Abstract
Enzymatic activity is crucial for various technological applications, yet the complex structures and limited stability of enzymes often hinder their use. Hence, de novo design of robust biocatalysts that are much simpler than their natural counterparts and possess enhanced catalytic activity has long been a goal in biotechnology. Here, we present evidence for the ability of a single amino acid to self-assemble into a potent and stable catalytic structural entity. Spontaneously, phenylalanine (F) molecules coordinate with zinc ions to form a robust, layered, supramolecular amyloid-like ordered architecture (F–Zn(ii)) and exhibit remarkable carbonic anhydrase-like catalytic activity. Notably, amongst the reported artificial biomolecular hydrolases, F–Zn(ii) displays the lowest molecular mass and highest catalytic efficiency, in addition to reusability, thermal stability, substrate specificity, stereoselectivity and rapid catalytic CO2 hydration ability. Thus, this report provides a rational path towards future de novo design of minimalistic biocatalysts for biotechnological and industrial applications.
Enzymes attain unique three-dimensionally folded protein structures encoded by the precise amino-acid sequence to execute highly specific and efficient catalytic reactions1–3. The tremendous advantages of enzymatic reactions, for both biological in vivo activity and technological applications, include high efficiency, a high degree of selectivity (regio-, chemo- and enantio-) and eco-friendly reaction conditions4. Enzymes thus provide great opportunities to carry out important chemical reactions in numerous research and industrial fields, including food, health, cosmetics, agriculture, energy and applied enzymology, an important branch of industrial biotechnology5. Despite the great potential of enzymes in biotechnology, in most cases practical applications have been hampered mainly due to the difference between the cellular environment and the industrial setting6. Amongst the pool of enzymes, carbonic anhydrases (CAs, EC 4.2.1.1) are some of the fastest known metalloenzymes, potentially having global implications for biotechnology (Fig. 1a)7. CAs are the most ubiquitous enzymes present in various organisms and they participate in diverse physiological functions, such as pH homeostasis, calcification, bone resorption and photosynthesis. CA is the principal protein constituent of erythrocytes, along with haemoglobin, and plays a central role in the transport of carbon dioxide via its reversible catalytic hydration into bicarbonate (HCO3−) and protons (H+). In addition, CA exhibits efficient esterase activity. This unique catalytic ability of CAs has attracted considerable interest in biotechnology, carbon sequestration systems, biofuels and calcite production8. However, due to critical drawbacks in CAs, such as lack of thermal and long-term stability, high production costs and difficulties associated with recovery and reusability, the search for new, robust CA mimetic biomaterials is still ongoing. Consequently, considerable effort has been made towards fabrication of artificial CA mimics9–17. However, the length of the genetically coded amino-acid sequences, the complexity of the folding process and the stability of the protein pose tremendous challenges. The X-ray crystallographic structure of CA II reveals a Zn(ii) ion in the active site, which coordinates three histidine side chains extending from adjacent, antiparallel β-strands (Fig. 1a). Consequently, numerous approaches are currently being developed to try to mimic the catalytic activity of CA II, including organometallic complexes, polymers, micelles and gels. However, these artificial organic systems are not common in native enzymes and biological systems. In this regard, biomolecular assemblies are an ideal supramolecular framework for constructing artificial enzymes because the building blocks and the driving forces for self-assembly are non-covalent interactions, which share many characteristics with natural proteins. Recent studies identified a library of short Zn(ii)-coordinated peptide amyloid assemblies showing catalytic activity for ester bond hydrolysis, a typical model reaction for CAs11,13,18–20. Therefore, the Zn(ii) ion acts as an important cofactor and the amyloid cross-β-sheet folding represents an optimal structural scaffold for the design of artificial CA mimetics. Specifically, aromatic dipeptides and metabolites are potential minimalistic mimics of amyloid assemblies21–27. In addition, we have demonstrated that the single amino acid, phenylalanine (F), can spontaneously self-assemble into hydrogen-bonded supramolecular cross-β-sheet secondary structures with amyloid characteristics24,25. Despite the success of initial studies on amyloid-associated catalysis11,13,20,28–35, so far, single amino-acid assemblies have not been examined for biocatalytic applications. Therefore, considering the amyloid-like and highly ordered assembly characteristics of phenylalanine, we set out to explore the possible CA-like catalytic activity of phenylalanine assemblies in the presence of zinc ions as a cofactor.
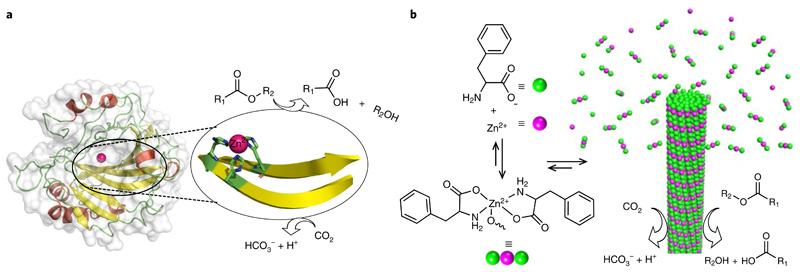
Fig. 1. Design of a minimalistic F–Zn(ii) biocatalyst through bioinspiration from structural insights of CA II.
a, Structure of human carbonic anhydrase (CA II; PDB entry 1CA2), which displays a zinc-coordinated cross-β-sheet catalytic centre that is involved in the catalysis of ester hydrolysis and interconversion of carbon dioxide to bicarbonate and proton. b, Chemical structure of phenylalanine and the monomeric 2:1 (F:Zn(ii)) complex and schematic of the nucleation and growth mechanism for rapid F–Zn(ii) co-assembly with CA-like catalytic activity.
Here, we present the rapid, single-crystal formation of zinc-bound phenylalanine (F–Zn(ii)) supramolecular amyloid-like cross-β-sheet assemblies. A combination of X-ray diffraction and spectroscopic methods was applied to characterize the molecular packing interactions among the phenylalanine and zinc ions. Remarkably, F–Zn(ii) crystals displayed CA-like biocatalytic activity towards both ester hydrolysis and hydration of carbon dioxide in an aqueous medium (Fig. 1b). This very small amino acid-based enzyme mimetic could, potentially, lead to a better understanding of hydrolase enzyme, as well as provide a model for de novo design of minimalistic and robust enzyme mimetics.
Results
Synthesis and structural insights of F–Zn(ii) self-assembly
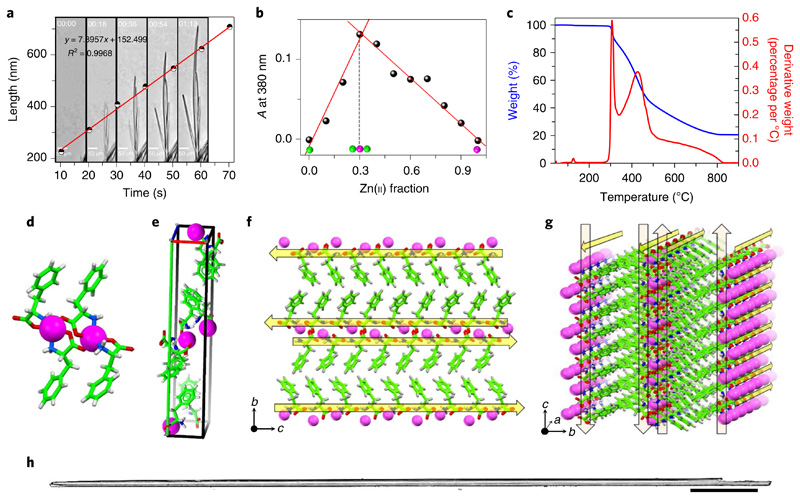
Aiming to generate zinc-incorporated structures, we first examined the self-assembly process of phenylalanine in the presence of zinc ions. Under alkaline and mild heating conditions, the phenylalanine and zinc ions spontaneously self-assembled into needle-like F–Zn(ii) crystals that were several micrometres long. Next, to estimate the rate at which the phenylalanine molecules and Zn(ii) ions self-organize in solution to form the ordered crystals, we monitored the crystallization process in real time (Fig. 2a). Briefly, a quartz cuvette was filled with a freshly prepared alkaline solution of phenylalanine and ZnCl2 building blocks (in a 2:1 ratio) and immediately capped to prevent evaporation and concentration changes. The crystallization process inside the cuvette was monitored using light microscopy, and the images were captured at one-second intervals (Supplementary Video 1). Spontaneously formed, small nucleation seeds were observed in fractions of seconds, and unidirectional progressive growth was sustained for several minutes. Figure 2a shows five consecutive snapshots of crystal growth at different time points. The plot of the representative single-structure growth as a function of time exhibited an excellent linear fit (R2 ≤ 1.00) with an elongation rate of 7.90 nm s−1 (Fig. 2a). For this specific structure, which was 16.66 μm in width, this rate translates into a volume increase of ~1.72 μm3 s−1. On the basis of a crystal structure unit cell volume of 1,668.94 Å3 (Supplementary Table 1), which contains a total of 12 molecules (eight phenylalanine and four Zn(ii)), we can estimate that 8.24 × 109 phenylalanine and 4.12 × 109 Zn(ii) building-block molecules organize together into an ordered structure in each second. To gain more insight into the nucleation step that initiates the spontaneous crystallization, in-situ absorption spectral measurements were performed during the crystallization process in alkaline medium. Addition of Zn(ii) ions to an alkaline phenylalanine solution resulted in the appearance of a new, broad absorption band centred at a wavelength of λmax = 380 nm, which can be attributed to the metal-to-ligand charge transfer coupled with ligand-to-ligand charge transfer transition of the F–Zn(ii) coordinated complex (Supplementary Fig. 1).36 The Job plot generated at the charge transfer band (λmax = 380 nm) versus the fraction of Zn(ii) ions in a fixed total concentration ([F+Zn(ii)] = 2 mM) clearly showed an inflection point at a 0.32 Zn(ii) fraction, corresponding to a 2:1 (F:Zn(ii)) binding stoichiometry (Fig. 2b). No crystallization was observed when the fraction of Zn(ii) was below 0.3 under similar conditions. Fourier transform infrared spectroscopy supported the coordination of phenylalanine carboxylic and amino groups to Zn(ii) by displaying a new metal–oxygen stretching band at 1,620 cm−1 and a significant red shift in the symmetric stretching band of NH, from 3,122 cm−1 to 3,249 cm−1 (Δν ≅ 127 cm−1, where Δν is the difference in frequency values), compared with those of phenylalanine alone (Supplementary Fig. 1b). Thus, the crystallization was mainly nucleated by a 2:1 phenylalanine to Zn(ii) complex formation (Fig. 1b). To increase the surface area, the crystals were ground to a fine powder and thoroughly characterized. The powder X-ray diffraction spectra of the ground crystals completely matched those of the single-crystal structure simulated powder pattern, thus confirming that the crystal structure within the ground powder form is identical to that of the single crystals (Supplementary Fig. 1c). In addition, energy dispersive X-ray spectroscopy measurements identified the presence of Zn along with the carbon (C) and oxygen (O) atoms (Supplementary Fig. 1d). Moreover, thermal gravimetric analysis experiments displayed a high thermal stability for the complex, with three thermal decomposition temperatures of 307 °C (14.6%), 428 °C (45.8%) and 584 °C (18.4%) (Fig. 2c).
Fig. 2. Characterization of F–Zn(ii).
a, Optical microscopy snapshots taken from Supplementary Video 1 that monitor the assembly kinetics of the F–Zn(ii) needle-like architectures captured every 18 s which are graphical representations of the elongation rate of a single structure. The calculated average measured elongation rate is 7.89 nm s−1. b, A UV–visible Job plot that monitors the absorption (A) at 380 nm as a function of the fraction of Zn(ii) ions. The inflection point at 0.3 corresponds to a stable 2:1 (F:Zn(ii)) binding stoichiometry. c, Thermogravimetric analysis of F–Zn(ii) crystals (blue curve) that reveals the high thermal stability (≤300 °C). The weight losses are depicted by the first derivative curve (red). d–g, Crystallographic insights of F–Zn(ii) (CCDC 1850564). d, Zinc-coordinated phenylalanine assembly that exhibits a 2:1 (F:Zn(ii)) primary complex structure. e, Unit cell that displays eight phenylalanine molecules and four Zn(ii) ions. f,g, Supramolecular cross-β-sheet layered organization along the crystallographic b–c (f) and a–b (g) planes. Yellow arrows direct from the the N to the C terminus of F. h, Optical microscope images of needle-like, long, single crystals of F–Zn(ii). Scale bar, 500 μm.
To gain better insight into the assembly process, we further performed single-crystal X-ray diffraction analysis (Fig. 2d–g). The determined structure revealed molecular packing of a monoclinic crystal system with a P21 space group containing eight phenylalanine molecules and four zinc ions per unit cell (Fig. 2e). Each zinc ion was coordinated with two phenylalanine residues through amine (NH2) and carboxylate (−OOC) groups (Fig. 2d). The coordination sphere is completed by one of the adjacent zinc–phenylalaninate carboxyl oxygens bonding to Zn(ii) on the axial position to form a distorted trigonal bipyramidal geometry. This primary complex further propagated into a layered structure that is stabilized by alternating hydrophobic (aromatic environment) and hydrophilic (both hydrogen bonding and Zn(ii) coordination) interactions. Each layer was composed of two continuous rows of phenylalanine molecules held together by Zn(ii) coordination and hydrogen bonds along the crystallographic a and c axes (Fig. 2f,g). These layers were further stabilized by interlayer edge-to-face aromatic phenyl ring interactions along the b axis. Collectively, the Zn(ii) coordination and hydrogen bonding, within and between the layers, in combination with the aromatic stacking, resulted in a strong set of interactions to hold this well-ordered, three-dimensional complex structure within the crystal lattice. More interestingly, the hydrogen-bonded phenylalanine molecular orientation (N and C termini) within the layers (along with the b–c and a–c crystal planes), and the aromatic interactions between the layers (along with the b axis), showed a significant resemblance to the β-sheet secondary structure of an amyloid-forming peptide37,38. The yellow arrows in Fig. 2f,g depict the direction of the N to C terminus-ordered phenylalanine molecules held together by hydrogen bonding and Zn(ii) coordination interactions, demonstrating the supramolecular cross-β-sheet, amyloid-like, hydrogen-bonded secondary structure. These superstructures further propagate along the crystallographic long axis, thereby forming needle-like single crystals (Fig. 2h). In contrast, another amino acid, glycine (G), coordinates with Zn(ii) (G–Zn(ii)) to form a closed tetramer ring organization39. This result clearly demonstrates the significance of stacking interactions gained by the aromatic phenyl moieties in the amyloid-like supramolecular assembly of F–Zn(ii), rather than there being mere coordination sites to zinc. Therefore, aromatic moieties play a central role by providing an energetic contribution as well as order and directionality to the assembly.
F–Zn(ii) catalytic esterase activity
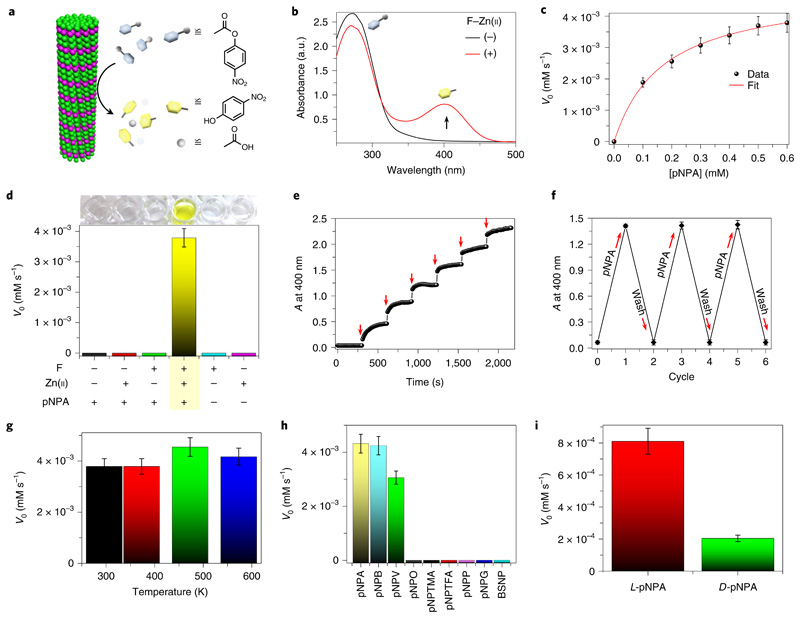
After demonstrating the minimalistic abiotic structural model, we sought to validate the CA-like catalytic activity of F–Zn(ii). The ester hydrolysis activity of CA has been widely assayed and standardized using a simple chromogenic substrate, p-nitrophenyl acetate (pNPA), by monitoring the appearance of the yellow hydrolysed product, p-nitrophenolate (pNP) (Fig. 3a). We therefore tested F–Zn(ii) hydrolase activity towards pNPA in deionized (DI) water using UV–visible absorption spectroscopy (Fig. 3b). In the presence of F–Zn(ii), the colourless pNPA aqueous solution turned yellow (Supplementary Videos 2 and 3), which was further evident from the appearance of an intense product absorption band at 400 nm (Fig. 3b). The absorption kinetic profile monitored at 400 nm exhibited an enzyme-like fast esterase trend in the presence of F–Zn(ii) (Supplementary Fig. 2). In contrast, under similar conditions, the control substrate solution without the catalyst did not show any visible colour change, nor an absorption band at 400 nm, even after weeks of incubation. This suggests that no background reaction took place. In addition, a control experiment (Supplementary Fig. 2) using G–Zn(ii) as a catalyst showed approximately fivefold less activity than F–Zn(ii), revealing the fact that the differences in the microenvironment at the active sites of F–Zn(ii) and G–Zn(ii) may account for the major difference in catalytic activity. Michaelis–Menten kinetics is a well-known model of enzyme kinetics that distinguishes enzymes from other chemical catalysts. To determine whether the F–Zn(ii) catalyst follows characteristic enzyme kinetics, the initial catalytic hydrolysis rates were determined as a function of various substrate concentrations. A smooth Michaelis–Menten model fitting (R2 = 0.998) plot verified the enzyme-like characteristics of F–Zn(ii), allowing us to extract values for the Michaelis constant (KM) = 0.169 mM and the maximum rate (Vmax) = 4.85 × 10−3 mM s−1 (Fig. 3c). Remarkably, the calculated catalytic efficiency (kcat/KM, where kcat is the catalytic rate constant) was found to be 76.54 M−1 s−1, which is the highest efficiency value among the reported artificial biomolecular hydrolases shown in Table 1 (refs.11–17). Moreover, this is the smallest known artificial hydrolase, about ~175-fold smaller (molecular mass of F, 165.08 Da) than CA II (~29,000 Da). Therefore, based on the molecular mass, the activity of F–Zn(ii) (kcat/KM = 46 × 10−2 (g l−1)−1 s−1) is eight times greater than that of the native CA II enzyme (pNPA hydrolysis at pH 7, kcat/KM = 5.7 × 10−2 (g l−1)−1 s−1). Furthermore, control experiments using all the possible combinations of catalyst, such as phenylalanine (F), Zn(ii) and pNPA, revealed the hydrolysis activity to be strictly F–Zn(ii) catalyst dependent, with no significant above-background activity observed in the absence of any of the catalytic building blocks (Fig. 3d). Moreover, the in-situ microscopy (Supplementary Video 3) and filtration experiments (Supplementary Fig. 4) provide further evidence of the catalytic function of F–Zn(ii) single crystals, and excludes the contribution from dissolved or non-assembled structures. In addition, the catalytic activity was pH dependent and increased with the pH of the reaction medium (Supplementary Fig. 6b).
Fig. 3. Catalytic esterase activity of F–Zn(ii).
a, Schematic illustration of ester hydrolysis catalysed by F–Zn(ii). b, UV–visible absorption spectra of a pNPA (10−3 M) aqueous solution with ((+), red curve) or without ((−), black curve) F–Zn(ii) catalyst (0.1 mg ml−1); a.u., absorbance units. The black arrow shows the change in the peak following addition of F–Zn(ii). c, Initial rate (V0) of hydrolysis catalysed by F–Zn(ii) as a function of substrate concentration, with the smooth Michaelis–Menten curve fitting. d, The control pNPA hydrolysis experiments showing catalysis by F–Zn(ii). No hydrolysis is observed when one of F or Zn(ii) is omitted from the system. e, Overlaid spectra of absorbance (A) changes at 400 nm as a function of time with sequential addition of F–Zn(ii) (0.02 mg ml−1, red arrows) to constant pNPA aqueous solution (10−3 M). The sharp jumps in absorbance are characteristic of F–Zn(ii)-catalysed fast product formation. f, Changes in absorbance at 400 nm for the chromogenic pNPA hydrolysis reaction during the F–Zn(ii) catalytic recycling and reuse process. g, The thermal catalytic stability of F–Zn(ii). The catalyst was annealed at different temperatures (298.15, 373.15, 473.15 and 573.15 K) and evaluated with the initial rate (V0) of pNPA hydrolysis measured at room temperature. h, The initial rate (V0) of hydrolysis for a series of different substrates in the presence of F–Zn(ii). The graph exhibits a clear substrate-specific (ester hydrolysis) catalytic activity. i, The initial hydrolysis rate (V0) of two enantiomers of Boc-phenylalanine 4-nitrophenyl ester (l/d-pNPA) catalysed by F–Zn(ii). Error bars in all graphs represent standard deviations of at least three independent measurements.
Table 1. Biomolecular artificial hydrolases.
| Amino-acid sequence | Molecular mass (Da) | Substrate | kcat/Km (M−1 s−1) | kcat/Km × 10−2 ((g l−1)−1 s−1) | Refs. |
|---|---|---|---|---|---|
| F | 165 | pNPA | 76.54a and 10.62b (pH 7.0) | ~46a and ~6.4b | This study |
| Carbonic anhydrase (CA II) | ~29,000 | pNPA | 1,670 (pH 7.0) | ~5.7 | 46 |
| Ac-IHIHIYI-NH2 | 949 | pNPA | 355 (pH 8.0) | ~37 | 18 |
| Ac-IHIHIQI-NH2 | 914 | pNPA | 62 (pH 8.0) and 15.76 (pH 7.3) | ~6.7 and ~1.7 | 11,13 |
| HKH-LLLAAA(K)-palmitoyl | 1,338 | DNPAc | 19.76 (pH 7.4) | ~1.4 | 12 |
| HSGVKVKVKVKVDPPTKVKVKVKV-NH2 | 2,510 | pNPA | 19.18 (pH 9.0) | ~0.76 | 14 |
| Fmoc-FFH-NH2 + Fmoc-FFR-NH2 | 671 + 689 | pNPA | 1.82 (pH 7.5) | ~0.13 | 16 |
| Ac-GLKALEEKCKALEEKLKALEEKHKALEEKG-NH2 | 3,432 | pNPA | 1.38 (pH 7.5) | ~0.04 | 10 |
| RSGQQKFQFQFEQQ- NH2 + HSGQQKFQFQFEQQ- NH2 | 1,785 + 1,766 | pNPA | 0.15 (pH 7.4) | 4.2×10−5 | 17 |
In deionized water.
In Tris–HCl (25 mM) buffer solution.
2,4-dinitrophenyl acetate (DNPA).
Furthermore, the catalytic rate is greatly influenced by the size of the F–Zn(ii) catalyst, with the ground powder (average mean size 250 nm) showing two times higher efficiency than the large crystals (>300 μm) (Supplementary Fig. 5), which was attributed to the increased catalytic surface area with a decrease in particle size. However, the F–Zn(ii) catalytic rate of pNPA hydrolysis is greatly hampered in Tris–HCl-buffered solution (25 mM, pH 7), suggesting occupation of the H2O-binding site with anionic species present in the buffer solution (Supplementary Fig. 2d). Having established efficient esterase activity, we proceeded to investigate the catalytic response by monitoring the product absorbance band at 400 nm as a function of time, with addition of small and equal amounts of F–Zn(ii) at regular intervals (Fig. 3e). Interestingly, a sharp jump in absorbance was displayed with each successive addition of F–Zn(ii). Such instantaneous changes provide strong evidence for a quick and effective enzyme-like catalytic response. An important aspect of a catalyst is its reusability. We, therefore, evaluated the reusability of the F-Zn(ii) catalyst by employing it in consecutive cycles of pNPA hydrolysis. Figure 3f demonstrates the reuse capacity of F–Zn(ii) for more than five cycles and, as shown, F–Zn(ii) maintained 99% of its activity even after five cycles of reuse. One of the notable features of the F–Zn(ii) catalyst is its high thermal structural stability, as confirmed by thermogravimetric analysis (Fig. 2c). To analyse the thermal catalytic stability, F–Zn(ii) was incubated at 373.15, 473.15 and 573.15 K for 2 h, and the pNPA esterase activity was measured in aqueous solution at room temperature (Fig. 3g). Interestingly, all the heated F–Zn(ii) samples displayed a similar catalytic rate, equal to the unincubated control, demonstrating an extremely high thermal catalytic stability. Next, to determine the temperature effect on reaction rates, we further performed the esterase activity at different reaction temperatures (295 K to 323 K). Interestingly, a gradual increase in the rate of pNPA hydrolysis was observed with a rise in temperature, providing valuable information on temperature-controlled catalytic function (Supplementary Fig. 6d). On the other hand, the catalytic activity of native CA II is completely lost on heating to 363.15 K. Therefore, the significant higher thermal catalytic stability of F–Zn(ii) is potentially useful for various industrial applications. This remarkable catalytic stability is attributed to the strong Zn(ii)-coordinated, amyloid-like structural integrity of F–Zn(ii). Substrate specificity and stereoselectivity are important characteristics of enzymatic catalysis. To evaluate the substrate specificity of F–Zn(ii), the hydrolysis reaction kinetics were analysed using a series of nitrophenyl esters with different side-chain lengths (4-nitrophenyl acetate (pNPA), 4-nitrophenyl butyrate (pNPB), 4-nitrophenyl valerate (pNPV), 4-nitrophenyl octanoate (pNPO)), bulky side chains having electron releasing (−CH3; 4-nitrophenyl trimethylacetate (pNPTMA)) and electronwithdrawing (–F; 4-nitrophenyl trifluoroacetate (pNPTFA)) groups and chemically distinct substrates (p-nitrophenyl phosphate (pNPP), p-nitrophenyl-β-d-glucopyranoside (pNPG) and benzene-sulfonic acid 4-nitrophenyl ester (BSNP)) (Fig. 3h). Among these, F–Zn(ii) could selectively catalyse the ester hydrolysis with a side-chain length-dependent rate, validating the substrate-specific catalytic action. The longer side chain led to a reduced hydrolysis rate. However, no catalytic hydrolysis was observed on the bulky side chain (pNPTMA and pNPTFA) and chemically distinct substrates (pNPP, pNPG and BSNP). Next, the stereoselectivity was tested using l- and d-Boc-phenylalanine 4-nitrophenyl ester (l/d-pNPA) enantiomers as substrates (Fig. 3i). Interestingly, the l-pNPA hydrolysis rate is fourfold higher than that of the d-pNPA enantiomer, suggesting a stereospecific catalytic function. This can be attributed to the asymmetric organization (chiral space group P21) of phenylalanine and Zn(ii) molecules within the crystal lattice promoting the stereospecific catalytic function.
Reaction pathway for F–Zn(ii) catalytic esterase reaction
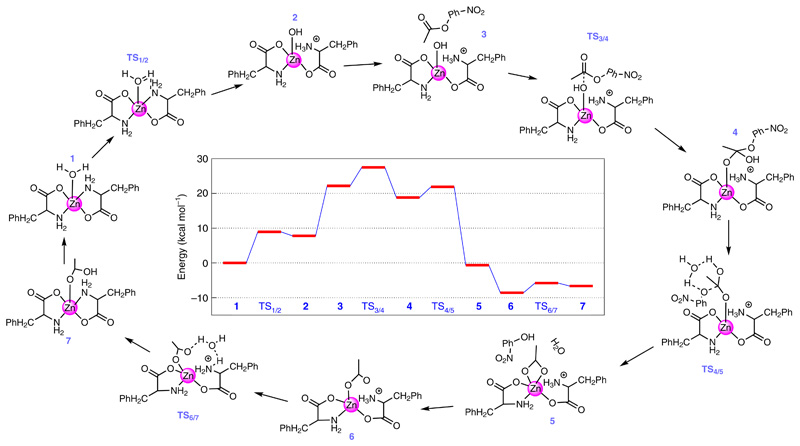
Following the successful experimental demonstration of the catalytic esterase activity of F–Zn(ii), we performed density functional theory calculations at the M06–2X/6-31 + g(d,p) level of theory to understand the step-by-step reaction mechanism. For the simulation, we considered a monomer unit of the F–Zn(ii) catalyst, and the initial geometry for the monomer unit was obtained from the experimental X-ray diffraction structure. The solvent medium was modelled using the polarizable continuum model. To better describe the solvent–monomer interactions, we also added five explicit water molecules near the F–Zn(ii) unit. As shown in Fig. 4, the monomer unit including the surrounding water molecules, complex 1, is the starting point of the entire reaction. In complex 1, one of the water molecules is bound to the zinc atom (Zn–O distance = 2.095 Å). The bound water molecule loses a proton to one of the NH2 groups (bound to the Zn) to generate the OH− nucleophile complex 2. The transition state for the proton transfer, complex TS1/2, involves the formation of a proton bridge between the Zn-bound H2O and the NH2 through another water molecule surrounding the complex. This proton transfer is an endothermic reaction with an energy difference (ΔG0) of 7.77 kcal mol−1 between complexes 1 and 2 and an energy barrier (ΔG≠) of 8.92 kcal mol−1. The calculated energies, ΔG0 and ΔG≠, between the complexes 1 and 2 are close to the energies (ΔG0 = 4.31 kcal mol−1 and ΔG≠ = 11.03 kcal mol−1) calculated for the first two steps of the pNPA hydrolysis in the presence of self-assembled short peptides40. Complex 3 is formed once pNPA approaches the hydroxide ion of complex 2. The subsequent bond formation between the carbon atom of the carbonyl group of pNPA and the oxygen atom of the hydroxide ion (bound to the Zn atom) leads to the formation of complex 4. This nucleophilic attack of OH− goes through the transition state, complex TS3/4, where the carbon atom of the carbonyl group is in close proximity to the oxygen atom of the hydroxide ion. The formation of 4 from 3 is an exothermic reaction with ΔG0 = −3.36 kcal mol−1 and ΔG≠ = 5.32 kcal mol−1. It is important to note that, in complex 4, the oxygen atom binding to the Zn belongs to the carbonyl group of pNPA and not the oxygen atom of the hydroxide ion. From complex 3 to complex 4, the distance between the Zn atom and the oxygen atom of the hydroxide ion (carbonyl group) increases (decreases) from 1.91 Å to 3.05 Å (3.39 Å to 1.91 Å). Also, the ester bond (the bond between the carbon of the carbonyl group and the oxygen of the phenoxide group of the pNPA) increases from 1.36 Å to 1.45 Å. This elongated ester bond dissociates further to form complex 5 through TS4/5. In 5, one of the products, namely, p-nitrophenol is formed and the remaining acetate ion is bound to the Zn complex. The conversion of complex 4 to 5 is highly exothermic, with ΔG0 = −22.53 kcal mol−1 and with a small barrier of ΔG≠ = 3.095 kcal mol−1. Complex 6 is formed after releasing the pNP from the active site. Finally, the transfer of a proton from the NH3 moiety to the acetate ion through the proton bridge (TS6/7) leads to the formation of complex 7, where the other product of acetic acid is formed, and the original complex is restored. The ΔG0 and ΔG≠ values for this step are 1.89 kcal mol−1 and 2.74 kcal mol−1, respectively. The release of the acetic acid molecule from the active site and the subsequent binding of a new water molecule to the active site re-initiates the entire process. It is important to note that when the proton transfers from the H2O to NH2 (that is, steps for TS1/2 → 2), the NH2 group moves away from the Zn site, and the reverse is observed when the proton transfers from the NH3 moiety to the acetate ion (steps for TS6/7 → 7).
Fig. 4. F–Zn(ii) catalytic esterase reaction mechanism and the chemical structures along the reaction pathway.
The entire reaction starts with the binding of a water molecule (from the solvent) to the central zinc atom of the monomeric F–Zn(ii) catalytic complex (1). The calculated free energy profile is shown in the centre. The optimized three-dimensional geometries of all the steps involved in the reaction are shown in Supplementary Fig. 9.
To further address the effect of aggregation on the catalytic reaction kinetics, we performed several additional calculations with a dimer configuration. In these calculations, we explored various possible sites for the proton transfer and considered different paths for the pNPA molecule while it is approaching the Zn-bound hydroxide ion. A detailed description of these additional calculations is given in the Supplementary Discussion. Briefly, as we move from the monomer to the dimer, the energy barrier between complex 1 and TS3/4 reduces (by ~8.5 kcal mol−1) and the exothermicity of the reaction from complex 3 to 4 increases (by ~8.2 kcal mol−1). Due to the combination of these effects, an aggregation shows enhanced catalytic activity for pNPA hydrolysis compared to the monomer and this result was demonstrated through our experiments.
F–Zn(ii) catalytic CO2 hydration reaction
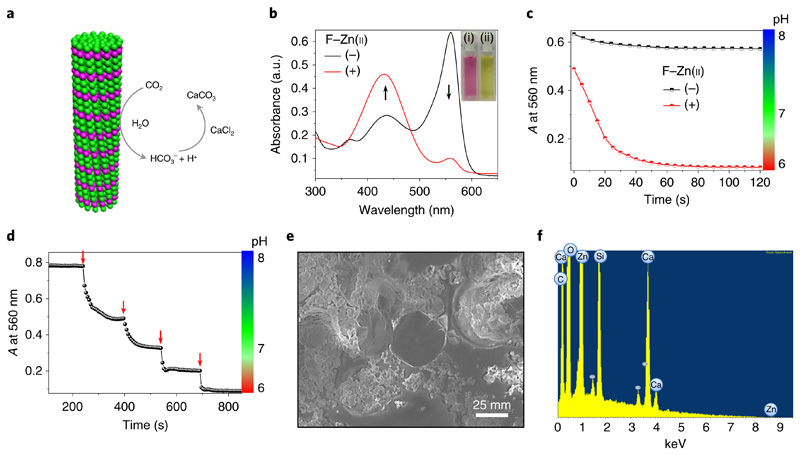
The native catalytic reaction of CA is CO2 hydration. In the presence of water, CA reversibly converts CO2 into bicarbonate and a proton, resulting in a drop in overall pH. Therefore, we have evaluated whether our designed F–Zn(ii) crystals can catalyse a similar reaction (Fig. 5a). The CO2 hydration experiments were carried out by mixing CO2-saturated deionized water into Tris buffer (25 mM, pH 8) solution in the presence or absence of the F–Zn(ii) catalyst. A colorimetric method using the pH indicator phenol red was applied to measure the shift in pH during the reaction. In the absence of F–Zn(ii), the solution was pink and displayed an intense absorption band at 560 nm and a less intense band at 430 nm, which is characteristic of an alkaline (pH 8) medium (Fig. 5b). Interestingly, on addition of F–Zn(ii) the solution turned pale yellow and exhibited minimal absorption at 560 nm, accompanied by an increased absorption band at 430 nm, which is characteristic of an acidic environment. Furthermore, the decrease in absorbance at 560 nm was followed over time (Fig. 5c). In the presence of F–Zn(ii), a rapid (within 60 s) decrease in absorbance (ΔA ≅ 0.54) corresponding to an acidic pH of 6 was observed. However, without the F–Zn(ii) catalyst, the solution showed an alkaline environment (pH 8) with a minimal change in absorbance (ΔA ≅ 0.06). These observations clearly suggest that the F–Zn(ii)-catalysed CO2 hydration induced a pH switch from alkaline to acidic. The CO2- hydration kinetic parameters in the presence of F–Zn(ii) were found to be kcat = 7.8 s−1, kcat/KM = 962 M−1s−1 and KM = 8.1 mM. However, the background, uncatalysed reaction (kcat = 0.48 s−1, kcat/KM = 100 M−1s−1 and KM= 4.8 mM) is ninefold slower than the F–Zn(ii)-catalysed reaction. Next, the stepwise catalytic function was monitored on addition of small amounts of saturated CO2 aqueous solution (10 μl) at regular intervals (Fig. 5d). A sharp and instantaneous drop in absorbance (or pH) with each successive addition provided strong evidence of the CO2 hydration reaction catalysed by F–Zn(ii). Furthermore, the hydrated CO2 was sequestered in the form of CaCO3 through a reaction with CaCl2 (10 mM) in Tris buffer at pH 8. A white precipitate was indeed obtained on addition of the CaCl2 solution to the catalyst-treated aqueous solution located in a CO2 closed chamber. In contrast, no visible precipitate was obtained in the absence of F–Zn(ii). A field emission scanning electron microscopy image of the precipitate showed plate-like crystalline microstructures (Fig. 5e). Energy dispersive X-ray spectroscopy analysis indicated the presence of calcium (Ca) element along with carbon (C), oxygen (O) and zinc (Zn), with the elementary compositions (Supplementary Fig. 9a) being close to the ratio of CaCO3, thus further confirming the catalytic sequestration of CO2 (Fig. 5f). In addition, Fourier transform infrared spectroscopy analysis supported the formation of CaCO3 by displaying characteristic absorbance bands at 1,407, 845 and 670 cm−1 (Supplementary Fig. 9b).
Fig. 5. F–Zn(ii) catalytic carbon dioxide hydration and sequestration.
a, Schematic representation of F–Zn(ii) catalytic CO2 hydration and sequestration on reaction with CaCl2 to produce solid CaCO3. b, UV–visible absorption spectral changes of the pH indicator phenol red (5 × 10−5 M) in a mixture containing CO2-treated deionized water in Tris buffer (pH 8) with (+) and without (−) the F–Zn(ii) (3.7 × 10−4 M) catalyst. The black arrows show the changes in the peaks following addition of F-Zn(ii). Inset: images of the corresponding solutions without (left) and with (right) F–Zn(ii). c, The time-dependent absorbance at 560 nm and the corresponding pH profile of phenol red in CO2-treated deionized water, Tris buffer (pH 8) in the absence (−) and presence (+) of F–Zn(ii). d, Stepwise drop in absorbance at 560 nm and respective pH by sequential addition of CO2-treated deionized water (10 μl, red arrows) into the Tris buffer (25 mM, pH 8) containing F–Zn(ii). e, Scanning electron micrograph of the solid precipitate after sequestration of CO2-treated F–Zn(ii) aqueous solution with CaCl2. f, Energy dispersive X-ray spectrum of the precipitate showing calcium (Ca), along with carbon (C), oxygen (O) and zinc (Zn) elements.
Discussion
In summary, we describe a minimalistic de novo enzyme design strategy involving a simple amino acid metallo-supramolecular structure to mimic the CA hydrolase enzyme. In contrast to other biomolecular CA mimics, F–Zn(ii) demonstrated rapid single-crystal growth kinetics with high thermal stability. Importantly, the F–Zn(ii) single crystals compellingly exhibited two CA catalytic activities, both as esterase towards the pNPA substrate and in CO2 hydration. A remarkable esterase catalytic efficiency of 76.54 M−1 s−1 (46 × 10−2 (g l−1)−1 s−1) was found, which is the highest amongst reported artificial biomolecular-based CA mimetics, and is even greater than that of native CA II enzyme on the basis of molecular mass. Furthermore, the efficient catalytic reusability for several catalytic cycles, the thermal catalytic stability over a wide range of high temperatures and the substrate specificity, stereoselectivity and rapid catalytic CO2 hydration properties, are promising in the context of industrial applications. The combination of experimental and computational results suggests that the extraordinary activity of F–Zn(ii) is engendered by its amyloid-like supramolecular structural framework, along with the dense ordered array of zinc ions on the surface allows easy access of substrates to the highly Lewis-acidic Zn(ii) catalytic centre. Given these exciting results, computational insights suggest that the incorporation of proton accepting groups at amine or aromatic rings is likely to be a working strategy to further improve the catalytic activity of F–Zn(ii). Thus, the enzyme-like catalytic activity exhibited by simple, zinc-bound amino-acid assemblies may provide the link between amino acids and the emergence of primitive precursor enzymes during evolution. We envisage that this minimalistic strategy, with advantages of rich structural diversity and metal chelation of natural amino acids, will provide the basis for de novo design of various other enzyme mimetics for industrial and biotechnological applications.
Methods
Preparation of F–Zn(ii) single crystals
One equivalent of ZnCl2 aqueous solution was slowly added to two equivalents of an l-phenylalanine alkaline solution containing two equivalents of NaOH under heating (60 °C). Spontaneously, needlelike white crystals started growing at the liquid–air interface. The crystals were filtered off, washed with deionized water and ethanol several times and used for the catalytic studies.
X-ray data collection and processing
A single needle crystal (250 × 5 × 5 μm3) was mounted at the end of a loop (Dual-Thickness MicroLoop LD 20, MiTeGen). The crystal was cryo-protected by a quick dip in a mixture of 65% mother liquor and 35% glycerol, and then cryo-cooled to 100 K in a nitrogen stream for X-ray diffraction data collection. Data were collected at the Advanced Photon Source (Argonne National Laboratory), beamline 24-ID-C, using a Pilatus-6MF pixel array detector. A 10-μm aperture was used and the crystal was translated along the length of the needle to expose fresh crystal during data collection. A total of 720 images were collected using an oscillation of 0.5° and wavelength of 0.8266. Data reduction and scaling were performed using XDS41. Data processing statistics indicate strong diffraction (I/σ > 9) to 0.75 Å resolution, but completeness diminished between resolutions of 0.90 Å to 0.75 Å due to the limits of the data collection geometry. The data were 91% complete at a resolution of 0.90 Å and 81% complete at a resolution of 0.75 Å (36% complete in the shell of 0.77–0.75 Å). Data collection and refinement statistics are reported in Supplementary Table 1.
The crystal structure was determined using direct methods with the program SHELXT42. The model was refined using REFMAC5 (ref. 43) and SHELXL44. After each refinement step, the model was visually inspected in Coot45 using both 2Fo − Fc and Fo − Fc difference maps. Here Fo and Fc are the observed structure factors and the calculated structure factors respectively.
Esterase activity
The esterase activity of the prepared samples was measured using a 96-well UV-Star UV transparent plate (Greiner BioOne) with a flat bottom using of pNPA as the substrate and F–Zn(ii) single crystals as the catalyst in deionized water. Reaction progress was monitored using a Biotek Synergy HT plate reader (Biotek) at 22 °C and continuous stirring mode. The pH-dependence studies were performed in Tris–HCl buffer solution (25 mM).
Kinetic experiments
The kinetic experiments were performed as described above. To increase the catalytic rate, F–Zn(ii) single crystals were mechanically ground into a fine powder form using a mortar and pestle and were dispersed in NMP solution. Separately, 0.1 M substrate (pNPA) stock solution was prepared in acetonitrile. The reaction was initiated by addition of the catalyst (3.7 × 10−4 M) to the substrate solution at the appropriate concentration (prepared from dilution of stock) in deionized water (200 μl total reaction volume containing 1% of NMP and 1% acetonitrile). The progress of the reaction was monitored at the product (pNP) absorbance wavelength (λmax = 400 nm) as a function of time. The linear part of the absorbance versus time plot of esterase activity was fitted with an extinction coefficient for the product (pNP) (ε = 264 M−1 cm−1, obtained experimentally from the slope of absorbance (λmax = 400 nm) versus the concentration of pNP under similar conditions (Supplementary Fig. 2)) and attained the initial reaction rate (V0) at various substrate concentrations. The average of the V0 values was plotted as a function of substrate concentration and fitted to Michaelis–Menten kinetics to obtain the Vmax and KM values. All measurements were replicated at least three times and averaged for accuracy.
To analyse the thermal stability, 100 mg of catalyst was inserted into a closed glass vial and annealed in an oil bath at various temperatures, ranging from 298.18 K to 573.15 K. After reaching room temperature, the catalytic activity was measured as described above. To study the reusability, 2 mg of catalyst was inserted into a 1.5-ml Eppendorf tube containing 1 ml of pNPA (1 mM) in deionized water. The reaction mixture was vortexed for 5 min and centrifuged to precipitate the catalyst. The supernatant was separated and the UV–visible absorbance spectra were recorded. After each reaction cycle, the catalyst was precipitated, washed with deionized water and reused for the next cycle of reaction.
CO2 hydration and in vitro crystallization of CaCO3
Carbon dioxide-saturated water was prepared by bubbling CO2 gas into 100 ml deionized water in a round-bottomed flask using a porous gas bubbler (flow rate of 600 cm3 min−1) under ice-cold conditions for 1 h. Then, 50 μl of CO2-treated water was added to the pH indicator phenol red (50 μM)–Tris buffer (20 mM, pH 8, 150 μl) solution in the presence or absence of the F–Zn(ii) catalyst, and the absorbance spectra were recorded. The CO2 hydration reaction was monitored as the change in the 560 nm absorbance band and the total pH change was extracted. In vitro CaCO3 crystallization was carried out by slow diffusion of CO2 evaporated from the decomposition of (NH4)2CO3 (for 24 h at 25 °C) into a crystallization system containing droplets of aqueous CaCl2 (7.5 mM) solution and F–Zn(ii) (2 mg ml−1). Control experiments were carried out without the catalyst.
Supplementary Material
Supplementary information is available for this paper at 10.1038/s41929-019-0348-x.
Acknowledgements
This work was partially supported by a grant from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (BISON, Advanced ERC grant, no. 694426) (to E.G.). P.M. gratefully acknowledges the Center for Nanoscience and Nanotechnology of Tel Aviv University for a postdoctoral fellowship, C. M. Dobson, University of Cambridge, and B. Rosen, Tel Aviv University, for stimulating discussions. S.S.R.K.C.Y. and B.M.W. acknowledge the support of the US Army Research Office under grant no. W911NF-17-1-0340 and the National Science Foundation for the use of supercomputing resources through the Extreme Science and Engineering Discovery Environment (XSEDE), project no. TG-ENG160024. D.S.E and M.R.S. acknowledge the Northeastern Collaborative Access Team beamline 24-ID-C, which is funded by the National Institute of General Medical Sciences from the National Institutes of Health (grant no. P41 GM103403) and uses resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility, operated under contract no. DE-AC02-06CH11357. The Pilatus 6M detector is funded by an NIH-ORIP HEI grant (no. S10 RR029205). We also thank S. Rencus-Lazar for linguistic editing and all the members of the Gazit laboratories for helpful discussions.
Footnotes
Kai Tao: 0000-0003-3899-5181
Linda J. W. Shimon: 0000-0002-7861-9247
Bryan M. Wong: 0000-0002-3477-8043
Ehud Gazit: 0000-0001-5764-1720
Data availability
The X-ray crystallographic coordinates for the structure reported in this study have been deposited at the Cambridge Crystallographic Data Centre (CCDC), under deposition 1850564. Other data that support the plots within this paper and other finding of this study are available from the corresponding author upon reasonable request.
Author contributions
P.M. and E.G. conceived and designed the experiments. P.M. grew the single crystals of catalytic complex and performed all experiments. P.M. and K.T. conducted the Fourier transform infrared and scanning electron microscopy measurements. D.S.E., M.R.S. and L.J.W.S. collected the single-crystal diffraction data and solved the crystal structure. S.S.R.K.C.Y. and B.M.W. performed the computational studies. P.M. and E.G. wrote and edited the manuscript. All authors discussed and commented on the manuscript.
Competing interests
The authors declare no competing interests.
Reprints and permissions information is available at www.nature.com/reprints.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vendruscolo M, Dobson CM. Structural biology: dynamic visions of enzymatic reactions. Science. 2006;313:1586–1587. doi: 10.1126/science.1132851. [DOI] [PubMed] [Google Scholar]
- 2.DeGrado W, Wasserman Z, Lear J. Protein design, a minimalist approach. Science. 1989;243:622–628. doi: 10.1126/science.2464850. [DOI] [PubMed] [Google Scholar]
- 3.Benkovic SJ, Hammes-Schiffer S. A perspective on enzyme catalysis. Science. 2003;301:1196–1202. doi: 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]
- 4.Aldridge S. Industry backs biocatalysis for greener manufacturing. Nat Biotechnol. 2013;31:95–96. doi: 10.1038/nbt0213-95. [DOI] [PubMed] [Google Scholar]
- 5.Schmid A, et al. Industrial biocatalysis today and tomorrow. Nature. 2001;409:258–268. doi: 10.1038/35051736. [DOI] [PubMed] [Google Scholar]
- 6.Arnold FH. Combinatorial and computational challenges for biocatalyst design. Nature. 2001;409:253–257. doi: 10.1038/35051731. [DOI] [PubMed] [Google Scholar]
- 7.Christianson DW, Fierke CA. Carbonic anhydrase: evolution of the zinc binding site by nature and by design. Acc Chem Res. 1996;29:331–339. [Google Scholar]
- 8.Boone C, Habibzadegan A, Gill S, McKenna R. Carbonic anhydrases and their biotechnological applications. Biomolecules. 2013;3:553–562. doi: 10.3390/biom3030553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song WJ, Tezcan FA. A designed supramolecular protein assembly with in vivo enzymatic activity. Science. 2014;346:1525–1528. doi: 10.1126/science.1259680. [DOI] [PubMed] [Google Scholar]
- 10.Zastrow ML, Peacock AFA, Stuckey JA, Pecoraro VL. Hydrolytic catalysis and structural stabilization in a designed metalloprotein. Nat Chem. 2012;4:118–123. doi: 10.1038/nchem.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rufo CM, et al. Short peptides self-assemble to produce catalytic amyloids. Nat Chem. 2014;6:303–309. doi: 10.1038/nchem.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guler MO, Stupp SI. A self-assembled nanofiber catalyst for ester hydrolysis. J Am Chem Soc. 2007;129:12082–12083. doi: 10.1021/ja075044n. [DOI] [PubMed] [Google Scholar]
- 13.Friedmann MP, et al. Towards prebiotic catalytic amyloids using high throughput screening. PLoS ONE. 2015;10:e0143948. doi: 10.1371/journal.pone.0143948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C, et al. Switchable hydrolase based on reversible formation of supramolecular catalytic site using a self-assembling peptide. Angew Chem Int Ed Engl. 2017;56:14511–14515. doi: 10.1002/anie.201708036. [DOI] [PubMed] [Google Scholar]
- 15.Singh N, Conte MP, Ulijn RV, Miravet JF, Escuder B. Insight into the esterase like activity demonstrated by an imidazole appended self-assembling hydrogelator. Chem Commun. 2015;51:13213–13216. doi: 10.1039/c5cc04281j. [DOI] [PubMed] [Google Scholar]
- 16.Huang Z, et al. Self-assembly of amphiphilic peptides into bio-functionalized nanotubes: a novel hydrolase model. J Mater Chem B. 2013;1:2297–2304. doi: 10.1039/c3tb20156b. [DOI] [PubMed] [Google Scholar]
- 17.Zhang C, et al. Self-assembled peptide nanofibers designed as biological enzymes for catalyzing ester hydrolysis. ACS Nano. 2014;8:11715–11723. doi: 10.1021/nn5051344. [DOI] [PubMed] [Google Scholar]
- 18.Al-Garawi ZS, et al. The amyloid architecture provides a scaffold for enzyme-like catalysts. Nanoscale. 2017;9:10773–10783. doi: 10.1039/c7nr02675g. [DOI] [PubMed] [Google Scholar]
- 19.Lee M, et al. Zinc-binding structure of a catalytic amyloid from solid-state NMR. Proc Natl Acad Sci USA. 2017;114:6191–6196. doi: 10.1073/pnas.1706179114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lengyel Z, Rufo CM, Moroz YS, Makhlynets OV, Korendovych IV. Copper-containing catalytic amyloids promote phosphoester hydrolysis and tandem reactions. ACS Catal. 2018;8:59–62. doi: 10.1021/acscatal.7b03323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gazit E. Self-assembled peptide nanostructures: the design of molecular building blocks and their technological utilization. Chem Soc Rev. 2007;36:1263–1269. doi: 10.1039/b605536m. [DOI] [PubMed] [Google Scholar]
- 22.Tao K, Makam P, Aizen R, Gazit E. Self-assembling peptide semiconductors. Science. 2017;358 doi: 10.1126/science.aam9756. eaam9756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makam P, Gazit E. Minimalistic peptide supramolecular co-assembly: expanding the conformational space for nanotechnology. Chem Soc Rev. 2018;47:3406–3420. doi: 10.1039/c7cs00827a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adler-Abramovich L, et al. Phenylalanine assembly into toxic fibrils suggests amyloid etiology in phenylketonuria. Nat Chem Biol. 2012;8:701–706. doi: 10.1038/nchembio.1002. [DOI] [PubMed] [Google Scholar]
- 25.Mossou E, et al. The self-assembling zwitterionic form of l-phenylalanine at neutral pH. Acta Crystallogr C. 2014;70:326–331. doi: 10.1107/S2053229614002563. [DOI] [PubMed] [Google Scholar]
- 26.Shaham-Niv S, Adler-Abramovich L, Schnaider L, Gazit E. Extension of the generic amyloid hypothesis to nonproteinaceous metabolite assemblies. Sci Adv. 2015;1:e1500137. doi: 10.1126/sciadv.1500137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aizen R, Tao K, Rencus-Lazar S, Gazit E. Functional metabolite assemblies—a review. J Nanopart Res. 2018;20:125. doi: 10.1007/s11051-018-4217-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carny O, Gazit E. A model for the role of short self-assembled peptides in the very early stages of the origin of life. FASEB J. 2005;19:1051–1055. doi: 10.1096/fj.04-3256hyp. [DOI] [PubMed] [Google Scholar]
- 30.Rout SK, Friedmann MP, Riek R, Greenwald J. A prebiotic template-directed peptide synthesis based on amyloids. Nat Commun. 2018;9 doi: 10.1038/s41467-017-02742-3. 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenwald J, Riek R. On the possible amyloid origin of protein folds. J Mol Biol. 2012;421:417–426. doi: 10.1016/j.jmb.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 32.Omosun TO, et al. Catalytic diversity in self-propagating peptide assemblies. Nat Chem. 2017;9:805–809. doi: 10.1038/nchem.2738. [DOI] [PubMed] [Google Scholar]
- 33.Zozulia O, Dolan MA, Korendovych IV. Catalytic peptide assemblies. Chem Soc Rev. 2018;47:3621–3639. doi: 10.1039/c8cs00080h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh N, Kumar M, Miravet JF, Ulijn RV, Escuder B. Peptide-based molecular hydrogels as supramolecular protein mimics. Chem Eur J. 2017;23:981–993. doi: 10.1002/chem.201602624. [DOI] [PubMed] [Google Scholar]
- 35.Greenwald J, Kwiatkowski W, Riek R. Peptide amyloids in the origin of life. J Mol Biol. 2018;430:3735–3750. doi: 10.1016/j.jmb.2018.05.046. [DOI] [PubMed] [Google Scholar]
- 36.Hossain M Elius, Hasan M Mahmudul, Halim ME, Ehsan MQ, Halim MA. Interaction between transition metals and phenylalanine: a combined experimental and computational study. Spectrochim Acta A. 2015;138:499–508. doi: 10.1016/j.saa.2014.11.084. [DOI] [PubMed] [Google Scholar]
- 37.Makin OS, Atkins E, Sikorski P, Johansson J, Serpell LC. Molecular basis for amyloid fibril formation and stability. Proc Natl Acad Sci USA. 2005;102:315–320. doi: 10.1073/pnas.0406847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riek R, Eisenberg DS. The activities of amyloids from a structural perspective. Nature. 2016;539:227–235. doi: 10.1038/nature20416. [DOI] [PubMed] [Google Scholar]
- 39.Konar S, et al. Structural determination and characterization of copper and zinc bis-glycinates with X-ray crystallography and mass spectrometry. J Coord Chem. 2010;63:3335–3347. [Google Scholar]
- 40.Song R, et al. Principles governing catalytic activity of self-assembled short peptides. J Am Chem Soc. 2019;141:223–231. doi: 10.1021/jacs.8b08893. [DOI] [PubMed] [Google Scholar]
- 41.Kabsch W. XDS. Acta Crystallogr D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheldrick GM. SHELXT – integrated space-group and crystal-structure determination. Acta Crystallogr A. 2015;71:3–8. doi: 10.1107/S2053273314026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 44.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verpoorte JA, Mehta S, Edsall JT. Esterase activities of human carbonic anhydrases B and C. J Biol Chem. 1967;242:4221–4229. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.