Abstract
Carotenoids play many crucial roles in organisms. Recently, the de novo synthesis of carotenoids has been reported in pea aphid (Acyrthosiphon pisum) through horizontally transferred genes. However, their upstream pathway in the pea aphid is poorly understood. Geranylgeranyl diphosphate synthase (GGPPS) is the functional enzyme in the synthesis of geranylgeranyl diphosphate (GGPP) which is a precursor for the biosynthesis of many biological metabolites, including carotenoid synthesis. In this study, we performed a series of experiments to characterize GGPPS gene and its association with carotenoid biosynthesis. (1) determining the transcript abundance and carotenoid content in two geographical strain with red and green morphs, and (2) examining the abundance of carotenoid related genes and carotenoid levels after silencing of GGPPS in both red and green morphs. We observed that GGPPS was more highly expressed in the green morph than in the red morph of two strains of the pea aphid. The total level of carotenoids was also higher in green morphs than in red morphs in both strains. In addition to the total carotenoid difference, the carotenoids found in the two morphs also differed. There were α-carotene, β-carotene, and γ-carotene in the green morphs, but three additional carotenoids, including cis-torulene∗, trans-torulene∗, and 3,4-didehydrolycopene∗, were present in the red morphs. Silencing the GGPPS by RNAi in both the red and green morphs decreased the expression of some carotenoid biosynthesis-related genes, including carotenoid synthase/cyclase genes and carotenoid desaturase genes in green morphs. Carotenoid levels were decreased in both green and red morphs. However, the specific carotenoids present were not changed after silencing GGPPS. These results demonstrated that GGPPS may act as the upstream enzyme to influence the synthesis of the total amount of carotenoids. The present study provided important molecular evidence for the conserved roles of GGPPS associated with carotenoids biosynthesis and will enhance further investigation on the mechanisms of carotenoid biosynthesis in pea aphid.
Keywords: geranylgeranyl diphosphate synthase, carotenoid biosynthesis, horizontal gene transfer, RNAi, aphid
Introduction
Carotenoid is a generic term for a natural pigment, common in animals, higher plants, fungi, algae, and bacteria. Carotenoids play many crucial roles in organisms, including photosynthesis (Niedzwiedzki et al., 2017), protection of photo-oxidation (Frank and Brudvig, 2004), diapause (Bryon et al., 2017), and biological regulation (Heath et al., 2013). There are three ways that insects obtain carotenoids. These include dietary ingestion (Shimizu et al., 1981), endosymbionts (Sloan and Moran, 2012), and de novo synthesis. Synthesis has been reported in piercing-sucking pests, such as aphids, mosquitoes, and mites, through functional carotenoid biosynthetic genes, which were originally horizontally transferred from fungi (Moran and Jarvik, 2010; Altincicek et al., 2012; Cobbs et al., 2013). In the pea aphid, Acyrthosiphon pisum, carotenogenic genes are derived from fungi via horizontal gene transfer and through a series of duplication induced three Carotenoid Synthase or Cyclase genes (CscA-C) and four Carotenoid Desaturase genes (CdeA-D) (Moran and Jarvik, 2010; Supplementary Figure S1). In plants, bacteria, fungi, and algae, GGPP is synthesized by GGPPS and used as a precursor for the biosynthesis of carotenoids (Misawa et al., 1990; Hundle et al., 1994; Bartley and Scolnik, 1995; Mende et al., 1997; Cazzonelli and Pogson, 2010; Yang et al., 2016). However, the upstream pathway of de novo carotenoid synthesis in the pea aphid is poorly understood.
Geranylgeranyl diphosphate (GGPP) is a 20C organic compound synthesized by geranylgeranyl diphosphate synthase (GGPPS) through the head-to-tail condensation of three isopentenyl pyrophosphate (IPP) groups to the allyl head of dimethylallyl pyrophosphate (DMAPP). Some GGPPS can alternatively use geranyl pyrophosphate (GPP) or farnesyl pyrophosphate (FPP) as a substrate to produce GGPP (Sagami et al., 1993; Zhang and Li, 2014). Generally, GGPPS contains five highly conserved motifs, and the first and second aspartate-rich (FARM and SARM) motifs DDxx(xx)D are deemed to be the binding and catalysis sites in prenyltransferases (Quondam et al., 1997; Chang et al., 2006). According to the amino acid residues of the fourth and fifth positions before the first DDxx(xx)D, GGPPS is divided into three types: type-I (archaea), type II (plants and bacteria), and type III (yeasts and animals) (Barbar et al., 2013). The GGPPS in insects belong to type III GGPPS and they have an extra motif VI (GxNP) (Zhang and Li, 2014; Yang et al., 2016).
The number of GGPPS varies among species, and different GGPPS may produce GGPP using different metabolic pathways (Okada et al., 2000). For example, 12 GGPPS have been identified and 10 are functional that can synthesize GGPP from Arabidopsis thaliana. These genes are distributed in different subcellular compartments and display different expression patterns and most GGPPSs are distributed in plastids and expressed in the specific root or seed tissues (Lange and Ghassemian, 2003; Beck et al., 2013). However, GGPPS11 (At4g36810) is widely expressed in photosynthetic tissues to provide GGPP for the biosynthesis of chlorophylls, carotenoids, or plastoquinones, which are vital for photosynthesis in A. thaliana (Okada et al., 2000; Beck et al., 2013; Coman et al., 2014). Two types of GGPPS (the first type is encoded GGPP for primary metabolism and the second one is responsible for secondary metabolism) have been isolated in Gibberella fujikuroi (Bettina, 2005; Singkaravanit et al., 2010). Only one GGPPS is found in most insects, such as the fruit fly Drosophila melanogaster (Lai et al., 1998), bumblebee, Bombus terrestris (Prchalová et al., 2016), spruce budworm, Choristoneura fumiferana (Barbar et al., 2013), and cotton aphid, Aphis gossypii (Zhang and Li, 2014).
Geranylgeranyl diphosphate synthase acts as a rate-limiting enzyme from IPP transformed to GGPP, and its enzyme activity not only affects the biosynthesis of carotenoids and chlorophylls, but it also affects the synthesis of other products derived from the isoprenoid pathway (Zhang and Li, 2014; Zhou et al., 2017). Targeting the isoprenoid pathway has been widely used in clinical agents (Swanson and Hohl, 2006) and the inhibitor of GGPPS could provide an alternative way in dealing with the diseases involving geranylgeranylation (Wiemer et al., 2011; Varney et al., 2018; Waller et al., 2019). In D. melanogaster, the geranylgeranyl pyrophosphate synthase-encoding gene, quemao, is critical in isoprenoids biosynthesis and heart development (Lai et al., 1998; Yi et al., 2006). GGPP synthesized by GGPPS is used to produce a diterpene in Reticulitermes speratus (Hojo et al., 2011). Silencing of GGPPS influences the biosynthesis of alarm pheromone (E)-β-farnesene (EβF) in A. gossypii (Sun and Li, 2018).
In this study, we report the identification of a single GGPPS in A. pisum and GGPPS expression and carotenoid content in two strains (NY and GS) of A. pisum. The expression patterns of GGPPS, carotenoid synthase/cyclase and desaturase genes, and carotenoid content were evaluated upon silencing of GGPPs in A. pisum. These results provide a new approach to the molecular regulation of carotenoid and promote the understanding of carotenoid biosynthesis in aphids.
Materials and Methods
Insects
To clarify the potential role of GGPPS in pea aphid, two population of A. pisum was used in the present study. The New York (NY) strain of A. pisum was originally collected from an alfalfa field in New York City, United States, in 2012. The Gansu (GS) strain was collected from an alfalfa field in Gansu Province, China, in 2016. These two strain kept in the lab for more than 3 years, without any exposure to pesticides and were fed separately on broad bean (Vicia faba) under 22 ± 1°C, 60 ± 10% relative humidity, and a 16:8 h (light: dark) photoperiod for more than 30 generations before applied in this study. All of the aphids were reared at a relatively low density (less than 30 individuals per seedling) in a continuous culture before being used in experiments. All of the progeny were produced by asexual reproduction.
cDNA Synthesis and GGPPS Sequence Confirmation
Total RNAs used for GGPPS sequence confirmation and expression profiles were extracted using the TRIzol kit (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. DNase I (Promega, Madison, WI, United States) and PrimeScript RT Reagent Kit (Takara, Dalian, China) were used to removing possible genomic DNA contamination and synthesize the first-strand cDNA. The synthesized cDNA was stored at −20°C until use.
The GGPPS sequence was retrieved from both the genome database of A. pisum (AphidBase)1 and the NCBI database and the sequence was confirmed by sequencing the RT-PCR products by Sanger sequencing (Supplementary Table S1). Briefly, the PCR reactions were performed in a C1000TM thermal cycler by an initial denaturation for 3 min at 95°C, followed by 35 cycles of 95°C for 30 s, 55°C to 60°C for 30 s, 72°C extension for 1 min, and a final extension for 10 min at 72°C. The reaction mixtures contained 10 × PCR buffer (Mg2+ free), 2.5 mM Mg2+, 2.5 mM dNTP mix, each specific primer (10 mM), and rTaq (Takara). The PCR products were purified and then ligated into a pGEM-T easy vector (Promega), which were then sequenced on an ABI Model 3100 automated sequencer (Invitrogen Life Technologies, Shanghai, China) to verify the GGPPs sequence.
Phylogenetic Analysis
To illustrate the evolutionary positions of GGPPS in the pea aphid, the relative homologies from different representative classifications, including insect, vertebrate, yeast, bacterium, and plant were used to construct the phylogenetic tree. All of the sequences were obtained from NCBI2. The full-length amino acid sequences were aligned using MEGA 5.05 with the default settings in ClustalW with the maximum likelihood method. Bootstrap values were calculated based on 1000 replicates (Tamura et al., 2011). Sequence alignment of GGPPS with the pea aphid and several other species were produced with JalView 2.9 (Waterhouse et al., 2009).
Spatiotemporal Expression and Sample Preparation
To detect the expression of GGPPS in two color types of an aphid, we collected red and green adults (within 12 h after the fourth instar nymph molt) from the NY and GS strains. In each group, 10 aphids were pooled as one biological replicate, and 4 biological replicates were included.
For determining the expression patterns of GGPPS at different developmental stages and tissues, we collected different developmental stages, namely, first, second, third, and fourth instar nymphs, and adults from the green morph of NY strain. For the nymphs, each stage was collected at three time points, namely, the early, middle, and late stages of each instar. In adults, four age time points were collected, namely, 8, 9, 12, and 15 days aphids (Supplementary Figure S2). Thirty insects were pooled as one sample. Tissues from the brain (200 individuals), stylet (200 individuals), integument (50 individuals), muscle (100 individuals), gut (100 individuals), fat body (100 individuals), and embryo (50 individuals) were excised from adults (within 12 h after the fourth instar nymph molt). Four biological replicates were performed for each group. All of the RNA samples were extracted by TRIzol and stored at −80°C until use.
Quantitative Real-Time PCR (RT-qPCR)
RT-qPCR primers were designed using PRIMER 3.03 (Supplementary Table S1). The RT-qPCR reaction mixtures contained nuclease-free water, each primer (0.2 mM), cDNA template, and NovoStart SYBR qPCR SuperMix (Novoprotein, Shanghai, China). The cDNA template used in the RT-qPCR reaction was diluted from the originally synthesized cDNA above with nuclease-free water to a final concentration of approximately 500 ng/μL. The reaction was performed on a Bio-Rad CFX Connect Real-Time System (Bio-Rad, Hercules, CA, United States) by following: 95°C for 120 s, 40 cycles of 95°C for 15 s, and 60°C for 30 s, and a final cycle of 60°C for 5 s and melting curve data were collected every 0.5°C until 95°C. A standard curve was established for each primer pair to determine the amplification efficiency. The reference genes elongation factor-1 alpha (EF1α) (GenBank accession number: AY219737.1) and ribosomal protein S20 (RPS20) (GenBank accession number: NM_001162819.2) were used to normalize the gene expression levels (Chen et al., 2016). The relative gene expression levels, based on two reference genes, were calculated using qBASE (Hellemans et al., 2007).
Carotenoid Extraction and HPLC Analysis
In parallel with part 2.4, we also collected red and green newly emerged adults within 12 h for determining the carotenoid content from the NY and GS strains. Twenty adults were pooled as a biological replicate, and four biological replicates were included in each group.
Carotenoids were extracted by following a standard protocol with slight modifications (Valmalette et al., 2012). Briefly, aphids were first ground in 200 μL of extraction solvent (acetone/hexane, 1:1 v/v, containing 0.1% of BHT as an antioxidant). Then, the extraction solvent was added to 1 mL and vortexed for 30 min. The extracts were centrifuged at 12,000 × g at 4°C for 10 min. The organic phases were dried under anhydrous sodium sulfate, filtered, and concentrated to dryness with a vacuum concentrator. The residue was dissolved in 50 μL methyl tert-butyl ether (MTBE) (containing 0.1% of BHT as an antioxidant). Samples were placed in amber vials before high-performance liquid chromatography (HPLC) analysis.
Carotenoids were separated along a C30 column (250 × 4.6 mm, 5 μm particle size) (YMC Co., Ltd., Kyoto, Japan) (Zhang et al., 2009; Valmalette et al., 2012). The mobile phases were acetonitrile/methanol (3:1 v/v, containing 0.1% of BHT as antioxidant) as eluent A, and MTBE (containing 0.1% of BHT as antioxidant) as eluent B. Flow rate was fixed at 1 mL/min and the column temperature was set at 30°C. A gradient program was performed: 0–10 min, 95% A/5% B, isocratic elution: 10–19 min, 86% A/14% B; 19–29 min, 75% A/25% B; 29–54 min, 50% A/50% B; 54–66 min, 26% A/74% B; 67–67 min, back to the initial conditions for re-equilibration. The injection volume was 20 μL and the detection was monitored at 450 nm. The α-carotene, β-carotene, and γ-carotene were identified and quantitatively determined by the standards using Agilent 1260 LC (Agilent Technologies, Santa Clara, CA, United States). The cis-torulene∗, all trans-torulene∗, and 3, 4-didehydrolycopene∗ were identified according to previous studies and calculated based on the β-carotene equivalent (Tsuchida et al., 2010; Valmalette et al., 2012).
Serial dilutions of carotenoid standards were used to establish the standard curves. For α-carotene standard (Sigma, St. Louis, MO, United States), 4, 2, 1, 0.8, 0.6, and 0.4 μg/mL were included. For β-carotene standard (Sigma), 4, 2, 1, 0.8, and 0.6 μg/mL were included and for γ-carotene standard (Sigma), concentrations were 1, 0.8, 0.6, 0.4, and 0.2 μg/mL. The calibration curve and coefficients were listed in Supplementary Figures S3–S5.
RNAi Bioassays
The double-strand RNA (dsRNA) was synthesized using a Transcript Aid T7 High Yield Transcription Kit (Thermo Scientific, Wilmington, DE, United States) according to the manufacturer’s instructions (Supplementary Table S1). The dsRNA was diluted with Nuclease-free water to a final concentration of 5,000 ng/μL.
Newly emerged green and red form adults (≤12 h old) from the NY strain were used in the RNAi experiments. The dsRNA injection was accomplished as in a previous study (Ye et al., 2019). Briefly, 600 ng of dsGGPPS and dsGFP (negative control) were injected into the aphids using an M3301 micromanipulator (World Precision Instruments, Sarasota, FL, United States). In each treatment, 25 aphids were injected as one biological replicate and 4 biological replicates were completed. After injection, the aphids were moved to broad bean leaves. After 36 h, in each biological replicate, 5 aphids were collected to detect gene expression levels and 20 aphids were collected for analyzing carotenoids content.
Statistical Analysis
Significant differences in carotenoid content in red and green morphs from the two strains and transcript levels of GGPPS in tissues were tested by one-way analysis of variance (ANOVA) followed by Tukey’s honestly significant difference test (Tukey’s HSD) for multiple samples comparison (P < 0.05). Significant differences in transcript levels of GGPPS in red and green morphs from the two strains were tested by Student’s t-test. Student’s t-tests were used to analyze the significance of gene expression level and carotenoid content between the dsGGPPS treatment and control (dsGFP). All of the statistical analyses were performed using SPSS version 20.0 software (IBM, Armonk, NY, United States).
Results
Phylogenetic Analysis of GGPPS
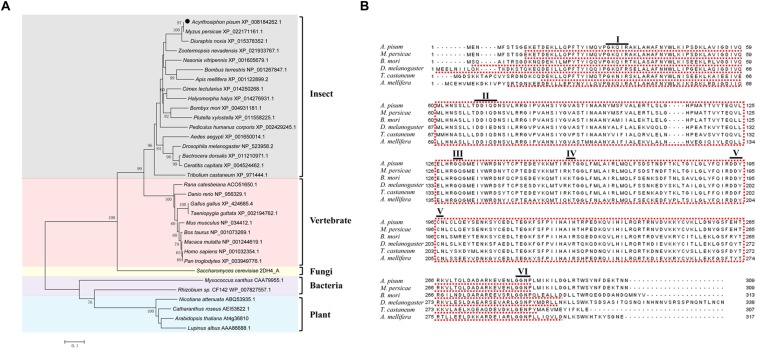
The open reading frame of GGPPS in A. pisum contains 930 bp and encodes 309 amino acid residues. The predicted isoelectric point of GGPPS is 6.32 and the molecular weight is 35 kDa. To investigate the evolutionary relationships of the GGPPS gene among different organisms, the full-length amino acid of GGPPS from seven insects, nine vertebrates, one fungus, two bacteria, and four plants were used to construct a phylogenetic tree. The GGPPS of insects and vertebrates belong to a large branch, close to fungal GGPPS (Saccharomyces cerevisiae) with a high value. GGPPS from A. pisum was the closest to the other two aphid species, Myzus persicae and Diuraphis noxia. GGPPS from bacteria and plants were clustered into another branch (Figure 1A).
FIGURE 1.
Phylogenetic tree and sequence alignments of GGPPS. (A) Phylogenetic tree of GGPPS generated by MEGA 5.05. (B) Alignment of the amino acid sequence of Acyrthosiphon pisum GGPPS with other insect GGPPS. A. pisum, Acyrthosiphon pisum; M. persicae, Myzus persicae; B. mori, Bombyx mori; D. melanogaster, Drosophila melanogaster; T. castaneum, Tribolium castaneum; A. mellifera, Apis mellifera. Conserved motifs are underlined, and the first and second aspartate-rich motifs (FARM: motif II and SARM: motif V, respectively) are DDIQD and DDYCN.
Sequence analysis showed that GGPPS from A. pisum and other insects were conserved and mainly include six conserved motifs: motif I (GKxxR), motif II (DDIQ/ED), motif III (GQ), motif IV (KT), motif V (DDYCN), and motif VI (GxNP). The first aspartate-rich motif (FARM) of A. pisum GGPPS was DDIQD, while in the second aspartate-rich motif (SARM), the last residue was an asparagine (DDYCN) instead of aspartate (Figure 1B and Supplementary Figure S6).
Expression Patterns of GGPPS in Green and Red Morphs
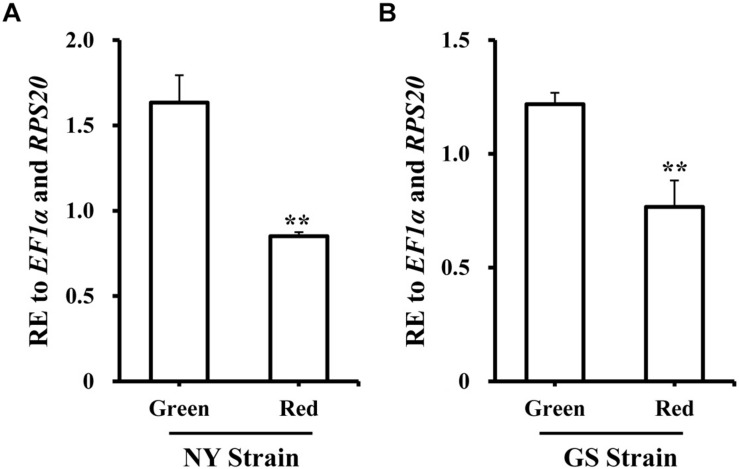
To better improve our understanding of the role in pea aphid, the expression level of GGPPS was analyzed in spatiotemporal dynamics and different body-color. Firstly, samples from the different developmental stages and tissues from the NY strain green morph were collected. GGPPS was consistently expressed during all of the tested stages and tissues. From the 1st to the 3rd instar nymphs, GGPPS expression slightly decreased and then increased along with aphid development (Supplementary Figure S7A). GGPPS were significantly higher in the brain and embryo tissues compared to other tissues, including stylet, integument, muscle, gut, and fat body (Supplementary Figure S7B). Secondly, the expression patterns of GGPPS were determined in red and green morphs of A. pisum from the NY and GS strains. In both strains, GGPPS was more highly expressed in the green morph compared to the red morph (Figure 2).
FIGURE 2.
Expression profiles of GGPPS in green and red morphs of the NY and GS strains of Acyrthosiphon pisum. (A) Expression patterns of GGPPS in red and green morphs of NY strain. (B) Expression patterns of GGPPS in red and green morphs of GS strain. A significant difference between red and green morphs is indicated by asterisks (∗∗P < 0.01).
Carotenoid Contents in Green and Red Morphs
Next, we determined the carotenoid contents of red and green morphs. In green morphs, only α-carotene, β-carotene, and γ-carotene were detected (Table 1). In red morphs, three additional carotenoids were detected, including cis-torulene∗, all trans-torulene∗, and 3,4-didehydrolycopene∗ (Table 1). The content of α-carotene, β-carotene, and total carotenoids was higher in the green morph compared to red morph in the NY and GS strains. No differences in compositions of individual carotenoids and the total content were observed between morphs of the NY and GS strains with the same body color (Table 1).
TABLE 1.
Carotenoid content of green and red morphs from NY and GS strains of Acyrthosiphon pisum.
| NY strain | GS strain | |||
| Green | Red | Green | Red | |
| α-carotene | 0.4920 ± 0.0202 b | 0.1846 ± 0.0123 a | 0.4592 ± 0.0273 b | 0.1798 ± 0.0147 a |
| β-carotene | 3.9132 ± 0.1498 b | 0.7828 ± 0.1569 a | 3.7664 ± 0.2266 b | 0.6316 ± 0.0831 a |
| γ-carotene | 0.1066 ± 0.0102 | 0.1150 ± 0.0343 | 0.0737 ± 0.0115 | 0.0980 ± 0.0140 |
| cis-torulene∗ | ND a | 0.8189 ± 0.2115 b | ND a | 0.7188 ± 0.0796 b |
| All-trans-torulene∗ | ND a | 0.5111 ± 0.0925 b | ND a | 0.4781 ± 0.0292 b |
| 3, 4-didehydrolycopene∗ | ND a | 0.3595 ± 0.0642 b | ND a | 0.3308 ± 0.0145 b |
| total | 4.5118 ± 0.1776 b | 2.7718 ± 0.5618 a | 4.2993 ± 0.2630 b | 2.4371 ± 0.2241 a |
∗ represents that the carotenoids were identified according to previous studies and calculated based on the β-carotene equivalent (Tsuchida et al., 2010; Valmalette et al., 2012). ND represents not detected. Lowercase letters (a and b) indicate significant differences in the content of the same carotenoid composition among different morphs or strains were tested by one-way ANOVA followed with Tukey’s honestly significant difference test for multiple samples comparison (P < 0.05).
Expression Profile of Carotenoids Associated Genes and Carotenoid Content Upon Silencing of GGPPS
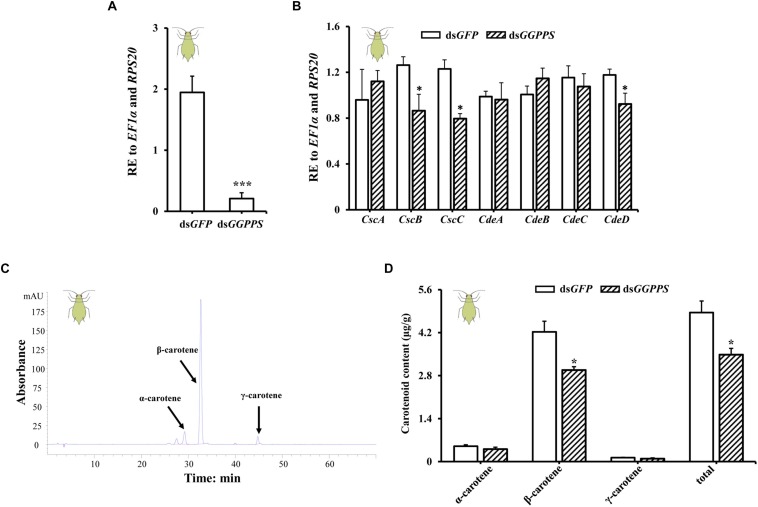
To future study the function of GGPPS in association with carotenoid biosynthesis, we examined the expression patterns of carotenoid biosynthesis-associated genes (including carotenoid synthase/cyclase genes and carotenoid desaturase genes obtained from fungi through horizontal gene transfer) and the carotenoid content in green and red morphs after RNAi of GGPPS. In green morphs, the expression level of GGPPS was significantly reduced by 89.4% after dsGGPPS injection (Figure 3A), and this decreased the expression levels of two carotenoid synthase/cyclase genes (CscB and CscC) and one carotenoid desaturase gene (CdeD). No significant effects on other carotenoid biosynthesis-related genes were observed (Figure 3B). The total carotenoid content was significantly decreased after silencing of GGPPS and the content of β-carotene was significantly reduced. Levels of α-carotene and γ-carotene were not significantly influenced and only slightly decreased (Figures 3C,D).
FIGURE 3.
Expression patterns of GGPPS, carotenoid biosynthesis-related genes, and carotenoids content upon silencing of GGPPS in the green morph of Acyrthosiphon pisum. (A) RNAi efficiency of GGPPS. (B) Expression patterns of carotenoid biosynthesis-related genes after RNAi of GGPPS. (C) The prominent peak retention time of green morph by HPLC. Black arrows represent different carotenoids. (D) Carotenoid content of the green morph after silencing of GGPPS. Significant differences between dsGFP and dsGGPPS are indicated by asterisks (∗P < 0.05; ∗∗∗P < 0.001).
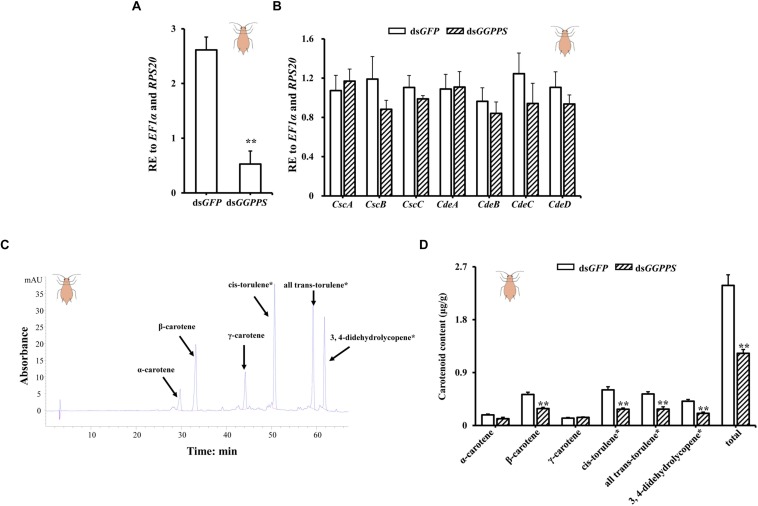
In the red morph, silencing produced an 80% reduction of GGPPS (Figure 4A), but there were no significant differences in the carotenoid biosynthesis-related genes. However, the expression of two carotenoid synthase/cyclase genes (CscB and CscC) and three carotenoid desaturase genes (CdeB, CdeC, and CdeD) decreased (Figure 4B). Similarly, the total carotenoid content was significantly reduced. Contents of β-carotene, cis-torulene∗, all trans-torulene∗, and 3,4-didehydrolycopene∗ were all significantly decreased, and α-carotene content decreased but there was no obvious effect on the content of γ-carotene (Figures 4C,D).
FIGURE 4.
Expression patterns of GGPPS, carotenoid biosynthesis-related genes, and carotenoids content upon silencing of GGPPS in the red morph of Acyrthosiphon pisum. (A) RNAi efficiency of GGPPS. (B) Expression patterns of carotenoid biosynthesis-related genes after RNAi of GGPPS. (C) The prominent peak retention time of red morph by HPLC. Black arrows represent different carotenoids. (D) Carotenoid content of the red morph after silencing of GGPPS. Significant differences between dsGFP and dsGGPPS are indicated by asterisks (∗∗P < 0.01).
Discussion
Geranylgeranyl diphosphate is a precursor for the biosynthesis of carotenoids and has been studied in organisms that synthesize carotenoids. Some insect species, such as aphids, can synthesize carotenoids via carotenoid biosynthesis-related genes obtained from fungi through horizontal gene transfer (Moran and Jarvik, 2010; Nováková and Moran, 2012). However, whether GGPPS is responsible for carotenoids biosynthesis in A. pisum, similar to reports in other organisms, has not been determined.
Based on the genome dataset of A. pisum, a single GGPPS in the isoprenoid biosynthesis pathway was identified. Sequence alignments of GGPPS with different insects showed that GGPPS are highly conserved, and the first five conserved motifs occurred in all trans-prenyltransferases. These results implied that GGPPS perform conserved functions in insects (Zhang and Li, 2014). GGPPS among animals (such as insects and vertebrates) and fungi group together and are referred to as type III GGPPS. Since fungi were the donors of the aphids horizontally transferred carotenoid-associated genes, it is reasonable to predict that GGPPS is the upstream enzyme determining the synthesis of aphid carotenoids. In addition, GGPPS could be categorized into three groups based on amino acid residues at the fourth or fifth location before the FARM or the insertion within the FARM. The pea aphid GGPPS was similar to type III GGPPS (Barbar et al., 2013), which is preferred for accepting FPP, compared to DMAPP, as an allylic substrate (Yang et al., 2014).
In plants, GGPPS are a small gene family and all homologs have tissue- and organelle-specific expression. GGPPS4 (At2g23800) is highly expressed in flower, GGPPS6 (At3g14530) is highly expressed in root and siliques, while GGPPS11 (At4g36810) is presented high abundance in all organs and tissues and is the one mainly responsible for most metabolites in A. thaliana (Beck et al., 2013). However, we identified only one ortholog of GGPPS in pea aphid and the transcript was more abundant in the green morph than in the red morph. This indicates the association of GGPPS and carotenoid synthesis. Bioinformatics analysis has revealed the carotenoid-associated genes in the pea aphid genome (Moran and Jarvik, 2010; Nováková and Moran, 2012). However, the biological and ecological functions of these genes are unclear. In addition, the original horizontal gene transfer has also undergone a series of duplications and there are now seven carotenoid associated genes in pea aphids (Moran and Jarvik, 2010). This suggests that these genes have provided a selective advantage to aphids possessing them. Similarly, sap-feeding spider mites, such as Tetranychus urticae and Panonychus citri, obtained carotenoid biosynthesis-related genes through horizontal gene transfer, and one carotenoid desaturase gene is involved in carotenoid accumulation and diapause (Altincicek et al., 2012; Bryon et al., 2017). Upon silencing of GGPPS by RNAi, the expression level of some carotenoid biosynthesis-related genes was decreased, indicating these horizontally transferred genes are functional and linked with GGPPS in the pea aphid. In A. thaliana, a GGPPS11 (At4g36810) knockdown strain (ggpps11-5) showed significantly reduced carotenoid levels compared to wild-type, while the lines of overexpression of GGPS11 alone do not accumulate higher amounts of carotenoids (Ruiz-Sola et al., 2016; Camagna et al., 2019). In Xanthophyllomyces dendrorhous, an increase in the levels of GGPPS resulted in higher carotenoid content (Breitenbach et al., 2011). Red and green morphs of A. pisum were significantly different in their relative expression levels of GGPPS and this may be associated with the total levels of carotenoids in the two-color forms. Silencing of GGPPS decreased the carotenoid content rather than changing the carotenoid composition. These data are consistent with the finding that carotenoid content and compositions are different in the two-color morphs (green morphs contain higher carotenoids content than that in red morphs, while red contains a number of different carotenoids than that in green morphs) (Moran and Jarvik, 2010; Tsuchida et al., 2010). The body-color polyphenism (red and green) may be related to carotenoids composition rather than the level of total carotenoids content (Moran and Jarvik, 2010; Tsuchida, 2016). The differences of total carotenoids in the two-color forms of A. pisum are probably associated with biological and ecological adaptations, such as host location, energy reserves, and probing behavior (Tsuchida, 2016; Zhang et al., 2016), which will require further study.
Besides contributing to carotenoids biosynthesis, GGPPS may have multiple physiological functions. Silencing of GGPPS increased the EβF amount but didn’t change the body length, body width, emitting cornicle droplets, fecundity, and survival in A. gossypii (Sun and Li, 2018). The duplication of the GGPPSs might be involved in the evolution of the chemical defense in Nasutitermes takasagoensis (Hojo et al., 2019). The GGPPS (qm) mutant in Drosophila showed a “broken-hearted” phenotype in embryos (Yi et al., 2006).
In total, our results indicate there might be an association with higher expression of GGPPS in the green morph and a relatively higher carotenoid content. Although direct silencing of GGPPS decreased the carotenoid contents, this suggests that GGPP may act as the substrate for carotenoid synthesis by downstream horizontally transferred genes regardless of red or green aphids. Does this mean carotenoids synthesis is a case of the substrate (GGPP) the concentration-dependent manner in green aphids or GGPPS plays a regulatory role in carotenoids synthesis? Besides, in this study, we focused on the very upstream gene GGPPS (non-horizontal transferred gene) in the evaluation of its involvement of carotenoids biosynthesis, and further association with the seven horizontally transferred carotenoids associated genes (carotenoid synthase/cyclase genes and carotenoid desaturase genes). It still requires intensive studies on further investigation of these seven genes in involvement with host adaptation, specifically, why aphids need duplicated carotenoids genes (Moran and Jarvik, 2010; Nováková and Moran, 2012)? Which are absent in most insects. This provides a nice model to study how horizontal transferred genes in facilitating the better adaptation of insects. So far, this is not yet clear any aphid species that are lacking these horizontally transferred genes, thus it is not possible to perform a comparative study between aphids with carotenoids synthesis genes and aphids without carotenoids synthesis genes. Further studies may be helpful in using tools such as genome editing approach to analyze the association between GGPPS and carotenoids synthesis in aphids (Le Trionnaire et al., 2019).
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
The research project was conducted on invertebrate species that are not subject to specific ethical issues and legislation.
Author Contributions
B-YD, JN, and J-JW conceived and designed the experiments. B-YD performed all of the experiments with the help of FS, LY, and T-YC. JN and J-JW provided the materials. B-YD, JN, and FS analyzed data. B-YD, JN, FS, and J-JW wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This research was supported in part by the China Postdoctoral Science Foundation (2018M640894), and the project was funded by Chongqing Special Postdoctoral Science Foundation (XmT2018063) and by the earmarked fund for Modern Agro-industry (Citrus) Technology Research System of China (CARS-26).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.01398/full#supplementary-material
The suppositional carotenoid biosynthesis pathway in the pea aphid. [According to Moran and Jarvik (2010)].
Schematic diagram of samples collected for different developmental stages. N1-1: Newly enclosed nymph in 4 h; N1-2: middle of 1st instar nymph; N1-3: late stage of 1st instar nymph; N2-1: early stage of 2nd instar nymph; N2-2: middle stage of 2nd instar nymph; N2-3: late stage of 2nd instar nymph; N3-1: early stage of 3rd instar nymph; N3-2: middle stage of 3rd instar nymph; N3-3: late stage of 3rd instar nymph; N4-1: early stage of 4th instar nymph; N4-2: middle stage of 4th instar nymph; N4-3: late stage of 4th instar nymph; AD-1: 8 days old adult; AD-2: 9 days old; AD-3: 12 days old; AD-4: 15 days old.
Standard curve of α-carotene obtained by HPLC. The linear regression equation was generated using 0.08, 0.04, 0.02, 0.016, 0.012, and 0.008 μg concentrations.
Standard curve of β-carotene obtained by HPLC. The linear regression equation was generated using 0.08, 0.04, 0.02, 0.016, and 0.012 μg concentrations.
Standard curve of γ-carotene obtained by HPLC. The linear regression equation was generated using 0.02, 0.016, 0.012, 0.008, and 0.004 μg concentrations.
Alignment of the amino acid sequence of Acyrthosiphon pisum GGPPS with GGPPS from other species. Conserved motifs are indicated by the red box with dotted lines.
Expression patterns of GGPPS during different developmental stages and tissues in the green morph of Acyrthosiphon pisum, NY strain. (A) The expression patterns of GGPPS in different developmental stages. N1: 1st instar nymph, N2: 2nd instar nymph, N3: 3rd instar nymph, N4: 4th instar nymph, AD: adult. (B) Expression patterns of GGPPS in tissues. BR, brain; ST, stylet; IN, integument; MU, muscle; GU, gut; FB, fat body; EM, embryo. Mean (±SE) expression level was based on four biological replicates. Lowercase letters (a and b) above each bar indicate significant differences among tissues (One-way ANOVA followed by Tukey’s honestly significant difference multiple comparison test; P < 0.05).
Primer sequences used for sequence confirmation, quantitative real-time PCR (qRT-PCR) and double stranded RNA (dsRNA) synthesis.
References
- Altincicek B., Kovacs J. L., Gerardo N. M. (2012). Horizontally transferred fungal carotenoid genes in the two-spotted spider mite Tetranychus urticae. Biol. Lett. 8 253–257. 10.1098/rsbl.2011.0704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbar A., Couture M., Sen S. E., Béliveau C., Nisole A., Bipfubusa M., et al. (2013). Cloning, expression and characterization of an insect geranylgeranyl diphosphate synthase from Choristoneura fumiferana. Insect. Biochem. Mol. Biol. 43 947–958. 10.1016/j.ibmb.2013.07.004 [DOI] [PubMed] [Google Scholar]
- Bartley G. E., Scolnik P. A. (1995). Plant carotenoids: pigments for photoprotection, visual attraction, and human health. Plant Cell 7 1027–1038. 10.1105/tpc.7.7.1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck G., Coman D., Herren E., Ruiz-Sola M. A., Rodríguez-Concepción M., Gruissem W., et al. (2013). Characterization of the synthase gene family in Arabidopsis thaliana. Plant Mol. Biol. 82 393–416. 10.1007/s11103-013-0070-z [DOI] [PubMed] [Google Scholar]
- Bettina T. (2005). Gibberellin biosynthesis in fungi: genes, enzymes, evolution, and impact on biotechnology. Appl. Microbiol. Biotechnol. 66 597–611. 10.1007/s00253-004-1805-1 [DOI] [PubMed] [Google Scholar]
- Breitenbach J., Visser H., Verdoes J. C., Ooyen A. J. J. V., Sandmann G. (2011). Engineering of geranylgeranyl pyrophosphate synthase levels and physiological conditions for enhanced carotenoid and astaxanthin synthesis in xanthophyllomyces dendrorhous. Biotechnol. Lett. 33 755–761. 10.1007/s10529-010-0495-2 [DOI] [PubMed] [Google Scholar]
- Bryon A., Kurlovs A. H., Dermauw W., Greenha R., Riga M., Grbic M., et al. (2017). Disruption of a horizontally transferred phytoene desaturase abolishes carotenoid accumulation and diapause in Tetranychus urticae. Proc. Natl. Acad. Sci. U.S.A. 114:E5871. 10.1073/pnas.1706865114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camagna M., Grundmann A., Bär C., Koschmieder J., Beyer P., Welsch R. (2019). Enzyme fusion removes competition for geranylgeranyl diphosphate in carotenogenesis. Plant Physiol. 179 1013–1027. 10.1104/pp.18.01026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzonelli C. I., Pogson B. J. (2010). Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 15 266–274. 10.1016/j.tplants.2010.02.003 [DOI] [PubMed] [Google Scholar]
- Chang T. H., Guo R. T., Ko T. P., Wang A. H. J., Liang P. H. (2006). Crystal structure of type-III geranylgeranyl pyrophosphate synthase from Saccharomyces cerevisiae and the mechanism of product chain length determination. J. Biol. Chem. 281 14991–15000. 10.1074/jbc.m512886200 [DOI] [PubMed] [Google Scholar]
- Chen N., Fan Y. L., Bai Y., Li X. D., Zhang Z. F., Liu T. X. (2016). Cytochrome P450 gene, CYP4G51, modulates hydrocarbon production in the pea aphid, Acyrthosiphon pisum. Insect Biochem. Mol. Biol. 76 84–94. 10.1016/j.ibmb.2016.07.006 [DOI] [PubMed] [Google Scholar]
- Cobbs C., Heath J., Stireman J. O., Abbot P. (2013). Carotenoids in unexpected places: gall midges, lateral gene transfer, and carotenoid biosynthesis in animals. Mol. Phylogenet. Evol. 68 221–228. 10.1016/j.ympev.2013.03.012 [DOI] [PubMed] [Google Scholar]
- Coman D., Altenhoff A., Zoller S., Gruissem W., Vranová E. (2014). Distinct evolutionary strategies in the GGPPS family from plants. Front. Plant Sci. 5:230. 10.3389/fpls.2014.00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank H. A., Brudvig G. W. (2004). Redox functions of carotenoids in photosynthesis. Biochemistry 43 8607–8615. 10.1021/bi0492096 [DOI] [PubMed] [Google Scholar]
- Heath J. J., Cipollini D. F., Stireman J. O., III (2013). The role of carotenoids and their derivatives in mediating interactions between insects and their environment. Arthropod Plant Interact. 7 1–20. 10.1007/s11829-012-9239-7 [DOI] [Google Scholar]
- Hellemans J., Mortier G., Paepe A. D., Speleman F., Vandesompele J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8:R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojo M., Shigenobu S., Maekawa K., Miura T., Tokuda G. (2019). Duplication and soldier-specific expression of geranylgeranyl diphosphate synthase genes in a nasute termite Nasutitermes takasagoensis. Insect Biochem. Mol. Biol. 111:103177. 10.1016/j.ibmb.2019.103177 [DOI] [PubMed] [Google Scholar]
- Hojo M., Toga K., Dai W., Yamamoto T., Maekawa K. (2011). High-level expression of the geranylgeranyl diphosphate synthase gene in the frontal gland of soldiers in Reticulitermes speratus (Isoptera: Rhinotermitidae). Arch. Insect Biochem. Physiol. 77 17–31. 10.1002/arch.20415 [DOI] [PubMed] [Google Scholar]
- Hundle B., Alberti M., Nievelstein V., Beyer P., Kleinig H., Armstrong G. A., et al. (1994). Functional assignment of Erwinia herbicola Eho10 carotenoid genes expressed in Escherichia coli. Mol. Gen. Genet. 245 406–416. 10.1007/bf00302252 [DOI] [PubMed] [Google Scholar]
- Lai C., Mcmahon R., Young C., Mackay T. F., Langley C. H. (1998). quemao, a Drosophila bristle locus, encodes geranylgeranyl pyrophosphate synthase. Genetics 149 1051–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange B. M., Ghassemian M. (2003). Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol. Biol. 51 925–948. [DOI] [PubMed] [Google Scholar]
- Le Trionnaire G., Tanguy S., Hudaverdian S., Gleonnec F., Richard G., Cayrol B., et al. (2019). An integrated protocol for targeted mutagenesis with CRISPR-Cas9 system in the pea aphid. Insect Biochem. Mol. Biol. 110 34–44. 10.1016/j.ibmb.2019.04.016 [DOI] [PubMed] [Google Scholar]
- Mende K., Homann V., Tudzynski B. (1997). The geranylgeranyl diphosphate synthase gene of Gibberella fujikuroi: isolation and expression. Mol. Gen. Genet. 255 96–105. 10.1007/s004380050477 [DOI] [PubMed] [Google Scholar]
- Misawa N., Nakagawa M., Kobayashi K., Yamano S., Izawa Y., Nakamura K., et al. (1990). Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J. Bacteriol. 172 6704–6712. 10.1128/jb.172.12.6704-6712.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran N. A., Jarvik T. (2010). Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science 328 624–627. 10.1126/science.1187113 [DOI] [PubMed] [Google Scholar]
- Niedzwiedzki D. M., Dilbeck P. L., Tang Q., Martin E. C., Bocian D. F., Hunter C. N., et al. (2017). New insights into the photochemistry of carotenoid spheroidenone in light-harvesting complex 2 from the purple bacterium Rhodobacter sphaeroides. Photosynth. Res. 131 291–304. 10.1007/s11120-016-0322-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková E., Moran N. A. (2012). Diversification of genes for carotenoid biosynthesis in aphids following an ancient transfer from a fungus. Mol. Biol. Evol. 29 313–323. 10.1093/molbev/msr206 [DOI] [PubMed] [Google Scholar]
- Okada K., Saito T., Nakagawa T., Kawamukai M., Kamiya Y. (2000). Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant Physiol. 122 1045–1056. 10.1104/pp.122.4.1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prchalová D., Buèek A., Brabcová J., Žáèek P., Kindl J., Valterová I., et al. (2016). Regulation of isoprenoid pheromone biosynthesis in bumblebee males. Chembiochem 17 260–267. 10.1002/cbic.201500415 [DOI] [PubMed] [Google Scholar]
- Quondam M., Barbato C., Pickford A., Helmercitterich M., Macino G. (1997). Homology modeling of Neurospora crassa geranylgeranyl pyrophosphate synthase: structural interpretation of mutant phenotypes. Protein Eng. Des. Sel. 10 1047–1055. 10.1093/protein/10.9.1047 [DOI] [PubMed] [Google Scholar]
- Ruiz-Sola M. A., Coman D., Beck G., Barji M. V., Colinas M., Graf A., et al. (2016). Arabidopsis geranylgeranyl diphosphate synthase 11 is a hub isozyme required for the production of most photosynthesis-related isoprenoids. New Phytol. 209 252–264. 10.1111/nph.13580 [DOI] [PubMed] [Google Scholar]
- Sagami H., Korenaga T., Ogura K. (1993). Geranylgeranyl diphosphate synthase catalyzing the single condensation between isopentenyl diphosphate and farnesyl diphosphate. J Biochem. 114 118–121. 10.1093/oxfordjournals.jbchem.a124125 [DOI] [PubMed] [Google Scholar]
- Shimizu I., Kitabatake S., Kato M. (1981). Effect of carotenoid deficiency on photosensitivities in the silkworm, Bombyx mori. J. Insect Physiol. 27 593–599. 10.1016/0022-1910(81)90106-2 [DOI] [Google Scholar]
- Singkaravanit S., Kinoshita H., Ihara F., Nihira T. (2010). Geranylgeranyl diphosphate synthase genes in entomopathogenic fungi. Appl. Microbiol. Biotechnol. 85 1463–1472. 10.1007/s00253-009-2171-9 [DOI] [PubMed] [Google Scholar]
- Sloan D. B., Moran N. A. (2012). Endosymbiotic bacteria as a source of carotenoids in whiteflies. Biol. Lett. 8 986–989. 10.1098/rsbl.2012.0664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z. J., Li Z. X. (2018). The terpenoid backbone biosynthesis pathway directly affects the biosynthesis of alarm pheromone in the aphid. Insect Mol. Biol. 27 824–834. 10.1111/imb.12521 [DOI] [PubMed] [Google Scholar]
- Swanson K. M., Hohl R. J. (2006). Anti-cancer therapy: targeting the mevalonate pathway. Curr. Cancer Drug Targets 6 15–37. 10.2174/156800906775471743 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T. (2016). Molecular basis and ecological relevance of aphid body colors. Curr. Opin. Insect Sci. 17 74–80. 10.1016/j.cois.2016.07.005 [DOI] [PubMed] [Google Scholar]
- Tsuchida T., Koga R., Horikawa M., Tsunoda T., Maoka T., Matsumoto S., et al. (2010). Symbiotic bacterium modifies aphid body color. Science 330 1102–1104. 10.1126/science.1195463 [DOI] [PubMed] [Google Scholar]
- Valmalette J. C., Dombrovsky A., Brat P., Mertz C., Capovilla M., Robichon A. (2012). Light-induced electron transfer and ATP synthesis in a carotene synthesizing insect. Sci. Rep. 2:sre00579. 10.1038/srep00579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varney M. L., Goetz D. B., Wiemer D. F., Holstein S. A. (2018). Isoprenoid amide bisphosphonates as a novel class of geranylgeranyl diphosphate synthase inhibitors. Blood 132:4679 10.1182/blood-2018-99-112965 [DOI] [Google Scholar]
- Waller D. D., Park J., Tsantrizos Y. S. (2019). Inhibition of farnesyl pyrophosphate (FPP) and/or geranylgeranyl pyrophosphate (GGPP) biosynthesis and its implication in the treatment of cancers. Crit. Rev. Biochem. Mol. 54 41–60. 10.1080/10409238.2019.1568964 [DOI] [PubMed] [Google Scholar]
- Waterhouse A. M., Procter J. B., Martin D. M. A., Clamp M., Barton G. J. (2009). Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25 1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemer A. J., Wiemer D. F., Hohl R. J. (2011). Geranylgeranyl diphosphate synthase: an emerging therapeutic target. Clin. Pharmacol. Ther. 90 804–812. 10.1038/clpt.2011.215 [DOI] [PubMed] [Google Scholar]
- Yang C., Pan H., Liu Y., Zhou X. (2014). Selection of reference genes for expression analysis using quantitative real-time PCR in the pea aphid, Acyrthosiphon pisum (Harris) (Hemiptera, Aphidiae). PLoS One 9:e110454. 10.1371/journal.pone.0110454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L. E., Huang X. Q., Lu Q. Q., Zhu J. Y., Lu S. (2016). Cloning and characterization of the geranylgeranyl diphosphate synthase (GGPS) responsible for carotenoid biosynthesis in Pyropia umbilicalis. J. Appl. Phycol. 28 671–678. 10.1007/s10811-015-0593-6 [DOI] [Google Scholar]
- Ye C., An X., Jiang Y. D., Ding B. Y., Shang F., Christiaens O., et al. (2019). Induction of RNAi core machinery’s gene expression by exogenous dsRNA and the effects of pre-exposure to dsRNA on the gene silencing efficiency in the pea aphid (Acyrthosiphon pisum). Front. Physiol. 9:1906. 10.3389/fphys.2018.01906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P., Han Z., Li X., Olson E. N. (2006). The mevalonate pathway controls heart formation in drosophila by isoprenylation of Gγ1. Science 313 1301–1303. 10.1126/science.1127704 [DOI] [PubMed] [Google Scholar]
- Zhang H., Li Z. X. (2014). A type-III insect geranylgeranyl diphosphate synthase with a novel catalytic property. Protein Pept. Lett. 21 615–623. 10.2174/0929866521666140214123942 [DOI] [PubMed] [Google Scholar]
- Zhang J. C., Tao N. G., Xu Q., Zhou W. J., Cao H. B., Xu J., et al. (2009). Functional characterization of citrus PSY gene in Hongkong kumquat (Fortunella hindsii swingle). Plant Cell Rep. 28 1737–1746. 10.1007/s00299-009-0774-3 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang X. X., Zhu J. Y., Zhang Z. F., Tian H. G., Liu T. X. (2016). Strategies used by two apterous strains of the pea aphid Acyrthosiphon pisum for passive disperal. Biol. Open 5 1535–1544. 10.1242/bio.018903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Wang C. Y., Gutensohn M., Jiang L., Zhang P., Zhang D., et al. (2017). A recruiting protein of geranylgeranyl diphosphate synthase controls metabolic flux toward chlorphyll biosynthesis in rice. Proc. Natl. Acad. Sci. U.S.A. 114 6866–6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The suppositional carotenoid biosynthesis pathway in the pea aphid. [According to Moran and Jarvik (2010)].
Schematic diagram of samples collected for different developmental stages. N1-1: Newly enclosed nymph in 4 h; N1-2: middle of 1st instar nymph; N1-3: late stage of 1st instar nymph; N2-1: early stage of 2nd instar nymph; N2-2: middle stage of 2nd instar nymph; N2-3: late stage of 2nd instar nymph; N3-1: early stage of 3rd instar nymph; N3-2: middle stage of 3rd instar nymph; N3-3: late stage of 3rd instar nymph; N4-1: early stage of 4th instar nymph; N4-2: middle stage of 4th instar nymph; N4-3: late stage of 4th instar nymph; AD-1: 8 days old adult; AD-2: 9 days old; AD-3: 12 days old; AD-4: 15 days old.
Standard curve of α-carotene obtained by HPLC. The linear regression equation was generated using 0.08, 0.04, 0.02, 0.016, 0.012, and 0.008 μg concentrations.
Standard curve of β-carotene obtained by HPLC. The linear regression equation was generated using 0.08, 0.04, 0.02, 0.016, and 0.012 μg concentrations.
Standard curve of γ-carotene obtained by HPLC. The linear regression equation was generated using 0.02, 0.016, 0.012, 0.008, and 0.004 μg concentrations.
Alignment of the amino acid sequence of Acyrthosiphon pisum GGPPS with GGPPS from other species. Conserved motifs are indicated by the red box with dotted lines.
Expression patterns of GGPPS during different developmental stages and tissues in the green morph of Acyrthosiphon pisum, NY strain. (A) The expression patterns of GGPPS in different developmental stages. N1: 1st instar nymph, N2: 2nd instar nymph, N3: 3rd instar nymph, N4: 4th instar nymph, AD: adult. (B) Expression patterns of GGPPS in tissues. BR, brain; ST, stylet; IN, integument; MU, muscle; GU, gut; FB, fat body; EM, embryo. Mean (±SE) expression level was based on four biological replicates. Lowercase letters (a and b) above each bar indicate significant differences among tissues (One-way ANOVA followed by Tukey’s honestly significant difference multiple comparison test; P < 0.05).
Primer sequences used for sequence confirmation, quantitative real-time PCR (qRT-PCR) and double stranded RNA (dsRNA) synthesis.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.