Abstract
Transcriptomics and candidate gene/protein expression studies have indicated several biological processes modulated by methylphenidate (MPH), widely used in attention-deficit/hyperactivity disorder (ADHD) treatment. However, the lack of a differential proteomic profiling of MPH treatment limits the understanding of the most relevant mechanisms by which MPH exerts its pharmacological effects at the molecular level. Therefore, our aim is to investigate the MPH-induced proteomic alterations using an experimental design integrated with a pharmacogenomic analysis in a translational perspective. Proteomic analysis was performed using the cortices of Wistar-Kyoto rats, which were treated by gavage with MPH (2 mg/kg) or saline for two weeks (n = 6/group). After functional enrichment analysis of the differentially expressed proteins (DEP) in rats, the significant biological pathways were tested for association with MPH response in adults with ADHD (n = 189) using genome-wide data. Following MPH treatment in rats, 98 DEPs were found (P < 0.05 and FC < −1.0 or > 1.0). The functional enrichment analysis of the DEPs revealed 18 significant biological pathways (gene-sets) modulated by MPH, including some with recognized biological plausibility, such as those related to synaptic transmission. The pharmacogenomic analysis in the clinical sample evaluating these pathways revealed nominal associations for gene-sets related to neurotransmitter release and GABA transmission. Our results, which integrate proteomics and pharmacogenomics, revealed putative molecular effects of MPH on several biological processes, including oxidative stress, cellular respiration, and metabolism, and extended the results involving synaptic transmission pathways to a clinical sample. These findings shed light on the molecular signatures of MPH effects and possible biological sources of treatment response variability.
Subject terms: Pharmacogenomics, ADHD
Introduction
Methylphenidate (MPH) is a widely used stimulant for pharmacological treatment of attention-deficit/hyperactivity disorder (ADHD). Its safety and efficacy for ADHD treatment in both adults and children are supported by meta-analyses; however, a considerable proportion of patients do not present a satisfactory response1–3. The main direct targets of MPH are the dopamine and norepinephrine transporters (DAT and NET, respectively). In addition, the interactions of MPH with other proteins of these neurotransmission systems, such as dopaminergic, adrenergic, and AMPA receptors4–6, as well as the modulation of signaling of other neurotransmitters, including serotonin, glutamate, and GABA7–9, are suggested to be involved in its pharmacological actions. Moreover, several other pathways such as those involved in neuronal plasticity, energy metabolism, cell differentiation, circadian rhythm, and ubiquitin-dependent protein degradation are also affected by MPH, as evidenced by transcriptomics and candidate gene/protein expression studies10–17. Although these studies used different methodologies, including in vivo and in vitro treatments, the overall evidence indicates that MPH actions involve a complex biological scenario, but the most relevant mechanisms for treatment response are still unclear.
Neuroimaging studies can complement the elucidation of MPH actions by pointing out the most relevant regions involved, and they suggest that MPH attenuates some ADHD-related structure alterations (e.g., reduced volumes of gray matter), mainly in basal ganglia and anterior cingulate cortex regions18,19. Functional studies reported that MPH mitigates ADHD dysfunctions in the prefrontal cortex, anterior cingulate cortex, and striatum. Of note, MPH treatment influences ventral frontostriatal functional connectivity without tasks20, as well as activates prefrontal cortex, basal ganglia, and cerebellum during tasks21,22. This set of evidence suggests that MPH effects involve mainly frontal and striatal regions, and a comprehensive investigation of the changes induced by MPH in such regions can provide a better understanding of its pharmacological effects.
Proteomic technology is a promising tool to investigate the molecular basis of ADHD, and can also help to clarify the existing gaps regarding MPH actions. This high-throughput hypothesis-free technique provides broad coverage of the brain profile, and the differential global protein expression under different conditions can shed light on specific-state biological underpinnings23. For example, proteomic molecular signatures of psychostimulants, including cocaine and methamphetamine, have suggested alterations in metabolism, oxidative stress, degenerative process, mitochondrial dysfunction, synaptic proteins, and plasticity24–26. Even though previous studies have explored MPH-induced effects in the expression of particular proteins, we are not aware of investigations using a hypothesis-free approach to characterize the proteomic profile according to MPH treatment. In addition, pharmacogenomic studies of MPH are scarce, especially in adults, precluding further insights on the molecular basis of its effects.
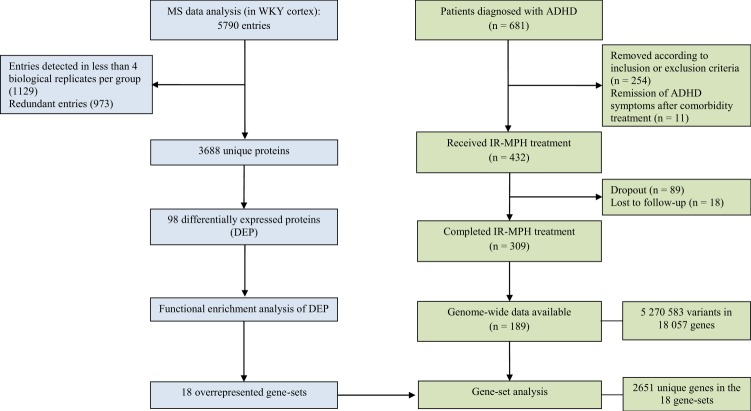
Therefore, this study aims to identify the MPH-induced alterations in the proteome of cortex tissue of Wistar-Kyoto (WKY) rats. Functional enrichment of the differentially expressed proteins (DEPs) into biological pathways was performed to provide an overview of the systems affected by MPH treatment. Moreover, from a translational perspective, the proteomic results were used to generate hypotheses to be tested for association with MPH treatment response in a clinical sample of adults with ADHD, using genome-wide data. The rationale of our study design and the details on the proteomics and pharmacogenomics analyses are shown in Fig. 1. This integrative approach, in addition to providing general insights for biological pathways modulated by MPH treatment, points out which of these pathways may also influence MPH treatment response variability.
Fig. 1. Flowchart of data analysis for proteomics (left) and pharmacogenomics (right).
ADHD attention-deficit/hyperactivity disorder, IR-MPH immediate-release methylphenidate, MS mass spectrometry, WKY Wistar-Kyoto.
Materials and methods
Differential proteomics of rats treated with MPH
Animals
WKY rats, males, and adults (90–120 days) weighing 260–320 g were used in this experiment. The rats were housed in 49 × 34 × 16 cm polypropylene cages (maximum of 4 per cage) at a temperature of 22 °C ± 2 °C on a 12-h light/dark cycle (lights on at 7 a.m.). Animals had ad libitum access to food and water. A period of 2 weeks for the stabilization and acclimation to the environment was provided to the animals prior to the initiation of the interventions. All the experimental design of this study was approved by the Institutional Committee for Animal Care and Use (GPPG-HCPA protocol no. 20160346) and performed in accordance with the Guide for the Care and Use of Laboratory Animals 8th edition (2011). The maintenance of the animals followed the law 11.794 (Brazil), which establishes procedures for the scientific use of animals. Vigorous attempts were made to minimize animal suffering, and decrease external sources of pain and discomfort, as well as to use the minimum number of animals required to produce reliable scientific data.
Drugs
Immediate-release methylphenidate (IR-MPH; Ritalin, Novartis, Brazil) was dissolved in 0.9% saline solution before the administration to the rats by gavage at a dose of 2 mg/kg. This dose results in plasma concentrations similar to what is considered clinically effective27–30.
Experimental design
WKY rats were randomly assigned to control (saline-treated) or MPH-treated groups. Each group was composed by six animals (biological replicates), which is in the range that usually provides relevant results among proteomic animal studies. The animals received IR-MPH or 0.9% saline solutions both by gavage twice a day for 14 days. Treatment was performed at the same time and by the same researcher (7 a.m. and 7 p.m. daily). Approximately 12 h after the last IR-MPH dose, animals were killed by decapitation and the brains were instantly removed for dissection. The cortices were used for this experiment, and they were immediately processed and prepared for protein extraction (the flowchart of the experimental design is provided in Supplementary Fig. 1).
Sample preparation and protein extraction for mass spectrometry
After dissection, the cerebral cortex tissues were individually homogenized and sonicated on cold HEPES/sucrose buffer containing protease and phosphatase inhibitors. The proteins were extracted using methanol and chloroform and stored at −80 °C until further processing. Protein extracts were resuspended with 8 M urea 50 mM ammonium bicarbonate, and protein concentration was determined using a bicinchoninic acid (BCA) protein assay (Thermo Scientific). Proteins were then digested, acidified with formic acid and subsequently desalted using AssayMap C18 cartridges (Agilent) mounted on an Agilent AssayMap BRAVO liquid handling system. More detailed procedures on the preparation of proteins prior to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis are described in the Supplementary Methods.
Mass spectrometry
Dried samples were reconstituted with 2% acetonitrile, 0.1% formic acid and analyzed by LC-MS/MS using a Proxeon EASY 1200 nanoLC system (Thermo Fisher Scientific) coupled to an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific). Peptide separation was performed on a 25 cm column with 75 µm inner diameter packed in-house with ethylene bridged hybrid (BEH) C18 1.7 µm resin (Waters) in a 180-min gradient of 6–45% of solvent B (80% acetonitrile, 0.1% formic acid) at a flow rate of 310 nL/min. The mass spectrometer was operated in positive data-dependent acquisition mode. MS1 spectra were measured with a resolution of 120,000, an automatic gain control (AGC) target of 1e6, a maximum injection time of 100 ms and a mass range from 350 to 1500 m/z. The instrument was set to run in top speed mode with 3 s cycles for the survey and the MS/MS scans. After a survey scan, tandem MS was performed on the most abundant precursors exhibiting a charge state from 2 to 8 of greater than 5e4 intensity by isolating them in the quadrupole at 0.8 Th. High-energy collision dissociation (HCD) fragmentation was applied with 30% collision energy, and resulting fragments were detected using the turbo scan rate in the ion trap. The AGC target for MS/MS was set to 1e4, and the maximum injection time limited to 15 ms. The dynamic exclusion was set to 20 s with a 10 ppm mass tolerance around the precursor and its isotopes. MS data are available via ProteomeXchange with identifier PXD015473.
Proteomics data analysis
The MaxQuant software version 1.5.5.1 was used to analyze the mass spectra. MS/MS spectra were searched against the Rattus norvegicus Uniprot protein sequence database (version January 2016) and the common Repository of Adventitious Proteins (cRAP) sequences (commonly known protein contaminants) from Global Proteome Machine database. Evidence table output from MaxQuant was used for label-free protein quantitative analysis. First, calculated peptide intensities were log base 2 transformed and normalized across samples to account for systematic errors. Following normalization, all non-razor peptide sequences were removed from the list, and protein-level quantification and testing for differential abundance were performed using MSstats Bioconductor package31. The model decomposes log-intensities into the effects of biological replicates, peptides, and statistical interactions. Specific settings used in MaxQuant and further details on the normalization procedure can be found in Supplementary Methods. For all these procedures performed in each biological replicate, no identification of group allocation was provided during the experiment.
Since MaxQuant cluster unresolved protein redundancy into protein groups, the entry with a higher number of peptides matched to the protein was selected to be included in the analysis. Proteins were excluded if not detected in at least 4 out of 6 animals. Proteins presenting a log base 2 transformed fold-change (Log2FC) higher than 1.0 (upregulated with MPH) or lower than −1.0 (downregulated with MPH) plus a nominal P-value < 0.05 were considered as significantly differentially expressed between the MPH-treated and saline-treated groups. The consideration of a cutoff for effect sizes (Log2FC) in addition to the P-value helps to reduce the probability of false-positive results without eliminating true-positive identifications that occur when applying traditional methods of multiple testing correction in quantitative proteomics32.
Functional enrichment analysis of DEPs
The DEPs (as gene names; Table 1) were analyzed using FUMA web application v1.3.3c33 to obtain information on the biological context in which these proteins are inserted. FUMA is an integrative web-based platform that provides pathway enrichment results by performing hypergeometric tests to evaluate whether genes of interest are overrepresented in any of the pre-defined sets from different categories. The categories of interest for this study in terms of biological mechanisms include Gene Ontology (GO) Biological Process, Kyoto Encyclopedia of Genes and Genomes (KEGG), and Reactome. False Discovery Rate (FDR) correction taking into account the total number of gene-sets in each category was used to determine the significantly overrepresented gene-sets, with an adjusted P-value cutoff of 0.05. The minimum number of overlapped genes was set as 2. Additional tools (DAVID and Enrichr) were used as a complementary approach to compare the results and outline the most consistently enriched pathways among the different software. DAVID test the significance of gene-term enrichment with a modified Fisher’s exact test (EASE score; set as 0.05), which compares the percentage of input genes that comprises a particular pathway to this proportion in the whole genome34. Enrichr retrieves P-values based on Fisher’s exact test, but it precomputes a background expected rank for each term in each gene-set category35.
Table 1.
Differentially expressed proteins with MPH treatment.
| Accession number | Protein names | Gene names | Log2FC (MPH/Saline) | P-value |
|---|---|---|---|---|
| Q3ZAU5a | DDHD domain-containing 1 | Ddhd1 | −11.053 | 0.0100 |
| A0A0G2K6R0a | Trinucleotide repeat-containing 6B | Tnrc6b | −10.937 | 0.0125 |
| Q63540a | Ataxin-1 | Atxn1 | −10.600 | 0.0125 |
| F1MAR6a | Proline dehydrogenase | Prodh1 | −10.133 | 0.0100 |
| P10818 | Cytochrome c oxidase subunit 6A1, mitochondrial | Cox6a1 | −3.693 | 0.0001 |
| Q6LED0 | Histone H3.1 | Hist1h3b | −2.876 | 0.0052 |
| D4A5X7 | Ganglioside-induced differentiation-associated-protein 1 | Gdap1 | −2.108 | 0.0040 |
| Q04940 | Neurogranin | Nrgn | 2.082 | 0.0185 |
| D4ADS4 | Microsomal glutathione S-transferase 3 | Mgst3 | −2.048 | 0.0088 |
| P14056 | Serine/threonine-protein kinase A-Raf | Araf | −1.982 | 0.0063 |
| B4F7E8 | Niban-like protein 1 | Fam129b | −1.864 | 0.0025 |
| F1MA59 | Collagen type IV alpha 1 chain | Col4a1 | −1.776 | 0.0038 |
| A0A0H2UHZ1 | Aquaporin-4 | Aqp4 | −1.734 | 0.0147 |
| Q62717 | Calcium-dependent secretion activator 1 | Cadps | −1.732 | 0.0032 |
| M0R3V7 | Sodium/calcium exchanger 1 | Slc8a1 | −1.731 | 0.0157 |
| Q0VGK0 | Gamma-aminobutyric acid receptor-associated protein-like 1 | Gabarapl1 | −1.713 | 0.0146 |
| A0A0U1RRQ1 | NADH:ubiquinone oxidoreductase subunit A1 | Ndufa1 | −1.656 | 0.0108 |
| F1M779 | Clathrin heavy chain;Clathrin heavy chain 1 | Cltc | −1.653 | 0.0182 |
| P30835 | ATP-dependent 6-phosphofructokinase, liver type | Pfkl | −1.651 | 0.0350 |
| Q812E9 | Neuronal membrane glycoprotein M6-a | Gpm6a | −1.630 | 0.0464 |
| P23978 | Sodium- and chloride-dependent GABA transporter 1 | Slc6a1 | −1.615 | 0.0417 |
| Q6MFX9 | Myelin-oligodendrocyte glycoprotein | Mog | −1.608 | 0.0176 |
| A0A0G2K4L6 | Similar to TSC22 domain family protein 2 | Tsc22d2 | 1.574 | 0.0084 |
| Q6P9Y4 | ADP/ATP translocase 1 | Slc25a4 | −1.540 | 0.0410 |
| F1LUT4 | Phospholipid-transporting ATPase | Atp8a1 | −1.501 | 0.0410 |
| A0A0G2JZK7 | Sodium/calcium exchanger 2 | Slc8a2 | −1.495 | 0.0384 |
| P11505 | Plasma membrane calcium-transporting ATPase 1 | Atp2b1 | −1.481 | 0.0310 |
| A0A0G2K757 | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit 2 | Rpn2 | −1.441 | 0.0222 |
| F1M110 | Glycylpeptide N-tetradecanoyltransferase | Nmt2 | −1.437 | 0.0032 |
| M0RDI5 | Mitochondrial calcium uniporter | Mcu | −1.410 | 0.0361 |
| A0A0G2JWE3 | Protein IWS1 homolog | Iws1 | −1.393 | 0.0126 |
| Q07984 | Translocon-associated protein subunit delta | Ssr4 | −1.388 | 0.0274 |
| P31647 | Sodium- and chloride-dependent GABA transporter 3 | Slc6a11 | −1.375 | 0.0489 |
| A0A0G2K6E2 | Thioredoxin reductase 2, mitochondrial | Txnrd2 | −1.368 | 0.0052 |
| F1LQB2 | Structural maintenance of chromosomes protein | Smc3 | −1.368 | 0.0408 |
| Q4V7D9 | Acid sphingomyelinase-like phosphodiesterase | Smpdl3b | −1.363 | 0.0065 |
| Q63584 | Transmembrane emp24 domain-containing protein 10 | Tmed10 | −1.354 | 0.0125 |
| Q63564 | Synaptic vesicle glycoprotein 2B | Sv2b | −1.316 | 0.0382 |
| Q68FW7 | Threonine–tRNA ligase, mitochondrial | Tars2 | −1.305 | 0.0136 |
| P00762 | Anionic trypsin-1 | Prss1 | 1.302 | 0.0019 |
| D4A106 | WD repeat domain 3 | Wdr3 | −1.298 | 0.0035 |
| A0A0G2K9V6 | Threonine–tRNA ligase, cytoplasmic | Tars | −1.298 | 0.0301 |
| D3ZEI4 | Hepatocyte cell adhesion molecule | Hepacam | −1.289 | 0.0177 |
| A0A0G2JSU4 | N-myc downstream regulated gene 2, isoform CRA_b | Ndrg2 | −1.285 | 0.0310 |
| P18088 | Glutamate decarboxylase 1 | Gad1 | −1.278 | 0.0145 |
| B0BN81 | 40S ribosomal protein S5 | Rps5 | −1.251 | 0.0461 |
| P06685 | Sodium/potassium-transporting ATPase subunit alpha-1 | Atp1a1 | −1.234 | 0.0333 |
| B0BNG3 | Lectin, mannose-binding 2 | Lman2 | −1.231 | 0.0040 |
| O35165 | Golgi SNAP receptor complex member 2 | Gosr2 | −1.223 | 0.0148 |
| Q02563 | Synaptic vesicle glycoprotein 2A | Sv2a | −1.220 | 0.0440 |
| D4ACM1 | Elongator complex protein 3 | Elp3 | −1.211 | 0.0303 |
| B2GUV7 | Eukaryotic translation initiation factor 5B | Eif5b | −1.208 | 0.0005 |
| Q566R4 | Leucine repeat adapter protein 2 | Fam89b | 1.206 | 0.0116 |
| D3ZYT2 | Mitochondrial ribosomal protein S5 | Mrps5 | 1.205 | 0.0349 |
| Q6AY18 | SAR1 gene homolog A (S. cerevisiae), isoform CRA_b | Sar1a | −1.190 | 0.0174 |
| B1PLB1 | CD34 antigen | Cd34 | 1.189 | 0.0259 |
| F1LQG0 | Huntingtin-associated protein 1 | Hap1 | 1.187 | 0.0075 |
| O89035 | Mitochondrial dicarboxylate carrier | Slc25a10 | −1.187 | 0.0434 |
| Q4V898 | RNA-binding motif protein, X chromosome | Rbmx | −1.184 | 0.0027 |
| F1LS72 | Ubiquitin-like modifier-activating enzyme 2 | Uba2 | −1.183 | 0.0182 |
| P16975 | SPARC | Sparc | 1.170 | 0.0073 |
| D3ZE85 | DOMON domain-containing protein FRRS1L | Frrs1l | −1.170 | 0.0019 |
| B4F774 | Ganglioside-induced differentiation-associated protein 1-like 1 | Gdap1l1 | −1.169 | 0.0383 |
| D3ZZ21 | NADH dehydrogenase (Ubiquinone) 1 beta subcomplex, 6 | Ndufb6 | −1.163 | 0.0072 |
| D4ABI7 | Very-long-chain (3 R)-3-hydroxyacyl-CoA dehydratase | Hacd3 | −1.150 | 0.0362 |
| G3V728 | 4-nitrophenylphosphatase domain and non-neuronal SNAP25-like protein homolog 1 (C. elegans), isoform CRA_b | Nipsnap1 | −1.145 | 0.0012 |
| D3ZQD3 | Oxoglutarate dehydrogenase-like | Ogdhl | −1.133 | 0.0207 |
| F1M471 | EPM2A-interacting protein 1 | Epm2aip1 | −1.130 | 0.0173 |
| D4A8N2 | Ferredoxin 2 | Fdx2 | 1.112 | 0.0170 |
| A1L1M0 | cAMP-dependent protein kinase catalytic subunit alpha | Prkaca | −1.117 | 0.0358 |
| G3V746 | Glutamate receptor ionotropic, NMDA 2B | Grin2b | −1.106 | 0.0364 |
| Q5XI78 | 2-oxoglutarate dehydrogenase, mitochondrial | Ogdh | −1.102 | 0.0280 |
| D3ZZN3 | Acetyl-coenzyme A synthetase | Acss1 | −1.101 | 0.0071 |
| F1LMR7 | Dipeptidyl aminopeptidase-like protein 6 | Dpp6 | −1.101 | 0.0146 |
| D4AD70 | 60S ribosomal protein L38 | Rpl38 | −1.097 | 0.0003 |
| D3ZIS5 | Cytochrome c oxidase assembly factor COX19 | Cox19 | 1.091 | 0.0161 |
| Q3SWS9 | Janus kinase and microtubule-interacting protein 1 | Jakmip1 | 1.073 | 0.0111 |
| Q5PPG6 | Nucleosome assembly protein 1-like 5 | Nap1l5 | 1.070 | 0.0081 |
| B4F7A9 | Casein kinase 2 alpha 2 | Csnk2a2 | −1.054 | 0.0031 |
| P84100 | 60S ribosomal protein L19 | Rpl19 | −1.052 | 0.0373 |
| Q4KM74 | Vesicle-trafficking protein SEC22b | Sec22b | −1.048 | 0.0462 |
| Q5XIJ4 | Protein FAM210A | Fam210a | −1.035 | 0.0177 |
| A0A0G2K4T7 | General transcription factor II-I | Gtf2i | −1.031 | 0.0020 |
| Q641Y2 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 2, mitochondrial | Ndufs2 | −1.029 | 0.0243 |
| P62332 | ADP-ribosylation factor 6 | Arf6 | −1.025 | 0.0081 |
| A0A0G2K490 | TRAF2 and NCK-interacting kinase | Tnik | −1.021 | 0.0419 |
| B2GV54 | Neutral cholesterol ester hydrolase 1 | Nceh1 | −1.019 | 0.0211 |
| Q9JIX3 | Bis(5-adenosyl)-triphosphatase | Fhit | −1.019 | 0.0008 |
| A0A0H2UHV9 | Coatomer subunit gamma-2 | Copg2 | −1.015 | 0.0028 |
| P00388 | NADPH–cytochrome P450 reductase | Por | −1.011 | 0.0321 |
| Q5XI38 | Lymphocyte cytosolic protein 1 | Lcp1 | −1.010 | 0.0488 |
| D4A4F9 | RCG20461 | Stum | −1.007 | 0.0223 |
| A0A0G2K9J0 | Tetratricopeptide repeat, ankyrin repeat and coiled-coil-containing 2 | Tanc2 | −1.006 | 0.0333 |
| D4A3D9 | Serine/threonine kinase 32C | Stk32c | −1.005 | 0.0134 |
| D4A193 | Receptor expression-enhancing protein | Reep1 | −1.004 | 0.0222 |
| A2RRU1 | Glycogen [starch] synthase, muscle | Gys1 | −1.003 | 0.0374 |
| Q4QQV3 | Protein FAM162A | Fam162a | −1.001 | 0.0055 |
| Q6PST4 | Atlastin-1 | Atl1 | −1.001 | 0.0484 |
MPH methylphenidate, Log2FC log base 2 transformed fold-change
aNot detected by the proteomic technique in any biological replicate from the MPH-treated group. The Log2FC and P-value are imputed values. The former was calculated as the average of Log2 protein intensity divided by 3.3, while imputed P-value was calculated as 0.05 divided by the number of replicates the specific protein was detected across replicates of the detected condition. The effects of MPH treatment on the expression of proteins are indicated by negative (downregulated) or positive (upregulated) Log2FC values
Gene-set analysis of MPH-treated adults with ADHD
In order to evaluate which of the gene-sets generated from functional enrichment analysis of the DEPs of the cortex of rats would also be involved with MPH treatment response in humans, a gene-set analysis using genome-wide data was performed for a clinical sample of adults with ADHD treated with IR-MPH. Considering the molecular targets of MPH are comparable in healthy or pathological conditions, this approach intends to translate the proteomic findings of the molecular effects of MPH in brain tissue of rats to a clinical perspective focused on the treatment response variability. Therefore, it is expected to reveal not only the molecular signatures of MPH but also in which pathways the genetic variability would be relevant for treatment response to MPH.
Sample
This sample comprised 189 individuals of European descent who were ascertained from the ADHD Outpatient Program in the adult division at Hospital de Clínicas de Porto Alegre (HCPA), Brazil. Diagnostic procedures for ADHD followed DSM-IV criteria36, and other lifetime psychiatric comorbidities were assessed using the Structured Clinical Interview for DSM-IV Axis I Disorders (SCID-I)37. The inclusion criteria were: (a) being white Brazilian of European descent; (b) aged 18 years or older; (c) fulfillment of the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, (DSM-IV) diagnostic criteria for ADHD;36 and (d) eligibility to IR-MPH treatment. The exclusion criteria were: (a) contraindication for IR-MPH use; (b) current stimulant treatment; (c) evidence of a clinically significant neurological disease that might affect cognition (for example, delirium, dementia, epilepsy, head trauma and multiple sclerosis), (d) current or past history of psychosis, and (e) estimated intelligence quotient score lower than 70. Detailed characteristics of the sample are shown in Supplementary Table 1. More detailed information on the sample has been described elsewhere38,39. This study was carried out in accordance with the Declaration of Helsinki, and all participants signed an informed consent form previously approved by the institutional review board of the hospital (No. 00000921).
Treatment and outcome measure
The pharmacological intervention followed the Brazilian guidelines for a more extensive study on clinical predictors of treatment response to IR-MPH40. Patients with unstable psychiatric comorbidities were treated with appropriate medications before initiation of treatment with IR-MPH, and they were later re-evaluated to confirm the ADHD diagnosis. After stabilization, they received IR-MPH standard initial daily dose of 10 mg, with doses increasing weekly until symptom control or occurrence of limiting adverse effects. At least 30 days of treatment were required to define the endpoint. The evaluation of treatment response was assessed considering the continuous variable of percentage reduction in severity scores of ADHD symptoms of the Portuguese version of the Swanson, Nolan and Pelham Rating Scale, version 4 (SNAP-IV) from baseline to endpoint. The quantitative measure of treatment response (percentage reduction in symptom severity) rather than a stratification of the sample into groups of responders versus non-responders was chosen since it provides higher statistical power to detect associations.
Genotyping
DNA was extracted from peripheral blood. The individuals were genotyped on the Infinium PsychArray-24 BeadChip (Illumina, San Diego, CA, USA). This microarray includes 265,000 SNPs from the Infinium Core-24 BeadChip, 245 000 markers from the Infinium Exome-24 BeadChip, and 50,000 additional markers previously associated with common psychiatric disorders. The data were processed at the Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard (Cambridge, MA, USA). Quality control (QC), principal components analysis, and genotype imputation procedures were implemented using the default values on RICOPILI following the PGC pipeline (https://sites.google.com/a/broadinstitute.org/ricopili/home). The European population of the 1000 Genomes Project Phase 1 was used as the reference panel. Postimputation QC was performed using the following settings: info score > 0.6, minor allele frequency > 1%, call rate > 98% and Hardy-Weinberg equilibrium test with P > 1e-06. The resulting dataset after QC consisted of 5,270,583 SNPs in 18,057 genes for 189 individuals.
Gene-set analysis
MAGMA v1.07b was used to perform the gene-set analysis following the basic steps of the software guidelines that include: annotation, gene analysis, and gene-set analysis41. For SNP annotation to genes, gene locations for build 37 (hg19) were used, setting a 2 kb upstream and 1 kb downstream window. For gene analysis, the principal components regression model was used. MAGMA’s gene analysis uses multiple regression to address linkage disequilibrium between genome-wide markers and to detect multi-marker effects41. Potential confounding variables were included in the analysis when associated with the outcome. Based on this, concomitant use of medication and baseline severity scores (assessed by the clinical global impression—severity (CGI-S) scale) were included as covariates, as well as the ten first principal components to control for population stratification. In the gene-set analysis, each of the 18 enriched pathways generated from the DEPs in rats was tested for association with MPH treatment response in the clinical sample (measured by the percentage change of symptoms between the baseline and the endpoint). The competitive testing uses the results from gene analysis to evaluate whether the combined effect of genes comprising the gene-sets is larger than all other genes in the genome (not included in the gene-sets) in relation to the outcome. FDR was applied for multiple testing correction considering all the gene-sets tested.
Results
Differential proteomics of rats treated with MPH
Identification of DEPs
According to our pre-defined criteria of Log2FC and P-value, 98 proteins showed differential expression with MPH treatment (86 proteins were considered downregulated, and 12 upregulated; Table 1). Among them, four proteins (DDHD1, TNRC6B, ATXN1, and PRODH1) were exclusively detected in the saline-treated group, while no protein was expressed only in the MPH-treated group. Few of these proteins showed substantial differential expression between groups with Log2FC higher than + 2.0 (NRGN) or lower than −2.0 (COX6A1, HIST1H3B, GDAP1, and MGST3), and most of them showed subtler changes with Log2FC of ± 1.5.
Functional enrichment analysis of DEPs
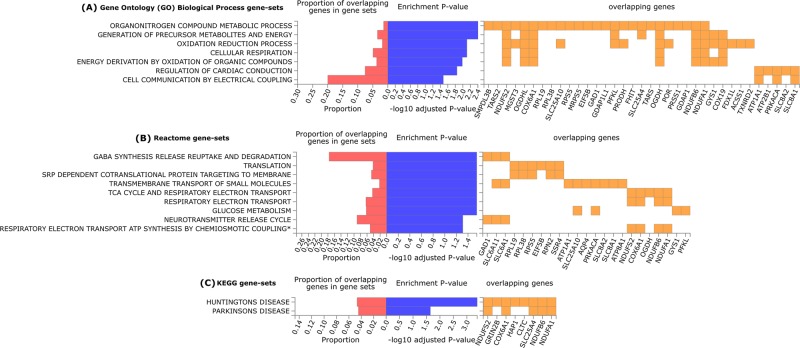
The enrichment analysis of the DEPs (entered as gene names) into functional categories using FUMA resulted in seven significantly overrepresented pathways (gene-sets) from GO Biological process (Fig. 2a), nine from Reactome (Fig. 2b) and two from KEGG (Fig. 2c) categories after FDR correction. This analysis was based on 97 genes, since the Stum gene (Uniprot protein accession code D4A4F9) was not recognized by FUMA, even under alternative names/codes. The complementary analyses using DAVID and Enrichr software can be found in Supplementary Tables 2 and 3, respectively. We observed differences in the association values retrieved by these tools. For example, although several pathways were overrepresented in DAVID with EASE cutoff of 0.05, only Reactome Muscle Contraction remains significant after FDR correction (Supplementary Table 2), while Enrichr retrieves several significantly enriched pathways from Reactome and KEGG categories after FDR correction, but none from the GO Biological Process category (Supplementary Table 3). Despite the differences in the statistical significance, in general, there is a relatively high concordance of the overrepresented pathways among the tools used, and we will focus the discussion on the principal results from FUMA, taking into account the most consistent findings among the three tools.
Fig. 2. FUMA results of the overrepresented gene-sets retrieved from the functional enrichment analysis of the 97 differentially expressed proteins with MPH treatment in the cortex of Wistar-Kyoto rats.
The blue bars represent the enrichment P-value (-log10 adjusted) after FDR correction considering the number of gene-sets tested per category. The red bars indicate the proportion of overlapping inputted genes according to the size (number of genes) of each gene-set. The orange squares show the inputted genes that are part of the enriched gene-sets. (a) Significantly enriched gene-sets from the Gene Ontology (GO)—Biological Process category. (b) Significantly enriched gene-sets from the Reactome category. (c) Significantly enriched gene-sets from the Kyoto Encyclopedia of Genes and Genomes (KEGG) category. *Respiratory electron transport ATP synthesis by chemiosmotic coupling and heat production by uncoupling proteins.
Gene-set analysis of MPH-treated adults with ADHD
All 18 gene-sets retrieved by FUMA from the three categories that were significantly overrepresented from the proteomic analysis were tested for association with IR-MPH treatment response in a clinical sample of adults with ADHD (Table 2). Gene-set analysis for the percentage change in severity scores of ADHD after IR-MPH treatment resulted in nominal associations for two gene-sets from the Reactome category: the GABA synthesis release reuptake and degradation, containing 17 genes (P = 0.013), and Neurotransmitter release cycle, containing 32 genes (P = 0.008). However, these associations did not remain significant after FDR correction (Pcorr = 0.117 and Pcorr = 0.117, respectively). In the analysis stratifying the sample by sex, the nominal associations observed for the whole sample remain only in men (P = 0.043 and Pcorr = 0.387; P = 0.009 and Pcorr = 0.162, respectively; Supplementary Table 4). No associations were found for the gene-sets from the GO Biological Process and KEGG categories.
Table 2.
Competitive gene-set analysis of the percentage change in symptoms of ADHD according to SNAP-IV scale after treatment with IR-MPH in adults with ADHD.
| Gene-set | Number of genes | Beta (SE) | P-value |
|---|---|---|---|
| Gene ontology (GO)—biological process | |||
| Organonitrogen compound metabolic process | 1685 | 0.007 (0.021) | 0.365 |
| Generation of precursor metabolites and energy | 272 | −0.005 (0.047) | 0.538 |
| Oxidation reduction process | 840 | 0.025 (0.029) | 0.191 |
| Cellular respiration | 134 | 0.025 (0.064) | 0.348 |
| Energy derivation by oxidation of organic compounds | 202 | −0.028 (0.054) | 0.694 |
| Regulation of cardiac conduction | 63 | −0.163 (0.105) | 0.939 |
| Cell communication by electrical coupling | 15 | 0.309 (0.201) | 0.063 |
| Reactome | |||
| GABA synthesis release reuptake and degradation | 17 | 0.455 (0.205) | 0.013a |
| Translation | 133 | 0.031 (0.067) | 0.322 |
| SRP dependent cotranslational protein targeting to membrane | 100 | 0.032 (0.075) | 0.335 |
| Transmembrane transport of small molecules | 391 | 0.009 (0.044) | 0.429 |
| TCA cycle and respiratory electron transport | 111 | 0.018 (0.071) | 0.401 |
| Respiratory electron transport | 62 | 0.029 (0.092) | 0.378 |
| Glucose metabolism | 59 | 0.066 (0.103) | 0.261 |
| Neurotransmitter release cycle | 32 | 0.375 (0.155) | 0.008b |
| Respiratory electron transport ATP synthesis by chemiosmotic coupling and heat production by uncoupling proteins | 78 | −0.007 (0.082) | 0.534 |
| Kyoto Encyclopedia of Genes and Genomes (KEGG) | |||
| Huntington’s disease | 165 | 0.024 (0.061) | 0.353 |
| Parkinson’s disease | 108 | −0.053 (0.074) | 0.763 |
ADHD attention-deficit/hyperactivity disorder, IR-MPH immediate-release methylphenidate, SNAP-IV Swanson, Nolan, and Pelham Rating Scale, version 4, SE standard
aFDR = 0.117
bFDR = 0.117
Discussion
This study investigated the global protein alterations induced by MPH in the cortex of rats. The 98 DEPs identified were analyzed in the context of biological pathways to provide a comprehensive overview of which systems are affected by MPH administration. The overall findings pointed out some pathways presenting biological plausibility, such as those related to synaptic neurotransmission, and others with a less established relationship that should be further explored, such as those involving oxidative stress, respiratory chain, and metabolic processes. The major relevance of this exploratory approach, besides unraveling putative molecular effects of MPH, is the perspective to translate the results to clinical samples. In this sense, we were also able to reinforce the relevance of some findings in a clinical sample of adults with ADHD, for which we found suggestive evidence on the association between MPH treatment response and genetic variability in pathways related to neurotransmitter release and GABA transmission.
Among the most interesting pathways overrepresented in the functional enrichment analysis of the DEPs with MPH treatment are those involved in synaptic neurotransmission, including gene-sets related to neurotransmitter release and transport, vesicle-mediated transport and GABA transmission (see Fig. 2 and Supplementary Tables 2 and 3). In line with these results, it has been reported that MPH induces alterations in synaptic vesicle-mediated neurotransmission in lysates of striatal synaptosomes of rats, where the content, transport, and release of dopamine were altered with MPH treatment42,43. Additionally, gene expression levels of proteins involved in neurotransmitter release were reduced with MPH treatment in PC12 cells10, and an enrichment analysis from transcriptomics studies also indicated the involvement of the synaptic transmission pathway in the MPH treatment15. Regarding the GABA neurotransmitter specifically, there are also evidences that MPH can modulate the GABAergic transmission44,45.
The involvement of pathways related to synaptic neurotransmission in MPH treatment response was reinforced by the gene-set analysis using a clinical sample of adults with ADHD, which was based on the biological pathways generated from the functional enrichment of the DEPs in WKY rats. We adopted this strategy considering that pathways modulated by MPH at the proteomic level are a good source of biological candidates to be investigated regarding treatment response. This translational approach suggests that among the pathways altered by MPH in rats, those involved in neurotransmitter release and GABA transmission are also important for treatment response to MPH. Although similar findings were observed only in men in the analysis separated by sex, this does not suggest an apparent sex-specific effect since the stratification did not improve the association values, indicating that the inclusion of women in the analysis may contribute for the association as well. Therefore, genetic variants in such biological pathways could be an important source of treatment response variability. Indeed, SNARE-related genetic variants, involved in neurotransmitter exocytosis, have been associated with treatment response to MPH in candidate gene association studies, in particular, SNAP-25 in children46 and SYT1 in adults39.
In line with this, a review of proteomic studies on multiple addictive drugs reported that synaptic proteins (comprising those related to the SNARE complex and the GABA receptor signaling) were also associated with exposure to one or more addictive drugs, including cocaine and/or amphetamine/methamphetamine47, suggesting an important role of these proteins for the effects of psychostimulants. Besides these proteins, most pathways modulated by MPH in our study overlap with those previously reported for other psychostimulants, such as alterations in metabolism, oxidative stress, and cell signaling26,47. On the other hand, the overrepresentation of pathways related to cardiac processes in our study is not shared with proteomic results for other psychostimulants26,47. Although MPH-induced alterations in the cardiovascular system, e.g., heart rate, is reasonable since they have been reported in patients with ADHD48, it is also possible that proteins comprising these pathways are indeed more related to other biological systems in this context. For example, a group of genes included in “regulation of cardiac conduction” overlaps with “muscle contraction”, which are more specific gene-sets, but they also comprise pathways involving cell communication by electrical coupling, calcium ion signaling, and ion homeostasis, which are very general signaling processes likely relevant for several systems, and more plausible for MPH effects in the brain (see Fig. 2 and Supplementary Tables 2 and 3).
Transcriptomics studies also suggest effects of MPH on synaptic transmission and cell signaling related pathways;15 however, they also point out other pathways, including axon guidance, nervous system development, Wnt signaling, cell/neuron differentiation, neuropeptide signaling, which were not observed in our study. Even though our methods differ from those used in these studies, previous investigations presenting comparable experimental designs also report a low concordance between proteomic and transcriptomic techniques. Such limited correlation between mRNA levels and protein abundance is a result of complex post-transcriptional and -translational regulatory mechanisms, such as transcript stability and protein degradation, that are reflected in the intrinsic differences captured in each step of the flow of genetic information49. Ideally, they should be interpreted as complementary approaches, but since proteomics detects the final product of genes, it can provide more direct insights on functional mechanisms and cellular activities.
Besides, the enrichment analysis of the DEPs in our study also retrieved interesting results from the KEGG category. Among them, the Huntington and Parkinson gene-sets might indicate an involvement of genes related to movement and cognitive processes in MPH actions. This might suggest that genetic variation related to traits underlying these diseases could be linking such pathways with MPH effects. Interestingly, the review mentioned above of proteomic studies involving the exposure to addictive drugs also pointed out Huntington’s disease signaling pathway47.
The functional enrichment results should be interpreted considering the limitation of this approach regarding the possibility of false-positive results in hypergeometric-based tests, which is a consequence of the gene overlap among the gene-sets (as exemplified above for the “regulation of cardiac conduction” gene-set). However, there is a high concordance among the pathways generated by the three tools used for analysis. Also, the associations observed for the clinical sample of adults with ADHD treated with MPH extended some of the proteomic findings from rats, at least in terms of treatment response. Unfortunately, we were not able to assess other system effects in the clinical sample, such as the cardiovascular and oxidative stress processes that have been reported for patients treated with MPH. Moreover, the sample size for the clinical sample is relatively small, and this could be limiting the detection of additional associations. Among the experimental limitations, we should mention we used WKY male rats, and the results might not be generalized to females. Also, we did not consider the behavioral effects of MPH treatment, and our results are reflecting MPH-induced molecular modifications independently of the presence of ADHD symptoms. Therefore, our results should be interpreted similarly to basic research and nonclinical drug development studies, for which the effects of a medication are evaluated without biological interference related to the disease at the first moment. For further interpretation of our results in the context of ADHD, they should be replicated using animal models for ADHD. The exclusive proteins of the saline-treated group (DDHD1, TNRC6B, ATXN1, and PRODH1) may raise the possibility of an artifact. However, since these proteins were consistently detected among the biological replicates in this condition, a possible suppressing effect of MPH in their expression is more likely. Although the mechanisms underlying these putative effects remain to be elucidated, their relevance in neurodevelopmental phenotypes has already been demonstrated50–57. Finally, considering the exploratory nature of our study design, cortex was the only brain region evaluated, and we did not perform methodological replication; however, the enrichment analysis and the extension of some results to a clinical sample allow us to make inferences on the most important biological pathways and candidates that should be further explored in future studies and in different brain regions.
In general, our exploratory study unraveled several pathways from different biological processes that are modulated by MPH treatment in rats. Importantly, the findings involving processes related to synaptic neurotransmission were extended to humans for MPH treatment response. The overall evidence from previous experimental studies combined with our findings suggests that MPH response involves a complex interaction of neurotransmitter systems that go beyond the widely acknowledged dopamine and norepinephrine, especially related to GABA transmission. This translational perspective, that integrates proteomics and pharmacogenomics, can shed light on the molecular signatures of MPH and the possible biological sources of treatment response variability.
Supplementary information
Acknowledgements
The authors would like to thank the ProDAH-A team and all the individuals with ADHD that participated in this study. The authors are also thankful to the following funding sources: Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, 476529/2012-3, 466722/2014-1 and 305592/2014-9), Hospital de Clínicas de Porto Alegre (FIPE-HCPA 100358, 08543 and 160346), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), European Community’s Horizon 2020 Programme (H2020/2014-2020) under grant agreement nº 667302 (CoCA), and NIH grants NIH/NIHGM P41 GM103533-22, NIH/NIMH 5 R01 MH067880-14, and P30 CA030199.
Conflict of interest
The author(s) declare the following potential conflict of interest with respect to the research, authorship and/or publication of this article: Dr. Grevet was on the speaker’s bureau for Novartis and Shire for the last 3 years. He also received travel awards (air tickets and hotel accommodations) for participating in two psychiatric meetings from Shire and Novartis. Dr. Rohde has been on the speakers’ bureau/advisory board and/or has acted as a consultant for Medice, Novartis/Sandoz and Shire/Takeda in the last three years. He receives authorship royalties from Oxford Press and ArtMed. He also received travel awards for taking part of 2016 AACAP meeting from Novartis and 2018 APA meeting from Shire. The ADHD and Juvenile Bipolar Disorder Outpatient Programs chaired by him received unrestricted educational and research support from the following pharmaceutical companies in the last three years: Janssen-Cilag, Medice, Novartis/Sandoz, and Shire/Takeda. All other authors have nothing to disclose.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Bruna S. da Silva, Douglas T. Leffa
Co-senior authors contributed equally to this work: Claiton H. D. Bau, Verônica Contini
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41398-019-0649-5).
References
- 1.Castells X, et al. Efficacy of methylphenidate for adults with attention-deficit hyperactivity disorder: a meta-regression analysis. CNS Drugs. 2011;25:157–169. doi: 10.2165/11539440-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 2.Catalá-López F, et al. The pharmacological and non-pharmacological treatment of attention deficit hyperactivity disorder in children and adolescents: a systematic review with network meta-analyses of randomised trials. PLoS ONE. 2017;12:e0180355. doi: 10.1371/journal.pone.0180355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cortese Samuele, Adamo Nicoletta, Del Giovane Cinzia, Mohr-Jensen Christina, Hayes Adrian J, Carucci Sara, Atkinson Lauren Z, Tessari Luca, Banaschewski Tobias, Coghill David, Hollis Chris, Simonoff Emily, Zuddas Alessandro, Barbui Corrado, Purgato Marianna, Steinhausen Hans-Christoph, Shokraneh Farhad, Xia Jun, Cipriani Andrea. Comparative efficacy and tolerability of medications for attention-deficit hyperactivity disorder in children, adolescents, and adults: a systematic review and network meta-analysis. The Lancet Psychiatry. 2018;5(9):727–738. doi: 10.1016/S2215-0366(18)30269-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gamo NJ, Wang M, Arnsten AFT. Methylphenidate and atomoxetine enhance prefrontal function through α2-adrenergic and dopamine D1 receptors. J. Am. Acad. Child Adolesc. Psychiatry. 2010;49:1011–1023. doi: 10.1016/j.jaac.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnsten AF, Dudley AG. Methylphenidate improves prefrontal cortical cognitive function through alpha2 adrenoceptor and dopamine D1 receptor actions: relevance to therapeutic effects in attention deficit hyperactivity disorder. Behav. Brain Funct. 2005;1:2. doi: 10.1186/1744-9081-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rozas C, et al. Methylphenidate amplifies long-term potentiation in rat hippocampus CA1 area involving the insertion of AMPA receptors by activation of β-adrenergic and D1/D5 receptors. Neuropharmacology. 2015;99:15–27. doi: 10.1016/j.neuropharm.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Daniali S, et al. Chronic Ritalin administration during adulthood increases serotonin pool in rat medial frontal cortex. Iran. Biomed. J. 2013;17:134–139. doi: 10.6091/ibj.1173.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller Erin M., Quintero Jorge E., Pomerleau Francois, Huettl Peter, Gerhardt Greg A., Glaser Paul E. A. Chronic Methylphenidate Alters Tonic and Phasic Glutamate Signaling in the Frontal Cortex of a Freely-Moving Rat Model of ADHD. Neurochemical Research. 2018;44(1):89–101. doi: 10.1007/s11064-018-2483-1. [DOI] [PubMed] [Google Scholar]
- 9.Solleveld MM, Schrantee A, Puts NAJ, Reneman L, Lucassen PJ. Age-dependent, lasting effects of methylphenidate on the GABAergic system of ADHD patients. NeuroImage Clin. 2017;15:812–818. doi: 10.1016/j.nicl.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartl J, et al. Effects of methylphenidate: the cellular point of view. ADHD Atten. Deficit Hyperact. Disord. 2010;2:225–232. doi: 10.1007/s12402-010-0039-6. [DOI] [PubMed] [Google Scholar]
- 11.Schwarz R, et al. A preliminary study on methylphenidate-regulated gene expression in lymphoblastoid cells of ADHD patients. World J. Biol. Psychiatry. 2015;16:180–189. doi: 10.3109/15622975.2014.948064. [DOI] [PubMed] [Google Scholar]
- 12.Quansah E, Sgamma T, Jaddoa E, Zetterström TSC. Chronic methylphenidate regulates genes and proteins mediating neuroplasticity in the juvenile rat brain. Neurosci. Lett. 2017;654:93–98. doi: 10.1016/j.neulet.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 13.dela Peña I, et al. Prefrontal cortical and striatal transcriptional responses to the reinforcing effect of repeated methylphenidate treatment in the spontaneously hypertensive rat, animal model of attention-deficit/hyperactivity disorder (ADHD) Behav. Brain Funct. 2014;10:17. doi: 10.1186/1744-9081-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sadasivan S, et al. Methylphenidate exposure induces dopamine neuron loss and activation of microglia in the basal ganglia of mice. PLoS ONE. 2012;7:e33693. doi: 10.1371/journal.pone.0033693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grünblatt E, Bartl J, Walitza S. Methylphenidate enhances neuronal differentiation and reduces proliferation concomitant to activation of Wnt signal transduction pathways. Transl. Psychiatry. 2018;8:51. doi: 10.1038/s41398-018-0096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong Q, et al. Increased locomotor activity and non-selective attention and impaired learning ability in SD rats after lentiviral vector-mediated RNA interference of Homer 1a in the brain. Int. J. Med. Sci. 2013;10:90–102. doi: 10.7150/ijms.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baird AL, Coogan AN, Kaufling J, Barrot M, Thome J. Daily methylphenidate and atomoxetine treatment impacts on clock gene protein expression in the mouse brain. Brain Res. 2013;1513:61–71. doi: 10.1016/j.brainres.2013.03.038. [DOI] [PubMed] [Google Scholar]
- 18.Nakao T, Radua J, Rubia K, Mataix-Cols D. Gray matter volume abnormalities in ADHD: voxel-based meta-analysis exploring the effects of age and stimulant medication. Am. J. Psychiatry. 2011;168:1154–1163. doi: 10.1176/appi.ajp.2011.11020281. [DOI] [PubMed] [Google Scholar]
- 19.Frodl T, Skokauskas N. Meta-analysis of structural MRI studies in children and adults with attention deficit hyperactivity disorder indicates treatment effects. Acta Psychiatr. Scand. 2012;125:114–126. doi: 10.1111/j.1600-0447.2011.01786.x. [DOI] [PubMed] [Google Scholar]
- 20.Hong S-B, et al. Functional dysconnectivity of corticostriatal circuitry and differential response to methylphenidate in youth with attention-deficit/hyperactivity disorder. J. Psychiatry Neurosci. 2015;40:46–57. doi: 10.1503/jpn.130290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubia K, et al. Effects of stimulants on brain function in attention-deficit/hyperactivity disorder: a systematic review and meta-analysis. Biol. Psychiatry. 2014;76:616–628. doi: 10.1016/j.biopsych.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spencer TJ, et al. Effect of psychostimulants on brain structure and function in ADHD. J. Clin. Psychiatry. 2013;74:902–917. doi: 10.4088/JCP.12r08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turck CW, Filiou MD. What have mass spectrometry-based proteomics and metabolomics (not) taught us about psychiatric disorders? Mol. Neuropsychiatry. 2015;1:69–75. doi: 10.1159/000381902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.del Castillo C, et al. Proteomic analysis of the nucleus accumbens of rats with different vulnerability to cocaine addiction. Neuropharmacology. 2009;57:41–48. doi: 10.1016/j.neuropharm.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Iwazaki T, McGregor IS, Matsumoto I. Protein expression profile in the striatum of acute methamphetamine-treated rats. Brain Res. 2006;1097:19–25. doi: 10.1016/j.brainres.2006.04.052. [DOI] [PubMed] [Google Scholar]
- 26.Kobeissy F, et al. Recent updates on drug abuse analyzed by neuroproteomics studies: Cocaine, Methamphetamine and MDMA. Transl. Proteom. 2014;3:38–52. doi: 10.1016/j.trprot.2014.04.001. [DOI] [Google Scholar]
- 27.Aoyama T, Kotaki H, Iga T. Dose-dependent kinetics of methylphenidate enantiomers after oral administration of racemic methylphenidate to rats. J. Pharmacobiodyn. 1990;13:647–652. doi: 10.1248/bpb1978.13.647. [DOI] [PubMed] [Google Scholar]
- 28.Kuczenski R, Segal DS. Exposure of adolescent rats to oral methylphenidate: preferential effects on extracellular norepinephrine and absence of sensitization and cross-sensitization to methamphetamine. J. Neurosci. 2002;22:7264–7271. doi: 10.1523/JNEUROSCI.22-16-07264.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wargin W, et al. Pharmacokinetics of methylphenidate in man, rat and monkey. J. Pharmacol. Exp. Ther. 1983;226:382–386. [PubMed] [Google Scholar]
- 30.Thanos PK, et al. A pharmacokinetic model of oral methylphenidate in the rat and effects on behavior. Pharmacol. Biochem. Behav. 2015;131:143. doi: 10.1016/j.pbb.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi M, et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics. 2014;30:2524–2526. doi: 10.1093/bioinformatics/btu305. [DOI] [PubMed] [Google Scholar]
- 32.Pascovici D, Handler DCL, Wu JX, Haynes PA. Multiple testing corrections in quantitative proteomics: a useful but blunt tool. Proteomics. 2016;16:2448–2453. doi: 10.1002/pmic.201600044. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 35.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinforma. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.APA, A. P. A. Diagnostic and statistical manual of mental disorders (DSM-IV). Am. Psychiatr. Assoc. 78–85 (1994).
- 37.First, M. B., Spitzer, R. L., Gibbon, M. L., & Williams, J. B. W. Structured clinical interview for DSM-IV-TR axis I disorders, Research Version, Patient Edition. (SCID-I/P). Biometrics Res. (2002).
- 38.Victor MM, et al. Severity but not comorbidities predicts response to methylphenidate in adults with attention-deficit/hyperactivity disorder: results from a naturalistic study. J. Clin. Psychopharmacol. 2014;34:212–217. doi: 10.1097/JCP.0000000000000091. [DOI] [PubMed] [Google Scholar]
- 39.da Silva BS, et al. Exocytosis-related genes and response to methylphenidate treatment in adults with ADHD. Mol. Psychiatry. 2018;23:1446–1452. doi: 10.1038/mp.2017.90. [DOI] [PubMed] [Google Scholar]
- 40.Victor MM, et al. Reasons for pretreatment attrition and dropout from methylphenidate in adults with attention-deficit/hyperactivity disorder: the role of comorbidities. J. Clin. Psychopharmacol. 2009;29:614–616. doi: 10.1097/JCP.0b013e3181c00b1e. [DOI] [PubMed] [Google Scholar]
- 41.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Volz TJ, Farnsworth SJ, Hanson GR, Fleckenstein AE. Methylphenidate-induced alterations in synaptic vesicle trafficking and activity. Ann. N. Y. Acad. Sci. 2008;1139:285–290. doi: 10.1196/annals.1432.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Volz Trent J., Farnsworth Sarah J., King Jill L., Riddle Evan L., Hanson Glen R., Fleckenstein Annette E. Methylphenidate Administration Alters Vesicular Monoamine Transporter-2 Function in Cytoplasmic and Membrane-Associated Vesicles. Journal of Pharmacology and Experimental Therapeutics. 2007;323(2):738–745. doi: 10.1124/jpet.107.126888. [DOI] [PubMed] [Google Scholar]
- 44.Goitia B, et al. Differential effects of methylphenidate and cocaine on GABA transmission in sensory thalamic nuclei. J. Neurochem. 2013;124:602–612. doi: 10.1111/jnc.12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quansah E, Ruiz-Rodado V, Grootveld M, Probert F, Zetterström TSC. 1H NMR-based metabolomics reveals neurochemical alterations in the brain of adolescent rats following acute methylphenidate administration. Neurochem. Int. 2017;108:109–120. doi: 10.1016/j.neuint.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 46.McGough J, et al. Pharmacogenetics of methylphenidate response in preschoolers with ADHD. J. Am. Acad. Child Adolesc. Psychiatry. 2006;45:1314–1322. doi: 10.1097/01.chi.0000235083.40285.08. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Yuan W, Li MD. Genes and pathways co-associated with the exposure to multiple drugs of abuse, including alcohol, amphetamine/methamphetamine, cocaine, marijuana, morphine, and/or nicotine: a review of proteomics analyses. Mol. Neurobiol. 2011;44:269–286. doi: 10.1007/s12035-011-8202-4. [DOI] [PubMed] [Google Scholar]
- 48.Liang EF, et al. The effect of methylphenidate and atomoxetine on heart rate and systolic blood pressure in young people and adults with attention-deficit hyperactivity disorder (ADHD): systematic review, meta-analysis, and meta-regression. Int. J. Environ. Res. Public Health. 2018;15:1789. doi: 10.3390/ijerph15081789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seliger B, et al. Combined analysis of transcriptome and proteome data as a tool for the identification of candidate biomarkers in renal cell carcinoma. Proteomics. 2009;9:1567–1581. doi: 10.1002/pmic.200700288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doan RN, et al. Recessive gene disruptions in autism spectrum disorder. Nat. Genet. 2019;51:1092–1098. doi: 10.1038/s41588-019-0433-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nord AS, et al. Reduced transcript expression of genes affected by inherited and de novo CNVs in autism. Eur. J. Hum. Genet. 2011;19:727–731. doi: 10.1038/ejhg.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Celestino-Soper PB, et al. Deletions in chromosome 6p22.3-p24.3, including ATXN1, are associated with developmental delay and autism spectrum disorders. Mol. Cytogenet. 2012;5:17. doi: 10.1186/1755-8166-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eising E, et al. A set of regulatory genes co-expressed in embryonic human brain is implicated in disrupted speech development. Mol. Psychiatry. 2019;24:1065–1078. doi: 10.1038/s41380-018-0020-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Koning MB, et al. PRODH rs450046 and proline x COMT Val158 Met interaction effects on intelligence and startle in adults with 22q11 deletion syndrome. Psychopharmacol. (Berl.). 2015;232:3111–3122. doi: 10.1007/s00213-015-3971-5. [DOI] [PubMed] [Google Scholar]
- 56.Barøy T, et al. Haploinsufficiency of two histone modifier genes on 6p22.3, ATXN1 and JARID2, is associated with intellectual disability. Orphanet J. Rare Dis. 2013;8:3. doi: 10.1186/1750-1172-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rizzi TS, et al. The ATXN1 and TRIM31 genes are related to intelligence in an ADHD background: evidence from a large collaborative study totaling 4,963 subjects. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2011;156:145–157. doi: 10.1002/ajmg.b.31149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.