Abstract
Dopamine and cAMP regulated phosphoprotein 32 kDa (DARPP-32) also known as phosphoprotein phosphatase-1 regulatory subunit 1B and encoded by the PPP1R1B gene is an inhibitor of protein phosphatase-1 and protein kinase A. DARPP-32 is expressed in a wide range of epithelial cells and some solid tumours; however, its role in breast cancer is only partially defined. DARPP-32 expression was determined using immunohistochemistry in two independent cohorts of early stage invasive breast cancer patients (discovery n = 1352; validation n = 1655), and 112 HER2 positive breast cancer patients treated with trastuzumab and adjuvant chemotherapy. PPP1R1B mRNA expression was assessed in the METABRIC cohort (n = 1980), using artificial neural network analysis to identify associated genes. In the discovery cohort, low nuclear expression of DARPP-32 was significantly associated with shorter survival (P = 0.041), which was independent of other prognostic variables (P = 0.019). In the validation cohort, low cytoplasmic and nuclear expression was significantly associated with shorter survival (both P = 0.002), with cytoplasmic expression independent of other prognostic variables (P = 0.023). Stronger associations with survival in oestrogen receptor (ER) positive disease were observed. In patients treated with trastuzumab, low nuclear expression was significantly associated with adverse progression-free survival (P = 0.031). In the METABRIC cohort, low PPP1R1B expression was associated with shortened survival of ER positive patients. Expression of CDC42 and GRB7, amongst others, were associated with PPP1R1B expression. This data suggests a role for DARPP-32 as a prognostic marker with clinical utility in breast cancer.
Subject terms: Breast cancer, Prognostic markers, Breast cancer, Tumour biomarkers
Introduction
Dopamine and cAMP regulated phosphoprotein 32 kDa (DARPP-32) also known as phosphoprotein phosphatase-1 regulatory subunit 1B and encoded by the PPP1R1B gene, was first described in 19831 and has been widely characterised as a signalling protein highly concentrated in brain regions rich in dopaminergic nerve terminals2–4. DARPP-32 was originally demonstrated to be a potent inhibitor of protein phosphatase-1 (PP-1) and a substrate of calcineurin5,6. Protein kinase A (PKA) phosphorylation of Thr34 allows DARPP-32 to inhibit protein phosphatase-1 (PP-1); cyclin dependent kinase (CDK)-5 phosphorylation of Thr75 allows DARPP-32 to inhibit PKA and enhance β-adducin Ser713 phosphorylation7. A truncated DARPP-32 isoform, t-DARPP, lacks the Thr-34 phosphorylation site and was originally identified in gastric cancer8. Interaction between DARPP-32, calcineurin and Bcl-2 assists with the anti-apoptotic function of Bcl-2 by preventing Ca2+ induced cell death through interaction with inositol 1,4,5-triphosphate receptor (InsP3R)9.
DARPP-32 activation is regulated by an array of neurotransmitters, such as dopamine, glutamate, serotonin and adenosine, but has also been shown to mediate the actions of multiple drugs of abuse, including cocaine, amphetamine, nicotine and caffeine (reviewed in10). DARPP-32 has been implicated in a number of psychiatric and neurological disorders, such as schizophrenia. In addition to the central nervous system, DARPP-32 is expressed in a wide range of epithelial cells11.
High levels of DARPP-32 in colorectal cancer are associated with survival of Dukes A and B patients12, and in glioblastoma, high DARPP-32/STAT3 and DARPP-32/STAT5B mRNA ratios are associated with longer progression free survival and overall survival13. In gastric cancer, DARPP-32 can promote cell invasion through CXCR4-mediated activation of the MT1-MMP/MMP-2 pathway14. A PPP1R1B-STARD3 fusion transcript has also been identified in gastric cancer, that increases in vitro cell proliferation through the phosphatidylinositol-3-kinase (PI3K)/AKT pathway15. DARPP-32 has been shown to influence breast epithelial cell migration; in MCF-7 and MDA-MB-231 cells this has been shown to be in a DDR1 dependent manner16. DARPP-32 phosphorylation, induced by Wnt-5a, has also been shown to inhibit MCF-7 cell migration in a CREB-dependent manner17.
The truncated splice variant, t-DARPP is present in gastric, breast, prostate, colon and stomach cancers8,11, and in models of murine tumourigenesis, DARPP-32 expression is expressed in normal mouse tissue and some breast tumours, with t-DARPP expressed only within tumours18. In breast cancer, t-DARPP mRNA is expressed in 36% of primary breast cancers (n = 36) relative to adjacent normal breast tissues (n = 18)19. Interestingly, the expression of t-DARPP has been implicated in resistance to the HER2 targeted agent, trastuzumab, in HER2 positive breast cancer cells via sustained signalling through phosphatidylinositol-4,5 bisphosphate 3-kinase (PI3K)/akt pathway and activation of PKA in vitro19–23, and in conferring a survival advantage to the HER2 targeted agent, lapatinib24. The role of DARPP-32 and t-DARPP in cancer is reviewed in25.
We sought to determine the frequency and importance of DARPP-32 expression in two large independent cohorts of early stage invasive breast cancer patients, including an additional cohort of HER2 positive breast cancer patients treated with trastuzumab to examine if DARPP-32 was associated with patient survival. In addition to protein expression, we sought to assess PPP1R1B mRNA expression in a large, well-annotated series of breast cancer patients, including artificial neural network analysis to identify genes associated with PPP1R1B expression.
Methods
Patient cohorts
This study is reported according to reporting recommendations for tumour marker prognostic studies (REMARK) criteria26. For protein expression three well-characterised patient cohorts were used; the discovery cohort, functioned as a discovery set; the validation cohort, functioned as a validation set and the HER2 cohort was used to assess DARPP-32 expression in HER2 positive patients treated with trastuzumab. Breast cancer specific survival was calculated as the time interval between primary surgery and death resultant from breast cancer. Progression-free survival was defined as the date of surgery to relapse (including local and regional relapse).
Discovery cohort
1352 early stage invasive breast cancer patients were available for assessment in the discovery cohort, with all patients treated at Nottingham University Hospitals between 1987 and 1998. All patients were managed in a standard manner, where all patients underwent a mastectomy or wide local excision, as decided by disease characteristics or patient choice, followed by radiotherapy if indicated. Patients received systemic adjuvant treatment on the basis of Nottingham Prognostic index (NPI), oestrogen receptor (ER), and menopausal status. Patients with an NPI score less than 3.4 did not receive adjuvant treatment and patients with an NPI score of 3.4 and above were candidates for CMF combination chemotherapy (cyclophosphamide, methotrexate and 5-fluorouracil) if they were ER negative or premenopausal; and hormonal therapy if they were ER positive. Median follow-up was 205 months determined using the reverse Kaplan-Meier method and clinicopathological information for this cohort is available in Table 1.
Table 1.
Associations between the cytoplasmic and nuclear expression of DARPP-32, determined in the discovery cohort and validation cohort using immunohistochemistry, with clinicopathological variables.
| Discovery cohort | Validation cohort | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cytoplasmic expression | Nuclear expression | Cytoplasmic expression | Nuclear expression | |||||||||
| Low | High | P value | Low | High | P value | Low | High | P value | Low | High | P value | |
| Age | ||||||||||||
| <50 years | 221 (16.4%) | 238 (17.6%) | 0.470 | 295 (21.8%) | 164 (12.1%) | 0.467 | 222 (13.5%) | 283 (17.3%) | 0.881 | 272 (16.6%) | 233 (14.2%) | 0.251 |
| ≥50 years | 448 (33.2%) | 444 (32.9%) | 591 (43.7%) | 301 (22.3%) | 494 (30.1%) | 640 (39.0%) | 576 (35.1%) | 558 (34.0%) | ||||
| Tumour size | ||||||||||||
| <20 mm | 414 (30.8%) | 397 (29.5%) | 0.188 | 594 (40.8%) | 262 (19.5%) | 0.052 | 422 (25.8%) | 589 (36.0%) | 0.048 | 513 (31.3%) | 334 (20.4%) | 0.320 |
| ³20 mm | 253 (18.8%) | 281 (20.9%) | 334 (24.8%) | 200 (14.9%) | 293 (17.9%) | 334 (20.4%) | 498 (30.4%) | 293 (17.9%) | ||||
| T stage | ||||||||||||
| 1 | 404 (30.0%) | 415 (30.9) | 0.210 | 536 (39.9%) | 283 (21.0%) | 0.500 | 424 (25.9%) | 608 (37.2%) | 0.016 | 501 (30.6%) | 531 (32.5%) | 0.001 |
| 2 | 215 (16.0%) | 197 (14.6%) | 277 (20.6%) | 135 (10.0%) | 208 (12.7%) | 237 (14.5%) | 244 (14.9%) | 201 (12.3%) | ||||
| 3 | 49 (3.6%) | 65 (4.8%) | 70 (5.2%) | 44 (3.3%) | 81 (5.0%) | 77 (4.7%) | 100 (6.1%) | 58 (3.5%) | ||||
| Tumour grade | ||||||||||||
| 1 | 104 (7.7%) | 124 (9.2%) | 0.165 | 153 (11.4%) | 75 (5.6%) | 0.391 | 84 (5.1%) | 179 (10.9%) | <0.001 | 105 (6.4%) | 158 (9.6%) | <0.001 |
| 2 | 238 (17.7%) | 212 (15.8%) | 304 (22.6%) | 146 (10.9%) | 300 (18.3%) | 358 (21.9%) | 349 (21.3%) | 309 (18.9%) | ||||
| 3 | 325 (24.2%) | 342 (25.4%) | 426 (31.7%) | 241 (17.9%) | 331 (20.2%) | 386 (23.6%) | 393 (24.0%) | 324 (19.8%) | ||||
| ER status | ||||||||||||
| Negative | 132 (10.1%) | 216 (16.5%) | <0.001 | 173 (13.2%) | 175 (13.4%) | <0.001 | 90 (5.5%) | 230 (14.0%) | <0.001 | 114 (7.0%) | 206 (12.6%) | <0.001 |
| Positive | 518 (39.5%) | 444 (33.9%) | 683 (52.1%) | 279 (21.3%) | 626 (38.2%) | 694 (42.3%) | 734 (44.8%) | 586 (35.7%) | ||||
| PgR status | ||||||||||||
| Negative | 247 (19.4%) | 301 (23.7%) | 0.007 | 319 (25.1%) | 229 (18.0%) | <0.001 | 246 (15.9%) | 405 (26.1%) | <0.001 | 299 (19.3%) | 352 (22.7%) | <0.001 |
| Positive | 381 (30.0%) | 341 (26.9%) | 513 (40.4%) | 249 (16.5%) | 425 (27.4%) | 473 (30.5%) | 502 (32.4%) | 396 (25.6%) | ||||
| NPI category | ||||||||||||
| Good (≤3.4) | 199 (14.8%) | 208 (15.5%) | 0.810 | 275 (20.5%) | 132 (9.8%) | 0.432 | 230 (14.1%) | 339 (20.7%) | 0.002 | 275 (16.8%) | 294 (18.0%) | <0.001 |
| Intermediate (3.41–5.4) | 348 (25.9%) | 342 (25.5%) | 452 (33.7%) | 238 (17.7%) | 347(21.2%) | 465 (28.5%) | 405 (24.8%) | 407 (24.9%) | ||||
| Poor (>5.4) | 119 (8.9%) | 127 (9.5%) | 154 (11.5%) | 92 (6.9%) | 135 (8.3%) | 118 (7.2%) | 164 (10.0%) | 89 (5.4%) | ||||
| Tubule formation | ||||||||||||
| 1 | 31 (2.4%) | 44 (3.4%) | 0.218 | 43 (3.3%) | 32 (2.5%) | 0.160 | 28 (1.7%) | 92 (5.7%) | <0.001 | 43 (2.6%) | 77 (4.7%) | <0.001 |
| 2 | 201 (15.5%) | 219 (16.9%) | 286 (22.1%) | 134 (10.3%) | 184 (11.3%) | 295 (18.2%) | 219 (13.5%) | 260 (16.0%) | ||||
| 3 | 408 (31.5%) | 394 (30.4%) | 519 (40.0%) | 283 (21.8%) | 494 (30.4%) | 531 (32.7%) | 573 (35.3%) | 452 (27.8%) | ||||
| Pleomorphism | ||||||||||||
| 1 | 9 (0.7%) | 17 (1.3%) | 0.179 | 14 (1.1%) | 12 (0.9%) | 0.031 | 5 (0.3%) | 19 (1.2%) | 0.016 | 10 (0.6%) | 14 (0.9%) | 0.081 |
| 2 | 267 (20.6%) | 252 (19.5%) | 360 (27.8%) | 159 (12.3%) | 200 (12.3%) | 294 (18.1%) | 236 (14.5%) | 258 (15.9%) | ||||
| 3 | 36 (28.0%) | 387 (29.9) | 473 (36.5%) | 277 (21.4%) | 501 (30.8%) | 605 (37.3%) | 589 (36.3%) | 517 (31.8%) | ||||
| Mitosis | ||||||||||||
| 1 | 213 (16.4%) | 239 (18.4%) | 0.093 | 293 (22.6%) | 159 (12.3%) | 0.241 | 336 (20.7%) | 469 (28.9%) | 0.034 | 392 (24.2%) | 413 (25.5%) | 0.007 |
| 2 | 138 (10.6%) | 111 (8.6%) | 174 (13.4%) | 75 (5.8%) | 153 (9.4%) | 152 (9.4%) | 181 (11.2%) | 124 (7.6%) | ||||
| 3 | 289 (22.3%) | 307 (23.7%) | 381 (29.4%) | 215 (16.6%) | 217 (13.4%) | 295 (18.2%) | 262 (16.2%) | 250 (15.4%) | ||||
| HER2 status | ||||||||||||
| Negative | 570 (43.4%) | 552 (42.0%) | 0.052 | 762 (58.0%) | 360 (27.4%) | <0.001 | 636 (41.2%) | 779 (50.5%) | 0.025 | 750 (48.6%) | 665 (43.1%) | 0.113 |
| Positive | 83 (6.3%) | 109 (8.3%) | 104 (7.9%) | 88 (6.7%) | 44 (2.9%) | 83 (5.4%) | 58 (3.8%) | 69 (4.5%) | ||||
| Triple negative disease | ||||||||||||
| Negative | 558 (42.6%) | 515 (39.3%) | 0.001 | 734 (56.0%) | 339 (25.9%) | <0.001 | 640 (39.7%) | 733 (45.5%) | <0.001 | 753 (46.7%) | 620 (38.5%) | <0.001 |
| Positive | 96 (7.3%) | 142 (10.8%) | 130 (9.9%) | 108 (8.2%) | 67 (4.2%) | 171 (10.6%) | 85 (5.3%) | 153 (9.5%) | ||||
| Vascular invasion | ||||||||||||
| Negative | 430 (32.2%) | 464 (34.7%) | 0.141 | 582 (43.5%) | 312 (23.3%) | 0.589 | 481 (29.4%) | 678 (41.4) | 0.007 | 568 (34.7%) | 591 (36.1%) | <0.001 |
| Positive | 232 (17.4%) | 211 (15.8%) | 295 (22.1%) | 148 (11.1%) | 233 (14.2%) | 245 (15.0%) | 278 (17.0%) | 200 (12.2%) | ||||
The P values are resultant from Pearson χ2 test of association and significant values (P < 0.05) are highlighted in bold. ER is oestrogen receptor and PgR is progesterone receptor.
Validation cohort
1655 early stage invasive breast cancer patients were available for assessment in the validation cohort, with all patients treated at Nottingham University Hospitals between 1998 and 2006. All patients were managed in a standard manner, as described for the discovery cohort. Median follow-up was 148 months determined using the reverse Kaplan-Meier method and clinicopathological information for this cohort is available in Table 1.
HER2 positive cohort
112 HER2 positive breast cancer patients were available for assessment in the HER2 positive cohort, with all patients treated at Nottingham University Hospitals between 2004 and 2012. Patients were treated according to local guidelines, with adjuvant therapy and trastuzumab following surgery. Adjuvant hormone therapy was received by 47% of patients (40/75), with 74% of patients receiving adjuvant radiotherapy (59/80). Trastuzumab was given on a 3-weekly regimen for 52 weeks, with patients receiving trastuzumab following six cycles of 3-weekly FEC chemotherapy (fluorouracil, epirubicin and cyclophosphamide) or patients receiving three cycles of FEC, followed by three cycles of taxane (FEC-T), to which, trastuzumab was frequently added from the second cycle of taxane onwards. Median follow-up was 50 months determined using the reverse Kaplan-Meier method and clinicopathological information for this cohort is available in Table 2.
Table 2.
Associations between the expression of DARPP-32 determined in HER2 positive breast cancer patients treated with trastuzumab and adjuvant chemotherapy and clinicopathological variables.
| Cytoplasmic | Nuclear | ||||||
|---|---|---|---|---|---|---|---|
| Low | High | P value | Low | High | P value | ||
| Age | ≤40 years | 6 (5.4%) | 9 (8.0%) | 0.710 | 6 (5.4%) | 9 (8.0%) | 0.651 |
| >40 years | 34 (30.4%) | 63 (56.3%) | 33 (29.5%) | 64 (57.1%) | |||
| Tumour size | ≤20 mm | 24 (21.6%) | 44 (39.6%) | 0.965 | 24 (21.6%) | 44 (39.6%) | 0.767 |
| >20 mm | 15 (13.5%) | 28 (25.2%) | 14 (12.6%) | 29 (26.1%) | |||
| Node status | Negative | 16 (14.3%) | 28 (25.0%) | 0.908 | 15 (13.4%) | 29 (25.9%) | 0.896 |
| Positive | 24 (21.4%) | 44 (39.3%) | 24 (21.4%) | 44 (39.3%) | |||
| T stage | 0 | 0 (0.0%) | 1 (0.9%) | 0.644 | 0 (0.0%) | 1 (0.9%) | 0.684 |
| 1 | 25 (22.3%) | 44 (39.3%) | 25 (22.3%) | 44 (39.3%) | |||
| 2 | 14 (12.5%) | 22 (19.6%) | 13 (11.6%) | 23 (20.5%) | |||
| 3 | 1 (0.9%) | 5 (4.5%) | 1 (0.9%) | 5 (4.5%) | |||
| Tumour grade | 1 | 0 (0.0%) | 3 (2.7%) | 0.424 | 0 (0.0%) | 3 (2.7%) | 0.430 |
| 2 | 13 (11.6%) | 22 (19.6%) | 13 (11.6%) | 22 (19.6%) | |||
| 3 | 27 (24.1%) | 47 (42.0%) | 26 (23.2%) | 48 (42.9%) | |||
| ER status | Negative | 14 (12.5%) | 36 (32.1%) | 0.126 | 14 (12.5%) | 36 (32.1%) | 0.174 |
| Positive | 26 (23.2%) | 36 (32.1%) | 25 (22.3%) | 37 (33.0%) | |||
| PgR status | Negative | 16 (17.2%) | 43 (46.2%) | 0.163 | 16 (17.2%) | 43 (46.2%) | 0.265 |
| Positive | 14 (15.1%) | 20 (21.5%) | 13 (14.0%) | 21 (22.6%) | |||
| NPI category | Good (≤3.4) | 1 (1.0%) | 6 (5.9%) | 0.417 | 2 (2.6%) | 5 (5.0%) | 0.385 |
| Intermediate (3.41–5.4) | 27 (26.7%) | 42 (41.6%) | 23 (22.8%) | 46 (45.5%) | |||
| Poor (>5.4) | 10 (9.9%) | 15 (14.9%) | 12 (11.9%) | 13 (12.9%) | |||
The P values are resultant from Pearson χ2 test of association and significant values (P < 0.05) are highlighted in bold. ER is oestrogen receptor and PgR is progesterone receptor.
METABRIC series
Details of the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) data set (n = 1980) data set have been published elsewhere27. Tumours were collected by five centres in the UK and Canada between 1977–2005 and almost all ER negative and lymph node positive patients received adjuvant chemotherapy, whereas ER negative and/or lymph node positive patient did not. No patients with HER2 overexpression received trastuzumab. Median follow-up was 141 months determined using the reverse Kaplan-Meier method. DNA and RNA were isolated from samples and hybridised to the Affymetrix SNP 6.0 and Illumina HT-12 v3 platforms for genomic and transcriptional profiling as described by Curtis et al. (2012)27. This cohort was used to assess the prognostic significance of DARPP-32 at the mRNA level and determine associations with other genes using artificial neural network analysis.
Immunohistochemistry
Immunohistochemistry was performed on tissue microarrays for the discovery cohort, validation cohort and the HER2 cohort, and were comprised of single 0.6 mm cores taken from a representative tumour area as assessed on Haematoxylin and Eosin stained sections by a specialist breast cancer histopathologist. Immunohostochemical staining was achieved using a Novolink Polymer Detection kit (Leica) according to the manufacturers’ instructions, the use of which has been described previously28–30. In brief, xylene was used to deparaffinise tissue, followed by rehydration in ethanol then water. Antigen retrieval was achieved in 0.01molL−1 sodium citrate buffer (pH6.0), heated in a microwave for 10 minutes at 750 W followed by 10 minutes at 450 W. Tissue was treated with Novolink Peroxidase Block, washed with Tris-buffered saline (TBS), and then treated with Novolink Protein Block solution. Rabbit polyclonal anti-DARPP-32 (Abcam ab40801) diluted 1:500 was used as primary antibody and was incubated on tissue for one hour at room temperature. Antibody specificity was confirmed by Western blotting on breast cancer cell lysates prior to use. Tissue was washed with TBS prior to the application of Novolink Post Primary solution, which was subsequently washed with TBS and then Novolink Polymer solution was applied. Immunohistochemical reactions were developed using 3,3′ diaminobenzidine as the chromogenic substrate and tissue was counterstained with haematoxylin. Tissue was dehydrated in ethanol and fixed in xylene prior to mounting using DPX. Positive and negative controls were included with each staining run and were comprised of breast tumour composite sections comprising grade 1 and 2 early stage invasive tumour; negative controls had primary antibody omitted from each staining run (Supplementary Information).
Statistical analyses
Slides were scanned using a Nanozoomer Digital Pathology Scanner (Hamamatsu Photonics) and staining was assessed at 200x magnification. Staining in the cytoplasm was assessed using a semi-quantitative immunohistochemical H score, where staining intensity within tumour cells was assessed as none (0), weak (1), medium (2) or strong (3) over the percentage area of each staining intensity. Staining in the nucleus was examined in a semi-quantitative manner, where the percentage of tumour cells that demonstrated any staining intensity was assessed. Greater than 30% of cores for each TMA were double assessed, with both assessors blinded to clinical outcome and each other’s scores. Single measure intraclass correlation coefficients were above 0.7, indicating good concordance between scorers.
Statistical analysis was performed using IBM SPSS Statistics (version 24). Cases were stratified based on breast cancer specific survival for both the discovery and validation cohorts and the METABRIC cohort, and relapse-free survival for the HER2 positive cohort, using X-Tile software31. All differences were deemed statistically significant at the level of P ≤ 0.05. The Pearson χ2 test of association was used to determine the relationship between categorised protein expression and clinicopathological variables. Survival curves were plotted according to the Kaplan-Meier method with significance determined using the log-rank test.
Identification of genes associated with DARPP-32 expression
A supervised artificial neural network was used to further understand the molecular function of PPP1R1B in breast cancer in the METABRIC series. PPP1B1R expression was used as the supervising variable as described by Abdel-Fatah et al.32. The artificial neural network was conducted with a constrained multi-layer perceptron architecture and sigmoidal transfer function, where weights were updated by a feed forward back propagation algorithm. Probes from the METABRIC data were ranked based on their root mean squared (RMS) error for predication of DARPP-32 expression as a continuous variable.
Results
DARPP-32 protein staining location and frequency
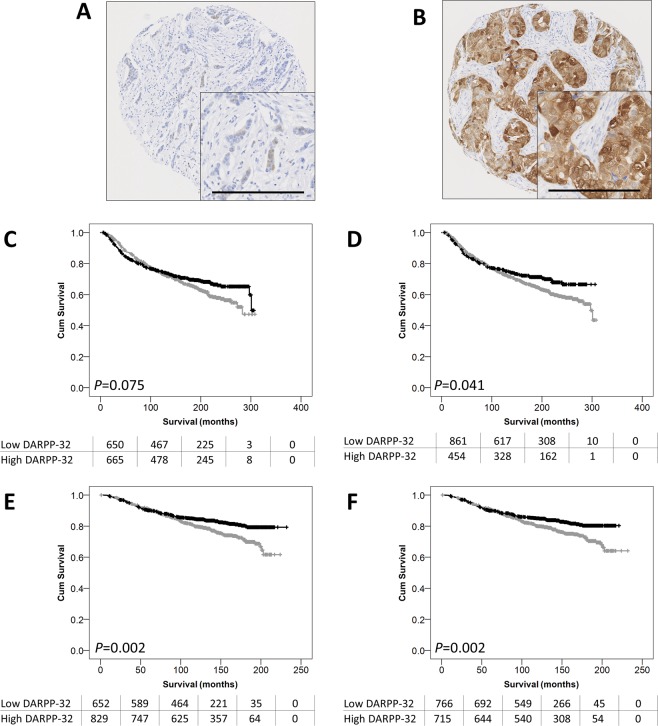
DARPP-32 protein expression was assessed in two large independent cohorts of early invasive breast cancer. Nuclear and cytoplasmic DARPP-32 expression was observed in all cohorts and varied from weak to intense, with heterogeneity observed between adjacent tumour cells. Representative photomicrographs are shown in Fig. 1. In the discovery cohort, 1352 patients were assessed, cytoplasmic DARPP-32 expression had a median H-score of 40, and ranged from 0 to 300. In the validation cohort, 1655 patients were assessed, cytoplasmic DARPP-32 expression had a median H score of 20 and ranged from 0 to 300. Nuclear DARPP-32 expression in the discovery cohort had a median H score of 0 and ranged from 0 to 100; in the validation cohort, the median DARPP-32 H-score was 5 and ranged from 0 to 100. X-tile was used to generate cut points for use in both cohorts based on breast cancer specific survival. In the discovery cohort, cytoplasmic DARPP-32 expression had a cut point of 35, with 49.6% of cases (670/1352) demonstrating low expression; in the validation cohort a cut point of 10 was used, with 43.6% of cases (722/1655) demonstrating low expression. In the discovery cohort, nuclear DARPP-32 expression had a cut point of 20, with 65.6% of cases (887/1352) demonstrating low expression; in the validation cohort, a cut point of 10 was used, with 51.5% of cases (853/1655) demonstrating low expression.
Figure 1.
Representative photomicrographs DARPP-32 staining. Photomicrographs of low DARPP-32 immunohistochemical staining (A), and high staining (B) are shown at 100x magnification with 200x magnification inset box where the scale bar represents 100 µm. Kaplan-Meier analysis of breast cancer specific survival showing the impact of low (grey line) and high (black line) DARPP-32 protein expression within the cytoplasm (C) or the nucleus (D) in the discovery cohort, and within the cytoplasm (E) or the nucleus (F) in the validation cohort. Significance was determined using the log-rank test. The numbers shown below the Kaplan-Meier survival curves are the number of patients at risk at the specified month.
Relationship between DARPP-32 protein expression and clinicopathological variables
In the discovery cohort, low cytoplasmic DARPP-32 expression was significantly associated with ER and PgR positive tumours (χ2 = 25.893, d.f. = 1, P < 0.001 and χ2 = 7.384, d.f. = 1, P = 0.007 respectively) and absence of triple negative disease (χ2 = 10.607, d.f. = 1, P = 0.001) (Table 1). In the validation cohort, low cytoplasmic DARPP-32 expression was significantly associated with larger tumour size (χ2 = 3.917, d.f. = 1, P = 0.048), higher tumour grade (χ2 = 17.517, d.f. = 2, P < 0.001), tubule formation (χ2 = 34.097, d.f. = 2, P < 0.001), pleomorphism (χ2 = 3.917, d.f. = 2, P = 0.016), lower mitosis (χ2 = 6.785, d.f. = 2, P = 0.034), increased tumour stage (χ2 = 8.215, d.f. = 2, P = 0.016), ER and PgR positive tumours (χ2 = 39.000, d.f. = 1, P < 0.001 and χ2 = 13.987, d.f. = 1, P < 0.001 respectively), belonging to a poor NPI prognostic group (χ2 = 12.386, d.f. = 2, P = 0.002) and HER2 status (χ2 = 5.017, d.f. = 1, P = 0.025), absence of triple negative disease (χ2 = 28.075, d.f. = 1, P < 0.001), and the presence of lymphovascular invasion (χ2 = 7.220, d.f. = 1, P = 0.007) (Table 1).
In the discovery cohort, low nuclear DARPP-32 expression was significantly associated with increased pleomorphism (χ2 = 6.943, d.f. = 2, P = 0.031), ER and PgR positive tumours (χ2 = 51.128, d.f. = 1, P < 0.001 and χ2 = 22.736, d.f. = 1, P < 0.001 respectively), HER2 negative tumours (χ2 = 13.790, d.f. = 1, P < 0.001), and the absence of triple negative disease (χ2 = 16.472, d.f. = 1, P < 0.001) (Table 1). In the validation cohort, low nuclear DARPP-32 expression was significantly associated with higher grade tumours (χ2 = 17.859, d.f. = 2, P < 0.001), tubule formation (χ2 = 26.145, d.f. = 2, P < 0.001), lower mitosis (χ2 = 10.070, d.f. = 2, P = 0.007), higher tumour stage (χ2 = 14.358, d.f. = 2, P = 0.001), ER and PgR positive tumours (χ2 = 41.180, d.f. = 1, P < 0.001 and χ2 = 15.031, d.f. = 1, P < 0.001 respectively), belonging to a poor NPI prognostic group (χ2 = 21.111, d.f. = 2, P < 0.001), the absence of triple negative disease (χ2 = 28.738, d.f. = 1, P < 0.001), and the presence of lymphovascular invasion (χ2 = 11.349, d.f. = 1, P = 0.001) (Table 1).
Association between DARPP-32 protein expression and survival
In the discovery cohort, low nuclear expression of DARPP-32 was significantly associated with adverse breast cancer specific survival (P = 0.041) (Fig. 1D). Low nuclear expression of DARPP-32 remained significantly associated with adverse survival (hazard ratio (HR): 0.766; 95% confidence interval (CI): 0.613–0.957; P = 0.019) when potentially confounding factors were included in multivariate assessment (including tumour size, stage, grade, NPI status, vascular invasion status, ER, PgR and HER2 receptor status (all with log-rank statistics of P < 0.001) (Table 2).
In the validation cohort, both low cytoplasmic and low nuclear DARPP-32 expression were significantly associated with survival (both P = 0.002) (Fig. 1E,F). Cytoplasmic expression of DARPP-32 remained significantly associated with adverse survival (HR: 0.744; 95% CI: 0.577–0.960; P = 0.023) when the potentially confounding factors were included in multivariate assessment (including tumour size, stage, grade, NPI status, vascular invasion status ER, PgR and HER2 receptor status (all with log rank statistics of P = 0.001). Nuclear expression of DARPP-32 did not remain significantly associated with survival in the validation cohort (HR: 0.786; 95% CI: 0.608–1.016; P = 0.066).
DARPP-32 expression in ER positive disease
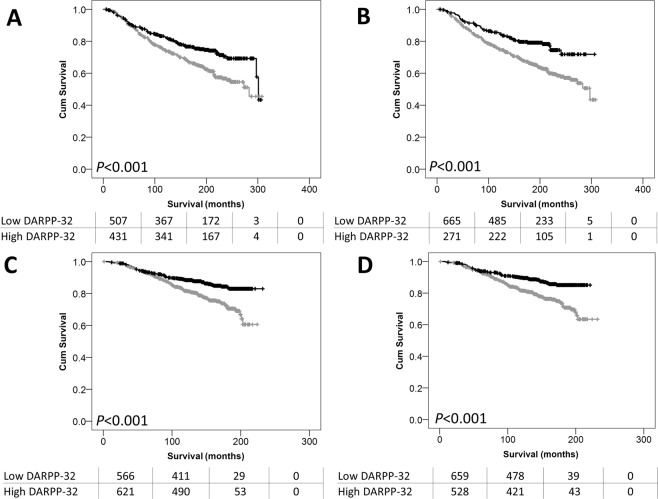
Low expression of DARPP-32 was particularly important in patients with ER positive disease. In the discovery cohort, low DARPP-32 cytoplasmic expression was significantly associated with adverse survival of ER positive patients (P < 0.001), but not ER negative patients (P = 0.099) (Fig. 2A). The same finding was observed when nuclear DARPP-32 expression was assessed, with low expression significantly associated with adverse survival of ER positive patients (P < 0.001), but not ER negative patients (P = 0.407) (Fig. 2B). In the validation cohort, low cytoplasmic expression of DARPP-32 associated with adverse survival of ER positive patients (P < 0.001), but not ER negative patients (P = 0.291) (Fig. 2C). Similar findings were for nuclear DARPP-32 expression were observed; with low expression associated with adverse survival of ER positive patients (P < 0.001), but not ER negative patients (P = 0.679) (Fig. 2D).
Figure 2.
DARPP-32 expression association with patient outcome. Kaplan-Meier analysis of breast cancer specific survival showing the impact of low (grey line) and high (black line) DARPP-32 expression within the cytoplasm in ER positive patients (A) and in the nucleus in ER positive patients (B) in the discovery cohort. DARPP-32 expression within the cytoplasm in ER positive patients (C) and in the nucleus in ER positive patients (D) in the validation cohort. Significance was determined using the log-rank test. The numbers shown below the Kaplan-Meier survival curves are the number of patients at risk at the specified month.
DARPP-32 expression in HER2 positive patients treated with trastuzumab
DARPP-32 was assessed in a cohort of 112 HER2 positive breast cancer patients treated with adjuvant chemotherapy and trastuzumab. A similar DARPP-32 expression pattern was observed to that in early breast cancer. In this cohort, cytoplasmic DARPP-32 had a median H score of 142.9 and ranged from 0 to 300; nuclear DARPP-32 expression had a median H score of 45.36 and ranged from 0 to 100. X-tile was used to generate a cut point for analysis based on relapse free survival; cytoplasmic DARPP-32 expression had a cut point of 20, with 35.7% of cases (40/112) demonstrating low expression, nuclear DARPP-32 expression had a cut point of 5, with 34.8% of cases (39/112) demonstrating low expression.
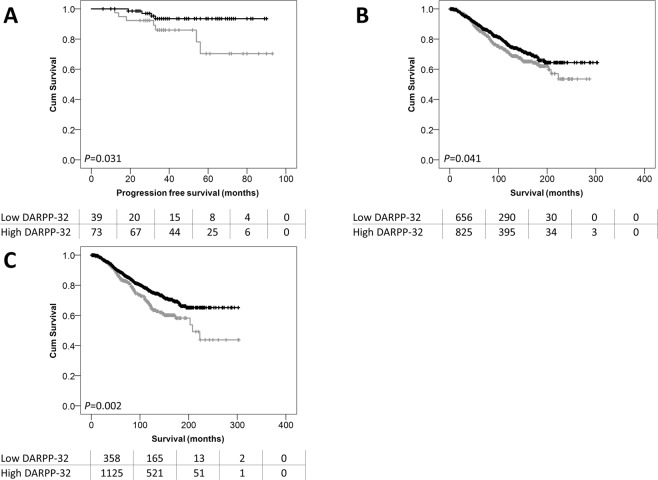
No associations were observed between DARPP-32 expression and clinicopathological criteria in this cohort (Table 2). Low nuclear DARPP-32 expression was significantly associated with adverse progression-free survival of HER2 positive breast cancer patients treated with trastuzumab (P = 0.031) (Fig. 3A); cytoplasmic DARPP-32 expression was not associated with progression-free survival (data not shown). Nuclear DARPP-32 expression did not remain associated with progression-free survival in multivariate Cox regression (HR = 0.387; 95% CI = 0.095–1.570; P = 0.184); when tumour size, lymph node status and NPI category were included (with individual Kaplan-Meier log rank significance of P = 0.012, P = 0.019 and P = 0.008 respectively).
Figure 3.
DARPP-32 expression association with HER2 positive patient outcome. Kaplan-Meier analysis of progression-free survival of HER2 positive breast cancer patients treated with adjuvant chemotherapy and trastuzumab showing the impact of high (black line) and low (grey line) DARPP-32 protein expression within the nucleus (A). Kaplan-Meier analysis of PPP1R1B probe 2 (B) and probe 3 (C) expression in ER positive breast cancer patients showing the impact of low expression (grey line) and high expression (black line) and breast cancer specific survival. Significance was determined using the log-rank test. The numbers shown below the Kaplan-Meier survival curves are the number of patients at risk at the specified month.
PPP1R1B mRNA expression and patient survival
Three PPP1R1B probes were available to assess mRNA expression in the METABRIC cohort and expression was categorised using the median gene expression value to assess for association with patient survival and to perform expression profiling. PPP1R1B probe 1 (ILMN_1690096) is located within a coding area that corresponds to the N-terminal region of DARPP-32; PPP1R1B probe 2 (ILMN_1759012) and PPP1R1B probe 3 (ILMN_2304495) were both located in untranslated regions (5′ and 3′ respectively). Probe 1 and 3 are located in areas found in the sequence for DARPP-32 (NM_032192), and probe 2 and 3 are located in areas found in the sequence for t-DARPP (NM_181505.3). No association was observed between the expression of the probes and disease specific survival. The gene expression data was analysed using an artificial neural network approach that uses a machine learning based data mining algorithm. A rank order of all the genes was produced based on the minimum average root mean squared error. The top 200 transcripts were selected for PPP1R1B probe 1, 2 and 3 and 73 common transcripts were identified (Table 3). The transcripts that were common to all three probes included CDC42, DKK1, GRB7, PNMT, and GPER amongst others.
Table 3.
Genes associated with two or more PPP1R1B probes in the METABRIC dataset identified by artificial neural network analysis. Those highlighted in bold were associated with expression of all three PPP1R1B probes.
| Gene | Illumina ID | Identity | Gene | Illumina ID | Identity |
|---|---|---|---|---|---|
| AFF3 | ILMN_1775235 | AF4/FMR2 family member 3 | NEDD9 | ILMN_1726164 | Cas scaffolding protein family member 2 |
| AMDHD1 | ILMN_1788239 | Amidohydrolase domain containing 1 | OAT | ILMN_1654441 | Ornithine aminotransferase |
| ATP13A5 | ILMN_1775285 | ATPase 13A5 | OAT | ILMN_2068747 | |
| BBOX 1 | ILMN_1734929 | Gamma-butyrobetaine hydrolase 1 | OBP2B | ILMN_1700666 | Odorant binding protein 2B |
| C22orf36 | ILMN_1737255 | Leucine rich repeat containing 75B | ORM2 | ILMN_1731785 | Orosomucoid 2 |
| CDC42 | ILMN_1696041 | Cell division cycle 42 | PAQR6 | ILMN_1689852 | Progestin and adipoQ receptor family member 6 |
| CEACAM1 | ILMN_1716815 | Carcinoembryonic antigen related cell adhesion molecule 1 | PDE4B | ILMN_1782922 | Phosphodiesterase 4B |
| CITED4 | ILMN_1787691 | Cbp/p300 interacting transactivator 4 | PNMT | ILMN_1710027 | Phenylethanolamine N-methyltransferase |
| CPA4 | ILMN_1784294 | Carboxypeptidase A4 | PPP1R1A | ILMN_2056606 | Protein phosphatase 1 regulatory inhibitor subunit 1A |
| CRABP1 | ILMN_1658040 | Cellular reinoic acid binding protein 1 | PROM1 | ILMN_1786720 | Prominin1 |
| CYP4Z1 | ILMN_1693594 | Cytochrome P450 family 4 subfamily Z member 1 | PVRL4 | ILMN_1749044 | Nectin cell adhesion molecule 4 |
| CYP4Z1 | ILMN_1728550 | RGMA | ILMN_1717636 | Repulsive guidance molecule BMP co-receptor A | |
| CYP4Z1 | ILMN_2359698 | RNF183 | ILMN_1692591 | Ring finger protein 183 | |
| CYP4Z2P | ILMN_1702829 | Putative inactive cytochrome P450 family member 4Z2 | S100A1 | ILMN_1653494 | S100 calcium binding protein A1 |
| DCD | ILMN_1722554 | Dercidin | S100A13 | ILMN_1738707 | S100 calcium binding protein A13 |
| DKK1 | ILMN_1773337 | Dickkopf WNT signalling pathway inhibitor 1 | SASH1 | ILMN_1712673 | SAM and SH3 domin containing 1 |
| ENPP3 | ILMN_1749131 | Ectonucleotide pyrophosphatase/phosphodiesterase 3 | SASH1 | ILMN_2185984 | |
| FAIM2 | ILMN_1803855 | Fas apoptotic inhibitory molecule 2 | SCGB2A2 | ILMN_1723333 | Secretoglobulin family 2 A member 2 |
| FAM134B | ILMN_1811330 | Reticulophagy regulator 1 | SLC22A15 | ILMN_1730639 | Solute carrier family 22 member 15 |
| FAM134B | ILMN_2283597 | SLC25A18 | ILMN_1754864 | Solute carrier family 25 member 18 | |
| FAM134B | ILMN_2387952 | SLC5A1 | ILMN_1681526 | Solute carrier family 5 member 1 | |
| FAT | ILMN_1754795 | FAT atypical cadherin 1 | SOX9 | ILMN_1805466 | SRY box 9 |
| FOLR1 | ILMN_2346339 | Folate receptor 1 | SPINK8 | ILMN_1728898 | Serine peptidase inhibitor, Kazal type 8 |
| GGT6 | ILMN_1788942 | Gamma-glutamyltransferase 6 | SPRY1 | ILMN_1691860 | Sprouty RTK signaling antagonist 1 |
| GPER | ILMN_1795298 | G protein-coupled oestrogen receptor 1 | ST6GAL1 | ILMN_1756501 | ST6 beta-galactosidase alpha-2,6-sialytransferase 1 |
| GRAMD2 | ILMN_1661443 | GRAM domain containing 2A | STAC2 | ILMN_1718295 | SH3 and cysteine rich domain 2 |
| GRB7 | ILMN_1740762 | Growth factor receptor bound protein 7 | TFAP2B | ILMN_1758404 | Transcription factor AP-2 beta |
| HOXA5 | ILMN_1753613 | Homeobox A5 | TFAP2B | ILMN_1853592 | |
| HSD17B2 | ILMN_1808713 | Hydroxysteroid 17 beta dehydrogenase 2 | TRPV6 | ILMN_1674533 | Epithelial calcium channel 2 |
| HSH2D | ILMN_1788017 | Hematopoietic SH2 domain containing | TSPAN6 | ILMN_1730998 | Tetraspannin 6 |
| ICAM1 | ILMN_1812226 | Intracellular adhesion molecule 1 | TTLL4 | ILMN_1746846 | Tubulin tyrosine ligase like 4 |
| IGSF9 | ILMN_1693941 | Immunoglobulin superfamily member 9 | UBE2E3 | ILMN_1669553 | Ubiquitin conjugating enzyme E2 E3 |
| KRT7 | ILMN_2163723 | Keratin 7 | UBE2E3 | ILMN_2390338 | |
| LOC340204 | ILMN_1789600 | VTCN1 | ILMN_1753101 | V-set containing T cell activation inhibitor 1 | |
| LOC646424 | ILMN_1661466 | ILMN_1854349_ | |||
| LOC730525 | ILMN_1651610 | ILMN_1889752_ | |||
| MAOB | ILMN_1727360 | Monoamine oxidase B | ILMN_1896906_ | ||
| MAST4 | ILMN_1738438 | Microtubule associated Ser/Thr kinase family member 4 | ILMN_1902123_ | ||
| MPZL2 | ILMN_1752932 | Myelin protein zero like 2 | ILMN_1904054_ |
All three PPP1R1B probes were further assessed in ER positive patients based on the observed DARPP-32 protein findings, with a cut point generated in this subgroup of patients using X-tile. PPP1R1B probe 1 expression was not associated with breast cancer specific survival of ER positive patients; however, low expression of PPP1R1B probe 2 and 3 were both associated with adverse survival of ER positive breast cancer patients (P = 0.041 and P = 0.002 respectively) (Fig. 3B,C).
Discussion
Low nuclear DARPP-32 expression was significantly associated with adverse survival of patients in two independent cohorts of patients treated at Nottingham University Hospitals (discovery cohort n = 1352 and validation cohort n = 1655). Furthermore, low cytoplasmic expression of DARPP-32 was associated with patient survival in the validation cohort. The epitope for the antibody used for immunohistochemistry is located within amino acids 0–30, meaning that only DARPP-32, and not t-DARPP expression was assessed in this study. There are limited reports of DARPP-32 expression and its association with patient survival in cancer. In oesophageal squamous cell carcinoma DARPP-32 is expressed after a phase of dysplasia, and low levels of DARPP-32 are associated with tumours that progress rapidly33. In colorectal cancer, lower expression of DARPP-32 is associated with improved overall survival and disease free survival12, and in non-small cell lung cancer high relative t-DARPP (in comparison to DARPP-32) is associated with tumour stage34. The expression of DARPP-32 in 230 breast cancer patients has been examined previously using N-terminal and C-terminal specific antibodies, to show that high N-terminal DARPP-32 expression is associated with adverse patient survival; this differs from the results presented here21. It is unclear why opposing results were observed; however, patient demographics, including clinicopathological variables, and patient treatment are not available in the previously published study, so cannot be directly compared with the current findings.
Interestingly, low DARPP-32 protein expression was particularly important in ER positive patients, in both cohorts of patients; there was also a strong association between DARPP-32 expression and ER receptor status. In addition to ER status, strong consistent associations were observed between DARPP-32 expression and PgR status and triple receptor negative disease in both patient cohorts.
There is accumulating evidence that DARPP-32, in particular t-DARPP, plays a role in response to trastuzumab; this study shows that low nuclear expression of DARPP-32 is significantly associated with adverse progression free survival in 112 HER2 positive patients treated with trastuzumab and adjuvant chemotherapy. This is agreement with published in vitro data, that indicates expression of DARPP-32 and t-DARPP expression in HER2 positive breast cancer is involved in resistance to trastuzumab22. Expression of t-DARPP has been shown to activate IGF-1R signalling in trastuzumab resistant breast cancer cells through increased glycolytic capacity35. In addition, DARPP-32 mRNA and protein levels have also been shown to fall in HER2 targeted agent, lapatinib, resistant breast cancer cell lines, with a t-DARPP mediated survival advantage observed24.
In models of murine tumourigenesis DARPP-32 expression is expressed in normal mouse tissue and some breast tumours, with t-DARPP expressed only within tumours18. It is interesting to hypothesise that the association between patient survival and low expression of DARPP-32 is observed with a corresponding shift to increased t-DARPP expression; this will be determined in future studies.
PPP1R1B expression was assessed in the METABRIC cohort, where three probes were available for assessment, probe 1 and 3 located in DARPP-32 and probe 2 and 3 located in t-DARPP. Artificial neural network analysis identified a number of genes associated with PPP1R1B expression. Artificial neural network analysis was performed using all three PPP1R1B probes and commonalities within the top 200 genes for each probe identified. Validation of these associations will be performed as part of future studies. Interestingly, CDC42 was identified from this analysis; cdc42 plays a role in filopodia formation and breast cancer cells expressing DARPP-32 have, in a study looking at Wnt-5A activation of DARPP-32, been shown to have lower cdc-42 activity17. In addition to CDC42, Dickkopf-1 (DKK1) was also identified and functions a wnt-5A pathway inhibitor. Furthermore, artificial neural network analysis identified an association between PPP1R1B with both GRB7 and PNMT, a link between PPP1R1B and these genes has already been described in upper gastrointestinal adenocarcinomas where DNA amplification at 17q is often detected (containing PPP1R1B, STARD3, TCAP, PNMT, PERLD1, ERBB2, C17orf37, and GRB7)36.
Conclusion
This study demonstrates that low DARPP-32 protein expression is associated with shorter survival in two large, independent, early stage invasive breast cancer patient cohorts, with a stronger association observed in ER positive disease. This finding was also observed at the mRNA level, with low PPP1R1B expression significantly associated with shorter survival of ER positive patients in the METABRIC cohort. Furthermore, low DARPP-32 expression was associated with shorter progression-free survival of HER2 positive patients treated with trastuzumab. This data suggests a potential role for DARPP-32 as a prognostic marker with clinical utility in breast cancer, requiring validation on samples from multiple institutions.
Compliance with ethical standards
Research involving human participants
Ethical approval for the discovery cohort, validation cohort and the HER 2 cohort was granted by Nottingham Research Ethics Committee 2, under the title ‘Development of a molecular genetic classification of breast cancer’ (C202313). METABRIC samples were collected by five centres in the UK and Canada and were acquired with appropriate consent from the respective institutional review boards27. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All samples collected from Nottingham used in this study were pseudo-anonymised; those collected prior to 2006 did not require informed patient consent under the Human Tissue Act, after 2006 informed consent was obtained from all individual participants included in the study.
Supplementary information
Acknowledgements
We thank the Nottingham Health Science Biobank and Breast Cancer Now for the provision of tissue samples. S.J.S. is funded through the University of Nottingham’s Research Vision as a Nottingham Research Fellow, and previously through the University of Nottingham Life Cycle 6 Campaign.
Author contributions
S.K., M.N.L., B.S. and S.J.S. performed the experiments. G.B. A.R.G. and S.J.S. performed the statistical analysis. P.M.M., S.Y.C., A.R.G., E.R. and I.O.E. provided patient TMAs and matched clinical data. S.G.M. and S.J.S. conceived and designed the study. S.J.S. wrote the manuscript. All authors reviewed the draft manuscript for critical comment.
Data availability
The METABRIC data is publically available https://www.ebi.ac.uk/ega/studies/EGAS00000000098. Immunohistochemistry datasets analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-53529-z.
References
- 1.Walaas SI, Aswad DW, Greengard P. A dopamine- and cyclic AMP-regulated phosphoprotein enriched in dopamine-innervated brain regions. Nature. 1983;301:69–71. doi: 10.1038/301069a0. [DOI] [PubMed] [Google Scholar]
- 2.Hemmings HC, Jr., Nairn AC, Aswad DW. & Greengard, P. DARPP-32, a dopamine- and adenosine 3′:5′-monophosphate-regulated phosphoprotein enriched in dopamine-innervated brain regions. II. Purification and characterization of the phosphoprotein from bovine caudate nucleus. J Neurosci. 1984;4:99–110. doi: 10.1523/JNEUROSCI.04-01-00099.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ouimet CC, Miller PE, Hemmings HC, Jr., Walaas SI, Greengard P. DARPP-32, a dopamine- and adenosine 3′:5′-monophosphate-regulated phosphoprotein enriched in dopamine-innervated brain regions. III. Immunocytochemical localization. J Neurosci. 1984;4:111–124. doi: 10.1523/JNEUROSCI.04-01-00111.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walaas SI, Greengard P. DARPP-32, a dopamine- and adenosine 3′:5′-monophosphate-regulated phosphoprotein enriched in dopamine-innervated brain regions. I. Regional and cellular distribution in the rat brain. J Neurosci. 1984;4:84–98. doi: 10.1523/JNEUROSCI.04-01-00084.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hemmings HC, Jr., Greengard P, Tung HY, Cohen P. DARPP-32, a dopamine-regulated neuronal phosphoprotein, is a potent inhibitor of protein phosphatase-1. Nature. 1984;310:503–505. doi: 10.1038/310503a0. [DOI] [PubMed] [Google Scholar]
- 6.King MM, et al. Mammalian brain phosphoproteins as substrates for calcineurin. J Biol Chem. 1984;259:8080–8083. [PubMed] [Google Scholar]
- 7.Engmann O, et al. DARPP-32 interaction with adducin may mediate rapid environmental effects on striatal neurons. Nat Commun. 2015;6:10099. doi: 10.1038/ncomms10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Rifai W, et al. Gastric cancers overexpress DARPP-32 and a novel isoform, t-DARPP. Cancer Res. 2002;62:4061–4064. [PubMed] [Google Scholar]
- 9.Chang MJ, et al. Feedback regulation mediated by Bcl-2 and DARPP-32 regulates inositol 1,4,5-trisphosphate receptor phosphorylation and promotes cell survival. Proc Natl Acad Sci USA. 2014;111:1186–1191. doi: 10.1073/pnas.1323098111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Svenningsson P, Nairn AC, Greengard P. DARPP-32 mediates the actions of multiple drugs of abuse. Aaps J. 2005;7:E353–360. doi: 10.1208/aapsj070235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beckler A, et al. Overexpression of the 32-kilodalton dopamine and cyclic adenosine 3′,5′-monophosphate-regulated phosphoprotein in common adenocarcinomas. Cancer. 2003;98:1547–1551. doi: 10.1002/cncr.11654. [DOI] [PubMed] [Google Scholar]
- 12.Kopljar M, et al. High Expression of DARPP-32 in Colorectal Cancer Is Associated With Liver Metastases and Predicts Survival for Dukes A and B Patients: Results of a Pilot Study. Int Surg. 2015;100:213–220. doi: 10.9738/INTSURG-D-14-00022.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Televantou D, et al. DARPP32, STAT5 and STAT3 mRNA expression ratios in glioblastomas are associated with patient outcome. Pathol Oncol Res. 2013;19:329–343. doi: 10.1007/s12253-012-9588-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu S, et al. Regulation of CXCR4-mediated invasion by DARPP-32 in gastric cancer cells. Mol Cancer Res. 2013;11:86–94. doi: 10.1158/1541-7786.MCR-12-0243-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yun SM, et al. PPP1R1B-STARD3 chimeric fusion transcript in human gastric cancer promotes tumorigenesis through activation of PI3K/AKT signaling. Oncogene. 2014;33:5341–5347. doi: 10.1038/onc.2013.472. [DOI] [PubMed] [Google Scholar]
- 16.Hansen C, Greengard P, Nairn AC, Andersson T, Vogel WF. Phosphorylation of DARPP-32 regulates breast cancer cell migration downstream of the receptor tyrosine kinase DDR1. Exp Cell Res. 2006;312:4011–4018. doi: 10.1016/j.yexcr.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 17.Hansen C, et al. Wnt-5a-induced phosphorylation of DARPP-32 inhibits breast cancer cell migration in a CREB-dependent manner. J Biol Chem. 2009;284:27533–27543. doi: 10.1074/jbc.M109.048884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christenson JL, Kane SE. Darpp-32 and t-Darpp are differentially expressed in normal and malignant mouse mammary tissue. Mol Cancer. 2014;13:192. doi: 10.1186/1476-4598-13-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vangamudi B, et al. t-DARPP regulates phosphatidylinositol-3-kinase-dependent cell growth in breast cancer. Mol Cancer. 2010;9:240. doi: 10.1186/1476-4598-9-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belkhiri A, et al. Expression of t-DARPP mediates trastuzumab resistance in breast cancer cells. Clin Cancer Res. 2008;14:4564–4571. doi: 10.1158/1078-0432.CCR-08-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamel S, et al. Both t-Darpp and DARPP-32 can cause resistance to trastuzumab in breast cancer cells and are frequently expressed in primary breast cancers. Breast Cancer Res Treat. 2010;120:47–57. doi: 10.1007/s10549-009-0364-7. [DOI] [PubMed] [Google Scholar]
- 22.Gu L, Waliany S, Kane SE. Darpp-32 and its truncated variant t-Darpp have antagonistic effects on breast cancer cell growth and herceptin resistance. PLoS One. 2009;4:e6220. doi: 10.1371/journal.pone.0006220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theile D, Geng S, Denny EC, Momand J, Kane SE. t-Darpp stimulates protein kinase A activity by forming a complex with its RI regulatory subunit. Cell Signal. 2017;40:53–61. doi: 10.1016/j.cellsig.2017.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christenson JL, Denny EC, Kane SE. t-Darpp overexpression in HER2-positive breast cancer confers a survival advantage in lapatinib. Oncotarget. 2015;6:33134–33145. doi: 10.18632/oncotarget.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avanes A, Lenz G, Momand J. Darpp-32 and t-Darpp protein products of PPP1R1B: Old dogs with new tricks. Biochem Pharmacol. 2019;160:71–79. doi: 10.1016/j.bcp.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McShane LM, et al. REporting recommendations for tumour MARKer prognostic studies (REMARK) Br J Cancer. 2005;93:387–391. doi: 10.1038/sj.bjc.6602678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curtis JP, et al. The pre-hospital electrocardiogram and time to reperfusion in patients with acute myocardial infarction, 2000-2002: findings from the National Registry of Myocardial Infarction-4. J Am Coll Cardiol. 2006;47:1544–1552. doi: 10.1016/j.jacc.2005.10.077. [DOI] [PubMed] [Google Scholar]
- 28.Davis J, et al. Low calpain-9 is associated with adverse disease-specific survival following endocrine therapy in breast cancer. BMC Cancer. 2014;14:995. doi: 10.1186/1471-2407-14-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storr SJ, et al. The calpain system is associated with survival of breast cancer patients with large but operable inflammatory and non-inflammatory tumours treated with neoadjuvant chemotherapy. Oncotarget. 2016;7:47927–47937. doi: 10.18632/oncotarget.10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin SG, et al. Low expression of G protein-coupled oestrogen receptor 1 (GPER) is associated with adverse survival of breast cancer patients. Oncotarget. 2018;9:25946–25956. doi: 10.18632/oncotarget.25408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 32.Abdel-Fatah TMA, et al. SPAG5 as a prognostic biomarker and chemotherapy sensitivity predictor in breast cancer: a retrospective, integrated genomic, transcriptomic, and protein analysis. Lancet Oncol. 2016;17:1004–1018. doi: 10.1016/S1470-2045(16)00174-1. [DOI] [PubMed] [Google Scholar]
- 33.Ebihara Y, et al. DARPP-32 expression arises after a phase of dysplasia in oesophageal squamous cell carcinoma. Br J Cancer. 2004;91:119–123. doi: 10.1038/sj.bjc.6601899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alam, S. K. et al. DARPP-32 and t-DARPP promote non-small cell lung cancer growth through regulation of IKKalpha-dependent cell migration. Commun Biol1, 10.1038/s42003-018-0050-6 (2018). [DOI] [PMC free article] [PubMed]
- 35.Lenz G, et al. t-Darpp Activates IGF-1R Signaling to Regulate Glucose Metabolism in Trastuzumab-Resistant Breast Cancer Cells. Clin Cancer Res. 2018;24:1216–1226. doi: 10.1158/1078-0432.CCR-17-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maqani N, et al. Molecular dissection of 17q12 amplicon in upper gastrointestinal adenocarcinomas. Mol Cancer Res. 2006;4:449–455. doi: 10.1158/1541-7786.MCR-06-0058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The METABRIC data is publically available https://www.ebi.ac.uk/ega/studies/EGAS00000000098. Immunohistochemistry datasets analysed during the current study are available from the corresponding author on reasonable request.