Abstract
Acute lung injury (ALI) is a pulmonary disorder that causes acute respiratory failure, thus leading to relative high mortality worldwide. However, the molecular mechanisms of ALI remain largely unknown. MicroRNA (miRNA)-dependent control of gene expression at a post-transcriptional level has been recently reported. Herein, we identify a candidate miRNA, miR-495, that affects the progression of ALI. Alveolar macrophages (NR8383) were treated with 1 μg/mL lipopolysaccharide (LPS) to establish a cell-injury model. Combined with the data from western blot, methylation-specific PCR, methylated DNA immunoprecipitation, and chromatin immunoprecipitation assays, NLRP3 inflammasome activation and methylation-dependent repression of miR-495 were found in LPS-exposed NR8383 cells. Dual-luciferase reporter gene assay and miR-495 gain-of-function experiments confirmed that NLRP3 was a target of miR-495. Next, the expression of miR-495 and NLRP3 was overexpressed or silenced to assess their effects on NLRP3 inflammasome activation, alveolar macrophage inflammation, and pyroptosis in vitro. As demonstrated, overexpressed miR-495 alleviated alveolar macrophage inflammation and pyroptosis and inhibited NLRP3 inflammasome activation by negatively regulating the NLRP3 gene. Consistently, elevated miR-495 alleviated lung injury and reduced the neutrophil infiltration and inflammation in rat models of LPS-induced ALI. Taken together, the data in our study demonstrated that methylation of the miR-495 promoter could downregulate miR-495, whose elevation could attenuate the activation of the NLRP3 inflammasome to protect against ALI, which provides novel therapeutic targets for ALI treatment.
Keywords: acute lung injury, microRNA-495, NOD-like receptor family pyrin domain containing 3, methylation, inflammasome
Introduction
Acute lung injury (ALI) is a prevalent and life-threatening disease resulting from an acute injury such as pneumonia, shock, sepsis, or aspiration.1 ALI causes lung injuries characterized by edema and injury to the lung tissues; impaired gas exchange; and severe inflammatory responses leading to diffuse alveolar injury, serious hypoxemia, and sub-par lung compliance.2,3 Owing to insufficient management and intricate pathogenesis, ALI accounts for high morbidity and mortality rates associated with acute respiratory failure worldwide.4,5 Although ALI and acute respiratory distress syndrome (ARDS) have been extensively explored for decades, several efforts in exploring therapeutic strategies and measures have ended up in failure.6 Moreover, patients who survive ALI are frequently confronted with long-term physical impairments.7 All in all, it is significant to develop novel and effective therapeutic targets for ALI to raise the quality of life of ALI-plagued patients.
MicroRNAs (miRNAs) are a group of short non-coding RNAs that participate in the processes of many lung diseases, including ALI.8 For instance, lung inflammation, vascular permeability, and neutrophil infiltration in ALI induced by lipopolysaccharide (LPS).9 In addition, Ai et al.10 indicated that miR-495 acts as a tumor suppressor, serving as a promising target for the treatment of lung cancer. Furthermore, NOD-like receptor family pyrin domain containing 3 (NLRP3) has also been highlighted as a putative target of miR-495 by bioinformatic prediction. NLRP3 is regarded as an intracellular signaling molecule that is implicated in the inflammatory response and development of lung diseases.11 It was further revealed that NLRP3 depletion influences the innate immune system, thus modulating intestinal inflammation and maintaining intestinal homeostasis.12 Another interesting topic, the NLRP3 inflammasome is a complex that consists of NLRP3, apoptosis-associated speck-like protein containing a caspase activation and recruitment domain (ASC), and caspase-1. Another study also reported that activation of the NLRP3 inflammasome may contribute to the progression of ALI.13,14 Based on these findings, we speculated whether the miR-495/NLRP3 axis could regulate the development of ALI. Therefore, in the present study, we aimed to explore the specific mechanism by which miR-495 participates in the processes of ALI by mediating the expression of NLRP3 using both in vitro and in vivo assays based on cell and rat models of LPS-induced ALI.
Results
NLRP3 Silencing Suppresses LPS-Induced Alveolar Macrophage Inflammation
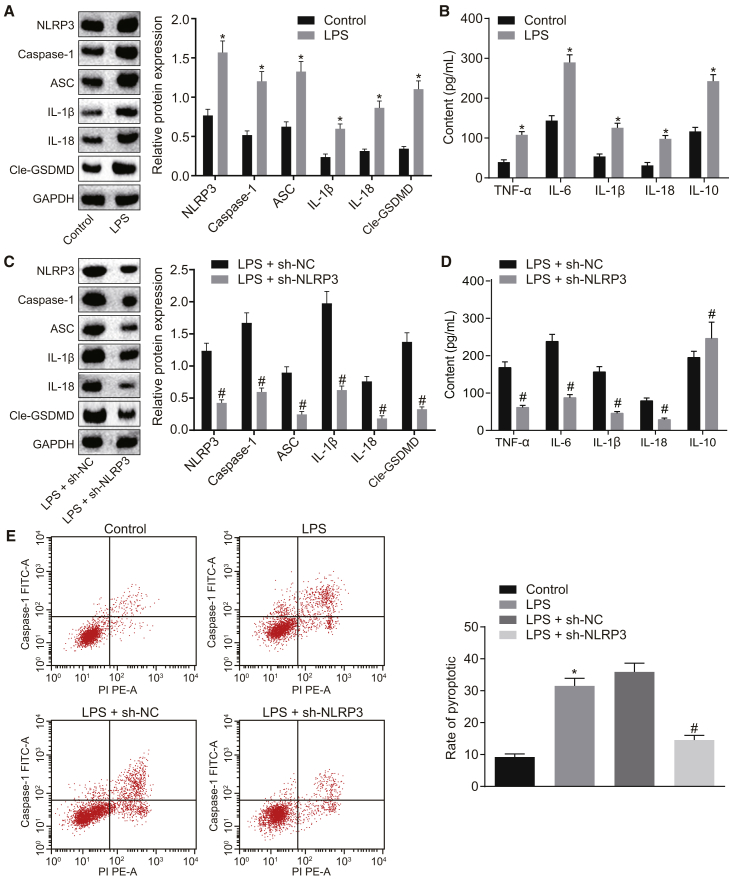
After establishing the cell models of LPS-induced injury, western blot analysis was applied to measure the protein expression levels of NLRP3, caspase-1, ASC, interleukin (IL)-1β, IL-18, and cleaved-gasdermin D (Cle-GSDMD) in NR8383 cells, which were all determined to be higher in NR8383 cells treated with LPS compared to NR8383 cells treated with F-12K culture medium (p < 0.05) (Figure 1A). Next, the levels of proinflammatory factors tumor necrosis factor α (TNF-α), IL-6, IL-1β, and IL-18 and of anti-inflammatory factor IL-10 in NR8383 cells were detected using ELISA, and the results revealed that LPS treatment increased the levels of the aforementioned inflammatory factors compared to F-12K culture medium treatment (Figure 1B), suggesting that LPS could successfully induce ALI in alveolar macrophages and that NLRP3 was upregulated in ALI. Subsequently, further assays were performed to identify the effects of NLRP3 on the expression levels of inflammation-related factors on NLRP3 silencing. As shown by western blot analysis, the protein expression levels of NLRP3, caspase-1, ASC, IL-1β, IL-18, and Cle-GSDMD in LPS-exposed NR8383 cells were noted to be reduced as a response to treatment with short-hairpin-RNA (shRNA)-targeting NLRP3 (sh-NLRP3) when compared to treatment with shRNA-negative control (sh-NC; p < 0.05) (Figure 1C). Additionally, the results of the ELISA revealed that LPS-exposed NR8383 cells transfected with short hairpin (sh)-NLRP3 exhibited downregulated expression levels of proinflammatory factors but upregulated expression of the anti-inflammatory factor IL-10 in comparison to the LPS-exposed NR8383 cells transfected with sh-NC (all p values < 0.05) (Figure 1D). Moreover, flow-cytometric analysis was performed to measure the pyroptosis of NR8383 cells, which demonstrated that LPS treatment enhanced pyroptosis in NR8383 cells versus the treatment with F-12K culture medium; however, pyroptosis was attenuated following treatment of LPS and sh-NLRP3 versus treatment of LPS and sh-NC (both p values < 0.05) (Figure 1E). These data revealed that NLRP3 inflammasome activation was detected in LPS-exposed NR8383 cells and that depletion of NLRP3 could ameliorate LPS-induced alveolar macrophage inflammation.
Figure 1.
LPS-Induced ALI Exhibits Activated NLRP3 Inflammasome and Inflammation of Alveolar Macrophages in LPS-Induced ALI Is Inhibited with the Silencing of NLRP3
(A) Protein expression levels of NLRP3, caspase-1, ASC, IL-1β, IL-18, and Cle-GSDMD in NR8383 cells treated with F-12K culture medium or LPS as detected by western blot analysis. (B) Expression levels of proinflammatory factors (TNF-α, IL-6, IL-1β, and IL-18) and anti-inflammatory factor IL-10 in NR8383 cells treated with F-12K culture medium or LPS as detected by ELISA. (C) Protein expression levels of NLRP3, caspase-1, ASC, IL-1β, IL-18, and Cle-GSDMD in LPS-exposed NR8383 cells following transfection with sh-NLRP3 or sh-NC as measured by western blot analysis. (D) Expression levels of proinflammatory factors (TNF-α, IL-6, IL-1β, and IL-18) and anti-inflammatory factor IL-10 in LPS-exposed NR8383 cells following transfection with sh-NLRP3 or sh-NC as measured by ELISA. (E) Pyroptosis of LPS-exposed NR8383 cells following transfection with sh-NLRP3 or sh-NC, as determined by PI/Hoechst 33342 double staining. *p < 0.05 versus NR8383 cells treated with F-12K culture medium; #p < 0.05 versus LPS-exposed NR8383 cells transfected with sh-NC. An unpaired t test was used to analyze data expressed as mean ± SD between two groups if the data conformed to normal distribution and homogeneity of variance. Data among multiple groups were compared using one-way ANOVA followed by Tukey’s post hoc test. The experiment was repeated three times.
NLRP3 Is the Target Gene of miR-495
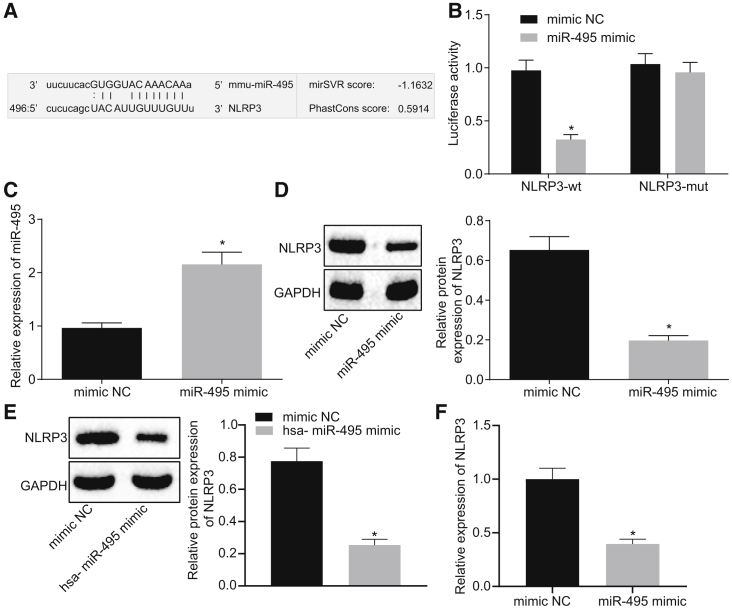
Bioinformatic prediction was performed in order to identify the upstream miRNA interacting with NLRP3. The prediction results revealed the presence of a specific binding site between the NLRP3 gene sequence and the miR-495 sequence, suggesting that NLRP3 was a putative target gene of miR-495 (Figure 2A) and that the miR-495 sequence was conservative. The relationship between miR-495 and NLRP3 was further verified using a dual-luciferase reporter gene assay. The results revealed that the luciferase activity was attenuated in response to co-transfection with the miR-495 mimic and NLRP3-wild-type (WT) 3′ UTR compared to co-transfection with mimic NC and NLRP3-WT (p < 0.05); however, the luciferase activity following co-transfection with miR-495 mimic and NLRP3 mutant type (Mut) was found to be not significantly different from that following co-transfection with mimic NC and NLRP3-Mut (p > 0.05) (Figure 2B). Moreover, as determined by qRT-PCR, the expression of miR-495 was increased in cells following miR-495 mimic transfection versus mimic NC transfection (p < 0.05) (Figure 2C). Furthermore, the results of western blot analysis indicated that cells transfected with miR-495 mimic presented with reduced expression of NLRP3 in comparison with cells transfected with mimic NC (p < 0.05) (Figure 2D). Subsequently, in order to investigate whether the interaction mechanism of miR-495 and NLRP3 was conservative in nature, we compared human and rat miR-495 sequences and found that miR-495 was highly conservative. Next, NR8383 cells were transfected with synthesized hsa-miR-495 mimic and its corresponding NC. Findings of western blot assay revealed that miR-495 mimic transfection led to a reduction in the expression levels of NLRP3 compared to mimic NC transfection (p < 0.05) (Figure 2E). Besides, the stability of NLRP3 mRNA was measured by determining its expression using qRT-PCR. It was revealed that NLRP3 expression was downregulated in miR-495-mimic-transfected cells versus the mimic-NC-transfected cells, suggesting that miR-495 could downregulate the expression of NLRP3 mRNA (Figure 2F). All in all, these findings suggested that miR-495 could modulate the expression of NLRP3 by influencing its stability.
Figure 2.
miR-495 Directly Targets NLRP3
(A) Binding site between miR-495 and NLRP3 3′ UTR predicted using the bioinformatic website microRNA.org. (B) Luciferase activity of NLRP3-WT and NLRP3-Mut following the co-transfection of miR-495 mimic or mimic NC detected by dual-luciferase reporter gene assay. (C) Expression of miR-495 in response to transfection with miR-495 mimic or mimic NC as measured by qRT-PCR. (D) Expression of NLRP3 in response to transfection with miR-495 mimic or mimic NC as measured by western blot analysis. (E) Protein expression of NLRP3 in response to transfection with hsa-miR-495 mimic or mimic NC as measured by western blot analysis. (F) Expression of NLRP3 in response to transfection with hsa-miR-495 mimic or mimic NC determined by qRT-PCR. *p < 0.05 versus NR8383 cells treated with mimic NC. Measurement data were expressed as mean ± SD. An unpaired t test was used to analyze data expressed as mean ± SD between two groups if the data conformed to normal distribution and homogeneity of variance. The experiment was repeated three times to obtain the mean value.
Methylation-Dependent Repression of miR-495 in LPS-Injured NR8383 Cells
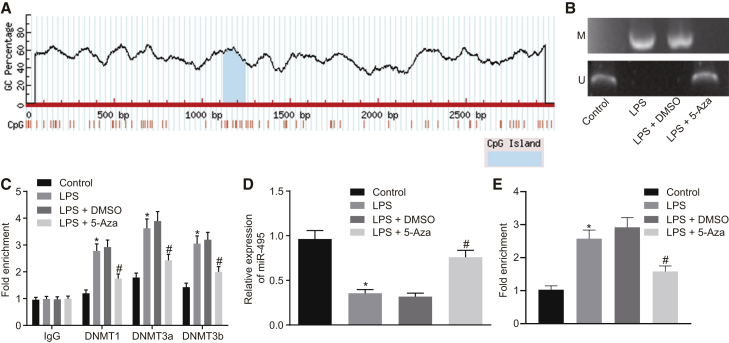
It is well known that methylation of miR-495 promoter downregulates the expression of miR-495.15 Thus, in order to further verify the role of miR-495 promoter methylation in the expression of miR-495 in LPS-induced ALI, we predicted the cytosine-phosphate-guanine (CpG) island in miR-495 promoter region using the MethPrimer website (https://www.urogene.org) and accordingly designed primers to perform a methylation-specific PCR (MSP) assay. The results revealed that the miR-495 promoter region site was methylated in NR8383 cells administered LPS compared to the NR8383 cells administered F-12K culture medium (Figure 3A); compared with LPS-exposed NR8383 cells treated with DMSO, the miR-495 promoter region site was found to be unmethylated in LPS-exposed NR8383 cells treated with 5-Aza (Figure 3B). Additionally, a chromatin immunoprecipitation (ChIP) assay was performed in order to test the enrichment of DNA methyltransferase 1 (DNMT1) and DNA methyltransferase 3a/b (DNMT3a/b) in the miR-495 promoter region, which revealed that the enrichment of DNMT1 and DNMT3a/b in the miR-495 promoter region was increased in the NR8383 cells exposed to LPS when compared to the NR8383 cells incubated with F-12K culture medium (p < 0.05) but was decreased in LPS-exposed NR8383 cells treated with 5-Aza relative to the LPS-exposed NR8383 cells treated with DMSO (p < 0.05) (Figure 3C). As suggested by qRT-PCR, the expression of miR-495 was noted to be reduced in LPS-treated NR8383 cells when compared with that in F-12K culture-medium-treated NR8383 cells (p < 0.05); when compared to the delivery of DMSO, the delivery of 5-Aza was found to elevate the expression of miR-495 in LPS-exposed NR8383 cells (p < 0.05) (Figure 3D). As detected by methylated DNA immunoprecipitation (MeDIP) assay, miR-495 promoter methylation levels in NR8383 cells manipulated with LPS were elevated compared to NR8383 cells manipulated with F-12K culture medium (p < 0.05); in comparison with DMSO treatment, treatment of 5-Aza resulted in reduced methylation levels of the miR-495 promoter in LPS-exposed NR8383 cells (p < 0.05) (Figure 3E). Taken together, the aforementioned results demonstrated that LPS induction increases methylation of miR-495 promoter and leads to the downregulation of miR-495 in LPS-induced ALI.
Figure 3.
Expression of miR-495 Is Reduced in LPS-Exposed NR8383 Cells due to the Methylation of miR-495 Promoter
(A) CpG island in the miR-495 gene promoter region predicted using the MethPrimer website. (B) Methylation of the miR-495 promoter in NR8383 cells treated with F-12K culture medium, LPS, combined LPS and DMSO, or combined LPS and 5-Aza as measured by MSP assay. (C) Enrichment of DNMT1 and DNMT3a/b in the miR-495 promoter region in NR8383 cells treated with F-12K culture medium, LPS, combined LPS and DMSO, or combined LPS and 5-Aza as measured by ChIP assay. (D) Expression of miR-495 in NR8383 cells treated with F-12K culture medium, LPS, combined LPS and DMSO, or combined LPS and 5-Aza as determined by qRT-PCR. (E) Methylation of miR-495 promoter in NR8383 cells treated with F-12K culture medium, LPS, combined LPS and DMSO, or combined LPS and 5-Aza detected by MeDIP assay. *p < 0.05 versus NR8383 cells treated with F-12K culture medium; #p < 0.05 versus NR8383 cells treated with LPS and DMSO. Measurement data were expressed as mean ± SD, and data among multiple groups were compared using one-way ANOVA and subjected to Tukey’s post hoc test. The experiment was repeated three times to obtain the mean value.
Restoring miR-495 Inhibits NLRP3 and Blocks LPS-Induced Inflammasome Activation
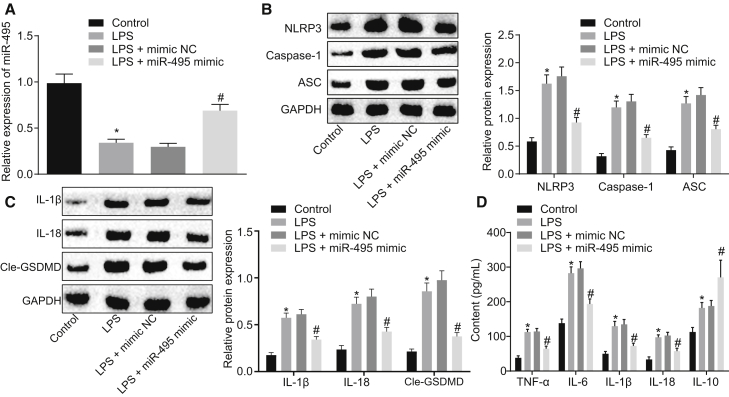
In the following experiments, we explored the effects of miR-495 on NLRP3 inflammasome activation by altering the expression of miR-495 with miR-495 mimic in NR8383 cells in LPS-induced injury models. qRT-PCR was performed to quantify miR-495 in NR8383 cells, and we found that LPS treatment downregulated the expression of miR-495 as the F-12K culture medium treatment for control (p < 0.05). In addition, compared with transfection with mimic NC, miR-495 mimic transfection was found to lead to restored expression of miR-495 in LPS-exposed NR8383 cells (p < 0.05) (Figure 4A). Next, the protein expression levels of NLRP3, ASC, and caspase-1 in NR8383 cells were measured using western blot analysis, whose findings revealed that LPS-exposed NR8383 cells presented with upregulated expression levels of NLRP3, ASC, and caspase-1 compared to NR8383 cells receiving F-12K culture medium (p < 0.05), which was opposite to the trends observed in the LPS-exposed NR8383 cells after transfection with miR-495 mimic (p < 0.05) (Figure 4B). Moreover, it was found that LPS treatment could elevate the expression levels of IL-1β, IL-18, and Cle-GSDMD when compared with the F-12K culture medium treatment (p < 0.05), whereas transfection with miR-495 mimic resulted in a decline in the expression levels of IL-1β, IL-18, and Cle-GSDMD in the LPS-exposed NR8383 cells (p < 0.05) (Figure 4C). As detected by ELISA, the levels of proinflammatory factors (TNF-α, IL-6, IL-1β, and IL-18) and anti-inflammatory factor IL-10 were noted to be elevated in response to LPS delivery (p < 0.05). However, the levels of pro-inflammatory factors were downregulated, and those of the anti-inflammatory factor were upregulated in LPS-exposed NR8383 cells following transfection with miR-495 mimic (p < 0.05) (Figure 4D). From these results, it could be inferred that upregulation of miR-495 could suppress the NLRP3 inflammasome activation in ALI.
Figure 4.
Activation of NLRP3 Inflammasome Triggered LPS Is Attenuated by Elevation of miR-495
(A) Expression of miR-495 in NR8383 cells treated with LPS, F-12K culture medium, combined LPS and mimic NC, or combined LPS and miR-495 mimic as detected by qRT-PCR. (B) Expression levels of NLRP3, ASC, and caspase-1 in NR8383 cells treated with LPS, F-12K culture medium, combined LPS and mimic NC, or combined LPS and miR-495 mimic as detected by western blot analysis. (C) Expression levels of IL-1β, IL-18, and Cle-GSDMD in NR8383 cells treated with LPS, F-12K culture medium, combined LPS and mimic NC, or combined LPS and miR-495 mimic as detected by western blot analysis. (D) Expression levels of proinflammatory factors (TNF-α, IL-6, IL-1β, and IL-18) and anti-inflammatory factor IL-10 in NR8383 cells treated with LPS, F-12K culture medium, combined LPS and mimic NC, or combined LPS and miR-495 mimic as detected by ELISA. *p < 0.05 versus NR8383 cells treated with F-12K culture medium; #p < 0.05 versus NR8383 cells treated with both LPS and mimic NC. Measurement data were expressed as mean ± SD, and data among multiple groups were compared using one-way ANOVA and subjected to Tukey’s post hoc test. The experiment was repeated three times to obtain the mean value.
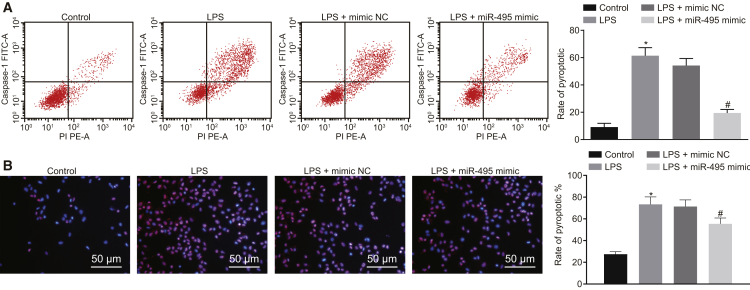
Restoring miR-495 Reduces LPS-Induced Pyroptosis of Alveolar Macrophages
After demonstration of the effect of miR-495 on the activation of the NLRP3 inflammasome in ALI, the focus of the experiment shifted to elucidating the role of miR-495 in the pyroptosis of alveolar macrophages. As detected by flow cytometry, the pyroptosis of NR8383 cells was noted to be promoted in response to LPS treatment compared with F-12K culture medium treatment; transfection with miR-495 mimic attenuated the pyroptosis of LPS-exposed NR8383 cells (p < 0.05) (Figure 5A). Pyroptosis was further measured using the propidium iodide (PI)/Hoechst 33342 double-staining method, which revealed that pyroptosis of NR8383 cells was enhanced following LPS delivery versus the delivery of F-12K culture medium; LPS-exposed NR8383 cells exhibited attenuated pyroptosis in response to transfection with miR-495 mimic (p < 0.05) (Figure 5B). To summarize, the aforementioned findings suggested that miR-495 elevation could inhibit pyroptosis of alveolar macrophages.
Figure 5.
Pyroptosis of Alveolar Macrophages Triggered by LPS Is Attenuated with the Elevation of miR-495
(A) Pyroptosis of LPS-exposed NR8383 cells determined by flow cytometry. (B) Pyroptosis of LPS-exposed NR8383 cells determined by PI/Hoechst 33342 double staining (200×). *p < 0.05, versus NR8383 cells treated with F-12K culture medium; #p < 0.05, versus LPS-exposed NR8383 cells transfected with mimic NC. Measurement data were expressed as mean ± SD, and data among multiple groups were compared using one-way ANOVA and subjected to Tukey’s post hoc test. The experiment was repeated three times to obtain the mean value.
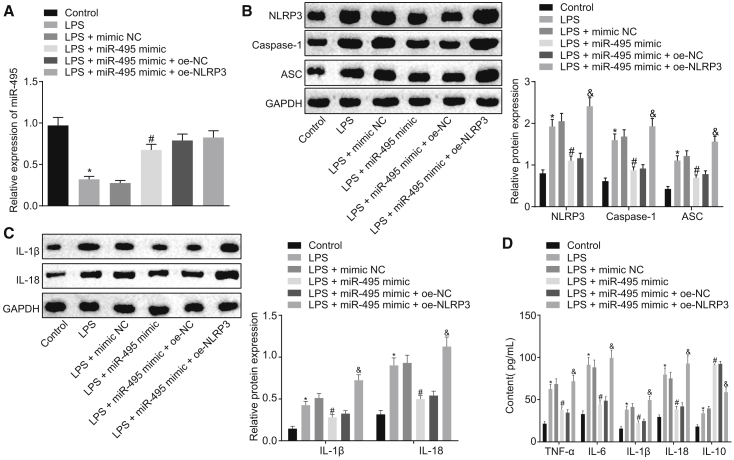
Restoring miR-495 Downregulates NLRP3 to Suppress LPS-Induced Inflammation and Pyroptosis In Vitro
Ad, we performed a series of assays to determine how miR-495 influences the LPS-induced injury in vitro by regulating the expression of NLRP3. LPS-exposed NR8383 cells were transfected with miR-495 mimic or co-transfected with both miR-495 mimic and oe-NLRP3 (NLRP3 overexpression plasmid) or their respective controls (mimic NC or oe-NC). The expression of miR-495 in NR8383 cells was measured using qRT-PCR, which revealed that NR8383 cells treated with LPS presented with reduced expression of miR-495 compared with NR8383 cells treated with F-12K culture medium (p < 0.05). Compared with LPS-exposed NR8383 cells transfected with mimic NC, LPS-exposed NR8383 cells transfected with miR-495 mimic exhibited increased expression of miR-495 (p < 0.05). However, compared with LPS-exposed NR8383 cells co-transfected with miR-495 mimic and oe-NC, there were no significant differences in the expression of miR-495 in LPS-exposed NR8383 cells co-transfected with miR-495 mimic and oe-NLRP3 (p > 0.05) (Figure 6A). Moreover, the expression levels of NLRP3, IL-1β, IL-18, ASC, and caspase-1 were measured using western blot analysis, and it was found that LPS-treated NR8383 cells presented with increased expression levels of these factors relative to the NR8383 cells treated with F-12K culture medium (p < 0.05). Transfection of miR-495 mimic was noted to downregulate the expression levels of NLRP3, ASC, caspase-1, IL-1β, and IL-18 in LPS-exposed NR8383 cells (p < 0.05), which were rescued by additional transfection with oe-NLRP3 (p < 0.05) (Figures 6B and 6C). As determined by ELISA, the levels of proinflammatory factors (TNF-α, IL-6, IL-1β, and IL-18) and anti-inflammatory factor IL-10 were found to be elevated in NR8383 cells administered treatment with LPS versus in NR8383 cells administered treatment with F-12K culture medium (p < 0.05). Relative to mimic NC transfection, miR-495 mimic transfection reduced the levels of pro-inflammatory factors and elevated those of the anti-inflammatory factor in LPS-exposed NR8383 cells (p < 0.05); however, transfection of oe-NLRP3 reversed the regulatory effects of miR-495 mimic on the aforementioned inflammatory factors (p < 0.05) (Figure 6D). Based on the aforementioned results, it could be concluded that elevated miR-495 attenuates LPS-induced inflammation and pyroptosis by negatively regulating NLRP3.
Figure 6.
Elevated Expression of miR-495 Depletes the Expression of NLRP3, Thus Hindering the LPS-Induced Inflammation In Vitro
(A) Expression of miR-495 in LPS-exposed NR8383 cells in response to transfection with plasmids including mimic NC, miR-495 mimic, oe-NC, and oe-NLRP3, as determined by qRT-PCR. (B) Expression levels of NLRP3, ASC, and caspase-1 in NR8383 cells in response to transfection with plasmids including mimic NC, miR-495 mimic, oe-NC, and oe-NLRP3, as determined by western blot analysis. (C) Expression levels of L-1β and IL-18 in NR8383 cells in response to transfection with plasmids including mimic NC, miR-495 mimic, oe-NC, and oe-NLRP3, as determined by western blot analysis. (D) Expression levels of proinflammatory factors (TNF-α, IL-6, IL-1β, and IL-18) and anti-inflammatory factor IL-10 in NR8383 cells in response to transfection with plasmids including mimic NC, miR-495 mimic, oe-NC, and oe-NLRP3, as determined by ELISA. *p < 0.05 versus NR8383 cells treated with F-12K culture medium; #p < 0.05, versus LPS-exposed NR8383 cells transfected with mimic NC; &p < 0.05 versus LPS-exposed NR8383 cells co-transfected with miR-495 mimic and oe-NC. Measurement data were expressed as mean ± SD, and data among multiple groups were compared using one-way ANOVA and subjected to Tukey’s post hoc test. The experiment was repeated three times to obtain the mean value.
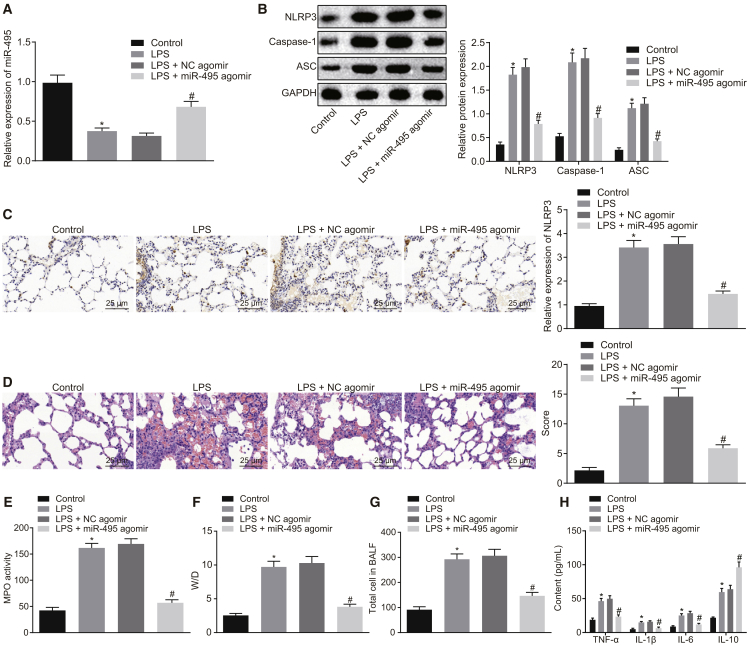
Restoring miR-495 Downregulates NLRP3 to Suppress the Development of ALI In Vivo
After uncovering the role of miR-495 in LPS-induced injury in vitro, we focused on investigating the function of miR-495 in the development of LPS-induced ALI in vivo. As measured by qRT-PCR, the expression of miR-495 was found to be reduced in LPS-administered rats compared with those administered normal saline (p < 0.05). The delivery of miR-495 agomir led to increased expression of miR-495 in LPS-treated rats relative to the delivery of NC agomir (p < 0.05) (Figure 7A). Moreover, the expression of NLRP3 in rat lung tissues was evaluated by western blot analysis and immunohistochemistry. The results demonstrated that the expression levels of NLRP3, ASC, and caspase-1 were higher in lung tissues of normal saline-treated rats than in lung tissues of LPS-treated rats (p < 0.05). The administration of miR-495 agomir led to reduced expression levels of NLRP3, ASC, and caspase-1 in LPS-treated rats (p < 0.05) (Figures 7B and 7C). In addition, pathological changes in the lung tissues were observed and detected using H&E staining. The results revealed that, in normal saline-treated rats, the lung tissues presented with complete structure, and a few macrophages could be observed within the alveolar space under the microscope. In comparison with the rats treated with normal saline, LPS-treated rats exhibited largely destroyed alveolar structure, accompanied with bleeding, massive neutrophil infiltration, thickened alveolar septa, and increased lung injury scores (p < 0.05). In contrast to the rats receiving both LPS and NC agomir, the injury of lung tissues in rats receiving both LPS and miR-495 agomir was greatly reduced, and lung injury scores were decreased (p < 0.05) (Figure 7D). Further, myeloperoxidase (MPO) activity was measured to assess neutrophil infiltration in lung tissues, which revealed that the MPO activity was increased in LPS-administered rats compared to the MPO activity normal saline-administered rats (p < 0.05), which was reduced by the delivery of miR-495 agomir in LPS-administered rats (p < 0.05) (Figure 7E). Furthermore, permeability of the lung tissues was assessed by testing the lung wet weight (W)/dry weight (D) ratio and protein content in bronchoalveolar fluid (BALF). The finding indicated that the lung W/D ratio and protein content in BALF were elevated following LPS treatment versus normal saline treatment (p < 0.05), both of which were reduced by the delivery of miR-495 agomir in LPS-administered rats (p < 0.05) (Figures 7F and 7G). As determined by ELISA, the levels of proinflammatory factors (TNF-α, IL-6, and IL-1β) and anti-inflammatory factor IL-10 were upregulated in response to LPS treatment versus normal saline treatment (p < 0.05); the expression levels of proinflammatory factors were downregulated, while that of the anti-inflammatory factor was upregulated following combined LPS and miR-495 agomir when in contrast to combined LPS and NC agomir (p < 0.05) (Figure 7H). All in all, the aforementioned results suggested that LPS could successfully induce ALI in rats and that increased miR-495 could hinder the development of ALI in vivo by negatively modulating the expression of NLRP3.
Figure 7.
Elevated Expression of miR-495 Depletes the Expression of NLRP3, Thus Protecting against ALI In Vivo
Rats were treated with normal saline or LPS, and LPS-treated rats were further introduced with NC agomir or miR-495 agomir. (A) Expression of miR-495 in lung tissues of rats as determined by qRT-PCR. (B) Expression levels of NLRP3, ASC, and caspase-1 in lung tissues of rats measured using western blot analysis. (C) Immunohistochemistry detection of NLRP3 contents in rat lung tissues (400×). (D) Pathological changes of lung tissues in rats observed using H&E staining (400×). (E) MPO activity in lung tissues of rats. (F) Detection of lung W/D ratio in rats. (G) Total protein content in BALF in rats. (H) Levels of proinflammatory factors (TNF-α, IL-6, and IL-1β) and anti-inflammatory factor IL-10 in the BALF of rats detected by ELISA. *p < 0.05 versus rats treated with normal saline; #p < 0.05 versus rats treated with LPS and NC agomir. Measurement data were expressed as mean ± SD, and data among multiple groups were compared using one-way ANOVA and subjected to Tukey’s post hoc test. The experiment was repeated three times to obtain the mean value.
Discussion
ALI is a severe lung disease that is usually accompanied by acute inflammatory responses, resulting in failure of the respiratory system.16 Despite tremendous efforts and research, ALI still presents with tremendous fatality and morbidity rates.17 Therefore, further studies focusing on novel and effective targets of ALI are needed to provide more insight into the management and treatment of ALI. The present study aimed to investigate the potential role of miR-495 in LPS-induced ALI. Our findings evidenced that overexpression of miR-495 could repress NLRP3 inflammasome activation to ultimately inhibit the development of LPS-induced ALI.
The present study revealed that the NLRP3 inflammasome was activated in LPS-induced ALI, while the expression of miR-495 was downregulated as a result of methylation of the miR-495 gene promoter. Numerous findings have indicated the involvement of the NLRP3 inflammasome in the inflammatory responses of ALI and suggested that its inhibition could result in the alleviation of ALI.18,19 In addition, Zhang et al.20 reported the activation of the NLRP3 inflammasome in LPS-induced ALI, accompanied by elevated expression levels of IL-1β and caspase-1, which is in accordance with our findings. Moreover, as illustrated by both Tian et al.21 and Liu et al.,22 NLRP3 inflammasome expression was downregulated in ALI in response to LPS treatment, and the inactivation of the NLRP3 inflammasome could contribute to the alleviation of LPS-induced ALI. IL-10, a member of the IL-10 cytokine family with numerous immunosuppressive effects, could be generated by diverse leukocyte subpopulations and is regulated by many signaling pathway and transcriptional networks.23 The IL-10 family is known to alleviate inflammatory response, improve innate immunity, and potentiate repairing mechanisms of tissues, thus maintaining tissue homeostasis in response to inflammation and infection.24 Similar to the present study, Ernst et al.25 and Barrett et al.26 noted upregulated levels of IL-10 expression following LPS stimulation. Furthermore, NLRP3 depletion was previously revealed to result in IL-10 elevation in preeclampsia,27 which was in line with our results. Zhu et al.,28 found that LPS treatment led to a downregulated level of IL-10 in rats with LPS-induced ALI, which was opposite to our finding, the increase of IL-10 was both determined in the cell and rat models after LPS stimulation. In addition, DNA methylation, an essential component of epigenetic modification, plays a crucial role in mediating multiple cellular processes by modulating gene expression,29 and hypermethylation of the miRNA gene promoter is constantly associated with the downregulation of miRNA.30,31 Matt et al.32 found that reduction of DNA methylation by zebularine could alleviate sickness and inflammation induced by LPS stimulation in the brain of mice. Furthermore, a previous study correlated abnormal methylation of genes in lungs with pathological and physiological processes of LPS-induced ALI.33 In line with our findings, Li et al.15,34 demonstrated that miR-495 promoter methylation could lead to the depletion of miR-495 in both gastric and breast cancers. In addition, downregulated expression of miR-495 was found in non-small-cell lung cancer cells and served as a tumor suppressor of this cancer.35
As expected, we uncovered that elevated miR-495 negatively regulates the NLRP3 gene to suppress NLRP3 inflammasome activation and alleviate alveolar macrophage inflammation in LPS-induced ALI. Consistently, accumulating evidence has indicated that NLRP3 is the target of miR-223, which inhibits activation of the NLRP3 inflammasome.36,37 Likewise, Zhou et al.38 demonstrated that miR-495 could negatively regulate expression of the NLRP3 inflammasome signaling pathway to attenuate injury and inflammation in cardiac microvascular endothelial cells. Additionally, elevation of miR-495 also contributed to decreased inflammation caused by high glucose content in cardiac fibrosis.39 Furthermore, another study demonstrated that the levels of proinflammatory factors (TNF-α, IL-6, and IL-1β) in alveolar macrophages were downregulated in the presence of miR-146a, suggesting that elevation of miR-146a alleviates the inflammatory response in LPS-induced ALI and could function as a promising target to prevent LPS-induced ALI.40 Moreover, the activation of NLRP3 inflammasome in alveolar macrophages was previously found to cause lung inflammation and injury induced by mechanical stretch, thus proposing NLRP3 as an underlying biomarker for treating ventilator-induced lung injury,41 which was identical to our results. Additionally, morin administration inhibited NLRP3 inflammasome activation, further protecting against LPS-induced ALI, accompanied by a reduced number of inflammatory cells in the BALF, downregulated expression of NLRP3 inflammasome, and attenuated MPO activity.42
In summary, the present study uncovered that methylation of the miR-495 promoter could downregulate the expression of miR-495 and that overexpression of miR-495 could inhibit the development of ALI by negatively regulating the NLRP3 gene, suggesting that upregulated miR-495 and downregulated NLRP3 could serve as promising therapeutic targets for the management of ALI. Moreover, the present study provides insights and a deeper understanding of the underlying mechanisms of ALI. However, further efforts are needed to confirm the efficacy of these biomarkers and targets in early detection, prediction, and prevention of lung injuries and diseases. Also, more research is warranted to elucidate the underlying mechanisms by which LPS activates DNMTs interacting with the miR-495 promoter in future studies.
Materials and Methods
Ethics Statement
All experimental protocols for the present study were approved by the Experimental Animal Ethics Committee of Shanghai 9th People’s Hospital, Shanghai Jiao Tong University School of Medicine. All animal experimentation strictly adhered to the principles to minimize the number, pain, suffering, and discomfort of the experimental animals.
Cell Culture and Treatment
Rat alveolar macrophages (NR8383 cells) purchased from the American Type Culture Collection (Manassas, VA, USA) were cultured in Ham’s F-12K complete medium (Thermo Fisher Scientific, Waltham, MA, USA) containing 1.5 g/L sodium bicarbonate, 15% fetal bovine serum, and 2 nM L-glutamine at 37°C with 5% CO2 in air. With the cell concentration adjusted to 4 × 105 cells per milliliter, the cells were added to a 96-well plate with 100 μL cells per well. Cells were continually cultured for 18 h with the addition of F-12K culture medium or LPS (final concentration, 1 μg/mL), and LPS-induced cell-injury models were established. Plasmids including mimic NC, miR-495 mimic, sh-NC, sh-NLRP3, oe-NC, and oe-NLRP3 were purchased from GenePharma (Shanghai, China) and were delivered into the LPS-administered cells according to the instructions for Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).43 Subsequently, the cells were treated with DMSO or 5-Aza (Sigma-Aldrich Chemical, St Louis, MO, USA).
Establishment of Rat Model of LPS-Induced ALI
Specific pathogen-free Sprague-Dawley rats (aged 8 weeks, weighing 200–250 g) were purchased from Hunan SJA Laboratory Animal (Changsha, Hunan, China). The rats were reared individually in a sterile environment with constant temperature (22°C), 50% relative humidity, and 12-h:12-h light/dark cycle. LPS (5 mg/kg) was instilled into the tracheas of the anesthetized rats to induce ALI.44 Subsequently, the rats were treated with normal saline or 5 mg/kg LPS. Rats administered with 5 mg/kg LPS were further manipulated with NC agomir or miR-495 agomir, which were both synthesized by Guangzhou RiboBio (Guangzhou, Guangdong, China). After 12 h of LPS treatment, all rats were euthanized, and the specimens were collected for further experimentation.
Western Blot Analysis
Cells were detached with trypsin and lysed using intensified radioimmunoprecipitation assay (RIPA) lysis buffer (Boster Biological Technology, Wuhan, Hubei, China) conjugated with a protease inhibitor. Subsequently, the protein concentration was measured using bicinchoninic acid (BCA) protein assay kits (Boster Biological Technology, Wuhan, Hubei, China). After protein separation using 10% SDS-PAGE, the proteins were transferred onto a polyvinylidene fluoride membrane, which was sealed with 5% BSA for 2 h. The membrane was then incubated overnight at 4°C with the addition of the following diluted primary antibodies: rabbit antibody to NLRP3 (ab214185, dilution ratio of 1:1,000), rabbit antibody to caspase-1 (ab62698, dilution ratio of 1:1,000), rabbit antibody to ASC (ab18193, dilution ratio of 1:1,000), IL-1β (ab200478, dilution ratio of 1:500), rabbit antibody to IL-18 (ab71495, dilution ratio of 1:300), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; ab181602, dilution ratio of 1:5,000). All aforementioned antibodies were purchased from Abcam (Cambridge, UK). Subsequently, the membrane was rinsed with PBS/Tween (PBST) three times and further incubated with horseradish peroxidase (HRP)-labeled secondary antibody of goat anti-rabbit (ab205719; dilution ratio of 1:2,000; Abcam, Cambridge, UK) at room temperature for 1 h, followed by development using enhanced chemiluminescence (ECL; Millipore, Bedford, MA, USA). The ImageJ software was used for quantification of the protein bands, with GAPDH serving as the internal control.
ELISA
Cell-culture medium in each well was centrifuged to collect the cell supernatant. According to the instructions of the ELISA kits, the levels of TNF-α (LC2062), IL-6 (JLC1236), IL-18 (JLC1702), IL-1β (JLC1704), IL-10 (JLC1689), and caspase-1 (JLC1702)—which were all provided by Shanghai Jingkang Bioengineering (Shanghai, China)—in the cell supernatant and alveolus lavage fluid were measured.
Dual-Luciferase Reporter Gene Assay
The interaction between miR-495 and NLRP3 was analyzed using the bioinformatic website http://www.microrna.org/microrna/home.do. Based on the findings of the predicted binding site, the synthesized NLRP3 mRNA 3′ UTR sequence and its mutant sequence were inserted into the pmirGLO vector (E1330; Promega, Madison, WI, USA) to generate NLRP3-WT and NLRP3-Mut reporter plasmids. The sequences are shown in Table 1. Next, NLRP3-WT and NLRP3-Mut reporter plasmids with correct sequences were co-transfected with miR-495 mimic or mimic NC into HEK-239T cells. After 48 h of culture, the luciferase activity in cells was detected based on the manufacturer’s protocols for dual-luciferase reporter gene assay kits (D0010, Beijing Solarbio Science and Technology, Beijing, China) provided by Genecopoeia (Rockville, MD, USA). Luminance was measured using the Glomax20/20 luminometer (E5311, Shaanxi Zhongmei Biotechnology, Xian, Shaanxi, China) provided by Promega (Madison, WI, USA).
Table 1.
The Sequences of NLRP3-WT, NLRP3-Mut, and miR-495 Promoter for Dual-Luciferase Reporter Assay
| Gene names | Sequence |
|---|---|
| NLRP3-WT | GCGTGGAAGCAGGACCACCAGGTGCCTCGGTCCTGCCCCAAGTCCTGCCCCAAGCCCCAGTGCGCACTGCTCTTCACTGCTATCAAGCCCTCCTTCACCATCAGGATCACAGCCGAGGCTCTTCTGGTATAGGGTCTGGAGCAAAGGCTTGTGTGGGACCAAATATTTTTCCTCACATCGATAACGTGAAACTGCCAGAGGCTGCCCTTCCCATCATATCCTCAGTGGGCAAGGTGTTCCCTCTTGGTGACTTCATGGAAACAGCTTCAAGAAAACGCCTTTTCTGTCCTCCCCCGCCCTCCTCTTACTCCTGCCCCTCCTCCTCCTCCTCCTCCCCTCCCCCCCCCTCCTCCTCCGCTTCTCCCCCCACCTGTCTTTCTCTCTCTGGGCCTCTGGTTTTTTGACCTTTGCCCATACCTTCAGTCTTGTCTTCCTGTTAACTGACCATCCCGCATAAGGAGCTGCCCGTGGGCTAGATGGAAGGTTTGTGGCAGCCTCTCAGCTACATTGTTTGTTTTTATTTTTTAATAGTTATGATTTCTCTTTAGCTACCTGAAAACTCAGAGATTTATAAAACCCATTTTTGTATTTATTGTATGTTTGTACTGCTTTCTTAATTTAAAAATGTATCTAGAATTCTTTTAAGTTATTTATCCAAACTACTAAAAATAAATCAGTTTACACATTTAAAA |
| NLRP3-Mut | GCGTGGAAGCAGGACCACCAGGTGCCTCGGTCCTGCCCCAAGTCCTGCCCCAAGCCCCAGTGCGCACTGCTCTTCACTGCTATCAAGCCCTCCTTCACCATCAGGATCACAGCCGAGGCTCTTCTGGTATAGGGTCTGGAGCAAAGGCTTGTGTGGGACCAAATATTTTTCCTCACATCGATAACGTGAAACTGCCAGAGGCTGCCCTTCCCATCATATCCTCAGTGGGCAAGGTGTTCCCTCTTGGTGACTTCATGGAAACAGCTTCAAGAAAACGCCTTTTCTGTCCTCCCCCGCCCTCCTCTTACTCCTGCCCCTCCTCCTCCTCCTCCTCCCCTCCCCCCCCCTCCTCCTCCGCTTCTCCCCCCACCTGTCTTTCTCTCTCTGGGCCTCTGGTTTTTTGACCTTTGCCCATACCTTCAGTCTTGTCTTCCTGTTAACTGACCATCCCGCATAAGGAGCTGCCCGTGGGCTAGATGGAAGGTTTGTGGCAGCCTCTCAGCGCACGGCGGGCGGGTTATTTTTTAATAGTTATGATTTCTCTTTAGCTACCTGAAAACTCAGAGATTTATAAAACCCATTTTTGTATTTATTGTATGTTTGTACTGCTTTCTTAATTTAAAAATGTATCTAGAATTCTTTTAAGTTATTTATCCAAACTACTAAAAATAAATCAGTTTACACATTTAAAA |
| miR-495 Promoter | ACTCAGAATCCAGCCGCTTCGTGGGCAGCGTTCAGTCCATGCTTGGGGAAAGGGCTTTGT |
| TGCCGTTGCTCATCTCTGTCTGTTGTTCAGCTCTGCTTGCCTTACGAGGCCCCAGATGCC | |
| CTGGATGCCCTCAGTGTCAGACTGCGGAAGATGTGTCCCGGCTGTGACACGCGCTTCAGG | |
| GGAGGGCTGTTGGCGTCAGGTCCATGGAGAGGTATGGGAGTGGACATCTTGTCTGAGAGC | |
| AGACATTCTATTTCGGGTGTGTGTCTTGGTCCGTTCATCCTCAGTCTCCCTCCCATACCT | |
| AGGATGGCTTGTGGAGAGAAGGCCTGCCCTGAGGGCTGAAGTTGGGACCCACAGCTCTCA | |
| CCCCGGCCTTGGTATGCTCCCATGGCTACGGCAGAGGCACAAGAAAGTAATTAGAAACGC | |
| AGAAACTCCTTAAGGCCAAGGCTGGGAACTGCTATATTTCTACCCACAGATCATTGACCA | |
| GAACAATTACACGGCCGAACCCAAGGCAAGGGGCGGGGAGGCCCACTCCACCCGGGAGGG | |
| GGTTGTGGTGAGGTGTGAATGCAGGAACCGGTGAGGATTTGCCACCACCAAGTCAGTTGT | |
| CCACGTGGACCACCTGAGAACTTGGGTTTTATTCATTGCTCATTCATCCCTCCCTAACCC | |
| GCAGCTTCCGGTCTGCCCCGGGGGCTCTTCTCCGGCTCAGGCTGATCGACAGTGGTGACG | |
| CTGTGCCAGGACCCCTGGTCATTCCATCTGTTTGAGGCAAATGTTAAAAAATCCCTTGCG | |
| CTACCCATTCTATGACGACTCCCTTGGCTCAGAGGCTCCATCAGGGCAGACACTTTGCTG | |
| TTTGTCCCTAAAGGGTGGGTGGTAGATCTCATCCATCTTATCTGGGGAGTTATTGTGACT | |
| CTCCTGGTCCCTCCTCATGTATCCTCAGGAGGACTTTTAGGATGCAAAATCACTGTTTCT | |
| GTGCCATTGTCATATTCTGTGCTACTGGAGGAGACTTTTTGTTATTGAGAAATTGAACAC | |
| ATCGTGCTCACAAGCAGCTCCAGGACACAATGCCACCTGTGCAGAGGGGGCAGACCTGTT | |
| AGATGTCTCCCAGGTCTTGAGGGGGTTGTATGCACGGCCATATCTTTGTCACCGGTTGCC | |
| CGGTGCGCATCAGGACCCATGTGCTCTCAGGAAGCCCGCAGGAGTGATACCAGCCGCGGA | |
| GAAGCAGTGTTGTCAACGGTGCTGACGTGCGGTACTTAATGAGAAGTTGCCCGTGTTTTT | |
| TTCGCTTTATTTGTGACGAAACATTCGCGGTGCACTTCTTTTTCAGTATCCTATTCTGCC | |
| TTGAAGACGTCTTGGTTTGGGTGCAACTTCAGGGAAGGCACATGGGCCCCTGTAGTGGAC | |
| GCTGATCAGGACCTCCCTACCCCATGTCTTTTCTCCTTCCTAAATTTCTCCTTGTTTTTT | |
| TTTTTTTTTTTTTTTTTTATGAAAGCTCCCGTCTTCCCGCCGTGTGCTTTGATTGCATTG | |
| ACCCTGCCCCACTTCCTCCACCAGCCCAGGCTTAAGCGAATCTCTTGATCTCTTCTCCTT | |
| TGCCACTGCAATTGCTTCCGGAATAGGCCCATGACCCCACTGGGCCAGTGATAGGCAAGG | |
| AGATGCTTGCTGCAGCTTCTGGGAAAGAAACTGTTTCTCCCTTTCCTGTGATAGCTGCCA | |
| GAGTAGACACTGTCTATTCCATTCAGCTGGGAATAGACAGTGAACTTTGACGTGAGCTTC | |
| GAAGGTCCCACAGCCGTTTAAGGACTGAGAGGAGAGGGCTTGGTGCCACGCAGACACAGC | |
| AGAGGGAGTCTGTACCATGAAAGGCATCTTGGAGACCTAGAGAAGAGCCTGGATGATATC | |
| ATTTGAGTCCCTAGATCAAACCCTGCCTGAAGCTTGCTCTGTCTCTGTACTTTTTAGTTA | |
| TATACGTCTATAAATCCCTTACTTGTTTAAGCCAGTTGGAGTTAGATCTTATCTTACTTG | |
| CAATACAAACAGATGATACCACAGTGTGTTTTCCAGTGTGACTGGGAAAGCCTGCATTTT | |
| TTTCAATGATGTGTTTCTCACGGTTTGCCTATCTATCTTCAGTGTCCTATTTAAGGCAGG | |
| GTTTTGGTGGAAGGTGACTTTTCTTATGGACTTATGCTTTCAAAGCTTTGTCTTAGTGAT | |
| TTAATTTATGGTTGACAAATGATGCACAATTACCGTGTTTTGTGTCCAGCAATGGGTCAC | |
| TGGGTCCTCTCTGGAAGGCCTGATGGATGGTGGGCCCCAGAACTTCTGCCCCGATTCACT | |
| CTCCCTCGAATCCACCTCTCTCTGGGAGGCTGCCCTGCAAGTGATGTGGACTTGGGGAAG | |
| CCGTTGTCCTTTCCTGGCCCCTGCTGTCCTTTGGCTCTCTGTGGGTTGGCTGTATTTGTC | |
| ACAACTCACACTTAACTGTCAGTGTTTTAGAACCAAGCCATGCCTCCAGGGAACCCGGAG | |
| CACTTTAGAGAGGAGGTGCAGTGGTCAGCACCATGCAAGGTGCACAGAGAGATGGGTCCA | |
| CCGAGAGCCGGGGCTGCCTTGGTGGTTTGTTTCTCCTGATTCGATATCTTTGTCTTCCCA | |
| TCTCCTTCCTGGAACCATGGGCTCATGGAGGAGATTCTATTTTTGGGTCCAGGACCATCC | |
| CATTTTCTCTGAAGTGATGCTCTCCATGAGCATTTGCAGGGTTGATAGTAAACATTCCGA | |
| AAGCCGTCTGCCCTGATTATTGAATTCTGCCCATGAAGACTTCCTGGGATCCCGAAGTAG | |
| AGGGAGTCTGGGATCGGTTTGCTTTATCCGTGATGACTGTCCGCCTCTGCTCAGTGTCAG | |
| CCCAGCCCTTCGAAGCATGTCCCCCACCTCCCCTGGGGTGAGCCAACTTACCTGATGCTT | |
| TTAGGCTTAATTGAGCATAAGTTTGATTGGCAGCGTCTGCCCGGCCCTGCCCTCACGCCC | |
| TGACCCTCAGTGTCCCTTCACGCCCAGGTGTGCCTCTGGCAGGGAGGACGTGCTCATCTC |
RNA Isolation and Quantification
Total RNA was extracted using TRIzol Reagent (catalog no. 15596026, Invitrogen, Carlsbad, CA, USA) and reverse transcribed into cDNA using NCode miRNA First-Strand cDNA Synthesis Kits (Thermo Fisher Scientific, Waltham, MA, USA). The synthesized cDNA was detected using Fast SYBR Green PCR Kits (Applied Biosystems, Carlsbad, CA, USA). The primers for qRT-PCR were synthesized by Sangon Biotech (Shanghai, China), and the sequences were as follows: reverse primer: 5′-GAGACTGCGGATGTATAGAACTTGA-3′; forward primer: 5′-AAACAAACATGGTGCACTTCTT-3′.45
MSP Assay
The methylation of the miR-495 promoter region was detected using the EZ DNA Methylation-Gold Kit (D5005, Zymo Research, Irvine, CA, USA). The primers (and sequences) for methylated reaction in MSP amplification were miR-495-MD (5′-TAGAGAGATGGGTTTATCGAGAGTC-3″) and miR-495-MR (5′-AGAGATGGGTTTATTGAGAGTTGG-3″), and the primers and sequences for unmethylated reaction in MSP amplification were miR-495-UD (5′-TTGGTGAAGTTGGGTG-3′) and miR-495-UR (5′-AAATTCAATAATCAAAACAAACAAC-3′). All aforementioned primers were synthesized by Sangon Biotech (Shanghai, China). Purified RNA was added to the CT conversion reagent for denaturation and bisulfate conversion. Next, the purified RNA was subjected to PCR. The PCR products were subjected to agarose gel electrophoresis, and the obtained images were analyzed using gel electrophoresis imaging and an image analysis system.15
MeDIP Assay
MeDIP assay was carried out in accordance with the instructions of the MeDIP kits. Briefly, genomic DNA was extracted from the cells and purified following standard procedures. Next, the obtained DNA was sheared into 200–1,000 bp fragments by ultrasonication. The DNA fragments were then denaturized at 95°C to obtain single-chain DNA fragments. Subsequently, the single-chain DNA fragments were co-incubated with the antibody 5-mC to obtain a DNA-5-mC complex, which was captured by magnetic beads. The DNA pulled down was purified with phenol/chloroform extraction and then subjected to qPCR.
ChIP Assay
Cells were treated with 4% formaldehyde with a final concentration of 1%, and the collected cells were crushed ultrasonically. Antibodies to DNMT3b (ab2851, dilution ratio of 1:50, Abcam, Cambridge, UK), DNMT1 (ab13537, dilution ratio of 1:50, Abcam, Cambridge, UK), and DNMT3a (ab2850, dilution ratio of 1:50, Abcam, Cambridge, UK) were bound to the miR-495 gene promoter, and protein A agarose/salmon sperm DNA was bound to the gene promoter complex, which was precipitated. The precipitated complex was then rinsed to remove the non-specific binding. Subsequently, the enriched miR-495 promoter complex was cross-linked and purified for qRT-PCR.46
Flow Cytometry
Pyroptosis was measured using flow cytometry in strict accordance with the instructions of the FAM-FLICA Caspase Assay Kit (ImmunoChemistry Technologies, Bloomington, MN, USA).47
PI/Hoechst 33342 Double Staining
The cells were detached with 0.125% trypsin, with PI and Hoechst 33342 added, for double staining for 10 min. Next, the cells were observed and photographed under an inverted fluorescence microscope (Carl Zeiss, Oberkochen, Germany). PI could enter the nucleus through the membrane of apoptotic cells to exhibit red fluorescence. The apoptotic rate was calculated as follows: (the number of apoptotic cells ÷ the total number of cells) × 100%.
Detection of MPO Activity
Lung tissue homogenates were mixed with thiobarbituric acid, and the MPO activity in the supernatant was detected using a spectrophotometer by measuring the absorbance changes of lung tissues per gram.48
H&E Staining
The right-side lungs of rats whose alveoli were not lavaged were fixed with formalin and paraffin-sectioned for H&E staining. Subsequently, the pathological changes in the tissue sections were observed under an optical microscope.49
Detection of Pulmonary Edema-Related Parameters
The left-side lungs without alveolar lavage were extracted from the rats, and the bloodstained water on the lung surface was sucked and dried to measure the wet weight (W) of the lungs. After that, the lungs were dried at 80°C for 48 h, followed by measurement of dry weight (D). Next, the W/D ratio and lung water content were calculated.
Immunohistochemistry
Tissue specimens were paraffin-sectioned, deparaffinized into water, dehydrated using gradient alcohol, and washed with distilled water for 2 min. Subsequently, the sections were treated with 3% H2O2 for 20 min, washed with distilled water for 2 min, and rinsed with 0.1 M PBS for 3 min, followed by antigen retrieval in a water bath. Next, the sections were added with normal goat serum blocking solution (C-0005, Shanghai Haoran Bio Technologies, Shanghai, China) at room temperature for 20 min. Following that, the sections were incubated overnight at 4°C with the primary antibody rabbit NLRP3 (ab214185, dilution ratio of 1:500), and incubated with the secondary antibody of goat anti-rabbit immunoglobulin G (IgG; ab6785, dilution ratio of 1:1,000) at 37°C for 20 min. The aforementioned antibodies were purchased from Abcam (Cambridge, UK). The sections were added with HRP-labeled streptavidin protein working solution (0343-10000U, Imunbio Biotechnology, Beijing, China), incubated at 37°C for 20 min, and developed using diaminobenzidine (ST033, Guangzhou Weijia Technology, Guangzhou, China). Subsequently, the sections were counterstained with hematoxylin (PT001, Shanghai Bogoo Bioechnology, Shanghai, China) for 1 min and returned to blue coloration using 1% ammonia. The staining was observed under a microscope.
Statistical Analysis
Statistical analyses were performed using the SPSS v21.0 software (IBM, Armonk, NY, USA). Measurement data were presented as mean ± SD. An unpaired t test was used to analyze the unpaired data between two groups conforming to normal distribution and homogeneity of variance. Comparisons among multiple groups were assessed using one-way ANOVA and subjected to Tukey’s post hoc test. Comparisons among multiple groups at different time points were assessed using repeated-measurement ANOVA and subjected to a Bonferroni post hoc test. The factor relevance was analyzed using Pearson’s correlation analysis. The survival rates of rats were calculated using the Kaplan-Meier method and single-factor analysis was performed using the log-rank test. A value of p < 0.05 was considered to be statistically significant.
Author Contributions
Y.Y. and Y.M. designed the study. Y.M. and M.Y. collated the data, designed and developed the database, carried out data analyses, and produced the initial draft of the manuscript. Y.Y. and M.Y. contributed to drafting the manuscript. All authors have read and approved the final submitted manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The authors thank the reviewers for their helpful comments.
References
- 1.Lemos-Filho L.B., Mikkelsen M.E., Martin G.S., Dabbagh O., Adesanya A., Gentile N., Esper A., Gajic O., Gong M.N., US Critical Illness and Injury Trials Group: Lung Injury Prevention Study Investigators (USCIITG-LIPS) Sex, race, and the development of acute lung injury. Chest. 2013;143:901–909. doi: 10.1378/chest.12-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fang X., Bai C., Wang X. Bioinformatics insights into acute lung injury/acute respiratory distress syndrome. Clin. Transl. Med. 2012;1:9. doi: 10.1186/2001-1326-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dushianthan A., Grocott M.P., Postle A.D., Cusack R. Acute respiratory distress syndrome and acute lung injury. Postgrad. Med. J. 2011;87:612–622. doi: 10.1136/pgmj.2011.118398. [DOI] [PubMed] [Google Scholar]
- 4.Czyzewski A.M., McCaig L.M., Dohm M.T., Broering L.A., Yao L.J., Brown N.J., Didwania M.K., Lin J.S., Lewis J.F., Veldhuizen R., Barron A.E. Effective in vivo treatment of acute lung injury with helical, amphipathic peptoid mimics of pulmonary surfactant proteins. Sci. Rep. 2018;8:6795. doi: 10.1038/s41598-018-25009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butt Y., Kurdowska A., Allen T.C. Acute lung injury: a clinical and molecular review. Arch. Pathol. Lab. Med. 2016;140:345–350. doi: 10.5858/arpa.2015-0519-RA. [DOI] [PubMed] [Google Scholar]
- 6.Hayes M., Curley G., Ansari B., Laffey J.G. Clinical review: Stem cell therapies for acute lung injury/acute respiratory distress syndrome—hope or hype? Crit. Care. 2012;16:205. doi: 10.1186/cc10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Needham D.M., Wozniak A.W., Hough C.L., Morris P.E., Dinglas V.D., Jackson J.C., Mendez-Tellez P.A., Shanholtz C., Ely E.W., Colantuoni E. Risk factors for physical impairment after acute lung injury in a national, multicenter study. Am. J. Respir. Crit. Care Med. 2014;189:1214–1224. doi: 10.1164/rccm.201401-0158OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pattarayan D., Thimmulappa R.K., Ravikumar V., Rajasekaran S. Diagnostic potential of extracellular microrna in respiratory diseases. Clin. Rev. Allergy Immunol. 2018;54:480–492. doi: 10.1007/s12016-016-8589-9. [DOI] [PubMed] [Google Scholar]
- 9.Fang Y., Gao F., Hao J., Liu Z. microRNA-1246 mediates lipopolysaccharide-induced pulmonary endothelial cell apoptosis and acute lung injury by targeting angiotensin-converting enzyme 2. Am. J. Transl. Res. 2017;9:1287–1296. [PMC free article] [PubMed] [Google Scholar]
- 10.Ai C., Jiang R., Fu L., Chen Y. MicroRNA-495 mimics delivery inhibits lung tumor progression. Tumour Biol. 2015;36:729–735. doi: 10.1007/s13277-014-2687-1. [DOI] [PubMed] [Google Scholar]
- 11.Coll R.C., Robertson A.A., Chae J.J., Higgins S.C., Muñoz-Planillo R., Inserra M.C., Vetter I., Dungan L.S., Monks B.G., Stutz A. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat. Med. 2015;21:248–255. doi: 10.1038/nm.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirota S.A., Ng J., Lueng A., Khajah M., Parhar K., Li Y., Lam V., Potentier M.S., Ng K., Bawa M. NLRP3 inflammasome plays a key role in the regulation of intestinal homeostasis. Inflamm. Bowel Dis. 2011;17:1359–1372. doi: 10.1002/ibd.21478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin N., Peng Z., Li B., Xia J., Wang Z., Yuan J., Fang L., Lu X. Isoflurane attenuates lipopolysaccharide-induced acute lung injury by inhibiting ROS-mediated NLRP3 inflammasome activation. Am. J. Transl. Res. 2016;8:2033–2046. [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang L., Zhang L., Kang K., Fei D., Gong R., Cao Y., Pan S., Zhao M., Zhao M. Resveratrol ameliorates LPS-induced acute lung injury via NLRP3 inflammasome modulation. Biomed. Pharmacother. 2016;84:130–138. doi: 10.1016/j.biopha.2016.09.020. [DOI] [PubMed] [Google Scholar]
- 15.Li Z., Zhang G., Li D., Jie Z., Chen H., Xiong J., Liu Y., Cao Y., Jiang M., Le Z., Tan S. Methylation-associated silencing of miR-495 inhibit the migration and invasion of human gastric cancer cells by directly targeting PRL-3. Biochem. Biophys. Res. Commun. 2015;456:344–350. doi: 10.1016/j.bbrc.2014.11.083. [DOI] [PubMed] [Google Scholar]
- 16.Leung W.S., Yang M.L., Lee S.S., Kuo C.W., Ho Y.C., Huang-Liu R., Lin H.W., Kuan Y.H. Protective effect of zerumbone reduces lipopolysaccharide-induced acute lung injury via antioxidative enzymes and Nrf2/HO-1 pathway. Int. Immunopharmacol. 2017;46:194–200. doi: 10.1016/j.intimp.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Cross L.J., Matthay M.A. Biomarkers in acute lung injury: insights into the pathogenesis of acute lung injury. Crit. Care Clin. 2011;27:355–377. doi: 10.1016/j.ccc.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang F., Fan K., Wang K., Bian C. Atractylodin attenuates lipopolysaccharide-induced acute lung injury by inhibiting NLRP3 inflammasome and TLR4 pathways. J. Pharmacol. Sci. 2018;136:203–211. doi: 10.1016/j.jphs.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Wang S., Zhao J., Wang H., Liang Y., Yang N., Huang Y. Blockage of P2X7 attenuates acute lung injury in mice by inhibiting NLRP3 inflammasome. Int. Immunopharmacol. 2015;27:38–45. doi: 10.1016/j.intimp.2015.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Li X., Grailer J.J., Wang N., Wang M., Yao J., Zhong R., Gao G.F., Ward P.A., Tan D.X., Li X. Melatonin alleviates acute lung injury through inhibiting the NLRP3 inflammasome. J. Pineal Res. 2016;60:405–414. doi: 10.1111/jpi.12322. [DOI] [PubMed] [Google Scholar]
- 21.Tian L., Li W., Wang T. Therapeutic effects of silibinin on LPS-induced acute lung injury by inhibiting NLRP3 and NF-κB signaling pathways. Microb. Pathog. 2017;108:104–108. doi: 10.1016/j.micpath.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 22.Liu Q., Ci X., Wen Z., Peng L. Diosmetin alleviates lipopolysaccharide-induced acute lung injury through activating the nrf2 pathway and inhibiting the nlrp3 inflammasome. Biomol. Ther. (Seoul) 2018;26:157–166. doi: 10.4062/biomolther.2016.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X., Wong K., Ouyang W., Rutz S. Targeting il-10 family cytokines for the treatment of human diseases. Cold Spring Harb. Perspect. Biol. 2019;11:a028548. doi: 10.1101/cshperspect.a028548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ouyang W., O’Garra A. Il-10 family cytokines il-10 and il-22: From basic science to clinical translation. Immunity. 2019;50:871–891. doi: 10.1016/j.immuni.2019.03.020. [DOI] [PubMed] [Google Scholar]
- 25.Ernst O., Glucksam-Galnoy Y., Athamna M., Ben-Dror I., Ben-Arosh H., Levy-Rimler G., Fraser I.D.C., Zor T. The camp pathway amplifies early myd88-dependent and type i interferon-independent lps-induced interleukin-10 expression in mouse macrophages. Mediators Inflamm. 2019;2019:3451461. doi: 10.1155/2019/3451461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrett J.P., Henry R.J., Villapol S., Stoica B.A., Kumar A., Burns M.P., Faden A.I., Loane D.J. NOX2 deficiency alters macrophage phenotype through an IL-10/STAT3 dependent mechanism: implications for traumatic brain injury. J. Neuroinflammation. 2017;14:65. doi: 10.1186/s12974-017-0843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Z., Zhao X., Shan H., Gao H., Wang P. microRNA-520c-3p suppresses NLRP3 inflammasome activation and inflammatory cascade in preeclampsia by downregulating NLRP3. Inflamm. Res. 2019;68:643–654. doi: 10.1007/s00011-019-01246-8. [DOI] [PubMed] [Google Scholar]
- 28.Zhu W.D., Xu J., Zhang M., Zhu T.M., Zhang Y.H., Sun K. MicroRNA-21 inhibits lipopolysaccharide-induced acute lung injury by targeting nuclear factor-κB. Exp. Ther. Med. 2018;16:4616–4622. doi: 10.3892/etm.2018.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y., Shi J.X., Pan X.F., Feng J., Zhao H. DNA microarray-based screening of differentially expressed genes related to acute lung injury and functional analysis. Eur. Rev. Med. Pharmacol. Sci. 2013;17:1044–1050. [PubMed] [Google Scholar]
- 30.He X.X., Kuang S.Z., Liao J.Z., Xu C.R., Chang Y., Wu Y.L., Gong J., Tian D.A., Guo A.Y., Lin J.S. The regulation of microRNA expression by DNA methylation in hepatocellular carcinoma. Mol. Biosyst. 2015;11:532–539. doi: 10.1039/c4mb00563e. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki H., Maruyama R., Yamamoto E., Kai M. DNA methylation and microRNA dysregulation in cancer. Mol. Oncol. 2012;6:567–578. doi: 10.1016/j.molonc.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matt S.M., Zimmerman J.D., Lawson M.A., Bustamante A.C., Uddin M., Johnson R.W. Inhibition of DNA methylation with zebularine alters lipopolysaccharide-induced sickness behavior and neuroinflammation in mice. Front. Neurosci. 2018;12:636. doi: 10.3389/fnins.2018.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X.Q., Lv C.J., Liu X.Y., Hao D., Qin J., Tian H.H., Li Y., Wang X.Z. Genome-wide analysis of DNA methylation in rat lungs with lipopolysaccharide-induced acute lung injury. Mol. Med. Rep. 2013;7:1417–1424. doi: 10.3892/mmr.2013.1405. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y., Luo D., Tian W., Li Z., Zhang X. Demethylation of miR-495 inhibits cell proliferation, migration and promotes apoptosis by targeting STAT-3 in breast cancer. Oncol. Rep. 2017;37:3581–3589. doi: 10.3892/or.2017.5621. [DOI] [PubMed] [Google Scholar]
- 35.Chu H., Chen X., Wang H., Du Y., Wang Y., Zang W., Li P., Li J., Chang J., Zhao G., Zhang G. MiR-495 regulates proliferation and migration in NSCLC by targeting MTA3. Tumour Biol. 2014;35:3487–3494. doi: 10.1007/s13277-013-1460-1. [DOI] [PubMed] [Google Scholar]
- 36.Haneklaus M., Gerlic M., Kurowska-Stolarska M., Rainey A.A., Pich D., McInnes I.B., Hammerschmidt W., O’Neill L.A., Masters S.L. Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1β production. J. Immunol. 2012;189:3795–3799. doi: 10.4049/jimmunol.1200312. [DOI] [PubMed] [Google Scholar]
- 37.Bauernfeind F., Rieger A., Schildberg F.A., Knolle P.A., Schmid-Burgk J.L., Hornung V. NLRP3 inflammasome activity is negatively controlled by miR-223. J. Immunol. 2012;189:4175–4181. doi: 10.4049/jimmunol.1201516. [DOI] [PubMed] [Google Scholar]
- 38.Zhou T., Xiang D.K., Li S.N., Yang L.H., Gao L.F., Feng C. Microrna-495 ameliorates cardiac microvascular endothelial cell injury and inflammatory reaction by suppressing the nlrp3 inflammasome signaling pathway. Cell. Physiol. Biochem. 2018;49:798–815. doi: 10.1159/000493042. [DOI] [PubMed] [Google Scholar]
- 39.Wang X., Jin H., Jiang S., Xu Y. MicroRNA-495 inhibits the high glucose-induced inflammation, differentiation and extracellular matrix accumulation of cardiac fibroblasts through downregulation of NOD1. Cell. Mol. Biol. Lett. 2018;23:23. doi: 10.1186/s11658-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeng Z., Gong H., Li Y., Jie K., Ding C., Shao Q., Liu F., Zhan Y., Nie C., Zhu W., Qian K. Upregulation of miR-146a contributes to the suppression of inflammatory responses in LPS-induced acute lung injury. Exp. Lung Res. 2013;39:275–282. doi: 10.3109/01902148.2013.808285. [DOI] [PubMed] [Google Scholar]
- 41.Wu J., Yan Z., Schwartz D.E., Yu J., Malik A.B., Hu G. Activation of NLRP3 inflammasome in alveolar macrophages contributes to mechanical stretch-induced lung inflammation and injury. J. Immunol. 2013;190:3590–3599. doi: 10.4049/jimmunol.1200860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tianzhu Z., Shihai Y., Juan D. The effects of morin on lipopolysaccharide-induced acute lung injury by suppressing the lung NLRP3 inflammasome. Inflammation. 2014;37:1976–1983. doi: 10.1007/s10753-014-9930-1. [DOI] [PubMed] [Google Scholar]
- 43.Zheng H.E., Wang G., Song J., Liu Y., Li Y.M., Du W.P. MicroRNA-495 inhibits the progression of non-small-cell lung cancer by targeting TCF4 and inactivating Wnt/β-catenin pathway. Eur. Rev. Med. Pharmacol. Sci. 2018;22:7750–7759. doi: 10.26355/eurrev_201811_16398. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Q., Nie J., Chen S.J., Li Q. Protective effects of ethyl gallate and pentagalloylglucose, the active components of Qingwen Baidu Decoction, against lipopolysaccharide-induced acute lung injury in rats. Drug Des. Devel. Ther. 2018;13:71–77. doi: 10.2147/DDDT.S186029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie Z., Tang Y., Su X., Cao J., Zhang Y., Li H. PAX3-FOXO1 escapes miR-495 regulation during muscle differentiation. RNA Biol. 2019;16:144–153. doi: 10.1080/15476286.2018.1564464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson J.D., Denisenko O., Sova P., Bomsztyk K. Fast chromatin immunoprecipitation assay. Nucleic Acids Res. 2006;34:e2. doi: 10.1093/nar/gnj004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hou L., Yang Z., Wang Z., Zhang X., Zhao Y., Yang H., Zheng B., Tian W., Wang S., He Z., Wang X. NLRP3/ASC-mediated alveolar macrophage pyroptosis enhances HMGB1 secretion in acute lung injury induced by cardiopulmonary bypass. Lab. Invest. 2018;98:1052–1064. doi: 10.1038/s41374-018-0073-0. [DOI] [PubMed] [Google Scholar]
- 48.Chen Y.T., Du Y., Zhao B., Gan L.X., Yu K.K., Sun L., Wang J., Qian F. Costunolide alleviates HKSA-induced acute lung injury via inhibition of macrophage activation. Acta Pharmacol. Sin. 2019;40:1040–1048. doi: 10.1038/s41401-018-0192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao H., Chen H., Xiaoyin M., Yang G., Hu Y., Xie K., Yu Y. Autophagy activation improves lung injury and inflammation in sepsis. Inflammation. 2019;42:426–439. doi: 10.1007/s10753-018-00952-5. [DOI] [PubMed] [Google Scholar]