Abstract
Horizontal gene transfer of integrative and conjugative elements (ICE) in bacterial pathogens of the bovine respiratory disease (BRD) complex has emerged as a significant cause of antimicrobial resistance (AMR) and therapeutic failure and mortalities in cattle. The aim of this study was to assess an AMR ICE occurring in Pasteurella multocida from a case of BRD, designated ICEMh1PM22 for its structure and host genome insertion site, and to identify consequences for host fitness and antimicrobial therapy. The modular structure of ICEMh1-like elements found in several related livestock pathogens was compared to ICEMh1PM22, and the repertoire of cargo genes in variable ICE modules was functionally categorized. AMR genes were identified as frequent additions to the variable modules of ICEMh1-like elements. Random PCR-based mapping of ICEMh1PM22-genome junctions in transconjugants provided evidence that ICEMh1PM22 integrates into the tRNA-leu for the UUG codon, and not into tRNA-leu for other codons. This was separately confirmed in the genomes of ICEMh1-like-harboring livestock pathogens. Bacterial genera harboring receptive tRNA-leuUUG were identified to establish the potential host range of ICEMh1-like elements. ICEMh1PM22-carrying transconjugants in P. multocida and Mannheimia haemolytica were less fit than isogenic strains without the ICE when grown without antimicrobial selection. This fitness cost was abrogated in the presence of subinhibitory concentrations of antimicrobials. Despite this cost, ICEMh1PM22 was retained in transconjugants in extended culture. To identify possible therapeutic efficiencies, antimicrobial combinations were screened for synergistic interactions against AMR ICEMh1PM22-carrying transconjugants. No antimicrobial combination tested exhibited synergistic interactions against AMR P. multocida or M. haemolytica harboring ICEMh1PM22. In conclusion, this study provided information on the structural variation of ICEMh1-like elements, refined the ICE insertion site and potential host range, and demonstrated the risk and consequences for AMR following horizontal transfer of ICE into BRD pathogens.
Keywords: bovine respiratory disease, Pasteurella multocida, Mannheimia haemolytica, antimicrobial resistance, synergy, integrative and conjugative element, conjugative transposon, cattle
Introduction
Integrative and conjugative elements (ICEs) are mobile genetic elements that can transfer autonomously by conjugation in bacteria. In general, ICEs can excise from the host chromosome to form a circular extrachromosomal intermediate which can be transferred and integrated into a receptive host (Johnson and Grossman, 2015). These widespread elements have increasingly been detected in bovine pathogens comprising the bacterial component of the bovine respiratory disease (BRD) complex (Michael et al., 2011b; Eidam et al., 2014). BRD bacteria are typically opportunistic pathogens which harmlessly reside in the nasopharynx, but upon exposure of cattle to stressors (e.g., weaning, transportation, feed changes) these bacteria can invade the respiratory tract and cause infection in conjunction with a variety of viral agents (Mosier, 2015). Serious BRD infections often lead to bovine pneumonia mortalities in calves. As a cause of significant economic loss to the cattle industry, BRD is typically prevented or treated with antimicrobials, including the use of in-feed antimicrobial prophylaxis (Cameron and McAllister, 2016). ICEs tend to have a modular structure, comprising modules for recombination and conjugation (Johnson and Grossman, 2015), but ICEs may also carry ‘cargo’ gene modules that are often advantageous to the recipient cell in terms of altering virulence, metabolic, and antimicrobial resistance capabilities. The first comprehensive reports of ICEs in BRD Pasteurellaceae described ICEPmu1 and ICEMh1, in Pasteurella multocida strain 36950 (Michael et al., 2011a, b) and Mannheimia haemolytica strain 42548, respectively (Eidam et al., 2014). Recently, related ICE sequences have been described in other BRD Pasteurellaceae isolates (Klima et al., 2016; Beker et al., 2018). Of particular concern, these ICEs can harbor multiple resistance genes, including determinants capable of conferring resistance to most veterinary antimicrobials that are approved for the treatment of BRD (FDA, 2016). In addition to those Pasteurellaceae, ICE ICEPmu1- and ICEMh1-like sequences have also been detected in mortality-associated Histophilus somni (Bhatt et al., 2018), as well as in disease-associated Pasteurellaceae from swine suffering from respiratory infection (M. J. Hauglund et al., USDA, unpublished data). Furthermore, resistance genes found in these ICE are often highly related to AMR genes from other human and veterinary bacteria (Michael et al., 2011a, b; Eidam et al., 2014; Cameron et al., 2018).
With the increasing availability of BRD-associated bacterial genome sequences, it is now evident that related ICEs are widespread in the Pasteurellaceae, which differ primarily in their ‘cargo’(Beker et al., 2018). ICEPmu1, the first BRD ICE characterized in P. multocida, was shown to integrate into a copy of the leucine tRNA (tRNA-leu) (Michael et al., 2011b). The chromosome-integrated form of ICEPmu1 was flanked by 13 bp direct repeats (5′-GATTTTGAATCAA-3′), which corresponded to the attB attachment site for site-specific recombination. In the circular form of ICEPmu1, the direct repeat sequence is flanked by left and right ‘attachment’ sites, attL and attR, which are thought to be the recombination sites for a XerCD-like pair of tyrosine recombinases (Castillo et al., 2017). In the integrated form, the direct repeat and either attL or attR comprise the left and right terminals of the ICE. ICEMh1 also targets tRNA-leu, but is flanked by a shorter, but otherwise identical 11 bp direct repeat (5′-GATTTTGAATC-3′) (Eidam et al., 2014). Thus, ICEPmu1 and ICEMh1 seem to have evolved from a common ancestor, and a number of genes (particularly those for recombination and conjugation) are syntenous (Eidam et al., 2014). Both ICEs also harbor two separate cargo regions named ‘resistance region 1′ and ‘resistance region 2′ because these regions characteristically harbor genes for antimicrobial resistance. ICEPmu1 has 12 resistance genes: strA and strB (conferring resistance to streptomycin), aphA1 (gentamicin), sul2 (sulfonamides), tet(H) (tetracyclines), floR (phenicols), erm(42) (macrolides and lincosamides), aadB (gentamicin), aadA15 (streptomycin and spectinomycin), blaOXA–2 (β-lactams), and msr(E) and mph(E) (macrolides) (Michael et al., 2011a, b). ICEMh1 has only 5 resistance genes: strA, strB, aphA1, sul2, tet(H) (Eidam et al., 2014). Both ICEs are fully mobilizable and proven to integrate into P. multocida, M. haemolytica, and E. coli recipients (Klima et al., 2014).
The observation that ICEPmu1 and ICEMh1 can easily move between genera and species highlights the extreme risk for rapid acquisition of AMR in BRD bacteria via horizontal gene transfer. The acquisition of these AMR genes poses an obvious threat to effective antimicrobial therapy, and could exacerbate economic losses associated with cattle mortalities. Greater knowledge of basic ICE biology is necessary to understand their potential host range, including acquisition by human pathogens, and the threat they pose to the efficacy of antimicrobial therapy. We explored these attributes using isogenic transconjugants of an ICEMh1-like element (ICEMh1PM22) previously identified in a bovine isolate (P. multocida PM22) from a BRD mortality (Klima et al., 2014). The objectives of this study were to (I) refine the insertion site of ICEMh1-like elements, (II) predict host range based on the insertion site, (III) explore the effect of ICEMh1PM22 acquisition on host fitness, and (IV) identify any synergistic antimicrobial combinations effective against bacteria harboring ICEMh1PM22. We tested for synergy using antimicrobials against which PM22 was both non-susceptible and susceptible to identify any combination capable of restoring susceptibility. We hypothesized that this information could be used to ensure prudent antimicrobial usage, inform future BRD mitigation strategies, and identify the potential risk in other areas of human and veterinary medicine.
Materials and Methods
Bacterial Strains and Growth Conditions
P. multocida PM22 was initially isolated following lung tissue necropsy of a beef cattle BRD mortality (acute fibrinous pneumonia) in a Texas feedlot (Klima et al., 2014). Pasteurellaceae were routinely grown on tryptic soy agar (TSA) blood agar (Dalynn Biologicals, Calgary, AB) or Brain Heart Infusion (BHI) agar (BD Difco, Mississauga, ON). E. coli were maintained with Luria-Bertani (LB) or Mueller-Hinton II (MH; cation-adjusted) agar/broth (BD Difco) supplemented with antibiotics where appropriate. All susceptibility testing was performed in MH broth. Spontaneous rifampin-resistant (RifR) mutants of P. multocida CCUG 17976 and M. haemolytica ATCC 33396 were obtained by plating OD600 2.0 on BHI supplemented with 50 mg/L rifampin (rifampicin; MilliporeSigma, Oakville, ON, United States), followed by incubation for ∼48 h at 37°C. To generate luciferase-expressing E. coli DH5α, the pAKlux2 plasmid (Karsi and Lawrence, 2007) was electroporated into E. coli DH5α and transformants were selected on and routinely maintained on LB agar supplemented with 100 mg/L of ampicillin (MilliporeSigma).
Conjugation and Mapping of ICE Insertion Junctions by Random PCR
Bacterial conjugation assays were performed as previously described (Klima et al., 2014), with minor modifications. Briefly, in three independent experiments, OD600 1.0 equivalents of P. multocida PM22 and RifR P. multocida CCUG 17976 or M. haemolytica ATCC 33396 were mated in 100 μL of BHI broth spotted on TSA blood agar, incubated aerobically at 37°C for 4 h, harvested and then plated on BHI agar supplemented with rifampin (50 mg/L), tetracycline (10 mg/L; MilliporeSigma), and spectinomycin (100 mg/L; MilliporeSigma). Six colonies were selected from each mating (18 in total for each species) and verified for ICEMh1PM22 acquisition by susceptibility testing and PCR screening (Klima et al., 2014). To further ensure that P. multocida transconjugants were not spontaneous, RifR mutants of P. multocida PM22, transconjugants were also tested for ceftiofur MIC (the cephalosporin resistance phenotype is not transferred to transconjugants). To map the ICEMh1PM22 insertion site, transconjugant DNA was purified (DNeasy Blood and Tissue kit, Qiagen, Montreal, QC) and amplified using the CEKG set of nested and random (i.e., containing degenerate sequence) oligonucleotides in combination with nested ICEMh1PM22-specific oligonucleotides: PM22-A1: 5′-CACCTTTAGTTGAAGCAATAG-3′; PM22-A2, 5′-TGGTAA AAGGTTTGTTTGTAC-3′; PM22-B1, 5′-ATCGTAGTAAGTG TGTATTTG-3′; and PM22-B2 5′-ATTATTTGAACAGTTC TACGC-3′ (Eurofins Genomics, Toronto, ON) in tandem rounds of PCR as described by Salama et al. (2004). Resulting amplicons were gel-purified (Zymoclean Gel DNA Recovery Kit, Zymo Research, Irvine, CA, United States) and sequenced (Eurofins Genomics).
Growth Curves and Co-culture Competitions
Growth curves of RifR P. multocida CCUG 17976 and M. haemolytica ATCC 33396 and their isogenic ICEMh1PM22 transconjugants were performed in 200 μL volumes of MH broth inoculated at an OD600 of 0.005 in 96-well plates (Nunc) sealed with gas-permeable film (Breathe-Easy sealing membrane, MilliporeSigma). Bacterial growth, with shaking at 37°C, was monitored continuously for 12 h in a plate-reader (Biotek HT Synergy). For 2-strain co-culture experiments, luciferase-expressing E. coli DH5α pAKlux2 and test strains (including DH5α without pAKlux2 as a control) were each inoculated at an OD600 of 0.0025 (i.e., total OD600 of ∼0.005) into black clear bottom 96-well plates (Nunc, Thermo-Fisher Scientific, Ottawa, ON, Canada). Bacterial growth was monitored as above, with the addition of luminometry. The maximum light produced (in relative light units, RLU) in each competition was recorded, and used to generate a competitive index (i.e., RLUTeststrains vs. DH5α pAKlux2/RLUDH5α vs. DH5α pAKlux2). Thus, values <1 indicated that test E. coli DH5α pAKlux2 was outperformed compared to an E. coli-only control. For long-term repeated passage experiments to assess if ICEMh1PM22 could be lost from the host strain, ICEMh1PM22 transconjugants were inoculated into 2 mL of MH broth or MH broth supplemented with 0.5 MIC (subinhibitory for the susceptible WT) oxytetracycline (0.125 mg/L), spectinomycin (32 mg/L), or tylosin (16 mg/L) in a sterile 96-well block (Greiner Bio-One, Monroe, NC), and sub-cultured (1/100) every ∼3 d into fresh media supplemented with the same antimicrobials for 150 d. To assess for loss of ICEMh1PM22, CFU from OD600 0.1-equivalents from each culture were enumerated in parallel on MH agar (total count) and MH agar supplemented with each antimicrobial at concentrations selective for ICEMh1PM22: oxytetracycline (10 mg/L); spectinomycin (512 mg/L); or tylosin (64 mg/L). Likewise, the effect of subinhibitory concentrations on co-culture competitions with luciferase-expressing E. coli was assessed with 0.5 MICs (for WT strains) of oxytetracycline (0.125 mg/L) and spectinomycin (32 mg/L) as above for co-cultures. Tylosin was not tested because E. coli is intrinsically resistant to macrolides.
MIC Determination and Checkerboard Assay Antimicrobial Synergy Screening
Minimum inhibitory concentrations (MICs) were determined for the RifR and isogenic ICEMh1PM22 P. multocida CCUG 17976 and M. haemolytica ATCC 33396 transconjugants according to the CLSI approved standard M07 - Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically (CLSI, 2018). Escherichia coli ATCC 25922 and E. faecalis ATCC 29212 were used as quality control organisms for susceptibility testing. Susceptibility designations for P. multocida PM22 were previously assigned (Klima et al., 2014) in accordance with the CLSI approved standard M31-A3 – Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria collected from animals (CLSI, 2008). Antimicrobials tested were purchased from MilliporeSigma (oxytetracycline hydrochloride, chlortetracycline, tilmicosin, gamithromycin, spectinomycin dihydrochloride pentahydrate, penicillin G sodium salt, ampicillin, ceftiofur hydrochloride, sulfamethoxazole, enrofloxacin, tiamulin fumarate, clindamycin hydrochloride and florfenicol) or from AlfaAesar (Haverhill, MA; tylosin tartrate, neomycin sulfate hydrate and sulfamethazine). Briefly, antimicrobial stocks were prepared immediately prior to testing with respect to potency in the appropriate solvent and filter sterilized where appropriate. Broth microdilution assays were performed in triplicate in 96-well plates in a total volume of 100 μL of MH broth containing 2-fold dilution series of each antimicrobial and inoculated with 5 μL of each strain tested (total inoculum of ∼5 × 104 CFU). Plates were grown aerobically at 37°C for 20 h and MIC was registered as the lowest concentration inhibiting visually detectable growth. For checkerboard synergy assays, two-dimensional arrays of serial concentrations (8 × 8 wells; 2-fold dilution series) of antimicrobials were created for two experiments: (I) all combinations of 5 antimicrobials (i.e., oxytetracycline, spectinomycin, tilmicosin, tylosin, and sulfamethazine) representing major antimicrobial classes to which transconjugants were non-susceptible, and (II) selected combinations of ‘non-susceptible’ antimicrobials (i.e., oxytetracycline, spectinomycin, and tylosin) with antimicrobials to which ICEMh1PM22 transconjugants were susceptible (i.e., penicillin G, enrofloxacin, florfenicol, ceftiofur, and chlortetracycline). Growth and sterility controls were included in each checkerboard assay, which were performed in triplicate. MICs were interpreted by eye and OD600 was measured in a plate reader to facilitate calculation of a fractional inhibitory concentration index (FICI). For discrepancies between replicates, the highest antimicrobial concentration determined the MIC. The FIC for each antimicrobial (A or B) was determined by the MIC of the antimicrobial in combination and divided by the MIC of the drug alone, according to:
Interactions were conservatively interpreted as ‘synergistic’ (FICI ≤ 0.5) or ‘no interaction’ (FICI > 0.5–4.0) in line with synergy testing guidelines in the Journal of Antimicrobial Chemotherapy (Odds, 2003).
Bioinformatics and Statistical Analyses
Statistical tests and graphing were performed with Sigmaplot 13.0 (Systat Software Inc.), with error bars indicating standard error of the mean (SEM). ICEMh1-like-containing Pasteurellaceae genomes were identified by default BLAST alignment to ICEMh1PM22, using only results with >80% sequence identity and >50% query coverage and eliminating isogenic or partial sequences. Sequence manipulations and alignments were performed in Geneious 8.1.9 using default MUSCLE parameters or progressive Mauve alignment (Darling et al., 2010) for identification of conserved and variable regions (and genes) in ICEMh1-like sequences. Gene Annotations in 41 ICEMh1-like sequences were standardized by re-annotation with PROKKA (Seemann, 2014) and manually scrutinized for putative function. Trees were constructed with PhyML (Jukes-Cantor substitution model). Sequence logos were produced with WebLogo (Crooks et al., 2004). Sequence similarity between ICEMh1-like sequences was visualized with Circos (Krzywinski et al., 2009). Predicted tRNA structure and identification was performed with tRNAscan-SE (Chan and Lowe, 2019). To identify bacteria with tRNA-leuUUG containing the conserved direct repeat and palindrome sequences, a BLAST search was performed using the trnL2 (tRNA-leuUUG) sequence from M. haemolytica M42458 with parameters: Program, blastn; Word size, 11; Expect value, 10; Hitlist size, 20,000; Match/Mismatch scores, 2/-3; Gapcosts, 5/2. This resulted in 9604 hits/alignments, which were filtered for those containing the ICEMh1-associated direct repeat (i.e., attB) and palindrome (with 100% identity). From this, a taxonomic report was generated to show bacterial genera.
Results
General Structure of the ICEMh1-like Variant ICEMh1PM22 From a BRD-Associated P. multocida Isolate
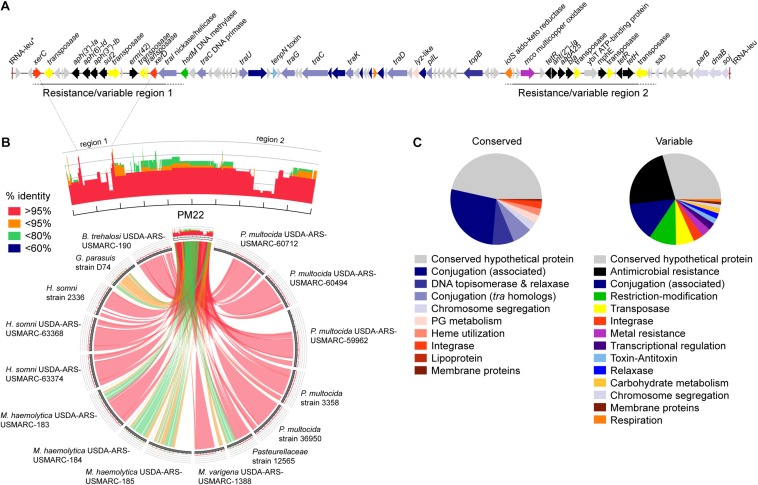
ICEMh1PM22 is a 77,786 bp element in P. multocida PM22 (accession # CP045724) bound by two copies of a tRNA for leucine (tRNA-leu), one of which is disrupted and potentially non-functional (on left, Figure 1A) whereas the second copy is full-length and likely functional (on right, Figure 1A). There is also a third disrupted copy of tRNA-leu upstream of the start codon of the full-length functional tRNA-leu (not shown). The ICEMh1PM22 sequence may encode for ∼94 proteins that have been annotated with putative functions or as conserved hypothetical proteins. Similar to the ICEMh1 prototype in M. haemolytica strain 42548 (Eidam et al., 2014) and ICEPmu1 in P. multocida strain 36950 (Michael et al., 2011b), and other Pasteurellaceae ICE (Beker et al., 2018), ICEMh1PM22 harbors multiple antimicrobial resistance genes/gene cassettes interspersed with numerous putative transposases in two discrete ‘resistance regions’. The first (left) region—resistance region 1, encodes aminoglycoside (aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib), sulfonamide (sul2), and macrolide (erm42) resistance genes, and the second—resistance region 2—encodes genes for tetracycline (tetH, tetR), aminoglycoside (ant(2″)-Ia, aadA25), macrolide (mphE) resistance, and a potential extended-spectrum β-lactamase (blaOXA–2) previously shown to be phenotypically non-functional in transconjugants (Klima et al., 2014). This two-region structure appears to be widely present in other ICEMh1-like elements found in 41 Pasteurellaceae genome sequences, encompassing the livestock pathogens P. multocida, M. haemolytica, Histophilus somni, Bibersteinia trehalosi, and Glaesserella parasuis (Figure 1B). Also routinely present in ICEMh1-like sequences are putative tyrosine recombinases (integrases), sometimes annotated as XerC and XerD, members of which are thought to work in tandem by mediating site-specific recombination (i.e., insertion and excision of the ICE) using two recognition sites which separately constitute the attL and attR sites in integrated ICE, and which are found on either side of a central attachment site involved in crossover (Michael et al., 2011b; Johnson and Grossman, 2015; Castillo et al., 2017). Alignment of xerC and xerD sequences found in 41 ICEMh1-like elements indicates a high degree of similarity (alignment identity: xerC, 77.9% identity; xerD 82.4% identity). XerC and XerD are likely part of the core or conserved backbone of ICEMh1-like elements, as are genes annotated as part of the conjugation machinery, such as the tra genes (Johnson and Grossman, 2015). Most of these were usually present in the central region of ICEMh1-like elements with >95% identity. We further examined the gene content in conserved and variable regions of 41 ICEMh1-like elements by extracting gene annotations from MAUVE-aligned sequences, defining ‘conserved’ as alignments found in >50% of ICEMh1-like elements with >80% sequence identity. ‘Conserved’ alignments harbored the majority of the conjugation machinery, such as tra and conjugation-associated proteins of unknown function, and DNA topoisomerases, integrases, and chromosome segregation factors potentially involved in ICE mobilization (Figure 1C). ‘Variable’ alignments contained more functionally diverse genes, including genes for antimicrobial and metal resistance, transposition, transcriptional regulation, potentially expanded metabolic functions (e.g., carbohydrate utilization and other respiratory genes), and toxin-antitoxin systems.
FIGURE 1.
Genomic characterization of the ICEMh1-like variant ICEMh1PM22. (A) Gene schematic of ICEMh1PM22 with genes shown as arrows and colored by functional characteristics, as shown in panel (C). (B) Comparison of ICEMh1PM22 with selected Pasteurellaceae genomic ICE sequences. In some cases, the ICE region cannot be delimited by the identification of the flanking direct repeat and includes sequences likely external to the ICE. Histograms represent the frequency and sequence identity (color) of alignments to ICEMh1PM22: red, >95% sequence identity; orange, >95%; green, <80%; blue, <60%. (C) Frequencies of annotated genes by functional characteristics found in conserved (left) and variable (right) regions in 41 ICEMh1-like-containing Pasteurellaceae (with >80% sequence identity and >50% query coverage alignment to ICEMh1PM22).
ICEMh1-like Elements Insert Into the tRNA-leu for the UUG Codon
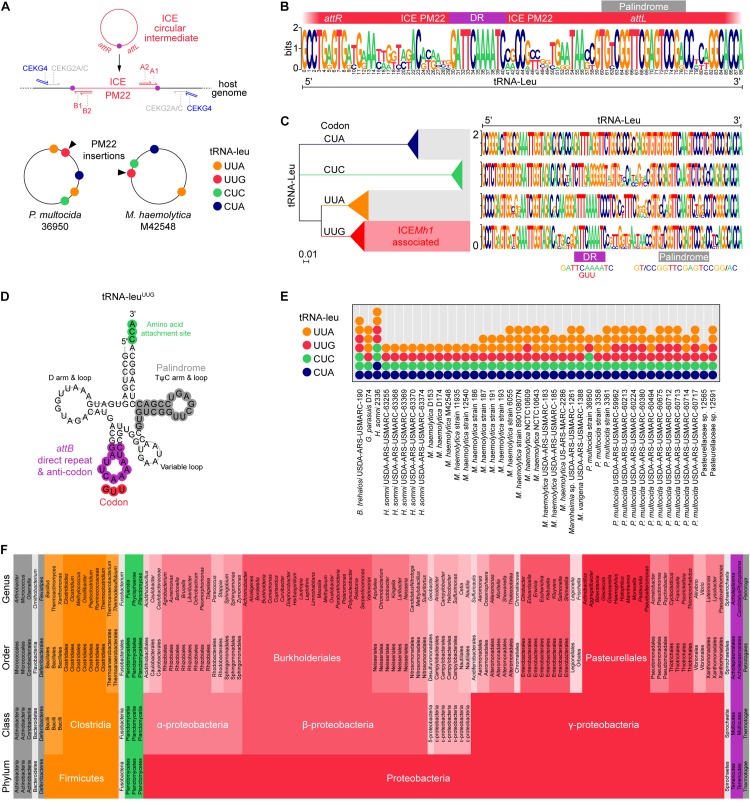
It was previously shown that ICEMh1-like elements insert into tRNA-leu genes in P. multocida and M. haemolytica (Michael et al., 2011b; Eidam et al., 2014). The terminal ends of ICEMh1 (and the cognate attB genome attachment site) were identified as a direct repeat (5′-GATTCAAAATC-3′) (Eidam et al., 2014). Given that there are multiple copies of tRNA-leu in most bacterial genomes, we initially thought that any could be receptive to ICEMh1-like elements. To determine this, we mapped ICEMh1PM22 junctions in transconjugants using degenerate (random) PCR oligonucleotides (Salama et al., 2004) with oligonucleotides specific for the terminal ends of ICEMh1PM22 (Figure 2A). Following sequencing and alignment of PCR-amplified junctions, ICEMh1PM22 transconjugants in P. multocida CCUG 17976 and M. haemolytica ATCC 33396 were found to have identical insertion sites, which we identified as a single tRNA-leu in each strain as shown mapped to the known genome sequences of P. multocida 36950 and M. haemolytica M42548 (Figure 2A). Furthermore, alignment of the left and right junctions in 41 ICEMh1-like-containing genomes indicated that those ICEs had exclusively inserted into tRNA-leu sequences in which both the direct repeat and an imperfect palindrome (5′-CGGTTCGAGTCCG-3′) were present (Figure 2B). To characterize this further, we examined all the tRNA-leu present in the 41 ICEMh1-like-containing genomes and found that these sequences could be categorized by multiple alignment phylogeny into four types in both P. multocida and M. haemolytica; representing 4 of the 6 possible anticodons for leucine (Figure 2C). This was consistent with data from the GtRNAdb tRNA databases (Chan and Lowe, 2015) which indicates that these bacteria typically encode at least 4 tRNAs for the leucine codons CUA, CUC, UUA, and UUG. The other two tRNA-leu codons, CUG and CUU, are absent in currently known P. multocida and M. haemolytica genomes. Only tRNA-leuUUG was integrated with ICEMh1-like elements in the 41 genomes studied. The predicted structure of tRNA-leuUUG suggests that the ICE direct repeat constitutes the tRNA anticodon loop, and the palindrome is the basis of the T-loop (or TψC loop) ribosome recognition site (Figure 2D). Our analysis of ICEMh1PM22 junctions also suggested that ICE insertion leaves one functional and one partial (disrupted) copy of tRNA-leuUUG (not shown). In the 41 ICEMh1-like-containing genomes, at least 1 functional copy of each tRNA was present, although sometimes full-length tRNA-leuUUG was present twice (Figure 1E). In all cases where two functional copies of tRNA-leuUUG were present, the copies were located directly on either side of the ICE insertion. We also observed that one terminus (i.e., direct repeat) of the ICE was absent in some ICEMh1-like-containing genomes, potentially indicating ICEs incapable of mobilization (not shown).
FIGURE 2.
Identification of tRNA-leuUUG as the putative attB insertion site of ICEMh1-like elements and predicted host range. (A) Schematic for ICE orientation and random PCR mapping of ICEMh1PM22 junctions in 18 Mannheimia haemolytica and 18 Pasteurella multocida transconjugants. ICEMh1PM22 insertions in every transconjugant were identical and aligned to a specific tRNA-leu in the genomes of P. multocida strain 36950 and M. haemolytica M42548. (B) Sequence logo of left and right MUSCLE-aligned junctions identified in 41 Pasteurellaceae harboring ICEMh1-like sequences. The left and right ends of ICEMh1-like variants contain a conserved direct repeat (DR; 5′-GATTCAAAATC-3′) attL conserves the tRNA TψC loop’s imperfect palindrome (5′-CGGTTCGAGTCCG-3′). The attL and attR sites are designated with respect to ICEPmu1 in Michael et al. (2011b). (C) Neighbor-joining tree of all tRNA-leu from 41 Pasteurellaceae harboring ICEMh1-like sequences. All ICEMh1-like insertions were associated with tRNA-leuUUG. (D) Predicted structure of tRNA-leuUUG showing presumptive ICEMh1 attB attachment site (also encoding the tRNA anticodon) and palindrome. (E) Frequencies of tRNA-leu codon types in 41 Pasteurellaceae harboring ICEMh1-like sequences. (F) Predicted host range of ICEMh1-like elements based on tRNA-leuUUG alignment by BLAST and strict (100% identity) conservation of direct repeat and palindrome. Colors are arbitrary.
Predicted Host Range of ICEMh1-like Elements
Given the conservation of the tRNA-leuUUG direct repeat and palindrome in the terminal ends of ICEMh1-like elements, we used these as criteria for in silico prediction of potential host range. The full-length M. haemolytica tRNA-leuUUG sequence was used as the basis for a blastn nr database search optimized for short input sequences. Following retrieval of 9,604 BLAST alignments, tRNA-leu containing the exact direct repeat and palindrome sequence were filtered, and a taxonomic report showing bacterial genera with members with those attributes was constructed (Figure 2F). As expected, the phylum with the greatest number of genera harboring similar tRNA-leu were the Proteobacteria, specifically γ-Proteobacteria, of which Pasteurellales were most abundant (an Order which includes P. multocida and M. haemolytica, and some human pathogens, including Haemophilus spp.). Enterobacterales were also frequently represented, and included many well-known food-borne and cattle-associated pathogens such as Escherichia coli and Salmonella spp. For E. coli, only some strains harbored the receptive tRNA-leu sequence, suggesting that only some members of any genus might be capable of transconjugation. Burkholderiales (β-proteobacteria) also harbored similar tRNA-leu, suggesting the potential for ICEMh1-like horizontal gene transfer to Burkholderia spp. and Bordetella spp. Likewise, bacteria from α-, δ-, and ε-Proteobacteria were also represented and may be receptive to ICEMh1, such that cattle-associated food-borne bacteria like Campylobacter could conjugate with ICE-bearing Pasteurellaceae. Some Gram-positive Firmicutes, specifically Clostridia, were also host to ICE-receptive tRNA-leu. Thus, if the tRNA-leuUUG sequence was the only requirement for mobilization and integration, ICEMh1 might be capable of crossing into these Gram-positive organisms. At present, it has only been experimentally confirmed that ICEMh1PM22 can transfer into RifR P. multocida and M. haemolytica (Klima et al., 2014), and E. coli strains K-12 and DH5α (not shown).
ICEMh1PM22 Acquisition Confers a Fitness Cost in Laboratory Conditions
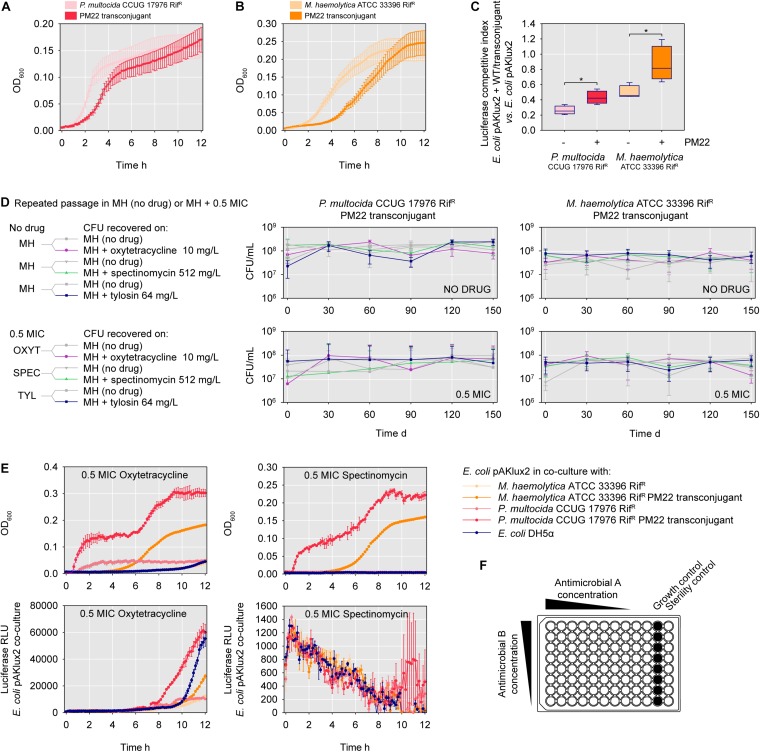
M. haemolytica ATCC 33396 transconjugants harboring ICEMh1PM22 were noticeably slower-growing than the isogenic WT on agar media supplemented with selective concentrations of antimicrobials. To assess the effect of ICE acquisition on growth parameters, we performed OD600-based growth curves for RifR P. multocida CCUG 17976 and M. haemolytica ATCC 33396, and their respective ICEMh1PM22 transconjugants. The transconjugant of RifR P. multocida CCUG 17976 exhibited both an extended lag phase and decreased growth rate compared to the ICE-minus strain (Figure 3A). Likewise, the transconjugant of the RifR M. haemolytica ATCC 33396 strain exhibited an extended lag phase, but did not have a significantly different growth rate (Figure 3B). To further explore the fitness costs of ICEMh1PM22 integration, we competed the isogenic RifR WT and transconjugants against E. coli DH5α expressing luciferase, and monitored light production to assess for E. coli fitness against all four strains (Figure 3C). In all cases, the Pasteurellaceae inhibited E. coli light production. However, competitions with strains harboring ICEMh1PM22 resulted in increased light production relative to the isogenic WT strains, also suggesting that ICEMh1PM22 confers a fitness cost on the host.
FIGURE 3.
ICEMh1PM22 fitness costs, addiction, and consequences for antimicrobial resistance in transconjugants. (A) Growth curves of P. multocida CCUG 17976 (spontaneous rifampin-resistant mutant) and isogenic ICEMh1PM22 transconjugant. Mean of 4 biological replicates with SEM. (B) Growth curves of M. haemolytica ATCC 33396 (spontaneous rifampin-resistant mutant) and isogenic ICEMh1PM22 transconjugant. Mean of 4 biological replicates with SEM. (C) Luciferase-based competition index from co-cultures of ICEMh1PM22 transconjugants and E. coli DH5α harboring the luciferase expression plasmid pAKlux2. Mean of 4 biological replicates with SEM; t-test, ∗P ≤ 0.05. (D) Long-term repeated passage of ICEMh1PM22 transconjugants. Transconjugants were grown in MH (no drug) or MH + 0.5 MIC (subinhibitory; MIC for susceptible P. multocida CCUG 17976 or M. haemolytica ATCC 33396 WT) for 150 days and plated on media with or without selective concentrations of indicated antimicrobials (i.e., 0.5 MIC for non-susceptible isogenic transconjugants). Mean of 3 biological replicates with SEM. (E) Upper panel: growth curves of co-cultures of E. coli DH5α pAKlux2 and E. coli DH5α, P. multocida CCUG 17976 ICEMh1PM22, or M. haemolytica ATCC 33396 ICEMh1PM22 in subinhibitory (0.5 MIC for susceptible P. multocida CCUG 17976 or M. haemolytica ATCC 33396) concentrations of oxytetracycline (left) or spectinomycin (right). Mean of 3 biological replicates with SEM. Lower panel: detection of E. coli luciferase production in co-cultures (as above) with either oxytetracycline (left) or spectinomycin (right). (F) Schematic of checkerboard synergy assay for drug interaction screening.
ICEMh1PM22 Was Retained in Transconjugants Following Repeated Laboratory Passage
Given the apparent cost of ICEMh1PM22 carriage, we hypothesized that hosts might lose the ICE following extended passage in non-selective conditions (i.e., without antimicrobials). To test this, the ICEMh1PM22 transconjugants for RifR P. multocida CCUG 17976 and M. haemolytica ATCC 33396 were repeatedly passaged over 150 days in media without antimicrobials or supplemented with sub-inhibitory concentrations of oxytetracycline, spectinomycin, or tylosin. The chosen concentrations were sub-inhibitory (0.5 MIC) to the parental WT strains. Every 30 days, CFU from each repeated passage were enumerated on MH agar (total CFU) and MH agar supplemented with each antimicrobial at a selective concentration (i.e., at a concentration where non-susceptibility was conferred by the ICEMh1PM22). The presence of ICEMh1PM22 was also confirmed in selected isolated colonies by PCR (not shown). This showed that repeated passage of ICEMh1PM22-carrying strains did not result in any measurable loss of the ICE (Figure 3D). In all cases there was no statistical difference between total and non-susceptible CFU counts in any repeated culture experiment. Thus, despite the fitness cost, ICEMh1PM22 was retained in the absence of selection in these experiments.
The Presence of ICEMh1PM22 Altered Collective Resistance to Sub-Inhibitory Antimicrobial Concentrations
Given that previous work had shown that ICEMh1PM22 could transfer into E. coli, we attempted to monitor transconjugation dynamics using luciferase-expressing E. coli in co-culture with RifR WT or ICEMh1PM22 transconjugants under antimicrobial pressure. We thought that increased luciferase production might indicate conjugation events. Therefore, oxytetracycline (0.125 mg/L) and spectinomycin (32 mg/L) were selected as bacteriostatic antimicrobials and used at 0.5 MIC for both P. multocida and M. haemolytica RifR WT. At these concentrations, oxytetracycline was sub-inhibitory to E. coli DH5α pAKlux2 (MIC: ∼1 mg/L), but spectinomycin was inhibitory (MIC: ∼16 mg/L). Co-cultures of the transconjugants or RifR WT with E. coli DH5α pAKlux2 were monitored for OD600 and luciferase production over 12 h. For oxytetracycline, increased luciferase production was observed for both P. multocida and M. haemolytica ICEMh1PM22 co-cultures compared to the ICE-minus RifR WT (Figure 3E, left). For spectinomycin, luciferase production was observed only in co-cultures of E. coli and the ICEMh1PM22 P. multocida transconjugant (Figure 3E, right). However, increased luciferase production was not due to E. coli acquisition of ICEMh1PM22, as non-susceptible E. coli could not be recovered on media supplemented with either tetracycline or spectinomycin at concentrations known to be selective for ICEMh1PM22 E. coli transconjugants (not shown). These findings suggest that the presence of ICEMh1PM22 negatively affected the efficacy of these antimicrobials against E. coli in co-culture, without horizontal gene transfer.
Antimicrobials Used to Treat BRD Exhibit No Synergistic Interactions Against ICEMh1PM22-Bearing P. multocida and M. haemolytica
Lastly, because ICEMh1PM22 harbors an extensive repertoire of antimicrobial resistance genes for drugs in frequent and common use for BRD, we investigated whether or not combination antimicrobial therapies were effective against P. multocida or M. haemolytica ICEMh1PM22 transconjugants. Checkerboard antimicrobial synergy assays (Figure 3F) were performed to test for (I) synergism between combinations of two antimicrobials against which ICEMh1PM22 confers non-susceptibility (i.e., oxytetracycline, spectinomycin, tilmicosin, tylosin, and sulfamethazine; Table 1), and (II) synergism between selected combinations of antimicrobials against which ICEMh1PM22 confers non-susceptibility (i.e., oxytetracycline, spectinomycin, and tylosin) and those that it does not (i.e., penicillin G, enrofloxacin, florfenicol, ceftiofur, and chlortetracycline). Following calculation and strict interpretation of the FICI scores for each combination, we found that no pairwise combination of oxytetracycline, spectinomycin, tilmicosin, tylosin, and sulfamethazine demonstrated synergism (Table 2). Likewise, no combination of a non-effective antimicrobial with any effective antimicrobial tested exhibited synergy (Table 3), and when present, susceptibility was due to the action of only the effective antimicrobial.
TABLE 1.
MIC changes in transconjugants.
| Antimicrobial | MICa | |||
| M. haemolytica ATCC 33396b | M. haemolytica ATCC 33396b × PM22 transconjugantc | P. multocida CCUG 17976b | P. multocida CCUG 17976b × PM22 transconjugantc | |
| Tetracyclines | ||||
| Oxytetracycline | 0.250 | 16 | 0.125 | 16 |
| Chlortetracycline | 0.125 | 2 | 0.125 | 1 |
| Macrolides | ||||
| Tylosin | 32 | 256 | 32 | 256 |
| Tilmicosin | 4 | 64 | 2 | 64 |
| Gamithromycin | 2 | 64 | 1 | 64 |
| Aminoglycosides | ||||
| Spectinomycin | 64 | 1024 | 64 | 1024 |
| Neomycin | 16 | 256 | 4 | 256 |
| β-lactams | ||||
| Penicillin G | 0.125 | 0.125 | 0.125 | 0.125 |
| Ampicillin | 0.250 | 0.250 | 0.125 | 0.125 |
| Ceftiofur | 0.125 | 0.125 | 0.125 | 0.125 |
| Sulfonamides | ||||
| Sulfamethazine | 128 | 1024 | 128 | 1024 |
| Sulfamethoxazole | 128 | 512 | 128 | 1024 |
| Others | ||||
| Enrofloxacin | 0.250 | 0.250 | 0.250 | 0.250 |
| Tiamulin | 8 | 8 | 4 | 8 |
| Clindamycin | 2 | 1024 | 4 | 1024 |
| Florfenicol | 1 | 1 | 1 | 1 |
aMinimum inhibitory concentration (MIC). The highest MIC resulting from three experiments is given. bMIC for spontaneous rifampicin-resistant mutant (Mutant MIC was equivalent to WT for all antimicrobials tested). cTransconjugants were selected for with 10 mg/L of tetracycline.
TABLE 2.
Results of checkerboard synergy testing (FICIs) with non-susceptible antimicrobial combinations.
| Antimicrobial | Rangea (mg/L) | MICALONEb (mg/L) | Combination FICIMEAN ± SD (FICILOW)c | ||||
| Spectinomycin | Tilmicosin | Tylosin | Sulfamethazine | Synergy (FICIMEAN ≤0.5) | |||
| Mannheimia haemolytica ATCC 33396 × PM22 transconjugant | |||||||
| Oxytetracycline | 1–64 | 16 | 0.84 ± 0.23 (0.63) | 0.57 ± 0.24 (0.31) | 0.98 ± 0.22 (0.63) | 0.99 ± 0.17 (0.75) | No interactions |
| Spectinomycin | 64–4096 | 1024 | 0.51 ± 0.30 (0.25) | 0.76 ± 0.63 (0.31) | 0.74 ± 0.65 (0.28) | No interactions | |
| Tilmicosin | 4–256 | 32 | 0.60 ± 0.28 (0.38) | 0.54 ± 0.09 (0.38) | No interactions | ||
| Tylosin | 16–1024 | 256 | 0.63 ± 0.09 (0.53) | No interactions | |||
| Sulfamethazine | 32–2048 | 1024 | No interactions | ||||
| Pasteurella multocida CCUG 17976 × PM22 transconjugant | |||||||
| Oxytetracycline | 1–64 | 16 | 1.23 ± 0.18 (1.06) | 0.58 ± 0.05 (0.50) | 6.59 ± 5.82 (2.00) | 1.21 ± 0.18 (1.03) | No interactions |
| Spectinomycin | 64–4096 | 1024 | 0.60 ± 0.28 (0.38) | 0.59 ± 0.28 (0.38) | 1.11 ± 0.09 (1.00) | No interactions | |
| Tilmicosin | 4–256 | 32 | 0.58 ± 0.28 (0.38) | 0.93 ± 0.25 (0.56) | No interactions | ||
| Tylosin | 16–1024 | 256 | 0.84 ± 0.23 (0.56) | No interactions | |||
| Sulfamethazine | 32–2048 | 1024 | No interactions | ||||
aRange of antimicrobial concentrations tested. bMinimum inhibitory concentration (MIC) of antimicrobial tested alone. cFractional inhibitory concentration index (FICI). The FICI values are given as the mean ± standard deviation resulting from three experiments. FICI values in brackets indicate the lowest value for the given combination. Synergism: ≤0.5; No interaction, ≥0.5.
TABLE 3.
Results of checkerboard synergy testing (FICIs) with non-susceptible and susceptible antimicrobial combinations.
| Antimicrobial | Rangea (mg/L) | MICALONEb (mg/L) | Combination FICIMEAN ± SD (FICILOW)c | |||
| Oxytetracycline | Spectinomycin | Tylosin | Synergy (FICIMEAN ≤0.5) | |||
| Mannheimia haemolytica ATCC 33396 × PM22 transconjugant | ||||||
| Penicillin G | 0.0078–0.5 | 0.125 | 1.11 ± 0.09 (1.00) | 0.59 ± 0.25 (0.25) | 0.65 ± 0.09 (0.56) | No interactions |
| Enrofloxacin | 0.0078–0.5 | 0.250 | 0.64 ± 0.30 (0.31) | 0.70 ± 0.29 (0.38) | 0.57 ± 0.23 (0.31) | No interactions |
| Florfenicol | 0.0625–4 | 1 | 0.68 ± 0.22 (0.50) | 0.61 ± 0.24 (0.38) | 0.81 ± 0.23 (0.56) | No interactions |
| Ceftiofur | 0.0313–2 | 0.125 | 1.28 ± 0.18 (1.06) | 1.28 ± 0.18 (1.06) | 3.29 ± 1.23 (1.50) | No interactions |
| Chlortetracycline | 0.0625–4 | 1 | 1.21 ± 0.18 (1.03) | 1.05 ± 0.51 (0.56) | 0.57 ± 0.24 (0.31) | No interactions |
| Pasteurella multocida CCUG 17976 × PM22 transconjugant | ||||||
| Penicillin G | 0.0078–0.5 | 0.125 | 0.93 ± 0.25 (0.56) | 0.65 ± 0.09 (0.56) | 1.13 ± 0.10 (1.00) | No interactions |
| Enrofloxacin | 0.0078–0.5 | 0.250 | 1.23 ± 0.18 (1.06) | 0.98 ± 0.22 (0.63) | 0.99 ± 0.20 (0.63) | No interactions |
| Florfenicol | 0.0625–4 | 1 | 0.74 ± 0.25 (0.50) | 0.53 ± 0.09 (0.56) | 0.61 ± 0.24 (0.38) | No interactions |
| Ceftiofur | 0.0313–2 | 0.125 | 1.14 ± 0.11 (1.00) | 1.28 ± 0.18 (1.06) | 1.14 ± 0.11 (1.00) | No interactions |
| Chlortetracycline | 0.0625–4 | 1 | 1.13 ± 0.10 (1.00) | 1.13 ± 0.10 (1.00) | 0.61 ± 0.24 (0.38) | No interactions |
aRange of antimicrobial concentrations tested. bMinimum inhibitory concentration (MIC) of antimicrobial tested alone. cFractional inhibitory concentration index (FICI). The FICI values are given as the mean ± standard deviation resulting from three experiments. FICI values in brackets indicate the lowest value for the given combination. Synergism, ≤0.5; No interaction, ≥0.5.
Discussion
Many ICEs site-specifically insert into tRNAs and, in general, are not able to change their location within the same host cell (Burrus et al., 2002; Michael et al., 2011b). Previous research has proven that ICEPmu1 and other ICEMh1-like elements integrate into identical or near identical tRNA-leu (Michael et al., 2011b; Eidam et al., 2014; Beker et al., 2018; Bhatt et al., 2018). Here, we confirmed these findings, and provided further explanation for the tRNA-leuUUG integration specificity of ICEMh1-like elements. The ICEMh1-like direct repeat is highly conserved because it is the basis for the tRNA-leuUUG anticodon loop. Likewise, the attL site is conserved because it forms the tRNA’s TψC loop. Unexpectedly, the attR site does not conserve the tRNA’s D arm and loop. We speculate that this is the reason that integration seems to result in one functional and one disrupted tRNA-leuUUG as the terminus with attL and the attL-associated direct repeat maintains the complete tRNA, but the attR terminus does not complete the second tRNA fragment. Examination of several genomes with ICEMh1-like elements suggests this is the most frequent outcome of ICE insertion, but there are also examples where the ICE is flanked by complete tRNA-leuUUG, as well as ICE insertions where a terminus (i.e., the direct repeat) cannot be identified. We speculate that these represent on-going recombination post-integration, and may be mechanisms through which ICEs become non-mobile, or acquire new genes from the host chromosome.
The highly conserved structural features of tRNAs evidently provide an excellent site for exploitation by ICEs and other mobile genetic elements. Many ICEs from different bacterial species integrate into various tRNAs (Burrus et al., 2002; Johnson and Grossman, 2015; Castillo et al., 2017). There are several possible advantages to this. Firstly, all known life uses tRNAs, and organisms encode multiple tRNAs for specific amino acids (i.e., the basis of redundancy in the genetic code). Although specific tRNAs may be absent or altered, they are still widely present in multiple bacterial species and provide for a broad host range. Secondly, tRNAs have redundancy, so ICE insertion into a tRNA need not be lethal even if the tRNA is disrupted. This would facilitate the transmission of the mobile genetic element without exerting a deleterious effect on the host, which might be expected if the insertion site was a highly conserved and a non-redundant housekeeping gene. However, although single tRNA deletions may have no appreciable phenotypic effects under certain conditions, some tRNAs for a particular amino acid are known to contribute differently to cellular fitness in challenging conditions (Bloom-Ackermann et al., 2014). Thirdly, the structural features of tRNAs provide for conserved recombination sites. Although relatively short sequences (<90 bp), tRNAs contain multiple stretches of complementary base pairs that provide tertiary structure, enabling interactions with amino acids, mRNA, and the ribosome. In the case of ICEMh1-like elements and tRNA-leuUUG, the 11 bp that constitute the ICE’s direct repeat and the tRNA’s anticodon loop were conserved with 100% identity in all ICEMh1-like-containing genomes. Exploiting the anticodon loop in this manner also likely enables a broader host range compared to more variable features of the tRNA. Likewise, in the case of tRNA-leuUUG, we speculate that other conserved bp in the tRNA provide for the attL and attR recognition and/or recombination sites of the putative XerCD tyrosine recombinases. Although we did not explore that possibility here, this would be consistent with known XerCD mechanisms (Castillo et al., 2017).
It is difficult to accurately assess the potential host range of ICEMh1-like elements. Here, we identified bacterial genera potentially receptive to the ICE based solely on the presence tRNA-leu, further refined by eliminating hits that did not contain the exact direct repeat and palindrome conserved at the attL terminus. As expected, related Pasteurellaceae and γ-Proteobacteria were most frequently represented, but genera from all Proteobacteria classes contained a receptive tRNA-leu. Most Gram-positive organisms may not be receptive to ICEMh1-like elements, but some Bacilli and Clostridia did harbor a receptive tRNA-leu. We attempted to transfer ICEMh1PM22 to Clostridium difficile, but did not successfully recover transconjugants (not shown). The host range of other conjugative elements in nature has been shown to be narrower than that observed in in vitro experiments (Rice, 1998). There are also other factors that could prevent ICEMh1-like element transmission to bacteria, despite the presence of a receptive tRNA-leu. These may include phage (Lin et al., 2011), certain metabolites (Cabezón et al., 2017), and CRISPR-Cas systems (Zhang et al., 2013). Furthermore, ICEs transfer to recipients relatively infrequently, remaining silent and integrated, with excision and horizontal transmission occurring at low frequencies within the population (Delavat et al., 2017). The putative host range of ICEMh1-like elements identified here includes many pathogens capable of causing significant human and veterinary disease. The transfer of AMR determinants into such bacteria could significantly compromise antimicrobial therapy.
Despite a fitness cost in the absence of antimicrobial selection, we also found that ICEMh1PM22 transconjugants do not lose the ICE after long-term passage. This is likely due to ‘addiction’ via the action of toxin-antitoxin genes common to most ICE (Harms et al., 2018). For practical purposes this implies that the acquisition of an AMR-carrying ICE is not reversible at the single-cell level. Given that ICEMh1-like elements may have a fitness cost to the host cell in the absence of antimicrobials, elimination of antimicrobial usage could help non-carriers to outcompete hosts. However, ICEMh1-like elements frequently contain metal-resistance and other genes conferring benefits under dynamic selective pressures in vivo, so elimination of antimicrobial pressure may not be effective at dampening ICE-carrying populations. We also used ICEMh1PM22 transconjugants to test for synergistic combinations of antimicrobials, hoping to identify combinations that might be effective against AMR BRD pathogens, potentially reducing antimicrobial usage and restoring drug effectiveness. Ultimately, we did not detect any synergistic interactions between the drugs tested using these organisms. This was not unexpected, and illustrates the relatively limited therapeutic options available to veterinarians for BRD treatment. The presence of multiple AMR determinants conferring resistance to most drug classes in these elements also underscores the dwindling effectiveness of antimicrobials. In conclusion, our results suggest that there is no easy solution to the emerging problem of AMR ICE in BRD pathogens using current antimicrobials and usage practices.
Data Availability Statement
The PM22 sequence was deposited in GenBank under accession number CP045724.
Author Contributions
AC and TM: conceptualization. AC and RZ: methodology, software, validation, formal analysis, and data curation. AC: investigation, visualization, writing, and original draft preparation. TM: resources, supervision, project administration, and funding acquisition. AC, RZ, and TM: writing, review, and editing.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Cassidy Klima for the initial isolation of the PM22 P. multocida isolate, and thank Reuben Ha, Cheyenne Conrad, Shaun Cook, and Wendi Smart for laboratory assistance and maintenance.
Footnotes
Funding. This research was funded by the Alberta Agriculture and Forestry Strategic Research and Development and the Beef Cattle Research Council (BCRC) – Agriculture and Agri-Food Canada Beef Cluster. AC was supported by an NSERC Postdoctoral Fellowship.
References
- Beker M., Rose S., Lykkebo C. A., Douthwaite S. (2018). Integrative and conjugative elements (ICEs) in Pasteurellaceae species and their detection by multiplex PCR. Front. Microbiol. 9:1329. 10.3389/fmicb.2018.01329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt K., Timsit E., Rawlyk N., Potter A., Liljebjelke K. (2018). Integrative conjugative element ICEHs1 encodes for antimicrobial resistance and metal tolerance in Histophilus somni. Front. Vet. Sci. 5:153. 10.3389/fvets.2018.00153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom-Ackermann Z., Navon S., Gingold H., Towers R., Pilpel Y., Dahan O. (2014). A comprehensive tRNA deletion library unravels the genetic architecture of the tRNA pool. PLoS Genet. 10:e1004084. 10.1371/journal.pgen.1004084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrus V., Pavlovic G., Decaris B., Guédon G. (2002). Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46 601–610. 10.1046/j.1365-2958.2002.03191.x [DOI] [PubMed] [Google Scholar]
- Cabezón E., De La Cruz F., Arechaga I. (2017). Conjugation inhibitors and their potential use to prevent dissemination of antibiotic resistance genes in bacteria. Front. Microbiol. 8:2329. 10.3389/fmicb.2017.02329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron A., McAllister T. A. (2016). Antimicrobial usage and resistance in beef production. J. Anim. Sci. Biotechnol. 7:68. 10.1186/s40104-016-0127-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron A., Klima C. L., Ha R., Gruninger R. J., Zaheer R., McAllister T. A. (2018). A novel aadA aminoglycoside resistance gene in bovine and porcine pathogens. mSphere 3:e00568-17. 10.1128/mSphere.00568-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo F., Benmohamed A., Szatmari G. (2017). Xer site specific recombination: double and single recombinase systems. Front. Microbiol. 8:453. 10.3389/fmicb.2017.00453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P. P., Lowe T. M. (2015). GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44 D184–D189. 10.1093/nar/gkv1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P. P., Lowe T. M. (2019). tRNAscan-SE: searching for tRNA Genes in Genomic Sequences. Methods Mol. Biol. 1962 1–14. 10.1007/978-1-4939-9173-0_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI, (2008). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Collected From Animals; Approved Standard. CLSI Document M31-A3. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- CLSI, (2018). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. CLSI Standard M07. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Crooks G. E., Hon G., Chandonia J.-M., Brenner S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14 1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A. E., Mau B., Perna N. T. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delavat F., Miyazaki R., Carraro N., Pradervand N., Van Der Meer J. R. (2017). The hidden life of integrative and conjugative elements. FEMS Microbiol. Rev. 41 512–537. 10.1093/femsre/fux008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidam C., Poehlein A., Leimbach A., Michael G. B., Kadlec K., Liesegang H., et al. (2014). Analysis and comparative genomics of ICE Mh1, a novel integrative and conjugative element (ICE) of Mannheimia haemolytica. J. Antimicrob. Chemother. 70 93–97. 10.1093/jac/dku361 [DOI] [PubMed] [Google Scholar]
- FDA, (2016). Approved Animal Drug Products (Green Book). Maryland, MA: Food and Drug Administration. [Google Scholar]
- Harms A., Brodersen D. E., Mitarai N., Gerdes K. (2018). Toxins, targets, and triggers: an overview of toxin-antitoxin biology. Mol. Cell. 70 768–784. 10.1016/j.molcel.2018.01.003 [DOI] [PubMed] [Google Scholar]
- Johnson C. M., Grossman A. D. (2015). Integrative and conjugative elements (ICEs): what they do and how they work. Annu. Rev. Genet. 49 577–601. 10.1146/annurev-genet-112414-055018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsi A., Lawrence M. L. (2007). Broad host range fluorescence and bioluminescence expression vectors for gram-negative bacteria. Plasmid 57 286–295. 10.1016/j.plasmid.2006.11.002 [DOI] [PubMed] [Google Scholar]
- Klima C. L., Cook S. R., Zaheer R., Laing C., Gannon V. P., Xu Y., et al. (2016). Comparative genomic analysis of Mannheimia haemolytica from bovine sources. PLoS One 11:e0149520. 10.1371/journal.pone.0149520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klima C. L., Zaheer R., Cook S. R., Booker C. W., Hendrick S., Alexander T. W., et al. (2014). Pathogens of bovine respiratory disease in North American feedlots conferring multidrug resistance via integrative conjugative elements. J. Clin. Microbiol. 52 438–448. 10.1128/JCM.02485-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A., Jimenez J., Derr J., Vera P., Manapat M. L., Esvelt K. M., et al. (2011). Inhibition of bacterial conjugation by phage M13 and its protein g3p: quantitative analysis and model. PLoS One 6:e19991. 10.1371/journal.pone.0019991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael G. B., Kadlec K., Sweeney M. T., Brzuszkiewicz E., Liesegang H., Daniel R., et al. (2011a). ICE Pmu1, an integrative conjugative element (ICE) of Pasteurella multocida: analysis of the regions that comprise 12 antimicrobial resistance genes. J. Antimicrob. Chemother. 67 84–90. 10.1093/jac/dkr406 [DOI] [PubMed] [Google Scholar]
- Michael G. B., Kadlec K., Sweeney M. T., Brzuszkiewicz E., Liesegang H., Daniel R., et al. (2011b). ICE Pmu1, an integrative conjugative element (ICE) of Pasteurella multocida: structure and transfer. J. Antimicrob. Chemother. 67 91–100. 10.1093/jac/dkr411 [DOI] [PubMed] [Google Scholar]
- Mosier D. (2015). Review of BRD pathogenesis: the old and the new. Anim. Health Res. Rev. 15 166–168. 10.1017/S1466252314000176 [DOI] [PubMed] [Google Scholar]
- Odds F. C. (2003). Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrob. Chemother. 52 1–1. 10.1093/jac/dkg301 [DOI] [PubMed] [Google Scholar]
- Rice L. B. (1998). Tn916 family conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob. Agents Chemother. 42 1871–1877. 10.1128/aac.42.8.1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama N. R., Shepherd B., Falkow S. (2004). Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J. Bacteriol. 186 7926–7935. 10.1128/jb.186.23.7926-7935.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Rho M., Tang H., Doak T. G., Ye Y. (2013). CRISPR-Cas systems target a diverse collection of invasive mobile genetic elements in human microbiomes. Genome Biol. 14:R40. 10.1186/gb-2013-14-4-r40 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The PM22 sequence was deposited in GenBank under accession number CP045724.