Abstract
Fusarium diseases are significant hindrances to food plant production and are very difficult to control, especially soilborne diseases caused by F. oxysporum. First I outline the Fusarium diseases and introduce examples of the recent outbreak of Fusarium diseases in Japan. Then I summarize my studies on (1) the control of Fusarium diseases by biological agents and by inducing resistance to diseases in plants, (2) the specific detection of forms and races in F. oxysporum using immunological measures and molecular measures based on phylogeny and pathogenicity-determining genes, and (3) molecular and genetic studies on Fusarium diseases, including evolutionary, genetic, and genomic analyses of the emergence and divergence of forms and races in F. oxysporum.
Keywords: Fusarium diseases, biological control, induced resistance, detection, molecular plant-microbe interactions, genome analyses
Introduction
For three decades, I have studied epidemics, control techniques such as biological control and inducing disease resistance in plants with chemical and physical stimuli, and specific detection techniques of Fusarium diseases, mainly of diseases caused by soilborne F. oxysporum. In addition, I have analyzed the mechanisms of pathogenic evolution and differentiation in F. oxysporum by molecular, genetic, and genomic methods.
In the 1980s, biochemical techniques, such as electrophoresis to separate proteins, and immunological techniques could be used for fungal studies. In the 1990s, we began using molecular techniques, for example, polymerase chain reaction (PCR), DNA sequencing, and gene targeting, to analyze the genes and their function in fungi. In the 2000s, project consortia sequenced the genomes of several fungal species, including Fusarium spp., and in the 2010s, the comparative analyses of genomes of Fusarium spp. revealed their genome and chromosomal structure and function. The finding of an accessory chromosome from F. oxysporum was one of the outcomes. For these several years, SMRT® sequencing has allowed us to analyze and compare the genomes of isolates of Fusarium spp. easily. Advances in technology have enabled us to try innovative methods to accelerate Fusarium researches.
1. Fusarium diseases
1.1. Fusarium spp.
Fusarium is a fungal genus belonging to Phylum Ascomycota, Subphylum Pezizomycotina, Class Sordariomycetes, Order Hypocreales, and Family Nectriaceae.1) Fusarium is a huge genus in which more than 1,500 species are estimated.
Fusarium spp. are ubiquitous in the environment, and several strains presenting pathogenicity to plants or animals and producing mycotoxins are reported. Fusarium oxysporum, F. solani, F. fujikuroi, and F. graminearum are the representative species known as plant-pathogenic Fusarium.
1.2. Fusarium oxysporum
Fusarium oxysporum produces a white, pale blue, purple, or light orange mycelial colony on standard media such as potato dextrose agar. Slightly arched macroconidia, usually with 1 to 4 septa, are mainly formed in sporodochia, and unicellular, colorless, and oval microconidia are formed in a false-headed manner on short conidiophores branching from the mycelium. The cell wall of mycelial cells and/or some macroconidial cells becomes thickened to form chlamydospores, which are durable and survive for several decades in soil.2) When a shake culture is performed in a liquid medium such as potato dextrose broth, a large number of yeast-like bud cells are formed, which are considered to be equivalent to the transfer form in plant vessels. The perfect stage of this fungus has not yet been found.
Fusarium oxysporum is also a ubiquitous fungus widely distributed in the environment. Although F. oxysporum was reported to be an airborne fungus in Shosoin in Nara, one of the oldest buildings in Japan built in the eighth century AD and one of the world heritages3) and as a pathogen of humans who have lost immunity,4) the fungus is famous as a soilborne pathogen of plants.
Soilborne plant diseases are those caused by infection of pathogens in soil via the roots. F. oxysporum is representative of soilborne pathogens. It inhabits the soil for a long time in the form of chlamydospores, penetrates the roots, extends in the tissues, colonizes and metastasizes in xylem vessels, and causes systemic yellowing, wilting, and death in plants.
1.3. Differentiation of forms and races in phytopathogenic F. oxysporum
Since F. oxysporum causes diseases in various plants, it is often thought to be versatile; however, the range of plant species that can host each isolate of F. oxysporum is strictly and clearly defined. More than 120 forms (formae speciales; ff. spp.) have been determined based on the range of plant species that can be hosts (Table 1). For example, the isolates that causes tomato wilt are determined to be f. sp. lycopersici, and this form never causes disease in the other plant species, such as cabbage and banana. Similarly, f. sp. cubense, the banana Panama pathogen, never causes disease in tomato or cabbage plants.
Table 1. Examples of forms (formae speciales) in plant pathogenic Fusarium oxysporum.
| Form (forma specialis) | Example of host plant |
|---|---|
| apii | Celery |
| cepae | Onion, Green onion |
| conglutinans | Cabbage |
| cubense | Banana |
| cucumerinum | Cucumber |
| lycopersici | Tomato |
| melongena | Egg plant |
| melonis | Melon |
| niveum | Water melon, Melon |
| pisi | Pea |
| raphani | Daikon radish |
| rapae | Brassica campestris |
| spinaceae | Spinach |
| tulipae | Tulipe |
Within a forma specialis, races are frequently distinguished by their specific pathogenicity to different cultivars. For example, three races (1–3) have been reported in f. sp. lycopersici, the tomato wilt fungus, which are classified based on their virulence on tomato cultivars carrying different resistance genes (I, I2, and I3; Table 2). The relationship between tomato cultivars and races of the tomato wilt pathogen can be explained by the “gene-for-gene theory” (Table 2).5) Original descriptions of races 1, 2, and 3 of f. sp. lycopersici appeared before 1895 in England, in 1939 in the USA, and in 1978 in Australia, respectively. In Japan, races 1, 2, and 3 were first reported, all in Fukuoka, in 1905, 1966, and 1997, respectively.6)
Table 2. Relationship between the tomato wilt fungus, Fusarium oxysporum f. sp. lycopersici (FOL) races and tomato cultivars.
| FOL race (AVR genes) | Tomato cultivar (R genes) | |||
|---|---|---|---|---|
| Ponderosa(i i2 i3) | Momotaro(I i2 i3) | Walter(I I2 i3) | Block(I I2 I3) | |
| 1 (AVR1 AVR2 AVR3) | S | R | R | R |
| 2 (— AVR2 AVR3) | S | S | R | R |
| 3 (— avr2 AVR3) | S | S | S | R |
S, compatible; R, incompatible
1.4. Examples of recent outbreaks of F. oxysporum diseases
An emergence of wilt caused devastating damage to tomato production in greenhouses in Hidaka, Kochi, Japan, in 2008. The genotype of the affected tomato cultivar was I I2 i3, suggesting the presence of F. oxysporum f. sp. lycopersici race 3 (Table 2), which had not previously been reported in Kochi. Certain characteristics of the pathogenic isolates from Kochi differed from those of reported Japanese race 3 isolates from Fukuoka, Hokkaido, and Kumamoto, and we determined the Kochi isolate to be a new biotype of race 3.6)
Panama disease caused by F. oxysporum f. sp. cubense is one of the most destructive diseases of banana plants and has affected production worldwide. Three races, 1, 2, and 4, have been reported. In the 1950s, production of the cultivar ‘Gros Michel’ in Central America dramatically decreased due to an epidemic of race 1. Because race 1 affected this cultivar severely, ‘Gros Michel’ has been replaced with the cultivar ‘Cavendish,’ which is resistant to race 1. Currently, ‘Cavendish’ accounts for about 99% of banana trading. Race 4, which is pathogenic to ‘Cavendish,’ was first reported in the 1960s in subtropical areas, such as Taiwan, and now it is designated subtropical race 4 (SR4). Since the late 1980s, devastating damage by F. oxysporum f. sp. cubense to the cultivar ‘Cavendish’ has been reported not only in subtropical areas but also in tropical areas. The pathogen of this severe epidemic was genetically distinct from SR4 and has been designated tropical race 4 (TR4). At present, TR4 is causing severe damage to banana plantations in Asia, such as the Philippines.
Although there had been no official record of Panama disease in Japan, in 2016, yellowing, browning, and wilt were observed on the cultivar ‘Shima-banana’ in Okinawa. We determined that the disease is caused by a Panama disease fungus, though fortunately not by TR4. This was the first official report of an occurrence of Panama disease in banana plants in Japan.7)
Although F. oxysporum is known as a soilborne pathogen, the pathogen is initially transmitted with seeds to the field and colonizes the soil. The pea wilt pathogen, F. oxysporum f. sp. pisi, is a famous seedborne disease occurring mainly in North America, Europe, Australia, and New Zealand and is monitored by the Plant Protection Station (PPS), Ministry of Agriculture, Forestry and Fisheries (MAFF), of the Japanese government as a potentially invasive disease. Since 2002, occurrences of the disease have been reported in several parts of Japan, and MAFF has been attempting to eradicate this disease under the Plant Protection Act. In cooperation with the PPS, we have established a specific detection system for f. sp. pisi, which will be described later.
F. oxysporum f. sp. batatas is the causal agent of sweet potato wilt disease. Wilt was a big issue in sweet potato production in the 1980s, and it was controlled with the use of tolerant cultivars and benzimidazole fungicides. Recently, as a result of repeated use of benzimidazole fungicides to control the fungus in the vine cutting at transplanting, benomyl-resistant strains of f. sp. batatas have emerged in several parts of Japan, and the disease is again attracting attention as an obstacle to sweet potato production.8)
2. Control of Fusarium diseases
2.1. Current general status of soilborne Fusarium disease control
Once soilborne fusaria pathogens spread in the field, their removal is very difficult. Currently, in order to control the diseases caused by soilborne pathogens, soil disinfection using fumigants, such as chloropicrin, hot water, or solarization, or using resistant cultivars is popular. However, the efficacy obtained with these treatments often is less sufficient than expected, or there is a need to reduce their usage because of adverse effects on the environment.
In order to solve these problems, research of other techniques to control fusarium diseases has been advanced. Here, studies on biological control and inducing disease resistance in plants as alternatives to control fusarium diseases are introduced.
2.2. Biological control of Fusarium diseases
Biological control is a way of controlling plant diseases using live microorganisms. Trichoderma lignorum was registered as a fungicide based on the Agricultural Chemicals Regulation Law in Japan in 1954 to control Rhizoctonia disease in tobacco. This was the first registered case of a biofungicide in the world.
In these 3 to 4 decades, many studies have been conducted, aiming to establish biocontrol of Fusarium diseases, including soilborne F. oxysporum diseases and rice ‘Bakanae disease caused by F. fujikuroi. A nonpathogenic strain of F. oxysporum was registered in 2002 as a biofungicide to control soilborne wilt of sweet potato plants caused by F. oxysporum f. sp. batatas.9) On the other hand, in 2003, Trichoderma atroviride was registered as a biofungicide to control rice ‘Bakanae’ by seed or nursery-box treatment, and this was followed by Talaromyces flavus in 2007. Both have been in the market.
We have isolated a nonpathogenic Fusarium sp. W5 from a rice plant tissue that carries biocontrol activity against rice ‘Bakanae.’ Using the isolate, we established a novel treatment method of biocontrol agent, spraying a spore suspension of W5 onto rice flowers, and demonstrated its strong biocontrol efficacy to reduce the seedborne ‘Bakanae’ (patents, Japan 6241001, Korea 10-1770656, Indonesia IDP000052506, United States 10264796 B2). We visualized the interaction between W5 and the ‘Bakanae’ pathogen using transformants expressing fluorescent proteins on/in rice plant tissues. W5 inhibited hyphal extension of the ‘Bakanae’ pathogen on/in rice flowers and seedlings, possibly by competition, and survived on/in rice seeds for at least six months.
Recently, we also found that soil-drench application of the spore suspension of W5 reduced the incidences of soilborne Fusarium diseases, such as tomato wilt and banana Panama disease.
2.3. Possible application of induced resistance to control soilborne diseases
During growth, plants are continuously challenged by a wide spectrum of environmental stimuli, including abiotic (e.g., extreme temperatures, drought, and mineral imbalance) and biotic (e.g., pests and microbes) ones. Plants recognize these stimuli and respond appropriately to them.
Plants usually protect themselves from microbes by activating defense reactions such as systemic acquired resistance (SAR) after recognizing microbial stimuli. However, pathogenic microbes can weaken or invalidate plant defenses against invasion. Therefore, improving the plant defenses prior to the approach of pathogenic microbes seems to be a good way to control the diseases.
Interestingly, plants’ response pathways to abiotic and biotic stimuli partly overlap. Thus, when plants are exposed to abiotic stimuli, the plants can acquire an improved defense by chance. For example, chemical stimuli, such as probenazole (PBZ), acibenzolar-S-methyl (ASM), tiadinil (TDL), and isotianil, have been used as plant activators that do not have direct antipathogen activity but can induce disease resistance in plants.
We found that foliar spray with validamycin A (VMA) effectively controls soilborne Fusarium diseases such as tomato wilt10) and banana Panama disease11) by inducing SAR.12)
Recently, we reported that ultrasound, which refers to frequencies above the limit of human hearing, is a candidate for inducing resistance to pathogens in plants.13) We revealed that an aerial ultrasound of 40.5 kHz could induce disease resistance in tomato and rice plants when they were irradiated with an ultrasound of ca. 100 dB for 2 weeks during nursery season, and it reduced the incidence of wilt and blast diseases, respectively, when plants were inoculated with a pathogen 0 or 1 week after terminating irradiation. Quantitative reverse transcription PCR (RT-qPCR) showed that SAR was induced in the ultrasound-irradiated tomato tissues.
3. Diagnose of Fusarium diseases
3.1. Importance of diagnosis in a Fusarium disease management program
Earlier detection of plant pathogens in the field is essential for plant health certification and appropriate disease management, for example, soil disinfestation, the use of disease-resistant cultivars, and the application of fungicides. However, there had been no practical procedure to detect Fusarium spp. except the isolation and inoculation test using test plants.
What needs to be differentiated in disease diagnosis? In F. oxysporum, for example, there are several forms, each of which has a different host plant species, as I wrote in 1.3. Moreover, there are races, each of which has different host cultivars within a form. The question usually posed by farmers is whether they can grow, for example, tomatoes and also cultivar A of the tomato in their fields. This indicates that we need to differentiate the forms and races of F. oxysporum in the soil.
3.2. Immunological detection
Immunological detection is based on the specific reaction between antibody and antigen. Immunology provides rapid, specific, and sensitive tools for the detection of pathogens. In fact, various immunological procedures have been used commonly against human pathogens and plant viruses. Improved procedures tailored to specific applications are needed for the diagnosis of plant diseases caused by various pathogens.
I developed a monoclonal antibody (MoAb) that reacts specifically to Fusarium spp. and studied on the application of the MoAb to detect F. oxysporum.14) A direct tissue-blotted immunobinding assay (DT-IBA) was applied to transverse sections from the stems or crowns of tomato and cucumber plants. In this assay, samples of plants infected with F. oxysporum showed a positive reaction within 4 hr, suggesting the possibility using the MoAb for disease diagnosis in the field and in quarantine.14) I also established a gel penetrate-blotted immunobinding assay (GP-IBA) to detect Fusarium spp. in soil using the MoAb.15) The mycelia of F. oxysporum in infected stems, crowns, and roots of tomato and Japanese honewort plants were effectively detected by immunofluorescence assay (IFA) with a combination of the MoAb and FITC-conjugated anti-mouse IgM-sheep IgG, indicating that the IFA is a useful tool to observe the behavior of Fusarium spp. in plant tissue.
Although detection using an antibody has advantages such as low cost and ease of use, the MoAb recognized all of the isolates of Fusarium spp., including nonpathogenic strains, and it turned out that the immunological detection methods cannot satisfy the needs of farmers.
3.3. Detection based on phylogeny
The evolution of technology in the 1990s has enabled molecular biological tools, such as PCR, to be used for the detection of specific plant pathogens. Because we supposed that specific pathogen types, such as forms and races, correlate with molecular phylogenetic relationships, we attempted to design PCR primer sets that recognize the differences in the nucleotide sequences of the genes. We compared the partial nucleotide sequences of endo-polygalacturonase gene (pg1) and exo-polygalacturonase (pgx4) genes from isolates of F. oxysporum ff. spp. lycopersici and radicis-lycopersici and designed specific primer sets based on the nucleotide differences that appeared among the pathogenic types. A combination of amplifications with these primer sets seemed to effectively differentiate the two forms and races in f. sp. lycopersici.6,16) Kashiwa (2016) successfully detected f. sp. conglutinans, the cabbage yellows pathogen, specifically and rapidly via PCR and real-time PCR using the primer and probe set designed on pg1 gene.17)
However, additional phylogenetic analyses using numerous isolates let us know that the forms and races of the pathogen may not always correlate with molecular phylogenetic relationships.18)
3.4. Detection based on pathogenicity-related genes
As I describe in 4.4, we recently identified the genes determining forms and races in F. oxysporum. Based on this information, we have designed primer sets that specifically determine race 119) and race 320) by loop-mediated isothermal amplification (LAMP) using special primers and probes sets designed at the SNPs in the avirulence genes determining races.
We also developed three primer sets targeting SIX6, SIX13, and pisatin demethylase (PDA) genes for the specific determination of F. oxysporum f. sp. pisi, the pea wilt pathogen (1.4), by PCR and LAMP. Almost all of the isolates of f. sp. pisi carry SIX6 and SIX13 genes sharing by several formae speciales other than f. sp. pisi. The PDA gene is possessed only by f. sp. pisi. A closely related species, F. solani f. sp. pisi, the root rot disease pathogen, carries PDA but not SIX6 or SIX13. Now we can specifically distinguish F. oxysporum f. sp. pisi from other F. oxysporum and F. solani f. sp. pisi by PCR and LAMP, and these techniques are useful for sanitary testing of seeds at plant quarantine facilities and for pathogen survival assessment after soil disinfection in the field.21)
4. Molecular and genetic studies on Fusarium diseases
4.1. From biochemical research to molecular biological research
Researchers have long attempted to understand the mechanisms by which Fusarium causes symptoms such as wilt in plants and presents host specificity such as forms (formae speciales) and races. Beckman (1983) suggested several pathogenicity-related factors, such as polygalacturonase produced by F. oxysporum.22) Biochemical and cytological approaches predominated in 1980s.
In the 1990s, molecular techniques, such as the cloning of genes, PCR amplification of DNA fragments, gene disruption, and gene expression analyses, were developed and became applicable to fungal study. In 1998, pg1 was reported as the first determined pathogenicity-related gene in F. oxysporum.23,24) After that, many studies were performed, focusing on the genes that relate to pathogenicity and host specificity in F. oxysporum.
4.2. Coevolution of the tomato wilt pathogen with the tomato plant
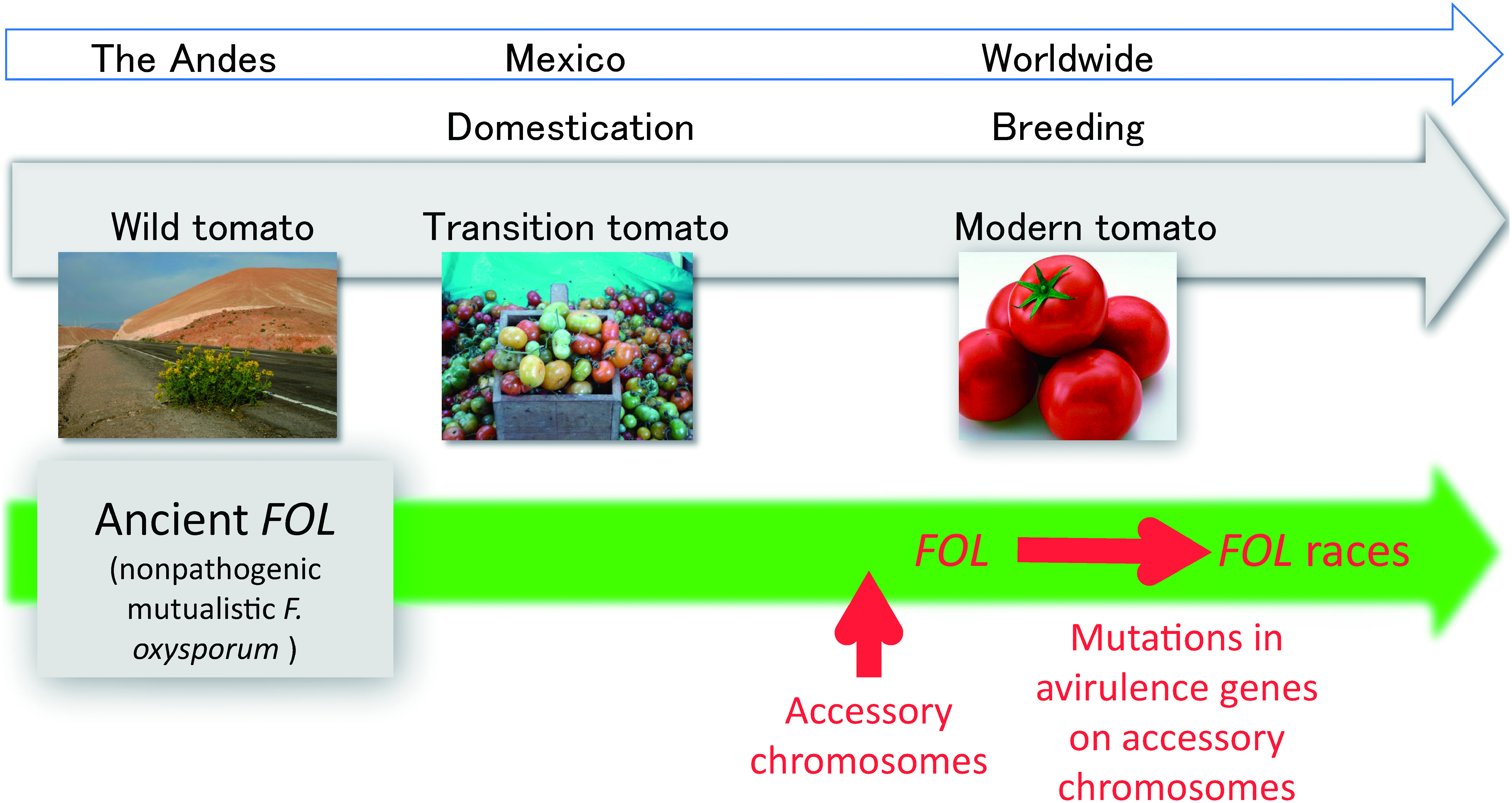
The tomato (Solanum lycopersicum) originated in Andean South America and spread prehistorically to Mexico, where it was domesticated. During the Spanish conquest in the 16th century, tomatoes were transmitted to Europe, where modern breeding of the tomato began. Today, the Andes continues to sustain wild Solanum species. Solanum lycopersicum var. cerasiforme, an apparent intermediate between wild and cultivated tomatoes, is currently found as a native-grown tomato in Mexico (transition tomato).
“When, where, and how did the tomato wilt pathogen (F. oxysporum f. sp. lycopersici; FOL) emerge?” This is a fundamental but difficult question to address. We hypothesized that FOL emerged from a mutualistic nonpathogenic F. oxysporum on the tomato during the tomato domestication and breeding processes. To verify the hypothesis, we collected more than 400 isolates of F. oxysporum from the tissues and rhizosphere of wild and transition tomatoes and compared them with FOL. They were all nonpathogenic to tomato cultivars tested. In the phylogenetic tree containing the F. oxysporum and FOL isolates, FOL isolates were found in three clades, which was consistent with the findings by Kawabe (2005).18) Several nonpathogenic isolates from the wild and transition tomatoes were grouped in FOL clades, whereas most of the F. oxysporum isolates were not, suggesting that these nonpathogenic isolates share common origins with FOL and that the origin of FOL existed with the wild tomato in the Andes.25)
4.3. Accessory chromosomes
Ma et al. (2010) found that FOL has a small chromosome (ca. 2.0 Mb) that is not necessary to survive.26) They also showed that a nonpathogenic F. oxysporum isolate gains virulence when it obtains the small chromosome from FOL. Now these small chromosomes are designated accessory chromosomes. They also found that the AVR genes determining races in FOL are located on the accessory chromosomes.
Interestingly, the FOL isolates in the three distinct clades (3.2)18) carry an identical set of AVR genes on accessory chromosomes. On the other hand, all of the nonpathogenic isolates from the wild and transition tomatoes in the same clades as FOL carry no AVR genes. Thus, the horizontal mobility of the accessory chromosome carrying pathogenicity-related genes has been hypothesized to be the driving force of the emergence of FOL in F. oxysporum (Fig. 1).
Fig. 1. Pictorial explanation of coevolution of the tomato wilt fungus, Fusarium oxysporum f. sp. lycopersici (FOL), and the tomato. The origin of FOL was the mutualistic nonpathogenic F. oxysporum that existed with the wild tomato in the Andes. During breeding, FOL emerged from the mutualistic nonpathogenic F. oxysporum by gaining accessory chromosomes carrying pathogenicity-related genes. The divergence of races in FOL occurred via mutations in avirulence genes (Table 2).
Other forms of F. oxysporum have an accessory chromosome(s) that carries pathogenicity-related genes in its own manner.27)
4.4. Mechanisms of race diversity
The relationship between the three races in FOL and tomato cultivars is explained in 1.3 (Table 2). The races are differentiated by the possession of AVR genes, and it was suggested that FOL races emerged as follows: race 1 (AVR1 AVR2 AVR3) lost the AVR1 locus and became race 2 (avr1null AVR2 AVR3), which escaped recognition by the I gene; a nucleotide substitution in race 2 AVR2 resulted in emergence of race 3 (avr1null avr2 AVR3), which evaded recognition by both I and I2.28) The arms race, or the “boom-and-bust” cycle between tomato and tomato wilt pathogen (FOL), might be continuing.
4.5. Evolution of races in the field
The 2008 outbreak of tomato wilt in Kochi was caused by a new biovar of race 3 of FOL (1.3). Inami (2012) found that although the pathogen was race 3, it has an AVR1 gene that is truncated by the insertion of a transposon hormin, which belongs to the hAT family.29) This provides evidence that mobile genetic elements may be one of the driving forces underlying race evolution and proposes an additional path for the emergence of FOL races: race 2 emerged from race 1 by transposon insertion into AVR1, not by deletion of the AVR1 locus; then a point mutation in race 2 AVR2 resulted in the emergence of the new biovar of race 3 (avr1th380 avr2G121A AVR3).
An additional collection of isolates of races 2 and 3 of FOL obtained from fields in Japan from 2000 to 2017 was subjected to analysis of their AVR genes. Among the eight isolates analyzed, we found 4 kinds of mutations (avr1null, avr1th685, avr1tf-380, and avr15′-ND) in AVR1, and 5 kinds (avr2null, avr2G121A, avr2T122A, avr2G134C, and avr2C146T) in AVR2. None of the eight isolates had the same combination of mutations in AVR1 and AVR2, even though some of them are phylogenetically very close. This suggests that the race differentiation of FOL in the field is more diversified than expected. This may relate to the fact that the avirulence genes exist on accessory chromosomes.
Concluding remarks
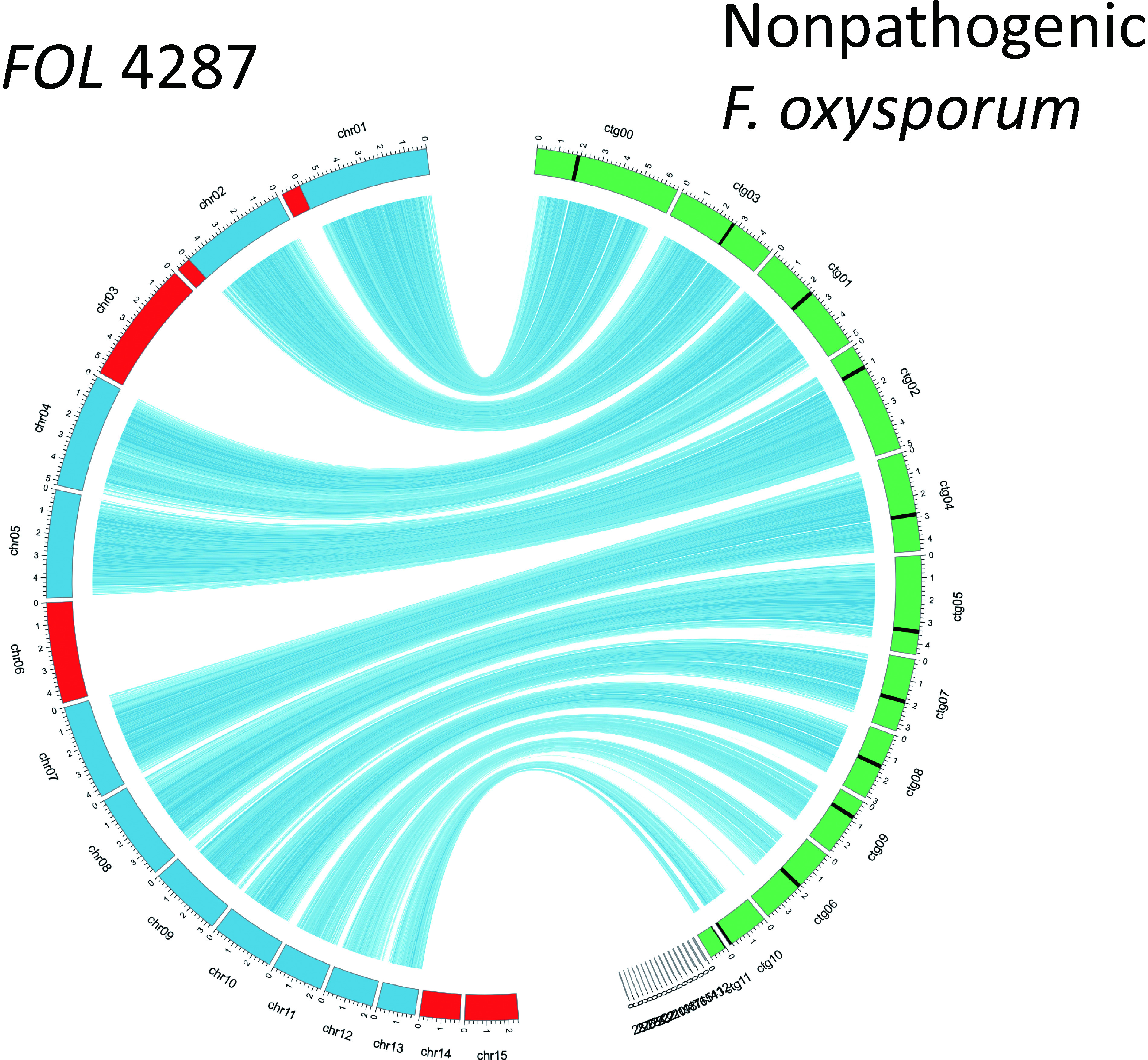
Era of genome analysis—In the 2000s, large consortial genome analysis projects for fungi were conducted, and the genome sequencing of several plant pathogenic fungi was achieved. One of them is the genome sequence project of Fusarium spp., and the genome data set of the tomato wilt fungus (F. oxysporum f. sp. lycopersici) has been opened to the public.26) Genomic information greatly contributes to clarifying the onset mechanism of Fusarium diseases and the host determination mechanism and to elucidating the substantial avirulence genes, discovering accessory chromosomes, and revealing the interaction with the host (Fig. 2).30,31) Additionally, in the last few years, with the remarkable progress of SMRT® sequencing technology, comparative genome analyses among strains have become easy, and even in a small laboratory like ours, genome sequencing can be used as a research tool. In fact, twelve supercontigs were obtained as a result of the SMRT® sequencing of a banana Panama disease fungus (F. oxysporum f. sp. cubense) genome, suggesting that it has become easy to to understand the essence of chromosomes.32) I predict that such innovation in research techniques will further accelerate Fusarium studies and reveal biologically significant findings.
Fig. 2. Comparison of genomes of the tomato wilt fungus, Fusarium oxysporum f. sp. lycopersici (FOL) 4287, and a nonpathogenic F. oxysporum isolate.31) The core chromosomes of FOL are presented in blue and accessory chromosomes in red. The chromosomes of a nonpathogenic F. oxysporum isolate are presented in green.
Acknowledgments
I am grateful to the Pesticide Science Society of Japan, which gave me an award. I would like to thank the members of Laboratory of Plant Pathology, Tokyo University of Agriculture and Technology (TUAT): Dr. Tohru Teraoka, Dr. Ken Komatsu, Dr. Masato Kawabe, Dr. Keigo Inami, Dr. Takeshi Kashiwa, Dr. Yu Ayukawa, and contract members and students; RIKEN Institute: Dr. Isamu Yamaguchi, Dr. Katsuyoshi Yoneyama, Dr. Yutaka Arimoto, Dr. Takashi Kamakura, Dr. Makoto Kimura, Dr. Hideo Nakashita, Dr. Takayuki Motoyama, Dr. Shuta Asai, Ms. Yumiko Kobayashi, Ms. Shigeko Sekido, Mr. Toshiyuki Shimizu, and contract students; the University of Tokyo: the late Dr. Yoji Doi, Dr. Tuneo Tsuchizaki, the late Dr. Syuichi Yamashita, Dr. Shigeto Namba, and contract students; and many researchers in the institutes of prefectures, companies, and NARO, including Dr. Toshio Kijima, for their kind support. This study was mainly supported by grants from the JSPS and MAFF.
References
- 1).J. F. Leslie and B. A. Summerell: “The Fusarium Laboratory Manual,” Blackwell, Ames, US, 2006.
- 2).J. A. Lucas: “Plant Pathology and Plant Pathogens,” John Wiley & Sons, New York, 1998.
- 3).Y. Emoto: Sci. Conserv. 8, 81–86 (1972) (in Japanese). [Google Scholar]
- 4).M. C. Dignani and E. Anaissie: Clin. Microbiol. Infect. 10(Suppl 1), 67–75 (2004). [DOI] [PubMed] [Google Scholar]
- 5).H. H. Flor: Phytopathology 45, 680–685 (1955). [Google Scholar]
- 6).K. Inami, C. Yoshioka, Y. Hirano, M. Kawabe, S. Tsushima, T. Teraoka and T. Arie: J. Gen. Plant Pathol. 76, 116–121 (2010). [Google Scholar]
- 7).T. Nitani, K. Akai, R. Hasegawa, Y. Ayukawa, R. R. Garcia, A. Chitose, K. Komatsu, H. Kikuno, K. T. Natsuaki and T. Arie: J. Gen. Plant Pathol. 84, 165–168 (2018). [Google Scholar]
- 8).S. Shimada, T. Nishimiya and K. Watanabe: Jpn. J. Phytopathol. 85, 108–111 (2019) (abstract in Japanese). [Google Scholar]
- 9).K. Ogawa and H. Kodama: Ann. Phytopathol. Soc. Jpn. 52, 15–21 (1986). [Google Scholar]
- 10).R. Ishikawa, K. Shirouzu, H. Nakashita, H. Lee, T. Motoyama, I. Yamaguchi, T. Teraoka and T. Arie: Phytopathology 95, 1209–1216 (2005). [DOI] [PubMed] [Google Scholar]
- 11).R. Syofi, R. Ishikawa, H. Nakashita, T. Teraoka and T. Arie: in the abstract book of the annual meeting for the Pesticide Science Society of Japan (2005).
- 12).R. Ishikawa, K. Shirouzu, H. Nakashita, T. Teraoka and T. Arie: J. Pestic. Sci. 32, 83–88 (2007). [Google Scholar]
- 13).D. Kawakami, T. Yoshida, Y. Kanemaru, M. H. Huarhua Zaquinaula, M. Tomomichi, M. Arimoto, T. Shibata, A. Goto, Y. Enami, H. Amano, T. Teraoka, K. Komatsu and T. Arie: J. Pestic. Sci. 32, 83–88 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).T. Arie, Y. Hayashi, K. Yoneyama, A. Nagatani, M. Furuya and I. Yamaguchi: Ann. Phytopathol. Soc. Jpn. 61, 311–317 (1995). [Google Scholar]
- 15).T. Arie, Y. Hayashi, K. Yoneyama and I. Yamaguchi: J. Pestic. Sci. 22, 321–325 (1997). [Google Scholar]
- 16).Y. Hirano and T. Arie: J. Gen. Plant Pathol. 72, 272–283 (2006). [Google Scholar]
- 17).T. Kashiwa, K. Inami, T. Teraoka, K. Komatsu and T. Arie: J. Gen. Plant Pathol. 82, 240–247 (2016). [Google Scholar]
- 18).M. Kawabe, Y. Kobayashi, G. Okada, I. Yamaguchi, T. Teraoka and T. Arie: J. Gen. Plant Pathol. 71, 263–272 (2005). [Google Scholar]
- 19).Y. Ayukawa, K. Komatsu, T. Kashiwa, K. Akai, M. Yamada, T. Teraoka and T. Arie: Lett. Appl. Microbiol. 63, 202–209 (2016). [DOI] [PubMed] [Google Scholar]
- 20).Y. Ayukawa, S. Hanyuda, N. Fujita, K. Komatsu and T. Arie: Sci. Rep. 7, 4253 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).S. Kotera, M. Hishiike, K. Komatsu and T. Arie: Jpn. J. Phytopathol. 84, 92–93 (2018) (abstract in Japanese). [Google Scholar]
- 22).C. H. Beckman: “The Nature of Wilt Diseases of Plants,” Amer. Phytopathol. Soc., St. Paul, USA, 1987.
- 23).T. Arie, S. Gouthu, S. Shimazaki, T. Kamakura, M. Kimura, M. Inoue, K. Takio, A. Ozaki, K. Yoneyama and I. Yamaguchi: Ann. Phytopathol. Soc. Jpn. 64, 7–15 (1998). [Google Scholar]
- 24).A. Di Pietro, M. Isabel and G. Roncero: Mol. Plant Microbe Interact. 11, 91–98 (1998). [DOI] [PubMed] [Google Scholar]
- 25).K. Inami, T. Kashiwa, M. Kawabe, A. Onokubo-Okabe, N. Ishikawa, E. Rodriguez Perez, T. Hozumi, L. Aragon Caballero, F. Caceres De Baldarrago, M. Jimenez Roco, K. A. Madadi, T. L. Peever, T. Teraoka, M. Kodama and T. Arie: Microbes Environ. 29, 200–210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).R. J. Ma, H. C. van der Does, K. A. Borkovich, J. J. Coleman, M. J. Daboussi, A. Di Pietro, M. Dufresne, M. Freitag, M. Grabherr, B. Henrissat, P. M. Houterman, S. Kang, W. B. Shim, C. Woloshuk, X. Xie, J. R. Xu, J. Antoniw, S. E. Baker, B. H. Bluhm, A. Breakspear, D. W. Brown, R. A. Butchko, S. Chapman, R. Coulson, P. M. Coutinho, E. G. Danchin, A. Diener, L. R. Gale, D. M. Gardiner, S. Goff, K. E. Hammond-Kosack, K. Hilburn, A. Hua-Van, W. Jonkers, K. Kazan, C. D. Kodira, M. Koehrsen, L. Kumar, Y. H. Lee, L. Li, J. M. Manners, D. Miranda-Saavedra, M. Mukherjee, G. Park, J. Park, S. Y. Park, R. H. Proctor, A. Regev, M. C. Ruiz-Roldan, D. Sain, S. Sakthikumar, S. Sykes, D. C. Schwartz, B. G. Turgeon, I. Wapinski, O. Yoder, S. Young, Q. Zeng, S. Zhou, J. Galagan, C. A. Cuomo, H. C. Kistler and M. Rep: Nature 464, 367–373 (2010).20237561 [Google Scholar]
- 27).T. Kashiwa, K. Inami, M. Fujinaga, H. Ogiso, T. Yoshida, T. Teraoka and T. Arie: J. Gen. Plant Pathol. 79, 412–421 (2013). [Google Scholar]
- 28).P. M. Houterman, B. J. C. Cornelissen and M. Rep: PLoS Pathog. 4, e1000061 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).K. Inami, C. Yoshioka-Akiyama, Y. Morita, M. Yamasaki, T. Teraoka and T. Arie: PLoS One 7, e44101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).T. Kashiwa, T. Kozaki, K. Ishii, B. G. Turgeon, T. Teraoka, K. Komatsu and T. Arie: Fungal Genet. Biol. 98, 46–51 (2017). [DOI] [PubMed] [Google Scholar]
- 31).H. Saito, S. Asai, S. Kotera, K. Komatsu and T. Arie: Jpn. J. Phytopathol. 85, 49–50 (2019) (abstract in Japanese). [Google Scholar]
- 32).S. Asai, Y. Ayukawa, P. Gana, S. Masuda, K. Komatsu, K. Shirasu and T. Arie: Microbiol. Res. Announce. 8, e00654-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]


