Abstract
Purpose
Liver cancer has a high incidence of mortality. DNA replication and posttranscriptional modifications play important roles in the development of liver cancer. Pescadillo (PES1) is a nuclear protein that is involved in embryonic development, ribosome synthesis, DNA replication, and cell cycle progression. Recently, abnormal PES1 expression was reported in several tumors, including neuroblastoma, colon cancer, gastric cancer, and breast cancer. Based on bio-informatic analysis, cell experiments and animal models, the aim of this study is to investigate the expression patterns and specific roles of PES1 in liver cancer.
Patients and methods
PES1 expression was represented by boxplots. The correlation between PES1 expression and clinical features was assessed by the chi-squared test and Fisher’s exact tests. Kaplan–Meier curves compared overall survival between different levels of PES1 expression, and Cox analysis selected potential variables associated with overall survival. The MTT assay investigated the proliferation rate, the scratch assay assessed the migratory ability, and the Transwell assay evaluated the invasion capacity of tumor cells in vitro. Animal models were used to confirm the tumorigenic roles of PES1 in vivo. GSEA illustrated the molecular mechanisms that PES1 participated in.
Results
We found that PES1 was highly expressed in liver cancer tissues, served as a diagnostic marker, and correlated with poor overall survival (OS) and relapse-free survival (RFS) in patients. In vitro studies indicated that PES1 promoted tumor cell proliferation (P=0.0034), migration (P=0.0026), and invasion (P=0.0008), and this tumorigenic role was confirmed in animal models. GSEA further illuminated molecular mechanisms that PES1 participated in liver cancer occurrence and progression.
Conclusion
This study suggested that PES1 was upregulated in liver cancer and correlated with poor prognosis, by promoting tumor cell proliferation, migration, and invasion, and PES1 may be a novel diagnostic and prognostic bio-marker and a promising therapeutic target in liver cancer.
Keywords: liver cancer, PES1, diagnosis, tumorigenesis, prognosis
Introduction
Liver cancer has a high morbidity and mortality worldwide. Globally, it ranked sixth of all tumors for incidence and third for deaths in 2013, while in developing countries it ranked fifth for incidence and second for death.1 Despite rapid developments in medical technology, it remains an incurable disease that places a huge burden on patients and society.2 The poor prognosis of liver cancer can be attributed to the fact that most diagnoses currently take place in the late stage of disease progression.3 Therefore, the identification of a specific biomarker for early-stage diagnosis and prognosis has great clinical importance.
DNA replication and posttranscriptional modifications are necessary processes in tumor initiation and progression.4,5 Pescadillo (PES1) is a nuclear protein of 588 amino acids that mediates the protein transport process.6 It plays important roles in embryonic development, ribosome synthesis, DNA replication, and cell cycle progression.7 Recently, abnormal PES1 upregulation has been identified in some cancers, including neuroblastoma, colon cancer, gastric cancer, and breast cancer.7–10 The upregulation of PES1 in hepatocellular carcinoma (HCC) was reported by Fan et al in 201811 and Wang et al in 2019;12 however, the expression of PES1 in other liver cancer types, in distinct subgroups, and diagnostic values has not been revealed. Furthermore, as a tumorigenesis-related molecule, the role of PES1 in cell migration and invasion remain to be elucidated.
In this study, we examined the expression of PES1 in liver cancer and analyzed its relationship with patient prognosis. We also verified the tumorigenic function of PES1 in liver cancer in vitro and in vivo. Our results suggest that PES1 could be a novel diagnostic and prognostic biomarker in patients with liver cancer.
Materials And Methods
Bioinformatic Analysis
RNA sequencing and clinical data for liver cancer patients were downloaded from The Cancer Genome Atlas (TCGA) database using the RTCGAToolbox package in R (version 3.5.1).13,14 Gene Set Enrichment Analysis (GSEA) was performed using the GSEA 3.0 software and the enrichment score (ES) was calculated for the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways from the Molecular Signature Database.
Cell Lines And Culture
Hepa 1–6 cells were purchased from the American Type Culture Collection (USA). Cells were cultured in Dulbecco’s modified Eagle’s medium (Sigma) supplemented with 10% fetal bovine serum (FBS, Solarbio). Cells were incubated in a humidified incubator with 5% CO2.
Cell Transfection
The PES1 sequence was amplified and inserted into pCMV vector (Beyotime). The transfection of PES1 over-expression or control plasmid was performed using Lipofectamine 3000 (Invitrogen) following the manufacturer’s instructions. After 48 hrs, cells were collected and used for experiments.
RNA Isolation And PCR
RNA was extracted using Trizol Reagent (Invitrogen) from the transfected cells, according to the manufacturer’s instructions. cDNA was synthesized from 1000 ng of RNA using PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa). PCR was carried out according to the following procedures: 95°C 3 mins, followed by 35 cycles of 95°C 30 s, 58°C 30 s, and 72°C 2 mins. The primers used were listed as follows: PES1 forward primer: 5ʹ-ATGGGAGGTCTGGAGAAG-3ʹ; PES1 reverse primer: TCACACGGGCCTTGTCCTCTT.
Proliferation Assay
Hepa 1–6 cells were seeded in 96-well plates at a concentration of 1×104 cells per well and transfected with PES1 or EGFP plasmid. Forty-eight hrs after transfection, MTT (Sigma) was added to the cells and the absorption at 570 nm was measured 2 hrs later.
Migration Assay
Hepa 1–6 cells were grown in 6-well plates until confluent and transfected with PES1 or EGFP plasmid. Forty-eight hrs after transfection, cells were scratched with a p200 tip. Photographs were taken immediately and 24 hrs after scratching. The size of the scratches was measured by ImageJ software (NIH).
Invasion Assay
Transwell chambers (8 mm pore size) were coated with Matrigel (Corning) according to the manufacturer’s instructions. PES1- or EGFP-transfected cells were plated in the upper chambers in serum-free medium, while medium containing 10% FBS was used to fill the lower chambers. After 24 hrs incubation, cells were fixed with 4% paraformaldehyde and stained with Giemsa stain (Solarbio). The number of invading cells was counted in five random fields (×100 magnification) per insert.
In Vivo Experiments
The animal experiments were approved by the Institutional Animal Care and Use Committee of First Hospital of Jilin University and performed in accord with the institutional and NIH guidelines. 5×105 Hepa 1–6 liver cancer cells transfected with PES1 or EGFP control plasmid were injected subcutaneously into the flanks of C57BL/6 mice (8–12 weeks old, 18–22 g). Tumor volumes were measured every 2 days and calculated using the following formula: tumor volume = (length×width2)/2.
Histology
Animals were sacrificed and tumors were fixed in 4% paraformaldehyde (PFA) overnight and then embedded in paraffin. Sections at 4 μm were processed for Hematoxylin-Eosin (H&E) staining and images were photographed with a light microscope (Olympus).
Statistical Analysis
Differences between discrete variables were examined by box plots. The chi-squared test and Fisher’s exact tests measured the correlations between PES1 expression and liver cancer pathological features. Overall survival between different levels of PES1 expression was compared by Kaplan–Meier curves, with P-values calculated by the log rank test using the Survival package in R.15,16 Univariate Cox regression analysis was used to select potential variables associated with overall survival, while multivariate Cox analysis assessed the effects of PES1 expression on survival and other pathological features.
Data from in vitro and in vivo experiments were analyzed by the Student’s t-test (unpaired, two-tailed) using Graph Pad Prism Software. Results are shown as means±SD. P<0.05 was considered statistically significant.
Results
PES1 Is Upregulated In Liver Cancer
Patient data were downloaded from the TCGA database. Of the 373 patients, 121 (32.44%) were female and 252 (67.56%) were male. A total of 244 (66.12%) patients were older than 55 years while 125 (33.88%) were younger. Detailed patient information is shown in Table 1.
Table 1.
Clinical Characteristics Of The Included Patients With Liver Cancer
| Characteristics | Numbers Of Cases |
|---|---|
| PES1 | |
| High | 186 (49.87) |
| Low | 187 (50.13) |
| Vital status | |
| Survival | 243 (65.15) |
| Death | 130 (34.85) |
| T classification | |
| T1 | 182 (49.06) |
| T2 | 95 (25.61) |
| T3 | 80 (21.56) |
| T4 | 13 (3.5) |
| Tx | 1 (0.27) |
| NA | 2 |
| N classification | |
| N0 | 253 (68.01) |
| N1 | 4 (1.08) |
| Nx | 115 (30.91) |
| NA | 1 |
| M classification | |
| M0 | 267 (71.58) |
| M1 | 4 (1.07) |
| Mx | 102 (27.35) |
| Gender | |
| Female | 121 (32.44) |
| Male | 252 (67.56) |
| Radiation therapy | |
| No | 341 (97.43) |
| Yes | 9 (2.57) |
| NA | 23 |
| Histological type | |
| Fibrolamellar carcinoma | 3 (0.8) |
| Hepatocellular carcinoma | 363 (97.32) |
| Hepatocholangiocarcinoma (mixed) | 7 (1.88) |
| Residual tumor | |
| R0 | 326 (89.07) |
| R1 | 17 (4.64) |
| R2 | 1 (0.27) |
| Rx | 22 (6.01) |
| NA | 7 |
| Race | |
| American Indian or Alaska native | 2 (0.55) |
| Asian | 159 (43.8) |
| Black or African American | 17 (4.68) |
| White | 185 (50.96) |
| NA | 10 |
| Age | |
| ≤55 | 125 (33.88) |
| >55 | 244 (66.12) |
| NA | 4 |
| Stage | |
| I | 172 (49.28) |
| II | 87 (24.93) |
| III | 85 (24.36) |
| IV | 5 (1.43) |
| NA | 24 |
Abbreviation: NA, not applicable.
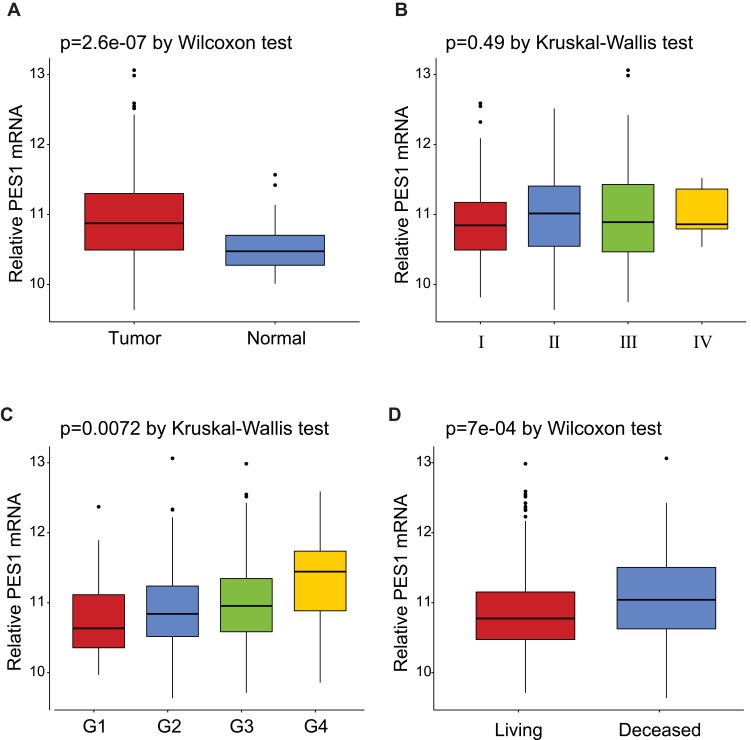
The expression of PES1 was compared between patients with liver cancer and healthy controls (Figure 1A), and shown to be significantly elevated in liver cancer patients (p=2.6e-07 by Wilcoxon test). Moreover, PES1 expression was distinct in patients of different subgroups sorted by clinical stage (p=0.49 by Kruskal–Wallis test), histological grade (p=0.0072 by Kruskal–Wallis test), and vital status (p=7e-04 by Wilcoxon test) (Figure 1B–D).
Figure 1.
Expression of PES1 mRNA in liver cancer based on analysis of TCGA data. (A) PES1 was significantly upregulated in liver cancer tissues when compared with normal liver tissues (p=2.6e-07 by Wilcoxon test). (B) The expression of PES1 in distinct stage of liver cancer (p=0.49 by Kruskal–Wallis test). (C) PES1 expression was increased as histologic grade progressing (p=0.0072 by Kruskal–Wallis test). (D) The deceased states of patients had higher PES1 expression level than living states (p=7e-04 by Wilcoxon test).
PES1 Presents Modest Diagnostic Value In Liver Cancer
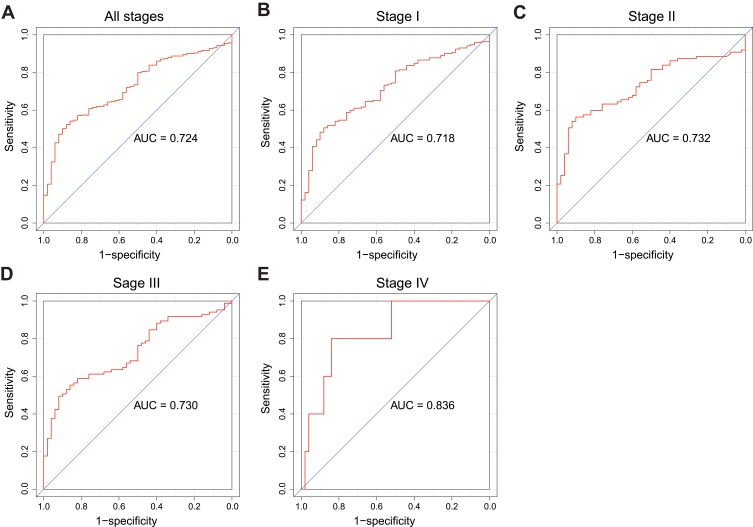
Receiver operating characteristic (ROC) curve was generated to evaluate the diagnostic value of PES1. The area under the curve (AUC) was 0.724, which indicated moderate diagnostic value (Figure 2A). Subgroup analysis further demonstrated the diagnostic value of PES1 expression in distinct stage of liver cancer, with AUC of 0.718 for stage I (Figure 2B), 0.732 for stage II (Figure 2C), 0.730 for stage III (Figure 2D), and 0.836 for stage IV (Figure 2E).
Figure 2.
The ROC curve of PES1 expression for liver cancer diagnosis. Non-tumor samples and tumor samples in all stages (A), stage I (B), stage II (C), stage III (D) and stage IV (E).
Correlation Between Clinical Characteristics And PES1 Expression In Liver Cancer
The relationship between clinical features and expression of PES1 in patients with liver cancer is summarized in Table 2. Vital status (P=0.002067), sex (P=0.03887), and race (P=0.008083) were significantly correlated with PES1 expression.
Table 2.
Correlation Between The Clinical Variables And PES1 Expression In Patients
| Features | Variables | N | PES1 (%) | χ2 | P | |
|---|---|---|---|---|---|---|
| High | Low | |||||
| Vital status | Survival | 243 | 107 (57.5) | 136 (72.7) | 9.489 | 0.002067 |
| Death | 130 | 79 (42.5) | 51 (27.3) | |||
| T classification | T1 | 182 | 85 (45.7) | 97 (51.9) | 2.579 | 0.76461 |
| T2 | 95 | 50 (26.9) | 45 (24.1) | |||
| T3 | 80 | 43 (23.1) | 37 (19.8) | |||
| T4 | 13 | 7 (3.8) | 6 (3.2) | |||
| Tx | 1 | 0 (0.0) | 1 (0.5) | |||
| N classification | N0 | 253 | 133 (71.5) | 120 (64.2) | 3.718 | 0.293629 |
| N1 | 4 | 1 (0.5) | 3 (1.6) | |||
| Nx | 115 | 52 (28.0) | 63 (33.7) | |||
| M classification | M0 | 267 | 135 (72.6) | 132 (70.6) | 0.188 | 0.910332 |
| M1 | 4 | 2 (1.1) | 2 (1.1) | |||
| Mx | 102 | 49 (26.3) | 53 (28.3) | |||
| Gender | Female | 121 | 51 (27.4) | 70 (37.4) | 4.267 | 0.03887 |
| Male | 252 | 135 (72.6) | 117 (62.6) | |||
| Radiation therapy | No | 341 | 166 (89.2) | 175 (93.6) | 2.322 | 0.3132 |
| Yes | 9 | 6 (3.2) | 3 (1.6) | |||
| Histological type | Fibrolamellar carcinoma | 3 | 0 (0.0) | 3 (1.6) | 3.143 | 0.207738 |
| Hepatocellular carcinoma | 363 | 182 (97.8) | 181 (96.8) | |||
| Hepatocholangio-carcinoma (mixed) | 7 | 4 (2.2) | 3 (1.6) | |||
| Residual tumor | R0 | 326 | 164 (88.2) | 162 (86.6) | 3.991 | 0.40729 |
| R1 | 17 | 8 (4.3) | 9 (4.8) | |||
| R2 | 1 | 1 (0.5) | 0 (0.0) | |||
| Rx | 22 | 8 (4.3) | 14 (7.5) | |||
| Race | American Indian or Alaska native | 2 | 2 (1.1) | 0 (0.0) | 13.765 | 0.008083 |
| Asian | 159 | 95 (51.1) | 64 (34.2) | |||
| Black or African American | 17 | 7 (3.8) | 10 (5.3) | |||
| White | 185 | 77 (41.4) | 108 (57.8) | |||
| Age | ≤55 | 125 | 64 (34.4) | 61 (32.6) | 4.332 | 0.114656 |
| >55 | 244 | 118 (63.4) | 126 (67.4) | |||
| Stage | I | 172 | 83 (44.6) | 89 (47.6) | 2.016 | 0.732797 |
| II | 87 | 48 (25.8) | 39 (20.9) | |||
| III | 85 | 43 (23.1) | 42 (22.5) | |||
| IV | 5 | 2 (1.1) | 3 (1.6) | |||
Note: Bold values indicate P<0.05 and are statistically significant.
Overexpression Of PES1 Is Associated With Poor Overall Survival (OS) In Patients With Liver Cancer
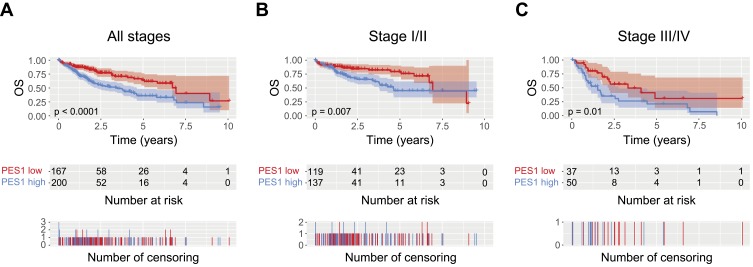
The role of PES1 expression in prognosis was analyzed by Kaplan–Meier analysis with the log rank test. Patients with higher PES1 expression had a significantly shorter survival time than those with lower expression (Figure 3A, P<0.0001). Subgroup analysis based on clinical stage found that PES1 upregulation significantly reduced OS in patients with both stage I/II (Figure 3B, P=0.007) and stage III/IV (Figure 3C, P=0.01). Univariate analysis revealed that poor OS was significantly associated with T classification (P=0.000), residual tumor (P=0.003), tumor stage (P=0.001), and PES1 expression (P=0.000). Multivariate tests using the Cox regression model revealed that high PES1 expression was an independent risk factor for patient prognosis (hazard ratio=2.09, P=0.000) (Table 3).
Figure 3.
Liver cancer patients with high PES1 expression were associated with poor overall survival (OS). Kaplan-Meier analysis of OS for liver cancer patients with all stages (A), stage I/II (B) and stage III/IV (C).
Table 3.
Association Between PES1 Expression And OS
| Characteristics | Hazard Ratio | 95% CI | P | Hazard Ratio | 95% CI | P |
|---|---|---|---|---|---|---|
| PES1 expression | 2.11 | 1.46–3.05 | 0.000 | 2.09 | 1.44–3.02 | 0.000 |
| T classification | 1.66 | 1.39–1.99 | 0.000 | 1.79 | 1.42–2.26 | 0.000 |
| N classification | 0.73 | 0.51–1.05 | 0.086 | |||
| M classification | 0.72 | 0.49–1.04 | 0.077 | |||
| Gender | 0.80 | 0.56–1.14 | 0.220 | |||
| Radiation therapy | 0.51 | 0.26–1.03 | 0.060 | |||
| Histological type | 0.99 | 0.27–3.66 | 0.986 | |||
| Residual tumor | 1.42 | 1.13–1.80 | 0.003 | 1.46 | 1.14–1.86 | 0.002 |
| Histological grade | 1.04 | 0.84–1.30 | 0.698 | |||
| Age | 1.00 | 0.69–1.45 | 0.997 | |||
| Stage | 1.38 | 1.15–1.66 | 0.001 | 0.88 | 0.71–1.09 | 0.250 |
Note: Bold values indicate P<0.05 and are statistically significant.
Abbreviation: CI, confidence interval.
Overexpression Of PES1 Is Associated With Poor Relapse-Free Survival (RFS) In Patients With Liver Cancer
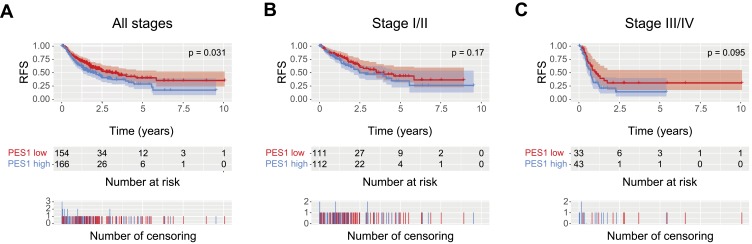
Kaplan-Meier analysis showed patients with higher PES1 expression presented shorter RFS than those with lower expression (Figure 4A, P=0.031). PES upregulation also impacted the RFS by subgroup analysis, such as patients in stage I/II (Figure 4B, P=0.17) and stage III/IV (Figure 4C, P=0.095). Univariate analysis revealed poor RFS was associated with T classification (P=0.000), residual tumor (P=0.042), stage (P=0.000), and PES1 expression (P=0.031). Multivariate analysis further demonstrated high PES1 expression was an independent risk factor for patient prognosis (hazard ratio=1.44, P=0.032) (Table 4).
Figure 4.
Liver cancer patients with high PES1 expression were associated with poor relapse-free survival (RFS). Kaplan-Meier analysis of RFS for liver cancer patients with all stages (A), stage I/II (B) and stage III/IV (C).
Table 4.
Association Between PES1 Expression And RFS
| Characteristics | Hazard Ratio | 95% CI | P | Hazard Ratio | 95% CI | P |
|---|---|---|---|---|---|---|
| PES1 expression | 1.44 | 1.03–2.01 | 0.031 | 1.44 | 1.03–2.02 | 0.032 |
| T classification | 1.78 | 1.49–2.12 | 0.000 | 1.60 | 1.22–2.09 | 0.001 |
| N classification | 0.97 | 0.67–1.40 | 0.874 | |||
| M classification | 1.17 | 0.79–1.74 | 0.432 | |||
| Gender | 0.99 | 0.70–1.41 | 0.966 | |||
| Radiation therapy | 0.74 | 0.26–2.16 | 0.584 | |||
| Histological type | 2.02 | 0.66–6.24 | 0.220 | |||
| Residual tumor | 1.28 | 1.01–1.61 | 0.042 | 1.36 | 1.07–1.71 | 0.011 |
| Histologic grade | 0.98 | 0.80–1.21 | 0.883 | |||
| Age | 0.90 | 0.63–1.28 | 0.550 | |||
| Stage | 1.66 | 1.38–1.99 | 0.000 | 1.16 | 0.89–1.50 | 0.273 |
Note: Bold values indicate P<0.05 and are statistically significant.
Abbreviation: CI, confidence interval.
PES1 Promotes Liver Cancer Cell Proliferation, Migration, And Invasion In Vitro
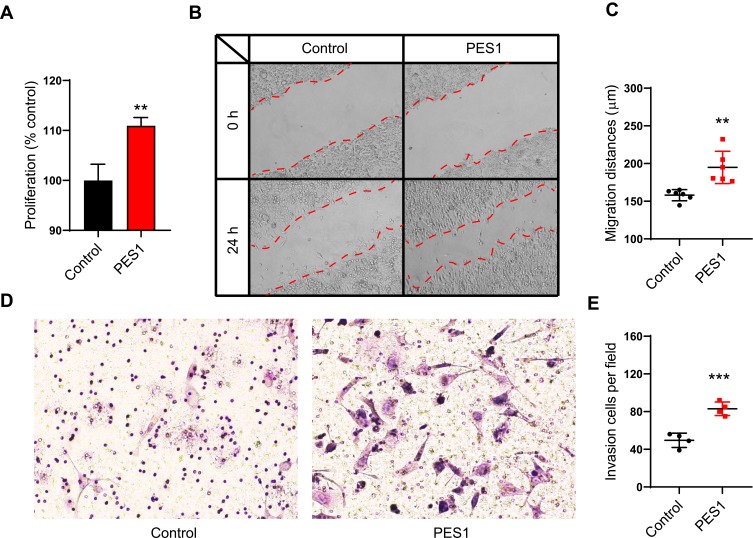
Because of the association between PES1 upregulation and poor prognosis in liver cancer, we speculated that PES1 could promote tumor progression. We therefore overexpressed PES1 in liver cancer Hepa 1–6 cell lines and investigated its effect on the cells. The MTT assay showed that PES1 upregulation significantly promoted Hepa 1–6 cell proliferation compared with control (P<0.01; Figure 5A). PES1 also increased the migratory distance of tumor cells in the scratch assay significantly (P<0.01; Figure 5B and C). Moreover, the invasion capacity was significantly increased in PES1-overexpressing cells (P<0.001; Figure 5D and E). Taken together, these results suggest that PES1 accelerated tumor progression by promoting tumor cell proliferation, migration, and invasion, and might be a therapeutic target for liver cancer.
Figure 5.
PES1 promoted liver cancer cell proliferation, migration and invasion. (A) MTT proliferation assay in PES1 or control plasmid transfected Hepa 1–6 cells. (B, C) Wound scratch assay showed PES1 overexpression promoted the migration of Hepa 1–6 cells. (D, E) Transwell invasion assay revealed PES1 overexpression cells had stronger invasion capacity than control. Error bars indicate the mean ± SD. **p < 0.01, ***p < 0.001.
PES1 Promotes Tumor Growth In Liver Cancer Models
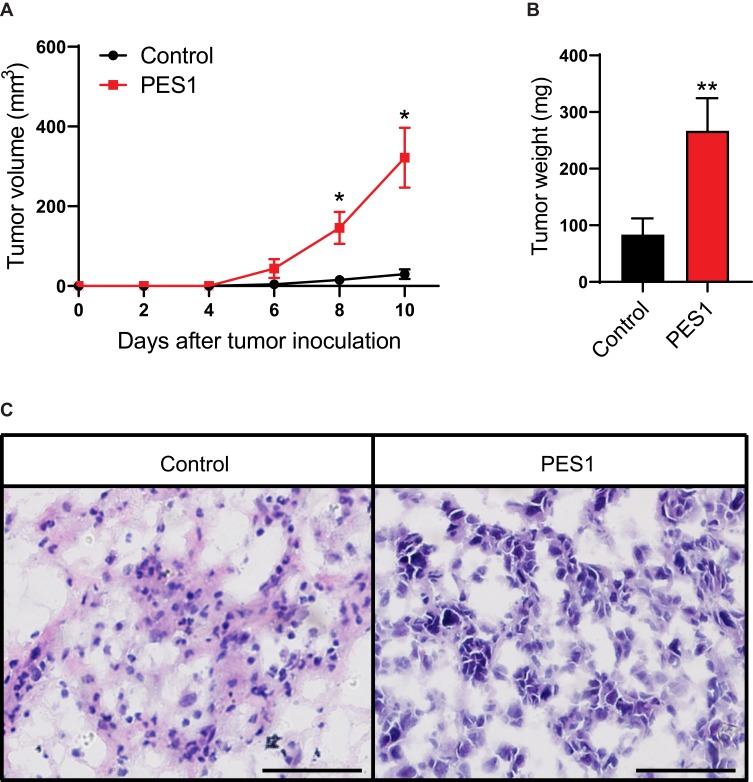
As we found PES1 presented tumorigenic roles in vitro, we further investigated the effect of PES1 upregulation on tumor growth of Hepa 1–6 cells in C57BL/6 mice. Results showed tumors from PES1-overexpressing cells had a faster growth rate than tumors from control cells (P<0.05; Figure 6A). Meanwhile, the tumor weight from PES1-overexpressing cells was also higher than from control cells (P<0.01; Figure 6B). Histologic analysis further presented advanced degree of malignancy in PES1 overexpression tumors (Figure 6C).
Figure 6.
PES1 promoted tumor growth in liver cancer models. Overexpression of PES1 increased tumor volume (A), tumor weight (B) and histological grade (C). Bar = 50 μm. Error bars indicate the mean ± SD. *p < 0.05, **p < 0.01.
Association Between KEGG Pathways And PES1 Expression
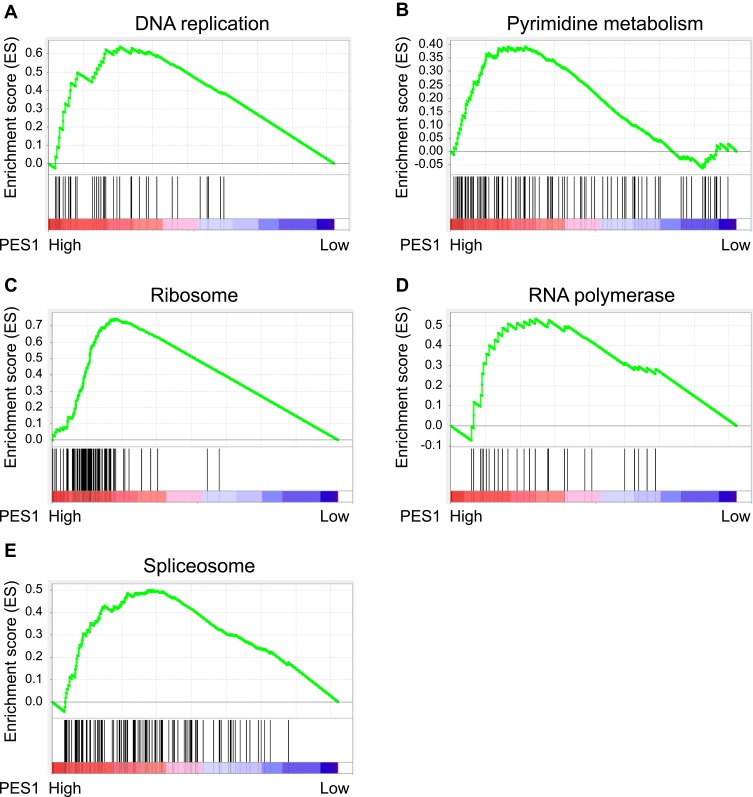
We explored and verified KEGG pathways which PES1 involved in the whole gene expression level using GSEA. As shown in Figure 7, five enriched KEGG pathways, including DNA replication, pyrimidine metabolism, ribosome, RNA polymerase, and spliceosome, were associated with high PES1 expression, which further illuminated molecular mechanisms that PES1 participated in liver cancer occurrence and progression.
Figure 7.
Gene set enrichment analysis (GSEA) indicated the associated Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. PES1 expression was correlated positively with gene signatures related to DNA replication (A), pyrimidine metabolism (B), ribosome (C), RNA polymerase (D) and spliceosome (E).
Discussion
In the present study, we determined the diagnostic and prognostic roles of PES1 expression in liver cancer patients and the functional consequences of PES1 in liver cancer cell line in vitro and in vivo. To our knowledge, no previous investigation has revealed the diagnostic value of the PES1 gene in human liver cancer, especially in subgroups, or the role of PES1 in migration and invasion of cells and RFS in patients. We concluded that PES1 was upregulated in liver cancer tissues, significantly associated with both OS and RFS. In vitro and in vivo experiments confirmed the tumorigenic effects of PES1, and GSEA also illustrated the molecular mechanisms that PES1 participated in.
A lack of diagnosis during early-stage disease contributes to the poor prognosis of liver cancer patients, so identifying a biomarker for early diagnosis would have notable benefits.3 DNA replication and posttranscriptional modifications play critical roles in tumorigenesis and tumor progression.4,5 As a nuclear protein, PES1 was first identified as essential for normal zebrafish embryonic development, and has been reported to play important roles in many other physiological processes including DNA replication, ribosome genesis, chromosomal stability, and cell proliferation.17–23 PES1 interacts with Bop1 and WDR12 to form the PeBoW-complex; this functions in the synthesis and maturation of ribosomes, which is a crucial process in cell proliferation.18 PES1 mutants or other disturbances to the complex can lead to p53 activation, which is regarded as the most common genetic alteration in human cancers, regulating numerous genes involved in the cell cycle, apoptosis, DNA repair, and metabolism.19,24
Several studies have reported the upregulation of PES1 in tumor tissues. Xie et al reported significantly higher PES1 expression in colon cancer tissues compared with adjacent tissues,7 while it was also highly up-regulated in gastric cancers.25 Our study observed increased PES1 expression in liver cancer, which is in accordance with previous work.11 Additionally, we documented differences in PES1 expression in distinct clinical stages, histological grade, and vital statuses of patients, which further divided patients into subgroups. ROC analysis further provided the diagnostic value of PES1 expression in liver cancer.
PES1 overexpression is correlated with tumor initiation and proliferation. Short hairpin RNA-mediated PES1 ablation previously inhibited colon cancer cell proliferation,7 and Li et al reported that PES1 silencing inhibited gastric cancer growth and altered the expression of proliferation-related genes.9 Moreover, Fan et al found that PES1 overexpression promoted liver cancer cell proliferation while its knockdown inhibited their growth.11 Based on these studies, we propose that the correlation between PES1 and the vital status of patients reflects the role of PES1 in promoting tumor proliferation.
Several articles have reported the adverse effects of PES1 in cancer prognosis. In neuroblastoma, high PES1 expression was associated with aggregation and a worse outcome, while PES1 knockdown suppressed tumor growth and induce apoptosis.8 Additionally, PES1 was shown to regulate the balance between estrogen receptor (ER)α and ERβ to promote the progression of breast cancer10,26,27 and ovarian cancer.28 Similar to earlier findings, our study found that higher PES1 expression was associated with worse outcomes in patients with liver cancer; however, we also reported heterogeneity in the prognostic role of PES1 because of distinct clinical stages, which provide the possibility of precision therapy for patients with liver cancer. Moreover, we showed that PES1 was associated with poor OS in stage Ⅰ/Ⅱ, suggesting that its prognostic evaluation role is effective even in the very early stages of the disease.
The MTT assay is one of the most common strategies to analyze cell proliferation. Our results showed that cells with upregulated PES1 exhibited a higher growth rate, suggesting that PES1 promotes tumor growth. The scratch assay and Transwell assay also revealed that PES1 upregulation increased cell migration and invasion, which further confirmed its strong correlation with tumor progression and metastasis. Finally, the tumorigenic roles of PES1 were verified in animal models, which made the theory more persuasive. Recently, two studies of the proliferative role of PES1 in hepatocellular carcinoma (HCC) obtained similar findings to our own research,11,12 which not only confirmed our results, but also suggested that PES1 was a research hotspot. Fan et al showed PES1 was upregulated in HCC, promoted HCC proliferation and glycolysis. Wang et al revealed upregulated PES1 promoted HCC proliferation via PI3K/AKT pathway, and correlated worse OS. In the present study, we demonstrated PES1 was upregulated in all liver cancer patients, rather than HCC, and the expression was distinct in patients of subgroups. We presented PES1 could be a diagnostic marker in liver cancer, and this value was different in each stage. Besides OS, we also analyzed the RFS of patients and showed PES1 was an independent risk factor for prognosis evaluation in both OS and RFS. In addition to proliferation, cell migration and invasion were important factors for tumorigenesis, which was also assessed in our study. Moreover, GSEA provided the pathways that PES1 participated in, which illuminated the molecular mechanisms of liver cancer occurrence and progression. However, all our data of patients were derived from the TCGA database; so, clinical samples are required to verify our findings in the future.
Conclusion
In conclusion, we found that PES1 was upregulated in liver cancer tissues and that its high expression was strongly correlated with clinical progression and poor survival in patients. In vitro experiments showed that PES1 promoted tumor growth, migration, and invasion, and animal models confirmed its tumorigenic roles. Our results suggest that PES1 could be a novel biomarker for the diagnosis and prognosis of patients with liver cancer and a promising target for tumor therapy.
Acknowledgments
We thank Sarah Williams, PhD, from Liwen Bianji, Edanz Group China, for editing the English text of a draft of this manuscript. This work was supported by the Project of Talent Development Funds of Jilin Province, China, No. 802160100428.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Fitzmaurice C, Dicker D, et al; Global Burden of Disease Cancer C. The global burden of cancer 2013. JAMA Oncol. 2015;1(4):505–527. doi: 10.1001/jamaoncol.2015.0735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asrani S, Devarbhavi H, Eaton J, Kamath P. Burden of liver diseases in the world. J Hepatol. 2018;70:151–171. [DOI] [PubMed] [Google Scholar]
- 3.Tsuchiya N, Sawada Y, Endo I, Saito K, Uemura Y, Nakatsura T. Biomarkers for the early diagnosis of hepatocellular carcinoma. World J Gastroenterol. 2015;21(37):10573–10583. doi: 10.3748/wjg.v21.i37.10573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macheret M, Halazonetis T. DNA replication stress as a hallmark of cancer. Annu Rev Pathol. 2015;10:425–448. doi: 10.1146/annurev-pathol-012414-040424 [DOI] [PubMed] [Google Scholar]
- 5.Nasr Z, Pelletier J. Tumor progression and metastasis: role of translational deregulation. Anticancer Res. 2012;32(8):3077–3084. [PubMed] [Google Scholar]
- 6.Li S, Wang M, Qu X, et al. SUMOylation of PES1 upregulates its stability and function via inhibiting its ubiquitination. Oncotarget. 2016;7(31):50522–50534. doi: 10.18632/oncotarget.10494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie W, Feng Q, Su Y, et al. Transcriptional regulation of PES1 expression by c-Jun in colon cancer. PLoS One. 2012;7(7):e42253. doi: 10.1371/journal.pone.0042253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakaguro M, Kiyonari S, Kishida S, et al. Nucleolar protein PES1 is a marker of neuroblastoma outcome and is associated with neuroblastoma differentiation. Cancer Sci. 2015;106(3):237–243. doi: 10.1111/cas.2015.106.issue-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Zhou X, Lan X, Zeng G, Jiang X, Huang Z. Repression of PES1 expression inhibits growth of gastric cancer. Tumour Biol. 2016;37(3):3043–3049. doi: 10.1007/s13277-015-4069-8 [DOI] [PubMed] [Google Scholar]
- 10.Cheng L, Li J, Han Y, et al. PES1 promotes breast cancer by differentially regulating ERalpha and ERbeta. J Clin Invest. 2012;122(8):2857–2870. doi: 10.1172/JCI62676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan P, Wang B, Meng Z, Zhao J, Jin X. PES1 is transcriptionally regulated by BRD4 and promotes cell proliferation and glycolysis in hepatocellular carcinoma. Int J Biochem Cell Biol. 2018;104:1–8. doi: 10.1016/j.biocel.2018.08.014 [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Sun J, Zhang N, et al. PES1 enhances proliferation and tumorigenesis in hepatocellular carcinoma via the PI3K/AKT pathway. Life Sci. 2019;219:182–189. doi: 10.1016/j.lfs.2018.12.054 [DOI] [PubMed] [Google Scholar]
- 13.Team RDC. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Computing 2012; 1:12–21. [Google Scholar]
- 14.Samur MK. RTCGAToolbox: a new tool for exporting TCGA firehose data. PLoS One. 2014;9(9):e106397. doi: 10.1371/journal.pone.0106397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Therneau TM. A Package for Survival Analysis in S. 2015.
- 16.Therneau TM, Grambsch PM. Modeling survival data: extending the cox model. Technometrics. 2000;97(457):353–354. [Google Scholar]
- 17.Allende ML, Amsterdam A, Becker T, Kawakami K, Gaiano N, Hopkins N. Insertional mutagenesis in zebrafish identifies two novel genes, pescadillo and dead eye, essential for embryonic development. Genes Dev. 1996;10(24):3141–3155. doi: 10.1101/gad.10.24.3141 [DOI] [PubMed] [Google Scholar]
- 18.Grimm T, Holzel M, Rohrmoser M, et al. Dominant-negative Pes1 mutants inhibit ribosomal RNA processing and cell proliferation via incorporation into the PeBoW-complex. Nucleic Acids Res. 2006;34(10):3030–3043. doi: 10.1093/nar/gkl378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holzel M, Grimm T, Rohrmoser M, et al. The BRCT domain of mammalian Pes1 is crucial for nucleolar localization and rRNA processing. Nucleic Acids Res. 2007;35(3):789–800. doi: 10.1093/nar/gkl1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holzel M, Rohrmoser M, Schlee M, et al. Mammalian WDR12 is a novel member of the Pes1-Bop1 complex and is required for ribosome biogenesis and cell proliferation. J Cell Biol. 2005;170(3):367–378. doi: 10.1083/jcb.200501141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lapik YR, Fernandes CJ, Lau LF, Pestov DG. Physical and functional interaction between Pes1 and Bop1 in mammalian ribosome biogenesis. Mol Cell. 2004;15(1):17–29. doi: 10.1016/j.molcel.2004.05.020 [DOI] [PubMed] [Google Scholar]
- 22.Prisco M, Maiorana A, Guerzoni C, et al. Role of pescadillo and upstream binding factor in the proliferation and differentiation of murine myeloid cells. Mol Cell Biol. 2004;24(12):5421–5433. doi: 10.1128/MCB.24.12.5421-5433.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rohrmoser M, Holzel M, Grimm T, et al. Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Mol Cell Biol. 2007;27(10):3682–3694. doi: 10.1128/MCB.00172-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parrales A, Iwakuma T. Targeting oncogenic mutant p53 for cancer therapy. Front Oncol. 2015;5:288. doi: 10.3389/fonc.2015.00288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim B, Bang S, Lee S, et al. Expression profiling and subtype-specific expression of stomach cancer. Cancer Res. 2003;63(23):8248–8255. [PubMed] [Google Scholar]
- 26.Thomas C, Gustafsson JA. Targeting PES1 for restoring the ERalpha/ERbeta ratio in breast cancer. J Clin Invest. 2012;122(8):2771–2773. doi: 10.1172/JCI65133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Yu L, Zhang H, et al. Down-regulation of pescadillo inhibits proliferation and tumorigenicity of breast cancer cells. Cancer Sci. 2009;100(12):2255–2260. doi: 10.1111/j.1349-7006.2009.01325.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Zhuang Q, Lan X, Zeng G, Jiang X, Huang Z. PES1 differentially regulates the expression of ERalpha and ERbeta in ovarian cancer. IUBMB Life. 2013;65(12):1017–1025. doi: 10.1002/iub.1228 [DOI] [PubMed] [Google Scholar]