Abstract
A hallmark of cancer cells is their ability to reprogram nutrient metabolism. Thus, disruption to this phenotype is a potential avenue for anti-cancer therapy. Herein we used a phenotypic chemical library screening approach to identify molecules that disrupted nutrient metabolism (by increasing cellular oxygen consumption rate) and were toxic to cancer cells. From this screen we discovered a 1,4-Naphthoquinone (referred to as BH10) that is toxic to a broad range of cancer cell types. BH10 has improved cancer-selective toxicity compared to doxorubicin, 17-AAG, vitamin K3, and other known anti-cancer quinones. BH10 increases glucose oxidation via both mitochondrial and pentose phosphate pathways, decreases glycolysis, lowers GSH:GSSG and NAPDH/NAPD+ ratios exclusively in cancer cells, and induces necrosis. BH10 targets mitochondrial redox defence as evidenced by increased mitochondrial peroxiredoxin 3 oxidation and decreased mitochondrial aconitase activity, without changes in markers of cytosolic or nuclear damage. Over-expression of mitochondria-targeted catalase protects cells from BH10-mediated toxicity, while the thioredoxin reductase inhibitor auranofin synergistically enhances BH10-induced peroxiredoxin 3 oxidation and cytotoxicity. Overall, BH10 represents a 1,4-Naphthoquinone with an improved cancer-selective cytotoxicity profile via its mitochondrial specificity.
Keywords: Cancer metabolism, Quinone, Peroxiredoxin, Mitochondria
Graphical abstract
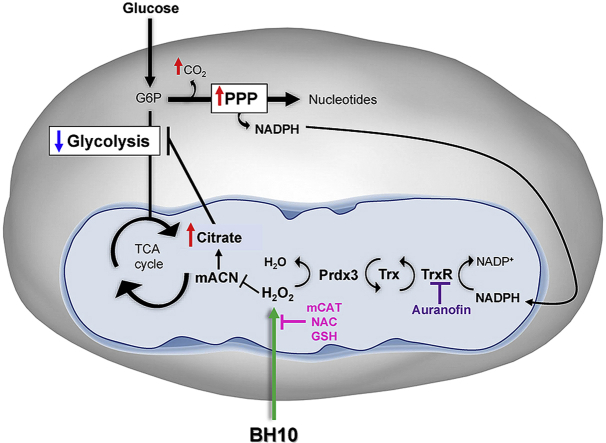
Schematic overview of BH10-induced effects on cancer cells. Treatment of cancer cells with BH10 induced mitochondrial oxidative stress. Activity of the redox-sensitive enzyme mitochondrial aconitase (mACN) was impaired leading to citrate accumulation and a decrease in glycolysis. BH10 increased pentose phosphate pathway activity to increase NADPH production for antioxidant defence. Mitochondrial oxidative stress is evidenced by oxidation of the peroxiredoxin/thioredoxin reductase system, including peroxiredoxin 3 dimerization (oxidation), and depletion of NADPH. BH10's cytotoxicity and effect on PRDX3 dimerization were synergistically enhanced with the thioredoxin reductase inhibitor auranofin, while over-expression of mitochondrial-targeted catalase (mCAT) or treatment with antioxidants N-acetylcysteine (NAC) and GSH protected cells from BH10-induced death.
Highlights
-
•
Phenotypic screen identifies a 1,4-Naphthoquinone BH10 with improved cancer-selective cytotoxicity
-
•
BH10 decreases glycolysis and increases glucose oxidation
-
•
BH10-mediated cytotoxicity is linked to mitochondrial oxidative stress
1. Introduction
The discovery of new anti-cancer agents has historically relied on target-based drug screens [1]. However, phenotypic drug screens are gaining renewed interest and are thought to identify drug leads/clinical candidates more likely to possess therapeutically-relevant molecular mechanisms of action [1]. Furthermore, phenotypic screens are proven to outperform target-based approaches leading to FDA-approved first-in-class small molecule drugs [2]. It has been reported that many target-based drug screens fail due to diverse biochemical mechanisms at the target [2]. In contrast, phenotypic screens prioritise a biological effect over a mechanism of action and avoid some hurdles faced by target-based models of drug discovery [1,2].
Most genetic aberrations that initiate and promote cancer development, such as oncogenic KRAS, BRAF, and MYC, activated Akt, and loss of tumour suppressors (e.g. LKB1, PTEN, p53), in one way or another impact cellular metabolism [3]. Not surprisingly, dysregulated metabolism is now recognised as a ‘hallmark’ of cancer [4,5]. Although reported many decades ago by Otto Warburg, the altered metabolism of glucose by tumour cells remains an area of intense investigation and potential avenue of drug discovery [6]. Increased glucose uptake and flux of glucose-derived carbons into lipids, nucleotides, and amino acids facilitates cancer cell anabolic growth, proliferation, anti-oxidant defence, metastasis, and immune system evasion, as reviewed [3]. However, it is not just metabolic pathways in the cytoplasm that provide cancer cells with the flexibility to thrive under diverse conditions. Cancer cells can also utilise different fuel sources through mitochondrial networks to aid tumour growth and progression [3]. Thus, the intrinsic ability of cancer cells to use both glycolysis and mitochondrial oxidative metabolism provides a selective advantage when exposed to stressors including anti-cancer therapies and/or hypoxia, allowing cancer cells to survive under these challenging conditions [[7], [8], [9]]. This highlights the importance of targeting diverse metabolic pathways for therapy to overcome the metabolic flexibility of cancer cells.
With these factors in mind, we conducted a phenotypic screen to identify small molecules that altered cell metabolism and were more toxic to cancer cells than normal cells. Specifically, we describe a cell-based phenotypic screen developed to detect small molecules that increase cellular oxygen consumption rate (primary screen) and showed greater toxicity to cancer cells than non-cancer cells (secondary screen). Screening for molecules that increase oxygen consumption is a gain-of-function screen that indicates increased nutrient oxidation and/or reactive oxygen species (ROS) production, which are two prominent mechanisms for disrupting cancer cell metabolism and viability [10,11].
2. Materials and methods
2.1. 1 Cell culture
Human non-cancerous cells; skin (CCD-25SK, obtained from ATCC), endometrium (hTERT-immortalised MAD11, from Dr Hui Li, University of Virginia, USA), breast (MCF10A, obtained from ATCC), mesothelium (MeT-5A, from Dr Glen Reid, University of Sydney, Australia), lung fibroblasts (WI-38, obtained from ATCC, MRC-5 from Dr. Joshua McCarroll, Children's Cancer Institute, Australia). Human cancer cells; mesothelioma (MSTO from Dr Glen Reid, Asbestos Disease Research Institute, University of Sydney, Australia), endometrial cancer (ISHIKAWA, KLE, MFE-296, MFE-319, HEC1A, RL95-2 and AN3-CA were from Dr Johnathan Lancaster, H. Lee Moffitt Cancer Centre and Research Institute, Florida, USA), liver cancer (HepG2 and HuH7, obtained from ATCC), colon cancer (HCT116 were obtained from ATCC, LIM2405, LIM1215 and LIM1899 were from Dr Antony Burgess, Walter and Eliza Hall Institute of Medical Research, Australia), ovarian cancer (SKOV-3 and A2780, obtained from ATCC), prostate cancer (LNCaP and PC3, obtained from ATCC), breast cancer (MDA-MB-231 and MCF7, obtained from ATCC), leukemia (K562, from Dr Maria Kavallaris, Children's Cancer Institute, Australia), melanoma (SKMEL-2, A375, SKMEL-28 and HT144, obtained from ATCC), lung cancer (CALU-1, from ATCC, and H441, H1650, and H1975 were from Dr Joshua McCarroll, Children's Cancer Institute, Australia). Primary murine hepatocytes were isolated as described [12]. Mouse cancer cells; liver (Hepa-1c1c7, obtained from ATCC) and melanoma (B16F1, from Dr Timothy Bullock, University of Virginia, USA). Mouse 3T3-L1 cells over-expressing mitochondrial catalase (or empty vector control) were a generous gift from Dr Daniel Fazakerley (Charles Perkins Centre, Sydney University). All cell lines were maintained as per instructions from the American Type Culture Collection. Cells were passaged 2–3 times per week with trypsin/EDTA and were kept in culture for a maximum of 4–5 weeks.
2.2. Chemicals
Chemicals were purchased from Sigma Aldrich or Cayman Chemical (Ann Arbor, Michigan USA), and were prepared in DMSO, sterile water, or PBS, and stored at appropriate temperatures, following manufacturer's instructions. BH10 was first purchased from TimTec (Newark, DE) and then synthesized by Dr Naresh Kumar as described in Section 2.14 below (UNSW, Sydney, Australia).
2.3. Chemical library screen
The ApexScreen 5040 small molecule diversity library was purchased from TimTec (Newark, DE) and compounds were screened for alterations in cellular O2 consumption using 96-well Becton Dickinson (BD) Oxygen Biosensor (OBS) microplates (Bedford, MA). Molecules were screened at a concentration of 5 μg/mL (TimTec Apex Screen) in rat L6 myoblasts, as described previously [13]. Hits were identified as those that increased O2 consumption over vehicle (DMSO) control.
2.4. Cytotoxicity assays
Cells (2500–5000/well) were seeded in 96-well plates and incubated at 37 °C overnight prior to drug treatment. Drugs diluted in cell culture medium were added to each well at the indicated concentrations. Cell viability was determined 48 h later by addition of thiazolyl blue tetrazolium bromide (MTT) reagent (3 h). Formazan crystals were solubilized in solvent (4 mM HCl, 0.1% NP-40 in isopropanol) and absorbance read at 590/620 nm using an EnSight Multimode plate reader (PerkinElmer). Viability of drug-treated cells is displayed as a percentage of control cells i.e. cells with equivalent concentrations of the appropriate vehicle. Synergy calculations were conducted according to the methods outlined by Chou and Talalay [14], using a non-constant drug ratio. Data were analysed in Compusyn (Combosyn, Inc.) whereby combination index (CI) values less than 1 indicate drug synergy.
2.5. Cell death assays
Cell death was measured using the Muse® Annexin V and Dead Cell Assay kit (Merck), according to manufacturer's instructions. Briefly, HEC1A cells were seeded in 6-well plates (100,000/well) and the next day treated for 22 h with BH10 (20 μM final), 3-BP (10 mM final, prepared in PBS and neutralised to pH 7.4), or vehicle (DMSO). Cells on a separate plate were exposed to UV irradiation (150,000 μJ/cm2). Non-attached and attached cells were harvested, centrifuged at 300×g for 5 min, and cell pellets resuspended in phenol red-free DMEM supplemented with 25 mM glucose, 4 mM l-glutamine, 1 mM sodium pyruvate, 1.5 g/L sodium bicarbonate and 1% FBS (2–3 x 105 cells/mL). Cells were diluted 1:1 with Muse reagent, incubated for 5 min at room temperature, and then run on the Muse™ Cell Analyzer (2000 counts/sample). Caspase activity was measured using Caspase-Glo® 3/7 assay reagent (Promega). HEC1A cells were seeded in 96-well plates at 5000 cells/well and treated for 6–22 h with BH10 (20 μM final) or vehicle (DMSO) prepared in media, or UV-irradiated in the presence or absence Q-VD-OPh hydrate (50 μM final, Merck). Caspase Glo® reagent was added to cells (1:1 ratio) and the lysed cell solution transferred to a white 96-well plate. Luminescent signal was detected using an EnSight Multimode plate reader (PerkinElmer).
2.6. Western blotting
Protein lysates (5–20 μg) were resolved on Any kD™ Mini-Protean TGX Precast gels (Bio-Rad) and electro-transferred to nitrocellulose membrane. Proteins were detected with rabbit antibodies; PARP (Cell Signalling), PRDX2, catalase (Abcam), PRDX3 (AbFrontier), and a mouse antibody; 14-3-3 (Santa Cruz). Primary antibodies were detected with donkey anti-mouse IgG (AlexaFluor790) or anti-rabbit IgG (AlexaFluor680) (Abcam) and membranes scanned on the LI-COR ODYSSEY System (LI-COR, Lincoln, NE, USA). For PRDX blotting; MAD11 (1 × 105/well) and HEC1A (2 × 105/well) cells were seeded in 6 well plates a day prior to drug treatments. Cells were treated with media or GSH (2 mM, pH neutralised) for 15 min prior to adding vehicle (DMSO), BH10, vitamin K3, auranofin, or BH10 + auranofin for 150 min. Cells were then rinsed and incubated with ice-cold catalase-treated PBS (1 mL/well) containing 100 mM NEM for 10 min on ice. Catalase-treated PBS was prepared by adding 5 μL pre-warmed (37 °C for 30 min) bovine catalase solution to 2.89 mL 50 mM pre-warmed potassium phosphate buffer ( ~1000 units/mL bovine catalase). This solution was diluted 1:10 into warm PBS (~100 units/mL) and incubated at 37 °C for another hour to remove H2O2. Following catalase treatment, cells were lysed in HES-SDS buffer [250 mM Sucrose, 20 mM HEPES (pH 7.4), 2% SDS (w/v), 1 mM EDTA] containing 100 mM NEM. Cell lysates were diluted with sample buffer without a reducing agent and run on gels according to methods described in manuscript. Densitometry was used to calculate the ratio of PRDX dimer to monomer for each condition.
2.7. Metabolomics
HEC1A cells were seeded in 15 cm dishes at 8 million cells per dish, with 4 replicate plates per treatment per time point. Scaling to 15 cm dishes with less volume per cell number required us to use 50 μM BH10 to kill >80% of cells at 24 h. Therefore, the metabolomics studies were performed with 50 μM BH10 and equivalent volumes of vehicle (DMSO). Cells were incubated for 5, 30, 90 and 150 min and then rinsed twice with ice-cold PBS. Cells were then scraped in ice-cold PBS and pelleted by centrifugation at 2000 rpm at 4 °C. PBS was removed, then cell pellets were frozen on dry ice and stored at −80 °C until shipment to Metabolon Inc. (Durham, NC, USA). An equivalent amount of material for each sample was extracted and run across the DiscoveryHD4™ global profiling platform. A recovery standard was added prior to the first step in the extraction process for QC purposes. Proteins were precipitated with methanol under vigorous shaking for 2 min (Glen Mills GenoGrinder 2000) followed by centrifugation. The resulting extract was divided into five fractions: one for analysis by UPLC-MS/MS with positive ion mode electrospray ionization, one for analysis by UPLC-MS/MS with negative ion mode electrospray ionization, one for LC polar platform, one for analysis by GC-MS, and one sample was reserved for backup. Samples were placed briefly on a TurboVap® (Zymark) to remove the organic solvent. For LC, the samples were stored overnight under nitrogen before preparation for analysis. For GC, each sample was dried under vacuum overnight before preparation for analysis. Raw data was extracted, peak-identified (using area-under-the-curve) and QC processed using Metabolon's hardware and software. Compounds were identified by comparison to library entries of purified standards or recurrent unknown entities. Overall, a total of 373 compounds of known identity (named biochemicals) were identified and analysed.
2.8. 14C- and 3H- tracer experiments
Sub-confluent cells were incubated in Krebs Ringer Phosphate (KRP) solution (120 mM NaCl, 600 μM Na2HPO4, 400 μM NaH2PO4, 6 mM KCl, 1.2 mM MgSO4, 12.5 mM HEPES pH 7.4, 1 mM CaCl2) with 20 μM BH10 or vehicle (DMSO) with nutrients, (5 mM glucose, 500 μM glutamine, 125 μM palmitate, 50 μM acetate, 1 mM l-carnitine, 0.25% fatty acid-free BSA) containing either 10 μCi/mL D-[3-3H]glucose, 5 μCi/mL D-[14C(U)] glucose, 5 μCi/mL D-[14C(1)] glucose, 5 μCi/mL D-[14C(6)] glucose, 1 μCi/mL L[14C(U)]-glutamic acid, or 2 μCi/mL[1–14C]-palmitic acid for 90 min in sealed wells. Substrate oxidation was measured by adding 2 M HClO4 (to halt the reaction) and evolved 14C–CO2 was captured in PCR tubes with 50 μL 1 M NaOH, and then transferred to scintillation vials. For measurements of glycolysis, reactions were stopped with 1 N HCl, and D-[3-3H] glucose was separated from tritiated [3H]2O (generated from dehydration at enolase in glycolytic reaction) by diffusion. Samples were counted using a LS 6500 Multi-purpose Scintillation Counter (Beckman Coulter) and substrate metabolism for each sample (pmol of substrate disintegrated per minute) normalised to protein content per well.
2.9. 13C-glucose tracer experiments
Polar metabolites were extracted from cells using a methanol-water-chloroform method [15]. HEC1A cells were seeded at 5 × 105/well in 6-well plates and treated with vehicle (DMSO) or BH10 (50 μM) for 90 min (isotopic steady-state was reached at this time point in labelled medium). Cells were washed and quenched using ice-cold isotonic (0.9% w/v) NaCl, and then scraped in 5 mL of 1:1 methanol: water solution at −30 °C. 1 nM of internal standard (chlorophenylalanine) was added to each well. Cell slurry was transferred to Falcon tubes containing 5 mL chloroform at −30 °C. Liquid-liquid extraction was performed by vortexing briefly and centrifuging at maximum speed for 10 min. The top aqueous phase was transferred into glass tubes and liquid evaporated without heat by SpeedVac (Savant). Dried samples were promptly derivatised by TBDMS-silylation method and analysed by GC-MS [16]. Isotopologue abundances have been corrected for natural enrichment.
2.10. Seahorse flux analysis
Cells (8000 cells MAD11/well and 15,000 HEC1A cells/well) were seeded in 96-well Seahorse tissue culture plates. The next day, cells were rinsed twice with PBS prior to addition of Seahorse media (160 μL/well) and then placed in a non-CO2 incubator at 37 °C for 1 h. Seahorse media contained phenol red-free DMEM (Sigma), 25 mM glucose, 4 mM l-glutamine and 1 mM sodium pyruvate (pH 7.4, filter sterilized). BH10 and vehicle (DMSO) were prepared in media (10x concentration) and loaded into port A. For thiol compound experiments, NAC and GSH were prepared at 20 mM (10x concentration) in Seahorse media, neutralised to pH 7.4, and loaded into port B. Basal OCR (pmoles O2/min) was measured using the Seahorse XFe96 Flux Analyzer (Seahorse Biosciences, North Billerica, MA). OCR rates were averaged over a minimum of 5 wells/plate/treatment condition and 3 independent plates and normalised to protein content per well.
2.11. NADPH and GSH assays
An NADP/NADPH-Glo assay (Promega) was used to measure NADPH/NADP+ levels, according to manufacturer's instructions. Briefly, cells were seeded in 6-well plates (1 × 105/well for MAD11 and 2 × 105/well for HEC1A) and treated for 2.5 h with BH10 (20 μM) or equivalent volumes of vehicle control (DMSO). Media was removed from wells prior to adding equal volumes (60 μL) of PBS then 1% dodecyltrimethylammonium bromide in 0.2 N NaOH. The remainder of the assay followed the protocol supplied with the kit. Luminescent signal was detected using an EnSight Multimode plate reader (PerkinElmer). GSH:GSSG ratios were measured using the Glutathione (GSH/GSSG/Total) Fluorometric Assay Kit (Biovision, K264). Cells were seeded in 10 cm dishes (1 × 106 for MAD11 and 2 × 106 for HEC1A) in 10 mL media. The following day, cells were treated with BH10 (20 μM) or equivalent volumes of vehicle (DMSO) for 2.5 h. Cells were rinsed with ice-cold PBS and then scraped from the dish in 800 μL ice-cold PBS. Cells were spun at 5000 rpm for 3 min at 4 °C, cell pellets resuspended in 100 μL glutathione assay buffer, and ice-cold perchloric acid (20 μL) was added to each sample (60 μL). GSH and GSSG levels were then measured according to manufacturer's instructions.
2.12. Aconitase assay
HEC1A cells were seeded and treated with vehicle or BH10 as per GSH assays. Mitochondrial fractions were extracted using a Cell Fractionation Kit (Abcam, ab109719) from cells pellets resuspended in 80 μL buffer A. Mitochondrial aconitase activity was measured using the Aconitase Enzyme Activity Microplate assay (Abcam, ab109712). Samples were diluted to 0.45 mg/mL (protein conc.), and 55 μL of each were added to 5 μL aconitase solution. Absorbance was read at 240 nm at multiple time points and the rate of enzyme activity was calculated in OD/min.
2.13. Oxidative DNA damage ELISA
Cells were seeded in 6-well plates (1 × 105/well MAD11 and 2 × 105/well HEC1A) and treated for 6 h with BH10 or vitamin K3 (20 μM) or equivalent volumes of vehicle (DMSO). Non-attached and attached cells were harvested, centrifuged at 300×g for 5 min, and pellets collected for DNA extraction using the GenElute Mammalian genomic DNA miniprep kit (Sigma). DNA was then concentrated by ethanol precipitation and DNA (3 μg in 20 μL) was then denatured by adding 2 μL of 0.5 M sodium acetate (pH 5.1), 0.2 μL of 1 M MgCl2 and heating samples to 100 °C for 5 min. Samples were cooled on ice (5 min) before adding Nuclease P1 (0.375 μL of 40 ng/μL stock, Sigma N8630) and heating at 50 °C for 1 h pH was adjusted to 7.5–8.5 with 1 M Tris (0.25 μL of 1 M Tris pH 10.5). Alkaline phosphatase (Sigma) was added to each sample (0.03U per 3 μg DNA) before heating to 37 °C for 30 min followed by 100 °C for 10 min. Samples were diluted to 0.04 ng/μL using buffer from the GenElute kit and placed on ice. Samples (equiv. of 2 μg DNA in 50 μL) were analysed using the DNA/RNA Oxidative Damage ELISA kit (Cayman Chemical), according to manufacturer's instructions.
2.14. Synthesis of BH10
BH10 was synthesized by a method modified from Brun et al. [17]. The appropriate naphthoquinone (1 equivalent) was dissolved in appropriate solvent (20 mL) and to the resulting mixture were added 1 equivalent of appropriate amine and 1 equivalent of Et3N. The resulting mixture was stirred at room temperature for 17–72 h. The resulting mixture was concentrated in vacuo, diluted in EtOAc and washed with water before the organic layer was dried over anhydrous Na2SO4. The solvent was evaporated, and the resulting residue was purified via flash column chromatography (when required) in the appropriate solvent system to afford the product. The general synthetic scheme was; 2,3-dichloro-[1,4]-naphthoquinone (0.51 g, 2.20 mM), Et2O (20 mL), 4-(2-aminoethyl)morpholine (0.30 mL, 2.20 mM), Et3N (0.31 mL, 2.20 mM). Reaction stirred for 18 h. Product eluted with EtOAc/n-hexane = 3:1 to afford a dark orange powder 1 (0.63 g, 89%): mp: 110–112 °C; 1H NMR (300 MHz, CDCl3): δ 8.15 (d, J = 9.0 Hz, 1H), 8.03 (d, J = 9.5 Hz, 1H), 7.75 (dd, J = 7.5 Hz, 1H), 7.65 (dd, J = 7.6 Hz, 1H), 6.88 (br s, 1H), 3.98 (dd, J = 17.1 Hz, 2H), 3.78 (t, J = 9.1 Hz, 4H), 2.71 (t, J = 11.0 Hz, 2H), 2.55 (t, J = 8.8 Hz, 4H); 13C NMR (CDCl3): δ 180.70, 176.80, 144.81, 135.03, 132.68, 132.51, 130.31, 126.92, 126.88, 67.03, 57.80, 53.20, 40.81.
2.15. Statistical analyses
Data with normal distributions were analysed using parametric tests and data with unequal variances were analysed using non-parametric tests (indicated within figure legends). All statistical analyses were performed in GraphPad Prism with the exception of the metabolomics experiments where following normalization to Bradford protein concentration, log transformation and imputation of missing values, if any, with the minimum observed value for each compound, Welch's two-sample t-test was used to identify biochemicals that differed significantly between experimental groups.
3. Results
3.1. Phenotypic screen to identify modulators of oxidative metabolism
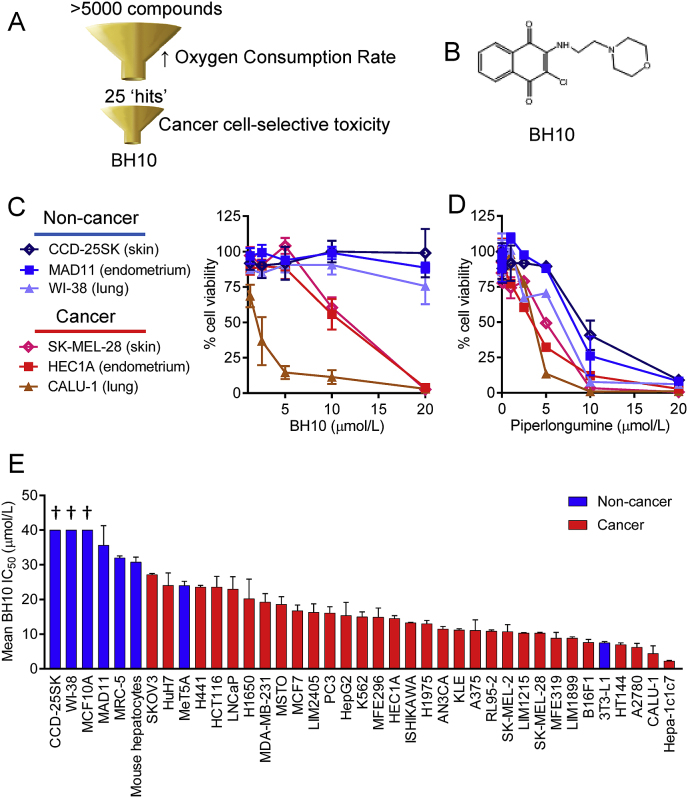
Herein we repurposed a diversity chemical library screen for cellular oxygen consumption rate (OCR), as described [13]. From a total of 5040 compounds screened, we selected the top 25 hits that increased OCR for secondary screening in a small panel of 3 cancer and 3 non-cancerous cells from skin, lung, and endometrium (Fig. 1A). Hit number 10 (2-chloro-3-[(2-morpholin-4-ylethyl)amino]naphthalene-1,4-dione), referred to as BH10 (Fig. 1B), had the best cancer-selective toxicity in this panel of cells and outperformed benchmarking to a control compound, piperlongumine, that is reported to have exceptional selectivity for killing cancer cells over non-cancer cells in a panel of 13 cancer cell lines and 6 non-cancer lines [18] (Fig. 1C–D) and works by inhibiting glutathione S-transferase pi 1 to induce oxidative stress [18].
Fig. 1.
Phenotypic drug screen identifies small molecule with better cancer cell-specific toxicity than piperlongumine. (A) Schematic overview of drug screen. (B) Chemical structure of BH10. Viability of cells (%, normalised to vehicle control) exposed to increasing doses of BH10 (C) and piperlongumine (D) for 48 h. Panel of non-cancerous and cancer cell lines are shown on the left. (E) Mean IC50 values for cells treated with BH10 for 48 h (IC50 values were calculated from dose response curves). Non-cancerous cells are displayed with blue bars and cancer cells are displayed with red bars. See Supp. Table 1 for IC50 values. n ≥ 3 for all experiments. Data represent mean ± SEM. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To better understand the anti-cancer properties of BH10, we screened BH10 across a much larger panel of 39 non-cancerous and cancer cells from different tissues of origin (Fig. 1E). The mean IC50 values for each cell (calculated from dose response curves) are shown in Fig. 1E (as listed in Supp. Table 1). From this screen we identified cells that were resistant and sensitive to BH10. Of the non-cancerous cells, 6 of 8 had IC50 values greater than 30 μM, whereas 19 of the 31 cancer cells had IC50 values less than 15 μM (Supp. Table 1).
3.2. BH10 has improved cancer-selective toxicity than other quinones and molecules that disrupt glucose metabolism
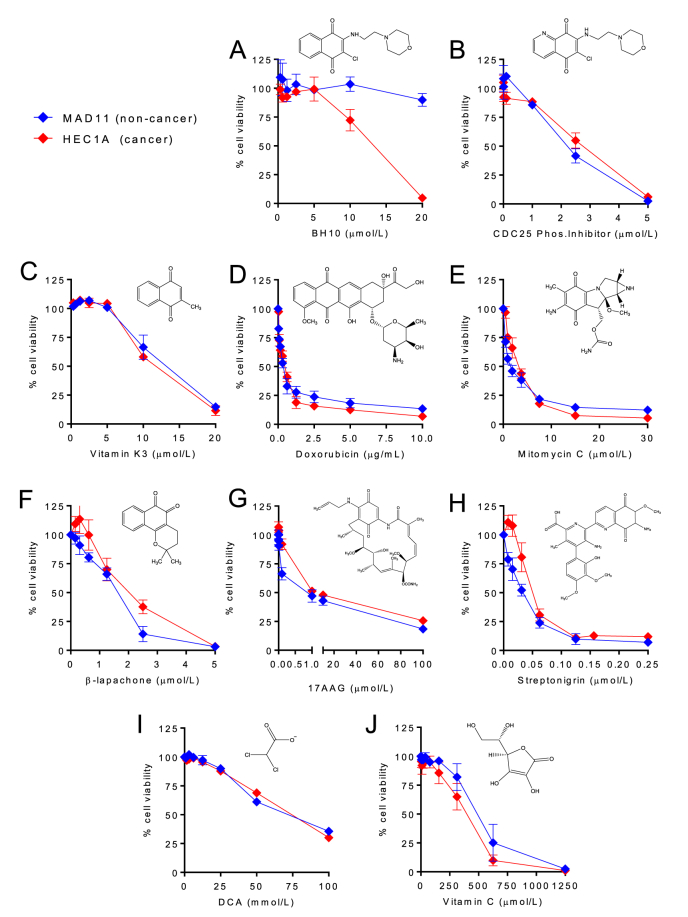
Quinones are an important class of anti-cancer molecules that have been used in the clinic for decades [19]. We therefore investigated whether BH10, which is a 1,4-naphthoquinone, had a different cytotoxicity profile than other quinones (Fig. 2). The HEC1A endometrial cancer cell line and the non-cancerous MAD11 endometrial cell line were tested for viability in response to a panel of known anti-cancer quinones and only BH10 showed selective toxicity towards the cancer cells (Fig. 2A) (see Supp. Table 2 for IC50 values). In contrast, the quinone molecules CDC25 phosphatase inhibitor II (Fig. 2B), vitamin K3 (Fig. 2C), doxorubicin (Fig. 2D), mitomycin C (Fig. 2E), β-lapachone (Fig. 2F), 17-AAG (Fig. 2G), and streptonigrin (Fig. 2H) did not show cancer-selective toxicity (see Supp. Table 2 for IC50 values). Dichloroacetate (DCA) was tested because it is thought to increase mitochondrial glucose oxidation by relieving pyruvate dehydrogenase kinase's inhibition of pyruvate dehydrogenase [20]; however, DCA was equally toxic to HEC1A and MAD11 cells and required high millimolar doses (Fig. 2I). Vitamin C was tested because it is shown to induce ROS, deplete glutathione, and target GAPDH [21]; however, Vitamin C had similar toxicity to HEC1A and MAD11 cells (Fig. 2J) (see Supp. Table 2 for IC50 values). Overall, these results indicate that BH10 has a distinct mechanism of action compared to other anti-cancer quinones or molecules that disrupt redox control and glucose metabolism.
Fig. 2.
BH10 shows improved cancer cell-selective toxicity compared to other anti-cancer quinones and molecules that disrupt glucose metabolism. Non-cancerous MAD11 (blue lines) and cancerous HEC1A cells (red lines) were exposed to increasing doses of (A) BH10, (B) CDC25 phosphatase inhibitor II, (C) vitamin K3, (D) doxorubicin, (E) mitomycin C, (F) β-lapachone, (G) 17-AAG, (H) streptonigrin, (I) dichloroacetate (DCA), and (J) vitamin C, for 48 h. Cell viability is shown as a percentage normalised to vehicle-treated cells. Each compound structure is shown with corresponding graphs. n ≥ 3 for all experiments. Data represent mean ± SEM. Statistical analyses comparing the IC50 values of MAD11 vs HEC1A for each drug are shown in Supp. Table 2. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. BH10 induces necrosis in cancer cells
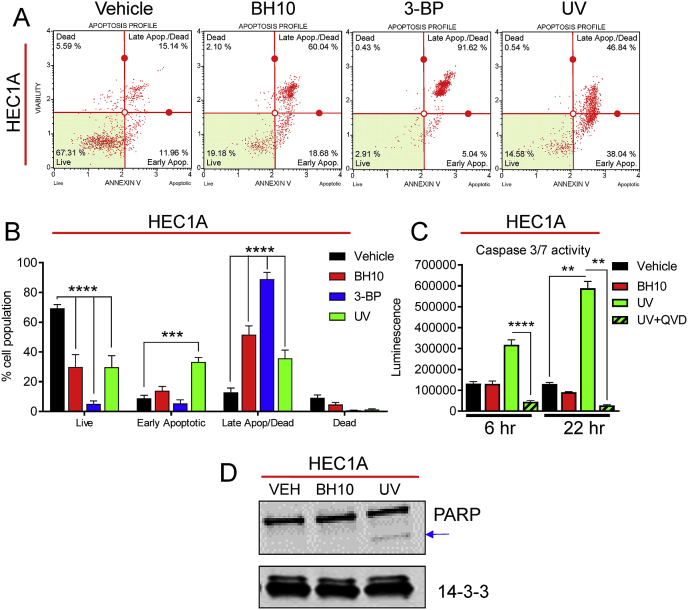
We next sought to determine how BH10 induced cancer cell death. Flow cytometry analyses showed that BH10 significantly increased the percentage of HEC1A cells in late apoptosis/necrosis (Fig. 3A–B). Ultraviolet radiation (UV) and 3-bromopyruvate (3-BP) were used as positive controls for apoptosis and necrosis, respectively [22]. UV treatment induced a significant increase in caspase 3/7 activity at 6 and 22 h, which was inhibited by the pan-caspase inhibitor Q-VD-OPH (QVD) (Fig. 3C). In contrast, BH10 did not increase caspase activity at either time point (Fig. 3C). UV treatment also induced PARP cleavage in HEC1A cells, while BH10 did not (Fig. 3D). Further analyses revealed that BH10-induced cancer cell death was not rescued by the necroptosis inhibitors necrostatin (RIPK1 inhibitor) or necrosulfanomide (MLKL inhibitor), the ferroptosis inhibitor ferrostatin, and BH10-induced cell death was also not altered by the PARP inhibitor, olaparib (not shown). These data support a mechanism for BH10-induced cancer cell death via necrosis.
Fig. 3.
BH10 induces necrosis in cancer cells. (A) Representative flow cytometry graphs from HEC1A cells treated with vehicle (DMSO), BH10 (20 μM), 3-bromopyruvate (3-BP, 10 mM), or UV-irradiated (22 h post treatment). (B) Quantification of flow cytometry data for the aforementioned conditions. (C) Caspase 3/7 activity in HEC1A cells following treatment with vehicle (DMSO), BH10 (20 μM), or UV-irradiation [(+/− the pan-caspase inhibitor, Q-VD-OPh hydrate (QVD)] (6 and 22 h post treatment). (D) Representative western blot of PARP (and cleaved PARP) protein in HEC1A cells treated with vehicle (DMSO), BH10 (20 μM), or UV-irradiated (6 h post treatment). 14-3-3 serves as a protein loading control. n ≥ 3 for all experiments. Data were analysed by 2-way ANOVA with Dunnet's multiple comparison test (B) and Kruskal-Wallis with uncorrected Dunn's test (C), where alpha = 0.05, ***p ≤ 0.001, ****p ≤ 0.0001. For B and C data represent mean ± SEM.
3.4. BH10 disrupts glucose, glutathione, nicotinamide, and mitochondrial metabolism
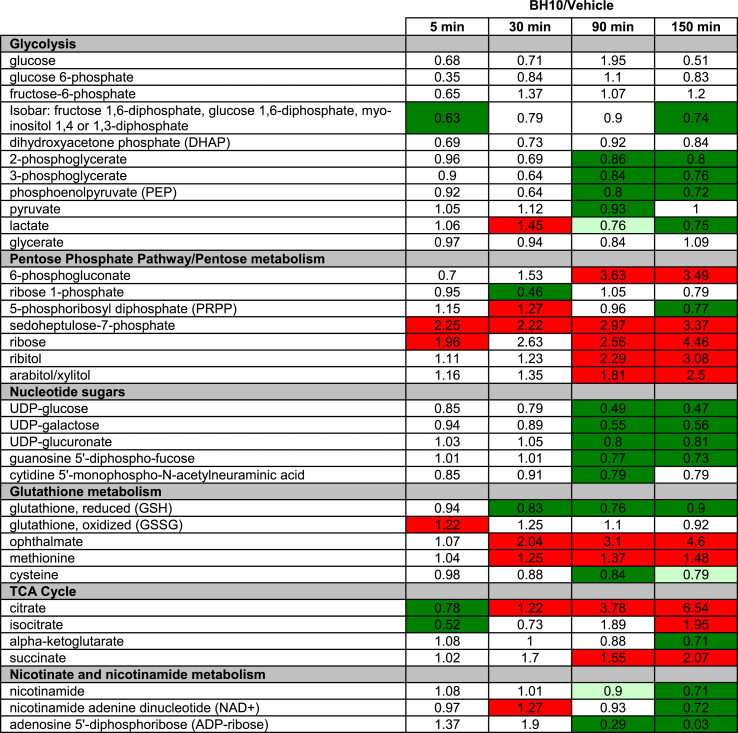
To expand our understanding of how BH10 influences cellular metabolism, metabolomics analyses were performed on HEC1A cancer cells treated with BH10 or vehicle control (DMSO) for 5, 30, 90 and 150 min (Table 1). Results showed that BH10 treatment disrupted glucose metabolism by decreasing several glycolytic intermediates that are metabolised downstream of phosphofructokinase, including lactate (Table 1). An increase in pentose phosphate pathway (PPP) intermediates, as well as 5-carbon sugar alcohols (ribose, ribitol, and arabitol/xylitol) were observed in cells treated with BH10 with accumulation increasing with time (Table 1). In addition, several activated nucleotide sugars (e.g., UDP-glucose, UDP-galactose) were decreased in BH10-treated cells at the later time points of 90 and 150 min (Table 1).
Table 1.
Metabolites altered in BH10-treated HEC1A cells. Metabolomics analysis of HEC1A cells treated with BH10 or vehicle control for the indicated time points. Dark red boxes indicate metabolites with mean values significantly increased in BH10-treated cells (p ≤ 0.05, fold change of ≥1.00), dark green boxes indicate metabolites with mean values significantly decreased in BH10-treated cells (p ≤ 0.05, fold change of <1.00), light green boxes indicate metabolites with mean values approaching significant decreases (0.05 < p < 0.1, fold change < 1.00), n = 4 for all time points. Replicate values for each metabolite and standard deviations for both BH10 and vehicle control, at each time point, are provided in Supp File 1.
Alterations in glutathione biosynthesis and degradation/recycling pathways were also observed in BH10-treated cells (Table 1). GSH was significantly lower (≥10% decrease) in BH10-treated cells from 30 to 150 min, with a brief but significant elevation in oxidized glutathione (GSSG) observed at 5 min (22% increase) (Table 1). A decline in GSH levels may be attributed to decreased synthesis, as accumulation of the amino acid precursor methionine, along with decreased availability of the rate-limiting biochemical for glutathione synthesis, cysteine, were observed in BH10-treated cells (Table 1). Pronounced elevations in ophthalamate, a glutathione-like biochemical that is synthesized by the same enzymes [gamma-glutamylcysteine synthetase (GCS) and glutathione synthetase (GS)] as glutathione, but with 2-aminobutyrate in place of cysteine, were observed in BH10-treated cells from 30 to 150 min (Table 1). These data suggest continued activity of glutathione synthetic machinery despite limited cysteine availability.
BH10-induced changes in glucose utilization also translated to alterations in tricarboxylic acid (TCA) cycle intermediates (Table 1). Accumulation of citrate was consistently observed after the 30-min time point and was the most-increased metabolite in BH10-treated cells (6.54-fold increase in BH10 vs vehicle-treated controls at 150 min, p < 0.05). Significant decreases in nicotinamide metabolites (nicotinamide and nicotinamide adenine dinucleotide, NAD+) were also observed while ADP-ribose was the most decreased metabolite (97% decrease in BH10 vs vehicle-treated controls at 150 min, p < 0.05) (Table 1).
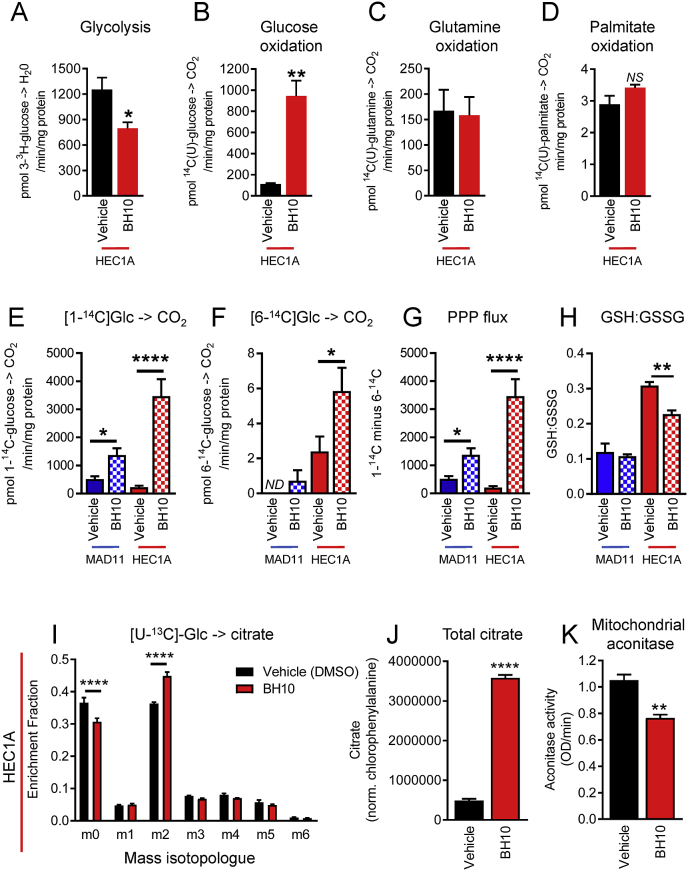
Using radiolabelled metabolic tracers, we confirmed that BH10 decreased glycolysis by 37% (p = 0.015, Fig. 4A) and increased glucose oxidation by 8-fold in HEC1A cancer cells (p = 0.022, Fig. 4B). In contrast, the oxidation of palmitate and glutamine were not significantly altered by BH10 (Fig. 4C–D). To distinguish mitochondrial glucose oxidation from pentose phosphate pathway (PPP) oxidation, we used 1–14C and 6–14C glucose tracers. 1-14C-glucose can be metabolised to 14CO2 in either the cytosol (via the PPP) or mitochondria (Fig. 4E), whereas 14CO2 released from 6-14C-glucose only occurs in mitochondria (Fig. 4F). Results show that BH10 treatment significantly increased CO2 generation from 1-14C-glucose in both non-cancerous MAD11 cells and cancerous HEC1A cells (p < 0.05, Fig. 4E). The effect of BH10 was greater in the HEC1A cells (15-fold increase, p < 0.0001) compared with MAD11 cells (2.7-fold increase, p = 0.019) (Fig. 4E). In contrast, BH10 treatment only increased CO2 generation from 6-14C-glucose in cancerous HEC1A cells (2.4-fold increase, p < 0.05, Fig. 4F). Measurements of PPP flux (obtained by subtracting values of 6–14C-glucose from 1-14C-glucose) showed that BH10 significantly increased flux of glucose to this pathway in MAD11 cells (2.7-fold increase, p = 0.019, Fig. 4G) but to a greater extent in HEC1A cancer cells (16.7-fold increase, p < 0.0001, Fig. 4G). We also confirmed that BH10 significantly decreased the ratio of GSH:GSSG in HEC1A cancer cells, but not in non-cancerous MAD11 cells (Fig. 4H). Using uniformly labelled stable isotope 13C-glucose we found that BH10 significantly decreased unlabelled citrate (m0) and increased the production of m2-labelled citrate in HEC1A cancer cells (Fig. 4I). These results demonstrate that glucose-derived carbons are the carbon source leading to increased levels of citrate, as opposed to reductive glutamine metabolism. Total citrate levels in this experiment were increased by 7.4-fold following 90 min of BH10 treatment (Fig. 4J), thereby validating the global metabolomics results in Table 1. We next investigated whether citrate accumulated because of decreased metabolism and found that BH10 caused a significant decrease (27.3%, p = 0.004) in the activity of mitochondrial aconitase (Fig. 4K), an enzyme that catalyses the isomeration of citrate to isocitrate.
Fig. 4.
BH10 alters glucose metabolism, and glutathione and citrate levels in cancer cells. Radiolabelled tracers were used to measure (A) glycolysis (uniformly-labelled 3H-glucose), (B) glucose oxidation (uniformly-labelled 14C-glucose), (C) glutamine oxidation (uniformly-labelled 14C-glutamine), (D) palmitate oxidation (uniformly-labelled 14C-palmitate), (E) 1C-glucose oxidation (1C-labelled 14C-glucose), (F) 6C-glucose oxidation (6C-labelled 14C-glucose), and (G) Pentose phosphate pathway (PPP) flux (1C-labelled 14C-glucose minus 6C-labelled 14C-glucose), in vehicle (DMSO) and BH10 (20 μM)- treated HEC1A and MAD11 cells (90 min treatment). (H) Ratio of GSH to GSSG in MAD11 and HEC1A cells treated with vehicle (DMSO) and BH10 (50 μM, the IC80 when tested in 15 cm2 plates) (150 min treatment). (I) Enrichment of 13C-glucose into citrate isotopologue in HEC1A cells treated with vehicle (DMSO) and BH10 (50 μM) (90 min treatment). (J) Total citrate levels in HEC1A cells treated with vehicle (DMSO) and BH10 (50 μM) (90 min treatment). (K) Mitochondrial aconitase activity in HEC1A cells treated with vehicle (DMSO) and BH10 (50 μM) (150 min treatment). n ≥ 3 for all experiments. Data from vehicle vs BH10-treated cells were analysed by unpaired student's t-tests (equal variances) or Mann-Whitney tests (unequal variances), where alpha = 0.05, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. Data represent mean ± SEM.
3.5. BH10-induced increases in OCR and cell death are rescued by antioxidants
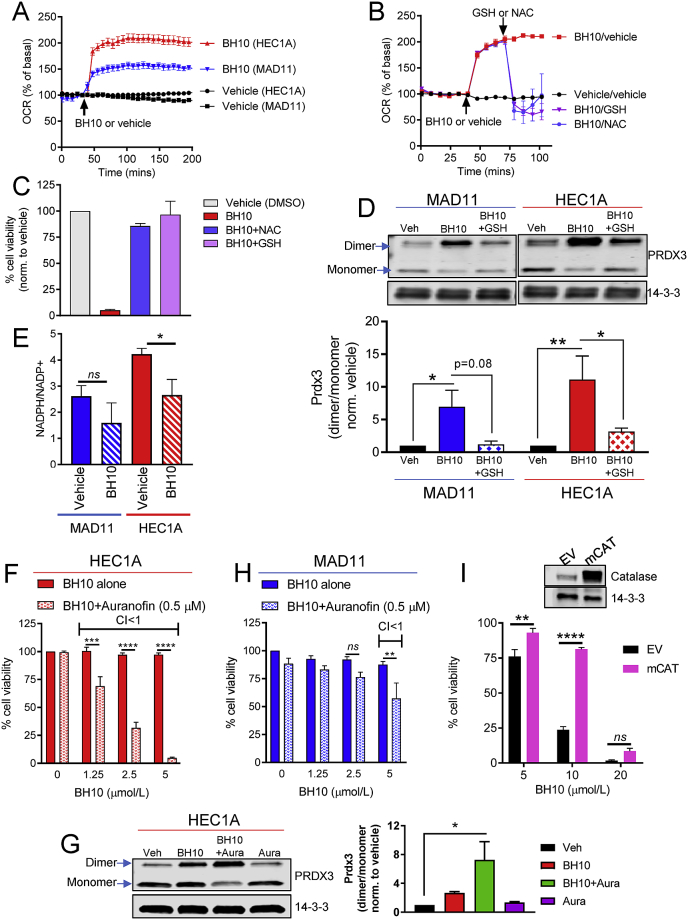
Aconitase is a redox-sensitive enzyme inhibited by oxidative damage, therefore we next investigated redox pathways. We first examined cellular oxygen consumption rate (OCR) in BH10-treated non-cancerous MAD11 cells and HEC1A cancer cells (Fig. 5A). BH10 increased OCR to ~200% in HEC1A cells, but only ~150% in MAD11 cells (Fig. 5A), suggesting that BH10 had a greater effect on OCR in cancer cells. Increases in OCR can be indicative of increased nutrient oxidation and/or increased reactive oxygen species (ROS) production. To investigate a role for ROS in the mechanism of BH10, we tested whether antioxidants were able to protect HEC1A cancer cells from BH10-induced increases in OCR and cell death. Indeed, both N-acetylcysteine (NAC) and GSH returned the level of OCR back to basal levels in BH10-treated cells (Fig. 5B) and rescued cells against BH10-induced cell death (Fig. 5C). These experiments suggest that BH10-induced cell death and increases in OCR are associated with oxidative stress.
Fig. 5.
BH10 increases mitochondrial oxidative stress. (A) Oxygen consumption rate (OCR) presented as a percentage of basal readings in MAD11 and HEC1A cells treated with vehicle (DMSO) or BH10 (20 μM). Injection of vehicle and BH10 indicated by arrow. (B) OCR presented as a percentage of basal readings in HEC1A cells treated with vehicle (DMSO) or BH10 (20 μM), followed by NAC or GSH (2 mM each). Injection of vehicle and BH10, or NAC or GSH, indicated by arrows. (C) Viability (%, normalised to vehicle control) of HEC1A cells treated with vehicle (DMSO) or BH10 (20 μM), alone or in combination with antioxidants NAC or GSH (2 mM each) for 48 h. (D) Representative western blot of peroxiredoxin 3 (PRDX3) dimerization in MAD11 and HEC1A cells treated with vehicle (DMSO), BH10 (BH10, 20 μM), or BH10 (BH10) in combination with GSH (150 min treatment). 14-3-3 serves as a protein loading control. Graph shows quantification of PRDX3 dimer to monomer (band densitometry) ratio for each condition shown in western blot. Viability (%, normalised to vehicle control) of HEC1A (F) and MAD11 (H) cells treated with BH10 alone or in combination with 0.5 μM auranofin. Synergy is indicated when combination index (CI) values are less than 1. (G) Representative western blot of peroxiredoxin 3 (PRDX3) dimerization in HEC1A cells treated with vehicle (Veh), BH10 (5 μM), BH10 (5 μM) in combination with auranofin (0.5 μM) (BH10 + Aura), or auranofin (0.5 μM) alone (Aura) for 150 min 14-3-3 serves as a protein loading control. Graph shows quantification of PRDX3 dimer to monomer (band densitometry) ratio for each condition shown in western blot. (I) Viability (%, normalised to vehicle control) of 3T3-L1 cells expressing mitochondrial catalase (mCAT) or empty vector (EV), treated with vehicle (DMSO) or BH10 (μM) for 48 h n ≥ 3 for all experiments. Data were analysed by 2-way ANOVA (D, F–I) and 1-way ANOVA (E), with multiple comparison tests, where alpha = 0.05, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, ns = not significant. Data represent mean ± SEM.
3.6. BH10 perturbs the mitochondrial peroxiredoxin/thioredoxin reductase system
We next investigated mitochondrial and cytoplasmic oxidative stress by measuring peroxiredoxin (PRDX) dimerization. PRDX proteins are involved in the cellular response to oxidative stress and exist in different subcellular compartments [23]. PRDX2 is found in the cytoplasm and nucleus, whereas PRDX3 is exclusively in mitochondria [23]. PRDX proteins detoxify hydrogen peroxide (H2O2) to water in a cycling reaction coupled with thioredoxin (TRX) and thioredoxin reductase, using NADPH as a primary electron donor [23]. We therefore examined whether BH10 altered the oxidation (dimerization) of PRDX2 or PRDX3. BH10 significantly increased PRDX3 dimerization, and this affect was greater in HEC1A cancer cells than MAD11 cells (Fig. 5D). GSH rescued PRDX3 dimer formation in both cell lines (Fig. 5D). In contrast, BH10 did not significantly alter PRDX2 dimerization (not shown) suggesting that BH10 may be selectively increasing mitochondrial oxidative stress, which is consistent with impaired mitochondrial aconitase activity (Fig. 4K). We also found that BH10 significantly decreased the NADPH/NADP+ ratio in HEC1A cancer cells, but not MAD11 cells (Fig. 5E).
To further explore the effect of BH10 on the PRDX/TRX system, we exposed cells to auranofin (a thioredoxin reductase inhibitor that impairs mitochondrial antioxidant defence [24]) in combination with BH10. Results showed that BH10 was highly synergistic (combination index values less than 1) with a non-toxic dose of auranofin (0.5 μM) in HEC1A cells. Cell death was induced with lower doses of BH10 (1.25–5 μM) compared with BH10 alone (Fig. 5F). In addition, PRDX3 dimerization was enhanced when BH10 (5 μM) was combined with auranofin (0.5 μM), compared to BH10 alone (Fig. 5G). These results suggest the synergistic cytotoxicity of these 2 drugs may be mediated through mitochondrial oxidative stress, as indicated by PRDX3 dimerization. In contrast to the results with the HEC1A cancer cells, BH10 had less synergy with auranofin in MAD11 cells (Fig. 5H). In addition, over-expression of mitochondrial-targeted catalase (an enzyme that detoxifies H2O2) partially protected cells against BH10-induced toxicity (Fig. 5I). Together, these data suggest that BH10-mediated cytotoxicity involves mitochondrial oxidative stress via H2O2.
3.7. Differences in oxidative stress between BH10 and vitamin K3
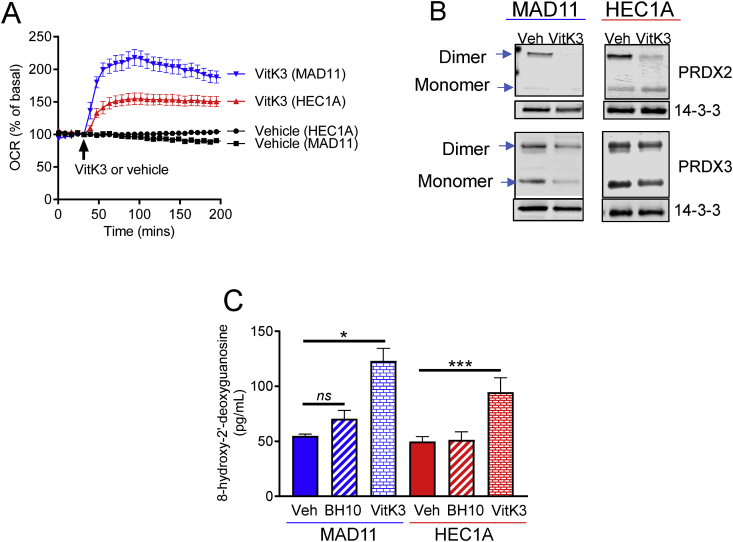
To further investigate mechanisms of cancer-selective toxicity, we compared BH10 to a closely related and well-known anti-cancer quinone, vitamin K3. Vitamin K3 is equipotent to BH10 in the context of cytotoxicity to cancer cells but is not selectively toxic to cancer cells (Fig. 2C) and increased OCR to greater levels in the non-cancerous cells (MAD11) than cancer cells (HEC1A) (Fig. 6A). Vitamin K3 was markedly different in its redox mechanism as evidenced by decreased total PRDX2 levels (monomer and dimer) in both cell lines and no effect on PRDX3 dimerization (Fig. 6B). Since nuclear PRDX2 protects cancer cells from death and DNA damage [25], we measured 8-hydroxy-2′-deoxyguanosine levels (an indicator of oxidative DNA damage [26,27]) in cells treated with BH10 and vitamin K3. Results showed that vitamin K3 significantly increased oxidative DNA damage in both MAD11 and HEC1A cells, whereas BH10 did not (Fig. 6C). Overall, these results demonstrate distinct mechanisms of oxidative stress between BH10 and vitamin K3, which may underlie the differences in cancer-selective toxicity between these two molecules.
Fig. 6.
Differences in oxidative stress between BH10 and vitamin K3. (A) Oxygen consumption rate (OCR) presented as a percentage of basal readings in MAD11 and HEC1A cells treated with vehicle (DMSO) or vitamin K3 (VitK3, 20 μM). Injection of vehicle and VitK3 indicated by arrow. (B) Representative western blots of peroxiredoxin 2 (PRDX2) and 3 (PRDX3) in MAD11 and HEC1A cells treated with vehicle (DMSO) and VitK3 (VK3, 20 μM) (150 min treatment). 14-3-3 serves as a protein loading control. (C) Oxidative DNA damage assay (measuring 8-hydroxy-2′-deoxyguanosine) performed on MAD11 and HEC1A cells treated with vehicle (DMSO), BH10 (20 μM) or VitK3 (20 μM) for 6 h. Data were analysed by 1-way ANOVA (C) with multiple comparison tests, where alpha = 0.05, *p ≤ 0.05, ***p ≤ 0.001, ns = not significant. Data in A and C represent mean ± SEM.
4. Discussion
Phenotypic screens for cancer drug discovery traditionally employ readouts of intracellular signalling and transcription, cell growth and death, or ‘classical’ phenotypes such as morphology, motility and differentiation [1]. In this study we selected molecules that were bioactive for increasing cellular oxygen consumption rate and screened them for cytotoxicity in a panel of cancer and non-cancerous cell lines. This approach led to the identification of the 1,4-Napthoquinone BH10.
1,4-Napthoquinones are known anti-cancer agents and structure-activity relationship (SAR) studies have reported anti-cancer potency of naphthoquinones similar to BH10 and including BH10. Brun et al. synthesized BH10 in an effort to make CDC25 inhibitors [17]. However, we found that CDC25 phosphatase inhibition is unlikely to be the BH10 mechanism of cytotoxicity because the CDC25 phosphatase inhibitor II did not have cancer-specific toxicity (Fig. 2B). Xu et al. also synthesized BH10 in an effort to make 20S protease inhibitors, but BH10 was not an effective inhibitor and had minimal effects on proliferation [28]. Hsu et al. also reported BH10 to be PT-262, which was described as an ERK, CDC2, and proliferation inhibitor [29] but an erratum later showed that PT-262 was a different molecule [30]. Although BH10 has been studied previously in the context of cancer, none of these studies compared the cytotoxicity of BH10 to cancer and non-cancerous cells, or identified mechanisms related to oxidative stress, mitochondria, or metabolism.
In this study, we enlisted a comprehensive set of metabolic studies which showed that treatment of cancer cells with BH10; 1) rapidly increased oxygen consumption rate, higher than that of non-cancerous cells, 2) increased glucose flux through the PPP, higher than in non-cancerous cells, 3) increased glucose flux into mitochondria and TCA cycle, and 4) increased citrate levels. This was coupled with a significant decrease in GSH:GSSG and NAPDH/NAPD+ ratios only in cancer cells and glycolysis/glycolytic intermediates. It is possible that the alterations in metabolism are secondary to oxidative stress occurring in mitochondria. For example, BH10 treatment led to a marked increase in citrate levels (~7-fold) that may be attributed to a decrease in the activity of mitochondrial aconitase; an enzyme that converts citrate to isocitrate, and is sensitive to oxidative stress and decreased GSH levels [31,32]. Citrate may be responsible for the decrease in glycolysis via allosteric inhibition of the rate-limiting glycolytic enzyme phosphofructokinase [33], which is supported by metabolomics data showing significant decreases in metabolites downstream of fructose-6-phosphate in BH10-treated cancer cells. The interconnection of these pathways is illustrated in the Graphical Abstract.
A BH10-mediated mitochondrial oxidative stress mechanism of cancer cell death is also supported by protection induced by mitochondria-targeted catalase and selective increases in PRDX3 oxidation in mitochondria. Furthermore, impairment of mitochondrial oxidative stress defence (with auranofin) demonstrated strong cytotoxic synergy with BH10 (Fig. 5F), which was associated with an increase in PRDX3 dimerization. These results point to a mitochondrial oxidative stress mechanism via increased H2O2 and activation of the mitochondrial peroxiredoxin/thioredoxin reductase system.
It is well established that ROS are elevated in cancer cells and, as an adaptive response, have higher levels of ROS-scavenging molecules such as GSH [34]. Consistent with this, we observed higher GSH:GSSG and NADPH/NADP+ ratios in cancer cells than normal cells (Fig. 4, Fig. 5E). GSH and NADPH are intricately linked, since NADPH is required for the conversion of GSSG to GSH via GSH reductase. This feature of cancer cells could explain why we observed significant decreases in both GSH:GSSG and NADPH/NADP + ratios in cancer cells upon exposure to BH10, but not in normal cells. Thus, higher levels of PRDX3 oxidation in cancer cells, coupled with significant decreases in GSH and NADPH specifically in cancer cells, may underlie cell death and selective toxicity to cancer cells. These results support the hypothesis that cancer cells may be more susceptible to the effects of oxidative stress than normal cells [10].
Perturbations in cell metabolism can induce cell death via different mechanisms, including apoptosis, necroptosis, ferroptosis and necrosis [35]. We found that BH10 did not induce caspase activity or PARP cleavage; indicating that cell death was not occurring via apoptosis. Inhibitors of necroptosis and ferroptosis also did not protect cancer cells from BH10-induced death, eliminating these pathways as potential mechanisms. These results point to a necrotic mechanism of cell death. In support of this, our metabolomics data showed BH10 treatment significantly decreased NAD+ and lactate levels at the 150-min time point prior to cell death. NAD+ is produced when pyruvate is converted to lactate by lactate dehydrogenase, and is an important substrate for PARP-mediated apoptosis [35]. BH10 treatment also significantly decreased ADP and ADP-ribose, while ribose was significantly increased, suggesting that BH10 treatment may disrupt poly(ADP-ribose) synthesis and/or degradation pathways required for apoptotic cell death. Interestingly, previous studies have shown that the synergistic cytotoxicity of β-lapachone combined with PARP inhibitors leads to tumour-selective apoptosis and is mediated via NQO1-dependent redox cycling [36]. In contrast, our results show that BH10 is not synergistic with the PARP inhibitor, olaparib. Furthermore, unlike β-lapachone, we propose that NQO1 is not involved in mediating BH10-induced cytotoxicity since the NQO1 inhibitor, ES936, did not protect against BH10-induced cell death (not shown). Another point of difference between these two quinones is that BH10 increases [U–13C]-glucose incorporation into m+2 citrate, while the opposite occurs for β-lapachone in an NQO1-dependent manner [37]. These results highlight different mechanisms of action and cell death between these quinones, which may underlie the differences in cytotoxicity between BH10 and β-lapachone in cancer vs non-cancer cells this study.
Differences in cancer cell selective toxicity between BH10 and other quinones, such vitamin K3, indicate BH10 has a distinct mechanism of action to these molecules. Examination of oxidative stress markers between vitamin K3 and BH10 showed that BH10 increased mitochondrial PRDX3 oxidation to higher levels in cancer cells, whereas vitamin K3 decreased PRDX2 abundance and induced oxidative DNA damage in both cancer and non-cancerous cells. These results suggest that BH10 and vitamin K3 induce oxidative stress in different subcellular locations, which may underpin their differences in cytotoxicity between cancer and non-cancerous cells. Further development of quinones as anti-cancer agents may be able to exploit this phenotype to improve cancer cell selective toxicity and decrease unwanted side effects.
5. Conclusion
Our phenotypic screen identified a 1,4-Naphthoquinone molecule (BH10) that has a unique mechanism of action and improved cancer-selective toxicity compared to other anti-cancer quinones, piperlongumine, and molecules that target glucose metabolism. Metabolic profiling revealed that BH10 targets multiple metabolic pathways that are often considered ‘hallmarks’ of cancer [5]. Indeed, BH10 reversed aspects of cancer metabolism by decreasing glycolysis while simultaneously increasing glucose oxidation via mitochondrial and pentose phosphate pathways. BH10 disrupted redox balance in mitochondria without inducing cytoplasmic PRDX2 oxidation or causing nuclear DNA oxidative damage.
While it is clear that mitochondrial oxidative stress is involved in the mechanism linking BH10 to cancer cell death, a limitation of this study is that the mechanism, or mechanisms, of BH10-mediated mitochondrial redox regulation remains unclear. Future studies are required to identify BH10 targets and molecular marker/s that predict sensitivity to this molecule. Proteomics-based BH10-pulldown studies were unable to validate a specific interaction (not shown) and we cannot exclude other possibilities including that BH10 is redox cycled by a mitochondrial enzyme. However, this study demonstrates the potential of phenotypic drug screens employing oxygen consumption rate as a gain-of-function primary readout for the discovery of novel anti-cancer agents.
Grant support
FLB was supported by a postdoctoral fellowship from the Hope Funds for Cancer Research (HFCR-14-06-04) and is currently supported by a Cancer Institute NSW ECF (2018/ECF003). Financial support was provided in part by a UNSW faculty collaboration grant to KLH and NK. JM is supported by a Cancer Institute NSW CDF. GEM was supported by an Australian Postgraduate Award. KGRQ is supported by a UNSW Sydney Scientia Fellowship.
Declaration of competing interest
The authors declare no conflicts of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101374.
Contributor Information
Frances L. Byrne, Email: frances.byrne@unsw.edu.au.
Kyle L. Hoehn, Email: k.hoehn@unsw.edu.au.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Moffat J.G., Rudolph J., Bailey D. Phenotypic screening in cancer drug discovery — past, present and future. Nat. Rev. Drug Discov. 2014;13:588. doi: 10.1038/nrd4366. [DOI] [PubMed] [Google Scholar]
- 2.Swinney D.C., Anthony J. How were new medicines discovered? Nat. Rev. Drug Discov. 2011;10:507. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 3.DeBerardinis R.J., Chandel N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016;2 doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell Metabol. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Hay N. Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Nat. Rev. Cancer. 2016;16:635. doi: 10.1038/nrc.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brognard J., Clark A.S., Ni Y., Dennis P.A. Akt/protein kinase B is constitutively active in non-small cell lung cancer cells and promotes cellular survival and resistance to chemotherapy and radiation. Cancer Res. 2001;61:3986–3997. [PubMed] [Google Scholar]
- 8.Lai S.Y., Fuller C.D., Bhattacharya P.K., Frank S.J. Metabolic imaging as a biomarker of early radiation response in tumors. Clin. Cancer Res. 2015;21:4996–4998. doi: 10.1158/1078-0432.CCR-15-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chae Y.C. Mitochondrial Akt regulation of hypoxic tumor reprogramming. Cancer Cell. 2016;30:257–272. doi: 10.1016/j.ccell.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panieri E., Santoro M.M. ROS homeostasis and metabolism: a dangerous liaison in cancer cells. Cell Death Dis. 2016;7:e2253. doi: 10.1038/cddis.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liberti M.V., Locasale J.W. The Warburg effect: how does it benefit cancer cells? Trends Biochem. Sci. 2016;41:211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chow J.D. Genetic inhibition of hepatic acetyl-CoA carboxylase activity increases liver fat and alters global protein acetylation. Mol. Metab. 2014;3:419–431. doi: 10.1016/j.molmet.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kenwood B.M. Identification of a novel mitochondrial uncoupler that does not depolarize the plasma membrane. Mol. Metab. 2014;3:114–123. doi: 10.1016/j.molmet.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou T.-C., Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzym. Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 15.Dietmair S., Timmins N.E., Gray P.P., Nielsen L.K., Kromer J.O. Towards quantitative metabolomics of mammalian cells: development of a metabolite extraction protocol. Anal. Biochem. 2010;404:155–164. doi: 10.1016/j.ab.2010.04.031. [DOI] [PubMed] [Google Scholar]
- 16.Quek L.E., Liu M., Joshi S., Turner N. Fast exchange fluxes around the pyruvate node: a leaky cell model to explain the gain and loss of unlabelled and labelled metabolites in a tracer experiment. Cancer Metabol. 2016;4:13. doi: 10.1186/s40170-016-0153-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brun M.-P. Design, synthesis, and biological evaluation of novel naphthoquinone derivatives with CDC25 phosphatase inhibitory activity. Bioorg. Med. Chem. 2005;13:4871–4879. doi: 10.1016/j.bmc.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Raj L. Selective killing of cancer cells by a small molecule targeting the stress response to ROS. Nature. 2011;475:231–234. doi: 10.1038/nature10167. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Wellington K.W. Understanding cancer and the anticancer activities of naphthoquinones - a review. RSC Adv. 2015;5:20309–20338. [Google Scholar]
- 20.Michelakis E.D., Webster L., Mackey J.R. Dichloroacetate (DCA) as a potential metabolic-targeting therapy for cancer. Br. J. Canc. 2008;99:989–994. doi: 10.1038/sj.bjc.6604554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yun J. Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science. 2015;350:1391–1396. doi: 10.1126/science.aaa5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Byrne F.L. Metabolic vulnerabilities in endometrial cancer. Cancer Res. 2014;74:5832–5845. doi: 10.1158/0008-5472.CAN-14-0254. [DOI] [PubMed] [Google Scholar]
- 23.Rhee S.G., Woo H.A., Kil I.S., Bae S.H. Peroxiredoxin functions as a peroxidase and a regulator and sensor of local peroxides. J. Biol. Chem. 2012;287:4403–4410. doi: 10.1074/jbc.R111.283432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pia Rigobello M. Gold complexes inhibit mitochondrial thioredoxin reductase: consequences on mitochondrial functions. J. Inorg. Biochem. 2004;98:1634–1641. doi: 10.1016/j.jinorgbio.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 25.Lee K.W. Peroxiredoxin II restrains DNA damage-induced death in cancer cells by positively regulating JNK-dependent DNA repair. J. Biol. Chem. 2011;286:8394–8404. doi: 10.1074/jbc.M110.179416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valavanidis A., Vlachogianni T., Fiotakis C. 8-hydroxy-2′ -deoxyguanosine (8-OHdG): a critical biomarker of oxidative stress and carcinogenesis. J. Environ. Sci. Health Part C. 2009;27:120–139. doi: 10.1080/10590500902885684. [DOI] [PubMed] [Google Scholar]
- 27.Li D. Oxidative DNA damage and 8-hydroxy-2-deoxyguanosine DNA glycosylase/apurinic lyase in human breast cancer. Mol. Carcinog. 2001;31:214–223. doi: 10.1002/mc.1056. [DOI] [PubMed] [Google Scholar]
- 28.Xu K. Design and synthesis of naphthoquinone derivatives as antiproliferative agents and 20S proteasome inhibitors. Bioorg. Med. Chem. Lett. 2012;22:2772–2774. doi: 10.1016/j.bmcl.2012.02.086. [DOI] [PubMed] [Google Scholar]
- 29.Hsu T.S. 7-Chloro-6-piperidin-1-yl-quinoline-5,8-dione (PT-262), a novel synthetic compound induces lung carcinoma cell death associated with inhibiting ERK and CDC2 phosphorylation via a p53-independent pathway. Cancer Chemother. Pharmacol. 2008;62:799–808. doi: 10.1007/s00280-007-0667-5. [DOI] [PubMed] [Google Scholar]
- 30.Hsu T.S. Erratum to: cancer chemother pharmacol. Cancer Chemother. Pharmacol. 2008;62 doi:10.1007/s00280-008-0718-6. [Google Scholar]
- 31.Raukas M. Mitochondrial oxidative stress index, activity of redox-sensitive aconitase and effects of endogenous anti- and pro-oxidants on its activity in control, Alzheimer's disease and Swedish Familial Alzheimer's disease brain. Free Radic. Res. 2012;46:1490–1495. doi: 10.3109/10715762.2012.728286. [DOI] [PubMed] [Google Scholar]
- 32.Yan L.J., Levine R.L., Sohal R.S. Oxidative damage during aging targets mitochondrial aconitase. Proc. Natl. Acad. Sci. U. S. A. 1997;94:11168–11172. doi: 10.1073/pnas.94.21.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kemp R.G., Fox R.W., Latshaw S.P. Amino acid sequence at the citrate allosteric site of rabbit muscle phosphofructokinase. Biochemistry. 1987;26:3443–3446. doi: 10.1021/bi00386a029. [DOI] [PubMed] [Google Scholar]
- 34.Bansal A., Simon M.C. Glutathione metabolism in cancer progression and treatment resistance. J. Cell Biol. 2018;217:2291–2298. doi: 10.1083/jcb.201804161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berghe T.V., Linkermann A., Jouan-Lanhouet S., Walczak H., Vandenabeele P. Regulated necrosis: the expanding network of non-apoptotic cell death pathways. Nat. Rev. Mol. Cell Biol. 2014;15:135. doi: 10.1038/nrm3737. [DOI] [PubMed] [Google Scholar]
- 36.Huang X. Leveraging an NQO1 bioactivatable drug for tumor-selective use of poly(ADP-ribose) polymerase inhibitors. Cancer Cell. 2016;30:940–952. doi: 10.1016/j.ccell.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silvers M.A. The NQO1 bioactivatable drug, β-lapachone, alters the redox state of NQO1+ pancreatic cancer cells, causing perturbation in central carbon metabolism. J. Biol. Chem. 2017;292:18203–18216. doi: 10.1074/jbc.M117.813923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.