Abstract
CG-NAP, also known as AKAP450, is an anchoring/adaptor protein that streamlines signal transduction in various cell types by localizing signaling proteins and enzymes with their substrates. Great efforts are being devoted to elucidating functional roles of this protein and associated macromolecular signaling complex. Increasing understanding of pathways involved in regulating T lymphocytes suggests that CG-NAP can facilitate dynamic interactions between kinases and their substrates and thus fine-tune T cell motility and effector functions. As a result, new binding partners of CG-NAP are continually being uncovered. Here, we review recent advances in CG-NAP research, focusing on its interactions with kinases in T cells with an emphasis on the possible role of this anchoring protein as a target for therapeutic intervention in immune-mediated diseases.
Keywords: adaptor protein, kinases, CG-NAP, AKAP450, T cell motility, immune synapse
Introduction
T lymphocytes play a central role in immune defense by mounting specific responses to eliminate infections and transformed cells. To perform an immunosurveillance function, T cells continuously circulate throughout the body until they encounter specific antigen on the surface of the antigen presenting cell (APC, Table 1). Such contact with an APC triggers an initial activation of the T cell, which rapidly reorients its organelles and mobilizes signaling proteins to the contact site. This process is accompanied by dynamic structural and cytoskeletal changes within the T cell. An activated T cell undergoes an episode of rapid proliferation, cytokine secretion, differentiation into effector subtypes, and site-specific recruitment. These functional processes in T lymphocytes are precisely regulated and are critical for mounting an effective immune response.
Table 1.
A list of abbreviations used.
| Abbreviations used | |
|---|---|
| AKAP | A-kinase anchoring protein |
| APC | Antigen presenting cell |
| cAMP | Cyclic adenosine monophosphate |
| CAMSAP2 | Calmodulin regulated spectrin associated protein family member 2 |
| CAMSAP3 | Calmodulin regulated spectrin associated protein family member 3 |
| CBFA2T3 | CBFA2/RUNX1 partner transcriptional co-repressor 3 |
| Cdk | Cyclin dependent kinase |
| CG-NAP | Centrosome- and Golgi-localized protein kinase N-associated protein |
| CHO | Chinese hamster ovary |
| CK1δ/ε | Casein kinase 1 delta/epsilon |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| EB1 | End-binding protein 1 |
| GM130 | 130 KDa cis-Golgi matrix protein |
| GTP | Guanosine triphosphate |
| HIV | Human immunodeficiency viruses |
| ICAM-1 | Intercellular adhesion molecule 1 |
| IL | Interleukin |
| IS | Immune synapse |
| JC virus | John Cunningham virus |
| KCNE1 | Potassium voltage-gated channel subfamily E regulatory subunit 1 |
| Kiz | Kizuna |
| LAT | Linker for activation of T cells |
| LFA-1 | Lymphocyte function-associated antigen 1 |
| MAPRE1 | Microtubule associated protein RP/EB family member 1 |
| MMG8 | Myomegalin variant 8 |
| MTCL1 | Microtubule cross-linking factor 1 |
| MTOC | Microtubule organizing center |
| NFAT | Nuclear factor of activated T cells |
| NFκB | Nuclear factor kappa B |
| PACT | Pericentrin-AKAP450 centrosomal targeting |
| PDE4D | Phosphodiesterase 4D |
| PKA | Protein kinase A |
| PKC | Protein kinase C |
| PKN | Protein kinase N |
| PLCγ1 | Phospholipase C gamma 1 |
| Plk1 | Polo-like kinase 1 |
| PP1 | Protein phosphatase 1 |
| PP2A | Protein phosphatase 2A |
| PR130 | Serine/Threonine-protein phosphatase 2a 72/130 KDa regulatory subunit B |
| RAPGEF2 | Rap guanine nucleotide exchange factor 2 |
| RUNX1T1 | RUNX1 partner transcriptional co-repressor 1 |
| siRNAs | Small interfering RNA |
| TCR | T cell receptor |
| TGFβ | Transforming growth factor beta |
| TUBGCP | Tubulin gamma complex associated protein |
| γ-TuRC | Gamma tubulin ring complex |
Multiple stages of T cell functions, such as activation, differentiation, conjugate formation with APCs, homing and motility are crucially regulated by protein kinases (1, 2). As members of the kinase superfamily are widely distributed within cells and often have broad substrate specificity, a crucial element in signal transduction is local control of substrate specificity (3, 4). How is an individual kinase directed to connect with a single substrate or multiple components of a pool of downstream substrates? In some cases, kinase specificity is achieved by influencing substrate recognition. In this context, a class of proteins collectively known as “adaptor, anchoring and scaffolding proteins,” have emerged as important mediators of signal transduction processes (5, 6). These proteins form specialized docking platforms that facilitate the formation of multicomponent signaling complexes, maintain static protein-protein interactions, position their kinase cargo in proximity to a subset of substrates, organize processes and components of protein kinase cascades and thus streamline cell signaling responses (6–13). In T lymphocytes, these signal-organizing proteins allow signals to be transduced with precision in response to molecular instructions from the cell surface (14–16). Most importantly, these anchoring/adaptor proteins facilitate the phosphorylation and dephosphorylation of protein kinases, including trans- and auto-phosphorylations, which are important for the kinases to gain catalytically competent conformation in order to respond to intra- and/or extra-cellular signals (16, 17). Anchoring/adaptor proteins thus control numerous cellular processes in T lymphocytes, including cell fate decisions, activation, differentiation and various stages of development and functions (16–19). Herein, we review the involvement of such an anchoring protein “Centrosome and Golgi localized protein kinase N (PKN)-Associated Protein” (CG-NAP), also known as A-Kinase Anchoring Protein 450 (AKAP450) (20–22), in the regulation of protein kinase dynamics and functional outcomes in T cells.
A-Kinase Anchoring Proteins (AKAPs)
AKAPs are a family of ubiquitously expressed structurally diverse signal-organizing proteins with tissue/cell-type specific expression patterns in human. So far, 41 AKAPs encoded by 41 genes have been experimentally validated in human cells and tissues (8, 23) (Table 2). Nine different AKAPs have been described in human T lymphocytes – AKAP1, AKAP2, AKAP5, AKAP8, AKAP9 (known as CG-NAP), AKAP11, Ezrin, RUNX1T1, and RUNX1T3 (24–26) and at least eight AKAPs with apparent molecular masses of 60, 75, 95, 120, 165, 190, 245, and 275 kDa were detected in mouse T lymphocytes (27); however, their involvements in the regulation of immune functions remain poorly understood.
Table 2.
A list of AKAP family proteins.
| S.N. | AKAPs | Name | Synonyms | HGNC ID (gene) | Chromosome |
|---|---|---|---|---|---|
| 1. | AKAP1 | A-kinase anchoring protein 1 | PRKA1, AKAP121, AKAP149, SAKAP84, S-AKAP84, AKAP84, D-AKAP1, PPP1R43, TDRD17 | HGNC:367 | 17q22 |
| 2. | AKAP2 | A-kinase anchoring protein 2 | PRKA2, AKAP-KL, KIAA0920, DKFZp564L0716, MISP2 | HGNC:372 | 9q31.3 |
| 3. | AKAP3 | A-kinase anchoring protein 3 | FSP95, SOB1, AKAP110, CT82 | HGNC:373 | 12p13.32 |
| 4. | AKAP4 | A-kinase anchoring protein 4 | p82, hAKAP82, AKAP82, Fsc1, HI, CT99 | HGNC:374 | Xp11.22 |
| 5. | AKAP5 | A-kinase anchoring protein 5 | AKAP75, AKAP79 | HGNC:375 | 14q23.3 |
| 6. | AKAP6 | A-kinase anchoring protein 6 | KIAA0311, mAKAP, AKAP100, PRKA6, ADAP6 | HGNC:376 | 14q12 |
| 7. | AKAP7 | A-kinase anchoring protein 7 | AKAP18, AKAP15 | HGNC:377 | 6q23.2 |
| 8. | AKAP8 | A-kinase anchoring protein 8 | AKAP95, DKFZp586B1222 | HGNC:378 | 19p13.12 |
| 9. | AKAP9 | A-kinase anchoring protein 9 | KIAA0803, AKAP350, AKAP450, CG-NAP, YOTIAO, HYPERION, PRKA9, MU-RMS-40.16A, PPP1R45, LQT11 | HGNC:379 | 7q21.2 |
| 10. | AKAP10 | A-kinase anchoring protein 10 | D-AKAP2, PRKA10, MGC9414 | HGNC:368 | 17p11.2 |
| 11. | AKAP11 | A-kinase anchoring protein 11 | KIAA0629, AKAP220, PRKA11, FLJ11304, DKFZp781I12161, PPP1R44 | HGNC:369 | 13q14.11 |
| 12. | AKAP12 | A-kinase anchoring protein 12 | AKAP250, SSeCKS, gravin | HGNC:370 | 6q25.1 |
| 13. | AKAP13 | A-kinase anchoring protein 13 | LBC, Ht31, BRX, AKAP-Lbc, c-lbc, PROTO-LB, HA-3, ARHGEF13 | HGNC:371 | 15q25.3 |
| 14. | AKAP14 | A-kinase anchoring protein 14 | AKAP28 | HGNC:24061 | Xq24 |
| 15. | AKAP17A | A-kinase anchoring protein 17A | CXYorf3, SFRS17A, XE7, XE7Y, DXYS155E, MGC39904, 721P, CCDC133 | HGNC:18783 | Xp22.33 and Yp11.32 |
| 16. | AKAP17BP | A-kinase anchoring protein 17B, pseudogene | AKAP16B, AKAP16BP | HGNC:38514 | Xq24 |
| 17. | ACBD3 | Acyl-CoA binding domain containing 3 | GOLPH1, GOCAP1, GCP60, PAP7 | HGNC:15453 | 1q42.12 |
| 18. | ARFGEF2 | ADP ribosylation factor guanine nucleotide exchange factor 2 | BIG2 | HGNC:15853 | 20q13.13 |
| 19. | CHD8 | Chromodomain Helicase DNA Binding Protein 8 | HELSNF1, Helicase with SNF2 Domain 1, AUTS18, Duplin, KIAA1564 | HGNC:20153 | 14q11.2 |
| 20. | CMYA5 | Cardiomyopathy associated 5 | C5orf10, SPRYD2, DKFZp451G223, TRIM76 | HGNC:14305 | 5q14.1 |
| 21. | C2orf88 | Chromosome 2 open reading frame 88 | MGC13057, smAKAP | HGNC:28191 | 2q32.2 |
| 22. | EZR | Ezrin | VIL2, Villin 2, P81, Cytovillin | HGNC:12691 | 6q25.3 |
| 23. | GSKIP | GSK3B Interacting Protein | C14orf129 | HGNC:20343 | 14q32.2 |
| 24. | ITGA4 | α4 integrin | CD49D | HGNC: 6140 | 2q31.3 |
| 25. | MAP2 | Microtubule associated protein 2 | MAP2A, MAP2B, MAP2C | HGNC:6839 | 2q34 |
| 26. | MSN | Moesin | IMD50, HEL70, Membrane-Organizing Extension Spike Protein | HGNC: 7373 | Xq12 |
| 27. | MYO7A | Myosin VIIA | USH1B, DFNB2, DFNA11, SRD2 | HGNC:7606 | 11q13.5 |
| 28. | MYRIP | Myosin VIIA and Rab interacting protein | DKFZp586F1018, exophilin-8, MyRIP, SLAC2-C, SLAC2C | HGNC:19156 | 3p22.1 |
| 29. | NBEA | Neurobeachin | KIAA1544, BCL8B, FLJ10197, LYST2 | HGNC:7648 | 13q13.3 |
| 30. | NF2 | Neurofibromin 2 | merlin, ACN, SCH, BANF | HGNC:7773 | 22q12.2 |
| 31. | OPA1 | Optic Atrophy Protein 1 | MyRIP, Optic Atrophy Protein 1, OPA1 Mitochondrial Dynamin Like GTPase, NTG, NPG, BERHS, LargeG, MTDPS14, KIAA0567, Dynamin-Like Guanosine Triphosphatase | HGNC: 8140 | 3q29 |
| 32. | PDE4DIP | Phosphodiesterase 4D Interacting Protein | MMGL, CMYA2, Myomegalin, Cardiomyopathy-Associated Protein | HGNC:15580 | 1q21.2 |
| 33. | PIK3CG | Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Gamma | P110γ, PI3Kγ, PI3CG, P120-PI3K | HGNC:8978 | 7q22.3 |
| 34. | RAB32 | Ras-Related Protein Rab-32 | Rab32 | HGNC:9772 | 6q24.3 |
| 35. | RSPH3 | Radial Spoke Head 3 | RSP3, RSHL2 | HGNC:21054 | 6q25.3 |
| 36. | RUNX1T1 | RUNX1 Partner Transcriptional Co-Repressor 1 | AML1T1, CBFA2T1, MTG8 | HGNC:1535 | 8q21.3 |
| 37. | RUNX1T3 | RUNX1 Partner Transcriptional Co-Repressor 3 | CBFA2T3, ETO2, HMTG16, MTG8-Related Protein 2, MTG16, MTGR2, ZMYND4 | HGNC:1537 | 16q24.3 |
| 38. | SPHKAP | SPHK1 interactor, AKAP domain containing | SKIP | HGNC:30619 | 2q36.3 |
| 39. | SYNM | Synemin | DMN, KIAA0353, SYN | HGNC:24466 | 15q26.3 |
| 40. | TNNT2 | Troponin T2 | c-troponinT, TnTC, CMPD2, LVNC6, CMD1D, CMH2 | HGNC:11949 | 1q32.1 |
| 41. | WASF1 | WAS protein family member 1 | WAVE1, SCAR1, KIAA0269, WAVE | HGNC:12732 | 6q21 |
Although members of the AKAP family differ greatly in their amino-acid sequences, structures, intracellular localizations and repertoire of protein binding partners, they all interact directly with the regulatory subunits of the protein kinase A (PKA) (28–33). However, the mechanism by which molecular interactions between specific AKAPs and PKA regulate normal and pathological signaling in human cells/tissues is just beginning to be understood.
AKAPs, through association with PKA, are involved in regulating T cell functions through the ubiquitous second messenger molecule cAMP (34–36), which controls cellular processes dictated by cell surface receptor-induced signaling (37–39). The interactions between AKAPs and PKA are complex as there are four distinct regulatory subunit isoforms of PKA – RIα, RIβ, RIIα, and RIIβ (7). These subunits differ in tissue distribution, cAMP sensitivity and AKAP-mediated localization, which fine-tune molecular signals depending on when and where PKA activity is applied (40). Most AKAPs bind to the RII isoform and a few dual-specific AKAPs can also interact with the RI isoform (33, 41). In addition, there are recent examples of RI selective AKAPs (42–44). Furthermore, most cell types simultaneously express multiple AKAPs (e.g., human T cells express at least 9 different AKAPs) (24–26).
It should be noted that the PKA-binding module of AKAPs denotes only one facet of their regulatory control. Apart from their interactions with PKA, AKAPs also interact with other downstream proteins and signaling enzymes, including protein kinase C (PKC) isoforms and PKN, protein phosphatases, phosphodiesterases, small GTPases (8) and substrates to integrate a diverse range of signals within distinct multivalent assemblies. The spatiotemporal interactions between enzymes and target substrates are important in the regulation of T cell functions as well as in the maintenance of T cell homeostasis (27).
CG-NAP: A Giant Member of the AKAP Protein Family
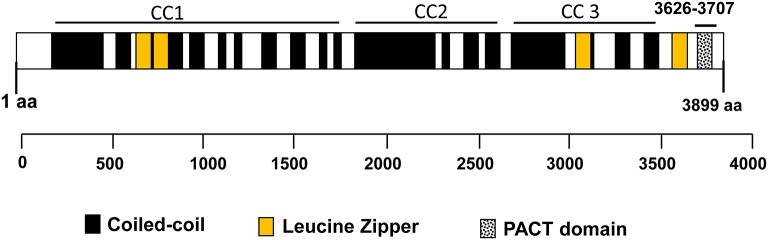
CG-NAP is a member of the AKAP family, prominently expressed in human T cells, in which this giant protein predominantly localizes to the centrosome (20). The human CG-NAP gene is located on the chromosome 7q21-22 and contains at least 50 exons (45–47). A total of 16 splice variants have been identified in the CG-NAP gene (Table 3). The cDNA derived from the CG-NAP gene contains 11.7 kb open reading frame coding the 3899 amino acid protein with a calculated molecular mass of 451.8 kDa (45). The CG-NAP protein has several stretches of coiled-coil structures and four leucine zipper-like motifs (Figure 1) and these structural motifs are involved in interactions with other signaling proteins (e.g., PKA, PKN and PKC isoforms) (45). Amino acid sequence comparison using BLAST analysis shows that regions of human CG-NAP share high homology with the rabbit AKAP120 and limited homology to the mouse pericentrin (48–50).
Table 3.
A list of 16 splice variants (transcripts) of the CG-NAP gene in human.
| S.N. | Name | Transcript ID | bp | Protein | Biotype | UniProt |
|---|---|---|---|---|---|---|
| 1. | AKAP9-201 | ENST00000356239.7 | 12471 | 3907aa | Protein coding | Q99996 |
| 2. | AKAP9-203 | ENST00000359028.6 | 12247 | 3910aa | Protein coding | A0A0A0MRF6 |
| 3. | AKAP9-202 | ENST00000358100.6 | 10309 | 3126aa | Protein coding | A0A0A0MRE9 |
| 4. | AKAP9-216 | ENST00000619023.4 | 6006 | 1637aa | Protein coding | A0A087WX84 |
| 5. | AKAP9-204 | ENST00000394534.6 | 5652 | 1769aa | Protein coding | H7BYL6 |
| 6. | AKAP9-206 | ENST00000435423.1 | 1904 | 318aa | Protein coding | H0Y6Q0 |
| 7. | AKAP9-213 | ENST00000491695.1 | 1389 | 282aa | Protein coding | A0A2R8Y590 |
| 8. | AKAP9-205 | ENST00000394564.5 | 1356 | 314aa | Protein coding | Q6PJH3 |
| 9. | AKAP9-207 | ENST00000438114.1 | 696 | 232aa | Protein coding | H7BZV6 |
| 10. | AKAP9-214 | ENST00000493453.1 | 6020 | No protein | Retained intron | – |
| 11. | AKAP9-211 | ENST00000487258.5 | 3983 | No protein | Retained intron | – |
| 12. | AKAP9-208 | ENST00000463118.1 | 891 | No protein | Retained intron | – |
| 13. | AKAP9-212 | ENST00000487692.1 | 869 | No protein | Retained intron | – |
| 14. | AKAP9-215 | ENST00000493976.1 | 724 | No protein | Retained intron | – |
| 15. | AKAP9-209 | ENST00000484815.1 | 671 | No protein | Retained intron | – |
| 16. | AKAP9-210 | ENST00000486313.1 | 584 | No protein | lncRNA | – |
The list was prepared from the Ensembl database using the HGNC ID HGNC:379.
Figure 1.
An illustration of the domain organization of the CG-NAP. Modular structures of coiled-coil (CC1, CC2, and CC3), 4 Leucine Zippers and the PACT domain are shown schematically.
CG-NAP/Protein Kinase Interactions in T Cells
The modular architecture of CG-NAP brings many protein kinases and their substrates in proximity within a cell and thus regulates the rate and magnitude of cytoplasmic catalysis. Since its initial characterization in 1999 (45) and the establishment of its role in regulating intracellular membrane trafficking and cell cycle progression (51), several interacting partners of CG-NAP have been identified in various cell types, including T lymphocytes (Figure 2).
Figure 2.
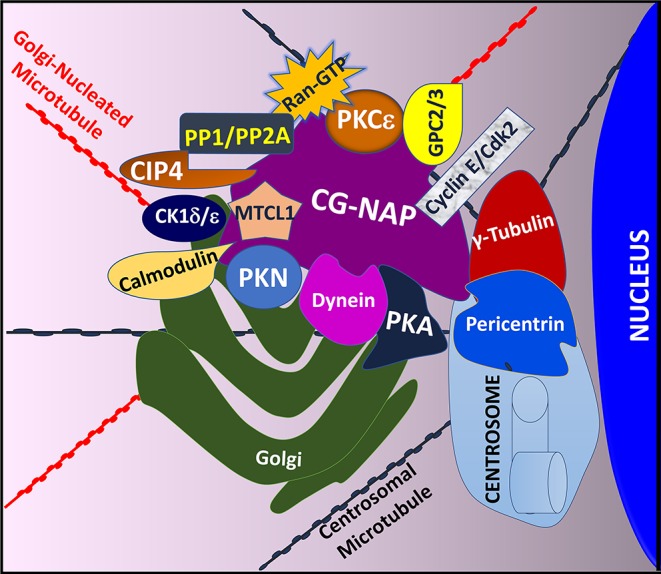
An illustration showing T cell CG-NAP as an anchoring protein that provides a docking platform for several kinases and their substrates. In particular, the Golgi-localized CG-NAP interacts with PKA, PKN, PKC isoforms, dynein, pericentrin, γ-tubulin, MTCL1, PP1, PP2A, CK1, calmodulin, CIP4, cyclin E, and Cdk2 and regulates T cell functions, including activation, proliferation, IS formation and migration.
Previous studies using co-immunoprecipitation approaches and deletion mutants to identify CG-NAP interacting partners revealed that CG-NAP functions as an anchoring molecular platform for protein kinases, including PKA (45). Using a yeast two-hybrid screening system, it has been demonstrated that CG-NAP interacts with the N-terminus of PKN (45). In addition, CG-NAP associates with the catalytic subunit of protein phosphatase 1 (PP1) (45), protein phosphatase 2A (PP2A) through its regulatory B subunit PR130 (45), casein kinase 1 delta and epsilon (CK1δ/ε) (52, 53), PKC isozymes (PKCβ, PKCδ, PKCε, PKCθ) (21, 54), calmodulin (55), the γ-tubulin ring complex (γ-TuRC) comprising of γ-tubulin, tubulin gamma complex associated proteins 2, 3, 4, 5 and 6 (TUBGCP2, TUBGCP3, TUBGCP4, TUBGCP5, and TUBGCP6) (55, 56), dynein/dynactin (57), Cdc42-interacting protein 4 (CIP4) (58), Ran (59), phosphodiesterase 4D (PDE4D) (45, 60), cyclin E/cyclin-dependent kinase (Cdk) 2 (61) and Golgin A2/GM130 (62) in various cell types (Figure 2). In cultured epithelial cells, CG-NAP forms a pericentrosomal complex with the EB1-binding-myomegalin protein complex and recruits calmodulin regulated spectrin-associated protein family member 2 and 3 (CAMSAP2 and CAMSAP3) and microtubule-associated protein RP/EB family member 1 (MAPRE1) to the Golgi (55, 63–66). Studies using various cell-types, including human T lymphocytes, further elucidate a role of CG-NAP in microtubule nucleation (20, 56, 67). These binding interactions with multiple proteins suggest a dynamic complexity in the active functions of CG-NAP.
In the context of T lymphocyte functions, crucial roles for CG-NAP have been demonstrated as (i) a component of the LFA-1-induced signaling complex, (ii) a mediator of T cell/APC immune synapse (IS) formation, (iii) an organizer of centrosomal re-localization, and iv) a facilitator of cytoskeletal rearrangement and motility (20–22). The LFA-1 β2 integrin plays multiple roles in the functioning of T lymphocytes, including migration to sites of inflammation/infection, proper functioning of the IS and functional programming for effector differentiation. In consequence, LFA-1-induced signals are critical in the pathogenesis of inflammatory diseases, such as psoriasis (68), and infectious diseases, including human immunodeficiency viruses (HIV) (69–71), Hepatitis C (72, 73) and John Cunningham (JC) virus (74). An active involvement of CG-NAP in mediating LFA-1 signaling suggests its potential implications for the above and other T cell-dependent diseases.
In prior studies, we have demonstrated that CG-NAP is an important component of the LFA-1 signaling complex for T lymphocyte migration (20, 21). These studies established that CG-NAP is expressed in T lymphocytes at the centrosome at rest and distributed both at the centrosome and along the trailing microtubules during migration (20, 21). CG-NAP co-immunoprecipitates with LFA-1 in activated migrating T cells (21). PKC isoforms, including PKCβ and PKCδ, also interact with CG-NAP in motile T cells (21). Hence, we concluded that the migratory signals in T lymphocytes induce the assembly of a multi-molecular protein complex involving CG-NAP, which serves as one of the docking platforms for PKCβ and PKCδ isoforms (21). PKCβ regulates LFA-1-mediated locomotion of activated T cells (75); whereas, PKCδ plays a critical role in TCR-induced negative regulation of IL-2 cytokine production and T cell proliferation (76).
Studies using cultured fibroblast cells demonstrated a direct association between CG-NAP and immature non- or hypo-phosphorylated PKCε at the Golgi and around the centrosome (54). Depending on environmental cues and upon phosphorylation, PKCε dissociates from the CG-NAP complex as a “mature” enzyme, which actively responds to second messenger signals (54). In human T cells, this PKC isoform regulates a diverse range of biological functions. In particular, PKCε modulates the TCR-associated signaling complex for T cell activation and cytokine secretion (77, 78), proliferation (79), sensitivity to TGFβ (79), development (80), gene expression (80), and survival (81, 82). PKCε directly activates the NFκB/NFAT/AP1 pathway in T cells leading to the up-regulation of IL-2 receptor expression and an increase in IL-2 production (83, 84). The LFA-1 signal for T cell migration activates PKCε, which phosphorylates the Rab GTPase Rab5a on Thr7, triggering a molecular cascade leading to the activation of the Rac1 protein and actin cytoskeletal rearrangements in motile T cells (85). Further studies are required to determine whether CG-NAP plays a role in the dynamic coordination of PKCε activities in human T cells.
CG-NAP/Kinase Interactions in T Cells at the Immune Synapse (IS)
Upon recognition of specific antigen on APCs and TCR engagement, a T cell undergoes a series of structural and molecular changes to form a flattened contact site, termed the “IS” (86). Within few seconds of T cell/APC contact, TCR signaling is triggered via an array of phosphorylation and de-phosphorylation cascades of membrane-proximal and -distal signaling elements. Within few minutes, the T lymphocyte rapidly reorients its cellular content to the intercellular contact zone. In particular, the stimulated T cell repositions its centromere from the uropod to the synapse at the contact site and dynamically orients cytoskeletal systems that allow asymmetric segregation of signaling and adhesive proteins toward the APC contact (87). This centrosomal polarization is important for the directional movement of recycling TCRs to the IS (88) and the positioning of the T cell secretory vesicles toward the APC (89). These molecular processes facilitate the polarized secretion of cytokines and cytolytic factors toward the bound target cell for effector immune responses (e.g., cell-mediated cytotoxicity and target cell destruction) (90), while preventing undesired bystander effects on neighboring cells. A single T lymphocyte is thus able to eliminate multiple target cells consecutively by integrin-mediated adhesion, rapid rearrangement of contacts and simultaneous formation of stimulatory and lytic synapses with defined central and peripheral signaling platforms. Moreover, the IS facilitates cell-to-cell communication between the T cell and the APC through exosomes and microvesicles (91, 92). After several hours of contact, T cell undergoes functional activation (93), and eventually differentiates to effector or memory T cells.
In the context of IS formation, CG-NAP coordinates dynamic interactions between protein kinases and their substrates at the centrosome in T cells. It colocalizes with a range of signaling molecules with implications for both the central supramolecular activation cluster (c-SMAC), which includes the TCR/CD3 complex and various costimulatory receptors, and the peripheral supramolecular activation cluster (p-SMAC) that incorporates LFA-1 (22). Functional consequences of CG-NAP loss in T cells during the IS formation, either by overexpression of a dominant-negative form or siRNA-mediated knockdown, include (i) impaired conformational activation and positioning of LFA-1 at the IS, (ii) defective segregation of LFA-1 at the p-SMAC ring, (iii) impaired LFA-1-associated signaling, (iv) reduced expression of the TCR CD3ϵ chain with decreased activation and clustering of TCR at the IS, (v) reduced phosphorylation of CD3ζ (Y83) in the TCR/CD3 complex, (vi) impaired recruitment of PKCθ to the IS, (vii) diminished phosphorylation of the phospholipase C gamma 1 (PLC-γ1), (viii) reduced activation of intracellular adaptor proteins, including the linker for activation of T cells (LAT) and Vav1, (ix) reduced phosphorylation of ERK1/2, (x) delocalization of the centrosome, (xi) defects in the translocation of microtubule organizing center (MTOC) toward the IS, and (xii) diminished production of IL-2 (22). The PKCθ isoform, PLC-γ1, ERK1/2, Vav1, and LAT play critical roles in TCR signaling. For example, activation of the TCR triggers PKCθ-mediated phosphorylation of the Rap guanine nucleotide exchange factor 2 (RAPGEF2) at Ser960, which regulates the adhesiveness of LFA-1 to its ligand ICAM-1 via Rap1 (94). Essential roles of PKCθ in regulating TCR-induced NFκB activation in mature thymocytes, inducible gene expression program in T cells, up-regulation and clustering of the LFA-1 on the T cell surface, adhesion capacity of T cells, effector T cell functions and protection from T cell-mediated autoimmune reactions have been documented (80, 95–97). An impaired PLC-γ1 activation in CG-NAP depleted T cells would impair diacylglycerol production, which is important for dynein function and MTOC translocation (22). TCR-induced phosphorylation of both LAT and Vav1 is critical for the functioning of the c-SMAC complex (22).
In the context of cytoskeletal reorganization at the IS, CG-NAP facilitates microtubule nucleation at the centrosome and non-centrosomal regions in human T cells (20). It coordinates PKA-mediated phosphorylation of dynein in motile T cells (20), which is crucial for centrosome repositioning at the IS (87, 98). Following APC/T cell contact, CG-NAP interacts with the kinase CK1δ that phosphorylates the microtubule plus-end binding protein EB1, which increases microtubule growth speeds (99) and has consequences for the IS. For example, T cell cytoskeletal remodeling elicits the APC to mobilize its intercellular adhesive molecules (ICAM-1 and−3) and subsequently the MHC-II molecules at the IS (100). Moreover, CG-NAP loss in human T cells impairs actin polymerization (22), which is crucial for the stabilization of APC/T cell contact at the IS (101).
CG-NAP mediates the activation of Aurora A protein kinase in human T cells (102), which is crucial for regulating signaling downstream of the TCR, such as activation of the Lck kinase and opening of the Ca2+ release-activated channels (CRAC)—both key signals involved in antigen-dependent T cell activation and in IS formation. Interestingly both knockdown and over-expression of CG-NAP significantly inhibit IL-2 secretion (22), suggesting multiple overlapping effects.
Thus, T cell CG-NAP contributes to the formation and maintenance of IS by serving as an intracellular scaffold for kinases and facilitating the organization and activation of receptor molecules.
CG-NAP/Kinase Interactions in T Cell Activation and Proliferation
The processes of T cell activation, proliferation and effector functions require several independent but coordinated molecular events initiated by TCR engagement (103). According to the clonal selection theory of adaptive immunity, the activation of a single lymphocyte clone provides sufficient function for an immediate immune response (i.e., proliferation of effector cells), as well as the regenerative capacity to maintain the selected lineage (i.e., development and differentiation of memory cells). In this context, CG-NAP is potentially involved in T cell proliferation and clonal expansion. While a direct role of CG-NAP in cell proliferation and cell cycle regulation has been identified in other cell types (51, 61, 104), further studies are required to dissect this role in T lymphocytes. Nonetheless some potential interactions may be inferred. For example, CG-NAP-depleted Chinese hamster ovary (CHO) cells and HeLa cells over-expressing C-terminus CG-NAP are arrested at the G1 stage of cell-cycle followed by the induction of apoptosis in these cells (51, 61, 104). It has been shown in CHO cells that CG-NAP, by anchoring cyclin E/Cdk2 to the centrosome, drives centrosomal amplification and cell cycle progression (61). Further studies are required to determine whether CG-NAP-cyclin/Cdk complexes are involved in T cell proliferation.
At the centrosome, CG-NAP interacts with the centrosomal protein 72 kDa (Cep72) (105) and recruits the Kizuna (Kiz) protein, which is phosphorylated by the polo-like kinase 1 (Plk1) (106). The phosphorylation of Kiz enhances its association with the CG-NAP interacting protein, pericentrin (106). This association galvanizes the pericentriolar material, facilitates microtubule nucleation on the centrosome and allows for tubulin polymerization at the plus-end of the microtubules (107). CG-NAP associates with the dynein/dynactin motor complex and together with kendrin/pericentrin, anchors γ-TuRC at the centrosome through binding to its TUBGCP2 and/or TUBGCP3 subunits at the amino terminal regions (55). It provides new nucleation sites for de novo microtubule polymerization (55, 67, 108), which is important for cell cycle progression and proliferation of T lymphocytes. While many components of these interactions have been identified in T cells, further studies are required to determine the precise mechanisms whereby CG-NAP can regulate proliferation in human T cells.
CG-NAP also plays a crucial role in the regulation of endosomal trafficking of the TCR and is required for the effective re-stimulation of T cells (109). CG-NAP-dependent signaling and endosomal trafficking are important for the retention of T cells at sites of inflammation in mice (109). However, the viability of CG-NAP-knockout mice and the normal T cell counts in mice with conditional deletion of CG-NAP (109) suggest that CG-NAP may largely be dispensable or redundant for the maintenance of the resting T cell complement in the mouse. Loss of CG-NAP function in T cells would thus impair their sustained activation and have immunological consequences (109).
It is now clear that repeated and transient contacts of effector T cells with APCs are needed to functionally activate T lymphocytes in tissues. This suggests that signal integration between successive contacts is necessary to achieve activation. In contrast, the interactions between naïve T cells and DCs in the lymph node are relatively less dynamic and, typically, such interactions last for several hours (110, 111). It has also been speculated that the mechanisms of T cell activation at inflammation sites may vary from the primary activation of naïve T cells in lymph nodes (109). These tissue-specific differences in T cell activation may explain why depletion of CG-NAP does not significantly affect baseline T cell presence and differentiation in lymphoid tissues but may significantly impact T cell re-activation under suboptimal antigen-presenting conditions, such as re-activation in inflamed non-lymphoid tissue (109). Nevertheless, it has now been established that CG-NAP interacts with kinases (such as PKCβ, PKCδ, PKCε, PKCθ) (21, 54) and protein phosphatases (such as PP1, PP2A) (45) and transduces important signals via TCR (22) to regulate tissue-specific T cell activation and proliferation.
CG-NAP/Kinase Interactions in T Cell Migration
The recruitment of T cells to the tissue sites of infection or inflammation is critical to an effective immune response. Stimulated T lymphocytes leave lymphatic tissue and search the periphery for infected or transformed cells. When a T lymphocyte encounters an APC or a transformed cell, it mounts a specific immune response in a highly controlled manner. Blockade of the multi-step process of T cell motility can impair immune reactions, while uncontrolled migration can contribute to the development of autoimmunity.
In one pathway, T cell motility is dependent on the interactions between the T cell integrin LFA-1 and its ligand ICAM-1, which is expressed on endothelial surfaces during inflammation (112). LFA-1 engagement triggers a plethora of signaling cascades causing dynamic phosphorylation/de-phosphorylation of substrates by kinases and phosphatases and formation of macromolecular signaling complexes that culminate in cytoskeletal remodeling and T cell motility (112). CG-NAP is an integral component of these LFA-1-induced multi-molecular complexes and can serve to link the centrosome, microtubules and kinases, critical to the polarization and migration of T cells (5, 20, 21).
In human T lymphocytes, CG-NAP predominantly localizes in close proximity to the centrosome and the Golgi (20). This Golgi localization of CG-NAP is disrupted by microtubule depolarization (20). In HeLa-Kyoto cells, CG-NAP was found to recruit the microtubule cross-linking factor 1 (MTCL1), a molecule which crosslinks and stabilizes non-centrosomal microtubules to the Golgi membranes (113). Hence, we hypothesize a potential role of CG-NAP/MTCL1 interactions in T cell motility, a process which requires further investigation. Furthermore, as an AKAP, CG-NAP interacts with PKA and this complex consequently phosphorylates centrosomal proteins pericentrin and dynein in motile T cells (20). Dynein plays a crucial role in MTOC repositioning, cytoskeletal organization and the movement and processes of signaling complexes during T cell activation and motility (114, 115). In addition to PKA, several other kinases and phosphatases, including CK1, PKC, PP1 and PP2A, phosphorylate and dephosphorylate dynein in various cell types (116, 117) and all these enzymes are known to be anchored to the CG-NAP (45, 52, 53).
The functional significance of CG-NAP/kinase interactions for T cell motility is further underscored by the finding that the association between CG-NAP and LFA-1-induced signaling complex is greatly reduced when T cells are maintained at low temperature conditions (21). These data suggest a potential link between CG-NAP/kinase interactions and metabolic pathways in motile T cells. T cells overexpressing the C-terminal (aa 3699-3796) mutant form of CG-NAP fail to polarize and migrate (21). This CG-NAP C-terminus region contains the PACT (pericentrin-AKAP450 centrosomal targeting) domain, which binds additional proteins (50). For example, calmodulin binds to the C-terminus of CG-NAP in a calcium-independent manner (50). In addition, CG-NAP interacts with PKCβ and PKCδ (21), a process critical for signal transduction in motile T cells (75, 118). While in vitro knockdown of CG-NAP in human T cells significantly inhibited T cell migration and chemotaxis (20), no major impact of CG-NAP depletion on T cell motility was observed in T cell-specific CG-NAP knockout mice (109). The discrepancy between these findings could be attributed to the different model systems and experimental conditions.
It has been shown that the intracellular distribution of CG-NAP in LFA-1-stimulated motile T cells is different from that in cells stimulated to migrate through interactions with fibronectin (21). These data suggest that CG-NAP plays a unique role in β2 integrin-mediated T cell motility and may not have a similar role in adhesion and motility induced via different integrin families, such as, the β1 integrin. It has also been demonstrated that LFA-1-induced macromolecular signaling assemblies bring together molecules involved in intracellular transport and secretion. For example, LFA-1-induced formation of CG-NAP/kinase interactome containing PKCβ is crucial for T cell IL-2 production (21).
Microtubules are prominent elements of the cytoskeleton and dynamic cytoskeletal remodeling is essential for T cell motility. In addition to the cellular microtubule arrays emanating from the centrosome or the MTOC, secondary networks exist, in which microtubules are not anchored to the centrosome. While non-centrosomal microtubules are known to be present in differentiated cells (e.g., neurons, skeletal muscles, and epithelial cells), a recent report has demonstrated non-centrosomal microtubules emanating from CG-NAP in motile T cells (20). GapmeR-mediated knockdown of CG-NAP (119) disrupted both the centrosomal and non-centrosomal microtubule nucleation and inhibited post-translational tyrosination and acetylation of tubulin, illustrating the complexity of CG-NAP's role in coordinating microtubule dynamics and stability in migrating T cells (20).
Prospects of Therapeutic Targeting of CG-NAP and Associated Challenges
Immune-mediated diseases caused by T cell dysfunction are an increasing cause of mortality worldwide. While available therapeutic agents target T cell trafficking and immune hyperactivity, such treatment modalities are often accompanied by significant side effects. For example, while blocking LFA-1/ICAM-1 interaction has been proven to be effective in treating immune diseases, such as psoriasis (120), such immunosuppression can trigger the activation of JC-1 virus in the central nervous system leading to the development of fatal progressive multifocal leukoencephalopathy (121, 122). In prior studies, we have demonstrated that pre-activation through the LFA-1 pathway also alters the T cell programme, such that these stimulated T cells become refractory to TGFβ-mediated suppression and exhibit increased IL-17 secretion (123). Further studies are required to delineate whether specific interactions between CG-NAP and its docking partners may mediate specific migratory or secretory signals impacting on immune effector mechanisms. Fine-tuning of these interactions can provide functional selectivity and may offer exciting therapeutic approaches for a wide array of immune-mediated diseases.
One such fine-tuning strategy could be to alter a specific CG-NAP/kinase interaction by targeting a single protein-protein interaction. This could be achieved by either developing inhibitors against a specific kinase interaction in the CG-NAP interactome or by targeting CG-NAP/kinase interacting domain by blocking peptides (124). Although more likely to serve as a research tool, blocking peptides may assist in designing and developing small molecules targeting CG-NAP/kinase interactions, representing an interesting area of research and drug discovery. Further structural modeling of CG-NAP/kinase interactions should also identify suitable targets for small molecule inhibitors.
Targeting CG-NAP in its entirety would be challenging mainly because (i) this adaptor protein is expressed as several alternatively spliced transcripts and (ii) the degree of complexity of CG-NAP's involvement in multiple aspects of T cell signaling makes it difficult to elucidate each of their individual roles. One plausible strategy to understand the role of CG-NAP in T cell functioning would be to selectively displace interacting kinases and their subtypes from the CG-NAP docking platform. This would require the development of isoform-specific disruptors and other molecular tools to dissect individual pathway and CG-NAP/kinase interactions with high specificity. Exciting tools are available to silence individual gene in T lymphocytes that can be used to study functional involvement of specific interaction between CG-NAP and an individual kinase. These include the use of antisense GapmeR (119), siRNAs, gene correction and CRISPR-Cas9 editing techniques, which can be employed to overcome immune-mediated pathologies.
Conclusion
It is evident from the past two decades of research that CG-NAP regulates a plethora of biological processes by organizing supramolecular complexes and facilitating dynamic interactions between many different kinases and their substrates. In T cells, CG-NAP plays an important role in motility and participates in multiple interdependent pathways of T cell activation and effector functions. Thus, systematic studies are warranted to shed light on common or distinct binding partners and functions of CG-NAP and clarify to what extent CG-NAP/kinase interactions regulate T cell functions.
There is growing interest in developing protein-protein interaction disruptors, which would open new opportunities for therapeutic targeting of individual interactions between CG-NAP and specific kinases. A better understanding of CG-NAP/kinase interactions in T lymphocytes and their functional perturbations in immune response regulation is likely to lead to new frontiers in the treatment of T cell-mediated diseases.
Author Contributions
NV and DK conceived the review idea. All authors contributed to the writing of this manuscript and approved the final manuscript version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported, in part, by the Lee Kong Chian School of Medicine (LKCMedicine), Nanyang Technological University (NTU) Singapore Start-Up Grant (L0412290), the Singapore Ministry of Education (MOE) under its Singapore MOE Academic Research Fund (AcRF) Tier 2 Grant (MOE2017-T2-2-004) to NV, and NIH grants (DK119186 and DK119192) to JS.
References
- 1.Finlay D, Cantrell D. The coordination of T-cell function by serine/threonine kinases. Cold Spring Harb Perspect Biol. (2011) 3:a002261. 10.1101/cshperspect.a002261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wells AD, Morawski PA. New roles for cyclin-dependent kinases in T cell biology: linking cell division and differentiation. Nat Rev Immunol. (2014) 14:261–70. 10.1038/nri3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pawson T. Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. (2004) 116:191–203. 10.1016/S0092-8674(03)01077-8 [DOI] [PubMed] [Google Scholar]
- 4.Li G, Qian H. Sensitivity and specificity amplification in signal transduction. Cell Biochem Biophys. (2003) 39:45–59. 10.1385/CBB:39:1:45 [DOI] [PubMed] [Google Scholar]
- 5.Verma NK, Kelleher D. Adaptor regulation of LFA-1 signaling in T lymphocyte migration: Potential druggable targets for immunotherapies? Eur J Immunol. (2014) 44:3484–99. 10.1002/eji.201344428 [DOI] [PubMed] [Google Scholar]
- 6.Flynn DC. Adaptor proteins. Oncogene. (2001) 20:6270–2. 10.1038/sj.onc.1204769 [DOI] [PubMed] [Google Scholar]
- 7.Smith FD, Esseltine JL, Nygren PJ, Veesler D, Byrne DP, Vonderach M, et al. Local protein kinase A action proceeds through intact holoenzymes. Science. (2017) 356:1288–93. 10.1126/science.aaj1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langeberg LK, Scott JD. Signalling scaffolds and local organization of cellular behaviour. Nat Rev Mol Cell Biol. (2015) 16:232–44. 10.1038/nrm3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott JD, Pawson T. Cell signaling in space and time: where proteins come together and when they're apart. Science. (2009) 326:1220–4. 10.1126/science.1175668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science. (2011) 332:680–6. 10.1126/science.1198701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeke A, Lukács M, Lim WA, Reményi A. Scaffolds: interaction platforms for cellular signalling circuits. Trends Cell Biol. (2009) 19:364–74. 10.1016/j.tcb.2009.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H, Jiang Z. The essential adaptors of innate immune signaling. Protein Cell. (2013) 4:27–39. 10.1007/s13238-012-2063-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vajjhala PR, Ve T, Bentham A, Stacey KJ, Kobe B. The molecular mechanisms of signaling by cooperative assembly formation in innate immunity pathways. Mol Immunol. (2017) 86:23–37. 10.1016/j.molimm.2017.02.012 [DOI] [PubMed] [Google Scholar]
- 14.Liu G, Dean A. Enhancer long-range contacts: the multi-adaptor protein LDB1 is the tie that binds. BBA Gene Regul. Mech. (2019) 1862:625–33. 10.1016/j.bbagrm.2019.04.003 [DOI] [PubMed] [Google Scholar]
- 15.Zhang Q, Ding S, Zhang H. Interactions between hematopoietic progenitor kinase 1 and its adaptor proteins (Review). Mol Med Rep. (2017) 16:6472–82. 10.3892/mmr.2017.7494 [DOI] [PubMed] [Google Scholar]
- 16.Rudd CE. Adaptors and molecular scaffolds in immune cell signaling. Cell. (1999) 96:5–8. 10.1016/S0092-8674(00)80953-8 [DOI] [PubMed] [Google Scholar]
- 17.Jordan MS, Singer AL, Koretzky GA. Adaptors as central mediators of signal transduction in immune cells. Nat Immunol. (2003) 4:110–6. 10.1038/ni0203-110 [DOI] [PubMed] [Google Scholar]
- 18.Koretzky GA, Myung PS. Positive and negative regulation of T-cell activation by adaptor proteins. Nat Rev Immunol. (2001) 1:95–107. 10.1038/35100523 [DOI] [PubMed] [Google Scholar]
- 19.Kong MS, Hashimoto-Tane A, Kawashima Y, Sakuma M, Yokosuka T, Kometani K, et al. Inhibition of T cell activation and function by the adaptor protein CIN85. Sci Signal. (2019) 12:567. 10.1126/scisignal.aav4373 [DOI] [PubMed] [Google Scholar]
- 20.Ong ST, Chalasani MLS, Fazil MHUT, Prasannan P, Kizhakeyil A, Wright GD, et al. Centrosome- and Golgi-localized protein kinase N-associated protein serves as a docking platform for protein kinase a signaling and microtubule nucleation in migrating T-cells. Front Immunol. (2018) 9:397. 10.3389/fimmu.2018.00397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El Din El Homasany BS, Volkov Y, Takahashi M, Ono Y, Keryer G, Delouvee A, et al. The scaffolding protein CG-NAP/AKAP450 is a critical integrating component of the LFA-1-induced signaling complex in migratory T cells. J Immunol. (2005) 175:7811–8. 10.4049/jimmunol.175.12.7811 [DOI] [PubMed] [Google Scholar]
- 22.Robles-Valero J, Martin-Cofreces NB, Lamana A, Macdonald S, Volkov Y, Sanchez-Madrid F. Integrin and CD3/TCR activation are regulated by the scaffold protein AKAP450. Blood. (2010) 115:74–4184. 10.1182/blood-2009-12-256222 [DOI] [PubMed] [Google Scholar]
- 23.Smith FD, Omar MH, Nygren PJ, Soughayer J, Hoshi N, Lau HT, et al. Single nucleotide polymorphisms alter kinase anchoring and the subcellular targeting of A-kinase anchoring proteins. Proc Natl Acad Sci USA. (2018) 115:E11465–74. 10.1073/pnas.1816614115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wehbi VL, Taskén K. Molecular mechanisms for cAMP-mediated immunoregulation in T cells - role of anchored protein kinase A signaling units. Front Immunol. (2016) 7:222. 10.3389/fimmu.2016.00222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukuyama T, Sueoka E, Sugio Y, Otsuka T, Niho Y, Akagi K, et al. MTG8 proto-oncoprotein interacts with the regulatory subunit of type II cyclic AMP-dependent protein kinase in lymphocytes. Oncogene. (2001) 20:6225–32. 10.1038/sj.onc.1204794 [DOI] [PubMed] [Google Scholar]
- 26.Schillace RV, Andrews SF, Liberty GA, Davey MP, Carr DW. Identification and characterization of myeloid translocation gene 16b as a novel a kinase anchoring protein in T lymphocytes. J Immunol. (2002) 168:1590–9. 10.4049/jimmunol.168.4.1590 [DOI] [PubMed] [Google Scholar]
- 27.Williams RO. Cutting edge: A-kinase anchor proteins are involved in maintaining resting T cells in an inactive state. J Immunol. (2002) 168:5392–6. 10.4049/jimmunol.168.11.5392 [DOI] [PubMed] [Google Scholar]
- 28.Colledge M, Scott JD. AKAPs: from structure to function. Trends Cell Biol. (1999) 9:216–21. 10.1016/S0962-8924(99)01558-5 [DOI] [PubMed] [Google Scholar]
- 29.Edwards AS, Scott JD. A-kinase anchoring proteins: protein kinase A and beyond. Curr Opin Cell Biol. (2000) 12:217–21. 10.1016/S0955-0674(99)00085-X [DOI] [PubMed] [Google Scholar]
- 30.Wong W, Scott JD. AKAP signalling complexes: focal points in space and time. Nat Rev Mol Cell Biol. (2004) 5:959–70. 10.1038/nrm1527 [DOI] [PubMed] [Google Scholar]
- 31.Carr DW1, Stofko-Hahn RE, Fraser ID, Bishop SM, Acott TS, Brennan RG, et al. Interaction of the regulatory subunit (RII) of cAMP-dependent protein kinase with RII-anchoring proteins occurs through an amphipathic helix binding motif. J Biol Chem. (1991) 266:14188–92. [PubMed] [Google Scholar]
- 32.Newlon MG1, Roy M, Morikis D, Carr DW, Westphal R, Scott JD, et al. A novel mechanism of PKA anchoring revealed by solution structures of anchoring complexes. EMBO J. (2001) 20:1651–62. 10.1093/emboj/20.7.1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gold MG, Lygren B, Dokurno P, Hoshi N, McConnachie G, Taskén K, et al. Molecular basis of AKAP specificity for PKA regulatory subunits. Mol Cell. (2006) 24:383–95. 10.1016/j.molcel.2006.09.006 [DOI] [PubMed] [Google Scholar]
- 34.Arumugham VB, Baldari CT. cAMP: a multifaceted modulator of immune synapse assembly and T cell activation. J Leukoc Biol. (2017) 101:1301–16. 10.1189/jlb.2RU1116-474R [DOI] [PubMed] [Google Scholar]
- 35.Taskén K, Stokka AJ. The molecular machinery for cAMP-dependent immunomodulation in T-cells. Biochem Soc Trans. (2006) 34:476–9. 10.1042/BST0340476 [DOI] [PubMed] [Google Scholar]
- 36.Kortum RL, Samelson LE. Priming the pump: adhesion enhances T cell antigen receptor-induced signaling. Immunity. (2009) 30:3–5. 10.1016/j.immuni.2008.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mosenden R, Taskén K. Cyclic AMP-mediated immune regulation–overview of mechanisms of action in T cells. Cell Signal. (2011) 23:1009–16. 10.1016/j.cellsig.2010.11.018 [DOI] [PubMed] [Google Scholar]
- 38.Grader-Beck T, van Puijenbroek AA, Nadler LM, Boussiotis VA. cAMP inhibits both Ras and Rap1 activation in primary human T lymphocytes, but only Ras inhibition correlates with blockade of cell cycle progression. Blood. (2003) 101:998–1006. 10.1182/blood-2002-06-1665 [DOI] [PubMed] [Google Scholar]
- 39.Ramstad C, Sundvold V, Johansen HK, Lea T. cAMP-dependent protein kinase (PKA) inhibits T cell activation by phosphorylating ser-43 of raf-1 in the MAPK/ERK pathway. Cell Signal. (2000) 12:57–563. 10.1016/S0898-6568(00)00097-8 [DOI] [PubMed] [Google Scholar]
- 40.Ould Amer Y, Hebert-Chatelain E. Mitochondrial cAMP-PKA signaling: What do we really know? BBA Bioenerg. (2018) 1859:68–877. 10.1016/j.bbabio.2018.04.005 [DOI] [PubMed] [Google Scholar]
- 41.Jarnaess E, Ruppelt A, Stokka AJ, Lygren B, Scott JD, Taskén K. Dual specificity A-kinase anchoring proteins (AKAPs) contain an additional binding region that enhances targeting of protein kinase A type I. J Biol Chem. (2008) 283:33708–18. 10.1074/jbc.M804807200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Means CK, Lygren B, Langeberg LK, Jain A, Dixon RE, Vega AL, et al. An entirely specific type I A-kinase anchoring protein that can sequester two molecules of protein kinase A at mitochondria. Proc Natl Acad Sci USA. (2011) 108:E1227–35. 10.1073/pnas.1107182108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burgers PP, Bruystens J, Burnley RJ, Nikolaev VO, Keshwani M, Wu J, et al. Structure of smAKAP and its regulation by PKA-mediated phosphorylation. FEBS J. (2016) 283:2132–48. 10.1111/febs.13726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burgers PP, Ma Y, Margarucci L, Mackey M, van der Heyden MA, Ellisman M, et al. A small novel A-kinase anchoring protein (AKAP) that localizes specifically protein kinase A-regulatory subunit I (PKA-RI) to the plasma membrane. J Biol Chem. (2012) 287:43789–97. 10.1074/jbc.M112.395970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi M, Shibata H, Shimakawa M, Miyamoto M, Mukai H, Ono Y. Characterization of a novel giant scaffolding protein, CG-NAP, that anchors multiple signaling enzymes to centrosome and the golgi apparatus. J Biol Chem. (1999) 274:17267–74. 10.1074/jbc.274.24.17267 [DOI] [PubMed] [Google Scholar]
- 46.Schmidt PH, Dransfield DT, Claudio JO, Hawley RG, Trotter KW, Milgram SL, Goldenring JR. AKAP350, a multiply spliced protein kinase A-anchoring protein associated with centrosomes. J Biol Chem. (1999) 274:3055–66. 10.1074/jbc.274.5.3055 [DOI] [PubMed] [Google Scholar]
- 47.Witczak O, Skalhegg BS, Keryer G, Bornens M, Tasken K, Jahnsen T, et al. Cloning and characterization of a cDNA encoding an A-kinase anchoring protein located in the centrosome, AKAP450. EMBO J. (1999) 18:1858–68. 10.1093/emboj/18.7.1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Westphal RS, Tavalin SJ, Lin JW, Alto NM, Fraser ID, Langeberg LK, et al. Regulation of NMDA receptors by an associated phosphatase-kinase signaling complex. Science. (1999) 285:93–6. 10.1126/science.285.5424.93 [DOI] [PubMed] [Google Scholar]
- 49.Dransfield DT, Yeh JL, Bradford AJ, Goldenring JR. Identification and characterization of a novel A-kinase-anchoring protein (AKAP120) from rabbit gastric parietal cells. Biochem J. (1997) 322:801–8. 10.1042/bj3220801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gillingham AK, Munro S. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. (2000) 1:524–9. 10.1093/embo-reports/kvd105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keryer G, Witczak O, Delouvée A, Kemmner WA, Rouillard D, Tasken K, et al. Dissociating the centrosomal matrix protein AKAP450 from centrioles impairs centriole duplication and cell cycle progression. Mol Biol Cell. (2003) 14:2436–46. 10.1091/mbc.e02-09-0614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sillibourne JE, Milne DM, Takahashi M, Ono Y, Meek DW. Centrosomal anchoring of the protein kinase CK1delta mediated by attachment to the large, coiled-coil scaffolding protein CG-NAP/AKAP450. J Mol Biol. (2002) 322:785–97. 10.1016/S0022-2836(02)00857-4 [DOI] [PubMed] [Google Scholar]
- 53.Giamas G, Hirner H, Shoshiashvili L, Grothey A, Gessert S, Kuhl M, et al. Phosphorylation of CK1delta: identification of Ser370 as the major phosphorylation site targeted by PKA in vitro and in vivo. Biochem J. (2007) 406:389–98. 10.1042/BJ20070091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takahashi M, Mukai H, Oishi K, Isagawa T, Ono Y. Association of immature hypophosphorylated protein kinase cepsilon with an anchoring protein CG-NAP. J Biol Chem. (2000) 275:34592–6. 10.1074/jbc.M005285200 [DOI] [PubMed] [Google Scholar]
- 55.Takahashi M, Yamagiwa A, Nishimura T, Mukai H, Ono Y. Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring gamma-tubulin ring complex. Mol Biol Cell. (2002) 13:3235–45. 10.1091/mbc.e02-02-0112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu J, de Heus C, Liu Q, Bouchet BP, Noordstra I, Jiang K, et al. Molecular pathway of microtubule organization at the Golgi apparatus. Dev Cell. (2016) 39:44–60. 10.1016/j.devcel.2016.08.009 [DOI] [PubMed] [Google Scholar]
- 57.Kim HS, Takahashi M, Matsuo K, Ono Y. Recruitment of CG-NAP to the Golgi apparatus through interaction with dynein-dynactin complex. Genes Cells. (2007) 12:421–34. 10.1111/j.1365-2443.2007.01055.x [DOI] [PubMed] [Google Scholar]
- 58.Larocca MC, Shanks RA, Tian L, Nelson DL, Stewart DM, Goldenring JR. AKAP350 interaction with cdc42 interacting protein 4 at the Golgi apparatus. Mol Bio Cell. (2004) 15:2771–81. 10.1091/mbc.e03-10-0757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keryer G, Di Fiore B, Celati C, Lechtreck KF, Mogensen M, Delouvee A, et al. Part of Ran is associated with AKAP450 at the centrosome: involvement in microtubule-organizing activity. Mol Biol Cell. (2003) 14:4260–71. 10.1091/mbc.e02-11-0773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Terrin A, Monterisi S, Stangherlin A, Zoccarato A, Koschinski A, Surdo NC, et al. PKA and PDE4D3 anchoring to AKAP9 provides distinct regulation of cAMP signals at the centrosome. J Cell Biol. (2012) 198:607–21. 10.1083/jcb.201201059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nishimura T, Takahashi M, Kim HS, Mukai H, Ono Y. Centrosome-targeting region of CG-NAP causes centrosome amplification by recruiting cyclin E-cdk2 complex. Genes Cells. (2005) 10:75–86. 10.1111/j.1365-2443.2005.00816.x [DOI] [PubMed] [Google Scholar]
- 62.Rivero S, Cardenas J, Bornens M, Rios RM. Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. EMBO J. (2009) 28:1016–28. 10.1038/emboj.2009.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Z, Zhang C, Qi RZ. A newly identified myomegalin isoform functions in Golgi microtubule organization and ER-Golgi transport. J Cell Sci. (2014) 127:4904–17. 10.1242/jcs.155408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J, Xu H, Jiang Y, Takahashi M, Takeichi M, Meng W. CAMSAP3-dependent microtubule dynamics regulates Golgi assembly in epithelial cells. J Genet Genomics. (2017) 44:39–49. 10.1016/j.jgg.2016.11.005 [DOI] [PubMed] [Google Scholar]
- 65.Yang C, Wu J, de Heus C, Grigoriev I, Liv N, Yao Y, et al. EB1 and EB3 regulate microtubule minus end organization and Golgi morphology. J Cell Biol. (2017) 216:3179–98. 10.1083/jcb.201701024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bouguenina H, Salaun D, Mangon A, Muller L, Baudelet E, Camoin L, et al. EB1-binding-myomegalin protein complex promotes centrosomal microtubules functions. Proc Natl Acad Sci USA. (2017) 114:E10687–96. 10.1073/pnas.1705682114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petry S, Vale RD. Microtubule nucleation at the centrosome and beyond. Nat Cell Biol. (2015) 17:1089–93. 10.1038/ncb3220 [DOI] [PubMed] [Google Scholar]
- 68.Lebwohl M, Tyring SK, Hamilton TK, Toth D, Glazer S, Tawfik NH, et al. Efalizumab study group. a novel targeted T-cell modulator, efalizumab, for plaque psoriasis. N Engl J Med. (2003) 349:2004–13. 10.1056/NEJMoa030002 [DOI] [PubMed] [Google Scholar]
- 69.Starling S, Jolly C. LFA-1 engagement triggers T cell oolarization at the HIV-1 virological synapse. J Virol. (2016) 90:9841–54. 10.1128/JVI.01152-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tardif MR, Tremblay MJ. Regulation of LFA-1 activity through cytoskeleton remodeling and signaling components modulates the efficiency of HIV type-1 entry in activated CD4+ T lymphocytes. J Immunol. (2005) 175:926–35. 10.4049/jimmunol.175.2.926 [DOI] [PubMed] [Google Scholar]
- 71.Hioe CE, Chien PC, Jr, Lu C, Springer TA, Wang XH, Bandres J, et al. LFA-1 expression on target cells promotes human immunodeficiency virus type 1 infection and transmission. J Virol. (2001) 75:1077–82. 10.1128/JVI.75.2.1077-1082.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Volkov Y, Long A, Freeley M, Golden-Mason L, O'Farrelly C, Murphy A, Kelleher D. The hepatitis C envelope 2 protein inhibits LFA-1-transduced protein kinase C signaling for T-lymphocyte migration. Gastroenterology. (2006) 130:482–92. 10.1053/j.gastro.2005.10.047 [DOI] [PubMed] [Google Scholar]
- 73.Petrovic D, Dempsey E, Doherty DG, Kelleher D, Long A. Hepatitis C virus–T-cell responses and viral escape mutations. Eur J Immunol. (2012) 42:17–26. 10.1002/eji.201141593 [DOI] [PubMed] [Google Scholar]
- 74.Schwab N, Ulzheimer JC, Fox RJ, Schneider-Hohendorf T, Kieseier BC, Monoranu CM, et al. Fatal PML associated with efalizumab therapy: insights into integrin αLβ2 in JC virus control. Neurology. (2012) 78:458–67. 10.1212/WNL.0b013e3182478d4b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Volkov Y, Long A, McGrath S, Ni Eidhin D, Kelleher D. Crucial importance of PKC-beta(I) in LFA-1-mediated locomotion of activated T cells. Nat Immunol. (2001) 2:508–14. 10.1038/88700 [DOI] [PubMed] [Google Scholar]
- 76.Gruber T, Barsig J, Pfeifhofer C, Ghaffari-Tabrizi N, Tinhofer I, Leitges M, et al. PKCdelta is involved in signal attenuation in CD3+ T cells. Immunol Lett. (2005) 96:291–3. 10.1016/j.imlet.2004.08.011 [DOI] [PubMed] [Google Scholar]
- 77.Keenan C, Volkov Y, Kelleher D, Long A. Subcellular localization and translocation of protein kinase C isoforms zeta and epsilon in human peripheral blood lymphocytes. Int Immunol. (1997) 9:1431–9. 10.1093/intimm/9.10.1431 [DOI] [PubMed] [Google Scholar]
- 78.Genot EM, Parker PJ, Cantrell DA. Analysis of the role of protein kinase C-alpha, -epsilon, and -zeta in T cell activation. J Biol Chem. (1995) 270:9833–9. 10.1074/jbc.270.17.9833 [DOI] [PubMed] [Google Scholar]
- 79.Mirandola P, Gobbi G, Masselli E, Micheloni C, Di Marcantonio D, Queirolo V, et al. Protein kinase Cε regulates proliferation and cell sensitivity to TGF-1β of CD4+ T lymphocytes: implications for Hashimoto thyroiditis. J Immunol. (2011) 187:4721–32. 10.4049/jimmunol.1003258 [DOI] [PubMed] [Google Scholar]
- 80.Lim PS, Sutton CR, Rao S. Protein kinase C in the immune system: from signalling to chromatin regulation. Immunology. (2015) 146:508–22. 10.1111/imm.12510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bertolotto C, Maulon L, Filippa N, Baier G, Auberger P. Protein kinase C theta and epsilon promote T-cell survival by a rsk-dependent phosphorylation and inactivation of BAD. J Biol Chem. (2000) 275:37246–50. 10.1074/jbc.M007732200 [DOI] [PubMed] [Google Scholar]
- 82.Pfeifhofer-Obermair C, Thuille N, Baier G. Involvement of distinct PKC gene products in T cell functions. Front Immunol. (2012) 3:220. 10.3389/fimmu.2012.00220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Szamel M, Appel A, Schwinzer R, Resch K. Different protein kinase C isoenzymes regulate IL-2 receptor expression or IL-2 synthesis in human lymphocytes stimulated via the TCR. J Immunol. (1998) 160:2207–14. [PubMed] [Google Scholar]
- 84.Simon AK, Auphan N, Pophillat M, Boyer C, Ghosh S, Rincón M, et al. The lack of NF-κB transactivation and PKC epsilon expression in CD4+CD8+ thymocytes correlates with negative selection. Cell Death Differ. (2000) 7:1253–62. 10.1038/sj.cdd.4400760 [DOI] [PubMed] [Google Scholar]
- 85.Ong ST, Freeley M, Skubis-Zegadło J, Fazil MHUT, Kelleher D, Fresser F, et al. Phosphorylation of Rab5a protein by protein kinase Cϵ is crucial for T-cell migration. J Biol Chem. (2014) 289:19420–34. 10.1074/jbc.M113.545863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bromley SK, Burack WR, Johnson KG, Somersalo K, Sims TN, Sumen C, et al. The immunological synapse. Annu Rev Immunol. (2001) 19:375–96. 10.1146/annurev.immunol.19.1.375 [DOI] [PubMed] [Google Scholar]
- 87.Yi J, Wu X, Chung AH, Chen JK, Kapoor TM, Hammer JA. Centrosome repositioning in T cells is biphasic and driven by microtubule end-on capture-shrinkage. J Cell Biol. (2013) 202:779–92. 10.1083/jcb.201301004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cesari F. TCR–CD3 recycling to the synapse. Nat Rev Immunol. (2009) 9:820–1. 10.1038/nri2680 [DOI] [Google Scholar]
- 89.Stinchcombe JC, Majorovits E, Bossi G, Fuller S, Griffiths GM. Centrosome polarization delivers secretory granules to the immunological synapse. Nature. (2006) 443:462–5. 10.1038/nature05071 [DOI] [PubMed] [Google Scholar]
- 90.Stinchcombe JC, Salio M, Cerundolo V, Pende D, Arico M, Griffiths GM. Centriole polarisation to the immunological synapse directs secretion from cytolytic cells of both the innate and adaptive immune systems. BMC Biol. (2011) 9:45. 10.1186/1741-7007-9-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. (2011) 2:282. 10.1038/ncomms1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Choudhuri K, Llodra J, Roth EW, Tsai J, Gordo S, Wucherpfennig KW, et al. Polarized release of T-cell-receptor-enriched microvesicles at the immunological synapse. Nature. (2014) 507:118–23. 10.1038/nature12951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Iezzi G, Karjalainen K, Lanzavecchia A. The duration of antigenic stimulation determines the fate of naive and effector T cells. Immunity. (1998) 8:89–95. 10.1016/S1074-7613(00)80461-6 [DOI] [PubMed] [Google Scholar]
- 94.Letschka T, Kollmann V, Pfeifhofer-Obermair C, Lutz-Nicoladoni C, Obermair GJ, Fresser F, et al. PKC-theta selectively controls the adhesion-stimulating molecule Rap1. Blood. (2008) 112:4617–27. 10.1182/blood-2007-11-121111 [DOI] [PubMed] [Google Scholar]
- 95.Sun Z, Arendt CW, Ellmeier W, Schaeffer EM, Sunshine MJ, Gandhi L, et al. PKC-theta is required for TCR-induced NF-kappaB activation in mature but not immature T lymphocytes. Nature. (2000) 404:402–7. 10.1038/35006090 [DOI] [PubMed] [Google Scholar]
- 96.Brezar V, Tu WJ, Seddiki N. PKC-theta in regulatory and effector T-cell functions. Front Immunol. (2015) 6:530. 10.3389/fimmu.2015.00530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Freeley M, Long A. Regulating the regulator: phosphorylation of PKC θ in T cells. Front Immunol. (2012) 6:227 10.3389/fimmu.2012.00227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hashimoto-Tane A, Yokosuka T, Sakata-Sogawa K, Sakuma M, Ishihara C, Tokunaga M, et al. Dynein-driven transport of T cell receptor microclusters regulates immune synapse formation and T cell activation. Immunity. (2011) 34:919–31. 10.1016/j.immuni.2011.05.012 [DOI] [PubMed] [Google Scholar]
- 99.Zyss D, Ebrahimi H, Gergely F. Casein kinase I delta controls centrosome positioning during T cell activation. J Cell Biol. (2011) 195:781–97. 10.1083/jcb.201106025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.de la Fuente H, Mittelbrunn M, Sánchez-Martín L, Vicente-Manzanares M, Lamana A, Pardi R, et al. Synaptic clusters of MHC class II molecules induced on DCs by adhesion molecule-mediated initial T-cell scanning. Mol Biol Cell. (2005) 16:3314–22. 10.1091/mbc.e05-01-0005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fuller CL, Braciale VL, Samelson LE. All roads lead to actin: the intimate relationship between TCR signaling and the cytoskeleton. Immunol Rev. (2003) 191:220–36. 10.1034/j.1600-065X.2003.00004.x [DOI] [PubMed] [Google Scholar]
- 102.Blas-Rus N, Bustos-Morán E, Martín-Cófreces NB, Sánchez-Madrid F. Aurora-A shines on T cell activation through the regulation of Lck. Bioessays. (2017) 39:1600156. 10.1002/bies.201600156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guy CS, Vignali DA. Organization of proximal signal initiation at the TCR:CD3 complex. Immunol Rev. (2009) 232:7–21. 10.1111/j.1600-065X.2009.00843.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mattaloni SM, Ferretti AC, Tonucci FM, Favre C, Goldenring JR, Larocca MC. Centrosomal AKAP350 modulates the G/S transition. Cell Logist. (2013) 3:e26331 10.4161/cl.26331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Oshimori N, Li X, Ohsugi M, Yamamoto T. Cep72 regulates the localization of key centrosomal proteins and proper bipolar spindle formation. EMBO J. (2009) 28:2066–76. 10.1038/emboj.2009.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fabbro M, Zhou BB, Takahashi M, Sarcevic B, Lal P, Graham ME, et al. Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev Cell. (2005) 9:477–88. 10.1016/j.devcel.2005.09.003 [DOI] [PubMed] [Google Scholar]
- 107.Paz J, Lüders J. Microtubule-organizing centers: towards a minimal parts list. Trends Cell Biol. (2018) 28:176–87. 10.1016/j.tcb.2017.10.005 [DOI] [PubMed] [Google Scholar]
- 108.Tovey CA, Conduit PT. Microtubule nucleation by γ-tubulin complexes and beyond. Essays Biochem. (2018) 62:765–80. 10.1042/EBC20180028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Herter JM, Grabie N, Cullere X, Azcutia V, Rosetti F, Bennett P, et al. AKAP9 regulates activation-induced retention of T lymphocytes at sites of inflammation. Nat Commun. (2015) 6:10182. 10.1038/ncomms10182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rothoeft T1, Balkow S, Krummen M, Beissert S, Varga G, Loser K, et al. Structure and duration of contact between dendritic cells and T cells are controlled by T cell activation state. Eur J Immunol. (2006) 36:3105–17. 10.1002/eji.200636145 [DOI] [PubMed] [Google Scholar]
- 111.Garcia Z, Pradelli E, Celli S, Beuneu H, Simon A, Bousso P. Competition for antigen determines the stability of T cell-dendritic cell interactions during clonal expansion. Proc Natl Acad Sci USA. (2007) 104:4553–8. 10.1073/pnas.0610019104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Smith A, Stanley P, Jones K, Svensson L, McDowall A, Hogg N. The role of the integrin LFA-1 in T-lymphocyte migration. Immunol Rev. (2007) 218:135–46. 10.1111/j.1600-065X.2007.00537.x [DOI] [PubMed] [Google Scholar]
- 113.Sato Y, Hayashi K, Amano Y, Takahashi M, Yonemura S, Hayashi I, et al. MTCL1 crosslinks and stabilizes non-centrosomal microtubules on the Golgi membrane. Nat Comm. (2014) 5:5266. 10.1038/ncomms6266 [DOI] [PubMed] [Google Scholar]
- 114.Lim WM, Ito Y, Sakata-Sogawa K, Tokunaga M. CLIP-170 is essential for MTOC repositioning during T cell activation by regulating dynein localisation on the cell surface. Sci Rep. (2018) 8:17447. 10.1038/s41598-018-35593-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Benzing C, Rossy J, Gaus K. Do signalling endosomes play a role in T cell activation? FEBS J. (2013) 280:5164–76. 10.1111/febs.12427 [DOI] [PubMed] [Google Scholar]
- 116.Carnegie GK, Means CK, Scott JD. A-kinase anchoring proteins: from protein complexes to physiology and disease. IUBMB Life. (2009) 61:394–406. 10.1002/iub.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wirschell M, Yamamoto R, Alford L, Gokhale A, Gaillard A, Sale WS. Regulation of ciliary motility: conserved protein kinases and phosphatases are targeted and anchored in the ciliary axoneme. Arch Biochem Biophys. (2011) 510:93–100. 10.1016/j.abb.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Quann EJ, Liu X, Altan-Bonnet G, Huse M. A cascade of protein kinase C isozymes promotes cytoskeletal polarization in T cells. Nat Immunol. (2011) 12:647–54. 10.1038/ni.2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fazil MHUT, Ong ST, Chalasani ML, Low JH, Kizhakeyil A, Mamidi A, et al. GapmeR cellular internalization by macropinocytosis induces sequence-specific gene silencing in human primary T-cells. Sci Rep. (2016) 6:37721. 10.1038/srep37721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li S, Wang H, Peng B, Zhang M, Zhang D, Hou S, et al. Efalizumab binding to the LFA-1 alphaL I domain blocks ICAM-1 binding via steric hindrance. Proc Natl Acad Sci USA. (2009) 106:4349–54. 10.1073/pnas.0810844106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Carson KR, Focosi D, Major EO, Petrini M, Richey EA, West DP, et al. Monoclonal antibody-associated progressive multifocal leucoencephalopathy in patients treated with rituximab, natalizumab, and efalizumab: a Review from the Research on Adverse Drug Events and Reports (RADAR) Project. Lancet Oncol. (2009) 10:816–24. 10.1016/S1470-2045(09)70161-5 [DOI] [PubMed] [Google Scholar]
- 122.Lin EJ, Reddy S, Shah VV, Wu JJ. A review of neurologic complications of biologic therapy in plaque psoriasis. Cutis. (2018) 101:57–60. [PubMed] [Google Scholar]
- 123.Verma NK, Dempsey E, Long A, Davies A, Barry SP, Fallon PG, et al. Leukocyte function-associated antigen-1/intercellular adhesion molecule-1 interaction induces a novel genetic signature resulting in T-cells refractory to transforming growth factor-β signaling. J Biol Chem. (2012) 287:27204–16. 10.1074/jbc.M112.376616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang Y, Ho TG, Bertinetti D, Neddermann M, Franz E, Mo GC, et al. Isoform-selective disruption of AKAP-localized PKA using hydrocarbon stapled peptides. ACS Chem Biol. (2014) 9:635–42. 10.1021/cb400900r [DOI] [PMC free article] [PubMed] [Google Scholar]