Abstract
Molecular docking has been applied to elucidate the binding of lamellarin analogues with HIV-1 integrase strand transfer complex (PDB ID: 5U1C). The results suggest hydrogen bond interaction with residue Glu92 is key, and stabilisation by π−π stacking interactions with DNA base is chiefly influential to strand transfer activity. Other residues involved in hydrogen bonding are Cys65, His67, Asp64, Asp116 and chelation with Mg2+ ion was seen for certain analogues. Furthermore, hydrophobic interactions can be accounted for several amino acids including Asp64, Cys65, Asp116, His67, Glu92, Tyr143, Phe121, Gly118, Pro142 and Val72, as well as the DNA base. The molecular docking results are in line with the reported literatures of other inhibitors and strand transfer activity observed previously by Faulkner. We further employed molecular docking simulation to virtually screen and identified 4 novel potential inhibitors of HIV-1 integrase strand transfer complex from a Chembridge diversity collection of 25,132 small molecule compounds; Chembridge ID compound codes: 22850303, 27553460, 24578440 and 27591056. The candidates clearly formed hydrogen bonding interactions with important residues: His67 and Glu92. In addition, hydrophobic interactions were seen with residues similar to interactions with lamellarin analogues. The calculated drug-like scores are suggestive of these compounds to have clinical potential and ADMET predictions implied of their acceptable pharmacokinetic and toxicity profiles.
Keywords: Pharmaceutical chemistry, Lamellarin, Molecular docking, Virtual screening, HIV, HIV-1 integrase, preADMET, ADMET
Pharmaceutical chemistry, Lamellarin; Molecular docking; Virtual Screening; HIV; HIV-1 integrase; preADMET; ADMET
1. Introduction
Acquired Immune Deficiency Syndrome (AIDS) is caused by an infection of the Human Immunodeficiency Virus (HIV) that damages the human immune system weakening the ability for the human host to combat against threatening infections. By 2018 people living with AIDS globally reached 37.9 million and the death toll of AIDS patients was estimated to be just under 800,000 [1]. Undoubtedly, AIDS is still a serious health concern to the global health community. At least 30 small molecules have been clinically approved between 1987 and 2019 for the treatment of AIDS [2]. Although effective in softening the pandemic, anti-HIV drug toxicity is a drawback, e.g., Stevens-Johnson syndrome [3] and mitochondrial toxicity that can lead to myopathy and hepatoxicity [4]. Additionally, mutant strains have emerged [5] placing a warrant on novel chemotherapeutic anti-HIV agents.
The development of small molecule-based anti-HIV drugs were heavily concentrated on targeting the catalytic and allosteric sites of reverse transcriptase (RT) and viral protease leading to the approval of a plethora of RT and viral protease inhibitors [5]. Rapid viral replication rate confers mutations when exposed to antiviral drugs, e.g., K65R and L74V mutants are commonly resistant against nucleoside RT inhibitors, and K103N and V106A are known for resistance to non-nucleoside RT inhibitors, thus, reducing the effectiveness of RT inhibitors [5]. Viral protease inhibitors suffered a similar fate and was affected largely by mutations at L90, M46 and V82 residues [5]. As a result, pharmaceutical scientists stretched beyond the development of RT and viral protease inhibitors, and developed novel classes of small molecule inhibitors, e.g., entry and maturation inhibitors [5, 6].
One class of anti-HIV agents that is less developed are strand transfer inhibitors that bind to retroviral integrase to modulate the integration of viral DNA into the host genome. Three drugs have been approved for the treatment of AIDS targeting HIV-1 integrase, namely raltegravir, dolutegravir and elvitegravir, and several scaffold inhibitors have been developed. Thus, searches for novel inhibitors of HIV-1 integrase with clinical potential is realistic. Previously, crystallisation of HIV-1 integrase strand transfer complexes (STCs) has proven to be challenging as a result of solubility issues and integrase aggregation resulting in mainly crystal structures with separate domains available on the Protein Data Bank (PDB) [7]. This limited structural and binding studies involving the full structure of the target and quite often the Prototype Foamy Virus (PFV) integrase/DNA complex was often used as a model. Recently, the full atomic structure of tetrameric HIV-1 integrase strand transfer complex (intasome) was modelled based on the X-ray crystal structures of separate individual domains and cryogenic electron microscopy imaging techniques (Fig. 1) [8]. The complete structure of the intasome provides the possibility to accurately elucidate the binding modes of compounds that target HIV-1 integrase STC.
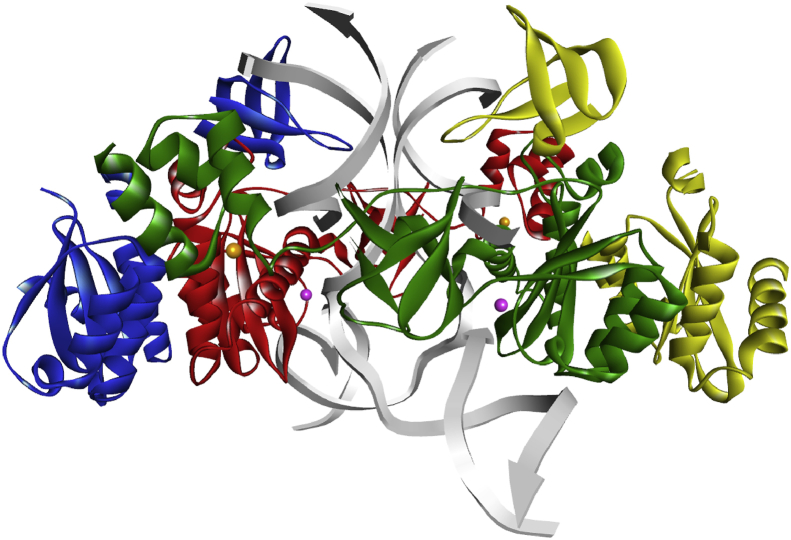
Fig. 1.
Structure of HIV-1 integrase STC intasome obtained from the Protein Data Bank (PDB code: 5U1C). Red and green structures depict the inner protomers, yellow and blue are the outer protomers, DNA strands are coloured grey, and metals ions are depicted as round spheres, orange and pink for Zn2+ and Mg2+ ions, respectively.
In this study, we applied a virtual screening approach using the Chembridge [9] diversity collection to search for potential inhibitors of HIV-1 integrase STC using the available structure. Chembridge Corporation is a company that supplies commercially available small molecules to drug discovery projects. Additionally, the computational binding interactions of lamellarin analogues with HIV-1 integrase STC was studied; Faulkner has previously reported strand transfer inhibition activities of lamellarin analogues [10, 11]. The lamellarins (see Fig. 2) are most notable for exerting cytotoxic effects against cancer cell lines with multiple mechanisms of action: topoisomerase [12, 13, 14, 15, 16], kinase inhibition [15] and mitochondrial damage [17]. Nevertheless, there has been a lack of study on the mechanistic binding action of these compounds with HIV-1 integrase.
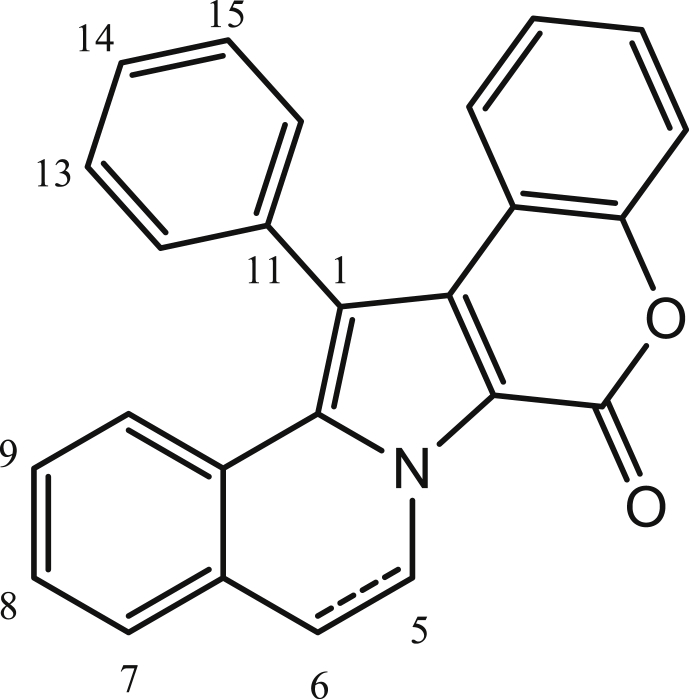
Fig. 2.
The chemical structure of the lamellarin scaffold.
2. Methodology
2.1. Ligand structure preparation
The molecular structures of lamellarin derivatives were drawn using the Scigress [18] version 2.8.1 software package. All sulfate groups were protonated for all lamellarin derivatives which replaced the counter ion, sodium. The drawn structures and the Chembridge diversity library in 3D format were subjected to steps of energy minimisation using the same Scigress 2.8.1 version software package. Molecular mechanics force-field MM2 [19] was applied followed by the semi-empirical PM6 [20] method.
2.2. Docking scaffold preparation
The structure of the intasome complex was obtained from the Protein Data Bank (PDB code: 5U1C) [8]. The structure is a tetramer complexed with integrated strands of viral and host target DNA. The outer protomers were removed as they lacked contacts with the DNA strands. The inner protomer with red ribbons (see Fig. 1) was chosen due to more contacts with the DNA strands. The DNA strands were included in the docking scaffold along with catalytic Mg2+ cofactor. The Zn2+ metal ions located at the N-terminal domains were removed. The Modeller [21] software version 9.2.0 was used to mutate Gln152 back to the wild-type Glu152 and the structure of the residue was refined.
2.3. Molecular docking
The Genetic Optimisation for Ligand Docking (GOLD) [22] version 5.6.2 was used as the molecular docking engine. GOLD has been established for its reliability and applicability in docking ligands with proteins and nucleic acids [23, 24, 25]. The semi-empirical ChemPLP [26] scoring function was implemented to validate the predicted binding modes and relative energies of the ligands. The centre of binding was defined at the catalytic and DNA interface site of HIV-1 integrase at coordinates (x, y, z) = (131.629, 111.574, 153.149) with 5 Å radius. All aspartic and glutamic acids were assumed to be deprotonated. The search efficiency was adjusted accordingly: 30% at 20 docking runs per ligand for pilot virtual screen, and 100% at 50 docking runs for the main virtual screen and lamellarin analogues. Search efficiency determines the search quality for complementary binding of ligands. The best three scoring runs were recorded.
2.4. Calculations of physicochemical properties
The Dragon 7.0 [27] software suite was used to calculate for the molecular descriptors: molecular weight (MW), MlogP, hydrogen bond donors (HDs) and acceptors (HAs), number of rotatable bonds (RBs) and total polar surface area (TPSA). Dragon 7.0 is well-known for its use in scientific research [23, 28].
2.5. Predictions of ADME and toxicity
The absorption, distribution, metabolism, excretion and toxicity were predicted for all compounds using the PreADMET webserver [29, 30]. The use of the webserver is well established in literature [31, 32].
3. Results and discussion
3.1. Molecular docking of lamellarin analogues
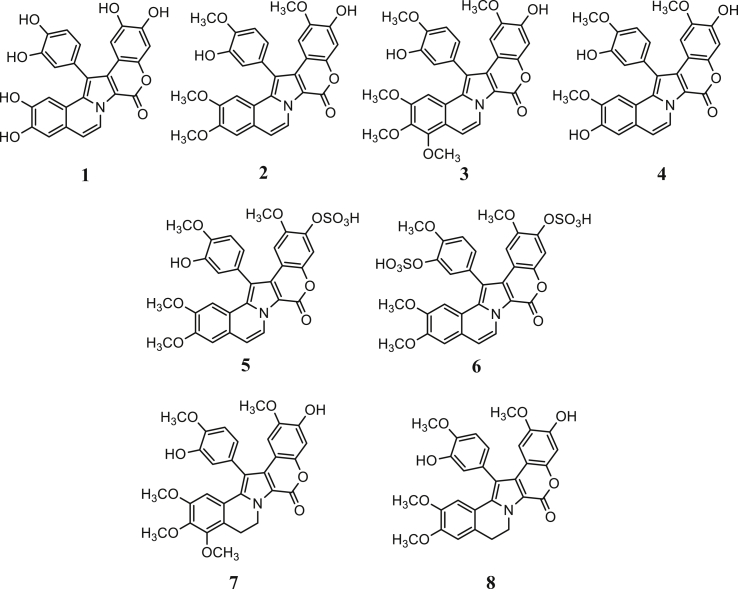
The lamellarins are mostly known for their cytotoxic effects and thus, are studied closely in the area of anticancer chemotherapeutics. They were extensively studied for their inhibition properties against topoisomerase, an enzyme that function to relieve DNA stress during replication. Reports suggest it binds to topoisomerase/DNA complex by interacting with the topoisomerase residues and planar intercalation to DNA [12, 14]. Regardless of their anticancer action, minimal progress was made following its reported anti-integrase activity against HIV by Faulkner [10, 11]. Taking into account of this work and its binding action with topoisomerase/DNA complex, it seemed possible that interaction can occur with both HIV-1 integrase and DNA. Considering the absence of co-crystallised ligands in the crystal structure, the COACH [33, 34] webserver was used to predict the ligand binding site of the HIV-1 integrase protomer to be at the catalytic site (see Table S1 and Figure S1). Molecular docking was applied to elucidate the binding action of 8 lamellarin analogues with known strand transfer inhibition activities: methoxy and hydroxyl substituted analogues 1 – 4, sulfated lamellarins 5 and 6, and methoxy and hydroxyl substituted with ring saturation analogues 7 and 8 (Fig. 3 and Table 1).
Fig. 3.
The chemical structures of 8 lamellarin analogues with anti-integrase strand transfer inhibition activities from references [10] and [11].
Table 1.
The best molecular docking scores of 8 lamellarin analogues and the strand transfer activities.
| Analogue | ChemPLP (with DNA) | ChemPLP (no DNA) | Strand transfer activity IC50 (μM)a |
|---|---|---|---|
| 1 | 58.01 | 38.29 | 1.3 |
| 2 | 36.08 | 26.93 | >1600 |
| 3 | 42.44 | 40.32 | 14 |
| 4 | 56.35 | 41.98 | 19 |
| 5 | 31.73 | 28.20 | 22 |
| 6 | 33.43 | 43.44 | 49 |
| 7 | 38.50 | 29.69 | 24 |
| 8 | 37.22 | 24.88 | 25 |
Activities from references [10] and [11].
The docking scores and strand transfer activities are shown in Table 1. From the docking scores, the binding of the ligands to the target with DNA is more favourable as seen from the higher docking scores for systems with DNA. This is partially in line with the suggested report that analogue 5 partially interact with just the catalytic domain, i.e., disintegration assay using purified catalytic domain resulted in IC50 = 64 μM compared to 7 μM for full length HIV-1 integrase [10]. Analogue 1 is the most potent and the highest scoring analogue. Analogues 3 and 4 are second and third most potent with scores of 42.44 and 56.35, respectively. Sulfated lamellarin analogues and ring saturated analogues were least scoring with scores ranging between 31 and 39. Analogue 2 is the least active analogue with docking score of 36.08.
The key residue interactions are shown in Table 2, and the graphical binding modes of the most active analogue 1 and a ring saturated analogue 8 are shown in Fig. 4. Molecular modelling suggests Glu92 to be one of the most important residues for hydrogen bonding interaction as this residue was observed to form hydrogen bonds with analogues 1–4 and 7–8, and hydrophobic interactions with all analogues. Occurrences of intermolecular interactions with Glu92 has been reported for compounds that target HIV-1 integrase [35, 36]. Additionally, mutations of Glu92 has been suggested in elvitegravir-resistant strains suggesting its importance in ligand binding [37]. Sulfated analogues 5 and 6 formed hydrogen bond interactions with the guanine base. The guanine base is predicted to be involved in forming hydrophobic and π−π stacking interactions with all analogues (see Fig. 4 and Figure S2). The π−π stacking interactions is predicted to contribute to the stability of the inhibitor-protein/DNA complex, which is seen in X-ray structures of approved drugs with viral intasome complexes [38, 39]. According to molecular docking, the sulfate groups are mostly exposed to the aqueous environment. Its presence in the lamellarin scaffold did not improve strand transfer activity which is in line with the low docking scores seen. This could possibly be attributed to the steric effects between the large sulfate groups and nucleotide base and lack of key hydrogen bonding amino acid residues. Other key hydrogen bonding residues are Cys65, His67 and catalytic residues Asp64 and Asp116 as seen for analogues 1 and 4. All residues have been reported to interact with inhibitors of HIV-1 integrase and are suggestive to have importance to antiviral activity [24, 35, 40, 41, 42]. Chelation with Mg2+ has been reported to be involved with integrase inhibition [24, 35, 43] and was seen with analogues 4, 6 and 7. All analogues pose π−π stacking interactions, however, analogues 7 and 8 contain one less unsaturated ring system and were less stabilised by the π−π stacking interactions that explains their lower scores compared to methoxy and hydroxyl substituted analogues.
Table 2.
Intermolecular bonding residues with lamellarin analogues. Chemical groups are shown to chelate with catalytic magnesium ion.
| Compound | H-bond residues | Hydrophobic interaction residues | Mg2+ chelation |
|---|---|---|---|
| 1 | Asp64, Cys65, His67, Glu92 | Asp64, Cys65, Asp116, His67, Glu92, guanine base | - |
| 2 | Glu92 | Cys65, His67, Val72, Glu92, Tyr143, guanine base | - |
| 3 | Glu92 | Cys65, His67, Asp116, Glu92, Tyr143, guanine base | - |
| 4 | Asp64, His67, Glu92, Asp116, guanine base | Asp64, His67, Glu92, Asp116, guanine base | 8-OH |
| 5 | Guanine base | Asp64, Cys65, His67, Glu92, Asp116, Phe121, guanine base | - |
| 6 | Guanine base | Asp64, Cys65, His67, Glu92, Asp116, Phe121, guanine base | 8-OMe |
| 7 | Glu92 | Asp64, Cys65, His67, Glu92, Asp116, Gly118, Pro142, Tyr143, guanine base | 9-OMe |
| 8 | Glu92 | Asp64, Cys65, His67, Val72, Glu92, guanine base | - |
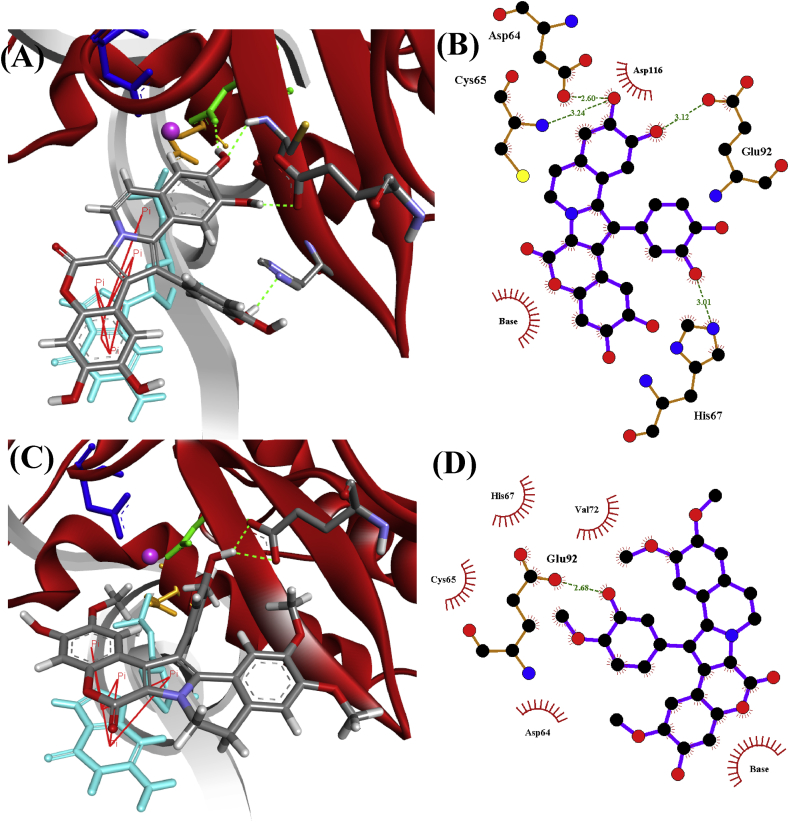
Fig. 4.
Graphical 2D and 3D representations of the binding modes of lamellarin analogues 1 (A and B) and 8 (C and D) with HIV-1 integrase STC. In the 3D representations (A and C), red ribbons depict HIV-1 integrase, grey strand is DNA, whereas green, blue and orange colours depict Asp64, Asp116 and Glu152, respectively, and round pink sphere represents Mg2+ metal ion cofactor. Bold red lines depict π−π stacking interactions, a guanine base in turquoise forming π−π stacking interactions with the lamellarin ligands and dotted green lines representing hydrogen bonds. The measured distances of the hydrogen bonds are shown with amino acids Cys65, Asp64, Glu92 and His67 to be 3.24, 2.60, 3.12 and 3.01 Å for analogue 1 (B), and 2.68 Å with Glu92 for analogue 8 (D). Hydrophobic interactions with residues and guanine base are depicted as red lashes in the 2D representations.
Glu92, His67 and guanine base were seen to be most important for hydrophobic interactions as they were able to interact with all analogues. Hydrophobic interactions with residues Cys65, Asp116 and Asp64 were also predicted to be important and interacted with most residues. Analogues 2, 3, and 7 were seen to form hydrophobic interactions with larger bulky amino acid Tyr143, and analogues 5 and 6 with Phe121. Hydrophobic interactions with smaller neutral amino acids Gly118 and Pro142 were seen with analogue 7, and Val72 with analogue 8.
3.2. Virtual screening for novel inhibitors of HIV-1 integrase STC
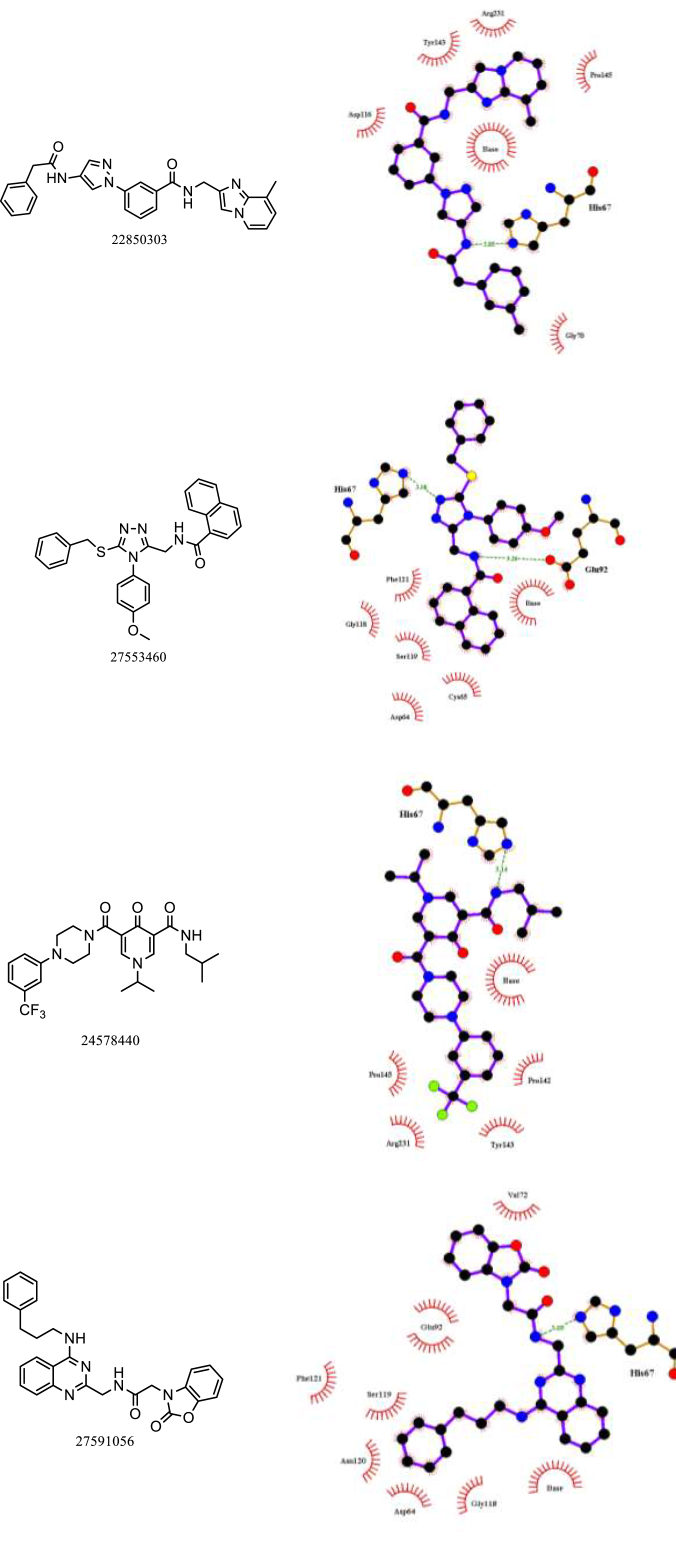
A total of 25,132 chemical entities from the Chembridge diversity collection were docked to the HIV-1 integrase/DNA interface. A pilot screen was initially conducted at 30% search efficiency with 20 docking runs. Compounds scoring more than 50 and hydrogen bond scores of greater than 1 were taken forward. A score of 50 was used as the cutoff on the basis that the most active lamellarin analogue 1 scored more than 50 and hydrogen bonds are considered crucial for ligand-target interactions. A total of 637 entities resulted and were docked to the interface site at 100% search efficiency with 50 docking runs. Compounds with scores of greater than 60 and hydrogen bonding score of 1 or greater were selected giving 50 compounds. The compounds were inspected visually for their plausible mode of binding, i.e., hydrogen bond formations with key amino acid interactions and formations of π−π stacking interactions with guanine: 18 compounds (see structures in Figure S3) were identified to form these intermolecular interactions. The 18 compounds consist of 9 that contain bicyclic heterocycle scaffolds that resemble closely to DNA purine core scaffold, 3 compounds that contain a pyridine-(1H)-one scaffold that mimics DNA pyrimidine core scaffold, 4 compounds with core pyrazole and triazole core scaffolds, and 2 compounds that are miscellaneous. The compounds were finally filtered based on chemical scaffold diversity, i.e., the best scoring compound from each scaffold category were selected resulting in a total of 4 potential candidates (see Fig. 5).
Fig. 5.
The chemical structures and Chembridge compound ID codes of four novel potential inhibitors of HIV-1 integrase STC identified from the Chembridge diversity collection by virtual screening, and the predicted binding interactions. The hydrogen bonds are shown in green dotted lines and the distances were measured in angstrom units. Hydrophobic interactions depicted in red lashes.
All four candidates were able to form π−π stacking interactions and hydrophobic interactions with the guanine base. Key hydrogen bonding with residue His67 was observed for all four candidates, and Glu92 with 27553460. Residues which formed hydrophobic interactions with lamellarin analogues were also seen amongst the four candidates, e.g., Asp116, His67, Tyr143, Asp64, Phe121, Gly118, Cys65, Glu92. Pro142, Val72 and guanine base. These interactions are highly suggestive of their strand transfer inhibition potential. Furthermore, additional residues were predicted to form hydrophobic interactions: Arg231, Gly70, Ser119, Asn12 and Pro145.
3.3. Drug-likeness, toxicity and ADMET properties of ligand candidates
The drug-like properties of the lamellarin analogues and virtual hit candidates were calculated (results in Table S3) using the Dragon 7.0 software to determine the drug-like scores of the compounds. The drug-like scores are shown in Table 3 and were calculated using a model developed by Eurtivong and Reynisson [44]: the model is based on the physicochemical properties of 1880 clinically approved drugs with the highest possible score of 6. Results revealed analogue 1 and sulfated lamellarin analogues 5 and 6 are less favourable as drug candidates, i.e., drug-like scores are most mediocre (scores of less than 4). The virtual hit candidates scored highly with drug-like scores >5 higher than the known drug, raltegravir. Analogues 2, 4 and 8 showed comparable drug-like scores to compound 27591056. Nonetheless, only analogues 4 and 8 showed promising evidences of strand transfer inhibition. Analogues 3 and 7 scored reasonably well with scores of 4.36. Regardless of only lamellarin analogues 1 and 4 which obeyed Lipinski's rules, most of these analogues scored favourably within the known drug chemical space. It was reported that the highest proportion of drugs (∼40%) scored between 4 and 5, and the second highest proportion of drugs (∼23%) scored between 5 and 6, which strongly suggests the potential for the lamellarin analogues and the virtual hits to be developed into future promising drug candidates [44].
Table 3.
Physicochemical properties and ADMET predictions of the lamellarin analogues and virtual hit candidates.
| Compound | Raltgravir | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 22850303 | 27553460 | 24578440 | 27591056 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Drug-like score | 4.13 | 3.76 | 4.80 | 4.36 | 4.81 | 3.82 | 2.90 | 4.36 | 4.80 | 5.35 | 5.01 | 5.10 | 4.80 |
| BBB | 0.043 | 0.288 | 0.035 | 0.026 | 0.168 | 0.050 | 0.052 | 0.073 | 0.029 | 0.027 | 0.800 | 0.045 | 0.038 |
| Buffer solubility (mg/L) | 16098.50 | 47.658 | 1.0799 | 0.9748 | 2.787 | 1.281 | 1.492 | 8.557 | 1.016 | 4.841 | 0.472 | 5.153 | 1.456 |
| Caco-2 | 20.04 | 16.556 | 25.123 | 27.999 | 18.939 | 2.269 | 1.887 | 28.36 | 25.48 | 29.10 | 50.66 | 27.45 | 25.68 |
| CYP2C19 inhibition | - | + | + | + | + | - | - | + | - | - | - | - | - |
| CYP2C9 inhibition | - | + | + | + | + | + | + | + | + | + | + | - | + |
| CYP2D6 inhibition | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CYP2D6 substrate | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CYP3A4 inhibition | + | + | + | + | + | + | + | + | + | - | - | + | - |
| CYP3A4 substrate | - | + | + | + | + | + | + | + | + | + | + | + | + |
| HIA (%) | 76.78 | 83.05 | 95.93 | 95.81 | 94.41 | 99.05 | 95.23 | 95.61 | 95.29 | 96.04 | 97.41 | 96.27 | 96.08 |
| Pgp inhibition | - | + | + | + | + | + | + | + | + | - | + | + | - |
| PPB (%) | 48.76 | 100.00 | 89.37 | 89.61 | 87.80 | 98.67 | 100.00 | 88.75 | 88.74 | 96.33 | 96.13 | 89.71 | 97.46 |
| PWS (mg/L) | 909.002 | 0.122 | 0.006 | 0.006 | 0.012 | 0.002 | 0.001 | 0.072 | 0.651 | 2.799 | 0.002 | 17.902 | 0.102 |
| carcino_Mouse | - | - | - | - | - | - | - | - | - | - | - | + | + |
| carcino_Rat | - | - | + | + | + | - | + | + | + | - | - | - | - |
| hERG_inhibition | Medium | Medium | Medium | Medium | Medium | Medium | Low | Medium | Medium | High | Medium | Medium | Ambiguous |
| TA100_10RLI | - | - | - | - | - | - | - | - | - | - | - | + | - |
| TA100_NA | + | - | - | - | - | - | - | - | - | - | - | - | - |
| TA1535_10RLI | - | - | - | - | - | - | - | - | - | - | - | + | - |
| TA1535_NA | - | - | - | - | - | + | - | - | - | - | - | - | - |
BBB = blood brain barrier penetration, Caco-2 = permeation of Caco-2 colorectal adenocarcinoma cells, CYP2C19 = cytochrome P4502C19, CYP2C9 = cytochrome P4502C9, CYP3A4 = cytochrome P4503A4, CYP2D6 = cytochrome P4502D6, Pgp = P-glycoprotein, HIA = human intestinal absorption, PPB = plasma binding protein, PWS = pure water solubility, TA100_10RLI/1535_10_RLI = TA100/TA1535 strain with metabolic activation by rat liver homogenate, TA100_NA/1535_NA = TA100/1535 strain with no metabolic activation.
Furthermore, the ADMET properties were predicted using the preADMET webserver. The blood brain barrier penetration is low and well below 0.1 for all compounds except for 27553460 with a value of 0.8. Buffer and pure water solubility needs to be optimised and improved compared to the known reference drug, raltegravir. Permeation into Caco-2 adenocarcinoma cells were predicted to be comparable to raltegravir for all compounds suggesting good intestinal absorption, which is in line with the calculated human intestinal absorption (HIA) predicting all compounds to have >80%, which is higher than raltegravir, HIA = 77%. All compounds were predicted to be free from binding to CYP2D6, whilst only lamellarin analogues inhibit CYP2C19. All compounds were predicted to bind to CYP3A4 either as inhibitors or substrates and only 24578440 and raltegravir were predicted to not inhibit CYP2C9. P-glycoprotein inhibition was predicted for all lamellarin analogues, 27553460 and 24578440 suggesting interference with the pharmacokinetic profiles of combination drugs. Plasma protein binding (PPB) exceeds 80% for all compounds, however, plasma protein binding is reversible and further pharmacological in vivo studies can justify a more accurate pharmacokinetic distribution.
Compounds 24578440 and 27591056 were predicted to be carcinogenic in mice, and lamellarin analogues 3–5 and 6–8 were predicted to be carcinogenic in rats. Only compound 22850303 was predicted to have a high risk of cardiotoxicity due to hERG inhibition, ambiguous for 27591056 and low/medium risks for raltegravir, lamellarin analogues, 22850303 and 27553460. Four Ames test were predicted for two strains TA100 and TA1535 strains with and without metabolic activation by rat liver 10% homogenate. Compound 24578440 and lamellarin analogue 5 were predicted to be mutagens. However, the reference known drug raltegravir was also predicted to have mutagenic potential from non-metabolic activated TA100 strain.
4. Conclusion
This work provides a simulative understanding in the way lamellarin analogues interact with HIV-1 integrase STC. The analogues showed intercalation to DNA bases and were stabilised by the π−π stacking interactions, as well as interacting with the surrounding amino acid residues at the catalytic/DNA interface of HIV-1 integrase STC. The scores calculated were in line with the experimental data conducted previously. Virtual screening identified 4 potential candidates from 4 different scaffold categories. The mode of binding of these compounds suggests DNA intercalation by π−π stacking interactions, hydrophobic interactions and hydrogen bonding with key residues such as Glu92 and His67 to be important for ligand binding. The drug-like properties of all compounds were calculated and the drug-like scores suggest favourable candidates for in vitro activity screening and further activity optimisation by means of medicinal chemistry. Furthermore, prediction of ADMET suggests good absorption and relatively acceptable toxicity profiles.
Declarations
Author contribution statement
Chatchakorn Eurtivong: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Kiattawee Choowongkomon, Poonsakdi Ploypradith: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Somsak Ruchirawat: Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by the Chulabhorn Royal Academy.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.World Health Organization https://www.who.int/news-room/fact-sheets/detail/hiv-aids
- 2.U.S. Food & Drug Administration https://www.fda.gov/patients/hiv-treatment/antiretroviral-drugs-used-treatment-hiv-infection
- 3.Metry D.W., Lahart C.J., Farmer K.L., Hebert A.A. Stevens-Johnson syndrome caused by the antiretroviral drug nevirapine. J. Am. Acad. Dermatol. 2001;44(2 Suppl):354–357. doi: 10.1067/mjd.2001.101885. [DOI] [PubMed] [Google Scholar]
- 4.Margolis A.M., Heverling H., Pham P.A., Stolbach A. A review of the toxicity of HIV medications. J. Med. Toxicol. 2014;10(1):26–39. doi: 10.1007/s13181-013-0325-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X. Anti-retroviral drugs: current state and development in the next decade. Acta Pharm. Sin. B. 2018;8(2):131–136. doi: 10.1016/j.apsb.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Clercq E. Anti-HIV drugs: 25 compounds approved within 25 years after the discovery of HIV. Int. J. Antimicrob. Agents. 2009;33(4):307–320. doi: 10.1016/j.ijantimicag.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 7.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Passos D.O., Li M., Yang R., Rebensburg S.V., Ghirlando R., Jeon Y., Shkriabai N., Kvaratskhelia M., Craigie R., Lyumkis D. Cryo-EM structures and atomic model of the HIV-1 strand transfer complex intasome. Science. 2017;355(6320):89–92. doi: 10.1126/science.aah5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.ChemBridge Corporation www.chembridge.com
- 10.Reddy M.V.R., Rao M.R., Rhodes D., Hansen M.S.T., Rubins K., Bushman F.D., Venkateswarlu Y., Faulkner D.J. Lamellarin α 20-sulfate, an inhibitor of HIV-1 integrase active against HIV-1 Virus in cell culture. J. Med. Chem. 1999;42(11):1901–1907. doi: 10.1021/jm9806650. [DOI] [PubMed] [Google Scholar]
- 11.Ridley C.P., Reddy M.V.R., Rocha G., Bushman F.D., Faulkner D.J. Total synthesis and evaluation of lamellarin α 20-Sulfate analogues. Bioorg. Med. Chem. 2002;10(10):3285–3290. doi: 10.1016/s0968-0896(02)00237-7. [DOI] [PubMed] [Google Scholar]
- 12.Cananzi S., Merlini L., Artali R., Beretta G.L., Zaffaroni N., Dallavalle S. Synthesis and topoisomerase I inhibitory activity of a novel diazaindeno[2,1-b]phenanthrene analogue of Lamellarin D. Bioorg. Med. Chem. 2011;19(16):4971–4984. doi: 10.1016/j.bmc.2011.06.056. [DOI] [PubMed] [Google Scholar]
- 13.Facompre M., Tardy C., Bal-Mahieu C., Colson P., Perez C., Manzanares I., Cuevas C., Bailly C. Lamellarin D: a novel potent inhibitor of topoisomerase I. Cancer Res. 2003;63(21):7392–7399. [PubMed] [Google Scholar]
- 14.Marco E., Laine W., Tardy C., Lansiaux A., Iwao M., Ishibashi F., Bailly C., Gago F. Molecular determinants of topoisomerase I poisoning by lamellarins: comparison with camptothecin and structure-activity relationships. J. Med. Chem. 2005;48(11):3796–3807. doi: 10.1021/jm049060w. [DOI] [PubMed] [Google Scholar]
- 15.Neagoie C., Vedrenne E., Buron F., Merour J.Y., Rosca S., Bourg S., Lozach O., Meijer L., Baldeyrou B., Lansiaux A., Routier S. Synthesis of chromeno[3,4-b]indoles as Lamellarin D analogues: a novel DYRK1A inhibitor class. Eur. J. Med. Chem. 2012;49:379–396. doi: 10.1016/j.ejmech.2012.01.040. [DOI] [PubMed] [Google Scholar]
- 16.Ohta T., Fukuda T., Ishibashi F., Iwao M. Design and synthesis of lamellarin D analogues targeting topoisomerase I. J. Org. Chem. 2009;74(21):8143–8153. doi: 10.1021/jo901589e. [DOI] [PubMed] [Google Scholar]
- 17.Kluza J., Gallego M.A., Loyens A., Beauvillain J.C., Sousa-Faro J.M., Cuevas C., Marchetti P., Bailly C. Cancer cell mitochondria are direct proapoptotic targets for the marine antitumor drug lamellarin. D. Cancer Res. 2006;66(6):3177–3187. doi: 10.1158/0008-5472.CAN-05-1929. [DOI] [PubMed] [Google Scholar]
- 18.Fujitsu Scigress. 2018. [Google Scholar]
- 19.Allinger N.L. Conformational analysis. 130. MM2. A hydrocarbon force field utilizing V1 and V2 torsional terms. J. Am. Chem. Soc. 1977;99(25):8127–8134. [Google Scholar]
- 20.Stewart J.J.P. Optimization of parameters for semiempirical methods V: modification of NDDO approximations and application to 70 elements. J. Mol. Model. 2007;13(12):1173–1213. doi: 10.1007/s00894-007-0233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics. 2016;54 doi: 10.1002/cpbi.3. 5.6.1-6.5.6.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CCDC Genetic Optimisation For Ligand Docking. 2019. [Google Scholar]
- 23.Leung E., Pilkington L.I., Naiya M.M., Barker D., Zafar A., Eurtivong C., Reynisson J. The cytotoxic potential of cationic triangulenes against tumour cells. MedChemComm. 2019 doi: 10.1039/c9md00305c. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vandurm P., Guiguen A., Cauvin C., Georges B., Le Van K., Michaux C., Cardona C., Mbemba G., Mouscadet J.F., Hevesi L., Van Lint C., Wouters J. Synthesis, biological evaluation and molecular modeling studies of quinolonyl diketo acid derivatives: new structural insight into the HIV-1 integrase inhibition. Eur. J. Med. Chem. 2011;46(5):1749–1756. doi: 10.1016/j.ejmech.2011.02.028. [DOI] [PubMed] [Google Scholar]
- 25.Li Y., Shen J., Sun X., Li W., Liu G., Tang Y. Accuracy assessment of protein-based docking programs against RNA targets. J. Chem. Inf. Model. 2010;50(6):1134–1146. doi: 10.1021/ci9004157. [DOI] [PubMed] [Google Scholar]
- 26.Korb O., Stutzle T., Exner T.E. Empirical scoring functions for advanced protein-ligand docking with PLANTS. J. Chem. Inf. Model. 2009;49(1):84–96. doi: 10.1021/ci800298z. [DOI] [PubMed] [Google Scholar]
- 27.Kode Cheminformatics, Dragon. 2018. [Google Scholar]
- 28.Kawczak P., Bober L., Bączek T. Activity evaluation of some psychoactive drugs with the application of QSAR/QSPR modeling methods. Med. Chem. Res. : an international journal for rapid communications on design and mechanisms of action of biologically active agents. 2018;27(10):2279–2286. doi: 10.1007/s00044-018-2234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S.K., Lee I.H., Kim H.J., Chang G.S., Chung J.E. EuroQSAR 2002 Designing Drugs and Crop Protectants: Processes, Problems and Solutions. Blackwell Publishing; Massachusetts, USA: 2003. The PreADME Approach: web-based program for rapid prediction of physico-chemical, drug absorption and drug-like properties; pp. 418–420. [Google Scholar]
- 30.Lee S.K., Chang G.S., Lee I.H., Chung J.E., Sung K.Y., No K.T. The PreADME: PC-Based Program for Batch Prediction of ADME Properties, EuroQSAR 2004, Istanbul, Turkey. 2004. Istanbul, Turkey. [Google Scholar]
- 31.Kovačević S.Z., Jevrić L.R., Podunavac Kuzmanović S.O., Lončar E.S. Prediction of in-silico ADME properties of 1,2-O-isopropylidene aldohexose derivatives. Iran. J. Pharm. Res. 2014;13(3):899–907. [PMC free article] [PubMed] [Google Scholar]
- 32.Wadapurkar R.M., Shilpa M.D., Katti A.K.S., Sulochana M.B. In silico drug design for Staphylococcus aureus and development of host-pathogen interaction network. Inform. Med. Unlocked. 2018;10:58–70. [Google Scholar]
- 33.Yang J., Roy A., Zhang Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics. 2013;29(20):2588–2595. doi: 10.1093/bioinformatics/btt447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J., Roy A., Zhang Y. BioLiP: a semi-manually curated database for biologically relevant ligand-protein interactions. Nucleic Acids Res. 2013;41(Database issue):D1096–D1103. doi: 10.1093/nar/gks966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vajragupta O., Boonchoong P., Morris G.M., Olson A.J. Active site binding modes of curcumin in HIV-1 protease and integrase. Bioorg. Med. Chem. Lett. 2005;15(14):3364–3368. doi: 10.1016/j.bmcl.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 36.Guenther S., Nair V. Binding modes of two novel dinucleotide inhibitors of HIV-1 integrase. Bioorg. Med. Chem. Lett. 2002;12(16):2233–2236. doi: 10.1016/s0960-894x(02)00353-0. [DOI] [PubMed] [Google Scholar]
- 37.Krishnan L., Li X., Naraharisetty H.L., Hare S., Cherepanov P., Engelman A. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc. Natl. Acad. Sci. 2010;107(36):15910–15915. doi: 10.1073/pnas.1002346107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hare S., Gupta S.S., Valkov E., Engelman A., Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464:232. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hare S., Smith S.J., Metifiot M., Jaxa-Chamiec A., Pommier Y., Hughes S.H., Cherepanov P. Structural and functional analyses of the second-generation integrase strand transfer inhibitor dolutegravir (S/GSK1349572) Mol. Pharmacol. 2011;80(4):565–572. doi: 10.1124/mol.111.073189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldgur Y., Craigie R., Cohen G.H., Fujiwara T., Yoshinaga T., Fujishita T., Sugimoto H., Endo T., Murai H., Davies D.R. Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design. Proc. Natl. Acad. Sci. 1999;96(23):13040–13043. doi: 10.1073/pnas.96.23.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ingale K.B., Bhatia M.S. HIV-1 integrase inhibitors: a review of their chemical development. Antivir. Chem. Chemother. 2011;22(3):95–105. doi: 10.3851/IMP1740. [DOI] [PubMed] [Google Scholar]
- 42.Huang M., Grant G.H., Richards W.G. Binding modes of diketo-acid inhibitors of HIV-1 integrase: a comparative molecular dynamics simulation study. J. Mol. Graph. Model. 2011;29(7):956–964. doi: 10.1016/j.jmgm.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta P., Roy N., Garg P. Docking-based 3D-QSAR study of HIV-1 integrase inhibitors. Eur. J. Med. Chem. 2009;44(11):4276–4287. doi: 10.1016/j.ejmech.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 44.Eurtivong C., Reynisson J. The development of a weighted index to optimise compound libraries for high throughput screening. Mol. Inform. 2019;38(3):1800068. doi: 10.1002/minf.201800068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.