Highlights
-
•
Radiomics features from QSM data have significant clinical value for the diagnosis of PD.
-
•
CNN features, which is better than Radiomics Features, are also useful in the diagnosis of PD.
-
•
The combination of radiomics features and CNN features can enhance the diagnostic accuracy.
Keywords: Parkinson's disease, QSM, Substantia nigra, Radiomics, Convolution neural network
Abstrat
Parkinson's disease is the second most common neurodegenerative disease in the elderly after Alzheimer's disease. The aetiology and pathogenesis of Parkinson's disease (PD) are still unclear, but the loss of dopaminergic cells and the excessive iron deposition in the substantia nigra (SN) are associated with the pathophysiology. As an imaging technique that can quantitatively reflect the amount of iron deposition, Quantitative Susceptibility Mapping (QSM) has been shown to be a promising modality for the diagnosis of PD. In the present work, we propose a hybrid feature extraction method for PD diagnosis using QSM images. First, we extract radiomics features from the SN using QSM and employ machine learning algorithms to classify PD and normal controls (NC). This approach allows us to investigate which features are most vulnerable to the effects of the disease. Along with this approach, we propose a Convolutional Neural Network (CNN) based method which can extract different features from the QSM image to further support the diagnosis of PD. Finally, we combine these two types of features and we find that the radiomics features and CNN features are complementary to each other, which helps further improve the classification (diagnostic) performance. We conclude that: (1) radiomics features from QSM data have significant clinical value for the diagnosis of PD; (2) CNN features are also useful in the diagnosis of PD; and (3) the combination of radiomics features and CNN features can enhance the diagnostic accuracy.
1. Introduction
Parkinson's disease (PD) is a progressive neurodegenerative disease that affects predominately dopamine-producing neurons in the substantia nigra (SN). It leads to shaking, stiffness, and difficulty with walking, balance, and coordination. In 2015, more than 6 million in the world had PD, causing significant economic loss to society (Vos et al., 2017). Although there are some treatments such as dopamine-related medication and deep brain stimulation, over time the patient still suffers a gradual increase in the severity of the symptoms. Therefore, early diagnosis of PD is crucial for treatment and disease management. Clinically, the physician makes the diagnosis of PD by reviewing the patient's history, symptoms, and drug reactions. However, it is often difficult to clearly differentiate idiopathic PD from other movement disorders. Thus, sensitive imaging biomarkers may help improve the diagnosis of PD.
Previous studies based on different neuroimaging modalities including structural or functional magnetic resonance imaging (MRI) and dopaminergic imaging methods using positron emission tomography (PET) have been used to diagnose PD with some success (Brooks and Pavese, 2011; Lorraine and Kalia, 2015). More recently, a great deal of attention has been paid to the role of increased iron deposition in the SN and its role in the development and progression of PD. Quantitative susceptibility mapping (QSM) is an MRI method used to quantify iron in vivo. Compared with these other imaging measures, the main advantage of QSM is that it provides a means to quantify iron content spatially in the deep gray matter and specifically in the SN (Haacke et al., 2015; Yan et al., 2018).
To validate the ability to measure iron content, QSM data has been compared with inductively coupled plasma mass spectrometry (ICP-MS) in cadaver brains and it has been shown that the magnetic susceptibility values in the deep brain nuclei are mainly derived from iron deposition (Langkammer et al., 2012). In addition, using X-ray fluorescence (XRF) imaging, magnetic susceptibility has proven to provide a reliable quantitative measurement of iron content (Zheng et al., 2013). Another study (Sun et al., 2015) also confirmed iron as the dominant QSM contrast in deep grey matter using a histologic iron-staining method.
Recently there has been a major interest in using iron-based contrast methods such as QSM to study the absence of the Nigrosome 1 (N1) territory as a biomarker for PD (Schwarz et al., 2018). In healthy controls, N1 appears hypointense in QSM, and the remaining parts of the SN appear hyperintense because of the normal age-related iron deposition in those areas. This visual determination of N1 is purely qualitative and does not take into account the shape or variable iron content of the SN. The feasibility to recognize and characterize the N1 territory is thus limited (Mahlknecht et al., 2017).
Our hypothesis is that AI methods such as radiomics and a Convolutional Neural Network (CNN) can successfully evaluate both iron increases and texture features to discriminate PD from normal controls (NC). Therefore, we propose to evaluate the QSM data in three ways: (1) using radiomics features from QSM to model the classification problem; (2) using a CNN to diagnose PD and obtain auto-encoded features from the QSM data; and (3) combining the radiomics features and CNN features to predict PD from NC.
2. Materials and methods
2.1. Data collection and ROI extraction
Eighty-seven cases of idiopathic PD (IPD) patients (41 males and 46 females, aged 60.9 ± 8.1 years old) and 53 normal controls (NC) (24 males and 29 females, aged 62.9 ± 7.1 years old) were collected at the Department of Radiology, Shanghai Ruijin Hospital. All patients were recruited from a local movement disorder clinic. The present study was approved by the local ethics committee and informed consent was signed by each subject. The data were collected between March 2012 and June 2015. The inclusion criteria were: (1) a diagnosis of idiopathic PD; (2) Mini-Mental State Exam (MMSE) score greater than or equal to 24; and (3) a Hoehn and Yahr (H&Y) scale of 1–3 but not higher since patients with higher scores would have more trouble staying still for the scan. The exclusion criteria were: (1) symptoms of secondary or atypical parkinsonism; (2) a history of cerebrovascular disease, seizures, brain surgery, brain tumor, moderate-to-severe head trauma or hydrocephalus; and (3) treatment with antipsychotic drugs or any other drug affecting clinical evaluation. Data were collected using a 16 echo, gradient echo imaging sequence on a 3T GE Signa HDxt from an eight-channel receive-only head coil with the following imaging parameters: TE1 = 2.69 ms with ΔTE = 2.87 ms, TR = 59.3 ms, pixel bandwidth = 488 Hz/pixel, flip angle = 12°, slice thickness = 1 mm, number of slices = 136, matrix size = 256 × 256, an in-plane resolution of 0.86 × 0.86 mm2, a parallel imaging factor of 2 with a total acquisition time of 10 min and 42 s.
Image reconstruction was performed as in (Li et al., 2011). Briefly, all phase images were averaged and filtered with the sophisticated harmonic artifact reduction for phase data (SHARP) and the susceptibility maps were derived from the frequency map using an improved LSQR (iLSQR) method (Li et al., 2011) (the regularization threshold for the Laplace filtering was set to 0.04). Units of susceptibility are given in parts per billion (ppb). Each SN was drawn by a clinician who is familiar with the anatomy of the midbrain and with this project (see Fig. 1).
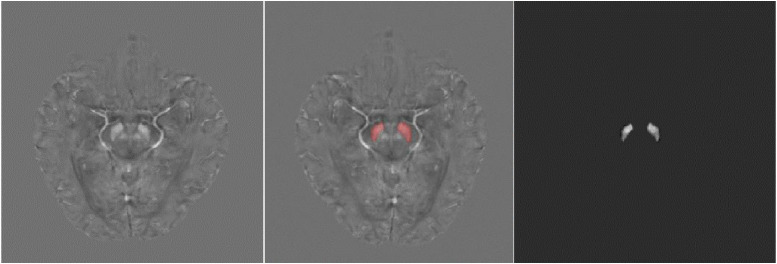
Fig 1.
Left: The original QSM image. Middle: The original QSM image with the mask of SN. Right: The SN extracted by the mask.
2.2. Classification with radiomics features
In order to verify the validity of radiomics features on the SN obtained from the QSM data, we extracted the radiomics features from the SN by using a process described in Section 2.2.1 and employed two machine methods: Logistic regression (LR) and support vector machine (SVM), to predict PD and NC as described in Section 2.2.2. The flow chart of the processing is shown in Fig. 2. All experiments were performed within the nested cross-validation framework: the inner loop was used to select the best model parameters and features, and the outer loop was used to evaluate the performance of the classifier.
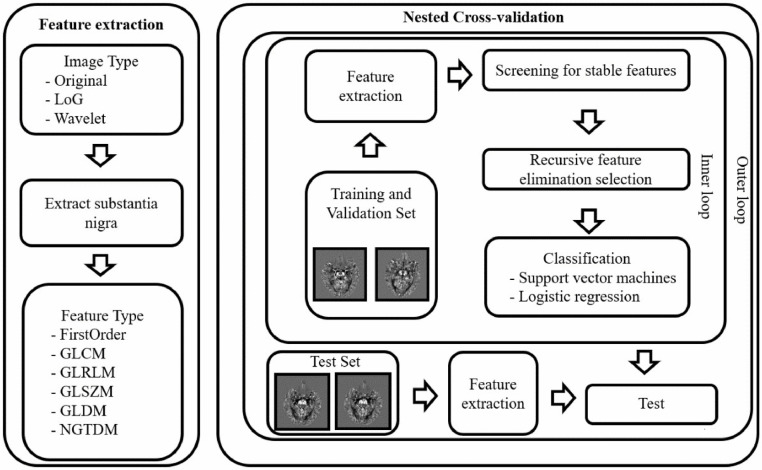
Fig 2.
Main flowchart. First, the QSM image is preprocessed into three different image types: Original, LoG, and Wavelet. Then, six types of radiomics features on SN are extracted from the different image types. The most stable and informative features are selected by the ICC and RFE processes. Then LR (or SVM) combines these selected features to distinguish PD and NC. The entire classification process is performed by a 7-fold nested cross-validation 5 times. Grey Level Co-occurrence Matrix (GLCM), Gray Level Run Length Matrix (GLRLM), Gray Level Size Zone Matrix (GLSZM), Neighboring Gray Tone Difference Matrix (NGTDM), Intraclass Correlation Coefficient (ICC), and Recursive Feature Elimination (RFE).
2.2.1. Image preprocessing and feature extraction
As discussed earlier, we first transform the QSM data into three different image types which can be used to extract features from:
-
(1)
Original: A copy of the original images.
-
(2)
Wavelet: Applies high (H) or low (L) pass filter in all of three dimensions in a 3D image. Theoretically, we have eight different filtering combinations: LLL, LLH, LHL, LHH, HLL, HLH, HHL, HHH. However, when the image rotates, the features extracted from the images processed by LLH, LHL, LHH, HLL, HLH and HHL filters will change too much, so we only preserve the images that are filtered by LLL and HHH.
-
(3)
LoG: Applies the Laplacian of Gaussian filter to the image to enhance edge information.
Then, we extract radiomics features from the QSM images. Specifically, by using the manual mask mentioned in Section 2.1, we first extracted two independent SN regions from the preprocessed images obtained in the previous step, as shown in Fig. 1. After that, we calculated the radiomics features of each connected region by Pyradiomics (https://pyradiomics.readthedocs.io/en/latest/). The extracted features contain six basic categories:(1) First-order statistical features; (2) Gray level co-occurrence matrix; (3) Gray level run length matrix; (4) Gray level size zone matrix; (5) Neighbouring gray tone difference matrix; and (6) Gray level dependence matrix. In this work, the feature set consisted of 2212 features for each case (i.e., the left substantia nigra and the right substantia nigra have 1106 features, respectively).
2.2.2. Feature selection and classification
Feature selection was performed to exclude the unstable factors in features and reduce the dimensionality of features. In order to evaluate the stability of the features extracted from the SN, we use the intraclass correlation coefficient (ICC) to screen out those features with high stability for manual mask. Here, ICC is a descriptive statistic that can be used to quantify the stability of a feature with respect to the accuracy of the mask. We first applied a small spatial deformation on the manually labeled mask to simulate a new mask set from a different doctor. Then, for each feature, we extracted the radiomics features based on the two mask sets and calculate the corresponding ICC. In our research, we chose features with ICC > 0.8 for the later steps. While the reduction in the number of features also makes our model more robust and interpretable, we used recursive feature elimination (RFE) for further feature selection to reduce the dimensionality of features. We chose the RFE method for feature selection based on the following considerations: (1) RFE can gradually eliminate useless features while keeping the robust features. (2) In the process of modeling, RFE makes it possible to combine logistic regression and SVM, so that the final classification model can decide which features are most useful, thereby integrating the learning and feature selection into the entire process. Given a classifier which can assign coefficients to features, the purpose of the RFE is to exclude the least important features from the current feature set. This procedure was recursively repeated until the number of remaining features reached a predetermined number. In order to avoid the influence of different feature scales, we removed the mean of the features on the training data and scaled them to unit variance before feature selection. Then, combined with an LR or SVC, we used RFE to filter out the most informative features for distinguishing PD from NC based on the feature set with high stability. After feature selection, we retrained an LR and an SVC model on the training and validation set with the selected features to predict whether a case is PD or NC.
To avoid data disclosure issues, all of the feature selection, hyperparameter selection and evaluation of the method were performed within a nested cross-validation framework. The nested cross-validation scheme involves two cross-validation routines: Outer cross-validation and inner cross-validation. We used a 7-fold outer cross-validation to test the generalization performance of the classification model and a 6-fold inner cross-validation to select features and optimize the hyperparameters. In each fold session, 100 examples were used for training, 20 for validation, and 20 for testing.
2.3. Classification with convolution neural network
Different from the radiomics based methods, we also proposed a convolution neural network (CNN) to distinguish PD patients from healthy controls. Since the features extracted by CNN are continuously optimized according to the classification accuracy on the training set, we can assume that these features can better solve the problem from the perspective of data driving, compared with the fixed feature extraction method of radiomics. Our CNN-based pipeline is shown in Fig. 3. Next, we introduce the CNN classification model from two aspects: data augmentation, and network construction and training.
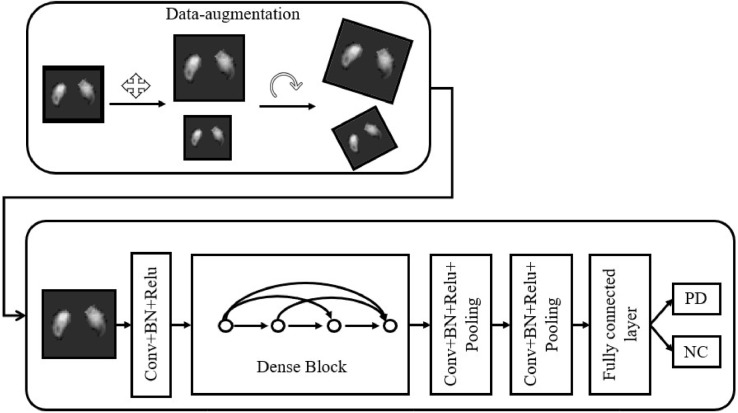
Fig 3.
The pipeline of the CNN. Data-augmentation, which helps prevent overfitting and provides more training data, is performed first, followed by a CNN classifier. The classifier will give a probability to PD and NC, respectively, according to the input image with SN.
2.3.1. Data augmentation
Previous studies show that, in the case of limited data, necessary data augmentation can help obtain a more robust classification model. Therefore, we performed data augmentation on all training data. First, we applied a dilation operation on the mask image. The dilated image can contain contextual information next to the SN. Both the dilated and the original mask images were used for ROI extraction. Then, we extracted a patch image containing the SN. The size of the patch image was fixed to 50 × 50 × 24. Before each training iteration, we randomly scale and rotate the patch image in a small range. The scaling coefficients were generated randomly from 0.95 to 1.05, and the rotation angles were generated randomly from −5 to +5°.
2.3.2. The construction and training of the CNN
Based on the most advanced deep learning classification network, i.e., Densenet (Huang et al., 2017), we constructed our classification network as summarized in Fig. 3.
After data augmentation, images with 1 channel (intensity channel) were passed into the convolution layer. Then, the data stream passes through a Convolution layer, Batch Normalization (BN) layer (Ioffe and Szegedy, 2015) and Relu layer successively. Here we call “Convolution layer + +BN layer + +Relu layer” as a “block” as indicated in Fig. 3. After block 1, which produced 6 feature maps, we added a “Dense block” which was introduced in (Huang et al., 2016). Specifically, the output of block1 generates a new 2-channel feature map through a new block. Then, two feature maps are concatenated into the channel dimension to form an 8-dimensional feature map. In our network, the same operation is repeated twice, resulting in a 10-channel feature map as the output of the “Dense block”. In the last two blocks, we added a “Pooling” layer to reduce the spatial dimension. After two blocks, we obtained a 4-dimension feature vector which encodes the input image in the semantic space. After introducing a fully connected layer and a Softmax layer, we obtained a classification probability for describing the confidence of illness.
The training of the CNN model is implemented by Pytorch, on a desktop equipped with a GTX 1080Ti GPU. For each experiment session, we divide the dataset into three parts: 100 subjects for training, 20 for validation, and 20 for testing same as described in 2.2.2. We employ Adam algorithm to optimize the CNN on the training set. For model selection, since we can get a model in each training step, we set inclusion criteria to prevent overfitting and enhance the generalization ability of the model: For each training step, only if the model had a classification accuracy larger than 90% on the training set and 80% on the validation set, was it recorded. After training, the model with the highest accuracy on the validation set was selected and used to evaluate the performance on the test set.
2.4. Hybrid features for diagnosis of PD
Radiomics features focus more on gray scale statistics and texture features (Jjm et al., 2017), while CNN pays more attention to specific higher-order semantic information (Zeiler and Fergus, 2014; Guorong et al., 2006). Therefore, we assume these two approaches may be complementarity for the classification task, as (Dong et al., 2016). Here, we combine them to look for an improved diagnosis of PD. We concatenate the radiomics features extracted in Section 2.2.2 and the 4-dimension feature vector extracted before the fully-connected layer in CNN as the new feature vector. By using this new feature vector, an LR or SVC is employed to model the classification task again. Except for the input features of the model, all the experimental steps were the same as in Section 2.2.
3. Results
3.1. Classification accuracy (PD/NC)
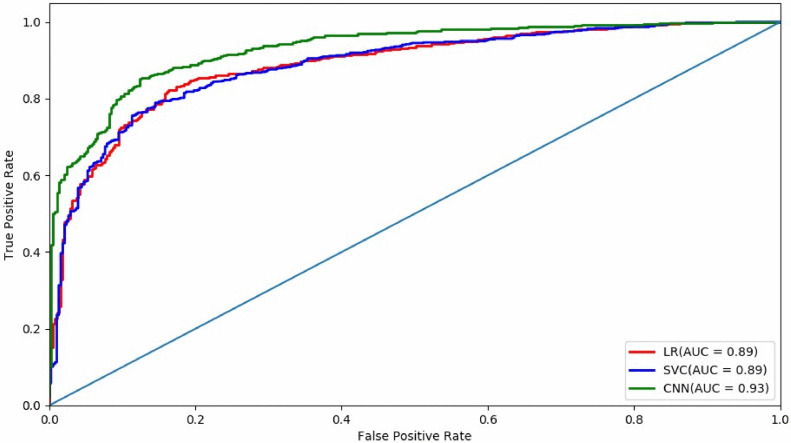
We performed fifty distinct 7-fold nested cross-validations on the dataset with three different machine learning algorithms: logistic regression (LR), support vector machine (SVM) and convolution neural network (CNN). In each cross-validation, six-seventh of the data was used as the training data (i.e., five-seventh for training, and one-seventh for validation in the CNN model), and the remaining one-seventh of the data was used as the test data. For each method, we put all the test cases in one set as the result. The AUC (Area Under the Curve), accuracy, sensitivity, and specificity for differentiating PD from NC were evaluated on the test set.
As summarised in the first three lines of Table 1, CNN gives the best classification performance: AUC = 0.93, Accuracy = 0.85, Sensitivity = =0.86, and Specificity = 0.83. Besides CNN, logistic regression and SVM also have comparable results. Even though these machine learning methods are not as good as CNN, they have better interpretability as the radiomics features provide more explanatory biomarkers.
Table 1.
PD vs. NC classification performance. Area under the receiver operating characteristics (AUC), accuracy (ACC), sensitivity, specificity, and BAC (= (sensitivity + specificity)/2) are reported with difference classifiers: SVM (support vector machine), LR (logistic regression), and our CNN (convolution neural network) method. The ‘Hybrid features’ means that both radiomics features and deep features extracted by CNN are used.
| Method | AUC | Accuracy | Sensitivity | Specificity | BAC |
| LR | 0.89 | 0.81 | 0.91 | 0.70 | 0.80 |
| SVC | 0.89 | 0.81 | 0.91 | 0.71 | 0.80 |
| CNN | 0.93 | 0.85 | 0.86 | 0.83 | 0.85 |
| LR (Hybrid features) | 0.96 | 0.88 | 0.97 | 0.78 | 0.88 |
| SVM (Hybrid features) | 0.96 | 0.90 | 0.93 | 0.86 | 0.89 |
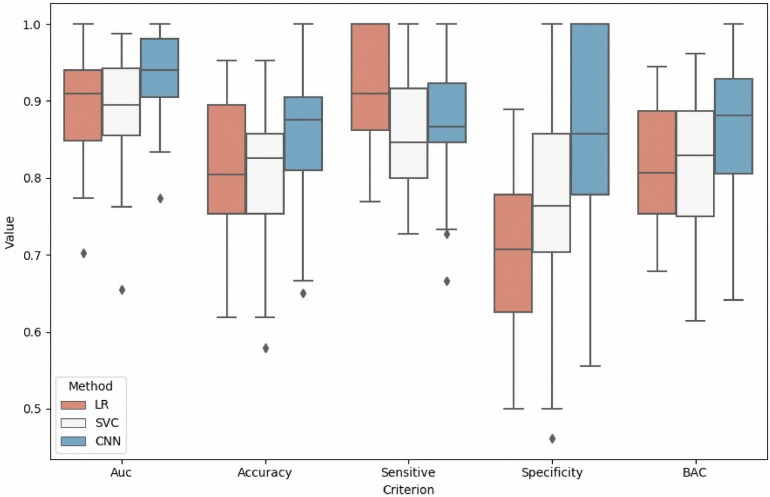
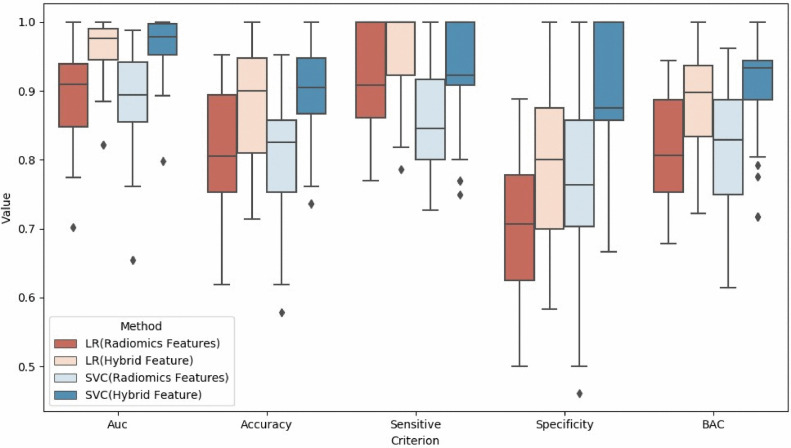
In addition to putting all the test results into one set for consideration, we calculated the results of each experiment and arranged them in Figs. 4 and 5. We can see that the CNN-based method not only obtains a better performance of almost all the classification indicators, but also is more stable than radiomics. Nevertheless, the radiomics features still get good classification results, which indicates that the gray scale and texture features of the SN can also distinguish PD very well.
Fig 4.
Comparison of results by LR (Logistic Regression), SVM (Support Vector Machine), and CNN (Convolution Neural Network). The box plot extends from the upper to lower quartile values of different criteria, while the box displays the quartile of values set. Points outside the box plot are outliers.
Fig 5.
The AUC curves of SVM (support vector machine), LR (logistic regression), and CNN (convolution neural network).
3.2. Combination of radiomics features and CNN features
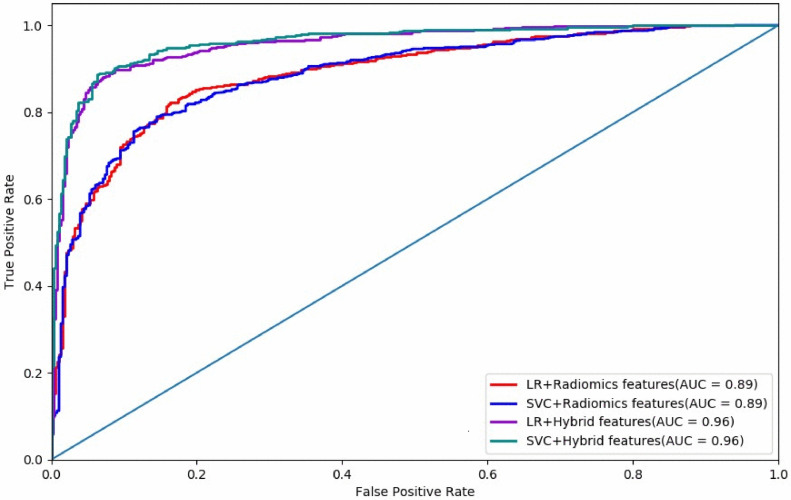
We combined the features extracted from the last convolution layer of CNN with the radiomics features extracted by RFE method and re-trained the LR and SVM models to get the classification results (see Table 1). It can be seen from Table 1 that the integration of the two features helps produce better performance in classification. With the help of radiomics features and deep features, we find that the SVM model gets the best results with the AUC comparable to that obtained from PET (Choi et al., 2017). Figs. 6 and 7 demonstrate that the combination of the radiomics features and CNN-based features improves both the classification accuracy and algorithm stability.
Fig 6.
Comparison of results by SVM (support vector machine), LR (logistic regression), and CNN (convolution neural network). The ‘Hybrid Features’ means that both radiomics features and deep features extracted by CNN are used.
Fig 7.
The AUC curves of SVM (support vector machine), LR (logistic regression), and CNN (convolution neural network).
3.3. Feature statistics and visualization
The high classification capacity shown by the radiomics features demonstrates the reliability of QSM in diagnosing PD. So, it is interesting to evaluate which features are chosen by the classification model. For each cross-validation round, we recorded which features were selected and counted them in Table 3. The clinical significance of these features will be further discussed in Section 4.
Table 3.
The radiomics features selected by logistic regression and SVM. We carried out nested cross-validation 50 times. In each cross-validation, logistic regression and SVM were combined with RFE, respectively, and selected a feature subset. We recorded the frequency of each feature selected and sorted out the features whose selected frequency is more than 20. In table, ‘Left’ and ‘Right’ denote the left and right hemispheres, respectively.
| Region | Image Type | Feature Class | Feature | Frequency |
| Right | Original | GLRLM | RunEntropy | 95 |
| Right | Original | GLCM | ClusterShade | 64 |
| Left | Wavelet-LLL | GLCM | IDN | 63 |
| Right | Log | GLCM | DifferenceEntropy | 59 |
| Right | Log | First-order | Median | 56 |
| Left | Log | GLDM | DependenceVariance | 54 |
| Right | Wavelet-LLL | GLCM | ClusterShade | 50 |
| Right | Log | First-order | Median | 49 |
| Left | Original | GLDM | DependenceEntropy | 48 |
| Right | Wavelet | GLDM | DependenceEntropy | 47 |
| Right | Original | GLDM | DependenceEntropy | 41 |
| Right | Log | First-order | Skewness | 36 |
| Right | Log | GLCM | DifferenceVariance | 36 |
| Right | Wavelet-LLL | GLRLM | RunEntropy | 31 |
| Right | Wavelet-LLL | GLCM | Correlation | 31 |
| Left | Wavelet-LLL | GLDM | DependenceEntropy | 27 |
| Right | Log | GLCM | IDN | 26 |
| Left | Log | GLRLM | LongRunEmphasis | 24 |
| Right | Log | First-order | Mean | 21 |
| Left | Log | First-order | Median | 20 |
| Left | Log | GLRLM | Median | 20 |
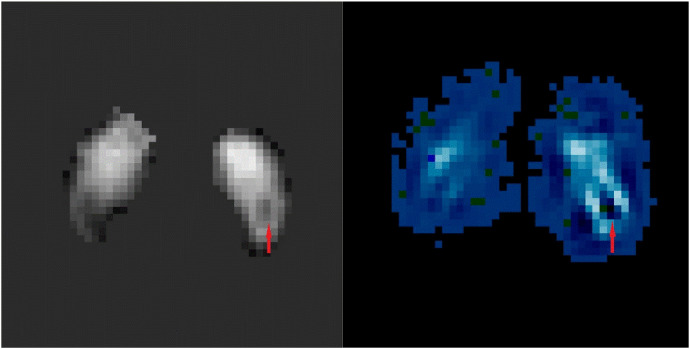
For the CNN-based method, although we have achieved the highest classification accuracy, there are still some problems. The hierarchical structure of CNN has the advantage of mining the characteristics of the data, but these features are often difficult to understand. In order to extract more information about the pattern of PD pathological changes from CNN, we employed the visualization technology Grad-Cam (Selvaraju et al., 2017) to highlight the most relevant region with the model's output through the CNN's parameters. The heat map visualized in Fig. 8 reveals pathophysiological changes consistent with current studies, some of which we will discuss in Section 4.
Fig 8.
The visualization of original image (left) and heat map (right) from Grad-Cam. The position pointed to by the red arrow is the 'Swallow tail’ representing the N1 territory, which usually appears in the caudal region of the substantia nigra in healthy people. According to the principle of grad-cam, if we reduce the intensity of the original image at the black area in the heat map, or increase the intensity of the original image at the white area in the heat map, the NC probability of CNN output will increase. In fact, no matter which kind of operation is carried out, the swallowtail sign will be more obvious. This is in complete agreement with our current understanding of the swallowtail sign.
4. Discussion
In this work, we found that CNN gave the best classification performance, and it was more stable than the radiomics features. Given that the radiomics features are easy to interpret and can still produce good classification results, we combined radiomics features and CNN-based features to further improve the classification performance, obtaining an AUC as high as 0.96.
Previous neuroimaging AI (e.g., ICCA, JFSS or SVM) studies on PD (see Table 2) were based on T1-weighted MR imaging. The classification accuracy of studies from T1 MR images ranged from 70.5% to 88% (Adeli et al., 2016; Amoroso et al., 2018; Liu et al., 2018). But T1 MR imaging can only show volumetric changes of brain structures from the whole brain data. However, the T1 image contrast in SN, which is a key structure pathologically involved in PD, is too low to show its contour. Therefore, it is difficult to determine which brain regions play an important role in the final diagnosis, because features come from different brain regions.
Table 2.
Classification accuracy from different methods. The method column is briefly described as the form of Method (medical image modality).
| Method | AUC | Accuracy | Sensitivity | Specificity |
| ICCA (MRI T1) Liu et al. | 0.71 | 0.71 | 0.625 | 0.786 |
| RLDA+JFSS (MRI T1) Adeli et al. | 0.84 | 0.82 | * | * |
| NF (MRI T1) Amoroso et al | 0.94 | 0.88 | 0.85 | 0.88 |
| CNN (MRI QSM) [Proposed] | 0.95 | 0.89 | 0.92 | 0.85 |
| SVM (MRI QSM) [Proposed] | 0.96 | 0.90 | 0.93 | 0.86 |
On the other hand, QSM is a recently developed MRI technique that is used to quantify iron in vivo. It provides both the quantitative information of the regional iron content and also its distribution within these nuclei. A previous QSM study on 21 PD patients and 21 normal controls by Murakami et al. (2015) showed a diagnostic performance accuracy of 88%, and the AUC for the ROC reached 0.9. Therefore, both radiomics features and CNN-based features in our work were performed on the QSM data of the SN structure to investigate the diagnostic performance.
The features used by CNN were extracted automatically by the back-propagation algorithm, which extracted the most discriminative features for the current classification task. Therefore, they have better discriminative power than radiomics features. As we can see from Table 3, most of the features selected in the LR and SVM are related to the brightness distribution in the region of the SN and are consistent with previous studies that have shown that excessive iron deposition occurs in the SN of PD patients (He et al., 2015; Morris and Edwardson, 1994; Sian‐Hülsmann et al., 2011; Wypijewska et al., 2010). From the features we selected, we can see that this phenomenon can be reflected not only in the mean of ROI gray, but also in other indicators of grayscale texture. For example, we can see that there are apparent anomalies in the median and 90 quantiles of grayscale in the region. Entropy is also frequently selected, and suggests that the distribution of the gray scale values is spatially cluttered. This indicates that the excessive deposition of iron does not occur uniformly in the SN but may be distributed in specific local regions.
The highlighted part of Fig. 8 from the CNN results for visualization represents a positive correlation with the judgment made by the network. This highlighted area is mainly located in the lateral aspects and the caudal portion of the SN. This is in agreement with previous work showing that there is a loss of dopaminergic neurons in the SN in PD patients in just these regions (Damier et al., 1999a, 1999b). So, these CNN-derived features appear to reflect the key pathophysiological change of PD as seen in other studies.
5. Conclusion
The findings from this work showed that hybrid features based on QSM of the SN from radiomics and CNN are consistent with previous histological and imaging studies, and provided reasonable diagnostic performance with an AUC as high as 0.96. The use of CNN and radiomics algorithms may provide a means by which to study diagnostic MR imaging biomarkers for PD.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported in part by grants from the National Key Research and Development Program of China (2018YFC0116400) for Feng Shi, Science and Technology Commission of Shanghai Municipality (17411952700) for Fuhua Yan and Naying He, the Shanghai Sailing Program (18YF1414700) for Naying He, a National Natural Science Foundation of china (81801652) for Naying He and Bin Xiao, and National Natural Science Foundation of china (81971576) for Fuhua Yan and Naying He.
Contributor Information
Qian Wang, Email: wang.qian@sjtu.edu.cn.
Fuhua Yan, Email: yfh11655@rjh.com.cn.
References
- Adeli E., Shi F., An L., Wee C.Y., Wu G., Wang T., Shen D. Joint feature-sample selection and robust diagnosis of Parkinson's disease from MRI data. Neuroimage. 2016;141:206–219. doi: 10.1016/j.neuroimage.2016.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoroso N., La R.M., Monaco A., Bellotti R., Tangaro S. Complex networks reveal early MRI markers of Parkinson's disease. Med. Image Anal. 2018;48:12–24. doi: 10.1016/j.media.2018.05.004. [DOI] [PubMed] [Google Scholar]
- Brooks D.J., Pavese N. Imaging biomarkers in Parkinson's disease. Prog. Neurobiol. 2011;95:614–628. doi: 10.1016/j.pneurobio.2011.08.009. [DOI] [PubMed] [Google Scholar]
- Choi H., Ha S., Im H.J., Paek S.H., Lee D.S. Refining diagnosis of Parkinson's disease with deep learning-based interpretation of dopamine transporter imaging. Neuroimage Clin. 2017;16:586–594. doi: 10.1016/j.nicl.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damier P., Hirsch E., Agid Y., Graybiel A.J.B. The substantia nigra of the human brain: I. Nigrosomes and the nigral matrix, a compartmental organization based on calbindin D28K immunohistochemistry. Brain. 1999;122:1421–1436. doi: 10.1093/brain/122.8.1421. [DOI] [PubMed] [Google Scholar]
- Damier P., Hirsch E.C., Agid Y., Graybiel A.M. The substantia nigra of the human brain. II. Patterns of loss of dopamine-containing neurons in Parkinson's disease. Brain. 1999;122(Pt 8):1437–1448. doi: 10.1093/brain/122.8.1437. [DOI] [PubMed] [Google Scholar]
- Dong N., Han Z., Ehsan A., Luyan L., Dinggang S. 3D Deep Learning for Multi-modal Imaging-Guided Survival Time Prediction of Brain Tumor Patients. MICCAI. 2016:212–220. doi: 10.1007/978-3-319-46723-8_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guorong W., Feihu Q., Dinggang S. Learning-Based Deformable Registration of MR Brain Images. IEEE Trans. Med. Imaging. 2006:1145–1157. doi: 10.1109/tmi.2006.879320. [DOI] [PubMed] [Google Scholar]
- Haacke E.M., Liu S., Buch S., Zheng W., Wu D., Ye Y. Quantitative susceptibility mapping: current status and future directions. Magn. Reson. Imaging. 2015;33:1–25. doi: 10.1016/j.mri.2014.09.004. [DOI] [PubMed] [Google Scholar]
- He N., Ling H., Ding B., Huang J., Zhang Y., Zhang Z., Liu C., Chen K., Yan F. Region-specific disturbed iron distribution in early idiopathic Parkinson's disease measured by quantitative susceptibility mapping. Hum. Brain Mapp. 2015;36:4407–4420. doi: 10.1002/hbm.22928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Liu Z., van der Maaten L., Weinberger K. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Best Paper Award. 2016. Densely Connected Convolutional Networks. [Google Scholar]
- Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Densely connected convolutional networks; pp. 4700–4708. [Google Scholar]
- Ioffe S., Szegedy C. International Conference on International Conference on Machine Learning JMLR.org; 2015. Batch normalization: accelerating deep network training by reducing internal covariate shift. [Google Scholar]
- Jjm V.G., Fedorov A., Parmar C., Hosny A., Aucoin N., Narayan V., Rgh B.T., Fillion-Robin J.C., Pieper S., Hjwl A.J.C.R. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langkammer C., Schweser F., Krebs N., Deistung A., Goessler W., Scheurer E., Sommer K., Reishofer G., Yen K., Fazekas F.J.N. Quantitative susceptibility mapping (QSM) as a means to measure brain iron? A post mortem validation study. Neuroimage. 2012;62:1593–1599. doi: 10.1016/j.neuroimage.2012.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Wu B., Liu C. Quantitative susceptibility mapping of human brain reflects spatial variation in tissue composition. Neuroimage. 2011;55:1645–1656. doi: 10.1016/j.neuroimage.2010.11.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorraine V.K., Anthony E.L. Parkinson's disease[J]. Lancet. 2015;386(9996):896–912. doi: 10.1016/S0140-6736(14)61393-3. [DOI] [PubMed] [Google Scholar]
- Mahlknecht P., Krismer F., Poewe W., Seppi K.J.M.D. Meta-analysis of dorsolateral nigral hyperintensity on magnetic resonance imaging as a marker for Parkinson's disease. Mov. Disord. 2017;32:619–623. doi: 10.1002/mds.26932. [DOI] [PubMed] [Google Scholar]
- Morris C.M., Edwardson J.A. Iron histochemistry of the substantia-nigra in Parkinsons-disease. Neurodegeneration. 1994;3:277–282. [PubMed] [Google Scholar]
- Murakami Y., Kakeda S., Watanabe K., Ueda I., Ogasawara A., Moriya J., Ide S., Futatsuya K., Sato T., Okada K., Uozumi T., Tsuji S., Liu T., Wang Y., Korogi Y. Usefulness of quantitative susceptibility mapping for the diagnosis of Parkinson disease. AJNR Am. J. Neuroradiol. 2015;36:1102–1108. doi: 10.3174/ajnr.A4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S.T., Mougin O., Xing Y., Blazejewska A., Bajaj N., Auer D.P., Gowland P.J.N.C. Parkinson's disease related signal change in the nigrosomes 1–5 and the substantia nigra using T2* weighted 7T MRI. NeuroImage Clin. 2018;19:683–689. doi: 10.1016/j.nicl.2018.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Proceedings of the IEEE International Conference on Computer Vision. 2017. Grad-cam: visual explanations from deep networks via gradient-based localization; pp. 618–626. [Google Scholar]
- Sian-Hülsmann J., Mandel S., Youdim M.B., Riederer P. The relevance of iron in the pathogenesis of Parkinson's disease. Ann. N.Y. Acad. Sci. 2011;118:939–957. doi: 10.1111/j.1471-4159.2010.07132.x. [DOI] [PubMed] [Google Scholar]
- Sun H., Walsh A.J., Lebel R.M., Blevins G., Catz I., Lu J.Q., Johnson E.S., Emery D.J., Warren K.G., Wilman A.H. Validation of quantitative susceptibility mapping with Perls' iron staining for subcortical gray matter. Neuroimage. 2015;105:486–492. doi: 10.1016/j.neuroimage.2014.11.010. [DOI] [PubMed] [Google Scholar]
- Vos T., Abajobir A.A., Abate K.H. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016[J] The Lancet. 2017;390(10100):1211–1259. doi: 10.1016/S0140-6736(17)32154-2. GB/T 7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wypijewska A., Galazka-Friedman J., Bauminger E.R., Wszolek Z.K., Schweitzer K.J., Dickson D.W., Jaklewicz A., Elbaum D., Friedman A. Iron and reactive oxygen species activity in Parkinsonian substantia nigra. Parkinsonism Relat. Disord. 2010;16:329–333. doi: 10.1016/j.parkreldis.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Yan F., He N., Lin H., Li R. Iron deposition quantification: applications in the brain and liver. J. Magn. Reson. Imaging. 2018;48:301–317. doi: 10.1002/jmri.26161. [DOI] [PubMed] [Google Scholar]
- Zeiler M.D., Fergus R. Proceedings of the European Conference on Computer Vision. Springer; 2014. Visualizing and understanding convolutional networks; pp. 818–833. [Google Scholar]
- Zheng W., Nichol H., Liu S., Cheng Y.C., Haacke E.M. Measuring iron in the brain using quantitative susceptibility mapping and X-ray fluorescence imaging. Neuroimage. 2013;78:68–74. doi: 10.1016/j.neuroimage.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]