Abstract
Catecholamines stimulate the first step of lipolysis by PKA-dependent release of the lipid droplet-associated protein ABHD5 from perilipin to co-activate the lipase ATGL. Here, we unmask a yet unrecognized proteolytic and cardioprotective function of ABHD5. ABHD5 acts in vivo and in vitro as a serine protease cleaving HDAC4. Through the production of an N-terminal polypeptide of HDAC4 (HDAC4-NT), ABHD5 inhibits MEF2-dependent gene expression and thereby controls glucose handling. ABHD5-deficiency leads to neutral lipid storage disease in mice. Cardiac-specific gene therapy of HDAC4-NT does not protect from intra-cardiomyocyte lipid accumulation but strikingly from heart failure, thereby challenging the concept of lipotoxicity-induced heart failure. ABHD5 levels are reduced in failing human hearts and murine transgenic ABHD5 expression protects from pressure-overload induced heart failure. These findings represent a conceptual advance by connecting lipid with glucose metabolism through HDAC4 proteolysis and enable new translational approaches to treat cardiometabolic disease.
Introduction
The effects of catecholamines through β-adrenergic receptors on cardiomyocytes and post-receptor signaling via protein kinase A (PKA) and Ca2+/Calmodulin-dependent kinase II (CaMKII) have been extensively studied with regard to physiological adaptation and pathological maladaptation.1,2 In the heart, most studies focused on the consequences of β1- and β2-adrenergic receptor activation on cellular processes such as Ca2+ handling, regulation of ion channels, sarcomere function or transcription.1,3 In adipocytes, the effects of catecholamines on lipid metabolism are well known but these have been less studied in cardiomyocytes.4–6 Such as, it has been demonstrated that the lipid droplet associated proteins perilipin 5 (PLIN5) and abhydrolase domain containing 5 (ABHD5), also known as comparative gene identification-58 (CGI-58), and adipose triglyceride lipase (ATGL) respond to PKA.7–10 PLIN5 and ABHD5 are both phosphorylated by PKA, resulting in dissociation of ABHD5 from PLIN5, leading to the activation of ATGL that initiates the first step of lipolysis, the transition from triacylglycerol (TAG) to diacylglycerol (DAG).11,12 Strikingly, human mutations of the genes encoding for ABHD5 and ATGL lead to a rare and deadly syndrome, termed neutral lipid storage disease (NLSD), that represents an unmet medical need.7,13,14 NLSD (at least in the case of ATGL) is among other symptoms characterized by severe cardiomyopathy and often requires heart transplantation. The patient’s phenotype could impressively be recapitulated in mice lacking ATGL or ABHD5, indicating that these two lipid droplet-associated proteins are essential for cardiac integrity and function.7,8,15 Based on the reported lipid accumulation in ABHD5 or ATGL deficient mice it is assumed that a lack of lipolysis-derived byproducts leads to a loss of mitochondrial oxidative capacity and by this drives the cardiomyopathic phenotype including contractile failure.7,8 However, the causal relationship between lipid accumulation and cardiac dysfunction is not formally proven.
Catecholaminergic stimulation of β-adrenergic receptors on cardiomyocytes leads not only to activation of PKA but also CaMKII.2 Short-term β-adrenergic stimulation during physiological adaptations favors activation of PKA, whereas sustained stimulation under pathological conditions leads to a shift towards CaMKII activation.16,17 Both kinases converge selectively on the epigenetic regulator histone deacetylase 4 (HDAC4).18–21 CaMKII binds and phosphorylates HDAC4 specifically at serine residue 632, thereby promoting cytosolic accumulation and dissociation from the transcriptional activator myocyte enhancer factor 2 (MEF2) as well as inhibiting proteolysis of HDAC4.18,21 The dissociation of the MEF2-HDAC4 complex permits MEF2 to activate a gene program that activates the hexosamine biosynthesis pathway and calcium mishandling, eventually leading to heart failure.20,22 Conversely, we could demonstrate that PKA triggers proteolytic processing of HDAC4 through a serine protease, resulting in an N-terminal HDAC4 polypeptide. This peptide was named HDAC4-NT and acts as a CaMKII-resistant selective MEF2 inhibitor, resulting in protective effects on cultured cardiomyocytes and diabetic hearts.19–21 But so far, the identity of the PKA-sensitive serine protease that cleaves HDAC4 has remained unknown (Fig. 1a).
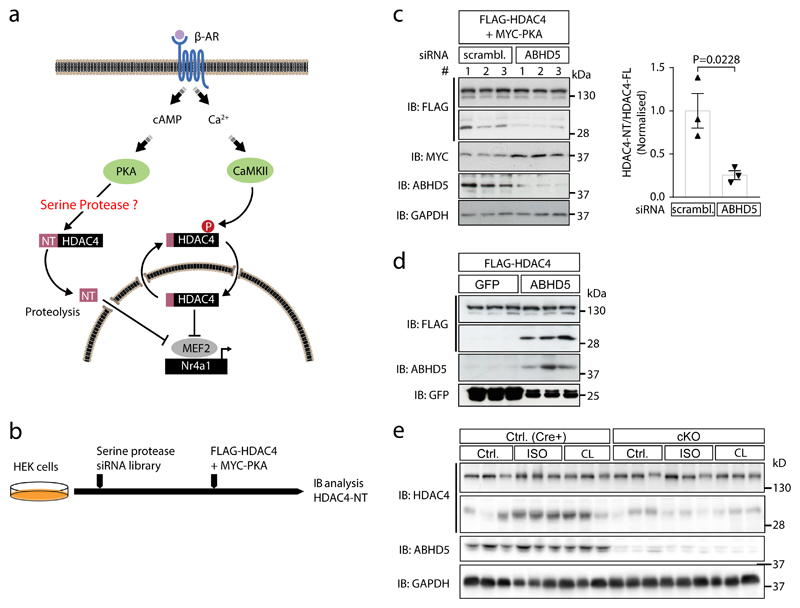
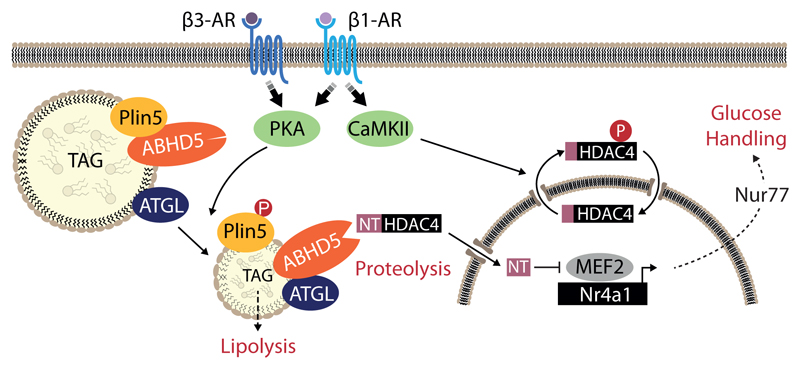
Fig. 1. ABHD5 is required for HDAC4 proteolysis.
(a) Current working model: CaMKII induces cytosolic accumulation of HDAC4 by phosphorylation, whereas PKA activates a so far unknown and here to be identified serine protease that cleaves off an N-terminal (NT) polypeptide of HDAC4. HDAC4-NT inhibits the transcription factor MEF2 in a CaMKII-resistant manner and represses genes such as Nr4a1 that is involved in the pathogenesis of heart failure. (b) siRNA-based protease screening strategy: Known and predicted serine proteases were silenced in HEK293 cells by transfection with a siRNA library. Subsequently, FLAG-HDAC4 and MYC-PKA were transfected to induce HDAC4-NT production. Cell lysates were processed for immunoblotting analysis to assess the degree of HDAC4 proteolysis and to detect MYC-PKA input. (c) HEK293 cells pre-transfected for 36 hrs either with three different siRNAs targeting ABHD5 or control scrambled siRNA (# 1, 2, 3) were transfected with expression plasmids encoding FLAG-HDAC4 and MYC-PKA. 24 hrs post plasmid transfection, cell lysates were prepared and analyzed by immunoblotting using antibodies directed against FLAG- and MYC-tags as well as endogenous ABHD5 and GAPDH as knockdown and loading control, respectively. Corresponding band intensities were quantified and the ratio between HDAC4-NT and HDAC4-FL was determined. Statistical analysis: Values are presented as Mean±SEM, n=3 (n represents independent samples); by unpaired two tailed t-test, P<0.05 considered as significant. (d) NRVMs were transduced with adenoviruses encoding FLAG-HDAC4 and ABHD5 plus GFP on a bicistronic promoter or GFP, and protein expression was detected using an antibody directed against FLAG, ABHD5 and GFP. The results show a representative blot of three independent experiments (e) Control mice (Cre) and cardiac-specific ABHD5-KO mice (cKO) were treated without or with isoproterenol (ISO) or the specific β3-adrenergic receptor agonist CL 316,243 (CL). Cardiac extracts were analyzed using antibodies directed against HDAC4 and ABHD5. An antibody against GAPDH was used as loading control. The results show a representative blot of two independent experiments.
Results
ABHD5 is required for HDAC4 proteolysis
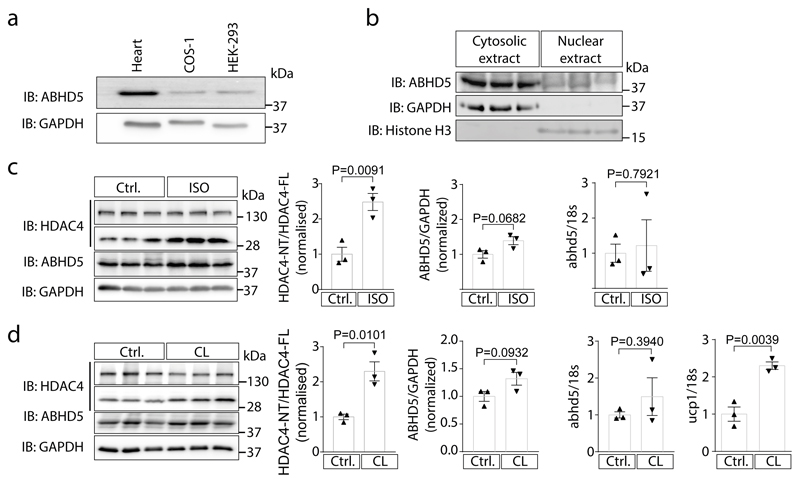
Our previous data using group-specific chemical protease inhibitors suggested that a serine protease is required for PKA-dependent HDAC4 proteolysis.19 Thus, we screened a human serine protease siRNA library that targets known and predicted serine proteases (Supplementary Data 1). We assayed for the level of HDAC4-NT production as a result of PKA and HDAC4 co-expression in the presence of different siRNAs (Fig. 1b). The screen and the subsequent validation identified ABHD5 to be required for PKA-dependent HDAC4 proteolysis in HEK cells (Fig. 1c). ABHD5 is broadly expressed in cell systems that have been used previously to study production of HDAC4-NT, but in relation we found also high levels in cardiac tissue (Extended Data Fig. 1a). ABHD5 localizes predominantly to the cytoplasm (Extended Data Fig. 1b) compared to the nucleus, suggesting that it could regulate cytosolic HDAC4 upon nucleo-cytoplasmic shuttling.19 Adenoviral gene delivery of ABHD5 and HDAC4 resulted also in HDAC4-NT production in neonatal rat ventricular myocytes (NRVMs) (Fig. 1d). In adipocytes, lipolysis is driven by activation of β3-adrenergic receptors.23 Thus, we tested whether β-adrenergic stimulation by isoproterenol (ISO) and the specific β3-adrenergic receptor agonist CL 316,243 results in production of endogenous HDAC4-NT in vivo, and we found indeed that both stimulation protocols induced HDAC4 proteolysis in the heart. (Extended Data Fig. 1c-d). Next, we stimulated adrenergic receptors in adult cardiac-specific ABHD5-deficient mice to test whether ABHD5 is required for HDAC4 proteolysis in vivo. Because mice with a global deletion of ABHD5 show early postnatal lethality,15 we generated an inducible cardiac-specific ABHD5 knockout model (cKO). Strikingly, both ISO and CL 316,243 induced HDAC4-NT production in Cre expressing control mice (Cre) but not in cKO, indicating that ABHD5 is required for stress-induced HDAC4 proteolysis (Fig. 1e).
ABHD5 possesses intrinsic serine protease activity
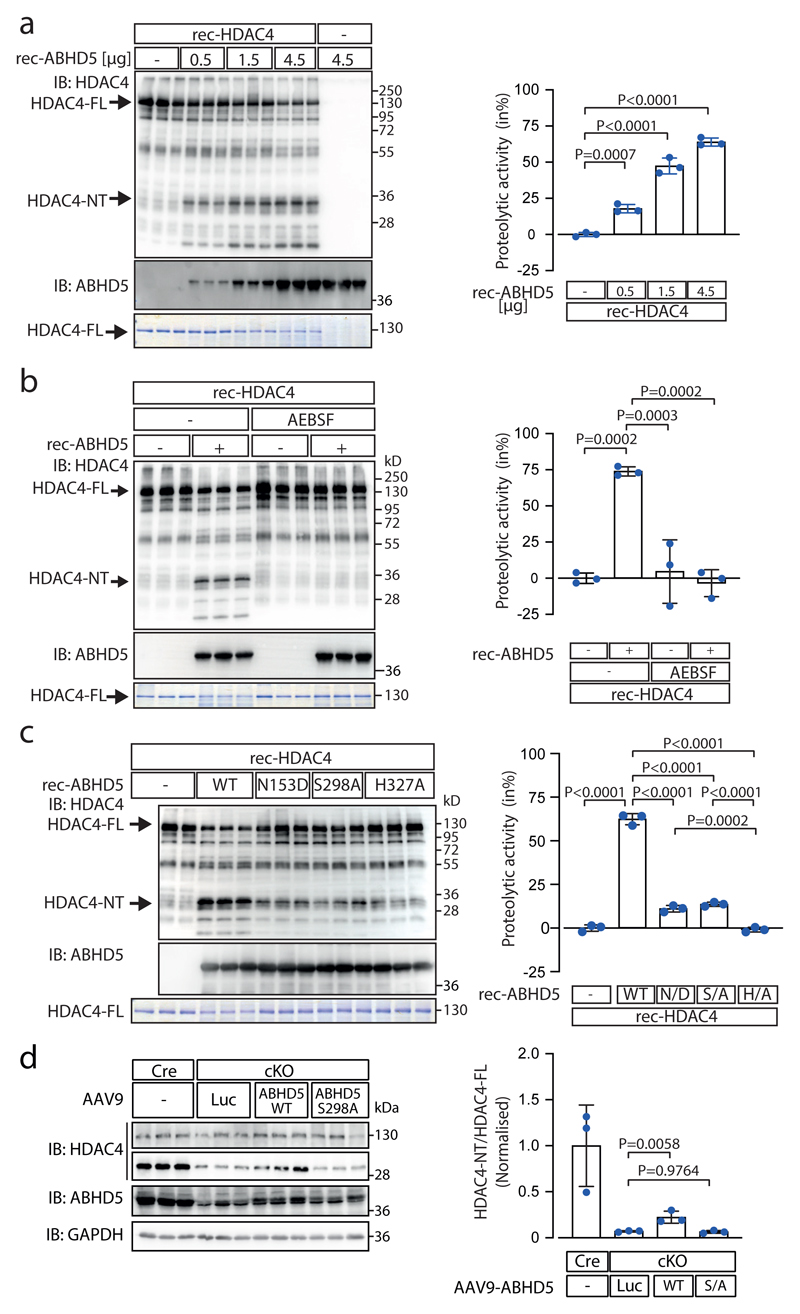
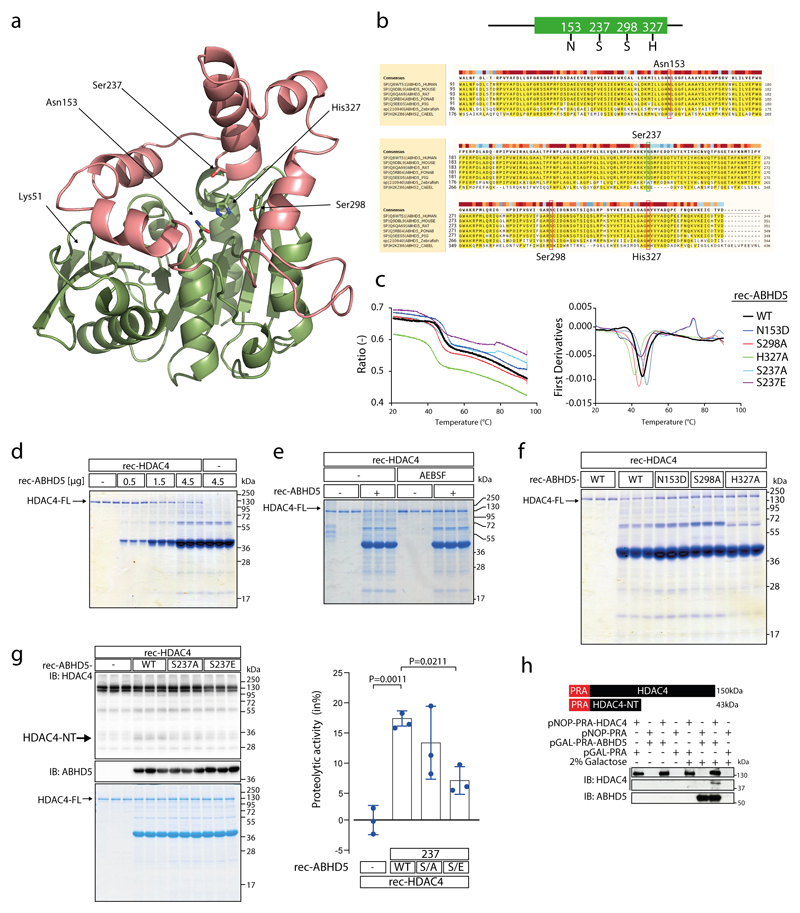
Based on the MEROPS peptidase database (https://www.ebi.ac.uk/merops/cgi-bin/pepsum?id=S33.975;type=peptidase) ABHD5 is predicted to act as a serine protease with amino acid (aa) residues Asn153, Ser298 and His327 building a catalytic triad. Based on a 3D model derived from the protein structure homology-modelling server SWISS-MODEL these aa residues form an arrangement somewhat reminiscent to a serine protease catalytic triad (Extended Data Fig. 2a; based on PDB file: Supplementary Data 2). Multiple sequence alignment of ABHD5 proteins from different species shows that Asn153, Ser298 and His327 and the surrounding aa residues are highly conserved in higher vertebrates (Extended Data Fig. 2b). Since the vertebrate stage the asparagine at 153 has replaced a serine within the canonical esterase/lipase motif ‘GXSXG’ of a related sub-family of proteins with intrinsic esterase/lipase/thioesterase activity,10,14 provoking the question whether with the loss of its intrinsic lipase activity ABHD5 may have gained serine protease activity. However, to the best of our knowledge this question has not been investigated yet experimentally. Therefore, we conducted a series of in vitro assays using recombinant WT and mutant ABHD5 (rec-ABHD5) and recombinant HDAC4 (rec-HDAC4) proteins to test whether ABHD5 possesses intrinsic serine protease activity. To assess similar protein folding, thermal stability of WT and mutant rec-ABHD5 was compared by nanoDSF (Tm: WT=47.4°C, Asn153Asp=49.8°C, Ser298Ala=45.4°C, His327Ala =42.9°C; Extended Data Fig. 2c). With increasing concentration of rec-ABHD5, full length rec-HDAC4 (HDAC4-FL) decreased and HDAC4-NT increased (Fig. 2a, Extended Data Fig. 2d). The relative loss of HDAC4-FL was used as a measure to quantify ABHD5’s protease activity. Strikingly, HDAC4 proteolysis was completely prevented by the serine protease inhibitor AEBSF, further corroborating that ABHD5 indeed possesses intrinsic serine protease activity (Fig. 2b, Extended Data Fig. 2e). Site directed mutagenesis of Asn153, Ser298 and His327 revealed that the aa of the predicted catalytic triad all contribute to ABHD5’s protease activity (Fig. 2c, Extended Data Fig. 2f). Whereas the histidine mutation abolished the protease activity almost completely, the asparagine and serine mutations reduced the protease activity strongly. Consistently, in vivo replacement of ABHD5 in cKO with ABHD5-WT but not ABHD5-S298A significantly increased HDAC4 proteolysis in vivo (Fig. 2d). Because it was reported that PKA phosphorylates mouse ABHD5 at Ser239 (homologous to human ABHD5 at Ser-237),6 we hypothesized that this site may affect ABHD5’s protease function in a PKA-dependent manner. Thus, we generated Ser-237 phospho-death (S237A) and phospho-mimetic (S237E) rec-ABHD5 mutants and found that both mutations reduced HDAC4-NT production (Extended Data Fig. 2g). We cannot rule out that the phospho-mimetic is not functional or properly folded (Tm: Ser237A=47.3°C, Ser235Glu=46.2°C) but the predicted 3D structure indicates that Ser237 (that is also highly conserved among vertebrates) lies in close proximity to Ser298 (Extended Data Fig. 2a-b), calling for further investigations whether Ser-237 may interfere with the proteolytic activity of ABHD5. For additional experimental validation, we co-expressed HDAC4 together with inducible ABHD5 in yeast. Induction of ABHD5 expression in the presence of constitutively expressed HDAC4 in yeast led again to HDAC4-NT production (Extended Data Fig. 2h). These data collectively support that ABHD5 possesses intrinsic protease activity but do not rule out that other domains or co-factors contribute to its full protease function. More studies (including crystallography) are needed to fully understand the features of the newly identified serine protease function of ABHD5.
Fig. 2. ABHD5 possesses intrinsic serine protease activity.
(a) Recombinant purified HDAC4 (rec-HDAC4) was incubated with increasing amounts of recombinant purified ABHD5 (rec-ABHD5) for 4 hrs at 37 °C. Proteolysis was analyzed by immunoblotting using antibodies directed against HDAC4 and by Coomassie staining, detecting absolute changes of full length HDAC4 (HDAC4-FL). rec-ABHD5 was detected by immunoblotting and Coomassie staining. The amounts of HDAC4-FL were determined in the Coomassie gel by densitometry. The ratio of HDAC4-FL in the presence of ABHD5 to HDAC4-FL in the absence of ABHD5 was subtracted from 1 and expressed as percent to calculate the proteolytic activity of ABHD5. (b) Similar assay as in ‘a’ was performed using rec-HDAC4 and rec-ABHD5 in the presence or absence of the serine protease inhibitor AEBSF. Proteolysis was analyzed by immunoblotting and Coomassie staining was used to quantify the proteolytic activity of ABHD5. (c) Similar assay as in ‘a’ and ‘b’ was performed using ABHD5 wild type (WT) and the ABHD5 mutants N153D (N/D), S298A (S/A) and H327A (H/A). Proteolysis was analyzed by immunoblotting, and Coomassie staining was used to quantify the proteolytic activity of ABHD5. (a-c) Statistical analysis: Values are presented as Mean±SEM, n=3 (n represents independent experiments); by one-way ANOVA, P<0.05 considered as significant. (d) ABHD5-WT, ABHD5-S298A and luciferase (Luc; control) were expressed with AAV9 vectors in cardiac-specific ABHD5 knockout mice (cKO) to replace endogenous ABHD5. HDAC4 and ABHD5 levels were analyzed by immunoblotting. Corresponding band intensities were quantified, and the ratio between HDAC4-NT and HDAC4-FL was determined as compared to Luc in cKO. Statistical analysis: Values are presented as Mean±SEM, n=3 (n represents different mice); by one-way ANOVA, P<0.05 considered as significant.
HDAC4-NT protects from heart failure in NLSD
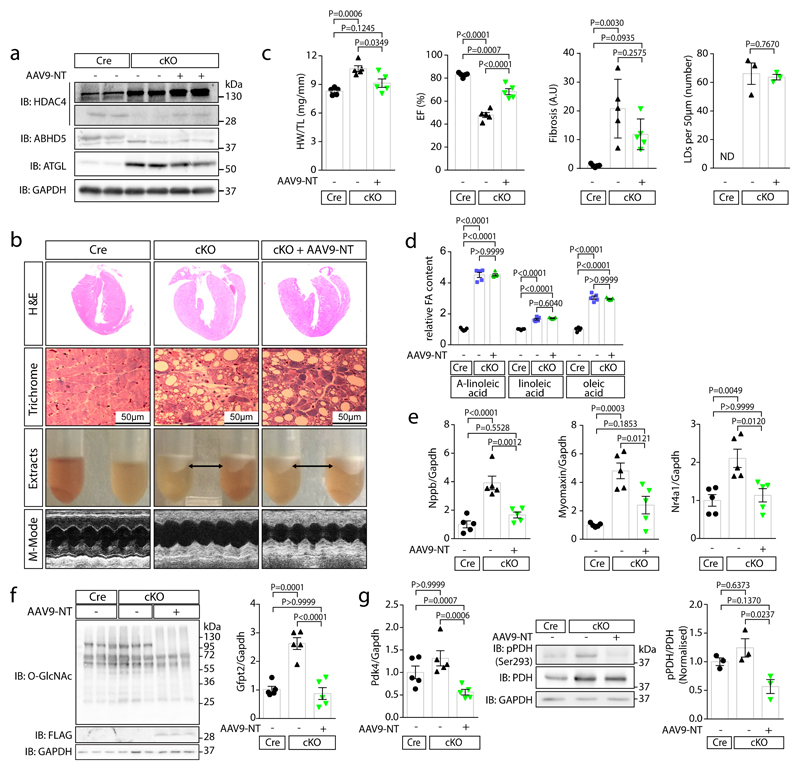
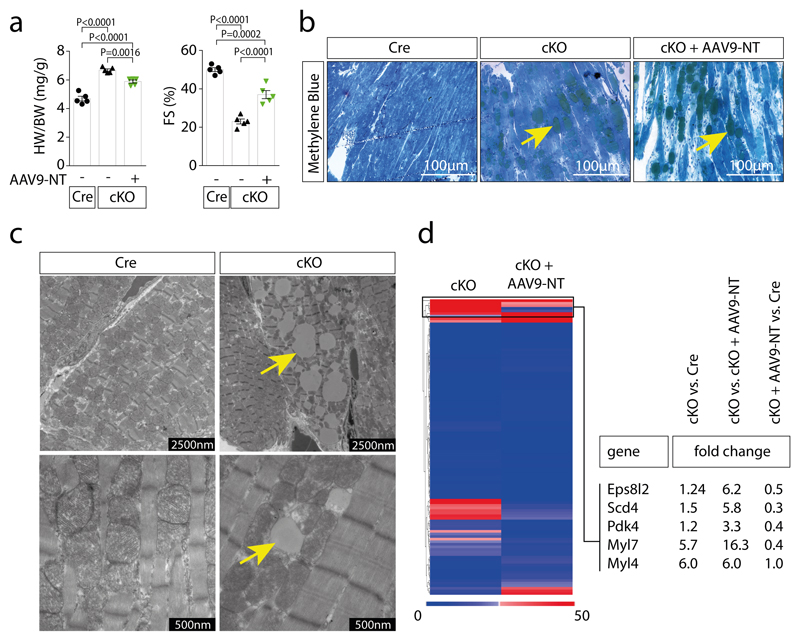
To investigate whether reduced HDAC4 proteolysis contributes to the NLSD cardiomyopathic phenotype upon ABHD5 deficiency, we used a rescue strategy. Gene delivery of HDAC4-NT using AAV9 resulted in cardiac HDAC4-NT levels close to its endogenous levels (Fig. 3a). Eight weeks after tamoxifen-induced cardiac ABHD5 deletion (cKO), we observed as compared to Cre expressing control mice (Cre) cardiac hypertrophy, dysfunction, fibrosis and massive intra-cardiomyocyte lipid accumulation (Fig. 3b-c; Extended Data Fig. 3a-b). AAV9-based HDAC4-NT gene delivery resulted in an attenuation of cardiac hypertrophy, an improvement in cardiac function and a trend towards less fibrosis despite unchanged lipid accumulation (Fig. 3b-d; Extended Data Fig. 3a-b). Interestingly, at the level of gene expression, HDAC4-NT gene delivery normalized not only the general marker for heart failure, Nppb, but also the specific MEF2 target genes Myomaxin and Nr4a1 (Fig. 3e), indicating that ABHD5 deficiency results in MEF2-dependent transcriptional activation and that re-expression of HDAC4-NT is sufficient to prevent activation of this pathway. These data challenge the current concept that lipotoxicity causes cardiac dysfunction at least in early ABHD5-deficiency. Accordingly, electron microscopy revealed that intracellular lipid droplets did not disturb the integrity of sarcomeres and mitochondria (Extended Data Fig. 3c). We have recently described that loss of HDAC4-NT results in MEF2-Nr4a1-Gfpt2-induced cardiac dysfunction.20 Likewise, we found here in cKO an induction of Gfpt2 (glutamine-fructose-6-phosphate transaminase 2), the rate-limiting enzyme of the hexosamine biosynthetic pathway, along with increased pan O-GlcNAcylation, which is normalized by the addition of HDAC4-NT (Fig. 3f). In addition, RNAseq and gene ontology analysis revealed that mainly metabolic processes were enriched (Suppl. Table 1). In particular, not only Nr4a1 and Gfpt2 as activators of O-GlcNAcylation were increased in cKO and normalized by HDAC4-NT, but also Pdk4 (pyruvate dehydrogenase kinase 4) was decreased by HDAC4-NT (Fig. 3g; Extended Data Fig. 3d; accession number: GSE135662). Accordingly, phosphorylation of pyruvate dehydrogenase (pPDH-Ser293) at the PDK4 site was normalized by HDAC4-NT in cKO (Fig. 3g). These data suggest that HDAC4-NT not only controls the glucose flux to the hexosamine biosynthesis pathway by regulating Gfpt2,20 but also by regulating pyruvate uptake into mitochondria through PDH, further establishing HDAC4-NT as a ‘gatekeeper' of glucose handling at different levels. This concept needs to be further validated in future experiments using glucose flux measurements.
Fig. 3. HDAC4-NT protects from heart failure in NLSD.
Cre-expressing control mice (Cre), cardiac-specific ABHD5-KO (cKO) and cKO with cardiac-specific HDAC4-NT gene delivery (cKO+AAV9-NT) were analyzed. (a) Immunoblotting for HDAC4, ABHD5, ATGL and GAPDH. (b) H&E and Masson’s-trichrome staining (Trichrome) of cardiac sections, a lipid layer after lysis and echocardiographic M-modes are shown. (a–b) Representative samples of one experiment with 5 mice. (c) Quantification of heart weight / tibia length ratios (HW/TL), ejection fraction (EF), fold change in fibrosis level in arbitrary units (A.U.) and number of lipid droplets in left ventricular myocardium (LDs, counted in trichrome sections are shown for the indicated groups). Statistical analysis for all except LDs: Values are presented as Mean±SEM, n=5 (n represents mice per indicated group); by one-way ANOVA, P<0.05 considered as significant. Statistical analysis for LDs: Values are presented as Mean±SEM, n=3 (n represents mice per indicated group); by unpaired two tailed t-test. (d) Relative quantification of indicated long chain fatty acids (FA) in three experimental groups (Cre, cKO and cKO+AAV9-NT). Statistical analysis: Values are presented as Mean±SEM, n=5 (n represents mice per indicated group); by one-way ANOVA, P<0.05 considered as significant. (e) Fold-change in mRNA-level of hypertrophic marker Nppb and specific MEF2 target genes, Myomaxin and Nr4a1, normalized to Gapdh in indicated groups. Statistical analysis: Values are presented as Mean±SEM, n=5 (n represents mice per indicated group); by one-way ANOVA, P<0.05 considered as significant. (f) Left: Immunoblotting of overall O-GlcNAcylation, FLAG-HDAC4-NT and GAPDH in Cre, cKO and cKO+AAV9-NT (n=3, n represents mice per indicated group). Right: Levels of Gfpt2 mRNA. Statistical analysis: Values are presented as Mean±SEM, n=5 (n represents mice per indicated group); by one-way ANOVA, P<0.05 considered as significant. (g) Left: Fold-change of Pdk4 mRNA normalized to Gapdh in indicated groups. Statistical analysis: Values are presented as Mean±SEM, n=5 (n represents mice per indicated group); by one-way ANOVA, P<0.05 considered as significant. Middle: immunoblotting of pyruvate dehydrogenase phosphorylation (pPDH-Ser293), pyruvate dehydrogenase (PDH) and GAPDH. Right: quantification of pPDH/PDH ratios. Statistical analysis: Values are presented as Mean±SEM, n=3 (n represents mice per indicated group); by one-way ANOVA, P<0.05 considered significant.
ABHD5 protects from heart failure
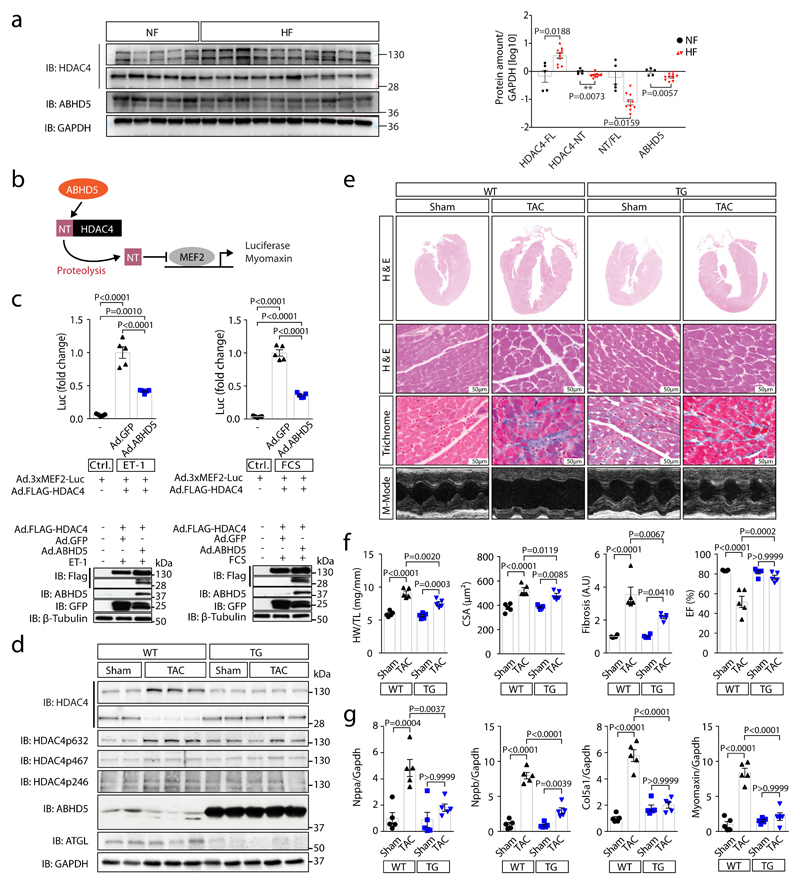
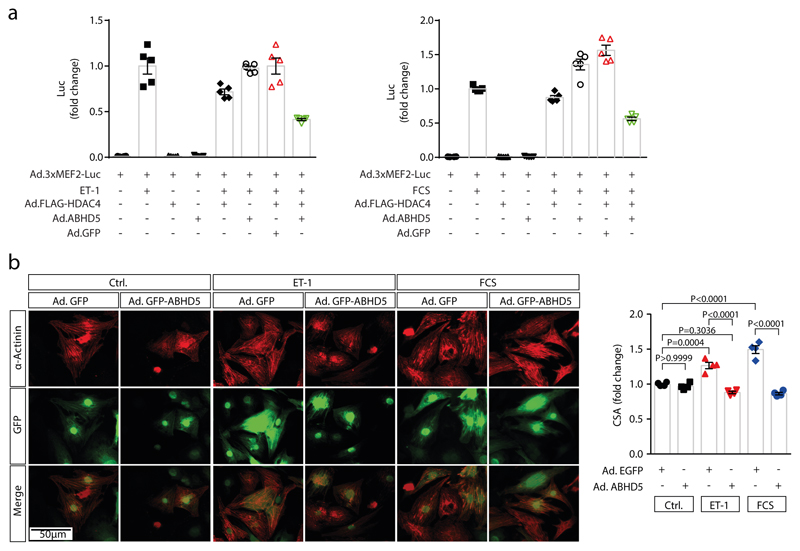
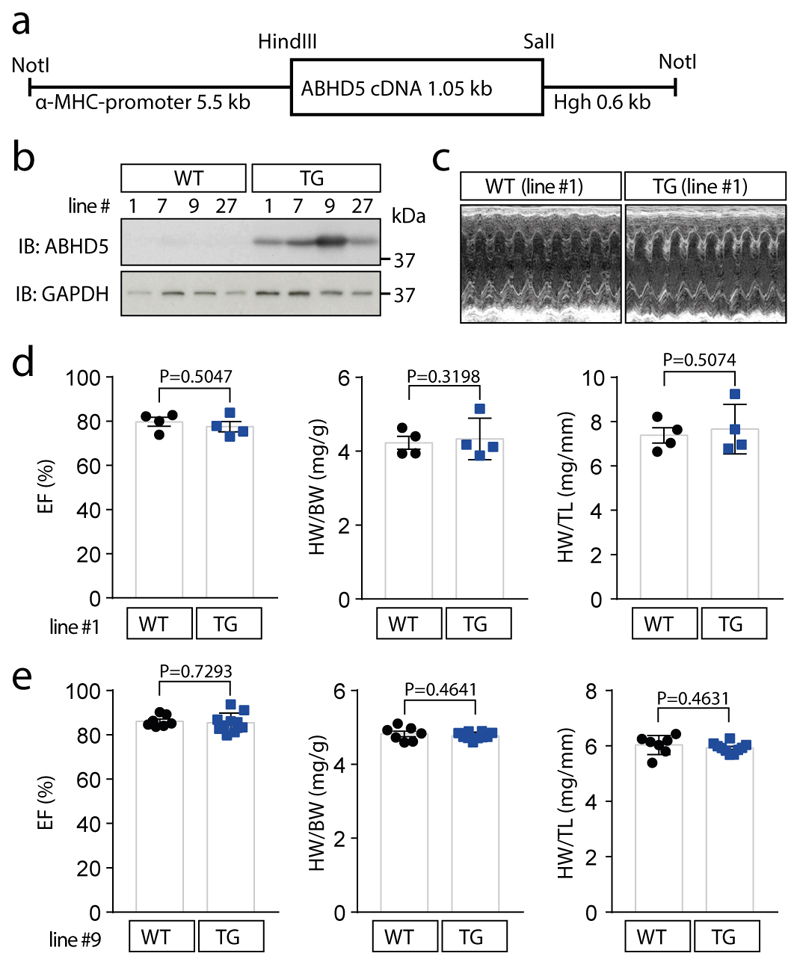
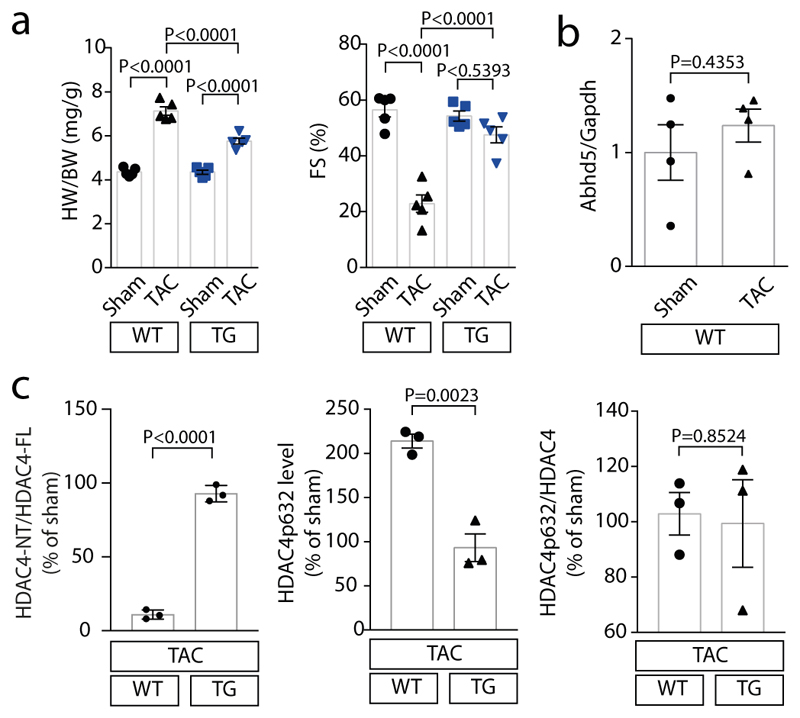
In human myocardial samples of patients with and without heart failure (from healthy donor hearts that could not be transplanted), we found that ABHD5 along with HDAC4-NT in relation to HDAC4-FL were down-regulated (Fig. 4a; Suppl. Table 2). Thus, we further explored the therapeutic potential of an ABHD5 gene addition strategy (granted patent: EP2956162B1). First, we tested whether ABHD5 expression in NRVMs would decrease MEF2 activity (as schematized in Fig. 4b), which induces heart failure-causing gene programs.20,22,24 Adenovirally expressed ABHD5 led to a significant repression of a MEF2 activity in the presence of HDAC4 upon stimulation with endothelin-1 (ET-1) or fetal calf serum (FCS) (Fig. 4c; Extended Data Fig. 4a). Moreover, adenoviral expression of ABHD5 in NRVMs prevented ET-1 and FCS-induced NRVM hypertrophy (Extended Data Fig. 4b). Next, we aimed to test whether ABHD5 exerts cardioprotective effects in vivo. Overexpression of ABHD5 in mice under the control of the cardiac-specific αMHC-promoter was achieved in four different transgenic mouse lines (Extended Data Fig. 5a-b). We found no obvious changes in cardiovascular parameters under unstressed conditions in αMHC-ABHD5 transgenic mouse line #1 and #9 (Extended Data Fig. 5c-e, Suppl. Table 3). Under conditions of pathological pressure overload induced by transverse aortic constriction (TAC), transgenic ABHD5 expression (we chose line #9, named ‘TG’) protected the heart from functional, structural and transcriptional remodeling. Immunoblot analysis showed that like in human myocardial samples, endogenous ABHD5 and HDAC4-NT were down-regulated upon TAC, which was prevented by transgenic ABHD5 expression (Fig. 4d, Extended Data Fig. 6b-c [left panel]). The association between ABHD5 and HDAC4-NT protein levels provides confirmatory evidence for the assumption that ABHD5 is required for HDAC4 proteolysis. In addition, we observed that ABHD5 overexpression affects the absolute but not relative phosphorylation levels of HDAC4 in particular at the specific CaMKII site Ser632 (Fig. 4d, Extended Data Fig. 6c [right]). In a recent article, we described that Ser632 phosphorylation of CaMKII can inhibit O-GlcNAcylation, which is required for HDAC4-NT production.21 This implies that ABHD5 might interfere with CaMKII binding to HDAC4. Thus, the interplay between these post-translational modifications and a potential regulatory role of ABHD5 needs to be investigated in the future. Importantly, in response to TAC, mice with cardiac ABHD5 overexpression showed a reduced hypertrophic response (Fig 4e-f, Extended Data Fig. 6a) and a blunted transcriptional up-regulation of the fetal genes Nppa and Nppb, the collagen Col5a1 and the specific MEF2 target gene Myomaxin (Fig. 4g), confirming that ABHD5 controls MEF2-dependent transcriptional pathways. Furthermore, interstitial fibrosis was prevented and different measures of cardiac function were improved (Fig. 4f, Extended Data Fig. 6a, Suppl. Table 4). Thus, myocardial overexpression of ABHD5 protects the heart from structural remodeling and failure.
Fig. 4. ABHD5 protects from heart failure.
(a) Left: immunoblotting of HDAC4-FL, HDAC4-NT, ABHD5 and GAPDH in non-failing (NF, lanes 1-5) and failing human myocardium (HF; lanes 6-10: dilated cardiomyopathy, DCM; lanes 11-15: ischemic cardiomyopathy, ICM). Right: Corresponding band intensities were normalized to GAPDH, and HDAC4-NT was normalized to HDAC4-FL. Statistical analysis: Values are presented as Mean±SEM, n=5 (NF) and n=10 (HF, n indicates separate human samples); by unpaired two tailed t-test, P<0.05 was considered as significant. (b) MEF2 activity was assessed by a luciferase reporter assay or Myomaxin expression in response to ABHD5-induced proteolysis of HDAC4. (c) Upper: MEF2 activity measured in lysates of endothelin-1 (ET-1) or fetal-calf-serum (FCS) stimulated NRVMs following transduction with indicated adenoviral constructs (Ad.3xMEF2C-Luc, Ad.FLAG-HDAC4, Ad.ABHD5 plus GFP on a bicistronic promoter or Ad.GFP). Statistical analysis: Values are presented as Mean±SEM, n=5 (n represents independent samples); by one-way ANOVA, P<0.05 considered as significant. Lower: Immunoblotting for FLAG-HDAC4, ABHD5, GFP and β-Tubulin (loading control) on luciferase assay lysates. Results shown represent at least three independent experiments. (d) Cardiac extracts from transgenic ABHD5 mice and wild type littermates (WT) three weeks after sham or TAC surgery were analyzed by immunoblotting. Expression of ABHD5 as well as HDAC4 and phosphorylated HDAC4, ATGL and GAPDH were detected. (e) Representative images of H&E staining, Masson’s-trichrome (Trichrome) staining, echocardiographic M-modes. (d-e) Representative samples of one experiment with 5 mice. (f) Quantification of heart weight / tibia length ratios (HW/TL), n=5, cardiomyocyte cross-sectional area (CSA), n=5, fibrosis in arbitrary units (A.U.), n=4 for sham WT and TG; and n=5 for TAC WT and TG, and ejection fraction (EF), n=5 (n represents mice per indicated group). Statistical analysis: Values are presented as Mean±SEM; by one-way ANOVA, P<0.05 considered as significant. (g) RT-PCR analysis of Natriuretic Peptide A (Nppa), Natriuretic Peptide B (Nppb), collagen 5a1 (Col5a1) and Myomaxin. Statistical analysis: Values are presented as Mean±SEM, n=5 (n represents mice per indicated group); by one-way ANOVA, P<0.05 considered as significant.
Discussion
Taken together, the data of this study show how the lipid droplet-associated protein ABHD5 controls cardiac function through an HDAC4-dependent mode of glucose handling regulation (Fig. 5). We unmasked a yet unanticipated proteolytic function of ABHD5. Besides its well-known PKA-dependent pro-lipolytic cofactor function of ATGL (without own intrinsic lipase activity due to the evolutionary loss of Ser-153), it also acts as a protease (with intrinsic serine protease function) that cleaves the signal-responsive epigenetic regulator HDAC4, resulting in CaMKII-resistant HDAC4-NT. HDAC4-NT in turn represses MEF2-dependent transcriptional programs driving cardiac dysfunction through metabolic control of calcium handling, but not affecting intra-cardiomyocyte lipid accumulation. Thus, this study demonstrates that lipid droplet associated signaling through ABHD5 represents a novel therapeutic target to treat cardiometabolic disease. For this study we used specifically HDAC4 as a substrate to identify ABHD5 as its protease. However, we cannot rule out that ABHD5 possesses additional substrates whose proteolytic products may also contribute to cardioprotection. Proteomic approaches are ongoing to identify potential other yet unknown substrates.
Fig. 5. Working model.
β3-adrenergic receptor stimulation leads to PKA-mediated phosphorylation of perilipin 5 (Plin5) and subsequent ABHD5 dissociation, which activates adipose triglyceride lipase (ATGL) catalyzing the first step of lipolysis. During this lipolytic process, ABHD5 not only activates ATGL but also acts as a serine protease on HDAC4. The resulting N-terminal HDAC4 polypeptide (HDAC4-NT) inhibits MEF2-dependent transcriptional programs including Nr4a1 expression, leading to changes in glucose handling (activation of the hexosamine biosynthesis pathway, O-GlcNAcylation of the calcium handling protein Stim1 and cardiac dysfunction).20 This pathway counteracts MEF2 activation that is driven by CaMKII-mediated phosphorylation of HDAC4 (see Fig. 1a). These findings represent therefore a yet unrecognized link between lipid metabolism and epigenetic control of cardiac function.
However and in particular, we demonstrate that HDAC4-NT gene therapy is sufficient to prevent severe cardiac dysfunction in a mouse model for NLSD.7,13,14 Further preclinical studies are currently ongoing to further establish HDAC4-NT gene therapy as a potential treatment strategy to reduce the mortality in NLSD models. Based on the data shown here, we also postulate that cardiac dysfunction in NLSD is rather driven by the protease function of ABHD5 instead of the ATGL (lipase) cofactor function, challenging the concept of lipotoxicity as cause for heart failure in cardiometabolic disease. Future studies also need to address the interplay between ABHD5 and ATGL towards HDAC4 proteolysis.
Our data also suggest that β3-adrenergic receptor agonists might be beneficial because they increase at least in mice endogenous levels of cardioprotective HDAC4-NT. In this regard, it will be interesting to await the results of an ongoing clinical study testing the therapeutic effects of the β3-adrenergic receptor agonist mirabegron in patients with structural heart disease.25
Finally, it will be also important to investigate the role of the here-described pathway for other cell types than cardiomyocytes. In this regard, lipid droplets are obviously the characteristic organelles of adipocytes and HDAC4 in white adipocytes was implied to play a role for beige adipocyte renaissance.26 It is therefore tempting to speculate that an HDAC4-dependent cross talk to lipid droplet-associated signaling might contribute to ‘beiging’.
In conclusion, the results of this study have broad implications on the understanding of fundamental cross talks between lipid and glucose metabolism in general, and on urgently needed new concepts for drug development strategies for cardiometabolic disease.
Methods
Human myocardial samples
Human myocardial samples were obtained from patients with end-stage dilated (DCM) or ischemic cardiomyopathy (ICM) at the time of cardiac transplantation (Suppl. Table 2). Non-failing (NF) human myocardial control samples were obtained from healthy donor hearts with no known history of cardiac disease which could not be transplanted for technical reasons. The study was approved by the Ethical Committee of the University of Göttingen (application numbers 14/9/11, 21/10/00, 31/9/00). We have obtained informed consent from all donors. For further details, refer to the reporting summary.
Mouse experiments
All experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committee at the Regierungspräsidium Karlsruhe, Germany. Mice were male littermates (except for in vivo proteolysis experiments, where mixed male and female mice were used [see below]) and age-matched - for all experiments. All genotypes are indicated in the described experiments.
Generation of ABHD5-transgenic mice
A cDNA encoding human ABHD5 was cloned into an expression plasmid containing α-MHC promoter and human growth hormone (hGH) poly-A (+) signal between SalI and HindIII. To exclude the bacterial expression component and linearize the rDNA plasmid for microinjection, the expression plasmid was digested by NotI (Extended Data Fig. 5a) and the rDNA of interest was purified for microinjection. Microinjection into fertilized one cell embryos was performed at the IBF facility of Heidelberg University. Genotyping was performed by PCR using genomic DNA isolated from the tail of founder mice. Genotyping primer sequences were presented as forward [cggcactcttagcaaaccttc] and reverse [gttgtgcccaagcaagatc]. Cardiac overexpression of ABHD5 in transgenic mice was evaluated by immunoblotting analysis using an anti-ABHD5 antibody. Transgenic mouse lines with different expression levels were identified and maintained in a C57BL/6N background.
Generation of ABHD5-deficienct mice
Inducible cardiomyocyte-specific ABHD5 knockout mice (cKO) were generated by mating α-MHC Mer-Cre-Mer transgenic mice27 to floxed ABHD5 mice15. 8-12-week old male Cre+ floxed and Cre+ non-floxed male mice were gavaged for 10 days (two days pause in between) with tamoxifen (80 mg/kg) to obtain cKO and Cre expressing control mice (Cre), respectively. Tamoxifen (10 mg/ml) was dissolved in regular sunflower oil solution containing 6% ethanol. Genotyping primer sequences were the following: Forward [taactgctgtggtatttgagg] and reverse [gactggaaggatttgagggg] to identify floxed ABHD5, and forward [aggttcgttcactcatgg] and reverse [tcgaccagtttagttaccc] to genotype the Mer-Cre-Mer transgenic mice. These mice were maintained in C57BL/6N background.
Transverse aortic constriction (TAC)
TAC surgery is an established surgical technique to induce pressure overload-induced heart failure.28,29 Mice were anesthetized by inhalation of 2% isoflurane and then intubated with a 20 gauge catheter and ventilated using a volume controlled rodent ventilator and Fluovac Anesthesia Systems (Hugo Sachs Elektronik, Harvard Apparatus GmbH, mouse ventilator minivent type 845) with a stroke volume of 0.2 ml oxygen and isoflurane mixture and a respiratory rate of 150 breaths/min. After shaving the hair on the chest, thoracotomy was performed to access the transverse aorta. The transverse aortic arch was then constricted between the innominate and left common carotid arteries with a 7-0 nylon suture tied 3 times firmly around a 27 gauge blunt needle for TAC. The needle was immediately withdrawn after the ligation. The operation procedure was performed on a clean and sterile operation field.
Experiments using adeno-associated virus vectors (AAV9)
HDAC4-NT (the first 201 aa of HDAC4) was cloned with an N-terminal FLAG-Tag into the AAV genome-plasmid pUF-CMVenh-MLC260.30 This plasmid contains a 260 bp spanning fragment of the cardiac-specific myosin light chain promoter (MLC260) and it is enhanced by a CMV enhancer. The use of cardiotrophic AAV9 vectors in addition to the use of a cardiac-specific promoter system allowed for cardiac-specific expression of HDAC4-NT.20 ABHD5 and ABHD5-S298A were cloned into a self-complementary (double-stranded) AAV genome-plasmid (pscTnT-miR122TS) containing the cardiac-specific troponin-T promoter.31 The respective control luciferase encoding plasmids were used pUF-CMVenh-MLC260-Luc and pscTnT-miR122TS-Luc.30,31 AAV9-NT, AAV9-ABHD5, AAV9-Luc and AAV9-ABHD5-S298A were produced in HEK293T cells (ATCC).32 In brief, each AAV genome plasmid was packaged into AAV9 capsids using Polyethylenimine (PEI) mediated co-transfection of 293T cells with pDP9rs and AAV genome-plasmid. 72 hrs post transfection, the cells as well as the supernatant were harvested followed by ammonium-sulfate precipitation for the supernatants and freeze- thaw cycles for generation of the cell lysates. For purification of the AAV vectors, iodixanol step gradients followed by ultracentrifugation were used. The final vector stocks were quantified using real-time PCR. For cardiac-specific gene delivery, 8–12-weeks old mice received the indicated AAV9 vectors through a single tail vein injection. To allow for expression of the above mentioned constructs, mice were kept for 6 weeks and then treated with tamoxifen for 12 days to induce the cKO. 14 weeks after tamoxifen treatment, mice were sacrificed. The dosages used for the experiments were 3x1011 genomic particles of AAV9-NT, 3x1011 genomic particles of AAV9-Luc (as control for AAV9-NT), 1x1012 genomic particles of ABHD5-WT, 1x1012 genomic particles of ABHD5-S298A and 1x1012 genomic particles of AAV9-Luc (as control for AAV9-ABHD5 and AAV9-ABHD5S298A).
Transthoracic echocardiography
Cardiac function was assessed by echocardiography using a Vevo 2100 high-resolution imaging system (Visual Sonics, Toronto, Canada) equipped with a 55 MHz transducer (MS550D).33 Briefly, mice were shaved on their chests and measurements were performed on conscious mice. M-mode and B-mode frames were recorded from the parasternal short axis and long axis. The operator and analyzer were blinded during the procedure.
Fibrosis Quantification in fatty cardiac tissue
For quantification of fibrosis area, the percentage of both fibrosis and fat tissue area was determined by using the point-counting-method, and the free fat tissue area calculated accordingly. The whole area of the heart section was analyzed.34 To obtain the final results, the percentage of free fat tissue area was set to 100%, and the related percentage of fibrosis was calculated. The obtained values were normalized to the Ctrl.
Cell culture
Neonatal rat ventricular myocytes (NRVMs) and HEK293cells (ATCC) were grown in Dulbecco's Modified Eagle's Medium (DMEM, Sigma [D5796]) supplemented with fetal calf serum (10%), l-glutamine (1%) and penicillin/streptomycin (1%). Expression plasmids encoding FLAG-HDAC4 (100 ng) and MYC-PKA (200 ng) were transfected using GeneJammer transfection reagent as suggested by the manufacturer (Agilent Technologies [204130]). NRVMs were transduced using adenovirus encoding FLAG-HDAC4, ABHD5 plus GFP on a bicistronic promoter and GFP as indicated. NRVMs were isolated from 1-to-3-day-old rats (Charles River). Briefly, hearts from neonatal rats were rapidly excised and washed in sterile PBS to remove blood and debris. Next, the ventricles were minced and dissociated into single cells by repeated pancreatin (Sigma [8049-47-6]) and collagenase II (Worthington [LS004177]) digestion with gentle stirring. The enzymatic digestion was inactivated in a complete medium and removed from dissociated cells by centrifugation. The obtained dissociated cells were resuspended in complete DMEM and then plated differentially for 2 hrs to eliminate fibroblasts. At the end, unattached cells were counted and plated accordingly.
Adult rat ventricular myocytes (ARVMs) were isolated following standard enzymatic digestion according to the Langendorff perfusion system. ARVMs were then maintained in Medium 199 (M199, Sigma[M7528]) supplemented with taurine (5 mM), carnitine (5 mM), creatine (4.4 mM), penicillin/streptomycin (1%), Insulin/Insuman® (Sanofi, 25 I.U), Mercaptopropionylglycine (4.9 mM). ARVMs were then fractionated into cytosolic-nuclear fractions. Briefly, ARVMs were first washed twice using ice cold PBS and then harvested in cytosolic buffer (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.05 % NP40 and protease inhibitor cocktail) followed by 10 min incubation on ice and centrifugation for 10 min at 3000 rpm at 4°C. The resultant supernatant was saved as cytosolic fraction and the pellet was further processed using nuclear fraction buffer (5 mM HEPES, pH 7.9, 1.5 mM MgCl2, 500 mM NaCl, 0.2 mM EDTA, 0.5 mM DTT and protease inhibitor cocktail). To obtain the nuclear fraction the pellet was washed once using ice cold PBS followed by resuspension in nuclear fraction buffer and incubation on ice for 30 min with 10 min interval vortexing before centrifugation at 14,000 rpm for 30 min at 4°C. Cytoplasmic and nuclear fractions of ARVMs were subjected to immunoblotting.
siRNA screening
HEK293 cells (ATCC) were transfected with a library of siRNAs (Ambion) directed against mRNA of 150 known or predicted human serine proteases (Supplementary Data 1). In the first step, each individual serine protease was targeted by two different siRNAs. Transfection mixes (100 μl) were prepared in opti-MEM medium (Invitrogen, [31985062]) by adding 5 μM siRNAs and 2 μl oligofectamine reagent (Invitrogen, [12252011]). The transfection mixes were then added to the HEK293 cells in the presence of 400 μl of serum free DMEM medium (Final volume=500 μl). After 4 hrs incubation, additional 500 μl of DMEM medium containing 20% FCS, 2% PenStrep (Penicillin Streptomycin) and 2% glutamine were added into the 500 μl serum free culture to constitute the complete DMEM medium. In a second step, the cells were transfected with FLAG-HDAC4 (100 ng) and MYC-PKA (200 ng) and as a control, with HDAC4 (100 ng) and an empty pcDNA 3.0 (200 ng) 36 hrs post siRNA transfection. Finally, cells were harvested and lysed in lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, Protease inhibitor cocktail tablets (Roche), 1 mM PMSF and 1% Triton X-100) for Western blotting analysis 24 hrs post expression plasmid transfection.
Proteolysis assay in a Saccharomyces cerevisiae system
To perform HDAC4 proteolysis assay in Saccharomyces cerevisiae, expression plasmids encoding HDAC4 (pNOP-HDAC4-Leu) and ABHD5 (pGAL-ABHD5-Trp) were transformed into Saccharomyces cerevisiae either together or with opposite backbone vectors and selected on selective plates lacking both leucine and tryptophan amino acids (SD-Leu-Trp). A single colony per transformation sample was picked and inoculated into a liquid culture of SC-Leu-Trp for expression. Expression of ABHD5 required induction by 2% galactose for 5 hrs. Yeast whole cell lysis was performed using glass beads in the presence of 20% Trichloroacetic acid (TCA) by a beat beater at 5,000 rpm (4x20sec, 5sec pause in between). The lysed homogenates were centrifuged at 13,000 rpm for 5 min at 4°C. Pellets were vacuum dried in a speed vac (30°C, 30-50 min [V-AQ]) and re-suspended in 1 M Tris pH 9.4 containing sample loading buffer (Laemmli) before boiling for 1 min at 95°C. Samples were resolved by 12% SDS-PAGE and subjected to Western blotting and protein detection using antibodies directed against the N-terminal region of HDAC4 (HDAC4 (N-18) [1:1000], Santa Cruz biotechnology) and full length ABHD5 (1:4000, Abnova). For further antibody details refer to the reporting summary.
Homology modeling
ABHD5 with uniprot ID_HumanQ8WTS1 was entered into the SWISS-MODEL automated protein structure homology-modeling server. The model searched for template library (SMTL version 2019-06-27, PDB release 2019-06-21) with blast and HHBlits for evolutionary related structures matching the ABHD5 sequence. To model ABHD5, the best fitting template with acceptable Qmean score was selected, in this case, proline iminopeptidase, was selected as a template to build a 3D model structure of ABHD5.35–38 The PDB file of the predicted ABHD5 model is provided in Supplementary Data 2.
Expression and purification of recombinant proteins
The expression and purification of recombinant HDAC4 protein (rec-HDAC4) was performed at the ‘Protein expression and Purification Core Facility’ of European Molecular Biology Laboratory (EMBL PEPCF) Heidelberg, Germany. Briefly, human full length HDAC4 (HDAC4-FL) was cloned into pFastBac HT A vector with an N-terminal TEV cleavable His6 tag. Recombinant bacmid was generated by transforming pFastBac HT A-His6-TEV-HDAC4 into E. coli DH10EMBacY. The second virus generation (V1) was used to infect Sf21 insect cells (~ 0.9 x 106 cells/ml) at a ratio of 1:100 in suspension culture using Sf-900 III serum free medium (Life Technologies). Protein expression was performed for 72 hrs at 27°C and 120 rpm. The cells were harvested by centrifugation for 30 min at 600 x g and 4°C. The cell pellet was re-suspended in buffer A (100 mM NaPO4, pH 7.5, 250 mM NaCl, 20 mM imidazole, 20 mM MgCl2), supplemented with 7U/ml Benzonase (Novagen, [70746]) and 1 x cOmplete™, EDTA-free Protease Inhibitor (Roche, [11697498001]). Insect cell lysis was accomplished using a 25 ml tissue grinder. The lysate was cleared by ultracentrifugation for 30 min at 125000 x g and 4°C. Next, the cleared lysate was loaded on a 5 ml Protino Ni-NTA column (MN), pre-equilibrated with buffer A using the Äkta Pure 25 system (GE). After extensive wash with buffer A, the bound protein was eluted with Buffer B (100 mM NaPO4, pH 7.5, 250 mM NaCl and 300 mM imidazole). Elution fractions containing HDAC4 were pooled and the His6 tag was cleaved with His6-TEV protease (in house produced, EMBL PEPCF). The cleavage was performed during dialysis overnight at 4°C against Buffer A. On the following day, the protein was applied again on a 5 ml Ni-NTA column, to remove the TEV protease and the un-cleaved HDAC4 protein. The flow through fractions containing the HDAC4 protein were collected, pooled together and concentrated to approximately 2mg/ml. The protein was aliquoted, flash frozen in liquid nitrogen and stored at -80°C. Purity of the protein was evaluated by SDS-PAGE and subsequent Coomassie staining. For the cleavage assay, HDAC4 was diluted 20-fold into 50 mM Tris-HCl, pH 7.5, 300 mM NaCl, 1 mM DTT, 10 % glycerol and concentrated to ~1 mg/ml.
For expression of N-terminal 6XHis-smt3-TEV tagged rec-ABHD5-WT (6XHis-smt3-TEV-mABHD5) in pET-28a was used.39 The putative ABHD5-catalytic mutants (6XHis-smt3-TEV ABHD5-N153D, S298A and H327A) as well as the putative PKA phospho-dead and –mimetic mutants (6XHis-smt3-TEV ABHD5-S237A and S237E) were generated as described below (site directed mutagenesis). The plasmids were then expressed in BL21-CodonPlus (DE3)-RIPL competent E.coli cells (Agilent technologies, 230280) in the presence 0.5 mM IPTG overnight at 25 °C, pelleted and re-suspended in lysis buffer (20 mM Tris-HCl pH 7.5, 500 mM NaCl, 0.1% NP-40, 30 mM imidazole and 1 mM DTT) and lysed using a French press. The samples were then cleared by centrifugation (12,000g for 30 min, 4 °C) and bound to a HisTrap HP column (GE health care, [17524701]). The unbound proteins were removed with 20 CVs (column volume) of lysis buffer and the protein was eluted with a series of gradient elutions in lysis buffer containing 300 mM imidazole (5 CV from 0-50%, 5 CV from 50-65%, 5 CV at 65% and 5 CV from 65-100%). Pure elution fractions containing His-smt3-TEV-ABHD5 were dialyzed (20 mM Tris-HCl pH 7.5, 300 mM NaCl, 10% glycerol and 1 mM DTT) and simultaneously TEV digested at 4 °C with slow agitation overnight. The protein was then concentrated using 30 kDa cut-off columns (Amicon Ultra-4, PLTK Ultracel-PL Membran, 30 kDa, [UFC803024]) and the final protein purity was assessed by SDS-PAGE. Similar thermal stability of rec-ABHD5-WT, -N153D, -S298A, -H327A, -S237A and -S237E mutant proteins was assessed by melting point determination with Differential Scanning Fluorimetry (DSF) at the EMBL Protein Expression and Purification Core Facility (PEPCF) on a Prometheus NT48 (NanoTemper technologies) by measuring the tryptophan fluorescence decrease and determination of the 330/350 nm ratio. Briefly, 10 μl of protein sample at 1 mg/ml was added to capillaries and sealed. The temperature was increased from 20 °C to 95 °C by a rate of 1 °C/min and the fluorescence emission at 330 nm and 350 nm were recorded after excitation with 280 nm.
In-vitro proteolysis assay
In-vitro proteolysis assay was performed by incubating rec-HDAC4 together with either rec-ABHD5 or indicated rec-ABHD5-mutant proteins in the presence of proteolysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 10 mM MgCl2 and 10 mM CaCl2) supplemented with or without 2.5 mM of the serine protease inhibitor AEBSF (Merck Millipore, [101500-100MG]) at 37 °C for 4 h with gentle agitation. As control cleavage reaction, only rec-HDAC4 or only rec-ABHD5 was used. The cleavage reaction was stopped by addition of 2X sample loading buffer and boiling at 95 °C. The samples were then separated on 12 % SDS- PAGE and subsequently transferred onto PVDF/Immobilon® membrane (Merck Millipore, [IPVH00019]) before probing the membranes with either anti-HDAC4 (Sigma, [H0163-200UL]) at 1:1000 dilution or anti-ABHD5 (Abcam, [ab59488]) at 1:1000 dilution for immunoblotting analysis. For additional antibody details refer to the reporting summary.
In vivo proteolysis assay
ABHD5 cKO, Cre expressing control and wildtype mice (C57BL/6N) at a age of 8–12 weeks (male and female) were starved for 4 hrs and then received a devided dose of two intraperitoneal injections (2 hrs between the first and second injection) of either isoproterenol (ISO, [sigma: I5627], 60 mg/kg body weight) or CL316,243 (CL [Santacruz, sc-203895], 20 mg/kg body weight). The control group mice received 0.9% NaCl. Organs were harvested 2 hrs after the second injection.
Expression plasmids and adenoviruses
Standard mammalian expression vectors for HDAC4 (FLAG tagged: FLAG-HDAC4) and the catalytic isoform of PKA (MYC tagged: MYC-PKA) were used.19 HDAC4 and ABHD5 were cloned into yeast expression plasmids bearing a constitutively active promoter [pNOP] and a galactose inducible promoter [pGAL] with or without FLAG or GST-Tag, respectively. Cloning primers are shown in Suppl. Table 5. The following adenoviral vectors encoding a MEF2 reporter luciferase, FLAG-HDAC4, ABHD5 and GFP were used: Ad.3xMEF2C-Luc (adenovirus expressing luciferase under the control of 3 MEF2 binding sites) and Ad.FLAG-HDAC419, Ad.ABHD5-GFP40 and Ad.GFP40. Ad.ABHD5-GFP encodes ABHD5 plus GFP on bicistronic promoter.
Quantitative RT-PCR
Total RNA was isolated and purified from cardiac tissue using trizol reagent procedure (Life technologies) and 1 μg of RNA was reverse transcribed to cDNA using cDNA synthesis kit (Thermo Scientific). 10 ng of cDNA was quantified using the SensiFAST Probe Lo-ROX (Bioline) kit employing Roche universal probe library (UPL). Primers used for quantitative RT-PCR analysis are shown in Suppl. Table 6.
Site-directed mutagenesis
Point mutations (ABHD5-N153D, -S298A, -H327A, -S237A, -S237E) were introduced using the PCR based quick change mutagenesis protocol following Phusion® High-Fidelity DNA polymerase (NEB, [M0530S]) and a DpnI enzyme (NEB, [R0176S]) digestion protocol using the primer listed, accordingly, in Suppl. Table 7. The resulting PCR and DpnI digestion products were then transformed into one shot™ TOP10 chemically competent E.coli cells (ThermoFisher Scientific, [C4040-06]) and the desired mutations were confirmed by Sanger sequencing.
Immunoblotting
HEK293 cells (ATCC) and NRVMs (isolated in house from Charles River bred rat pups) were harvested and lysed in standard lysis buffer with protease inhibitors (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, Protease inhibitor cocktail tablets (Roche), 1 mM PMSF and 1% Triton X-100). Murine ventricles were homogenized in lysis buffer with protease inhibitor (20 mM Tris, pH 7.4, 150 mM NaCl, protease inhibitor cocktail tablets (Roche), 1 mM PMSF and 0.5% NP-40). Human myocardial samples were grinded using mortar and pestle under liquid N2, and homogenized and lysed in lysis buffer (20 mM Tris, pH 7.4, 150 mM NaCl, protease inhibitor cocktail tablets (Roche), 1 mM PMSF and 0.5% NP-40). All samples were then analyzed by SDS-PAGE and transferred onto a PVDF membrane before probing with the indicated antibodies. The following antibodies were used for immunoblotting: anti-HDAC4 (N-18 [sc-5245], directed against the N-terminus) to detect HDAC4-NT, anti-HDAC4 (HDAC-144 [sc-56686], directed against the N-terminus AA 1-19 of HDAC4 human origin) to detect HDAC4-NT in human samples, anti-HDAC4 (H-92 [sc-11418], directed against AA 530-631) to detect full length HDAC4, anti-GFP (sc-53882) and anti-MYC (A-14 [sc-789]) were all obtained from Santa Cruz biotechnology and were used at 1:1000 dilution; and anti-HDAC4 (Sigma [H0163] directed against HDAC4 AA 1-19) to detect HDAC4-NT and was used 1:1000, anti-ABHD5 was obtained from Abnova (H00051099-M01) and was used at 1:4000 dilution; an additional anti-ABHD5 was obtained from Abcam (ab59488) and was used at 1:1000 dilution to detect wildtype and the mutant ABHD5-recombinant proteins employed in the in vitro proteolysis assay; anti-ATGL was obtained from Cell Signaling Technology (2138S) and was used at 1:1000 dilution, anti-GAPDH was obtained from EMD Millipore (MAB374) and was used at 1:5000 dilution, anti-β-tubulin was obtained from Sigma-Aldrich (T4026) and was used at 1:2000 dilution, anti-PDH was obtained from Cell Signaling Technology (2784s) and used at 1:1000 dilution, anti-pPDH(Ser-293) was obtained from Novus Biologicals (NB110-93479) and used at 1:1000 dilution, anti-O-GlcNAC was obtained from Cell Signaling Technology (9875s) and used at 1:1000 dilution, anti-Histone was obtained from Merck Millipore (04-298) and used at 1:1000 dilution. For more detailed information about antibodies used in this study, refer to the reporting summary
Histology and immunocytochemistry
Hearts were cut transversally for four chamber view, washed quickly in 0.9% NaCl and fixed in 4% PBS buffered paraformaldehyde (PFA). Samples were then embedded in paraffin, sectioned to the desired diameters (5 μm), and stained with either hematoxylin and eosin (H & E) or Trichrome. NRVMs [see above], were grown on gelatin-coated glass coverslips; fixed in 4% PBS buffered paraformaldehyde (PFA) and stained in PBS containing 5% goat serum and 0.1% triton-X-100. A primary antibody against anti-α-sarcomeric actinin (mouse monoclonal; Sigma [A7811]) was used at a dilution of 1:200. The secondary fluorochrome-conjugated (texas red) antibody (Vector Labs, TI-2000) was used at a dilution of 1:400.
Electronmicroscopy
Hearts were cut transversally for a four-chamber view, washed quickly in 0.9 % NaCl and fixed in Karnovsky solution (2 % PFA and 4 % glutaraldehyde buffered in PBS, and 0.2M cacodylate buffer), and then washed in cacodylate buffer to eliminate aldehyde. They were next incubated for 2 hrs post fixation in 1.5 % osmium-tetroxide and dehydrated in ascending percentages of ethanol (50 %, 70 %, 80 % and 96 %) followed by further dehydration in absolute ethanol and propylenoxide. Ethanol and propylenoxide were then replaced by ARALDITE (1:1) and propylenoxide (1:3). After overnight incubation in 100 % ARALDITE, samples were prepared in capsule and left for polymerization at 60 °C for two days. Finally, 70 nm sections were prepared with LEICA ULTRACUT UCT, contrasted with uranylacetate and lead citrate. Pictures of the samples were acquired with a ZEISS EM910 electron microscope.
Reporter assays
NRVMs [see above] were transduced with adenoviruses expressing luciferase reporter (under the control of 3 x MEF2 binding site upstream of TATA box), FLAG-HDAC4, ABHD5 plus GFP on a bicistronic promoter, and GFP alone. After adenoviral transduction, the NRVMs were cultured in a serum free DMEM (Starvation medium) for 24 hrs and were afterwards stimulated either with endothelin-1 (ET-1, 100 nM) or fetal calf serum (FCS, 10%). NRVMs were lysed and assayed for luminescence 24 hrs after stimulation.
Quantification of long chain fatty acids
Cardiac tissues were homogenized in Dulbecco's Phosphate-Buffered Saline pH7.4 or GST lysis buffer with protease inhibitor (20 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, Protease inhibitor cocktail tablets (Roche), 1 mM PMSF and 0.5 % NP-40) for analysis of long chain fatty acids (FAs) content41.
RNA sequencing (RNAseq)
Total RNA was extracted as described under ‘Quantitative RT-PCR’. Strand-specific TruSeq mRNA libraries were prepared at the Max Planck-Genome-Centre Cologne, Germany (http://mpgc.mpipz.mpg.de/home/) for polyadenylated transcript enrichment. All libraries were sequenced paired-end 2 x 150 nt on an Illumina HiSeq 3000 instrument.
Read processing and mapping
We used Flexbar to remove adapter sequences and low quality regions from FASTQ files.42 Reads greater 18 bp were retained and mapped against the murine 45S rRNA precursor sequence (BK000964.3) to remove rRNA contaminant reads. We used the mouse genome sequence and annotation (EnsEMBL 79) together with the splice-aware STAR read aligner (release 2.5.1b) to map our short reads.43 All subsequent transcriptome analyses were carried out with the cufflinks package44 and custom R scripts. A heat map was generated using a custom R script. RNAseq analysis code will be made available upon reasonable request
Gene set enrichment analysis
Gene set enrichment analyses were carried out using topGO45 defining all expressed genes (FPKM & gt; 1) with default parameters as a background group.
Statistics and Reproducibility
Values are presented as Mean±SEM (Standard Error of Mean). Statistical analysis was performed using GraphPad prism 8 (GraphPad Software, San Diego, California, USA). Statistical significance was evaluated either by analysis of variances (ANOVA) or t test. Differences among groups were tested by ordinary one-way ANOVA with post hoc comparisons by Bonferroni’s multiple comparison tests with each P value adjusted to account for multiple comparisons. Unpaired two tailed t-test was used where appropriate.
Extended Data
Extended Data Fig. 1. ABHD5 is required for HDAC4 proteolysis.
(a) Immunoblotting using an antibody directed against endogenous ABHD5 in cardiac and cellular COS as well as HEK293 extracts. GAPDH was used as loading control. (b) Cytosolic and nuclear fractions from adult rat ventricular myocytes were used for immunoblotting using an antibody directed against ABHD5, GAPDH and histone H3. (a-b) Representative results of two independent experiments. (c) Left panel: Immunoblotting analysis of cardiac extracts of control (Ctrl) or isoproterenol (ISO) treated mice using antibodies directed against HDAC4, ABHD5 and GAPDH as loading control. Middle left panel: Densitometric analysis of HDAC4-NT/FL ratio shown as fold change vs Ctrl. Middle-right panel and right panel: Quantification of cardiac ABHD5 protein normalized to GAPDH and abhd5-RNA levels normalized to 18S in unstressed mice and ISO-treated mice. Statistical analysis: Values are presented as Mean±SEM, n=3 (n represents mice per indicated group); by unpaired two tailed t-test, P<0.05 considered as significant. (d) Left panel: Immunoblotting analysis of cardiac extracts of control (Ctrl) or CL 316,243 (CL) treated mice using antibodies directed against HDAC4, ABHD5 and GAPDH as loading control. Middle-left panel: Densitometric analysis of HDAC4-NT/FL ratio shown as fold change vs Ctrl. Middle panel: Quantification of cardiac ABHD5 protein normalized to GAPDH and abhd5-RNA levels normalized to 18S in unstressed mice and CL-treated mice. Right panel: Quantification of ucp1 mRNA normalized to 18S in brown adipose tissue after treatment with CL as a marker of CL effective administration. Statistical analysis: Values are presented as Mean±SEM, n=3 (n represents mice per indicated group); by unpaired two tailed t-test, P<0.05 considered as significant.
Extended Data Fig. 2. ABHD5 possesses intrinsic serine protease activity.
(a) Prediction of 3D structure of ABHD5 using SWISS-MODEL. Highlighted are amino acids (aa) predicted to serve as putative catalytic triad (Asn153, Ser298 and His327) and the putative phosphorylation site Ser-237. Ser-237 and Ser-298 of ABHD5 lie in spatial proximity to each other. Lys51 represents the N-terminal starting aa of the peptide sequence used for modeling. Pink: cap domain; green: a/b hydrolase core. (b) Multiple sequence alignment of ABHD5 protein of different species as indicated by UniProt online tool (uniprot.org/align/). Red boxes denote aa N153, S298 and H327 of the predicted serine protease catalytic triad, green box denotes aa S237 as reported PKA phosphorylation site (aa numbering corresponds to human ABHD5 protein). (c) Tryptophan fluorescence decay plot upon thermal denaturation of rec-ABHD5-WT, -N153D, -S237A, -S237E -S298A and -H327A (1°C/min). Left: the ratio of fluorescence at 350/330 nm was plotted against the temperature. Right: Equivalent analysis to identify the melting temperature (Tm) of the rec-proteins. Measurements were performed once. (d) Coomassie staining after protease assay using recombinant purified HDAC4 (rec-HDAC4) and increasing amounts of recombinant purified ABHD5 (rec-ABHD5) as indicated. (e) Coomassie staining after protease assay using rec-HDAC4 and rec-ABHD5 in the presence or absence of serine protease inhibitor AEBSF as indicated. (f) Coomassie staining after protease assay using rec-HDAC4 and rec-ABHD5 wild type (WT) or putative catalytic triad mutants as indicated. The data in (d-f) represent results of three independent experiments. (g) Immunoblotting and Coomassie staining for HDAC4 and ABHD5 after in vitro proteolysis assays involving Rec-HDAC4, rec-ABHD5 wild type (WT) or Ser-237 phospho-death (S237A) and phospho-mimetic (S237E) mutants as indicated. Coomassie staining was used to quantify the proteolytic activity of ABHD5. Statistical analysis: Values are presented as Mean±SEM, n=3 (n represents independent experiments); by one-way ANOVA, P<0.05 considered as significant. (h) Immunoblotting for HDAC4 and ABHD5 in yeast cell extracts after proteolysis assay in a yeast expression system as indicated. Because HDAC4 contains the Protein-A (PRA; 15 kDa) tag, HDAC4-NT is expected to migrate at 43 kDa. The experiment was performed at least three times with similar results.
Extended Data Fig. 3. HDAC4-NT protects from ABHD5-deficiency induced cardiac dysfunction.
Cre expressing control mice (Cre), cardiac-specific ABHD5-KO (cKO) and cKO with HDAC4-NT gene delivery (cKO+AAV9-NT) were analyzed. (a) Heart weight/body weight ratios (HW/BW) and fractional shortening (FS) are shown for the indicated groups. Statistical analysis: Values are presented as Mean±SEM; n=5 (n represents mice per indicated group); by one-way ANOVA, P<0.05 considered as significant. (b) Representative images of Methylene Blue staining of cardiac thin-sections to qualitatively visualize lipid droplet content in the indicated groups. Arrows point to positive lipid stain. (c) Representative electron-microscopic images of the mitochondrial and sarcomeric structures of cardiomyocytes of control mice (Cre) and cKO mice; arrows point to the lipid accumulation on the mitochondrial and sarcomeric surface in cKO. (b–c) Representative samples of one experiment with 5 mice included. (d) RNAseq analysis of heart tissue of the three above mentioned groups. The left panel shows a heat map displaying expression differences in cKO vs. cKO+AAV9-NT. Right panel: Fold change in expression of selected genes in cKO vs. Cre, cKO vs cKO+AAV9-NT and cKO+AAV9-NT vs Cre.
Extended Data Fig. 4. ABHD5 inhibits MEF2 and cardiomyocyte hypertrophy.
(a) NRVMs were transduced with the Ad3xMEF2C-Luc reporter. Ad.FLAG-HDAC4, Ad.ABHD5 plus GFP on a bicistronic promoter and Ad.GFP (10 MOI) were then co-expressed as indicated and NRVMs were stimulated for 24 hrs with endothelin-1 (ET-1) or fetal calf serum (FCS); n=5 (n represents independent samples) and values are presented as Mean±SEM. (b) Right panel: Fluorescence microscopy of NRVMs expressing Ad.ABHD5 plus GFP on a bicistronic promoter or Ad.GFP (10 MOI each). NRVM hypertrophy was induced with 100 nM ET-1 or 10% FCS for 24 hrs. Shown are representative images of α-actinin staining (red) of NRVMs and GFP fluorescence to confirm adenoviral expression. Images are representative of at least three independent experiments. Scale bar=50 μm. Right panel: Quantification of NRVM cross sectional areas (CSA). Statistical analysis: Values are presented as Mean±SEM, n=4 (n represents independent samples); by one-way ANOVA, P<0.05 considered as significant.
Extended Data Fig. 5. Generation and baseline characterization of cardiac-specific ABHD5 transgenic mice (TG).
(a) Microinjected fragment of an expression plasmid containing hABHD5 cDNA flanked by α-MHC promoter and human growth hormone (hGH) poly-A (+) signal. (b) Cardiac overexpression of ABHD5 in mice of 4 different transgenic lines as compared to wild type littermates (WT) from the same lines was evaluated by immunoblotting analysis using an anti-ABHD5 antibody. (c–e) Baseline characterization of transgenic mice. (c) Representative echocardiography of a mouse from transgenic line #1 as compared to a WT littermate. The data in b are representative for 4 different mouse lines and the data in c are representative for 4 mice from line # 1 as well as 7 (WT) and 10 (TG) mice from line #9. (d) Quantification of ejection fraction (EF), heart weight/body weight ratio (HW/BW), heart weight/tibia length ratio (HW/TL) in WT littermates (n=4) and transgenic mice from line #1 (n=4) (e) and in WT littermates (n=7) and transgenic mice from line #9 (n=10, TG). (d, e) Statistical analysis: Values are presented as Mean±SEM; by unpaired two tailed t-test, P<0.05 considered as significant. ‘n’ represents mice per indicated group
Extended Data Fig. 6. ABHD5 attenuates cardiac hypertrophy upon TAC.
(a) Left panel: Quantification of HW/BW ratio of WT and TG mice three weeks after TAC or sham surgery. Right panel: Quantification of fractional shortening (FS) of WT and TG mice three weeks after TAC or sham surgery. Statistical analysis: Values are presented as Mean±SEM, n=5 (n represents mice per indicated group); by one-way ANOVA, P<0.05 considered as significant. (b) mRNA expression of Abhd5 in sham and TAC operated WT mice normalized to Gapdh. Statistical analysis: Values are presented as Mean±SEM, n=4 (n represents mice per indicated group); by unpaired two tailed t-test, P<0.05 are considered as significant. (c) Corresponding band intensities of experiment shown in Fig. 4d were quantified. Left panel: Ratio between HDAC4-NT and HDAC4-FL. Middle panel: HDAC4 P-632 levels were determined upon TAC in relation to sham in WT and TG mice and normalized to GAPDH. Right panel: Ratio between HDAC4 P-632 and total HDAC4. Statistical analysis: Values are presented as Mean±SEM, n=3 (n represents mice per indicated group); by unpaired two tailed t-test; P<0.05 are considered as significant.
Supplementary Material
Prediction of the 3D structure of ABHD5 extracted from an online protein structure homology-modelling server SWISS-MODEL (https://swissmodel.expasy.org)
Human-serine proteases siRNA library. List of siRNAs targeting known and predicted serine proteases (library was purchased from Ambion)
Acknowledgements
We thank Marco Hagenmüller for help with figures and manuscript editing, Qiang Sun, Michaela Oestringer, Jutta Krebs, Ulrike Oehl and Joshua Hartmann for technical help, and Regina Zahn, Kathryn Perez, Kim Remans and Jochen Baßler for help with purification of recombinant HDAC4 and ABHD5, and Sylvia Kaden for the help with electron microscopy. L.H.L. is recipient of the Clinician-Scientist Program (CSP) of the German Cardiac Society (DGK). M.O. is supported by the Austrian Science Fund (FWF): F73 SFB Lipid Hydrolysis. J.B. was supported by grants from the Deutsche Forschungsgemeinschaft (BA 2258/2-1BA; BA 2258/9-1; SFB 1118), the European Commission (FP7-Health-2010; MEDIA-261409) and the DZHK (Deutsches Zentrum für Herz-Kreislauf-Forschung - German Centre for Cardiovascular Research) and by the BMBF (German Ministry of Education and Research).
Footnotes
Data availability Statement
Raw data of experiments presented in Fig. 3F-G, Suppl. Table 1 and Extended Data Fig. 3d were deposited in the gene expression omnibus (GEO, GSE135662) with NCBI tracking system ID #20253819. Source Data provide all uncut gels of the immunoblotting figures in the manuscript, and all data supporting the findings of the manuscript could be obtained from the corresponding author upon request.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article
Author Contributions
J.B. and Z.H.J. designed the study. Z.H.J., S.K.D., J.T., M.D., C.X., F.S., D.S., X.G., L.W. B.C.W., G.F., and S.W.S. carried out experiments. Z.H.J., L.H.L., S.W.S., C.D, J.-H.G. and J.B. analyzed and interpreted data. T.F, O.J.M, S.S., P.M., C.D, C.M., M.O., G.H, H.A.K., J.T., and J.B. provided research support and conceptual advice. Z.H.J. J.T. and J.B. wrote the paper.
Competing Interests Statement
Z.H.J., L.H.L, O.J.M, H.A.K. and J.B. filed a patent on HDAC4-NT and ABHD5 gene therapy (US9914912B2). All other authors declare no conflict of interest.
References
- 1.Lohse MJ, Engelhardt S, Eschenhagen T. What is the role of beta-adrenergic signaling in heart failure? Circ Res. 2003;93:896–906. doi: 10.1161/01.RES.0000102042.83024.CA. [DOI] [PubMed] [Google Scholar]
- 2.Dewenter M, von der Lieth A, Katus HA, Backs J. Calcium Signaling and Transcriptional Regulation in Cardiomyocytes. Circ Res. 2017;121:1000–1020. doi: 10.1161/CIRCRESAHA.117.310355. [DOI] [PubMed] [Google Scholar]
- 3.Rockman HA, Koch WJ, Lefkowitz RJ. Cardiac function in genetically engineered mice with altered adrenergic receptor signaling. The American journal of physiology. 1997;272:H1553–1559. doi: 10.1152/ajpheart.1997.272.4.H1553. [DOI] [PubMed] [Google Scholar]
- 4.Wang H, et al. Unique regulation of adipose triglyceride lipase (ATGL) by perilipin 5, a lipid droplet-associated protein. J Biol Chem. 2011;286:15707–15715. doi: 10.1074/jbc.M110.207779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamaguchi T, et al. CGI-58 facilitates lipolysis on lipid droplets but is not involved in the vesiculation of lipid droplets caused by hormonal stimulation. J Lipid Res. 2007;48:1078–1089. doi: 10.1194/jlr.M600493-JLR200. [DOI] [PubMed] [Google Scholar]
- 6.Sahu-Osen A, et al. CGI-58/ABHD5 is phosphorylated on Ser239 by protein kinase A: control of subcellular localization. J Lipid Res. 2015;56:109–121. doi: 10.1194/jlr.M055004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haemmerle G, et al. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-alpha and PGC-1. Nat Med. 2011;17:1076–1085. doi: 10.1038/nm.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zierler KA, et al. Functional cardiac lipolysis in mice critically depends on comparative gene identification-58. J Biol Chem. 2013;288:9892–9904. doi: 10.1074/jbc.M112.420620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pollak NM, et al. The interplay of protein kinase A and perilipin 5 regulates cardiac lipolysis. J Biol Chem. 2015;290:1295–1306. doi: 10.1074/jbc.M114.604744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lass A, et al. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 2006;3:309–319. doi: 10.1016/j.cmet.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 11.Cerk IK, Wechselberger L, Oberer M. Adipose Triglyceride Lipase Regulation: An Overview. Curr Protein Pept Sci. 2018;19:221–233. doi: 10.2174/1389203718666170918160110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamaguchi T. Crucial role of CGI-58/alpha/beta hydrolase domain-containing protein 5 in lipid metabolism. Biol Pharm Bull. 2010;33:342–345. doi: 10.1248/bpb.33.342. [DOI] [PubMed] [Google Scholar]
- 13.Chanarin I, et al. Neutral-lipid storage disease: a new disorder of lipid metabolism. Br Med J. 1975;1:553–555. doi: 10.1136/bmj.1.5957.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lefevre C, et al. Mutations in CGI-58, the gene encoding a new protein of the esterase/lipase/thioesterase subfamily, in Chanarin-Dorfman syndrome. Am J Hum Genet. 2001;69:1002–1012. doi: 10.1086/324121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radner FP, et al. Growth retardation, impaired triacylglycerol catabolism, hepatic steatosis, and lethal skin barrier defect in mice lacking comparative gene identification-58 (CGI-58) J Biol Chem. 2010;285:7300–7311. doi: 10.1074/jbc.M109.081877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W, et al. Sustained beta1-adrenergic stimulation modulates cardiac contractility by Ca2+/calmodulin kinase signaling pathway. Circ Res. 2004;95:798–806. doi: 10.1161/01.RES.0000145361.50017.aa. [DOI] [PubMed] [Google Scholar]
- 17.Fischer TH, et al. Ca2+/calmodulin-dependent protein kinase II and protein kinase A differentially regulate sarcoplasmic reticulum Ca2+ leak in human cardiac pathology. Circulation. 2013;128:970–981. doi: 10.1161/CIRCULATIONAHA.113.001746. [DOI] [PubMed] [Google Scholar]
- 18.Backs J, Song K, Bezprozvannaya S, Chang S, Olson EN. CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J Clin Invest. 2006;116:1853–1864. doi: 10.1172/JCI27438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Backs J, et al. Selective repression of MEF2 activity by PKA-dependent proteolysis of HDAC4. J Cell Biol. 2011;195:403–415. doi: 10.1083/jcb.201105063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehmann LH, et al. A proteolytic fragment of histone deacetylase 4 protects the heart from failure by regulating the hexosamine biosynthetic pathway. Nat Med. 2018;24:62–72. doi: 10.1038/nm.4452. [DOI] [PubMed] [Google Scholar]
- 21.Kronlage M, et al. O-GlcNAcylation of Histone Deacetylase 4 Protects the Diabetic Heart from Failure. Circulation. 2019 doi: 10.1161/CIRCULATIONAHA.117.031942. [DOI] [PubMed] [Google Scholar]
- 22.Kim Y, et al. The MEF2D transcription factor mediates stress-dependent cardiac remodeling in mice. J Clin Invest. 2008;118:124–132. doi: 10.1172/JCI33255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang LK, Tao YX. Physiology and pathophysiology of the beta3-adrenergic receptor. Prog Mol Biol Transl Sci. 2019;161:91–112. doi: 10.1016/bs.pmbts.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Wei J, et al. Reversal of pathological cardiac hypertrophy via the MEF2-coregulator interface. JCI Insight. 2017;2 doi: 10.1172/jci.insight.91068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pouleur AC, et al. Rationale and design of a multicentre, randomized, placebo-controlled trial of mirabegron, a Beta3-adrenergic receptor agonist on left ventricular mass and diastolic function in patients with structural heart disease Beta3-left ventricular hypertrophy (Beta3-LVH) ESC Heart Fail. 2018;5:830–841. doi: 10.1002/ehf2.12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, et al. Adipocyte Liver Kinase b1 Suppresses Beige Adipocyte Renaissance Through Class IIa Histone Deacetylase 4. Diabetes. 2017;66:2952–2963. doi: 10.2337/db17-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sohal DS, et al. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res. 2001;89:20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- 28.deAlmeida AC, van Oort RJ, Wehrens XH. Transverse aortic constriction in mice. J Vis Exp. 2010 doi: 10.3791/1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreusser MM, et al. Cardiac CaM Kinase II genes delta and gamma contribute to adverse remodeling but redundantly inhibit calcineurin-induced myocardial hypertrophy. Circulation. 2014;130:1262–1273. doi: 10.1161/CIRCULATIONAHA.114.006185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muller OJ, Schinkel S, Kleinschmidt JA, Katus HA, Bekeredjian R. Augmentation of AAV-mediated cardiac gene transfer after systemic administration in adult rats. Gene Ther. 2008;15:1558–1565. doi: 10.1038/gt.2008.111. [DOI] [PubMed] [Google Scholar]
- 31.Geisler A, et al. microRNA122-regulated transgene expression increases specificity of cardiac gene transfer upon intravenous delivery of AAV9 vectors. Gene Ther. 2011;18:199–209. doi: 10.1038/gt.2010.141. [DOI] [PubMed] [Google Scholar]
- 32.Jungmann A, Leuchs B, Katus HA, Rommelaere J, Muller OJ. Protocol for efficient generation and characterization of adeno-associated viral (AAV) vectors. Hum Gene Ther Methods. 2017 doi: 10.1089/hgtb.2017.192. [DOI] [PubMed] [Google Scholar]
- 33.Lehmann LH, et al. Essential role of sympathetic endothelin A receptors for adverse cardiac remodeling. Proc Natl Acad Sci U S A. 2014;111:13499–13504. doi: 10.1073/pnas.1409026111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams J, et al. 13-cis retinoic acid inhibits development and progression of chronic allograft nephropathy. Am J Pathol. 2005;167:285–298. doi: 10.1016/S0002-9440(10)62973-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bienert S, et al. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017;45:D313–D319. doi: 10.1093/nar/gkw1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waterhouse A, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guex N, Peitsch MC, Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis. 2009;30(Suppl 1):S162–173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 39.Boeszoermenyi A, et al. Structure of a CGI-58 motif provides the molecular basis of lipid droplet anchoring. J Biol Chem. 2015;290:26361–26372. doi: 10.1074/jbc.M115.682203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Badin PM, et al. Regulation of skeletal muscle lipolysis and oxidative metabolism by the co-lipase CGI-58. J Lipid Res. 2012;53:839–848. doi: 10.1194/jlr.M019182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poschl JM, et al. Effects of dietary supplementation of saturated fatty acids and of n-6 or n-3 polyunsaturated fatty acids on plasma and red blood cell membrane phospholipids and deformability in weanling guinea pigs. Lipids. 1999;34:467–473. doi: 10.1007/s11745-999-0386-4. [DOI] [PubMed] [Google Scholar]
- 42.Dodt M, Roehr JT, Ahmed R, Dieterich C. FLEXBAR-Flexible Barcode and Adapter Processing for Next-Generation Sequencing Platforms. Biology (Basel) 2012;1:895–905. doi: 10.3390/biology1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Prediction of the 3D structure of ABHD5 extracted from an online protein structure homology-modelling server SWISS-MODEL (https://swissmodel.expasy.org)
Human-serine proteases siRNA library. List of siRNAs targeting known and predicted serine proteases (library was purchased from Ambion)