Abstract
Oligoarginine sequences conjugated to a short cancer-targeting peptide (CTP) selective for the prostate-specific membrane antigen (PSMA) receptor was developed for selective small interfering RNA (siRNA) delivery to a human metastatic/castration-resistant prostate cancer (PCa) cell line, which expresses PSMA on the surface. The PSMA-Rn (n = 6 and 9) peptides were synthesized by solid-phase peptide synthesis, characterized by liquid chromatography-mass spectrometry (LC-MS) and condensed with glucose-regulated protein (GRP)-silencing siRNAs. Native gels showed formation of stable CTP:siRNA ionic complexes. Furthermore, siRNA release was effected by heparin competition, supporting the peptides’ capabilities to act as condensing and releasing agents. However, dynamic light scattering (DLS) and transmission electron microscopy (TEM) studies revealed large anionic complexes that were prone to aggregation and limited cell uptake for RNAi activity. Taken together, these data support the notion that the development of efficient peptide-based siRNA delivery systems is in part contingent on the formulation of discrete nanoparticles that can effectively condense and release siRNA in cells.
Introduction
The prostate-specific membrane antigen (PSMA) is a type II transmembrane protein that functions as a surface carboxypeptidase enzyme, an exopeptidase with folate hydrolase activity, because it progressively liberates glutamates from glutamate-rich sources.1 Moreover, this receptor has been shown to have an internalization signal, which once activated allows for enzyme cell uptake via an endosome formed from clathrin-dependent endocytosis.2 In cancer, PSMA is expressed in all prostatic tissues, including primary prostate adenocarcinomas, metastatic prostate cancer, and in the tumor neovasculature of many solid tumors but not normal tissues.3,4 In prostate cancer (PCa), PSMA is highly expressed in poorly differentiated, highly metastatic prostatic cells and in castrate-resistant models.5 In fact, PSMA-based positron-emission tomography (PET) and computed tomography (CT) imaging is an emerging field in the diagnosis and treatment of advanced and resistant PCa, rendering PSMA a valuable biomarker for targeted forms of therapies.5
The phage display selection of PSMA binding peptides has resulted in the identification of three peptide sequences: GDHSPFT, SHFSVGS and EVPRLSLLAVFL, which are capable of targeting, binding, and internalizing within PSMA-expressing PCa cells.6 The selected PSMA binding peptides were based on the consensus sequences, SHSFSVGSGDHSPFT and GRFLTGGTGRLLRIS. These peptides were labeled with the 5-carboxyfluorescein (5-FAM) fluorophore and bound selectively to PSMA-expressing PCa cells. Moreover, cell uptake studies by fluorescence imaging revealed that the peptides accumulated intracellularly and were shown as disperse fluorescent punctuate regions found within the cells. Similarly, a separate phage display study was used to select another PSMA binding peptide.7 This 12-mer peptide, GTIQPYPFSWGY, was shown to: (1) have good binding affinities (8–9 μM) to PSMA+ LNCaP and C4-2 PCa cell lines; (2) facilitate cell surface staining for microscopy when fluorescently labeled; (3) enable the delivery of the D-(KLAKLAK)2 cytotoxic peptide to LNCaP cells to induce cell death; and (4) have favorable in vivo distribution where it selectively accumulated in prostate tissue of a C4-2 mouse xenograft with minimal uptake in any other major organ. These key lead peptides highlight the clinical potential in targeting this receptor as a diagnostic tool and for the specific delivery of therapeutics against PCa.
The oligoarginine and polyarginine peptides (Rn, where n = 6–16) are an important class of cell-penetrating peptides (CPPs) capable of small interfering RNA (siRNA) delivery in a wide range of cell types.8 These peptides are hydrophilic with a high polycationic charge density and have a higher cell-penetrating potential due to the strong affinity of the guanidinium group for the phospholipids in cell membranes. The charged side chains can infiltrate into the lipid bilayer and essentially create a pore within the membrane through which the CPP and its cargo can penetrate into the cells.8,9 For example, an R9 peptide complexed with siRNA has been used to silence EGFP expression in human gastric carcinoma cells.10 Moreover, an in vivo application of an R12 CPP-siRNA complex was found to reduce subcutaneous tumor growth in a mouse xenograft model via the silencing of the Her2 protein.11
However, a major limitation of using either cancer-targeting or cancer-penetrating peptides for siRNA transfection is respectively related to their limited cell permeability or nonspecific delivery. Here, we present a combination approach involving the use of a PSMA targeting peptide (PSMA-1, GRFLTGGTGRLLRIS) and oligoarginine penetrating peptides (Rn = 6 and 9) within a single peptide sequence for targeted delivery of glucose-regulated protein (GRP)-silencing siRNAs in PCa cells. The GRPs (GRP 75, 78, and 94) are chaperone proteins that serve as main sensors for misfolded proteins in the endoplasmic reticulum (ER) and trigger the unfolded protein response (UPR) under physiological and pathological cellular stress conditions.12 Importantly, GRPs are overexpressed in cancer, where they exhibit a variety of signaling pathways associated with cancer initiation, proliferation, adhesion, and invasion, which contributes to metastatic spread.13,14 Thus, GRPs have been classified as clinically proven therapeutic targets for specific cancer detection and treatment modalities. Toward this goal, we have developed GRP-silencing siRNA nanostructures15,16 and their bioconjugates17, 18, 19 for potentiating GRP knockdown and cell death in a panel of cancer cell lines. Building upon this work, we now report a peptide formulation capable of efficiently complexing and releasing siRNA, but with limited cell uptake due to aggregation into large, polydisperse anionic nanoparticles. Therefore, we emphasize the need for restricting particle size and charge distributions when developing peptide-based siRNA delivery formulations.
Results
Design and Synthesis of Peptide and siRNA Sequences
The PSMA binding peptide chosen for this work was based on the phage display selection reported by Shen et al.6 The peptide GRFLTGGTGRLLRIS (PSMA-1) was selected and polyarginine sequences R6 or R9 were added at the C terminus to generate the CTPs (Figure 1A), PSMA-1-Rn (n = 6 or 9), using classical Fmoc chemistry on the hydrophilic polyethylene glycol (PEG) resin (Scheme S1). Moreover, fluorescein isothiocyanate (FITC) labeling of the peptide with an aminohexanoic acid (Ahx) linker at the N terminus produced FITC-PSMA-1, to track PSMA binding on PCa cells, and FITC-PSMA-1-R9, to assess the influence of the nona-arginine sequence on siRNA delivery in PCa cells. Following synthesis, the peptides were cleaved and deprotected from the solid support and subjected to reversed-phase, ion-pairing (RP-IP) high-pressure liquid chromatography (HPLC) purification (>90%) (Figures S3, S4, S6, S8, and S9) and characterization by electrospray ionization (ESI)-MS (Table S1; Figures S5, S7, and S10).
Figure 1.
Rational Design of Peptide and siRNA Sequences Used in This Study for the Generation of the CTP:siRNA Complex
(A) Peptide and siRNA sequences used to generate the CTP:siRNA complex. (B) CTP:siRNA complex.
The GRP-silencing siRNAs adopting linear and higher order V-shape nanostructures were synthesized by automated solid-phase synthesis and isolated following our published procedures.15,16 For the synthesis of the V-shape siRNA, an orthogonally protected 5′-levulinyl 2′-monomethoxytrityl (5′-OLv 2′-OMMT) ribouridine phosphoramidite was used as branchpoint synthon to selectively extend the linear RNA sequence on solid phase into the desired V-shape RNA template (Figure 1A). Following solid-phase RNA synthesis, samples were cleaved and deprotected from the solid support, analyzed, and purified by RP-IP HPLC to obtain pure (>95%) single-stranded RNA. The linear and V-shape siRNA hybrids were developed in annealing buffer (10 mM Tris, 50 mM NaCl, 1 mM EDTA, pH 7.5–8.0) by mixing equimolar (250 pmol) quantities of the linear and V-shape RNA templates with their cRNA single strands. Samples were then heated briefly (95°C, 5 min), slowly cooled to room temperature (22°C), and stored overnight in a refrigerator (5°C) prior to analyses. Stable siRNA hybrids were then treated with CTP (PSMA-1-R6) to determine the optimal conditions for CTP:siRNA complex formation (Figure 1B).
PSMA-1-R6 Can Efficiently Complex and Release siRNA
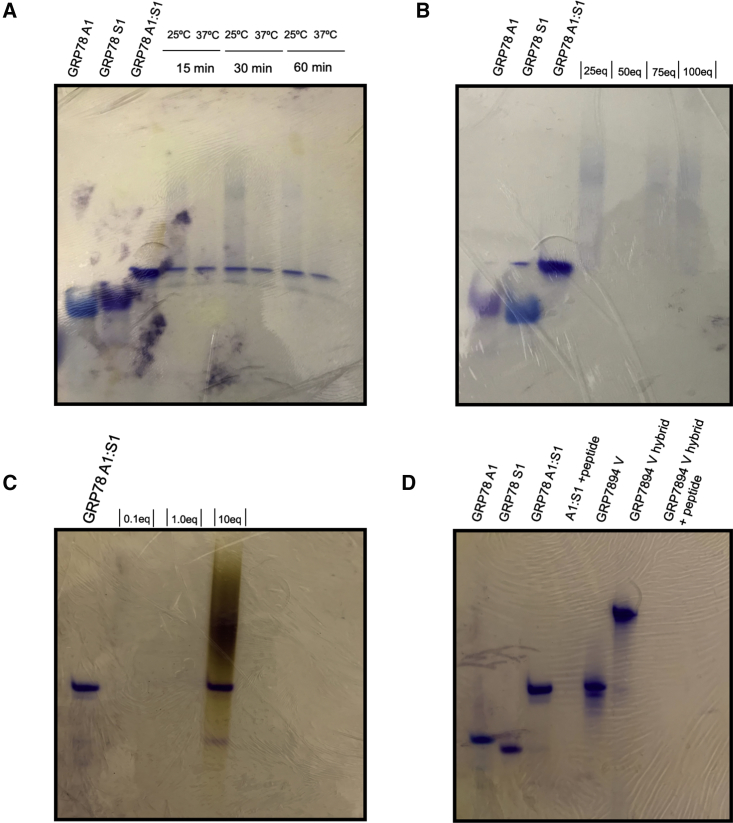
Native PAGE shift assays were then used to determine the extent of CTP:siRNA complexation by optimization of the stoichiometric mole ratios (Figure 2). The PSMA-1-R6 CTP was used to probe the extent of siRNA complexation as the R9 variant was rationalized to interact with the siRNAs in similar fashion. In order to optimize the stoichiometric ratios of the CTP:siRNA complexes, stoichiometric to excess amounts of CTP (1–100 equiv) were added to the hybrid siRNA (250 pmol) in annealing buffer (50 mM NaCl, 10 mM Tris, 1 mM EDTA, pH 7.3–7.5), followed by incubation at 37°C for 30 min prior to addition of the gel loading sucrose buffer for native PAGE analysis (Figure 2B). CTP:siRNA complex formation was identified by a shift to more retained bands on the gel with a decrease in intensity when compared to the untreated siRNA controls. It is expected that the CTP:siRNA formulation should move to a more retained higher order complex with slower electrophoretic mobility due to RNA charge masking, and with minimal signal detection due to the limited capabilities of the Stains-All dye to stain RNA in the CTP:siRNA complex form. These trends were also observed in the time and temperature optimization studies (Figure 2A), which indicates CTP:siRNA complex formation within 15 min at 25°C and 37°C. At 50 equiv of CTP, incubated at 37°C for 30 min, the most efficient CTP:siRNA complex was detected for the linear and V-shape siRNA (Figure 2D). Furthermore, the CTP:siRNA complex at these optimized conditions was challenged by a heparin-release assay to validate the bifunctional siRNA complex and release capabilities of the CTP. In this assay (Figure 2C), a concentration-dependent study (0.1–10 equiv) of heparin was used to determine the extent of CTP:siRNA complex stability and the optimal heparin concentration for siRNA displacement. Native PAGE analysis revealed that with excess heparin (10 equiv), siRNA (250 pmol) was displaced from the CTP (12.5 nmol) and stained as a dark violet band by the Stains-All dye solution. This result confirmed the reversibility of the CTP:siRNA complex formulation, which is critical for the prerequisite siRNA cell uptake and intracellular release requirements for RNAi activity.
Figure 2.
Optimization Conditions of the CTP:siRNA Complexes
(A) Time (0–60 min) and temperature (25°C and 37°C) dependence. (B) siRNA:CTP stoichiometric mole ratios (1:25–100 equiv). (C) Concentration-dependent (0.0163–1.63 mg/mL) heparin release assay at 37°C for 30 min. (D) CTP:siRNA complexes with linear and V-shape siRNA (250 pmol) in combination with PSMA-1-R6 (12.5 nmol).
Cell Uptake of the CTP:siRNA Complexes
Two PCa (PSMA+ LNCaP and a PSMA− PC3) cell lines were tested for FITC-PSMA-1 binding by flow cytometry (Figure S1). Surface expression of the PSMA receptor was confirmed on the LNCaP cells using a phycoerythrin (PE)-conjugated PSMA monoclonal antibody (GCP-05, Invitrogen). No antibody binding was observed on the PC3 cells, validating that this cell line is PSMA−. A distinct increase in the fluorescence intensity for the FITC-PSMA-1 peptide was detected on the LNCaP cells relative to the PC3 cells, indicating PSMA binding on the LNCaP cells with some small degree of nonspecific binding on the PC3 cells. The initial lead peptide, PSMA-1-R6, was probed for its ability to potentially function as a selective siRNA transfection vector within the LNCaP PSMA+ cells.
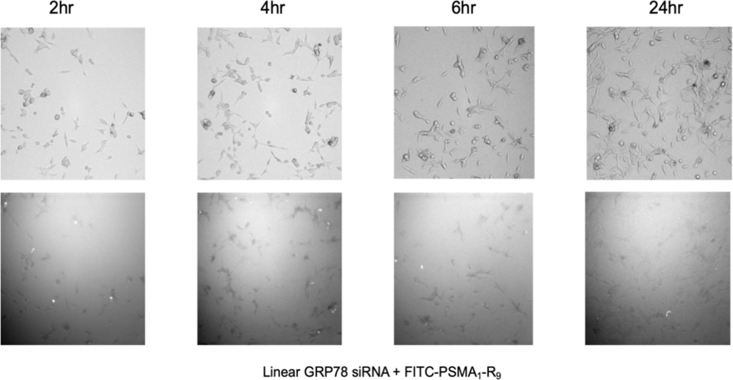
The FITC-labeled PSMA-1-R9 peptide:siRNA complex was evaluated for cell uptake using fluorescence microscopy. It was hypothesized that the lengthier poly(arginine), R9 versus R6, would benefit siRNA complex formation and cell uptake.10,11 Briefly, the PSMA-1-R9 peptide (1.25 μM) was incubated with siRNA (25 nM) to form the CTP:siRNA complex for 30 min at 37°C, followed by addition to a monolayer of LNCaP cells (∼60%–70% confluency) and imaged, using high content screening during a 24-h period. Little fluorescence was observed during the time course of this assay (2–24 h), indicating little (if any) cell binding and uptake of the CTP:siRNA complex (Figure 3).
Figure 3.
HCS Imaging of LNCaP Cells Incubated with FITC-PSMA-1-R9:siRNA Complexes Over Time
Top: brightfield channel. Bottom: LED filter (485 excitation/520 emission).
GRP78 Knockdown in LNCaP and PC3 Cells
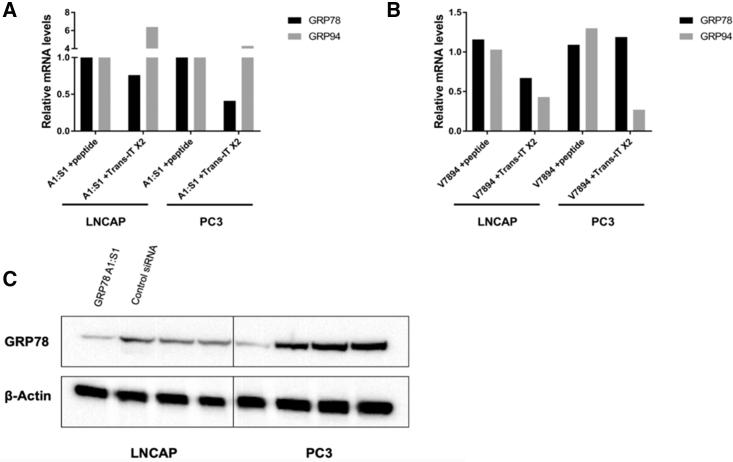
In order to evaluate whether any GRP knockdown activity would occur with the CTP:siRNA complex, in spite of its limited cell uptake, the CTP, PSMA-1-R6 (1.25 μM), was mixed with siRNA (25 nM) and incubated within a monolayer of either LNCaP or PC3 cells at ∼60%–70% confluency. Likewise, the siRNA was complexed with the Trans-IT X2 dynamic delivery system and transfected separately into the cells. The knockdown efficiency was monitored by qRT-PCR and western blot at 48 or 72 h post-transfection, respectively. No changes in the mRNA levels (Figure 4A) or the protein expression levels (Figure 4B) were observed when transfecting a linear siRNA against GRP78 into either cell line as compared to 30%–70% knockdown when using the Trans-IT X2 reagent. Similarly, peptide-based transfection of higher order V-shape siRNA against GRP78 and GRP94 into the LNCaP cells resulted in no knockdown of the target genes compared to siRNA transfection with the Trans-IT X2 dynamic delivery system.
Figure 4.
GRP Knockdown in the LNCaP or PC3 Cell Lines Using the PSMA-1-R6 Peptide or the Trans-IT X2 System as the Transfection Reagents
The cells were transfected with 25 nM siRNA with relative mRNA levels analyzed by qRT-PCR at 48 h and total protein levels analyzed at 72 h. All knockdown levels were normalized against a negative siRNA control. (A and B) mRNA knockdown comparison by qRT-PCR of the knockdown efficiency between the peptide or the Trans-IT X2 reagent of (A) linear siRNA against GRP78 and (B) V-shape siRNA against GRP78 and GRP94 is shown. (C) Protein expression change comparison between the peptide or the Trans-IT X2 reagent.
DLS and TEM Analysis of the CTP:siRNA Complexes
Dynamic light scattering (DLS) analysis and transmission electron microscopy (TEM) imaging were used to determine whether the PSMA-1-Rn (n = 6 or 9) peptides were capable of condensing siRNA into small, discrete, monodisperse nanoparticles, a requirement for efficient transfection.20 The sizes, shapes, and ionic charge distributions of the CTP:siRNA complexes were compared to the complexes generated with the siRNA:Trans-IT X2 dynamic delivery system functioning as control (Table S2). The siRNA:Trans-IT X2 complexes had uniform particle sizes ∼400 nm with approximate net neutral charges (0.77 ± 1.24 mV). In comparison, the CTP:siRNA complexes showed much larger particle sizes ranging from ∼800 to 1,000 nm with slight negative zeta potential measurements (−5.88 ± 0.25 for the PSMA-1-R6 complex and −2.16 ± 0.39 mV for the PSMA-1-R9 complex). Large aggregates (with effective diameters in the range of 1.5–2 μm) were also observed for the CTP:siRNA complexes, which compares to ∼4- to 5-fold the size of the siRNA:Trans-IT X2 complexes.
The TEM images (Figure S2) supported the DLS data and provided further insights into particle sizes and extent of aggregation. Interestingly, the CTP:siRNA complex with the more amphiphilic peptide sequence, PSMA-1-R9, generated large porous networks of peptide-bound siRNA, which may altogether restrict siRNA condensation for cell delivery. Conversely, the CTP:siRNA complexes with the PSMA-1-R6 peptide generated particles of similar morphology that were, on average, 200–300 nm larger than the siRNA:Trans-IT X2 complexes. In both cases, the CTP:siRNA complexes produced larger, polydisperse particles that were prone to aggregation when compared to the small, discrete, monodisperse nanoparticles generated by the siRNA:Trans-IT X2 dynamic delivery system. Furthermore, considering that surface charge density may play a critical role in cell binding and penetration, the more neutral siRNA:Trans-IT X2 dynamic delivery system may function to minimize repulsion at the cell surface for delivery. Taken together, size, shape, and charge are important factors for developing nanoparticle formulations for siRNA delivery.
Discussion
This study highlights the design, synthesis, characterization, and biological evaluation of a new class of PSMA-targeting peptides for siRNA delivery in PCa cells. The peptides were functionalized with poly(arginine) in order to improve siRNA complex formation for cell delivery. Native PAGE was used to optimize conditions for forming stable CTP:siRNA complexes that also resulted in siRNA release in a heparin displacement assay. The PSMA-targeting peptides were validated by flow cytometry, which revealed binding to the PSMA+ LNCaP PCa cells. Cell uptake studies by flow cytometry and high content screening of the FITC-labeled CTP:siRNA complexes failed to show cell binding and translocation, which translated into a lack of siRNA-mediated GRP knockdown at the mRNA and protein levels of expression. To account for this, DLS and TEM studies indicated large, polydisperse, anionic aggregates for the CTP:siRNA complexes when compared to the small, monodisperse neutral nanoparticles observed for the control transfection with the siRNA:Trans-IT X2 dynamic delivery system. These properties likely contributed to the divergent siRNA activities in PCa cells with the competing transfection reagents. These limitations are a current focus in our research work in order to improve siRNA cell uptake with peptide vectors. For example, incorporating PEG linkers or coatings in nanoparticle formulations have been shown to reduce aggregation and improve uptake and biodistribution in vivo.21,22 Ultimately, effective siRNA transfections will be based on nanoparticle formulations that are amenable to efficient cell uptake for siRNA activity inside cells. Toward this objective, we are currently exploring the effect of formulating siRNAs within stable metallic formulations that have been effectively used as biocompatible theranostic agents to condense siRNAs into small, discrete nanoparticles for detection in cell uptake studies and RNAi activity in PCa cells.23,24 Their successful applications provide the opportunity to further functionalize the nanoparticles with CTPs in order to develop cell-targeted delivery of siRNAs in cells and potentially also in tumor-bearing animal models. The latter is a current focus of our ongoing and future research work geared toward the development of effective cancer-targeting gene therapy strategies.
Materials and Methods
Materials
Amino acids for peptide synthesis were purchased from Novabiochem (San Diego, CA, USA). Peptide syntheses were conducted on a Rink Amide ChemMatrix (0.47 mmol/g) (Biotage, Charlotte NC, USA). 2-(6-Chloro-1H-benzotriazole-1-yl)-1,1,3,3-tetramethylaminium hexafluorophosphate (HCTU) was purchased from Advanced ChemTech (Louisville, KY, USA). FITC was purchased from Thermo Scientific (Rockford, IL, USA) as a single isomer and used in the dark to fluorescently label all peptides. Trifluoroacetic acid (TFA; Biograde) was purchased from VWR (Radnor, PA, USA); N-N-dimethylformamide (DMF), acetonitrile (MeCN), methanol, and dichloromethane (DCM) were all purchased from Macron (Center Valley, PA, USA). Piperidine, triethylsilane (TES; 98+%), and pyridine (ACS, 99%) were purchased form Alfa Aesar (Ward Hill, MA, USA). N-methylmorpholine (NMM) (99%) was purchased from Acros Organics (Pittsburg, PA, USA). Diethyl ether (99%, ACS), Et2O, used to precipitate peptides, was purchased from Sigma-Aldrich (St. Louis, MO, USA). Solid-phase oligonucleotide synthesis reagents and RNA phosphoramidites were obtained from ChemGenes (Wilmington, MA, USA) or Glen Research (Sterling, VA, USA).
siRNA Synthesis
RNA was synthesized on an automated ABI 3400 synthesizer. Reagents for the synthesis cycle were as follows: detritylation reagent (3% solution of dichloroacetic acid in DCM), coupling reagent (0.25 M ethylthiotetrazole in MeCN), capping reagent (cap A, 1:1:8 v/v/v acetic anhydride/pyridine/tetrahydrofuran; cap B, 16% N-methyl imidazole in tetrahdrofuran), oxidation reagent (0.02 M iodine in 75:20:5 v/v/v tetrahydrofuran/pyridine/water), and MeCN wash (biotech grade purchased from EMI). Oligonucleotide syntheses were performed on a 1-μmol scale using 1,000–2,000 Å long-chain alkylamine (LCAA) CPG. RNA phosphoramidites were prepared as 0.15 M solutions in anhydrous MeCN with coupling times of 10 min. For the synthesis of branch RNA, the branchpoint phosphoramidites were also prepared as 0.2 M solutions in MeCN and coupled with an extended reaction time of 20 min. RNA syntheses were performed using the following synthesis cycles: for detritylation, DCM wash (30 s) followed by delivery of 3% dichloroacetic acid in DCM (120 s); for coupling, delivery of RNA phosphoramidites with activator (0.25 M ethylthiotetrazole in MeCN); for capping, delivery of cap A and cap B (14 s); and for oxidation, delivery of oxidizing solution (0.02 M iodine in 75:20:5 v/v/v tetrahydrofuran/pyridine/water) for 20 s followed by a wait period of 20 s. Single-stranded RNA sequences were extended in the 3′→5′ orientation on solid phase until completion. V-shape RNAs were first grown in the 3′→5′ position followed by coupling of a branchpoint phosphoramidite. The branchpoint phosphoramidite was coupled for 20 min, followed by a manual decyanoethylation step (2:3 v/v trimethylamine/MeCN, 90 min) prior to the 2′-OMMT detritylation. The oligonucleotide-bound CPG was washed with MeCN (30 mL) and replaced on the synthesizer to complete 3′→5′ RNA synthesis of the subsequent sequence. The terminal 5′-OH was then capped with automated delivery of cap A and cap B (14 s). Cleavage and deprotection of the synthesized RNA was performed after drying the synthesizer columns under argon (Ar(g)) for 10 min. CPG-bound RNA was then transferred into autoclaved screw cap microtubes (1.5 mL) and treated with a 1-mL solution of 3:1 v/v ammonium hydroxide in absolute ethanol at 55°C for 16–20 h. RNA samples were then evaporated in vacuo and the CPG was washed twice with autoclaved distilled water (500 μL). Crude oligonucleotides were resuspended in a mixture of 1:1.5 v/v dimethylsulfoxide/triethylamine trihydrofluoride (150 μL) to complete the 2′-desilylation reaction at 65°C for 2 h. The crude RNA samples were subsequently precipitated from the reaction mixture with 3 M sodium acetate (NaOAc; 25 μL) in n-butanol (n-BuOH; 1 mL). Precipitation was completed in the freezer (−80°C) for 30 min prior to centrifugation (12,000 rpm), leaving the crude oligonucleotides as a solid white pellet. The collected RNA samples were dried in a SpeedVac concentrator and resuspended in autoclaved water (1 mL) for yield determination using UV absorbance measurements at 260 nm.
Peptide Synthesis
All peptides were synthesized by stepwise manual solid-phase peptide synthesis using Fmoc chemistry. Fmoc amino acids (3 equiv, 0.3 mmol) were coupled on a Rink amide linker PEG solid support (0.47 mmol/g, 0.1 mmol) for 1 h using HCTU (3 eq, 0.3 mmol) and NMM (6 equiv, 0.6 mmol) in DMF (3.5 mL). Fmoc deprotection was completed with a 20% piperidine in DMF solution (3 mL, twice for 10 min). Amino acid couplings and Fmoc deprotections were repeated until the desired sequences were completed. Upon completion of the PSMA-1 and PSMA-1-R9 sequences, the solid support was subjected to the addition of Fmoc-Ahx linker as previously described, followed by FITC labeling. A slurry of FITC (1.1 equiv, 0.33 mmol) in pyridine/DMF/DCM (12:7:5 v/v) was prepared and added to the reaction cartridge and left on a shaker for 16–18 h. After FITC-labeling was completed, the resin was washed with DMF (3 × 3 mL), MeOH (3 × 3 mL), and DCM (3 × 3 mL). Peptide cleavage and deprotection from the solid support was accomplished using a mixture of TFA/TES/H2O (95:2.5:2.5 v/v/v) for 4 h. Peptide samples were concentrated under nitrogen to a viscous oil, precipitated with cold Et2O, and centrifuged to a white pellet. The supernatant was decanted and the peptide pellets were dissolved in MeCN/H2O for liquid chromatography-mass spectrometry (LC-MS) analyses.
Peptide RP HPLC and ESI-MS
Sample analyses were performed on an Agilent 1100 series ESI/LC-MS with single quadrupole mass analyzer and LC conditions, which used an isocratic binary solvent system (85% MeOH/H2O, 0.1% formic acid (FA), 2 min) in positive mode. Analytical RP HPLC was performed using a Waters 2695 Symmetry C18 column (3.9 × 150 mm, 5-μm particle size) with a gradient of 20%–80% MeCN/H2O, 0.1% TFA, over 18 min at 25°C, with a 1 mL/min flow rate and detection at 220 or 480 nm for FITC-labeled samples. Purified peptides were lyophilized to a fine powder before being dissolved in TE buffer (10 mM Tris, 50 nM NaCl, 1 mM EDTA, pH 7.5–8.0) to confirm purity. Peptide sequences and purities can be found in Table S1.
Native PAGE Shift Assay
Linear GRP78 siRNA (250 pmol) was incubated with the PSMA-1-R6 peptide (25–100 equiv, 6.25–25 nmol) for 60 min at either 25°C or 37°C. Aliquots were taken at 15-min intervals, mixed with a 30% sucrose buffer, and loaded on an 18% native polyacrylamide gel. V-shape siRNA (250 pmol) was incubated separately with the PSMA-1-R6 peptide (50 equiv, 12.5 nmol) for 30 min at 37°C before being loaded onto the gel. The assay was run at 300 V, 100 mA, 12 W for 5 h followed by sample detection with a Stains-All solution. For the heparin titration assay, the linear siRNA (250 pmol) was incubated with the PSMA-1-R6 peptide (50 equiv, 12.5 nmol) for 30 min at 37°C and then cooled to room temperature (22°C). Heparin (0.1–10 equiv, 0.02–1.6 mg/mL) was titrated into the CTP:siRNA complex (250 pmol) and incubated for 30 min at 37°C before being loaded onto a separate 18% native polyacrylamide gel and run at 300 V, 100 mA, 12 W for 5 h followed by sample detection with a Stains-All solution.
Dynamic Light Scattering
A Malvern Zetasizer, Nano-ZS (Malvern Instruments, UK) employing a 173° scattering angle and a 4-mW incident He-Ne laser (633 nm) was used to measure the particle sizes (hydrodynamic diameter), size distributions, and zeta potentials of the siRNA:Trans-IT X2 dynamic delivery system control and CTP:siRNA complex. Samples were measured in triplicate at 25°C. All samples were loaded into folded capillary cells (DTS1070) equipped with electrodes on both sides to allow measurement of their zeta potentials and, by extension, the stability and degree of aggregation. Particle suspensions with highly positive or highly negative zeta potentials are considered stable because the electrical repulsion between the particles tends to counter the van der Waals forces that would otherwise result in aggregation and precipitation.
TEM Imaging
TEM analyses of the siRNA samples incubated with CTP or the Trans-IT X2 dynamic delivery system were performed with a JEOL 1200EX transmission electron microscope (JEOL, Japan) at an accelerating voltage of 80 kV. A mixture of 1:1 volume ratio 1% uranyl acetate as a sample suspension was prepared and 10 μL of this solution was placed on a TEM carbon-film-coated copper grid of 300 mesh (Electron Microscopy Sciences, Hatfield, PA). Each sample was allowed to sit for 5 min on the grid before wicking the excess liquid followed by storage for 1 week to allow the samples to dry. Images were taken with a SIA-L3C CCD camera (Scientific Instruments and Applications) using the software Maxim DL5 (Diffraction, Ottawa, ON, Canada).
Cell Culture
PC3 (bone metastatic PCa cell line, ATCC-CRL-1435) and LNCaP (clone FGC ATCC-CRL-1740) cells were purchased from ATCC. Cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS), 2.5 mM l-glutamine, and 1% penicillin/streptomycin at 37°C in a humidified tissue culture incubator containing 5% CO2.
siRNA Transfection
A monolayer of PC3 or LNCaP cells was grown in a 24-well culture plate in complete growth media until 60%–70% confluent to ensure expression of the PSMA receptor. Linear GRP78 siRNA (12.5 pmol) was incubated with either the PSMA-1-R6 or the FITC-PSMA-1-R9 peptides (625 pmol) for 30 min at 37°C in Opti-MEM before being added to the cells and diluted to a final siRNA concentration of 25 nM. siRNA was also complexed with the Trans-IT X2 transfection reagent in Opti-MEM according to the manufacturer’s protocol and added to the cells at a final concentration of 25 nM. The cells were then incubated at 37°C for 48–72 h.
qRT-PCR
Total mRNA was isolated following transfection (48 h) from TRIzol (Ambion)-preserved cells using a TriRNA Pure Kit (Geneaid), following the manufacturers’ instructions. The collected mRNA was then quantitated on a Qubit 3.0 fluorimeter using the Qubit broad range (BR) assay kit (Thermo Fisher Scientific), and mRNA (200 ng) was reversed transcribed into cDNA using a high-capacity cDNA kit (Applied Biosystems). RT-PCR was performed using pre-developed TaqMan gene expression primer probes for GRP78 (assay ID Hs99999174_m1), GRP94 (assay ID Hs00437665_g1), and GAPDH (Hs99999905_m1) and TaqMan fast advanced master mix. A qPCR fast assay was carried out on a StepOnePlus (Applied Biosystems). Fold changes were calculated with the ΔΔCt method using GAPDH as an endogenous control and the negative siRNA as the control sample.
Western Blot
Total protein was isolated from the cell cultures following transfection (72 h). Protein lysates were prepared by lysing the cells in ice-cold RIPA buffer (G-Biosciences) supplemented with protease and phosphatase inhibitors (MilliporeSigma), which were diluted 1:10 v/v as per the manufacturer’s recommendations. Cell debris was removed by centrifugation at 16,000 × g at 4°C, and protein concentrations were determined using a Pierce BCA kit (Thermo Fisher Scientific). A sample (20–35 mg) of the supernatant protein was mixed with LDS buffer and DTT, incubated at 70°C for 10 min and resolved on a 4%–12% Bis-Tris PAGE gradient gel before being transferred to a polyvinylidene fluoride (PVDF) membrane. Following transfer, the membrane was blocked in 5% skim milk for 1 h, washed, and incubated at 4°C overnight with a rabbit 1° monoclonal antibody against human GRP78, GRP94, GRP75, or β-actin (all purchased from Cell Signaling Technology) at a 1:1,000 dilution. The membrane was subsequently washed and incubated with an anti-rabbit horseradish peroxidase (HRP)-conjugated 2° Ab (Cell Signaling Technology) for 1 h at room temperature at a 1:2,000 dilution. The bands were visualized using a SignalFire enhanced chemiluminescence (ECL) reagent (Cell Signaling Technology) on a ProteinSimple FluorChem E imager.
Author Contributions
C.N.C., D.S., and J.Z. designed the study and wrote the manuscript. C.N.C. conducted the study in its entirety. S.S., G.A., and C.J.H. conducted the experiments. J.A.R. and U.S. provided facility use, training, and data analysis for DLS and TEM analysis. All authors reviewed and approved the manuscript.
Conflicts of Interest
All authors contributed equally to this work and declare no competing interests.
Acknowledgments
D.S. acknowledges financial support from the SHU URC award as well as Dr. Mark Hail and Kevin McCarl (Novatia Inc.) for MS data acquisition. U.S. acknowledges financial support from the PSC-CUNY Research Award and the Undergraduate Research and Mentoring Education (URME) program at Queens College as well as use of the Core Facility for Imaging, Cellular, and Molecular Biology at Queens College. This research was supported in part by funding from the Emerald Foundation, Inc. to J.Z.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.10.013.
Supplemental Information
References
- 1.Pinto J.T., Suffoletto B.P., Berzin T.M., Qiao C.H., Lin S., Tong W.P., May F., Mukherjee B., Heston W.D. Prostate-specific membrane antigen: a novel folate hydrolase in human prostatic carcinoma cells. Clin. Cancer Res. 1996;2:1445–1451. [PubMed] [Google Scholar]
- 2.Rajasekaran S.A., Anilkumar G., Oshima E., Bowie J.U., Liu H., Heston W., Bander N.H., Rajasekaran A.K. A novel cytoplasmic tail MXXXL motif mediates the internalization of prostate-specific membrane antigen. Mol. Biol. Cell. 2003;14:4835–4845. doi: 10.1091/mbc.E02-11-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross J.S., Sheehan C.E., Fisher H.A., Kaufman R.P., Jr., Kaur P., Gray K., Webb I., Gray G.S., Mosher R., Kallakury B.V. Correlation of primary tumor prostate-specific membrane antigen expression with disease recurrence in prostate cancer. Clin. Cancer Res. 2003;9:6357–6362. [PubMed] [Google Scholar]
- 4.Silver D.A., Pellicer I., Fair W.R., Heston W.D., Cordon-Cardo C. Prostate-specific membrane antigen expression in normal and malignant human tissues. Clin. Cancer Res. 1997;3:81–85. [PubMed] [Google Scholar]
- 5.von Eyben F.E., Baumann G.S., Baum R.P. PSMA diagnostics and treatments of prostate cancer become mature. Clin. Transl. Imaging. 2018;6:145–148. doi: 10.1007/s40336-018-0270-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen D., Xie F., Edwards B. Evaluation of phage display discovered peptides as ligands for prostate-specific membrane antigen (PSMA) PLoS ONE. 2013;8:e68339. doi: 10.1371/journal.pone.0068339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin W., Qin B., Chen Z., Liu H., Barve A., Cheng K. Discovery of PSMA-specific peptide ligands for targeted drug delivery. Int. J. Pharm. 2016;513:138–147. doi: 10.1016/j.ijpharm.2016.08.048. [DOI] [PubMed] [Google Scholar]
- 8.Tünnemann G., Ter-Avetisyan G., Martin R.M., Stöckl M., Herrmann A., Cardoso M.C. Live-cell analysis of cell penetration ability and toxicity of oligo-arginines. J. Pept. Sci. 2008;14:469–476. doi: 10.1002/psc.968. [DOI] [PubMed] [Google Scholar]
- 9.Futaki S., Suzuki T., Ohashi W., Yagami T., Tanaka S., Ueda K., Sugiura Y. Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J. Biol. Chem. 2001;276:5836–5840. doi: 10.1074/jbc.M007540200. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y.H., Hou Y.W., Lee H.J. An intracellular delivery method for siRNA by an arginine-rich peptide. J. Biochem. Biophys. Methods. 2007;70:579–586. doi: 10.1016/j.jbbm.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Kim S.W., Kim N.Y., Choi Y.B., Park S.H., Yang J.M., Shin S. RNA interference in vitro and in vivo using an arginine peptide/siRNA complex system. J. Control. Release. 2010;143:335–343. doi: 10.1016/j.jconrel.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 12.Lee A.S. Glucose-regulated proteins in cancer: molecular mechanisms and therapeutic potential. Nat. Rev. Cancer. 2014;14:263–276. doi: 10.1038/nrc3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cultrara C.N., Kozuch S.D., Ramasundaram P., Heller C.J., Shah S., Beck A.E., Sabatino D., Zilberberg J. GRP78 modulates cell adhesion markers in prostate Cancer and multiple myeloma cell lines. BMC Cancer. 2018;18:1263. doi: 10.1186/s12885-018-5178-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X.X., Li H.D., Zhao S., Zhao L., Song H.J., Wang G., Guo Q.J., Luan Z.D., Su R.J. The cell surface GRP78 facilitates the invasion of hepatocellular carcinoma cells. BioMed Res. Int. 2013;2013:917296. doi: 10.1155/2013/917296. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Maina A., Blackman B.A., Parronchi C.J., Morozko E., Bender M.E., Blake A.D., Sabatino D. Solid-phase synthesis, characterization and RNAi activity of branch and hyperbranch siRNAs. Bioorg. Med. Chem. Lett. 2013;23:5270–5274. doi: 10.1016/j.bmcl.2013.08.033. [DOI] [PubMed] [Google Scholar]
- 16.Patel M.R., Kozuch S.D., Cultrara C.N., Yadav R., Huang S., Samuni U., Koren J., 3rd, Chiosis G., Sabatino D. RNAi Screening of the glucose-regulated chaperones in cancer with self-assembled siRNA nanostructures. Nano Lett. 2016;16:6099–6108. doi: 10.1021/acs.nanolett.6b02274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah S.S., Cultrara C.N., Kozuch S.D., Patel M.R., Ramos J.A., Samuni U., Zilberberg J., Sabatino D. Direct transfection of fatty acid conjugated siRNAs and Knockdown of the glucose-regulated chaperones in prostate cancer cells. Bioconjug. Chem. 2018;29:3638–3648. doi: 10.1021/acs.bioconjchem.8b00580. [DOI] [PubMed] [Google Scholar]
- 18.Kozuch S.D., Cultrara C.N., Beck A.E., Heller C.J., Shah S., Patel M.R., Zilberberg J., Sabatino D. Enhanced cancer theranostics with self-assembled, multilabeled siRNAs. ACS Omega. 2018;3:12975–12984. doi: 10.1021/acsomega.8b01999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cultrara C.N., Shah S., Kozuch S.D., Patel M.R., Sabatino D. Solid phase synthesis and self-assembly of higher-order siRNAs and their bioconjugates. Chem. Biol. Drug Des. 2019;93:999–1010. doi: 10.1111/cbdd.13448. [DOI] [PubMed] [Google Scholar]
- 20.Zhang S., Li J., Lykotrafitis G., Bao G., Suresh S. Size-dependent endoytosis of nanoparticles. Adv. Mater. 2009;21:419–424. doi: 10.1002/adma.200801393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Braeckmans K., Buyens K., Bouquet W., Vervaet C., Joye P., De Vos F., Plawinski L., Doeuvre L., Angles-Cano E., Sanders N.N. Sizing nanomatter in biological fluids by fluorescence single particle tracking. Nano Lett. 2010;10:4435–4442. doi: 10.1021/nl103264u. [DOI] [PubMed] [Google Scholar]
- 22.Suk J.S., Xu Q., Kim N., Hanes J., Ensign L.M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 2016;99:28–51. doi: 10.1016/j.addr.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahme K., Guo J., Holmes J.D. Bioconjugated gold nanoparticles enhance siRNA delivery in prostate cancer cells. Methods Mol. Biol. 2019;1974:291–301. doi: 10.1007/978-1-4939-9220-1_21. [DOI] [PubMed] [Google Scholar]
- 24.Fitzgerald K.A., Rahme K., Guo J., Holmes J.D., O’Driscoll C.M. Anisamide-targeted gold nanoparticles for siRNA delivery in prostate cancer—synthesis, physicochemical characterization and in vitro evaluation. J. Mater. Chem. B Mater. Biol. Med. 2016;4:2242–2252. doi: 10.1039/c6tb00082g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.