Abstract
Zika virus (ZIKV) infection is an emerging pandemic threat to humans that can be fatal in newborns. Advances in digital health systems and nanoparticles can facilitate the development of sensitive and portable detection technologies for timely management of emerging viral infections. Here we report a nanomotor-based bead-motion cellphone (NBC) system for the immunological detection of ZIKV. The presence of virus in a testing sample results in the accumulation of platinum (Pt)-nanomotors on the surface of beads, causing their motion in H2O2 solution. Then the virus concentration is detected in correlation with the change in beads motion. The developed NBC system was capable of detecting ZIKV in samples with virus concentrations as low as 1 particle/μL. The NBC system allowed a highly specific detection of ZIKV in the presence of the closely related dengue virus and other neurotropic viruses, such as herpes simplex virus type 1 and human cytomegalovirus. The NBC platform technology has the potential to be used in the development of point-of-care diagnostics for pathogen detection and disease management in developed and developing countries.
Keywords: motion sensing, nanomotors, platinum nanoparticles, cellphone, Zika virus
Graphical Abstract
Zika virus (ZIKV) is raising significant global social and health concerns. Evidence of an increased incidence of microcephaly and other neurological complications such as Guillain–Barre syndrome during ZIKV outbreaks has been reported in infants.1–3 With the absence of specific medications and a preventive vaccine, sensitive and rapid diagnosis of ZIKV becomes critical and stays as an urgent public health demand.4,5 Recent developments in nanotechnology have further increased the ability to manipulate matter at the atomic and molecular scale to build a wide range of nanoparticle systems that possess versatile properties and applications.6–8 Catalytic properties of Au, Cu, Fe, and Pt nanoparticles are widely reported and have inspired a particularly interesting class of self-propelling catalytic motor-like micro/nanostructures.9,10 These structures are capable of converting chemical energy into mechanical motion, and three main propulsion mechanisms were reported to drive the motion of these materials, including self-electrophoresis, self-diffusiophoresis, and bubble thrust.11–14 Spherical particles, wires, rods, tubes, helices, and polymeric assemblies are among the most reported motor-like architectures.13–17 Their impressive autonomous, powerful, remotely controlled, and ultrafast motion is highly attractive with exciting applications in biomedicine and engineering.9,18 Furthermore, the catalytic motors possess a structure to function tenability. The addition of functional materials during the fabrication process of motors or modifying their surface allow multifunctions with broad applications.9,19,20 Self-propelling catalytic motor structures have recently been used in chemical and biological sensing,16,21 drug delivery,22–25 controlled transport and release of biomolecules,26,27 cell screening and manipulation,28–31 and waste treatment.32,33
Motion-based biosensing has attracted considerable attention owing to the fact that the motion as a signal readout is simple and potentially offers spatial resolution and real-time detections.34,35 Lately, numerous reports described the utilization of motion-based sensors for the detection of different nucleic acid and protein targets.26,28,35–39 Although those biosensors have demonstrated good performance and potential for target detection, most of them rely on using relatively sophisticated optical microscopy systems for tracking the changes in the motion and speed of motor structures in the presence of target analytes. Here we report an approach for virus detection through monitoring the catalytic-based motion of nanostructures under a cellphone optical system, which can have great potential to be used in the development of point-of-care diagnostics.
RESULTS AND DISCUSSION
NBC System Design and Development.
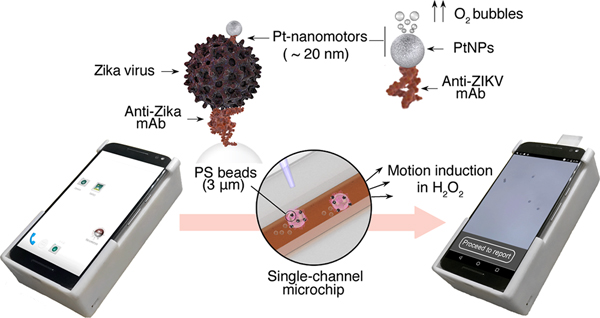
This work reports the development of a system for the immunological detection of ZIKV using Pt-nanomotors and bead motion monitoring under a cellphone optical system (Figure 1). In this assay, the applied Pt-nanomotors are specifically designed to interact with ZIKV captured on the surface of 3 μm polystyrene (PS) beads, forming a three-dimensional (3D) immunocomplex that moves in the presence of H2O2. While loaded on the surface of a single-channel microchip, the average motion velocity of the formed immunocomplexes (beads-virus-motors) is measured by a cellphone enabled with an optical attachment and a motion tracking cellphone application. The average motion velocity of the beads is then quantitatively correlated to the virus concentration in the tested sample. The Pt-nanomotors were mainly comprised of PtNPs conjugated with anti-Zika virus monoclonal antibody (anti-ZIKV mAb) specifically targeting the envelope protein. The motors move by catalyzing the decomposition of H2O2, and thus in the presence of ZIKV, an abundant number of motors accumulates on the surface of the beads and induces their motion. In contrast, in the absence of virus, the motors did not bind to the surface of beads and remain free in H2O2 solution, resulting in a significantly lower motion velocity of beads as compared to when target viruses were present in the sample. The cellphone setup used in this study comprises an android terminal modified with a cellphone application, a disposable microchip, and an optical cellphone attachment. We used a MotoX cellphone (Motorola, XT1575) in performing the experiments in this work. The optical cellphone attachment was designed using SolidWorks 2016 software and fabricated using a 3D printer (UltimakerII Extended) with Ultimaker PLA (polylactic acid) as printing material. The microchip was prepared with two main layers of glass slide and poly(methyl methacrylate) (PMMA) that were assembled together using a laser-machined double-sided adhesive (DSA) sheet to form a single longitudinal channel (Figure S1). The optical attachment includes an inexpensive acrylic lens for image magnification, electronics, and a LED light source (Figure S2). A slide holder was engraved on the cellphone attachment where the microchip can be inserted into the setup and imaged. A customized cellphone application was developed to specifically identify beads in the sample and track its movement to measure its velocity and calculate the virus concentration. The cellphone application can record videos, enumerate beads, automatically calculate their motion velocity, and report the results in ∼2 min (Figure S3). The cellphone application is enabled with a user-friendly interface to facilitate the testing process. The developed system was able to record videos of a sample at a rate of 30 frames per second (fps) with a maximum effective field-of-view (FOV) of 480 × 360 μm. The device was calibrated using a micrometer scale (Figure S4). The resolving power of the attachment was tested using micropolystyrene beads (3 μm). We observed that the system was able to visualize and detect the motion of the microbeads. The estimated material cost to fabricate the cellphone attachment and the used microchip was $3.59 and $0.38, respectively (Table S1).
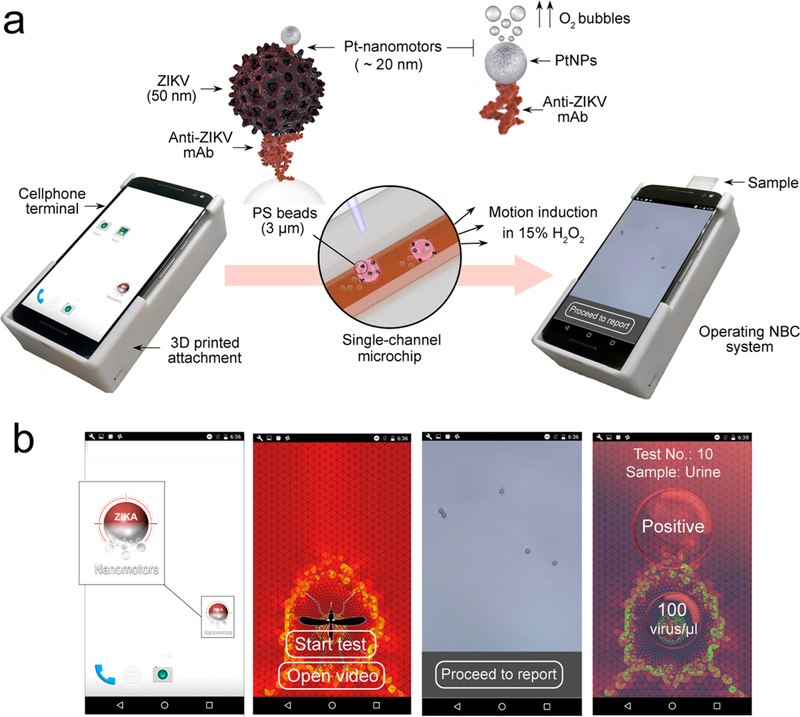
Figure 1.
Schematic of the NBC system for virus detection. (a) Operating the NBC system loaded with a sample. The operating NBC comprises three main components: (i) 3D printed optical attachment, (ii) single channel microchip for sample loading and visualization, and (iii) cellphone terminal. The detection protocol starts by capturing virus on the surface of 3 μm beads modified with anti-Zika virus monoclonal antibody (anti-ZIKV mAb) and assembling with Pt-nanomotors to form the Pt-ZIKV-bead complex. The Pt-nanomotors are prepared of spherical platinum nanoparticles (PtNPs) coupled with anti-ZIKV mAb. The presence of Pt-nanomotors on the surface of beads induces their motion in 15% hydrogen peroxide (H2O2) solution. The change in the beads velocity can be analyzed using the NBC system through a motion tracking cellphone application. (b) Motion tracking application interface and data processing. Based on the detected change in the velocity of the beads, the concentration of the virus in the tested samples is detected, and a report for ZIKV infection is generated.
Pt-Nanomotors Preparation and Characterization.
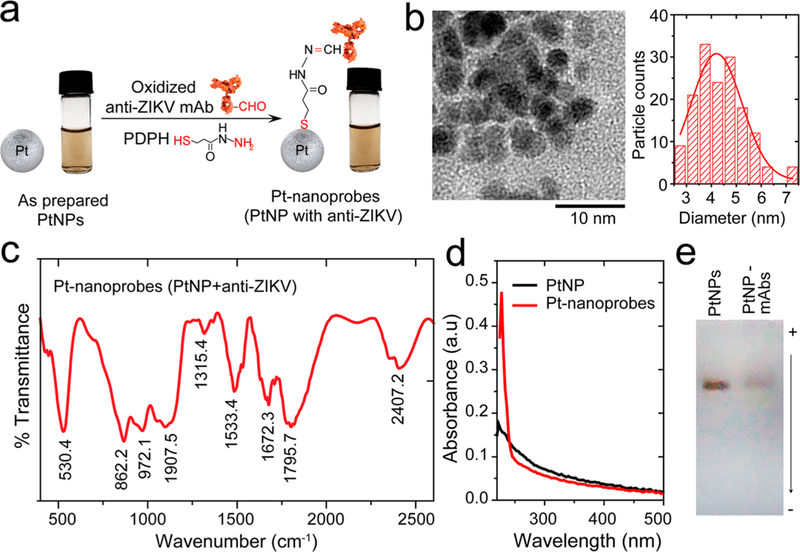
Pt-nanomotors were prepared from PtNPs functionalized with anti-ZIKV mAb following our surface chemistry protocol (Figure S5). This protocol relies on using a bifunctional cross-linker of 3-(2-pyridyldithio)propionyl hydrazide (PDPH) to bind the oxidized antibodies through their carbohydrate residues to the surface of nanoparticles (Figure 2a). Transmission electron microscopy (TEM) and the corresponding size distribution histogram indicate that the synthesized PtNPs are spherical in shape with an average diameter of 4.37 ± 0.986 nm (Figure 2b). We performed Fourier transform-infrared spectroscopy (FT-IR) to characterize the surface chemistry and antibody immobilization. The conjugation of mAb to PtNPs resulted in several peaks in FT-IR analysis that are characteristic for antibodies. Figure 2c shows FT-IR spectra of Pt-nanomotors with different bands appearing at 2407.2, 1672.3, 1533.4, 1315.4, 1907.5, and 862.2 cm−1, which can be assigned to C⩵O stretching, N–H bending, C–N stretching, C–C stretching, and S–metal bond, respectively.40,41 These bands correspond to the thiol–Pt bond formed by PDPH with the surface of PtNPs and to amid-I and -II characteristic of antibodies coupled to the surface of the PtNPs. Ultraviolet– visible (UV–vis) analysis of citrate-stabilized PtNPs and Pt-nanomotors (PtNPs modified with mAb) confirmed the stability of the synthesized nanomotors, and a strong absorption peak was observed at 223 nm, which is associated to the presence of the antibody as a protein structure (Figure 2d and Figure S6). On the other hand, the conjugation of antibodies to the surface of PtNPs caused retardation in the motion of the formed Pt-nanomotors (PtNPs-mAbs) compared to non-modified PtNPs when tested on agarose gel electrophoresis, which can be attributed to the difference in size and charge density value between PtNPs (no antibodies) and the formed nanomotors (PtNPs-antibody conjugates) (Figure 2e and Figures S7 and S8).42 The ratio of antibody molecules per nanoparticle was estimated to be 1.792 ± 0.693 antibody molecule/PtNP based on their corresponding absorption values at 223 nm (Figure S6 and Supporting Methods). Therefore, approximately 6.38% of the surface of Pt-nanomotor particle was covered with anti-ZIKV mAb, and 93.62% of the surface of PtNPs was available to interact with H2O2 for gas formation. In addition, this ratio of antibody surface coverage on Pt-nanomotors allows efficient labeling of the captured virus with minimum chance for the formation of large aggregates of beads, which can limit the motion of each complex and result in a false negative signal.
Figure 2.
Pt-nanomotor preparation and characterization. (a) Schematic presentation of the protocol used in the preparation of Pt-nanomotors. Oxidized anti-ZIKV mAb was conjugated to the surface of PtNPs using a bifuctional cross-linker of 3-(2-pyridyldithio)propionyl hydrazide (PDPH) that has a terminal thiol group bound to the metal surface of PtNPs on one side and a terminal hydrazide group bound to the free aldehyde group in the oxidized carbohydrate residue of antibody on the other side. The images show no color change or visible turbidity in the PtNP samples, indicating the stability of the prepared Pt-nanomotors. (b) TEM micrograph and particle size distribution histogram of the prepared PtNPs. (c) FT-IR spectrum of the prepared Pt-nanomotors showing several characteristic peaks for protein at 1795.7 cm−1, 1672.3 cm−1, and 1533.4 cm−1. (d) UV–vis absorption spectra of PtNPs and the prepared Pt-nanomotors. A strong absorption peak at 260 nm was observed with Pt-nanomotors, confirming the conjugation of antibodies to the surface of PtNPs. (e) Agarose gel electrophoresis of PtNPs and PtNP-mAbs (Pt-nanomotors).
Preparation and Characterization of ZIKV-Capturing Beads.
Beads coated with anti-ZIKV envelope mAb were used to allow specific formation of Pt-bead virus complexes by the accumulation of nanomotors on the surface of beads in the presence of ZIKV. Beads conjugated with anti-ZIKV mAb were prepared using a coupling protocol that allows the directional conjugation of antibodies to the surface of beads using adipic dihydrazide (Figure 3a and Supporting Methods). Carboxylated beads were initially activated with adipic acid using the well-known 1-ethyl-3-(3-(dimethylamino)propyl)carbodiimide hydrochloride(EDC)/sulfo-N-hydroxysuccinimide (sulfo-NHS) protocol. Then the free hydrazide groups on the surface of the beads were directly coupled to oxidized antibodies by sodium periodate and through their carbohydrate residue in FC region (Figure S9). The surface activation of beads with adipic acid was confirmed using ζ potential and FT-IR techniques. The ζ potential indicated a significant decrease to –10.2 ± 2.21 mV in the net negative surface charge of the beads after the activation with dihydrazide due to the presence of terminal amine with positive charge (Figure S10). FT-IR indicated the presence of 1720.5 and 1666.5 cm−1 vibrations that are characteristic for amide-I and -II groups of the adipic dihrydrazide existing on the surface of beads (Figure S11).40 For antibody conjugation, UV–vis spectroscopy indicated the presence of a strong peak at 223 nm that is specific for antibody used in our study (Figure S6). Using a standard curve prepared of different concentrations of anti-ZIKV antibody, the number of antibody molecules per each bead was ∼3 × 104 molecule (Figure 3b and Figure S6 and Supporting Methods). On the other hand, FT-IR analysis of bead-modified with antibodies showed the presence of a cluster of vibration bands at 1343.6, 1508.7, 1711.6, and 1920.2 cm−1 and two bands at 2907.9 and 2950.8 cm−1. The peaks at 1343.6, 1508.7, 1711.6, and 1920.2 cm−1 can be attributed to the amide-I and II characteristic to the antibody as protein structures, while the peaks at 2907.9 and 2950.8 cm−1 can be attributed to C=O of the carboxyl groups in the PDPH cross-linker used in antibody conjugation protocols (Figure 3c). Virus capture and PtNP-virus complex formation on the surface of beads were confirmed using sodium dodecyl sulfate polyacrylamide (SDS) gel electrophoresis, scanning electron microscopy (SEM), and inductively coupled plasma mass spectroscopy (ICP-MS) techniques. SDS gel electrophoresis analysis of the virus captured on beads indicated the presence of an intense band at 88.2 kDa that is characteristic to the non-structural protein 3 (NS3) of ZIKV and also appeared in the control sample of purified ZIKV in PBS (Figure 3d).43,44 SEM analysis of PtNP-virus assemblies on the surface of beads indicated that the formed assemblies were 80–100 nm in size and that there were approximately 1.2 PtNP-virus complexes per 1 μm2 (Figure 3e). The formed PtNP-virus-beads complexes were isolated by centrifugation, washed, and tested by ICP-MS technique to estimate the concentration of Pt metal on the surface of beads. The results confirmed the accumulation of Pt-nanomotors on the surface of beads in the presence of ZIKV at the rate of 1200 Pt-nanomotors per bead when 103 virus particles/μL were used in the reaction (Figure S12).
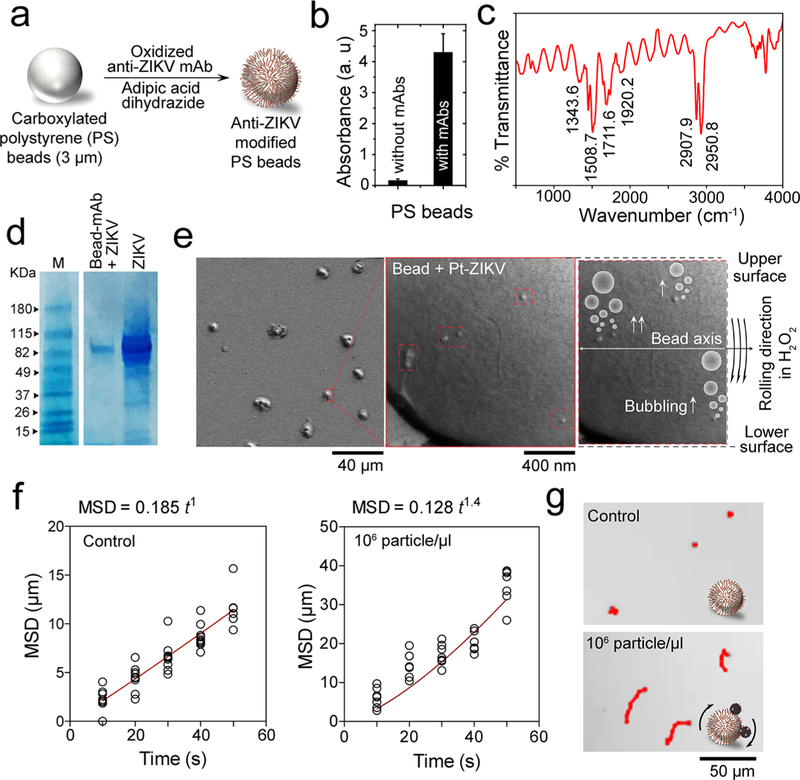
Figure 3.
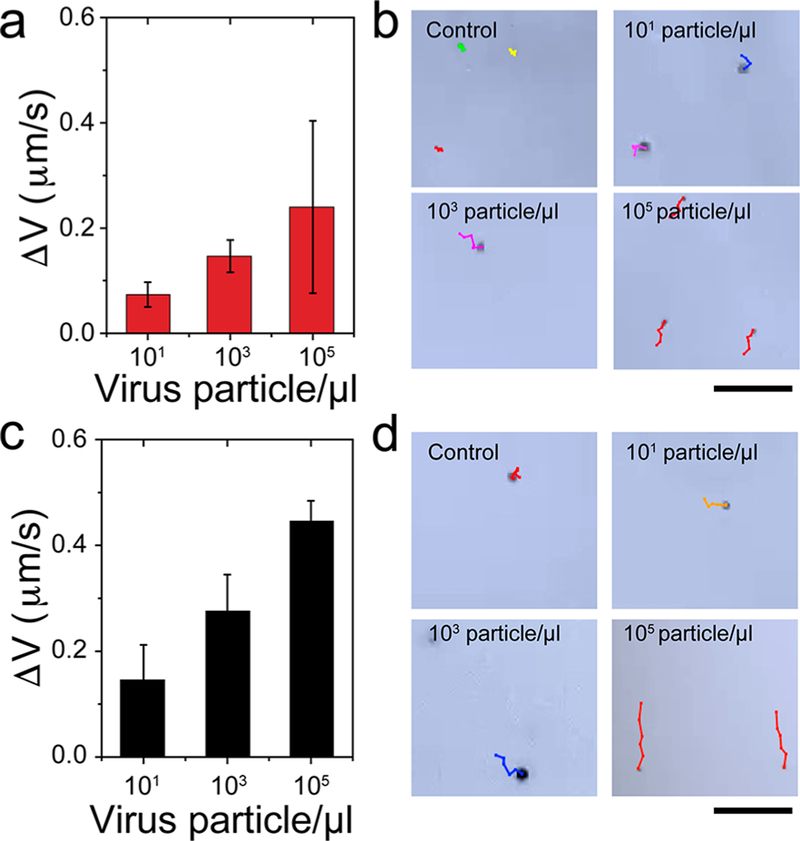
Virus capture and induction of beads motion by Pt-nanomotors. (a) Schematic presentation of surface modification of PS beads with anti-ZIKV mAb. (b) UV–vis absorption values at 223 nm for beads with and without antibody modification. (c) FT-IR spectrum of the prepared anti-ZIKV mAb-modified beads, showing several peaks characteristics for protein at 1711.6, 1508.7, 1343.6, and 2907.9 cm−1. (d) SDS gel electrophoresis for ZIKV captured on the surface of antibody-modified beads. M: protein marker; Bead-mAb + ZIKV: virus captured on beads modified with anti-Zika mAb; ZIKV: Zika virus prepared in phosphate buffer saline at 105 particles/μL. (e) SEM analysis of beads with Pt-virus complexes. (f) Motion analysis of beads in the presence and absence of the target ZIKV tested under bright-field light microscopy using 200× magnification. (g) Representative images of motion trajectories of beads in the presence and absence of ZIKV under light microscopy.
Validation of the NBC System Design for ZIKV Detection.
We initially tested the motion induction of PS beads in the presence of virus particles using bright-field microscopy. Aliquots of ZIKV-spiked phosphate buffer (PB) with virus concentrations of 106 particles/μL were added to antibody-modified beads followed by the addition of Pt-nanomotors to allow the formation of PtNP-bead-virus complexes (see Methods). The motion of the formed complexes was then tested in 10% H2O2 solution in a single channel microfluidic chip under light microscope and using ImageJ software. Figure 3f,g shows the difference between the motion of beads in the presence and absence of the target ZIKV. Using mean squared displacement (MSD) versus time t as a model to assess the motion of beads, we found that the MSD versus time did have a near linear dependence (α = 1.0), which is typical to the case for random diffusion where α = 1 in virus-free control samples. On the other hand, when 106 virus particles/μL was used in 10% H2O2, a power dependence with α = 1.4 was observed indicating that the bead motion is specifically correlated to the presence of ZIKV (Figure 3f and Movies S1 and S2). The beads in ZIKV-spiked samples were observed to move with an average velocity of 1.199 μm/sec (Figure S13), which was approximately 5.2-times higher than the average velocity of the beads in virus-free control samples. These results confirmed the ability of the virus to specifically induce the motion of beads when the bead-virus-PtNP complexes were formed. Trajectory images of the motions of beads in control ZIKV-spiked samples are shown in Figure 3f. Furthermore, we tested the change in the velocity of beads motion upon induction with different concentrations of ZIKV (from 0 particles/μL to 105 particles/μL). The results confirmed the ability of the virus to induce the motion of beads when assembled with Pt-nanomotors in 15% H2O2 solution. The velocity of beads motion increased significantly with the increase in virus concentrations tested (Figures S14 and S15 and Movies S3–S5). The ability of the developed NBC system to track the motion and measure the velocity of beads was then validated using samples with different concentrations of ZIKV (n = 40). The motion tracking cellphone application was optimized to have a correlation percentage of 89.11% with the measurements obtained by light microscopy and ImageJ software (Figure S16).
Evaluation of the NBC System in ZIKV Detection Using Spiked and Patient Samples.
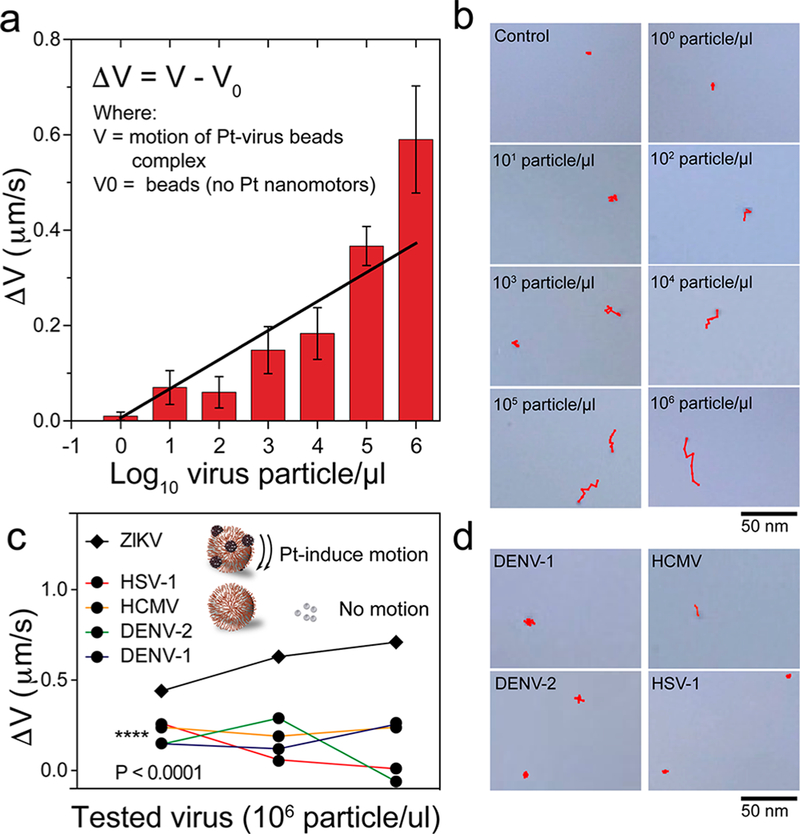
To evaluate the performance of the NBC assay in Zika detection, we used samples spiked with different ZIKV concentrations ranged from 100 particles/μL to 106 particles/μL and non-target viruses, including dengue types 1 (DENV-1) and 2 (DENV-2), human simplex virus type 1 (HSV-1), and human cytomegalovirus (HCMV). The antibody-modified beads were mixed with the samples for virus capture on beads and then incubated with Pt nanomotors to allow the formation of bead-virus-NPs complexes. The motion of the beads was then monitored in 15% H2O2 solution using the cellphone optical system. Figure 4a shows the change in the beads motion velocity (ΔV) caused by the addition of different concentrations of ZIKV. The results showed an excellent correlation (r2 = 0.89) between the virus concentration and the magnitude of the beads velocity change. Based on S/N = 2.0 and S/N = 3.0, the detection limit of the developed NBC assay for testing the target ZIKV in PB is down to 1 particle/μL and 10 particles/μL, respectively (Figure S17 and Movies S6–S8). Trajectory images of the motions of beads with different ZIKV concentrations as recorded by the NBC system are shown in Figure 4b. In addition, the detection specificity of the system was tested using the same concentration (106 particles/μL) of the target ZIKV and non-starget viruses including DENV-1, DENV-2, HSV-1, and HCMV. Figure 4c presents the change in beads velocity (ΔV) with ZIKV, DENV-1, DENV-2, HSV-1, and HCMV. The increase in the beads velocity caused by the target ZIKV was significant ( P < 0.0001) compared with the change in the velocity of beads caused by the addition of all the non-target viruses. Figure 4d shows representative digital images of the trajectories of beads in the presence of DENV-1, DENV-2, HSV-1, and HCMV. The images indicate a relatively slow motion of beads that is comparable to the random diffusion of beads in control samples (Figure 4d and Movie S9). To further confirm the potential of the NBC system for virus detection in biological samples, we tested the developed system using urine and saliva samples spiked with ZIKV (n = 9) at three different concentrations (i.e., 101 particles/μL, 10 3 particles/μL, and 105 particles/μL) ( Figure 5a–d). Figure 5a,b shows that the beads velocity change increased as the virus concentration in the sample increased and the cellphone system was able to detect ZIKV at concentrations as low as 101 particles/μL in urine samples. There was a variation in the motion velocity recorded for urine samples with relatively high virus concentration (105 particles/μL), which can be explained by the variation in the number of captured virus particles to different beads in the presence of high salt concentration and unfavorable pH conditions. However, we did not observe significant beads velocity change in virus-free control samples. These results confirmed the ability of the NBC system for target virus detection and particularly Zika virus in complex biological samples such as urine and saliva. Furthermore, we evaluated the performance of the NBC system in identifying the ZIKV-infected serum samples compared to the results obtained by the standard techniques currently approved by the U.S. Food and Drug Administration (FDA) and recommended by the Center for Disease Control (CDC) for qualitative detection of ZIKV, including CDC Zika MAC-ELISA (ZIKV IgM detection) and Aptima Zika virus assay (ZIKV RNA detection) (Table 1). The results indicated that the accuracy of the NBC system in classifying patient serum samples as positive (ZIKV-infected) and negative (non-infected) compared to CDC Zika MAC-ELISA was 100%, while having a correlation of 80% to Aptima Zika virus assay.
Figure 4.
Evaluation of the NBC system for ZIKV detection. Sensitivity testing of the NBC system for ZIKV detection: (a) the change in bead motion magnitude in the presence of different concentrations of ZIKV (from 100 particles/μL to 106 particles/μL); (b) trajectory images of the motion of beads in the control sample and samples with different concentrations of ZIKV. Specificity testing of the NBC system for ZIKV detection: (c) the change in bead motion magnitude due to ZIKV and non-target viruses, including herpes simplex virus type 1 (HSV-1), human cytomegalovirus (HCMV), and dengue virus (DENV) types 1 and 2 were measured. The tested virus concentration for samples in the specificity evaluation was 106 particles/μL; (d) trajectory images of the motion of beads in the presence of ZIKV and other non-target viruses. Error bars are standard deviations from a total of three independent measurements.
Figure 5.
ZIKV detection in biological samples using the NBC system. (a) Detection of ZIKV spiked in urine samples with virus concentrations of 101 particles/μL, 103 particles/μL, and 105 particles/μL. (b) Trajectory images of beads motion recorded for ZIKV-spiked urine samples. (c) Detection of ZIKV-spiked saliva samples with virus concentrations of 101 particles/μL, 103 particles/μL, and 105 particles/μL. (d) Trajectory images of beads motion recorded for ZIKV-spiked urine samples. Error bars are standard deviations from a total of three independent measurements.
Table 1.
NBC System Evaluation Using ZIKV-Infected Patient Serum Samples (n = 10)a
| CDC Zika MAC-ELISAb |
Aptima Zika virus assayc |
NBC systemd |
||||
|---|---|---|---|---|---|---|
| Patient no. | sample | ISR value | sample | S/Co value | sample | V (μm/s) |
| 1 | positive | 13.86 | positive | 18.51 | positive | 0.272 |
| 2 | positive | 14.96 | negative | 0.00 | positive | 0.253 |
| 3 | positive | 21.32 | positive | 31.91 | positive | 0.432 |
| 4 | positive | 3.78 | positive | 20.59 | positive | 0.208 |
| 5 | positive | 18.98 | positive | 32.91 | positive | 0.439 |
| 6 | positive | 17.25 | negative | 0.00 | positive | 0.372 |
| 7 | positive | 18.07 | positive | 33.09 | positive | 0.475 |
| 8 | positive | 14.14 | positive | 19.39 | positive | 0.246 |
| 9 | positive | 2.55 | positive | 17.18 | positive | 0.140 |
| 10 | negative | 0.60 | negative | 0.00 | negative | 0.065 |
The cellphone results were compared with the results obtained by the standard lab-based tests recommended for ZIKV detection.
Zika immune status ratio values (Zika ISR). Reading of ≥1.80 is positive and ≤1.60 is negative as defined by the manufacturer.
Signal-to-cutoff value (S/Co). Reading of >1.0 is positive and <1.0 is negative as defined by the manufacturer.
V is the velocity magnitude. Reading of ≥0.086 is positive and <0.086 is negative, considering S/N = 2 and the average velocity of control samples (no ZIKV) is 0.07 ± 0.008 μm/s.
This work has demonstrated the development of the NBC system for sensitive and specific detection of ZIKV by leveraging the advantage of catalytic properties of Pt-nano-motors that are prepared with PtNPs modified with antibodies to induce the motion of microbeads in the presence of target virus. This study integrates bead motions and cellphone for the detection of viruses by using specifically designed Pt-nano-motors. The high sensitivity (1 particles/μL, S/N = 2) of the NBC system is attributed to the efficient catalytic activity known for the PtNPs used in the preparation of Pt-nanomotors in our study. We specifically used PtNPs with ∼4.4 nm in diameter in the preparation of nanomotors to allow maximum accumulation of nanoparticles on the surface of virus particles captured on the beads, which leads to efficient induction of the motion of beads even at low concentrations of viruses. We further controlled the ratio of anti-ZIKV monoclonal antibody at ∼1.8 antibody molecules per nanomotor to preserve the catalytic activity of the motors without affecting their efficiency to interact with captured viruses on the surface of the beads. This optimum antibody concentration per PtNP further prevents the formation of aggregates during assay. Due to the limitation in visualizing nanomotors (<1000 nm in all dimensions) using cellphones even with advanced optics, beads that are micrometer in size are used in the NBC system to allow visualization of the motion change using a low-cost cellphone-based optical sensor. In our system, we used 3 μm PS beads with a density of 1.1 g/cm3 to minimize the effect of gravity forces on the beads and to increase the efficient detection time. Large beads can be easily observed using a cellphone with the aid of simple optical accessories. However, on the negative side, larger beads can experience larger hydrodynamic resistance in the solution, which demands a higher amount of nanomotors to cause a significant and detectable bead motion change. Also, it was necessary to use highly uniform beads that are within ∼0.16 μm variation in size to avoid any effect on the velocity of beads because of size variation (Figure S18). It is worth mentioning that the use of microparticles allows a highly specific motion-based detection because of the absence of background signal from samples. This can further help the expansion of our system for point-of-care testing by eliminating the need for nanomotors separation or washing before motion testing with a cellphone. Furthermore, the capture of targets on beads has been known for long time to allow direct sample testing without the need for pretesting sample preparation steps, making the NBC system advantageous over the standard polymerase chain reaction (PCR)-based techniques currently recommended for ZIKV testing. Monoclonal antibodies that target the surface envelope protein and can recognize different ZIKV strains (PRVACB59, H/PF/ 2013, and MR 766, Figure S19) were used in the preparation of both nanomotors and virus-capturing beads to allow highly specific detection of ZIKV. A combination of monoclonal and polyclonal antibodies is commonly used in capturing and labeling steps of immunoassays. Here, we used a monoclonal antibody in the preparation of nanomotors and beads to limit the formation of Pt-nanomotor bead complex in the presence of surface antigen of the virus and to improve the efficiency of the developed system for virus particle detection, which is critical for accurate detection of ZIKV infection (Figure S20). In addition, our antibody immobilization scheme allows a directional conjugation of antibodies to the surface of beads and nanoparticles through their FC region. The directional conjugation of antibodies helps to preserve the full activity of antibodies. It also allows highly specific interaction with the target with high avidity due to the full accessibility of Fab regions that interact with the virus on the surface of particles. The long-term shelf life and stability of the disposables used in antibody-based point-of-care diagnostics are also important. Freeze-drying the surface chemistry can prolong the stability and shelf life of the disposables. Others have demonstrated long-term shelf life for target detection on plastic chips and showed that antibodies immobilized on-chip were stable for more than 200 days.45
CONCLUSIONS
Mobile health technologies have the great potential to transform the current paradigm in the management of infectious diseases particularly in resource-limited settings.46–50 The work reported in this manuscript shows how nanotechnology, microfluidics, and cellphones can be potentially used to effectively address early detection of Zika virus infection at the point-of-care. We have demonstrated the feasibility and usefulness of a cellphone system for motion-based detection of target viruses using the catalytic activity of nanomotors. The results of this study indicate that the NBC system can allow rapid and simple viral load testing and could potentially be used in immunoassays that require simple and rapid identification of viruses. The NBC system has the potential to be used for broad applications in infectious disease diagnosis and treatment monitoring.
METHODS
Virus Culture and Isolation.
Zika virus PRVABC59 isolated by the U.S. Center for Disease Control (CDC) from a ZIKV-infected patient who traveled to Puerto Rico in 2015 (NCBI accession no. KU501215) was used in this study. Virus stock was received from CDC and propagated in the Vero cell line c6/36 following the standard protocols.51 Cells were grown until confluence was reached. Then the growth medium was discarded, and fresh media was added and warmed up to 33 °C. Virus was then added to the cells and incubated at 5° angle for 1 h in the incubator at 33 °C. DMEM-5 was again added and incubated for 6 days at a slant angle of 20° in an incubator at 33 °C. The virus was harvested by centrifuging the cell culture media at 4000 × g for 30 min at 4 °C. The supernatant was then collected and aliquoted into separate vials containing 500 μL each.
Virus Purification and Quantification.
Zika virus particles were purified by centrifugation on sucrose gradients.52 24 mL of virus supernatant was loaded into an ultracentrifuge tube, and 7 mL of 20% sucrose solution was slowly added to the bottom of the tube. The tubes were then centrifuged for 3.5 h at 100,000 × g and 4 °C. Then the formed virus pellet dried upside-down inside the biosafety cabinet at room temperature for 20 min. The virus was suspended in DMEM-30 and quantified by RT-PCR using a Zika Real Time RT-PCR Kit (MyBiosource, Inc., San Diego, CA, USA).
Microchip Fabrication.
The microfluidic device consists of three layers: PMMA (3.175 mm; McMaster-Carr, 8560K239) that contains the inlets and outlets of microchannels, double-sided adhesive (DSA) sheet (80 mm; 3M, 82603) that includes a single microfluidic channel, and a glass slide (25 × 75 mm; Globe Scientific, NJ, USA). The microchip design was initially prepared using the vector graphics editor CorelDraw X7 software. Then, the DSA and PMMA were cut using the VLS 2.30 CO2 laser cutter (Universal Laser systems AZ) with the laser power, speed, and pulse per inch of 93%, 2.3%, and 1000, respectively, for PMMA and 20%, 15%, 500, respectively, for DSA. All the materials used in the microchip preparation, including PMMA, DSA, and glass slides, were cleaned with ethanol, H2O2, and DI water using lint-free tissues. The DSA was then peeled off of one side and was applied to the clean side of the PMMA. After ensuring that the DSA was added properly, the other side of the DSA was peeled off and was stuck on to the precleaned glass slide.
Nanomotor Preparation and Characterization.
Platinum nanomotors that specifically recognize ZIKV were prepared of spherical platinum nanoparticles (PtNPs) modified with monoclonal anti-Zika virus (ZIKV-Env) antibody (EastCoast Bio, Inc. North Berwick, ME, USA, cat no. HM325). The synthesis protocol begins with PtNPs synthesis followed by antibody coupling to the surface of the PtNPs. PtNPs were synthesized using a modified method from literature.53 All glassware used were cleaned with aqua regia and ultrapure water. 36 mL of a 0.2% solution of chloroplatinic acid hexahydrate was mixed with 464 mL of boiling DI water. 11 mL of a solution containing 1% sodium citrate and 0.05% citric acid was added followed by a quick injection of 5.5 mL of a freshly prepared 0.08% sodium borohydrate solution, containing 1% sodium citrate and 0.05% citric acid. The reaction continued for 10 min, and the formed nanoparticles solution was gradually cooled down to room temperature. The formed PtNPs were modified with 3-(2-pyridyldithio)-propionyl hydrazide (PDPH) freshly reduced by 20 mM tris(2-carboxyethyl)phosphine (TCEP). For antibody coupling reaction, aliquots of 5 μL of antibody (7 mg/mL) were mixed with 10 mM of sodium metaperiodate and 0.1 M sodium acetate (pH 5.5) and incubated at 4 °C in the dark for 20 min. The oxidized antibody was washed by using filtration column unit (Amicon Ultra-15 Centrifugal Filter Unit, cat. no. UFC903008) and then added to PDPH activated PtNPs and allowed to react with the oxidized antibody for 1 h at room temperature. The formed Pt-nanomotors were washed by a dialysis membrane using phosphate buffer for 3 h with mild stirring at 4 °C. The prepared PtNPs and Pt-nanomotors were characterized using transmission electron microscopy (TEM), ultraviolet–visible (UV–vis) spectroscopy, Fourier transform-infrared spectroscopy (FT-IR), ζ potential, and dynamic light scattering (DLS).
Beads Modification and Characterization.
ZIKV was captured on the surface of 3 μm PS beads and labeled with Pt-nanomotors. The protocol used for this step involves three main reactions: (1) Polystyrene beads activation with adipic acid. In this reaction, 20 μL of Sperotech-SPHERO carboxyl beads with 1% w/v was diluted in 200 μL of 0.05 M 2-(N-morpholino)ethanesulfonic acid (MES) pH 5.0, then activated using EDC-NHS coupling reaction by adding 100× molar concentration of adipic acid dihydrazide. The reaction mixture was incubated at room temperature with agitation for 20 min. After the reaction, the activated beads were washed twice with MES buffer. (2) Anti-ZIKV monoclonal envelope antibody oxidation using sodium periodate following the described protocol in the previous section. (3) Oxidized coupling to the surface of hydrazide beads. The surface area of beads was calculated and the concentration of antibody was adjusted in a way that it covers 20%, 40%, 80%, and 100% of the beads. After optimization, the ratio of antibody covering the beads was optimized to be 40%. Antibody was added to the activated beads and was incubated for 2.5 h on the shaker 150 rpm. Excess antibody was washed twice. PBS was used as storage buffer for the modified beads and kept in dark at 4 °C. The prepared beads modified with anti-ZIKV antibody were characterized using UV–vis spectroscopy and FT-IR techniques.
Bead Motion Cellphone Assay.
The NBC system assay relies on the induction of the bead motion in the presence of target virus due to the formed bead-virus-PtNP complex. The working protocol comprises three main steps: (1) Virus capture on the surface of beads. 5 μL of the antibody-modified beads were added to a 1.5 mL centrifuge tube, 10 μL of ZIKV was added, and the final volume was made up to 100 μL with 100 mM phosphate buffer (pH 7.2). The sample was incubated for 20 min with mild shaking (150 rpm) at room temperature and washed twice with phosphate buffer to remove all non-captured viruses from the sample. (2) Pt-beads-virus complex formation. 20 μL of prepared Pt-nanomotors were added to the centrifuge tube and incubated for 20 min with mild mixing. The sample was washed 3 times using phosphate buffer to remove all free nanomotors. (3) Motion testing using the cellphone system. H2O2 solution (30%) was mixed with equal volume of the prepared Pt-bead-virus complex solution. 10 μL of the mixture was loaded on the microfluidic device, and the motion of the beads was measured using the developed cellphone system. The capture of ZIKV on the surface of beads was confirmed using SDS gel electrophoresis and SEM techniques. The induction of the beads motion in the presence of ZIKV was initially tested using bright-field light microscopy technique. Videos of virus-free control and ZIKV-spiked samples (106 particles/μL) were recorded under light microscopy using Snagit at 10 frames per second. Then videos were analyzed using ImageJ and MtrackJ plug-in to calculate the velocities of beads.
Detection Performance of the NBC System.
We evaluated the sensitivity of the NBC system using serially diluted ZIKV-spiked PB samples with virus concentrations ranging from 100 particles/μL to 106 particles/μL. 10 μL of each virus concentration was tested using our bead motion testing protocol, and 10 μL of the formed reaction mixture was loaded into the microchip and were immediately tested with the cellphone. This process was repeated for all of the samples with different virus concentrations. One positive control with ZIKV and without nanomotors was included in all of the three trials. The specificity of the developed NBC assay was tested using ZIKV and non-target viruses, including DENV-1, DENV −2, HCMV, and HSV-1 at 106 particles/μL using the same protocol.
Cellphone Optical System and Software.
The cellphone setup was designed using Solid Works 2015 software and 3D printed with a 3D printer (Ultimaker Extended II) using Ultimaker PLA (polylactic acid) as printing material. The setup was designed to record the videos using the cellphone rear camera. The optical cellphone attachment has an LED, electronics and switches, and lenses for image magnification. The electronic switch on the optical system is used to turn on and off the light source when needed. A Moto X smartphone (Motorola, XT1575) was used in this work. A microchip holder was engraved on the cellphone optical attachment for microchip manipulation and positioning. The cellphone application was designed using Android Studio. The cellphone application records a video of the sample for 2 min at 30 frames per second. The detection algorithm identifies the beads and tracks their motion to calculate the velocities. The virus concentration is calculated based on the bead motion change in the sample. The cellphone application is enabled with a user-friendly interface that can be operated by a lay user.
System Evaluation Using Spiked and ZIKV-Infected Patient Samples.
To evaluate the NBC system, we used ZIKV-spiked synthetic urine and artificial saliva samples with virus concentrations of 101 particles/μL, 103 particles/μL, and 105 particles/μL. ZIKV-infected serum patient samples (n = 10) purchased from Boca Biolistics, LLC (Pompano Beach, FL, USA) were also used for system evaluation. Each spiked sample was tested using our bead motion testing protocol for performance testing of NBC system.
Statistical Analyses.
Statistical analyses were performed using OriginPro 2015 (OriginLab Corporation, Northampton, USA) and GraphPad Prism software version 5.01 (GraphPad Software, Inc. La Jolla, CA, USA). Data were collected and analyzed using software, and each data point represents the average of a total of three independent measurements.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Donald Coen, Seamus McCarron, Sambeeta Das, and Francoise Giguel. Research reported in this publication was partially supported by the National Institute of Health under award numbers R01AI118502, R21HD092828, and P30ES000002; Harvard T.H. Chan School of Public Health, Harvard Center for Environmental Health through Harvard NIEHS grant; American Board of Obstetrics and Gynecology, American College of Obstetricians and Gynecol-ogists, American Society for Reproductive Medicine, Society for Reproductive Endocrinology and Infertility through ASRM award; and Harvard University Center for AIDS Research (CFAR) under award number 5P30AI060354–14.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.8b01515.
Detailed methods for UV–vis, TEM, SEM, FT-IR, ICP-MS, and gel electrophoresis analysis, microchip fabrication, optical attachment design and structure, Pt-nanomotors preparation protocols and characterization data, beads motion analysis, and material cost (PDF)
Movie S1 (AVI)
Movie S2 (AVI)
Movie S3 (AVI)
Movie S4 (AVI)
Movie S5 (AVI)
Movie S6 (AVI)
Movie S7 (AVI)
Movie S8 (AVI)
Movie S9 (AVI)
The authors declare no competing financial interest.
REFERENCES
- (1).Cugola FR; Fernandes IR; Russo FB; Freitas BC; Dias JLM; Guimarães KP; Benazzato C; Almeida N; Pignatari GC; Romero S; Polonio CM; Cunha I; Freitas CL; Brandão WN; Rossato C; Andrade DG; Faria DP; Garcez AT; Buchpigel CA; Braconi CT; et al. The Brazilian Zika Virus Strain Causes Birth Defects in Experimental Models. Nature 2016, 534, 267–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Rasmussen SA; Jamieson DJ; Honein MA; Petersen LR. Zika Virus and Birth Defects Reviewing the Evidence for Causality. N. Engl. J. Med 2016, 374, 1981–1987. [DOI] [PubMed] [Google Scholar]
- (3).Mlakar J; Korva M; Tul N; Popović M; Poljšak-Prijatelj M; Mraz J; Kolenc M; Resman Rus K; Vesnaver Vipotnik T; Fabjan Vodušek V; Vizjak A; Pižem J; Petrovec M; Županc TA. Zika Virus Associated with Microcephaly. N. Engl. J. Med 2016, 374, 951–958. [DOI] [PubMed] [Google Scholar]
- (4).Fauci AS; Morens DM. Zika Virus in the Americas Yet Another Arbovirus Threat. N. Engl. J. Med 2016, 374, 601–604. [DOI] [PubMed] [Google Scholar]
- (5).Saxena SK; Elahi A; Gadugu S; Prasad AK. Zika Virus Outbreak: An Overview of the Experimental Therapeutics and Treatment. VirusDisease 2016, 27, 111–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Riehemann K; Schneider SW; Luger TA; Godin B; Ferrari M; Fuchs H. Nanomedicine - Challenge and Perspectives. Angew. Chem., Int. Ed 2009, 48, 872–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Draz MS; Shafiee H. Applications of Gold Nanoparticles in Virus Detection. Theranostics 2018, 8, 1985–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Draz MS; Fang BA; Zhang P; Hu Z; Gu S; Weng KC; Gray JW; Chen FF. Nanoparticle-Mediated Systemic Delivery of siRNA for Treatment of Cancers and Viral Infections. Theranostics 2014, 4, 872–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Wang J. Nanomachines: Fundamentals and Applications; Wiley: New York, 2013. [Google Scholar]
- (10).Ismagilov RF; Schwartz A; Bowden N; Whitesides GM. Autonomous Movement and Self-Assembly. Angew. Chem., Int. Ed 2002, 41, 652–654. [Google Scholar]
- (11).Solovev AA; Mei Y; Ureña EB; Huang G; Schmidt OG. Catalytic Microtubular Jet Engines Self-Propelled by Accumulated Gas Bubbles. Small 2009, 5, 1688–1692. [DOI] [PubMed] [Google Scholar]
- (12).Lavrentovich OD; Lazo I; Pishnyak OP. Nonlinear Electrophoresis of Dielectric and Metal Spheres in a Nematic Liquid Crystal. Nature 2010, 467, 947–950. [DOI] [PubMed] [Google Scholar]
- (13).Wang W; Duan W; Ahmed S; Mallouk TE; Sen A. Small Power: Autonomous Nano- and Micromotors Propelled by Self-Generated Gradients. Nano Today 2013, 8, 531–554. [Google Scholar]
- (14).Illien P; Golestanian R; Sen A. Fuelled” Motion: Phoretic Motility and Collective Behaviour of Active Colloids. Chem. Soc. Rev 2017, 46, 5508–5518. [DOI] [PubMed] [Google Scholar]
- (15).Ceylan H; Giltinan J; Kozielski K; Sitti M. Mobile Microrobots for Bioengineering Applications. Lab Chip 2017, 17, 1705–1724. [DOI] [PubMed] [Google Scholar]
- (16).Chałupniak A; Morales-Narváez E; Merkoçi A. Micro and Nanomotors in Diagnostics. Adv. Drug Delivery Rev 2015, 95, 104–116. [DOI] [PubMed] [Google Scholar]
- (17).Fu X; Chen B; Tang J; Zewail AH. Photoinduced Nanobubble-Driven Superfast Diffusion of Nanoparticles Imaged by 4D Electron Microscopy. Sci. Adv 2017, 3, e1701160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Wang H; Zhao G; Pumera M. Beyond Platinum: Bubble-Propelled Micromotors Based on Ag and MnO 2 Catalysts. J. Am. Chem. Soc 2014, 136, 2719–2722. [DOI] [PubMed] [Google Scholar]
- (19).Zhang H; Duan W; Lu M; Zhao X; Shklyaev S; Liu L; Huang TJ; Sen A. Self-Powered Glucose-Responsive Micropumps. ACS Nano 2014, 8, 8537–8542. [DOI] [PubMed] [Google Scholar]
- (20).Dong B; Zhou T; Zhang H; Li CY. Directed Self-Assembly of Nanoparticles for Nanomotors. ACS Nano 2013, 7, 5192–5198. [DOI] [PubMed] [Google Scholar]
- (21).Wong F; Dey KK; Sen A. Synthetic Micro/Nanomotors and Pumps: Fabrication and Applications. Annu. Rev. Mater. Res 2016, 46, 407–432. [Google Scholar]
- (22).Gao W; Kagan D; Pak OS; Clawson C; Campuzano S; Chuluun-Erdene E; Shipton E; Fullerton EE; Zhang L; Lauga E; Wang J. Cargo-Towing Fuel-Free Magnetic Nanoswimmers for Targeted Drug Delivery. Small 2012, 8, 460–467. [DOI] [PubMed] [Google Scholar]
- (23).Gao W; Wang J. Synthetic Micro/nanomotors in Drug Delivery. Nanoscale 2014, 6, 10486–10494. [DOI] [PubMed] [Google Scholar]
- (24).Ma X; Zhao Y; Ng KW; Zhao Y. Integrated Hollow Mesoporous Silica Nanoparticles for Target drug/siRNA Co-Delivery. Chem. - Eur. J 2013, 19, 15593–15603. [DOI] [PubMed] [Google Scholar]
- (25).Patra D; Sengupta S; Duan W; Zhang H; Pavlick R; Sen A. Intelligent, Self-Powered, Drug Delivery Systems. Nanoscale 2013, 5, 1273–1283. [DOI] [PubMed] [Google Scholar]
- (26).Wu J; Balasubramanian S; Kagan D; Manesh KM; Campuzano S; Wang J. Motion-Based DNA Detection Using Catalytic Nanomotors. Nat. Commun 2010, 1, 36. [DOI] [PubMed] [Google Scholar]
- (27).Orozco J; Cortés A; Cheng G; Sattayasamitsathit S; Gao W; Feng X; Shen Y; Wang J. Molecularly Imprinted Polymer-Based Catalytic Micromotors for Selective Protein Transport. J. Am. Chem. Soc 2013, 135, 5336–5339. [DOI] [PubMed] [Google Scholar]
- (28).Kagan D; Campuzano S; Balasubramanian S; Kuralay F; Flechsig GU; Wang J. Functionalized Micromachines for Selective and Rapid Isolation of Nucleic Acid Targets from Complex Samples. Nano Lett. 2011, 11, 2083–2087. [DOI] [PubMed] [Google Scholar]
- (29).He Q; Shi J. Mesoporous Silica Nanoparticle Based Nano Drug Delivery Systems: Synthesis, Controlled Drug Release and Delivery, Pharmacokinetics and Biocompatibility. J. Mater. Chem 2011, 21, 5845. [Google Scholar]
- (30).Xu T; Soto F; Gao W; Dong R; Garcia-Gradilla V; Magaña E; Zhang X; Wang J. Reversible Swarming and Separation of Self-Propelled Chemically Powered Nanomotors under Acoustic Fields. J. Am. Chem. Soc 2015, 137, 2163–2166. [DOI] [PubMed] [Google Scholar]
- (31).Campuzano S; Orozco J; Kagan D; Guix M; Gao W; Sattayasamitsathit S; Claussen JC; Merkoçi A; Wang J. Bacterial Isolation by Lectin-Modified Microengines. Nano Lett. 2012, 12, 396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Balasubramanian S; Kagan D; Hu CMJ; Campuzano S; Lobo-Castañon MJ; Lim N; Kang DY; Zimmerman M; Zhang L; Wang J. Micromachine-Enabled Capture and Isolation of Cancer Cells in Complex Media. Angew. Chem., Int. Ed 2011, 50, 4161–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Ma X; Hahn K; Sanchez S. Catalytic Mesoporous Janus Nanomotors for Active Cargo Delivery. J. Am. Chem. Soc 2015, 137, 4976–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Campuzano S; Kagan D; Orozco J; Wang J. Motion-Driven Sensing and Biosensing Using Electrochemically Propelled Nano-motors. Analyst 2011, 136, 4621. [DOI] [PubMed] [Google Scholar]
- (35).Van Nguyen K; Minteer SD. DNA-Functionalized Pt Nanoparticles as Catalysts for Chemically Powered Micromotors: Toward Signal-on Motion-Based DNA Biosensor. Chem. Commun 2015, 51, 4782–4784. [DOI] [PubMed] [Google Scholar]
- (36).Yu X; Li Y; Wu J; Ju H. Motor-Based Autonomous Microsensor for Motion and Counting Immunoassay of Cancer Biomarker. Anal. Chem 2014, 86, 4501–4507. [DOI] [PubMed] [Google Scholar]
- (37).Vilela D; Orozco J; Cheng G; Sattayasamitsathit S; Galarnyk M; Kan C; Wang J; Escarpa A. Multiplexed Immunoassay Based on Micromotors and Microscale Tags. Lab Chip 2014, 14, 3505. [DOI] [PubMed] [Google Scholar]
- (38).Morales-Narváez E; Guix M; Medina-Sánchez M; Mayorga-Martinez CC; Merkoçi A. Micromotor Enhanced Microarray Technology for Protein Detection. Small 2014, 10, 2542–2548. [DOI] [PubMed] [Google Scholar]
- (39).García M; Orozco J; Guix M; Gao W; Sattayasamitsathit S; Escarpa A; Merkoçi A; Wang J. Micromotor-Based Lab-on-Chip Immunoassays. Nanoscale 2013, 5, 1325–1331. [DOI] [PubMed] [Google Scholar]
- (40).Bhardwaj SK; Bhardwaj N; Mohanta GC; Kumar P; Sharma AL; Kim KH; Deep A. Immunosensing of Atrazine with Antibody-Functionalized Cu-MOF Conducting Thin Films. ACS Appl. Mater. Interfaces 2015, 7, 26124–26130. [DOI] [PubMed] [Google Scholar]
- (41).Chandra S; Gupta LK. EPR, Mass, IR, Electronic, and Magnetic Studies on copper(II) Complexes of Semicarbazones and Thiosemicarbazones. Spectrochim. Acta, Part A 2005, 61, 269–275. [DOI] [PubMed] [Google Scholar]
- (42).Draz MS; Fang BA; Li L; Chen Z; Wang Y; Xu Y; Yang J; Killeen K; Chen FF. Hybrid Nanocluster Plasmonic Resonator for Immunological Detection of Hepatitis B Virus. ACS Nano 2012, 6, 7634–7643. [DOI] [PubMed] [Google Scholar]
- (43).Brecher M; Li Z; Liu B; Zhang J; Koetzner CA; Alifarag A; Jones SA; Lin Q; Kramer LD; Li H. A Conformational Switch High-Throughput Screening Assay and Allosteric Inhibition of the Flavivirus NS2B-NS3 Protease. PLoS Pathog. 2017, 13, e1006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Wong SSY; Poon RWS; Wong SCY. Zika Virus Infection-the next Wave after Dengue? J. Formosan Med. Assoc 2016, 115, 226–242. [DOI] [PubMed] [Google Scholar]
- (45).Chin CD; Laksanasopin T; Cheung YK; Steinmiller D; Linder V; Parsa H; Wang J; Moore H; Rouse R; Umviligihozo G; Karita E; Mwambarangwe L; Braunstein SL; van de Wijgert J; Sahabo R; Justman JE; El-Sadr W; Sia SK. Microfluidics-Based Diagnostics of Infectious Diseases in the Developing World. Nat. Med 2011, 17, 1015–1019. [DOI] [PubMed] [Google Scholar]
- (46).Romeo A; Leung TS; Sánchez S. Smart Biosensors for Multiplexed and Fully Integrated Point-of-Care Diagnostics. Lab Chip 2016, 16, 1957–1961. [DOI] [PubMed] [Google Scholar]
- (47).Mudanyali O; Mcleod E; Luo W; Greenbaum A; Coskun AF; Hennequin Y; Allier CP; Ozcan A. Wide-Field Optical Detection of Nanoparticles Using on-Chip Microscopy and Self-Assembled Nanolenses. Nat. Photonics 2013, 7, 247–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Laksanasopin T; Guo TW; Nayak S; Sridhara AA; Xie S; Olowookere OO; Cadinu P; Meng F; Chee NH; Kim J; Chin CD; Munyazesa E; Mugwaneza P; Rai AJ; Mugisha V; Castro AR; Steinmiller D; Linder V; Justman JE; Nsanzimana S; et al. A Smartphone Dongle for Diagnosis of Infectious Diseases at the Point of Care. Sci. Transl. Med 2015, 7, 273re1–273re1. [DOI] [PubMed] [Google Scholar]
- (49).Kanakasabapathy MK; Pandya HJ; Draz MS; Chug MK; Sadasivam M; Kumar S; Etemad B; Yogesh V; Safavieh M; Asghar W; Li JZ; Tsibris AM; Kuritzkes DR; Shafiee H. Rapid, Label-Free CD4 Testing Using a Smartphone Compatible Device. Lab Chip 2017, 17, 2910–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Kanakasabapathy MK; Sadasivam M; Singh A; Preston C; Thirumalaraju P; Venkataraman M; Bormann CL; Draz MS; Petrozza JC; Shafiee H. An Automated Smartphone-Based Diagnostic Assay for Point-of-Care Semen Analysis. Sci. Transl. Med 2017, 9, eaai7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Lanciotti RS; Lambert AJ; Holodniy M; Saavedra S; del Carmen Castillo Signor L. Phylogeny of Zika Virus in Western Hemisphere, 2015. Emerging Infect. Dis 2016, 22, 933–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Medina F; Medina JF; Colon C; Vergne E; Santiago GA; Munoz-Jordan JL. Dengue Virus: Isolation, Propagation, Quantification, and Storage. Curr. Protoc. Microbiol 2012, 27, 15D.2.1–15D.2.24. [DOI] [PubMed] [Google Scholar]
- (53).Bigall NC; Härtling T; Klose M; Simon P; Eng LM; Eychmuüller, A. Monodisperse Platinum Nanospheres with Adjustable Diameters from 10 to 100 Nm: Synthesis and Distinct Optical Properties. Nano Lett. 2008, 8, 4588–4592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.