Abstract
Blood and tissue contents of polyunsaturated fatty acid (PUFA) and long‐chain PUFA (LC‐PUFA) are related to numerous health outcomes including cardiovascular health, allergies, mental health and cognitive development. Evidence has accumulated to show that in addition to diet, common polymorphisms in the fatty acid desaturase (FADS) gene cluster have very marked effects on human PUFA and LC‐PUFA status. Recent results suggest that in addition to fatty acid desaturase 1 and fatty acid desaturase 2, the gene product of fatty acid desaturase 3 is associated with desaturating activity. New data have become available to show that FADS single nucleotide polymorphisms (SNPs) also modulate docosahexaenoic acid status in pregnancy as well as LC‐PUFA levels in children and in human milk. There are indications that FADS SNPs modulate the risk for allergic disorders and eczema, and the effect of breastfeeding on later cognitive development. Mechanisms by which FADS SNPs modulate PUFA levels in blood, breast milk and tissues should be explored further. More studies are required to explore the effects of FADS gene variants in populations with different ethnic backgrounds, lifestyles and dietary habits, and to investigate in greater depth the interaction of gene variants, diet and clinical end points, including immune response and developmental outcomes. Analyses of FADS gene variants should be included into all sizeable cohort and intervention studies addressing biological effects of PUFA and LC‐PUFA in order to consider these important confounders, and to enhance study sensitivity and precision.
Keywords: arachidonic acid, docosahexaenoic acid, FADS gene cluster, fatty acid desaturases, omega‐3 fatty acids, omega‐6 fatty acids
Introduction
The polyunsaturated fatty acid (PUFA) contents in blood and tissue lipids are closely related to numerous health outcomes including cardiovascular disease morbidity and mortality (Leaf 2006), immunological and inflammatory responses including allergies and atopic dermatitis (Trak‐Fellermeier et al. 2004; Kompauer et al. 2005), as well as mental health and psychiatric disorders (Muskiet & Kemperman 2006). Many of the physiological effects of PUFA are considered to be primarily mediated by tissue concentrations of long‐chain polyunsaturated fatty acids (LC‐PUFAs), in particular arachidonic acid (AA; 20:4n‐6), eicosapentaenoic acid (EPA; 20:5n‐3) and docosahexaenoic acid (DHA; 22:6n‐3) (3; 4) (Fig. 1). The availability of LC‐PUFA, particularly of AA and DHA, also plays a critical role during early growth and development during pregnancy and early childhood (Koletzko et al. 2008).
Figure 1.
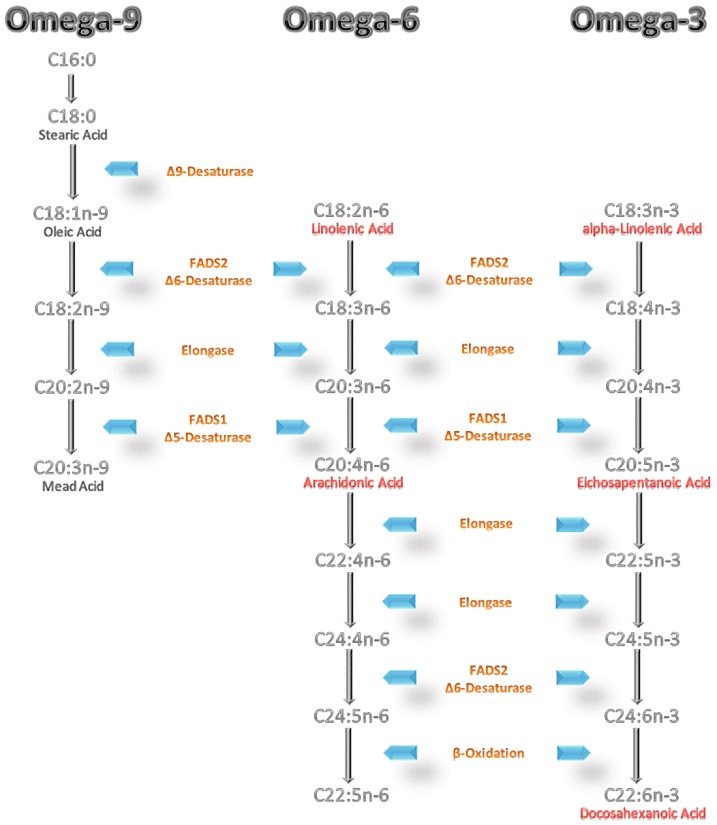
The omega‐9, omega‐6, and omega‐3 fatty acid metabolism pathways. FADS1, fatty acid desaturase 1; FADS2, fatty acid desaturase 2; SCD, stearoyl‐CoA desaturase; D5D, delta‐5 desaturase; D6D, delta‐6 desaturase; D8D, delta‐8 desaturase; D9D, delta‐9 desaturase.
LC‐PUFAs are provided by the diet, but can also be synthesized in human metabolism starting from the precursor essential fatty acids, linoleic acid (LA) and alpha‐linolenic acid (ALA) via consecutive desaturation and chain elongation (Fig. 1) (Sprecher & Baykousheva 1994; Sprecher et al. 1995; Park et al. 2009a). The delta‐5 desaturase (D5D) and delta‐6 desaturase (D6D) are thought to be the rate‐limiting enzymes in the formation of LC‐PUFA (Nakamura & Nara 2004). The human desaturase complementary DNAs were first cloned in 1999 (1999a, 1999b), and 1 year later were identified as fatty acid desaturase 1 (FADS1) and fatty acid desaturase 2 (FADS2) in the human genome (Marquardt et al. 2000). During the last 5 years, considerable evidence has accumulated to show that fatty acid desaturase (FADS) gene variants have marked effects on PUFA metabolism and on the contents of PUFA and LC‐PUFA in human blood and tissues, and first indications of effects on health end points have emerged. Here we review information on the role of FADS gene variants on PUFA metabolism and its potential biological importance.
Key messages
-
•
The availability of PUFA such as LA and ALA, and of LC‐PUFA, particularly AA and DHA, is essential for health and normal body function, modulates cardiovascular and immune health, and is of critical importance in pregnancy and early childhood for growth and development.
-
•
In addition to dietary PUFA supply, gene variants assessed by common SNPs in the FADS gene cluster markedly influence PUFA and LC‐PUFA status, including DHA levels in pregnancy and LC‐PUFA levels in human milk.
-
•
It appears highly likely that the gene product of FADS3 has desaturating activity.
-
•
Some studies indicate that FADS SNPs may modulate the risk for allergic disorders and eczema, and the effect of breastfeeding on later cognitive development.
-
•
More studies are required to explore the mechanisms by which FADS SNPs modulate PUFA levels in blood, breast milk and tissues, and to explore the effects of FADS gene variants in populations with different ethnic backgrounds, lifestyles and dietary habits, and to investigate in greater depth the interaction of FADS gene variants, diet and clinical end points, including immune response and developmental outcomes.
PUFA biochemistry and metabolism
Fatty acids are classified according to their length and degree of unsaturation of the hydrocarbon chain. Saturated fatty acids have no double bonds in the hydrocarbon chain, whereas unsaturated fatty acids contain at least one double bond. Fatty acids with at least two double bonds are referred to as PUFA, whereas PUFA with 20 or more carbon atoms are classified as LC‐PUFA. There are three PUFA families: the omega‐9, omega‐6 and omega‐3 series. Depending on the location of the last double bond, most distant from the carboxyl group (the alpha carbon atom), unsaturated fatty acids are classified as omega‐9 (or n‐9) [if the last double bond is located nine carbon atoms from the methyl end (omega carbon atom) of the molecule], omega‐6 (n‐6) or omega‐3 (n‐3) fatty acids. The parent fatty acids of these three families are oleic acid (omega‐9), LA (omega‐6) and ALA (omega‐3) (Fig. 1). These parent unsaturated fatty acids can be converted into fatty acids with longer chain length and higher degree of unsaturation by a series of alternating desaturation and chain elongation steps (Fig. 1). Current knowledge is that the three metabolic pathways share the same enzymes for desaturation and elongation.
The omega‐9 parent fatty acid, oleic acid, derives from the diet, but also can be synthesized endogenously. Palmitic acid (16:0) is elongated to stearic acid (18:0), which is further desaturated by the action of stearoyl‐CoA desaturase (SCD or delta‐9 desaturase) to oleic acid. An important product of the omega‐9 series is mead acid (C20:3n‐9). Mead acid is synthesized by D6 desaturation, elongation and D5 desaturation starting from oleic acid. In the case of LA or ALA deficiency, enhanced conversion of oleic acid to mead acid occurs, as the desaturase enzymes bind preferentially to ALA and LA. Therefore, increased concentrations of mead acid and an increased ratio of mead acid to AA in blood lipids are considered biomarkers of LA and ALA deficiency.
Dihomo‐gamma‐linolenic acid (DGLA, C20:3n‐6) can be formed by chain elongation of gamma‐linolenic acid (GLA) or by desaturation [delta‐8 desaturase (D8D)] of eicosadienoic acid (EDA). D5 desaturation of DGLA leads to AA. Further elongation and desaturation steps are possible.
An analogous pathway is used for omega‐3 fatty acids. ALA is converted to stearidonic acid by D6 desaturation, and stearidonic acid can be further elongated to eicosatetraenoic acid (ETA, 20:4n‐3). Alternatively, ALA can be elongated to eicosatrienoic acid (ETE, C20:3n‐3), which can be further desaturated (D8D) to ETA (Park et al. 2009a). The D5D product of ETA is EPA (C20:5n‐3), an important omega‐3 metabolite that serves as precursor of biologically potent eicosanoids. The major downstream product of the omega‐3 family is DHA (C22:6n‐3). There is no evidence for the previously assumed role of a delta‐4 desaturation in the conversion of EPA to DHA in mammals. A pathway in which DHA is synthesized by two chain elongations of EPA, followed by a D6 desaturation and a partial β‐oxidation was described by Sprecher (Sprecher 1999).
All desaturation and elongation steps take place in the endoplasmic reticulum. For the partial β‐oxidation, the last step, a compartmental translocation to peroxisomes is required (Sprecher 1999). The low activity of D6D and the compartmental translocation may explain the low conversion rate of omega‐3 docosapentaenoic acid (DPA, 22:5n‐3) to DHA in humans (Burdge 2004). There are indications for a higher conversion rate of ALA to EPA and DHA in women than in men, which is assumed to be due to oestrogen effects (Burdge 2006).
LA and ALA cannot be synthesized endogenously in mammals, and hence must be provided with the diet as essential nutrients. In typical current Western diets, up to 20% of dietary fat is comprised of PUFA. The most abundant dietary PUFA is LA. Together with ALA, LA contributes more than 95% of dietary PUFA intake (Calder 2008). Dietary sources of LA are vegetable oils such as corn, soybean, and sunflower oil and food products made with such oils. ALA is found in green plants, nuts and some common vegetable oils such as rapeseed (canola) or flax oil. The intakes of LC‐PUFA are much lower with most human diets than the intakes of LA and ALA. Dietary intakes of AA range typically between 50 and 500 mg day−1. High quantities of AA are found in, e.g. meat, eggs and offal. Oily fish and other seafood are the main dietary sources of the omega‐3 LC‐PUFA, EPA and DHA. Oily coldwater fish such as herring, mackerel, salmon, sardines, anchovy and tuna are rich sources of these fatty acids. Approximately 1.5–3.5 g of omega‐3 LC‐PUFA can be provided by one oily fishmeal. In the absence of oily fish consumption, omega‐3 LC‐PUFA intakes are typically very low and usually do not reach 100 mg day−1.
FADS
Key regulators of LC‐PUFA synthesis are the enzymes D5D and D6D (Nakamura & Nara 2004) (Fig. 1). Both desaturases are membrane‐bound enzymes produced in a majority of human tissues, with the highest activities found in liver. Significant activities of these enzymes are also found in adipose tissue (Sjogren et al. 2008), brain, heart and lung, whereas lesser amounts have been shown in placenta, skeletal muscle, kidney, pancreas and pregnant uterus (1999a, 1999b). The rat‐lactating mammary gland expresses D5D and D6D. Therefore, it has been suggested that the human mammary gland might also convert PUFA to LC‐PUFA, which subsequently can be incorporated into human milk (Rodriguez‐Cruz et al. 2006), but this hypothesis has not been confirmed by direct measurements.
D5D and D6D are encoded by the genes FADS1 and FADS2, respectively. They form a gene cluster jointly with the gene for fatty acid desaturase 3 (FADS3) on the human chromosome 11q12‐q13.1 (Fig. 2) (Marquardt et al. 2000; Nakamura & Nara 2004). FADS1 and FADS2 are found in a head‐to‐head fashion, whereas FADS2 and FADS3 have a tail‐to‐tail orientation. This cluster comprises 91.9 kb (Marquardt et al. 2000; Nakamura & Nara 2004). In the NCBI database (dbSNP build 130), around 500 single nucleotide polymorphisms (SNPs) are annotated for this region. The gene products of FADS1 and FADS2 have been characterized as D5D and D6D, respectively. In contrast, the function of the FADS3 gene product so far has not been characterized. Park et al. (2009b) identified several alternative splice forms of FADS3 and hypothesized that FADS3 might have a tissue‐ or PUFA‐specific role in LC‐PUFA synthesis.
Figure 2.
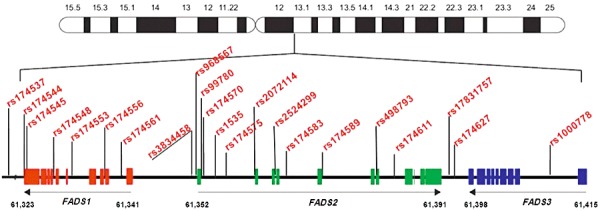
The human FADS gene cluster located on chromosome 11 with exon/intron organization and location of some published SNPs shown to be associated with fatty acid and phospholipid levels. SNP rs174537 is located 13.7 kb upstream of FADS1 (//indicates change of scaling). FADS, fatty acid desaturase; SNPs, single nucleotide polymorphisms; FADS1, fatty acid desaturase 1; FADS2, fatty acid desaturase 2; FADS3, fatty acid desaturase 3.
For the mammalian D6D, so far five substrates have been identified, i.e. C18:2n‐6, C24:6n‐6, C18:3n‐3, C24:5n‐3 and C16:0. The enzyme also has a D8D activity on 20:2n‐6 and 20:3n‐3. For D5D, only two substrates, namely 20:3n‐6 and 20:4n‐3, are known, indicating a higher substrate specificity of this enzyme (Park et al. 2009b). These two desaturases are thought to be rate limiting in the metabolic pathways of all three PUFA series.
Human FADS gene variants and PUFA metabolism
Given that D5D and D6D play important roles in the synthesis of LC‐PUFA, we hypothesized that polymorphisms of the desaturase encoding genes might affect desaturation activity, and thus PUFA concentrations in blood or tissue lipids. Our group therefore performed a first candidate gene study to explore whether FADS1 FADS2 gene cluster polymorphisms influence PUFA contents of serum phospholipids of human adults, which was published in the year 2006 (Schaeffer et al. 2006). In a cohort of 727 white adults (Erfurt, Germany) participating in the European Community Respiratory Health Survey I, 18 SNPs in the FADS1 FADS2 gene cluster and the fatty acid composition of serum phospholipids were analysed. FADS1 and FADS2 cluster SNPs were significantly associated with most omega‐6 and omega‐3 fatty acid levels except for omega‐6 DPA and omega‐3 DHA. Carriers of the minor alleles of 11 SNPs (rs174544, rs174553, rs174556, rs174561, rs3834458, rs968567, rs99780, rs174570, rs2072114, rs174583 and rs174589) had higher serum phospholipid contents of omega‐6 LA, EDA, DGLA, and of omega‐3 ALA, all of which are in the upper part of the pathway (cf. Fig. 1), and lower levels of the omega‐6 fatty acids GLA, AA, adrenic acid (ADA), and the omega‐3 fatty acids EPA and DPA, which are desaturation products. These data support the conclusion that the minor alleles lead to lesser expression of D6D and D5D. No association was found between genetic variants and DHA variance, which would support the concept that little DHA is synthesized endogenously, and DHA serum concentrations are primarily determined by the dietary supply of preformed DHA from fish and other sources. Analysis of reconstructed haplotypes indicated highly significant associations between haplotypes and fatty acid levels, which remained significant after correction for multiple testing and were in agreement with the findings of the SNP analysis. The effect size of SNPs was very high: genetic variants explained as much as 28.5% of the variation in serum AA contents in this cohort of free‐living individuals with considerable variation in lifestyle and dietary habits. The reconstructed haplotypes predicted some 12% and 10% of the variation of the AA precursors EDA and DGLA, respectively. The association between FADS polymorphisms and manifestations of respiratory and allergic disorders was also explored. There were no associations with total or specific immunoglobulin E (IgE) levels, but subjects carrying minor alleles of several SNPs had less than half the risk for allergic rhinitis and atopic eczema, even though statistical significance was lost after correcting for multiple testing.
The effects of human FADS gene variants on PUFA levels were replicated in several studies summarized in Table 1. Rzehak et al. genotyped three SNPs (rs174556, rs174561 and rs3834458) located in the FADS1 FADS2 gene cluster in a subgroup of Bavarian adults participating in the Bavarian Nutrition Survey II (Rzehak et al. 2009). Genotyping data were associated with fatty acid levels obtained from plasma phospholipids in 163 subjects and from erythrocyte membrane phospholipids in 535 subjects. This study confirmed the associations between variants in the FADS1 FADS2 gene cluster and serum phospholipid PUFA levels found by Schaeffer et al. (2006) in an independent cohort, and also replicated the 5‐loci haplotypes. Furthermore, minor allele haplotypes were found to influence PUFA levels in erythrocyte membranes, particularly AA and DGLA levels.
Table 1.
Studies exploring associations between FADS gene variants and fatty acid/phospholipid levels
| Study | Subjects | Significant SNP(s) | SNP location | Metabolites | Principal findings |
|---|---|---|---|---|---|
| Schaeffer et al. (2006) | n = 727 (GER) | rs174544, rs174553, rs174556, rs174561, rs3834458, rs968567, rs99780, rs174570, rs2072114, rs174583, rs174589 | FADS1/FADS2 | Fatty acids in serum phospholipids | Minor alleles are associated with: ↑LA, EDA, DGLA, ALA levels, ↓GLA, AA, ADA, EPA, DPA levels; AA – most significant associations, 28 % of variability genetically explained |
| Baylin et al. (2007) | n = 1694 (CRC, case) | rs3834458 | FADS1/FADS2 intergenic | Fatty acids in plasma (only in a subsample with n = 196) and in adipose tissue | Minor alleles are associated with: ↑EDA, ETE levels, ↓GLA, AA, EPA levels |
| Malerba et al. (2008) | n = 658 (ITA) | rs174545, rs174556, rs174561, rs3834458, rs174570, rs174583, rs174589, rs174611, rs174627 | FADS1/FADS2/FADS3 | Fatty acids in serum phospholipids and erythrocyte membranes | Minor alleles are associated with: ↑LA, EDA, ALA levels, ↓AA level |
| Martinelli et al. (2008) | n = 876 (ITA) | rs174545, rs174570, rs174583, rs1000778 | FADS1/FADS2/FADS3 | Fatty acids in serum phospholipids and erythrocyte membranes | Minor alleles are associated with: ↑LA level, ↓AA level, and AA/LA ratio |
| Xie and Innis (2008) | n = 69 (CAN, plasma/erythrocytes), n = 54 (CAN, breast milk) | rs174553, rs99780, rs174583 | FADS1 | Fatty acids in plasma phospholipids, erythrocyte glycerophospho‐ethanolamines, and breast milk | Minor alleles are associated with: ↑LA and ↓AA plasma and erythrocyte levels, ↓AA, EPA, DPA, DHA in breast milk |
| Rzehak et al. (2009) | n = 163 (GER, plasma), n = 535 (GER, erythrocytes) | rs174556, rs174561, rs3834458 | FADS1/FADS2 | Fatty acids in plasma and erythrocyte membrane phospholipids | Minor alleles are associated with: ↑DGLA levels, ↓AA levels; Replication of all haplotypes of the original five‐loci haplotypes from Schaeffer et al. (2006) |
| Tanaka et al. (2009) | n = 1210 (ITA, plasma), n = 1076 (USA, erythrocytes) | rs174537 | 5′ upstream region of FADS1 | Fatty acids in plasma and erythrocyte membranes | Minor alleles are associated with: ↑LA, ALA levels, ↓EDA, AA, EPA levels AA – 18.6% of variability genetically explained |
| Illig et al. (2010) | n = 1809 (GER) n = 422 (GBR) | rs174547 | FADS1 | Serum (lyso‐) phosphatidylcholines | Minor alleles are associated with: ↑PCaa36:3/PCaa36:4 ratio (≙↓D5D activity) |
| Molto‐Puigmarti et al. (2010) | n = 309 (NED) | rs174561, rs174575, rs3834458 | FADS1/FADS2 | Fatty acids in plasma phospholipids and in breast milk | Minor alleles are associated with: ↑LA, EDA, DGLA, ALA plasma levels, ↓GLA, AA, ADA, EPA, DPA, DHA plasma levels; Increasing fish/fish‐oil intake was associated with: →DHA plasma levels irrespective of the genotype, ↑DHA milk levels only in the major allele carriers |
| Bokor et al. (2010) | n = 1144 (EUR) | rs174546, rs968567, rs174570, rs174572, rs2072114, rs174589, rs174602, rs174611, rs174616 | FADS1/FADS2 | Fatty acids in serum phospholipids | Minor alleles are associated with: ↑LA levels, ↓AA levels, ↓D5D activity (estimated by AA/LA ratio) |
| Rzehak et al. (2010) | n = 333 (GER, serum) n = 546 (NED, plasma) | rs174545, rs174546, rs174556, rs174561, rs3834458 | FADS1/FADS2 | Fatty acids in serum glycerophospholipids and in plasma phospholipids | Minor alleles are associated with: ↑LA, ALA levels, ↓GLA, AA, ADA, EPA, DPA, DHA levels; genetically explained variability: LA = 5%, AA = 18%, DHA = 3%; SNPs are associated with eczema in GER but not in NED subjects, PUFA contents are not associated with eczema |
| Koletzko et al. (2011) | n = 4457 (GBR) | rs174548, rs174556, rs174561, rs3834458, rs968567, rs174570, rs174574, rs2727271, rs174576, rs174578, rs174579, rs174602, rs498793, rs526126, rs174448, rs174449, rs174455 | FADS1/FADS2/FADS3 | Fatty acids in erythrocyte phospholipids | Minor alleles are associated with: ↑ LA, ALA levels, ↓ AA, EPA, DHA levels, and AA/LA, EPA/ALA ratios |
| Lattka et al. (2011) | n = 769 (GER, 1.5 months) and n = 463 (GER, 6 months) | rs174547, rs174556, rs174602, rs498793, rs526126, rs174626, rs1000778, rs174455 | FADS1/FADS2/FADS3 | Fatty acids in breast milk | Minor alleles are associated with: ↓ AA, D5D activity (estimated by AA/DGLA ratio) FADS genotypes are associated with time course of fatty acid concentrations other than PUFA during lactation period |
ALA, alpha‐linolenic acid; AA, arachidonic acid; DGLA, dihomo‐gamma‐linolenic acid; D5D, delta‐5 desaturase; DHA, docosahexaenoic acid; EDA, eicosadienoic acid; EPA, eicosapentaenoic acid; FADS, fatty acid desaturase; GLA, gamma‐linolenic acid; LA, linoleic acid; SNP, single nucleotide polymorphisms; FADS1, fatty acid desaturase 1; FADS2, fatty acid desaturase 2; FADS3, fatty acid desaturase 3; ADA, ••; DPA, docosapentaenoic acid; ETE, eicosatrienoic acid; PUFA, polyunsaturated fatty acid.
Two further association studies analysed the fatty acid composition in both serum and red blood cell lipids. Malerba et al. (2008) analysed 13 SNPs located in the FADS gene cluster in 658 cardiovascular disease patients (Italian adults) participating in the Verona Heart Project (Malerba et al. 2008), and reported associations between SNPs and serum phospholipid and erythrocyte total fatty acids. Homozygote and heterozygote carriers of various minor alleles had higher levels of LA, EDA and ALA and lower levels of AA. For EPA and DHA no significant associations were observed. Only the associations between a constructed haplotype within the FADS1 FADS2 gene cluster and serum and erythrocyte AA levels were significant after adjustment for multiple testing. The same SNPs were examined by Martinelli et al. in 876 Italian adults participating in the Verona Heart Project, and the authors also explored an association with coronary artery disease (Martinelli et al. 2008). Among the studied subjects, 610 adults had angiographically documented coronary artery disease (cases), while the other 266 subjects (controls) did not. Cases had higher AA/LA and EPA/ALA ratios compared with controls, and the AA/LA ratio was a significant predictor of coronary artery disease using a multiple logistic regression model.
Baylin et al. (2007) also explored the association of FADS gene variants with cardiovascular disease. They performed a study aimed at identifying a common FADS2 gene promoter polymorphism (rs3834458) as a potential modulator of the effect of ALA on myocardial infarction and evaluated the effect of this polymorphism on PUFA concentrations in adipose tissue and the risk of myocardial infarction. Three thousand three hundred eighty‐eight Costa Rican subjects comprising 1694 controls and 1694 cases (men and women diagnosed as survivors of a first acute nonfatal myocardial infarction) were included. Fatty acids in plasma were analysed in a subsample of the cases (n = 196). The authors reported that a common deletion in the FADS2 promoter was associated with GLA, AA, EPA, ETA and EDA in plasma and adipose tissue. With increasing numbers of copies of the variant allele GLA, AA and EPA levels decreased, and levels of ETA and EDA increased. However, there was no significant association between this deletion and myocardial infarction, nor did this deletion modify the association between ALA and the risk of myocardial infarction.
Genome‐wide association studies (GWAS)
GWAS provided further evidence for the importance of FADS gene cluster polymorphisms in PUFA metabolism. In these studies, FADS polymorphisms are among the most significant hits.
The InCHIANTI study (Invecchiare in Chianti, ageing in the Chianti area, Tuscany, Italy) associated plasma levels of six PUFA (LA, EDA, AA, ALA, EPA and DHA) with genotyping data from 1210 Italian adults (Tanaka et al. 2009). Carriers homozygous for the minor allele of rs174537 near FADS1 had lower AA, EDA and EPA, and higher LA and ALA plasma levels than the major allele homozygotes. The most significant association was found between AA levels and rs174537, which accounted for 18.6% of the additive variance in AA concentrations. Tanaka et al. (2009) confirmed their findings in a second independent study cohort consisting of 1076 subjects participating in the Genetics of Lipid Lowering Drugs and Diet Network (white men and women from the USA) study. In this study, genotyping data were associated with levels of six PUFA (LA, AA, ALA, EPA, DPA and DHA) in erythrocytes (% of total fatty acids).
GWAS in subjects participating in the German Kooperative Gesundheitsforschung in der Region Augsburg study (KORA, Cooperative Health Research in the Augsburg Region, Germany) determined associations between the genotype of 284 males (55–79 years) with some 363 metabolites measured in serum samples of these participants (Gieger et al. 2008). Strong associations were found between the SNP rs174548, located on the FADS1 gene, with a number of plasma glycerophospholipid concentrations. Up to 12% of the observed variance of certain glycerophospholipid species, and up to 28% of the variance of certain glycerophospholipid ratios in plasma were explained by this SNP. The lowest levels of glycerophospholipid species containing PUFA with four and more double bonds were found for carriers of the minor allele of rs174548. Furthermore, AA was significantly reduced with increasing copy number of the minor allele. A positive association was shown between concentrations of glycerophospholipid species containing PUFA with three or fewer double bonds and the FASD1 genotype. The power of this GWAS was however limited because of the small number of participants. In 2010, the same group published GWAS using two cohorts of larger sample size. Genotyping data of 1809 participants from the KORA population were associated with 163 metabolites measured in fasting serum samples The findings were replicated in 422 participants of the TwinsUK (adult twin British registry) cohort (Illig et al. 2010). In both cohorts, the strongest association was observed for SNP rs174547 (FADS1) with the ratio between two glycerophosphocholines (PCaaC36:3/PCaaC36:4), considered a marker of D5D activity.
FADS gene variants in pregnant women
While all published studies agree on the strong association of FADS1 and FADS2 genotypes with several n‐6 and n‐3 fatty acids, reports on an association with DHA levels hardly exist. This question is of importance in particular with respect to pregnancy and early childhood because DHA is an omega‐3 LC‐PUFA with high incorporation into growing neural tissues with importance for brain and retina function (Larque et al. 2002). Only two studies reported an association of single FADS SNPs with DHA levels (Xie & Innis 2008; Molto‐Puigmarti et al. 2010); all other studies did not find such associations. Our group recently explored the relationship between FADS gene cluster polymorphisms and red blood cell (RBC) fatty acid (FA) levels in over 4000 pregnant women participating in the Avon Longitudinal Study of Parents and Children (Koletzko et al. 2011). Linear regression analysis of 17 SNPs in the FADS gene cluster was conducted with RBC phospholipid fatty acid data from 6711 samples of 4457 women obtained throughout pregnancy at a mean gestational age of 26.8 ± 8.2 weeks [mean ± standard deviation (SD)]. Independent of dietary effects, the minor alleles were consistently positively associated with the precursor FA, and negatively with LC‐PUFA as well as product/substrate ratios of the n‐6 (AA/LA ratio) and the n‐3 (EPA/ALA ratio) pathways (Table 2). In contrast to previous studies, we also found consistent significant inverse associations with DHA. Thus, FADS genotypes influence DHA levels in maternal RBC phospholipids and might affect the child's DHA supply during pregnancy. Similar, although weaker, associations were found for the FADS3 SNP rs174455. We thus conclude that it is highly likely that a gene product of FADS3 has desaturating activity.
Table 2.
Regression coefficients from analyses of long n‐6 and n‐3 fatty acid levels as a percentage of total fatty acid levels in red blood cell lipids on FADS SNPs in 6711 blood samples from 4457 pregnant women
| FADS1 | Intergenic | FADS2 | |||||
|---|---|---|---|---|---|---|---|
| rs174548 | rs174556 | rs174561 | rs3834458 | rs968567 | rs174570 | rs174574 | |
| Omega‐6 | |||||||
| 18:2 | 0.110 (0.020)*** | 0.112 (0.020)*** | 0.108 (0.020)*** | 0.106 (0.019)*** | 0.018 (0.024) | 0.150 (0.027)*** | 0.113 (0.019)*** |
| 20:4 | −0.154 (0.021)*** | −0.151 (0.021)*** | −0.155 (0.021)*** | −0.151 (0.020)*** | −0.131 (0.025)*** | −0.129 (0.029)*** | −0.147 (0.020)*** |
| Omega‐3 | |||||||
| 18:3 | 0.079 (0.021)*** | 0.084 (0.021)*** | 0.081 (0.021)*** | 0.075 (0.020)*** | 0.013 (0.025) | 0.071 (0.028)* | 0.073 (0.020)*** |
| 20:5 | −0.074 (0.021)*** | −0.069 (0.021)** | −0.074 (0.021)*** | −0.075 (0.021)*** | −0.091 (0.026)*** | −0.037 (0.029) | −0.068 (0.020)*** |
| 22:6 | −0.092 (0.021)*** | −0.092 (0.021)*** | −0.096 (0.021)*** | −0.097 (0.020)*** | −0.089 (0.025)*** | −0.073 (0.028)** | −0.092 (0.020)*** |
| Ratios | |||||||
| AA to LA | −0.262 (0.021)*** | −0.259 (0.021)*** | −0.262 (0.021)*** | −0.256 (0.020)*** | −0.175 (0.025)*** | −0.255 (0.028)*** | −0.254 (0.020)*** |
| EPA to ALA | −0.165 (0.020)*** | −0.165 (0.020)*** | −0.166 (0.020)*** | −0.161 (0.020)*** | −0.111 (0.025)*** | −0.117 (0.028)*** | −0.151 (0.019)*** |
| N | 6565 | 6524 | 6560 | 6558 | 6666 | 6583 | 6554 |
| FADS2 | |||||||
|---|---|---|---|---|---|---|---|
| rs2727271 | rs174576 | rs174578 | rs174579 | rs174602 | rs498793 | rs526126 | |
| Omega‐6 | |||||||
| 18:2 | 0.155 (0.028)*** | 0.108 (0.019)*** | 0.111 (0.019)*** | 0.053 (0.022)* | 0.080 (0.023)*** | −0.025 (0.018) | −0.011 (0.024) |
| 20:4 | −0.114 (0.030)*** | −0.144 (0.020)*** | −0.146 (0.020)*** | −0.120 (0.023)*** | −0.037 (0.024) | 0.041 (0.020)* | −0.079 (0.025)** |
| Omega‐3 | |||||||
| 18:3 | 0.123 (0.030)*** | 0.069 (0.020)*** | 0.070 (0.020)*** | 0.028 (0.023) | 0.063 (0.024)** | 0.007 (0.019) | −0.008 (0.025) |
| 20:5 | −0.017 (0.030) | −0.064 (0.021)** | −0.064 (0.020)** | −0.066 (0.023)** | −0.012 (0.024) | 0.031 (0.020) | −0.067 (0.025)** |
| 22:6 | −0.052 (0.030) | −0.091 (0.020)*** | −0.093 (0.020)*** | −0.088 (0.023)*** | −0.034 (0.024) | 0.045 (0.019)* | −0.092 (0.025)*** |
| Ratios | |||||||
| AA to LA | −0.241 (0.030)*** | −0.248 (0.020)*** | −0.252 (0.020)*** | −0.183 (0.023)*** | −0.096 (0.024)*** | 0.065 (0.020)*** | −0.093 (0.025)*** |
| EPA to ALA | −0.152 (0.029)*** | −0.144 (0.020)*** | −0.144 (0.019)*** | −0.101 (0.022)*** | −0.082 (0.023)*** | 0.026 (0.019) | −0.063 (0.024)** |
| N | 6569 | 6481 | 6574 | 6556 | 6591 | 6555 | 6595 |
| Intergenic | FADS3 | ||
|---|---|---|---|
| rs174448 | rs174449 | rs174455 | |
| Omega‐6 | |||
| 18:2 | 0.070 (0.019)*** | 0.065 (0.019)*** | 0.059 (0.019)** |
| 20:4 | −0.095 (0.020)*** | −0.103 (0.020)*** | −0.099 (0.020)*** |
| Omega‐3 | |||
| 18:3 | 0.043 (0.020)* | 0.043 (0.020)* | 0.032 (0.020) |
| 20:5 | −0.043 (0.020)* | −0.050 (0.020)* | −0.051 (0.021)* |
| 22:6 | −0.076 (0.020)*** | −0.082 (0.020)*** | −0.062 (0.020)** |
| Ratios | |||
| AA to LA | −0.164 (0.020)*** | −0.169 (0.020)*** | −0.160 (0.020)*** |
| EPA to ALA | −0.093 (0.019)*** | −0.101 (0.020)*** | −0.088 (0.020)*** |
| N | 6623 | 6583 | 6569 |
Modified from Koletzko et al. (2011). Fatty acids and ratios have been standardized to have a variance of one. Standard errors are reported in parentheses. *P < 0.05, **P < 0.01, ***P < 0.001. AA, arachidonic acid; EPA, eicosapentaenoic acid; FADS, fatty acid desaturase; LA, linoleic acid; SNPs, single nucleotide polymorphisms; FADS1, fatty acid desaturase 1; FADS2, fatty acid desaturase 2; FADS3, fatty acid desaturase 3; ALA, alpha‐linolenic acid.
FADS gene variants in adolescents and in children
A recently published study from Bokor et al. (2010) investigated whether associations between FADS gene cluster variants and fatty acid levels shown in adults are also present in adolescents. This study examined the relationship between 13 SNPs located in the FADS1 FADS2 gene cluster and serum phospholipid fatty acids in 1144 European adolescents (14.7 ± 1.4 years) participating in the Healthy Lifestyle in Europe by Nutrition in Adolescence cross‐sectional study. The findings were in line with results published in adults. Minor alleles of nine SNPs were associated with higher LA, lower AA and lower D5D activity, which were estimated from the AA/LA ratio.
Our group analysed associations between variants in the FADS gene cluster and blood fatty acid composition in two child cohort studies, and also explored the possible association with eczema (Rzehak et al. 2010). Data from two population‐based birth cohorts in the Netherlands and Germany (Kind, Ouders en gezondheid: Aandacht voor Leefstijl en Aanleg [KOALA], Inpluences of Lifestyle‐related factors on the immune system and the development of allergies in childhood [LISA]) were pooled (n = 879) and analysed by (logistic) regression regarding the mutual influence of SNPs in the 5 FADS gene cluster (rs174545, rs174546, rs174556, rs174561, rs3834458) on PUFA in blood and parent‐reported eczema until the age of 2 years. All SNPs were highly significantly associated with PUFA including DHA, also after correction for multiple testing, except for ALA and EPA. Genetic variation explained about 5% of the variation of blood LA content, 18% of AA and 3% of DHA content. All tested SNPs showed associations with eczema in the LISA study, but not in the KOALA study (Table 3), whereas blood PUFA content was not significantly associated with eczema either in the pooled cohort or in the analyses stratified by study cohort. These data suggest a possible link between inflammatory response, eczema and the metabolic pathway of LC‐PUFA formation. However, results are inconclusive between the two study populations of Dutch and German children, and further research is required to understand these discrepant results.
Table 3.
Association of FADS1 FADS2 variants with parental‐reported eczema in 879 children in the Dutch KOALA and German LISA studies
| SNP | KOALA study (the Netherlands) | LISA study (Germany) | ||||||
|---|---|---|---|---|---|---|---|---|
| n | OR | 95% CI | P‐value trend | n | OR | 95% CI | P‐value trend | |
| rs174545 | ||||||||
| C/C | 237 | 1.00 | – | 0.950 | 155 | 1.00 | – | 0.003 |
| C/G | 219 | 1.17 | 0.79–1.75 | 138 | 2.00 | 0.96–4.17 | ||
| G/G | 59 | 0.88 | 0.46–1.66 | 33 | 4.12 | 1.58–10.77 | ||
| rs174546 | ||||||||
| C/C | 238 | 1.00 | – | 0.998 | 156 | 1.00 | – | 0.005 |
| C/T | 218 | 1.16 | 0.78–1.73 | 138 | 1.87 | 0.91–3.84 | ||
| T/T | 60 | 0.86 | 0.46–1.63 | 33 | 3.84 | 1.48–9.93 | ||
| rs174556 | ||||||||
| C/C | 253 | 1.00 | – | 0.843 | 164 | 1.00 | – | 0.004 |
| C/T | 214 | 1.12 | 0.75–1.67 | 137 | 2.33 | 1.15–4.71 | ||
| T/T | 51 | 0.96 | 0.49–1.86 | 24 | 3.74 | 1.26–11.09 | ||
| rs174561 | ||||||||
| T/T | 256 | 1.00 | – | 0.953 | 164 | 1.00 | – | 0.004 |
| T/C | 212 | 1.09 | 0.73–1.62 | 137 | 2.33 | 1.15–4.71 | ||
| C/C | 52 | 0.93 | 0.48–1.80 | 24 | 3.74 | 1.26–11.09 | ||
| rs3834458 | ||||||||
| T/T | 237 | 1.00 | – | 0.818 | 152 | 1.00 | – | 0.004 |
| T/Z | 222 | 1.17 | 0.78–1.74 | 141 | 1.77 | 0.86–3.63 | ||
| Z/Z | 60 | 0.95 | 0.51–1.78 | 30 | 4.32 | 1.65–11.35 | ||
Note: OR of indicator‐coded SNPs (reference is homozygous major alleles genotype) on eczema are estimated by logistic regression. Adjustment comprises study cohort (indicator‐coded dummy variables: KOALA study conventional, KOALA study alternative recruitment group vs. LISA study as reference), sex, maternal education, maternal smoking during pregnancy and exclusive breastfeeding for at least 3 months. Modified from Rzehak et al. (2010).
FADS1, fatty acid desaturase 1; FADS2, fatty acid desaturase 2; SNPs, single nucleotide polymorphisms; OR, odds ratio; CI, confidence interval; KOALA, Kind, Ouders en gezondheid: Aandacht voor Leefstijl en Aanleg; LISA, Inpluences of Lifestyle‐related factors on the immune system and the development of allergies in childhood.
FADS gene variants and breastfeeding
The supply of LC‐PUFA to the fetus during gestation, particularly AA and DHA, during gestation to the fetus and post‐partum to the newborn is important for fetal growth, tissue development and early development of the nervous system (Larque et al. 2002). Breast milk contains relatively high amounts of AA and DHA, and breastfed infants have higher blood lipid concentrations of these two FA, as well as higher brain lipid content of DHA, than infants fed on formulas that do not provide LC‐PUFA (Farquharson et al. 1992; Makrides et al. 1994; Sauerwald et al. 2001). The contents of AA and DHA in breast milk are influenced by maternal dietary intake (Fidler et al. 2000; Yuhas et al. 2006). A further influencing factor appears to be the endogenous synthesis of these FA. A study from Rodriguez‐Cruz et al. (2006) showed that rat‐lactating mammary gland expresses D5D and D6D desaturases and thus has the capacity to synthesize LC‐PUFA.
In 1988, Koletzko et al. reported evidence for inter‐individual variations in LC‐PUFA metabolism and/or incorporation into breast milk lipids (Koletzko et al. 1988). They analysed the FA composition of human milk lipids in 24‐h collections of mature milk of 15 German women. No correlation was found between LC‐PUFA contents and their parent FAs, but the contents of omega‐6 and omega‐3 LC‐PUFA correlated significantly with each other. Because omega‐6 and omega‐3 LC‐PUFA originate from different dietary sources, our group assumed that some women had a higher ability to synthesize and secrete milk LC‐PUFA than others.
Xie and Innis analysed the association between genetic variants in the FADS1 FADS2 gene cluster and fatty acid levels in different blood compartments and in the breast milk of Canadian women (Xie & Innis 2008). Six SNPs (rs174553, rs174561, rs174575, rs174583, rs498793 and rs99780) were genotyped. For FA analyses in plasma phospholipids and erythrocyte glycerophosphoethanolamines, blood samples were collected from all 69 women at 16 weeks and again at 36 weeks of gestation. Furthermore, FA levels were analysed in breast milk collected from a subset of 54 women exclusively breastfeeding at 1 month post‐partum. Information on fat and FA intakes was assessed by an interview‐administered food frequency questionnaire at 16 and 36 weeks of gestation. For minor allele homozygotes of rs174553, rs174583 and rs99780, higher LA and lower AA levels were found in plasma phospholipids and erythrocyte glycerophosphoethanolamines. Minor alleles of the three SNPs were associated with lower levels of AA and EPA in breast milk. Women homozygous for the minor allele of rs174575 had lower levels of AA, EPA, DPA and DHA in breast milk.
The interaction of FADS gene variants with the effect of maternal fish and fish‐oil intake on DHA levels in plasma and human milk was investigated in 309 women participating in the Dutch KOALA birth cohort study (Molto‐Puigmarti et al. 2010). Three SNPs of the FADS1 FADS2 gene cluster (rs174561, rs174575 and rs3834458) were genotyped, and the fatty acid composition in plasma phospholipids collected at 36 weeks of gestation and in breast milk collected 1 month post‐partum was analysed. Fish and fish‐oil intake was assessed by a food frequency questionnaire at 34 weeks of gestation. Fatty fish and fish‐oil supplement intake were assessed by a short questionnaire during the week prior to breast milk sampling. Results of this study showed that DHA levels in plasma and breast milk were lower in women homozygous for the minor allele compared with women carrying the major allele. With increasing fish and fish‐oil intake, DHA levels increased in plasma phospholipids independently of the genotype, whereas DHA levels in breast milk increased only in lactating women carrying the major allele. The authors speculate that the incorporation of DHA into human milk is somehow reduced in women homozygous for the minor allele. These unexpected findings should be retested in another study. The reported maximum of fatty fish intake per week was three portions, and hence further studies should also evaluate whether higher amounts of fatty fish intake or omega‐3 LC‐PUFA supplementation might overcome the apparent limited incorporation of DHA into human milk of lactating women homozygous for the minor allele.
Our group investigated the influence of FADS SNPs on breast milk FA levels and their time course during lactation in the Ulm Birth Cohort study comprising 769 nursing mothers at 1.5 months after birth, and in a subset of 463 still breastfeeding mothers at 6 months post‐partum (Lattka et al. 2011). We conducted linear regression analysis of eight FADS SNPs with FA levels at both time points separately and assessed the genotype effect over time in a longitudinal analysis using a generalized estimating equation regression model. We observed significant associations of FADS genotypes with AA levels and the 20:4n‐6/20:3n‐6 ratio at both time points, but no association of FADS SNPs with the time course of AA levels. Longitudinal analysis of FAs other than LC‐PUFA by genotype over time showed associations for dodecanoic acid, cis‐15‐tetracosenoic acid and trans‐9‐octadecenoic acid. These data confirm that maternal FADS genotypes are associated with breast milk AA levels and therefore influence the child's supply of this FA.
The possible functional effects of the FADS genotype on breastfeeding effects have been investigated in two studies. Caspi et al. analysed the association between breastfeeding, two FADS SNPs and later IQ development in two independent birth cohorts (Caspi et al. 2007). They included 1037 children from the Dunedin Multidisciplinary Health and Development Study, New Zealand and 2232 infant twins from the Environmental Risk Longitudinal Twin Study in England and Wales. Genetic polymorphism in the FADS2 gene (rs174575) had no significant effect in the two total study populations, but the rs174575 polymorphisms interacted with breastfeeding in predicting the IQ in both cohorts. For breastfed children carrying the common C allele, a marked IQ advantage was found over children not breastfed. In children homozygous for the G allele, breastfeeding had no influence on the IQ. These effects remain significant even after adjustment for potential confounders such as intrauterine growth differences, social class differences and maternal cognitive ability.
Different results were reported by Steer et al. in 5934 children aged 8 years from the ALSPAC birth cohort (Steer et al. 2010). No genetic main effect on IQ was found for the FADS2 SNP rs174575. However, an interaction with this polymorphism was observed such that breastfed children homozygous for the minor allele (GG) performed better than their formula‐fed counterparts by an additional 5.8 points (1.4, 10.1) (interaction P = 0.0091). Interaction results were attenuated by about 10% after adjustment for seven factors. This study also investigated rs1535, another FADS2 polymorphism in linkage disequilibrium with rs174575, together with performance and verbal IQ, finding similar results, although effect sizes were generally reduced. Thus, this study did not replicate the findings of Caspi et al. (2007). In contrast to their study, GG children exhibited the greatest difference between feeding methods such that breastfed children performed similarly irrespective of child genotype, whereas formula‐fed GG children performed worse than other children on formula milk. Biologically, these results appear more plausible in that infants who have a lower ability to endogenously synthesize LC‐PUFA needed for incorporation into lipids of the rapidly growing brain would benefit more from a supply of preformed LC‐PUFA through breastfeeding. Further studies are required.
Conclusions
FADS gene variants have an important impact on PUFA metabolism and can influence PUFA and LC‐PUFA tissue availability. Several studies provide evidence that genetic variation in the FADS gene cluster can modify desaturase function. This effect was shown in several cohorts with different ethnic backgrounds and in adults, adolescents and children. FADS gene variants accounted for up to 28.5% of the variability in PUFA and LC‐PUFA levels in human tissues (Schaeffer et al. 2006; Gieger et al. 2008). Therefore, blood and tissue levels of the essential fatty acids LA and ALA and of their biologically active LC‐PUFA derivatives are influenced not only by diet, but to a large extent also by genetic variation. First results suggest marked effects of genetic variation in the FADS gene cluster on relevant clinical end points, including allergic rhinitis, eczema and cognitive development, with potentially major importance for public health. It appears necessary to better identify mechanisms by which FADS gene variants modulate PUFA levels in blood, breast milk and tissues. More studies are required to explore the effects of FADS gene variants in populations with different ethnic backgrounds, lifestyles and dietary habits, and to investigate in greater depth the interaction of FADS gene variants, diet and clinical end points, including immune response and developmental outcomes. Analyses of FADS gene variants should be included in all sizeable cohort and intervention studies addressing biological effects of PUFA and LC‐PUFA in order to consider these important confounders, and to enhance study sensitivity and precision.
Source of Funding
Financially supported in part by the Commission of the European Communities, specific RTD Programme ‘Quality of Life and Management of Living Resources’, within the 7th Framework Programme NUTRIMENTHE, FP7‐212652 and by the ‘Kompetenznetz Adipositas’ (‘Competence Network on Obesity’) funded by the Federal Ministry of Education and Research (FKZ: 01GI0826), and by the Munich Center of Health Sciences (MCHEALTH). BK is the recipient of a Freedom to Discover Award of the Bristol‐Myers‐Squibb Foundation, New York, NY, USA. Colin Steer was partly supported by NOAA, USA.
Conflicts of interest
Authors have no conflict of interest.
Acknowledgements
We are indebted to many fine colleagues and friends for fruitful collaboration and intellectual stimulation, in particular to Hermann Brenner, Tamás Decsi, Jean Golding, Joachim Heinrich, Thomas Illig and Dietrich Rothenbacher.
References
- Baylin A., Ruiz‐Narvaez E., Kraft P. & Campos H. (2007) Alpha‐linolenic acid, delta6‐desaturase gene polymorphism, and the risk of nonfatal myocardial infarction. The American Journal of Clinical Nutrition 85, 554–560. [DOI] [PubMed] [Google Scholar]
- Bokor S., Dumont J., Spinneker A., Gonzalez‐Gross M., Nova E., Widhalm K. et al (2010) Single nucleotide polymorphisms in the FADS gene cluster are associated with delta‐5 and delta‐6 desaturase activities estimated by serum fatty acid ratios. Journal of Lipid Research 51, 2325–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdge G.C. (2004) Alpha‐linolenic acid metabolism in men and women: nutritional and biological implications. Current Opinion in Clinical Nutrition and Metabolic Care 7, 137–144. [DOI] [PubMed] [Google Scholar]
- Burdge G.C. (2006) Metabolism of alpha‐linolenic acid in humans. Prostaglandins, Leukotrienes, and Essential Fatty Acids 75, 161–168. [DOI] [PubMed] [Google Scholar]
- Calder P.C. (2008) Polyunsaturated fatty acids, inflammatory processes and inflammatory bowel diseases. Molecular Nutrition & Food Research 52, 885–897. [DOI] [PubMed] [Google Scholar]
- Caspi A., Williams B., Kim‐Cohen J., Craig I.W., Milne B.J., Poulton R. et al (2007) Moderation of breastfeeding effects on the IQ by genetic variation in fatty acid metabolism. Proceedings of the National Academy of Sciences of the U S A 104, 18860–18865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.P., Nakamura M. & Clarke S.D. (1999a) Cloning, expression, and fatty acid regulation of the human delta‐5 desaturase. The Journal of Biological Chemistry 274, 37335–37339. [DOI] [PubMed] [Google Scholar]
- Cho H.P., Nakamura M.T. & Clarke S.D. (1999b) Cloning, expression, and nutritional regulation of the mammalian delta‐6 desaturase. Journal of Biological Chemistry 274, 471–477. [DOI] [PubMed] [Google Scholar]
- Farquharson J., Cockburn F., Patrick W.A., Jamieson E.C. & Logan R.W. (1992) Infant cerebral cortex phospholipid fatty‐acid composition and diet. Lancet 340, 810–813. [DOI] [PubMed] [Google Scholar]
- Fidler N., Sauerwald T., Pohl A., Demmelmair H. & Koletzko B. (2000) Docosahexaenoic acid transfer into human milk after dietary supplementation: a randomized clinical trial. Journal of Lipid Research 41, 1376–1383. [PubMed] [Google Scholar]
- Gieger C., Geistlinger L., Altmaier E., Hrabe de Angelis M., Kronenberg F., Meitinger T. et al (2008) Genetics meets metabolomics: a genome‐wide association study of metabolite profiles in human serum. PLoS Genetics 4, e1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illig T., Gieger C., Zhai G., Romisch‐Margl W., Wang‐Sattler R., Prehn C. et al (2010) A genome‐wide perspective of genetic variation in human metabolism. Nature Genetics 42, 137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koletzko B., Mrotzek M. & Bremer H.J. (1988) Fatty acid composition of mature human milk in Germany. The American Journal of Clinical Nutrition 47, 954–959. [DOI] [PubMed] [Google Scholar]
- Koletzko B., Lien E., Agostoni C., Bohles H., Campoy C., Cetin I. et al (2008) The roles of long‐chain polyunsaturated fatty acids in pregnancy, lactation and infancy: review of current knowledge and consensus recommendations. Journal of Perinatal Medicine 36, 5–14. [DOI] [PubMed] [Google Scholar]
- Koletzko B., Lattka E., Zeilinger S., Illig T. & Steer C. (2011) Genetic variants of the fatty acid desaturase gene cluster predict amounts of red blood cell docosahexaenoic and other polyunsaturated fatty acids in pregnant women: findings from the Avon Longitudinal Study of Parents and Children. The American Journal of Clinical Nutrition 93, 211–219. [DOI] [PubMed] [Google Scholar]
- Kompauer I., Demmelmair H., Koletzko B., Bolte G., Linseisen J. & Heinrich J. (2005) Association of fatty acids in serum phospholipids with hay fever, specific and total immunoglobulin E. The British Journal of Nutrition 93, 529–535. [DOI] [PubMed] [Google Scholar]
- Larque E., Demmelmair H. & Koletzko B. (2002) Perinatal supply and metabolism of long‐chain polyunsaturated fatty acids: importance for the early development of the nervous system. Annals of the New York Academy of Sciences 967, 299–310. [DOI] [PubMed] [Google Scholar]
- Lattka E., Rzehak P., Szabo E., Jakobik V., Weck M., Weyermann M. et al (2011) Genetic variants in the FADS gene cluster are associated with arachidonic acid concentrations of human breast milk at 1.5 and 6 mo postpartum and influence the course of milk dodecanoic, tetracosenoic, and trans‐9‐octadecenoic acid concentrations over the duration of lactation. The American Journal of Clinical Nutrition 93, 382–391. [DOI] [PubMed] [Google Scholar]
- Leaf A. (2006) Prevention of sudden cardiac death by n‐3 polyunsaturated fatty acids. Fundamental & Clinical Pharmacology 20, 525–538. [DOI] [PubMed] [Google Scholar]
- Makrides M., Neumann M.A., Byard R.W., Simmer K. & Gibson R.A. (1994) Fatty acid composition of brain, retina, and erythrocytes in breast‐ and formula‐fed infants. The American Journal of Clinical Nutrition 60, 189–194. [DOI] [PubMed] [Google Scholar]
- Malerba G., Schaeffer L., Xumerle L., Klopp N., Trabetti E., Biscuola M. et al (2008) SNPs of the FADS gene cluster are associated with polyunsaturated fatty acids in a cohort of patients with cardiovascular disease. Lipids 43, 289–299. [DOI] [PubMed] [Google Scholar]
- Marquardt A., Stohr H., White K. & Weber B.H. (2000) cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics 66, 175–183. [DOI] [PubMed] [Google Scholar]
- Martinelli N., Girelli D., Malerba G., Guarini P., Illig T., Trabetti E. et al (2008) FADS genotypes and desaturase activity estimated by the ratio of arachidonic acid to linoleic acid are associated with inflammation and coronary artery disease. The American Journal of Clinical Nutrition 88, 941–949. [DOI] [PubMed] [Google Scholar]
- Molto‐Puigmarti C., Plat J., Mensink R.P., Muller A., Jansen E., Zeegers M.P. et al (2010) FADS1 FADS2 gene variants modify the association between fish intake and the docosahexaenoic acid proportions in human milk. The American Journal of Clinical Nutrition 91, 1368–1376. [DOI] [PubMed] [Google Scholar]
- Muskiet F.A. & Kemperman R.F. (2006) Folate and long‐chain polyunsaturated fatty acids in psychiatric disease. The Journal of Nutritional Biochemistry 17, 717–727. [DOI] [PubMed] [Google Scholar]
- Nakamura M.T. & Nara T.Y. (2004) Structure, function, and dietary regulation of delta6, delta5, and delta9 desaturases. Annual Review of Nutrition 24, 345–376. [DOI] [PubMed] [Google Scholar]
- Park W.J., Kothapalli K.S., Lawrence P., Tyburczy C. & Brenna J.T. (2009a) An alternate pathway to long‐chain polyunsaturates: the FADS2 gene product delta8‐desaturates 20:2n‐6 and 20:3n‐3. Journal of Lipid Research 50, 1195–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W.J., Kothapalli K.S., Reardon H.T., Kim L.Y. & Brenna J.T. (2009b) Novel fatty acid desaturase 3 (FADS3) transcripts generated by alternative splicing. Gene 446, 28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Cruz M., Tovar A.R., Palacios‐Gonzalez B., Del Prado M. & Torres N. (2006) Synthesis of long‐chain polyunsaturated fatty acids in lactating mammary gland: role of delta5 and delta6 desaturases, SREBP‐1, PPARalpha, and PGC‐1. Journal of Lipid Research 47, 553–560. [DOI] [PubMed] [Google Scholar]
- Rzehak P., Heinrich J., Klopp N., Schaeffer L., Hoff S., Wolfram G. et al (2009) Evidence for an association between genetic variants of the fatty acid desaturase 1 fatty acid desaturase 2 (FADS1 FADS2) gene cluster and the fatty acid composition of erythrocyte membranes. The British Journal of Nutrition 101, 20–26. [DOI] [PubMed] [Google Scholar]
- Rzehak P., Thijs C., Standl M., Mommers M., Glaser C., Jansen E. et al (2010) Variants of the FADS1 FADS2 gene cluster, blood levels of polyunsaturated fatty acids and eczema in children within the first 2 years of life. PLoS One 5, e13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauerwald T.U., Demmelmair H. & Koletzko B. (2001) Polyunsaturated fatty acid supply with human milk. Lipids 36, 991–996. [DOI] [PubMed] [Google Scholar]
- Schaeffer L., Gohlke H., Muller M., Heid I.M., Palmer L.J., Kompauer I. et al (2006) Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Human Molecular Genetics 15, 1745–1756. [DOI] [PubMed] [Google Scholar]
- Sjogren P., Sierra‐Johnson J., Gertow K., Rosell M., Vessby B., de Faire U. et al (2008) Fatty acid desaturases in human adipose tissue: relationships between gene expression, desaturation indexes and insulin resistance. Diabetologia 51, 328–335. [DOI] [PubMed] [Google Scholar]
- Sprecher H. (1999) An update on the pathways of polyunsaturated fatty acid metabolism. Current Opinion in Clinical Nutrition and Metabolic Care 2, 135–138. [DOI] [PubMed] [Google Scholar]
- Sprecher H. & Baykousheva S. (1994) The role played by beta‐oxidation in unsaturated fatty acid biosynthesis. World Review of Nutrition and Dietetics 75, 26–29. [DOI] [PubMed] [Google Scholar]
- Sprecher H., Luthria D.L., Mohammed B.S. & Baykousheva S.P. (1995) Reevaluation of the pathways for the biosynthesis of polyunsaturated fatty acids. Journal of Lipid Research 36, 2471–2477. [PubMed] [Google Scholar]
- Steer C.D., Davey Smith G., Emmett P.M., Hibbeln J.R. & Golding J. (2010) FADS2 polymorphisms modify the effect of breastfeeding on child IQ. PLoS One 5, e11570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Shen J., Abecasis G.R., Kisialiou A., Ordovas J.M., Guralnik J.M. et al (2009) Genome‐wide association study of plasma polyunsaturated fatty acids in the InCHIANTI study. PLoS Genetics 5, e1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trak‐Fellermeier M.A., Brasche S., Winkler G., Koletzko B. & Heinrich J. (2004) Food and fatty acid intake and atopic disease in adults. The European Respiratory Journal 23, 575–582. [DOI] [PubMed] [Google Scholar]
- Xie L. & Innis S.M. (2008) Genetic variants of the FADS1 FADS2 gene cluster are associated with altered (n‐6) and (n‐3) essential fatty acids in plasma and erythrocyte phospholipids in women during pregnancy and in breast milk during lactation. The Journal of Nutrition 138, 2222–2228. [DOI] [PubMed] [Google Scholar]
- Yuhas R., Pramuk K. & Lien E.L. (2006) Human milk fatty acid composition from nine countries varies most in DHA. Lipids 41, 851–858. [DOI] [PubMed] [Google Scholar]


