Abstract
Fish scales are mineralized structures that play important roles in protection and mineral homeostasis. This tissue expresses multiple estrogen receptor subtypes and can be targeted by estrogens or estrogenic endocrine-disrupting compounds, but their effects are poorly explored. The transcriptome data here presented support the findings reported in the research article “Genistein and estradiol have common and specific impacts on the sea bass (Dicentrarchus labrax) skin-scale barrier” [1]. Juvenile sea bass were exposed to estradiol and the phytoestrogen genistein for 1 and 5 days, by intraperitoneal injections, and the effects on scale transcript expression were analysed by RNA-seq using an Illumina Hi-seq 1500. The raw reads of the 30 libraries produced have been deposited in the NCBI-SRA database with the project accession number SRP102504. Mapping of RNA-seq reads against the sea bass reference genome using the Cufflinks/TopHat package identified 371 genes that had significant (FDR<0.05) differential expression with the estradiol or genistein treatments in relation to the control scales at each exposure time, 254 of which presented more than a 2-fold change in expression. The identity of the differentially expressed genes was obtained using both automatic and manual annotations against multiple public sequence databases and they were grouped according to their patterns of expression using hierarchical clustering and heat-maps. The biological processes and KEGG pathways most significantly affected by the estradiol and/or genistein treatments were identified using Cytoscape/ClueGO enrichment analyses.
Keywords: Estrogen-responsive genes, Genistein-responsive genes, Fish scale, RNA-seq, Transcriptome
Specifications Table
| Subject area | Biology |
| More specific subject area | Aquaculture, Ecotoxicology, Environment, Marine fisheries |
| Type of data | Tables, figures |
| How data was acquired | Illumina Hi-Seq 1500 |
| Data format | Raw, metadata |
| Experimental factors | Marine cultured sea bass were exposed to estradiol and genistein by intraperitoneal injection and their scales collected after 1 or 5 days |
| Experimental features | RNA extraction, quality evaluation, RNA-seq library preparation, sequencing and bioinformatics analysis |
| Data source location | Faro, Algarve, Portugal (37° 1′ 0″ N;7° 56′ 0″W) |
| Data accessibility |
Data is available in this article and at the NCBI Sequence Read Archive (SRA), project SRP102504 (https://www.ncbi.nlm.nih.gov/sra/SRP102504). The links for each dataset of replicates for the six experimental groups are:https://www.ncbi.nlm.nih.gov/sra/SRX2673957[accn] for C1d,https://www.ncbi.nlm.nih.gov/sra/SRX2673962[accn] for E1d,https://www.ncbi.nlm.nih.gov/sra/SRX2674307[accn] for Gen1d,https://www.ncbi.nlm.nih.gov/sra/SRX2674405[accn] for C5d,https://www.ncbi.nlm.nih.gov/sra/SRX2674467[accn] for E5d andhttps://www.ncbi.nlm.nih.gov/sra/SRX2675383[accn] for Gen5d. |
| Related research article | “Genistein and estradiol have common and specific impacts on the sea bass (Dicentrarchus labrax) skin-scale barrier” [1]. |
Value of the Data
|
1. Data
Table 1 presents the detailed RNA-seq sequence statistics for each of the thirty replicate libraries that were constructed from sea bass scale RNA, grouped according to treatment: fish exposed to estradiol, E2 (E1d and E5d libraries, corresponding to 1 or 5 days of exposure respectively); fish exposed to genistein, Gen (Gen1d and Gen5d) and control fish (C1d and C5d). The transcriptome data released at the NCBI SRA database (Project SRP102504) contains the raw data files of the 30 RNA-seq libraries that were produced from the scales of E2 or Gen-exposed sea bass for each of the six experimental conditions: C1d (experiment with accession SRX2673957, n = 6 replicate libraries), E1d (SRX2673962, n = 5 libraries), Gen1d (SRX2674307, n = 5 libraries), C5d (SRX2674405, n = 5 libraries), E5d (SRX2674467, n = 5 libraries) and Gen5d (SRX2675383, n = 4 libraries).
Table 1.
Detailed RNA-seq statistics for each library, grouped by treatment. Individual libraries (n = 4–6 from individual fish) were prepared from RNA extracted from scales of fish sampled 1 day after treatment with the vehicle (C1d), estradiol (E1d) or genistein (Gen1d) and scales sampled five days after each treatment (C5d, E5d and Gen5d, respectively). The number of raw and filtered reads (in millions), percentage of mapped reads (for each individual library or on average) and the total numbers of reads produced in the study are presented.
| Treatment | Lib name | Raw reads (millions) | Filtered reads (millions) | Mapped reads (%) |
|---|---|---|---|---|
| C1d | C1d_1 | 36.2 | 33.0 | 89.3% |
| C1d_2 | 32.6 | 32.6 | 84.2% | |
| C1d_3 | 35.7 | 35.7 | 85.6% | |
| C1d_4 | 32.0 | 32.0 | 85.1% | |
| C1d_5 | 32.1 | 32.1 | 86.4% | |
| C1d_6 | 36.1 | 36.1 | 86.2% | |
| E1d | E1d_1 | 31.5 | 31.5 | 87.2% |
| E1d_2 | 37.6 | 37.6 | 84.6% | |
| E1d_3 | 37.6 | 37.6 | 84.8% | |
| E1d_4 | 39.2 | 39.2 | 84.1% | |
| E1d_5 | 31.6 | 28.3 | 83.1% | |
| Gen1d | Gen1d_1 | 41.6 | 41.6 | 84.0% |
| Gen1d_2 | 38.5 | 35.1 | 85.4% | |
| Gen1d_3 | 44.4 | 44.4 | 86.1% | |
| Gen1d_4 | 30.4 | 30.4 | 83.7% | |
| Gen1d_5 | 39.2 | 38.0 | 84.5% | |
| C5d | C5d_1 | 40.9 | 40.9 | 84.6% |
| C5d_2 | 30.7 | 30.7 | 87.3% | |
| C5d_3 | 32.1 | 32.1 | 86.6% | |
| C5d_4 | 29.3 | 29.3 | 88.3% | |
| C5d_5 | 45.8 | 45.8 | 85.6% | |
| E5d | E5d_1 | 35.4 | 35.4 | 85.6% |
| E5d_2 | 36.1 | 36.1 | 85.9% | |
| E5d_3 | 38.8 | 26.2 | 87.0% | |
| E5d_4 | 51.0 | 49.0 | 88.9% | |
| E5d_5 | 44.0 | 40.8 | 91.4% | |
| Gen5d | Gen5d_1 | 44.0 | 44.0 | 84.5% |
| Gen5d_2 | 40.9 | 40.9 | 86.1% | |
| Gen5d_3 | 42.0 | 42.0 | 85.9% | |
| Gen5d_4 | 44.3 | 44.3 | 85.7% | |
| Average | 37.7 | 36.8 | ||
| Total | 1131.7 | 1102.8 |
A total of 749 differentially expressed (DE) genes were identified. DE genes in the scale transcriptomes of E2-treated or Gen-treated sea bass were identified by comparison with the corresponding controls at each sampling time and genes changing expression in the control groups when comparing 1 day vs 5 days of exposure. The expression levels, fold changes and identities of the 332 DE genes in sea bass scales, that had more than a two-fold change in expression (“DE ≥ 2 FC”), are listed in Supplementary Table 1. A short version of this table (containing only the first 20 genes) is displayed in Table 2. Supplementary Table 2 contains the remaining 417 DE genes (FDR < 0.05 and < 2-fold changes) and an abbreviated version is presented in Table 3. Table 4 presents the detailed results from the annotation of the total list of 749 DE genes (that were used for global enrichment analyses), as well as for the selection of the 332 DE genes with ≥ 2-fold change. Fig. 1 summarizes the preference order strategy used to automatically annotate the 332 selected DE genes using multiple databases, which was followed by careful manual curation. More details about the annotation strategy can be found in the methods below and in the methods and results of the associated JSBMB manuscript [1]. Fig. 2 summarizes the number of genes that were DE in response to the treatments (E2 or Gen) or changed expression between sampling times, when considering different stringency levels: False Discovery Rate (FDR) < 0.05 and < or ≥ 2-fold change in expression. Fig. 3 presents a heatmap showing the grouping of the six treatment groups, according to the identified transcriptome changes in sea bass scales.
Table 2.
Selected list (first 20 genes) of the genes differentially expressed with a ≥2-fold difference in sea bass scales from treated versus control animals. The complete list with the expression and annotation details for the 332 genes with significant differential expression (FDR < 0.05 and FC ≥ 2) in each comparison can be consulted in Supplementary Table 1. Normalized expression levels in each experimental group are presented in fragments per kilobase of exon model per million reads mapped (fpkm) and significant differential expression for each comparison is presented in Log2 fold change (FC). The final annotation reached using the preference order strategy (Fig. 1 and Table 4) and revised by manual curation is presented, with the accession numbers from Swiss-Prot, Genbank or from the sea bass genome.
| Gene ID |
Relative expression levels (fpkm) |
Log2FC of E2 or Gen effects vs control at 1d |
Log2FC of E2 or Gen effects vs control at 5d |
Log2FC of C5d vs C1d |
Final gene annotation |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cufflinks# | C1d | E1d | Gen1d | C5d | E5d | Gen5d | up in E2 | Down in E2 | Up in Gen | Down in Gen | E2 up | E2 down | Gen up | Gen down | C5d up | C5d down | Accession | Hit description | Symbol |
| XLOC_019555 | 5.4 | 14.8 | 37.7 | 33.7 | 5.3 | 34.0 | 1.4 | – | 2.8 | – | – | −2.7 | – | – | 2.6 | – | O75443 | Alpha-tectorin | TECTA |
| XLOC_019129 | 34.2 | 225.7 | 761.3 | 1735.6 | 22.0 | 1398.8 | 2.7 | – | 4.5 | – | – | −6.3 | – | – | 5.7 | – | DLAgn_00248120 | Mhc class ii antigen beta chain | HLADPB1 |
| XLOC_020058 | 0.5 | 3.8 | 4.0 | 3.1 | 6.2 | 6.0 | 2.9 | – | 2.9 | – | – | – | – | – | 2.6 | – | Q5W7F1 | Neutral ceramidase or N-acylsphingosine amidohydrolase 2 | ASAH2 |
| XLOC_019225 | 25.2 | 63.2 | 50.8 | 25.0 | 17.3 | 22.9 | 1.3 | – | 1.0 | – | – | – | – | – | – | – | Q02817 | Mucin-2 | MUC2 |
| XLOC_019447 | 10.8 | 22.6 | 22.4 | 12.0 | 13.6 | 11.7 | 1.1 | – | 1.0 | – | – | – | – | – | – | – | XP_019109670.1 | Integumentary mucin C.1-like | MUCC1 |
| XLOC_019084 | 20.6 | 47.4 | 38.1 | 15.9 | 14.5 | 40.0 | 1.2 | – | – | – | – | – | 1.3 | – | – | – | G8HTB6 | ZP domain-containing protein / CUB and zona pellucida like domains 1 | CUZD1 |
| XLOC_019425 | 437.4 | 1926.9 | 1162.0 | 1718.5 | 2043.4 | 2161.0 | 2.1 | – | – | – | – | – | – | – | – | – | P28064 | Proteasome subunit beta type-8 | PSMB8 |
| XLOC_021846 | 92.1 | 215.6 | 136.4 | 206.0 | 144.3 | 113.2 | 1.2 | – | – | – | – | – | – | – | 1.2 | – | – | No hit found | – |
| XLOC_020866 | 5.0 | 19.1 | 5.9 | 0.6 | 0.4 | 1.1 | 1.9 | – | – | – | – | – | – | – | – | – | DLAgn_00214300 | Protein fam111a-like | FAM111A |
| XLOC_009298 | 49.5 | 135.9 | 60.4 | 73.6 | 55.8 | 70.0 | 1.5 | – | – | – | – | – | – | – | – | – | DLAgn_00110020 | Uncharacterized protein loc101483146 | hyp_loc101483146 |
| XLOC_020865 | 260.9 | 1274.3 | 269.3 | 12.9 | 5.6 | 19.7 | 2.0 | – | – | – | – | – | – | – | – | – | DLAgn_00214300 | Protein fam111a-like | FAM111A |
| XLOC_018302 | 1.6 | 3.9 | 2.2 | 1.6 | 3.5 | 1.5 | 1.3 | – | – | – | – | – | – | – | – | – | XP_006805317.1 | Uncharacterized protein LOC102781223 | hyp_LOC102781223 |
| XLOC_001638 | 3.9 | 4.2 | 9.2 | 3.2 | 6.8 | 7.9 | – | – | 1.2 | – | 1.1 | – | 1.3 | – | – | – | P35448 | Thrombospondin-1 | THBS1 |
| XLOC_009985 | 1.1 | 0.6 | 2.8 | 0.6 | 1.9 | 2.9 | – | – | 1.4 | – | – | – | 2.2 | – | – | – | P43300 | Early growth response protein 3 | EGR3 |
| XLOC_009459 | 2.8 | 2.5 | 5.8 | 2.4 | 4.4 | 6.1 | – | – | 1.0 | – | – | – | 1.4 | – | – | – | Q9ET55 | Nocturnin | CCRN4L |
| XLOC_015202 | 6.1 | 6.1 | 19.3 | 3.5 | 5.0 | 8.4 | – | – | 1.7 | – | – | – | 1.3 | – | – | – | Q16690 | Dual specificity protein phosphatase 5 | DUSP5 |
| XLOC_009646 | 8.0 | 5.6 | 19.0 | 3.0 | 7.6 | 13.2 | – | – | 1.2 | – | – | – | 2.2 | – | – | −1.4 | Q20A00 | DNA damage-inducible transcript 4-like protein | DDIT4L |
| XLOC_018946 | 261.3 | 451.9 | 567.7 | 482.9 | 396.7 | 190.1 | – | – | 1.1 | – | – | – | – | −1.3 | – | – | DLAgn_00241010 | Complement c1q-like protein 4 precursor | C1QL4 |
| XLOC_011902 | 6.6 | 8.2 | 13.4 | 20.8 | 25.4 | 9.1 | – | – | 1.0 | – | – | – | – | −1.2 | 1.7 | – | XP_005946242.1 | RNA-directed DNA polymerase from mobile element jockey-like | pred_pol413 |
| XLOC_002574 | 0.8 | 1.4 | 2.0 | 0.8 | 0.9 | 1.0 | – | – | 1.3 | – | – | – | – | – | – | – | Q61391 | Neprilysin / membrane metallo-endopeptidase | MME |
Table 3.
Selected list (first 20 genes) of the genes differentially expressed at FDR < 0.05 and at < 2-fold change difference, in sea bass scales from treated versus control animals. The complete list with the expression and annotation for the 417 genes with differential expression under these limits (FDR < 0.05 and FC < 2) can be consulted in Supplementary Table 2. Normalized expression levels in each experimental group are presented in fragments per kilobase of exon model per million reads mapped (fpkm) and significant differential expression for each comparison is presented in Log2 fold change (FC). The final annotation reached using the preference order strategy (Fig. 1 and Table 4) is presented, with the accession numbers from Swiss-Prot, Genbank or from the sea bass genome.
| Gene ID |
Relative expression levels (fpkm) |
Log2FC of E2 or Gen effects vs control at 1d |
Log2FC of E2 or Gen effects vs control at 5d |
Log2FC of C5d vs C1d |
Final gene annotation |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cufflinks# | C1d | E1d | Gen1d | C5d | E5d | Gen5d | E2 up | E2 down | Gen up | Gen down | E2 up | E2 down | Gen up | Gen down | C5d up | C5d down | Hit description | Symbol |
| XLOC_017886 | 20.5 | 32.3 | 35.2 | 21.7 | 28.2 | 26.6 | 0.7 | – | 0.8 | – | – | – | – | – | – | – | sp|P51890|LUM_CHICK Lumican OS=Gallus gallus | LUM |
| XLOC_009066 | 20.1 | 32.2 | 23.6 | 14.9 | 18.8 | 16.6 | 0.7 | – | – | – | – | – | – | – | – | – | sp|Q6NVM0|H10_XENTR Histone H1.0 | H1F0 |
| XLOC_000057 | 39.9 | 63.9 | 54.8 | 51.8 | 66.1 | 52.3 | 0.7 | – | – | – | – | – | – | – | – | – | sp|Q04857|CO6A1_MOUSE Collagen alpha-1(VI) chain | COL6A1 |
| XLOC_014484 | 17.5 | 28.2 | 25.8 | 20.4 | 14.9 | 17.2 | 0.7 | – | – | – | – | – | – | – | – | – | sp|Q53RD9|FBLN7_HUMAN Fibulin-7 | FBLN7 |
| XLOC_019995 | 42.4 | 68.5 | 41.0 | 32.9 | 42.9 | 22.8 | 0.7 | – | – | – | – | – | – | – | – | – | sp|P55918|MFAP4_BOVIN Microfibril-associated glycoprotein 4 | MFAP4 |
| XLOC_006221 | 7.3 | 11.8 | 7.0 | 7.3 | 10.5 | 6.6 | 0.7 | – | – | – | – | – | – | – | – | – | sp|Q4R6P7|SESN1_MACFA Sestrin-1 | SESN1 |
| XLOC_002145 | 58.5 | 95.2 | 82.9 | 108.5 | 94.2 | 73.3 | 0.7 | – | – | – | – | – | – | – | 0.9 | – | sp|O95428|PPN_HUMAN Papilin | PAPLN |
| XLOC_022309 | 58.4 | 96.0 | 62.0 | 45.3 | 81.2 | 35.5 | 0.7 | – | – | – | 0.8 | – | – | – | – | – | sp|Q9U8W8|TL5A_TACTR Techylectin-5A | – |
| XLOC_003581 | 10.8 | 18.6 | 11.2 | 12.6 | 8.8 | 6.9 | 0.8 | – | – | – | – | – | – | −0.9 | – | – | sp|Q802Y8|ZB16A_DANRE Zinc finger and BTB domain-containing protein 16-A | ZBTB16 |
| XLOC_011142 | 1033.0 | 1793.0 | 979.6 | 1063.6 | 1387.2 | 709.6 | 0.8 | – | – | – | – | – | – | – | – | – | sp|Q66S03|LECG_THANI Galactose-specific lectin nattectin | LADD |
| XLOC_013333 | 40.4 | 70.4 | 59.7 | 50.9 | 50.7 | 48.9 | 0.8 | – | – | – | – | – | – | – | – | – | sp|Q90611|MMP2_CHICK 72 kDa type IV collagenase | MMP2 |
| XLOC_020568 | 323.2 | 571.3 | 392.8 | 275.6 | 447.7 | 280.9 | 0.8 | – | – | – | – | – | – | – | – | – | gap junction epsilon-1 | – |
| XLOC_016220 | 5.0 | 8.9 | 6.1 | 6.5 | 5.6 | 5.4 | 0.8 | – | – | – | – | – | – | – | – | – | sp|Q568Y7|NOE2_RAT Noelin-2 | OLFM2 |
| XLOC_006639 | 175.0 | 315.2 | 258.5 | 221.0 | 268.7 | 245.3 | 0.8 | – | – | – | – | – | – | – | – | – | sp|Q01584|LIPO_BUFMA Lipocalin | LCN1 |
| XLOC_015159 | 46.9 | 88.4 | 67.0 | 84.6 | 64.2 | 95.1 | 0.9 | – | – | – | – | – | – | – | 0.9 | – | sp|P27590|UROM_RAT Uromodulin | UMOD |
| XLOC_019689 | 26.0 | 49.4 | 26.6 | 17.3 | 18.0 | 17.4 | 0.9 | – | – | – | – | – | – | – | – | – | No hit found | – |
| XLOC_014167 | 3.2 | 6.1 | 5.1 | 3.4 | 2.6 | 2.7 | 1.0 | – | – | – | – | – | – | – | – | – | sp|Q5E9P5|PAMR1_BOVIN Inactive serine protease PAMR1 OS=Bos taur... 413 e-114 | PAMR1 |
| XLOC_022211 | 101.7 | 197.8 | 148.2 | 85.2 | 76.3 | 155.6 | 1.0 | – | – | – | – | – | – | – | – | – | No hit found | – |
| XLOC_008732 | 3.6 | 6.9 | 6.7 | 5.9 | 4.9 | 5.3 | 1.0 | – | 0.9 | – | – | – | – | – | – | – | sp|Q9BQB4|SOST_HUMAN Sclerostin | SOST |
| XLOC_018728 | 132.5 | 258.4 | 160.0 | 121.5 | 235.3 | 99.0 | 1.0 | – | – | – | 1.0 | – | – | – | – | – | No hit found | – |
Table 4.
Detailed results from the annotation. Annotation results (in number of genes or percentage from total) are shown for the 749 genes differentially expressed with q < 0.05 (columns “DE all”) and for the selection of 332 genes differentially expressed at q < 0.05 with a minimum 2-fold change (columns “DE ≥ 2 FC”).
| DE all |
DE ≥ 2 FC |
|||
|---|---|---|---|---|
| Number | % | Number | % | |
| Annotation to different databases: | ||||
| Genes with BlastX hit to SwissProt | 593 | 79 | 229 | 68 |
| Genes assigned to predicted genes in genome | 600 | 80 | 275 | 83 |
| Genes with BlastX hit to GenBank | 676 | 90 | 283 | 85 |
| Final annotation using preference order: | ||||
| Genes annotated via SwissProt | 593 | 79 | 229 | 68 |
| Genes annotated via the sea bass genome | 92 | 12 | 61 | 19 |
| Genes annotated via GenBank | 34 | 5 | 24 | 7 |
| Annotated | 719 | 96 | 314 | 95 |
| Non-annotated | 30 | 4 | 18 | 5 |
| Total number of DE genes | 749 | 100 | 332 | 100 |
| Summary of annotation: | ||||
| Annotation to known proteins | 667 | 89 | 280 | 84 |
| Annotation to predicted proteins | 21 | 3 | 12 | 4 |
| Annotation to hypothetical/uncharacterized proteins | 20 | 3 | 14 | 4 |
| Mapping to non-annotated genes | 11 | 1 | 8 | 2 |
Fig. 1.
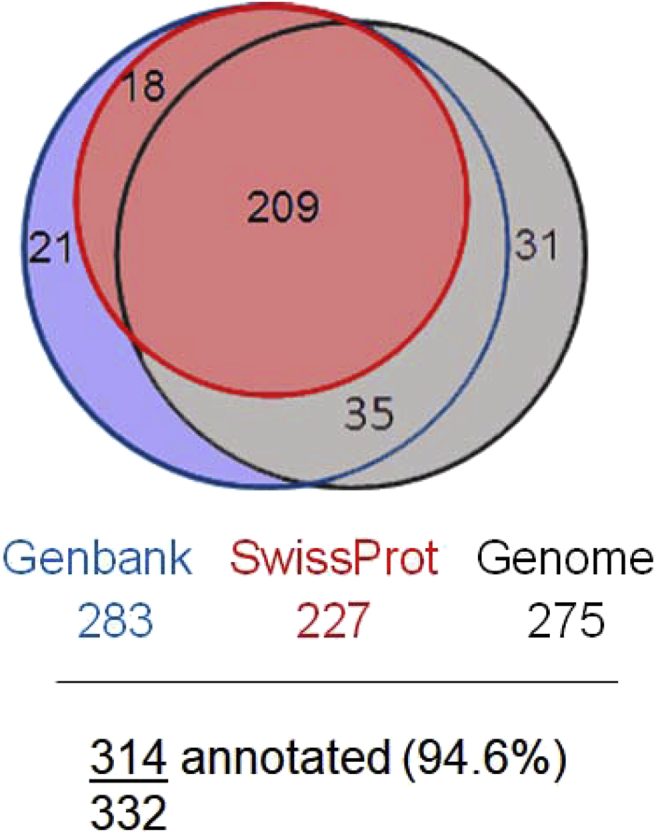
Annotation to different databases of the 332 genes found to be ≥ 2-fold differentially expressed. Venn diagrams indicate the number of genes with a significant match to the sea bass genome, Swiss-Prot protein database or GeneBank protein database, the number of genes annotated by more than one database are inside the intersecting areas. The annotation was carried out in order of preference; 1) matches to Swiss-Prot, 2) matches to the sea bass genome and 3) Genbank matches as indicated by the colour shading. The areas of each sphere are proportional to the number of genes annotated.
Fig. 2.
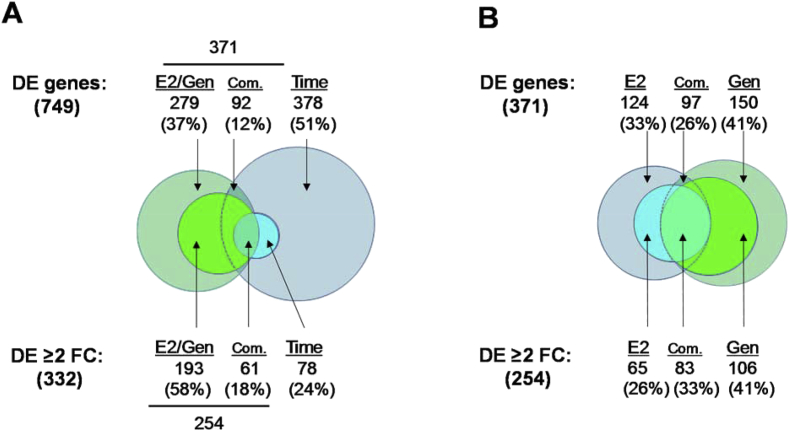
Proportion of differentially expressed (DE) genes identified in the present study. A. Venn diagrams representing common (Com.) and specific genes differentially expressed in sea bass scales in response to the treatments (17β-estradiol, E2, and/or genistein, Gen, compared to the corresponding controls at each sampling time 1 day and 5 days), compared to the differential expression in the control groups over time. The number of genes by treatment or time and their respective percentage are shown for two levels of stringency. “DE genes” above the diagram corresponds to the 749 genes differentially expressed at an FDR <0.05. Below the diagram the DE genes (332) differentially expressed with a minimum of 2-fold change are considered (“DE ≥ 2 FC” and FDR < 0.05). 51% of the “DE genes” changed expression only in control scales over time but these changes were of low magnitude (average fold change of 1.9-fold between C1d and C5d) and when the analysis stringency was increased to a minimum of 2-fold change, only 24% of these 332 genes changed expression in the control scales over time. The 254 genes (76%) significantly regulated by E2 or Gen were selected for further analyses. B. Shows the comparison of common/specific DE genes between E2 and Gen at the two stringency levels, FDR <0.05 or ≥2 FC and FDR <0.05 (irrespective of the sampling times). For the number of genes regulated by E2 and/or Gen at each sampling time see Fig. 2B of the associated paper in JSBMB [1].
Fig. 3.
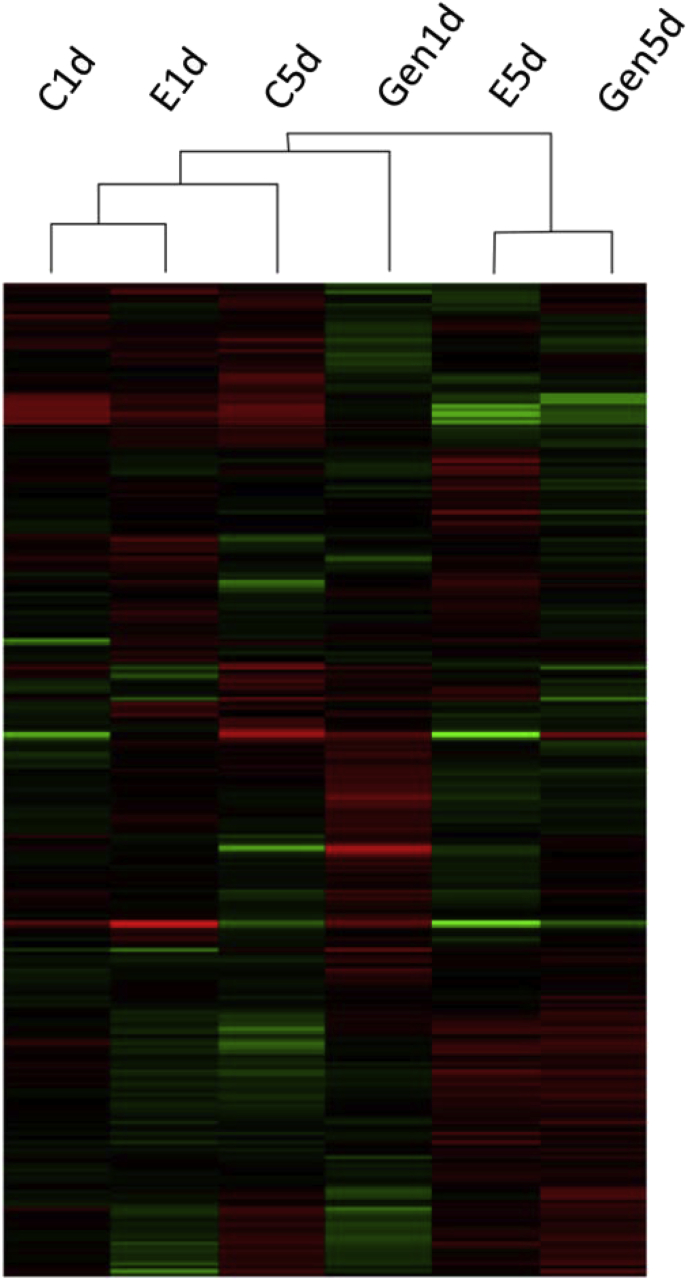
Heatmap of clustered DE genes identified in sea bass scales after treatment with E2 or Gen. The tree in the upper panel shows the hierarchical clustering of the DE genes (one gene/line) identified in the scales of the six treatment groups [injections with estradiol (E), genistein (G) or vehicle only (control, C) at 1 or 5 days (1d or 5d)]. The red gradient indicates high abundance, the green gradient indicates low abundance and black indicates equal abundance for each gene and condition relative to the average. 1 day after treatment the DE genes of the E2 group clustered more closely to the control than Gen1d. 5 days after treatment both E2-and Gen-treated scales clearly separated from the control and clustered together, suggesting a similar response at this time point.
Table 5, Table 6 present the significantly enriched GO Biological Processes (GO-BP) and KEGG pathways, respectively, of all genes that presented significant changes in expression in response to the treatments E2 and/or Gen (analysis “All”); Table 7, Table 8 present the significantly enriched GO-BP and KEGGs when directly comparing the E2-or Gen-responsive genes, irrespective of the sampling time (analyses “E2” vs “Gen”); Table 9, Table 10 list the significantly enriched GO-BP and KEGGs when comparing responsive gene lists between 1 day and 5 days (analyses “1d” vs “5d”).
Table 5.
Enrichment of GO Biological Processes (GO-BP) using all genes found to be differentially expressed in response to E2 and/or Gen (analysis “All”). Significantly enriched biological processes (FDR < 0.05) were identified by ID and term description and grouped into 23 functionally related networks (GO group) obtained by ClueGO analysis. Each group is named after its most significant term (lowest FDR) and highlighted in bold, which was chosen for GOTerm representation in Fig. 3 of the associated MS in JSBMB [1]. Functionally related groups are sorted by highest enrichment score, calculated as [ -Log2 (group FDR)].
| GOID | GOTerm | Term FDR | Group FDR | Enrichment Score | GO group | % Associated Genes | Nr. Genes |
|---|---|---|---|---|---|---|---|
| GO:0043207 | response to external biotic stimulus | 7.2E-10 | 1.8E-10 | 32.4 | 18 | 8.54 | 21.00 |
| GO:0051707 | response to other organism | 7.2E-10 | 1.8E-10 | 32.4 | 18 | 8.54 | 21.00 |
| GO:0009615 | response to virus | 3.8E-05 | 1.8E-10 | 32.4 | 18 | 12.96 | 7.00 |
| GO:0009617 | response to bacterium | 1.5E-03 | 1.8E-10 | 32.4 | 18 | 5.20 | 9.00 |
| GO:0051607 | defense response to virus | 2.1E-02 | 1.8E-10 | 32.4 | 18 | 7.14 | 3.00 |
| GO:0043207 | response to external biotic stimulus | 7.2E-10 | 5.9E-09 | 27.3 | 20 | 8.54 | 21.00 |
| GO:0051707 | response to other organism | 7.2E-10 | 5.9E-09 | 27.3 | 20 | 8.54 | 21.00 |
| GO:0051591 | response to cAMP | 4.8E-05 | 5.9E-09 | 27.3 | 20 | 75.00 | 3.00 |
| GO:0009617 | response to bacterium | 1.5E-03 | 5.9E-09 | 27.3 | 20 | 5.20 | 9.00 |
| GO:0046683 | response to organophosphorus | 2.3E-03 | 5.9E-09 | 27.3 | 20 | 20.00 | 3.00 |
| GO:0014074 | response to purine-containing compound | 2.3E-03 | 5.9E-09 | 27.3 | 20 | 20.00 | 3.00 |
| GO:0032496 | response to lipopolysaccharide | 2.7E-03 | 5.9E-09 | 27.3 | 20 | 8.33 | 5.00 |
| GO:0002237 | response to molecule of bacterial origin | 3.1E-03 | 5.9E-09 | 27.3 | 20 | 7.94 | 5.00 |
| GO:0034097 | response to cytokine | 3.5E-03 | 5.9E-09 | 27.3 | 20 | 4.29 | 9.00 |
| GO:0006954 | inflammatory response | 8.2E-03 | 5.9E-09 | 27.3 | 20 | 4.32 | 7.00 |
| GO:0042493 | response to drug | 1.2E-02 | 5.9E-09 | 27.3 | 20 | 9.38 | 3.00 |
| GO:0009612 | response to mechanical stimulus | 2.5E-02 | 5.9E-09 | 27.3 | 20 | 6.67 | 3.00 |
| GO:0016126 | sterol biosynthetic process | 2.9E-09 | 3.0E-07 | 21.7 | 22 | 31.03 | 9.00 |
| GO:0006694 | steroid biosynthetic process | 4.0E-09 | 3.0E-07 | 21.7 | 22 | 19.30 | 11.00 |
| GO:0016125 | sterol metabolic process | 9.6E-09 | 3.0E-07 | 21.7 | 22 | 20.83 | 10.00 |
| GO:0008202 | steroid metabolic process | 1.0E-08 | 3.0E-07 | 21.7 | 22 | 14.63 | 12.00 |
| GO:1901617 | organic hydroxy compound biosynthetic process | 8.7E-07 | 3.0E-07 | 21.7 | 22 | 15.25 | 9.00 |
| GO:0008203 | cholesterol metabolic process | 2.6E-06 | 3.0E-07 | 21.7 | 22 | 20.59 | 7.00 |
| GO:1902652 | secondary alcohol metabolic process | 4.3E-06 | 3.0E-07 | 21.7 | 22 | 18.92 | 7.00 |
| GO:0006695 | cholesterol biosynthetic process | 1.1E-05 | 3.0E-07 | 21.7 | 22 | 31.25 | 5.00 |
| GO:1902653 | secondary alcohol biosynthetic process | 1.8E-05 | 3.0E-07 | 21.7 | 22 | 27.78 | 5.00 |
| GO:1901615 | organic hydroxy compound metabolic process | 4.4E-05 | 3.0E-07 | 21.7 | 22 | 7.69 | 10.00 |
| GO:0044283 | small molecule biosynthetic process | 8.0E-05 | 3.0E-07 | 21.7 | 22 | 5.85 | 12.00 |
| GO:0046165 | alcohol biosynthetic process | 1.3E-04 | 3.0E-07 | 21.7 | 22 | 17.86 | 5.00 |
| GO:0008610 | lipid biosynthetic process | 4.1E-04 | 3.0E-07 | 21.7 | 22 | 4.86 | 12.00 |
| GO:0006066 | alcohol metabolic process | 4.5E-04 | 3.0E-07 | 21.7 | 22 | 8.33 | 7.00 |
| GO:0016053 | organic acid biosynthetic process | 1.3E-03 | 3.0E-07 | 21.7 | 22 | 5.97 | 8.00 |
| GO:0046394 | carboxylic acid biosynthetic process | 1.3E-03 | 3.0E-07 | 21.7 | 22 | 5.97 | 8.00 |
| GO:1901607 | alpha-amino acid biosynthetic process | 1.4E-03 | 3.0E-07 | 21.7 | 22 | 10.42 | 5.00 |
| GO:0008652 | cellular amino acid biosynthetic process | 1.6E-03 | 3.0E-07 | 21.7 | 22 | 9.80 | 5.00 |
| GO:0031099 | regeneration | 3.0E-03 | 3.0E-07 | 21.7 | 22 | 4.94 | 8.00 |
| GO:0031329 | regulation of cellular catabolic process | 3.0E-03 | 3.0E-07 | 21.7 | 22 | 5.51 | 7.00 |
| GO:0009894 | regulation of catabolic process | 3.5E-03 | 3.0E-07 | 21.7 | 22 | 5.22 | 7.00 |
| GO:0006520 | cellular amino acid metabolic process | 6.5E-03 | 3.0E-07 | 21.7 | 22 | 4.15 | 8.00 |
| GO:0022600 | digestive system process | 6.9E-03 | 3.0E-07 | 21.7 | 22 | 12.00 | 3.00 |
| GO:1901605 | alpha-amino acid metabolic process | 7.2E-03 | 3.0E-07 | 21.7 | 22 | 5.08 | 6.00 |
| GO:0007586 | digestion | 1.2E-02 | 3.0E-07 | 21.7 | 22 | 9.38 | 3.00 |
| GO:0072330 | monocarboxylic acid biosynthetic process | 1.5E-02 | 3.0E-07 | 21.7 | 22 | 5.97 | 4.00 |
| GO:0031331 | positive regulation of cellular catabolic process | 1.7E-02 | 3.0E-07 | 21.7 | 22 | 5.71 | 4.00 |
| GO:0009896 | positive regulation of catabolic process | 1.9E-02 | 3.0E-07 | 21.7 | 22 | 5.48 | 4.00 |
| GO:0097164 | ammonium ion metabolic process | 2.9E-02 | 3.0E-07 | 21.7 | 22 | 6.25 | 3.00 |
| GO:0015748 | organophosphate ester transport | 3.5E-02 | 3.0E-07 | 21.7 | 22 | 5.77 | 3.00 |
| GO:0006633 | fatty acid biosynthetic process | 3.5E-02 | 3.0E-07 | 21.7 | 22 | 5.77 | 3.00 |
| GO:0001878 | response to yeast | 4.6E-06 | 8.8E-06 | 16.8 | 12 | 25.00 | 6.00 |
| GO:0009620 | response to fungus | 1.2E-05 | 8.8E-06 | 16.8 | 12 | 20.69 | 6.00 |
| GO:0031099 | regeneration | 3.0E-03 | 2.7E-04 | 11.9 | 19 | 4.94 | 8.00 |
| GO:0009611 | response to wounding | 3.3E-03 | 2.7E-04 | 11.9 | 19 | 4.33 | 9.00 |
| GO:0042060 | wound healing | 3.4E-03 | 2.7E-04 | 11.9 | 19 | 4.68 | 8.00 |
| GO:0022600 | digestive system process | 6.9E-03 | 2.7E-04 | 11.9 | 19 | 12.00 | 3.00 |
| GO:0007586 | digestion | 1.2E-02 | 2.7E-04 | 11.9 | 19 | 9.38 | 3.00 |
| GO:0042246 | tissue regeneration | 4.3E-02 | 2.7E-04 | 11.9 | 19 | 4.08 | 4.00 |
| GO:0007599 | hemostasis | 4.5E-02 | 2.7E-04 | 11.9 | 19 | 5.08 | 3.00 |
| GO:0007596 | blood coagulation | 4.5E-02 | 2.7E-04 | 11.9 | 19 | 5.08 | 3.00 |
| GO:0050817 | coagulation | 4.9E-02 | 2.7E-04 | 11.9 | 19 | 4.84 | 3.00 |
| GO:0030162 | regulation of proteolysis | 3.4E-03 | 5.6E-03 | 7.5 | 11 | 4.02 | 10.00 |
| GO:0045861 | negative regulation of proteolysis | 1.7E-02 | 5.6E-03 | 7.5 | 11 | 4.03 | 6.00 |
| GO:0006270 | DNA replication initiation | 6.9E-03 | 1.2E-02 | 6.4 | 0 | 12.00 | 3.00 |
| GO:0048593 | camera-type eye morphogenesis | 6.4E-03 | 1.4E-02 | 6.2 | 17 | 4.64 | 7.00 |
| GO:0048592 | eye morphogenesis | 7.3E-03 | 1.4E-02 | 6.2 | 17 | 4.04 | 8.00 |
| GO:0042462 | eye photoreceptor cell development | 1.9E-02 | 1.4E-02 | 6.2 | 17 | 7.50 | 3.00 |
| GO:0031076 | embryonic camera-type eye development | 3.1E-02 | 1.4E-02 | 6.2 | 17 | 6.12 | 3.00 |
| GO:0001754 | eye photoreceptor cell differentiation | 4.8E-02 | 1.4E-02 | 6.2 | 17 | 4.92 | 3.00 |
| GO:0006979 | response to oxidative stress | 3.7E-03 | 1.4E-02 | 6.2 | 16 | 7.25 | 5.00 |
| GO:1990748 | cellular detoxification | 3.1E-02 | 1.4E-02 | 6.2 | 16 | 6.12 | 3.00 |
| GO:0098869 | cellular oxidant detoxification | 3.1E-02 | 1.4E-02 | 6.2 | 16 | 6.12 | 3.00 |
| GO:0098754 | detoxification | 3.5E-02 | 1.4E-02 | 6.2 | 16 | 5.77 | 3.00 |
| GO:0031589 | cell-substrate adhesion | 9.6E-03 | 1.5E-02 | 6.1 | 10 | 7.02 | 4.00 |
| GO:0007160 | cell-matrix adhesion | 3.2E-02 | 1.5E-02 | 6.1 | 10 | 6.00 | 3.00 |
| GO:0006730 | one-carbon metabolic process | 1.4E-02 | 1.8E-02 | 5.8 | 4 | 8.57 | 3.00 |
| GO:0051241 | negative regulation of multicellular organismal process | 1.3E-02 | 1.8E-02 | 5.8 | 3 | 4.32 | 6.00 |
| GO:0009266 | response to temperature stimulus | 1.7E-02 | 2.1E-02 | 5.6 | 8 | 7.89 | 3.00 |
| GO:0072527 | pyrimidine-containing compound metabolic process | 1.9E-02 | 2.3E-02 | 5.4 | 5 | 7.50 | 3.00 |
| GO:1901136 | carbohydrate derivative catabolic process | 3.2E-02 | 3.9E-02 | 4.7 | 6 | 6.00 | 3.00 |
| GO:0009410 | response to xenobiotic stimulus | 3.5E-02 | 4.0E-02 | 4.6 | 9 | 5.77 | 3.00 |
| GO:0001945 | lymph vessel development | 3.8E-02 | 4.2E-02 | 4.6 | 2 | 5.56 | 3.00 |
| GO:0048864 | stem cell development | 4.1E-02 | 4.3E-02 | 4.5 | 13 | 4.17 | 4.00 |
| GO:0014031 | mesenchymal cell development | 4.1E-02 | 4.3E-02 | 4.5 | 13 | 4.17 | 4.00 |
| GO:0014032 | neural crest cell development | 4.1E-02 | 4.3E-02 | 4.5 | 13 | 4.17 | 4.00 |
| GO:0000188 | inactivation of MAPK activity | 3.4E-04 | 4.6E-02 | 4.4 | 21 | 22.22 | 4.00 |
| GO:0043407 | negative regulation of MAP kinase activity | 5.6E-04 | 4.6E-02 | 4.4 | 21 | 19.05 | 4.00 |
| GO:0043409 | negative regulation of MAPK cascade | 1.4E-03 | 4.6E-02 | 4.4 | 21 | 14.29 | 4.00 |
| GO:0043405 | regulation of MAP kinase activity | 2.0E-03 | 4.6E-02 | 4.4 | 21 | 7.32 | 6.00 |
| GO:0071901 | negative regulation of protein serine/threonine kinase activity | 2.1E-03 | 4.6E-02 | 4.4 | 21 | 12.50 | 4.00 |
| GO:0033673 | negative regulation of kinase activity | 2.7E-03 | 4.6E-02 | 4.4 | 21 | 6.67 | 6.00 |
| GO:0006469 | negative regulation of protein kinase activity | 2.7E-03 | 4.6E-02 | 4.4 | 21 | 6.74 | 6.00 |
| GO:0051348 | negative regulation of transferase activity | 3.3E-03 | 4.6E-02 | 4.4 | 21 | 6.25 | 6.00 |
| GO:0001933 | negative regulation of protein phosphorylation | 3.4E-03 | 4.6E-02 | 4.4 | 21 | 6.12 | 6.00 |
| GO:0042326 | negative regulation of phosphorylation | 3.6E-03 | 4.6E-02 | 4.4 | 21 | 6.00 | 6.00 |
| GO:1902532 | negative regulation of intracellular signal transduction | 8.0E-03 | 4.6E-02 | 4.4 | 21 | 4.96 | 6.00 |
| GO:0071900 | regulation of protein serine/threonine kinase activity | 8.2E-03 | 4.6E-02 | 4.4 | 21 | 4.92 | 6.00 |
| GO:0010563 | negative regulation of phosphorus metabolic process | 9.6E-03 | 4.6E-02 | 4.4 | 21 | 4.72 | 6.00 |
| GO:0045936 | negative regulation of phosphate metabolic process | 9.6E-03 | 4.6E-02 | 4.4 | 21 | 4.72 | 6.00 |
| GO:0031400 | negative regulation of protein modification process | 9.6E-03 | 4.6E-02 | 4.4 | 21 | 4.72 | 6.00 |
| GO:0001706 | endoderm formation | 1.6E-02 | 4.6E-02 | 4.4 | 21 | 8.11 | 3.00 |
| GO:0001704 | formation of primary germ layer | 1.9E-02 | 4.6E-02 | 4.4 | 21 | 5.48 | 4.00 |
| GO:0001666 | response to hypoxia | 4.4E-02 | 4.6E-02 | 4.4 | 15 | 5.17 | 3.00 |
| GO:0070482 | response to oxygen levels | 4.6E-02 | 4.6E-02 | 4.4 | 15 | 5.00 | 3.00 |
| GO:0036293 | response to decreased oxygen levels | 4.6E-02 | 4.6E-02 | 4.4 | 15 | 5.00 | 3.00 |
| GO:0009142 | nucleoside triphosphate biosynthetic process | 4.5E-02 | 4.7E-02 | 4.4 | 7 | 5.08 | 3.00 |
| GO:0033334 | fin morphogenesis | 4.5E-02 | 4.7E-02 | 4.4 | 1 | 5.08 | 3.00 |
| GO:0042541 | hemoglobin biosynthetic process | 1.7E-03 | 6.2E-02 | 4.0 | 14 | 23.08 | 3.00 |
| GO:0020027 | hemoglobin metabolic process | 2.0E-03 | 6.2E-02 | 4.0 | 14 | 21.43 | 3.00 |
| GO:0055076 | transition metal ion homeostasis | 4.9E-02 | 6.2E-02 | 4.0 | 14 | 4.84 | 3.00 |
Table 6.
Enrichment of KEGG pathways using all genes found to be differentially expressed in response to E2 and/or Gen (analysis “All”). Significantly enriched pathways (FDR < 0.05) were identified by their KEGG identifier and have been grouped according to 7 functionally related groups obtained by ClueGO analysis. Each group is named after its most significant term (lowest FDR), which is indicated in bold. Functionally related groups are sorted by highest enrichment score, calculated as [- Log2 (group FDR)].
| KEGG Identifier | KEGG ID FDR | Group FDR | Enrichment Score | Groups | % Associated Genes | Nr. Genes |
|---|---|---|---|---|---|---|
| Steroid biosynthesis | 240.0E-9 | 210.0E-9 | 22.2 | 0 | 33.33 | 7.00 |
| ECM-receptor interaction | 1.5E-3 | 1.3E-3 | 9.6 | 5 | 8.64 | 7.00 |
| Alanine, aspartate and glutamate metabolism | 8.7E-3 | 3.0E-3 | 8.4 | 6 | 10.00 | 4.00 |
| Arginine biosynthesis | 10.0E-3 | 3.0E-3 | 8.4 | 6 | 12.00 | 3.00 |
| DNA replication | 9.6E-3 | 6.3E-3 | 7.3 | 2 | 10.53 | 4.00 |
| p53 signaling pathway | 12.0E-3 | 11.0E-3 | 6.5 | 4 | 6.76 | 5.00 |
| Arachidonic acid metabolism | 11.0E-3 | 12.0E-3 | 6.4 | 1 | 7.84 | 4.00 |
| Proteasome | 14.0E-3 | 14.0E-3 | 6.2 | 3 | 7.14 | 4.00 |
Table 7.
Enrichment of GO Biological Processes (GO-BP) by the genes differentially expressed in response to E2 or to Gen (analyses “E2” vs “Gen”). Significantly enriched biological processes (FDR < 0.05), identified by their ID and term description, are grouped into 12 functionally related networks (GO group) obtained by ClueGO analysis. Each group is named after its most significant term (lowest FDR), highlighted in bold, which was chosen for group representation in Fig. 3 of the associated MS in JSBMB [1]. Functionally related groups are sorted by the highest enrichment score, calculated as [- Log2 (group FDR)]. The classification of each term as specifically enriched in response to the E2 or Gen treatments (or both), obtained by ClueGO cluster analysis, is also shown. For groups including terms enriched in more than one treatment, the classification of the leading term was adopted for the group.
| GOID | GOTerm | Term FDR | Group FDR | Enrichment Score | GOGroups | % Associated Genes | Nr. Genes | Treatment | %Genes Specific for E2 | %Genes Specific for Gen |
|---|---|---|---|---|---|---|---|---|---|---|
| GO:0043207 | Response to external biotic stimulus | 540.0E-12 | 100.0E-12 | 33.2 | 7 | 8.54 | 21.00 | Specific for Gen | 51.67 | 73.19 |
| GO:0051707 | response to other organism | 540.0E-12 | 100.0E-12 | 33.2 | 7 | 8.54 | 21.00 | Specific for Gen | 51.67 | 73.19 |
| GO:0009615 | response to virus | 28.0E-6 | 100.0E-12 | 33.2 | 7 | 12.96 | 7.00 | Specific for Gen | 50.61 | 63.26 |
| GO:0009617 | response to bacterium | 1.1E-3 | 100.0E-12 | 33.2 | 7 | 5.20 | 9.00 | Specific for Gen | 55.51 | 74.01 |
| GO:0043207 | Response to external biotic stimulus | 540.0E-12 | 8.9E-9 | 26.7 | 9 | 8.54 | 21.00 | Specific for Gen | 51.67 | 73.19 |
| GO:0051707 | response to other organism | 540.0E-12 | 8.9E-9 | 26.7 | 9 | 8.54 | 21.00 | Specific for Gen | 51.67 | 73.19 |
| GO:0051591 | response to cAMP | 36.0E-6 | 8.9E-9 | 26.7 | 9 | 75.00 | 3.00 | Specific for Gen | 45.51 | 68.26 |
| GO:0009617 | response to bacterium | 1.1E-3 | 8.9E-9 | 26.7 | 9 | 5.20 | 9.00 | Specific for Gen | 55.51 | 74.01 |
| GO:0046683 | response to organophosphorus | 1.8E-3 | 8.9E-9 | 26.7 | 9 | 20.00 | 3.00 | Specific for Gen | 45.51 | 68.26 |
| GO:0014074 | response to purine-containing compound | 1.8E-3 | 8.9E-9 | 26.7 | 9 | 20.00 | 3.00 | Specific for Gen | 45.51 | 68.26 |
| GO:0032496 | response to lipopolysaccharide | 2.0E-3 | 8.9E-9 | 26.7 | 9 | 8.33 | 5.00 | Specific for Gen | 49.63 | 66.17 |
| GO:0002237 | response to molecule of bacterial origin | 2.3E-3 | 8.9E-9 | 26.7 | 9 | 7.94 | 5.00 | Specific for Gen | 49.63 | 66.17 |
| GO:0042493 | response to drug | 10.0E-3 | 8.9E-9 | 26.7 | 9 | 9.38 | 3.00 | Specific for Gen | 45.51 | 68.26 |
| GO:0009612 | response to mechanical stimulus | 23.0E-3 | 8.9E-9 | 26.7 | 9 | 6.67 | 3.00 | Specific for Gen | 45.51 | 68.26 |
| GO:0016126 | Sterol biosynthetic process | 2.2E-9 | 1.3E-6 | 19.6 | 11 | 31.03 | 9.00 | Specific for Gen | 20.36 | 91.63 |
| GO:0006694 | steroid biosynthetic process | 3.0E-9 | 1.3E-6 | 19.6 | 11 | 19.30 | 11.00 | Specific for Gen | 17.00 | 93.49 |
| GO:0016125 | sterol metabolic process | 7.3E-9 | 1.3E-6 | 19.6 | 11 | 20.83 | 10.00 | Specific for Gen | 18.53 | 92.66 |
| GO:0008202 | steroid metabolic process | 7.7E-9 | 1.3E-6 | 19.6 | 11 | 14.63 | 12.00 | Specific for Gen | 15.69 | 94.16 |
| GO:1901617 | organic hydroxy compound biosynthetic process | 660.0E-9 | 1.3E-6 | 19.6 | 11 | 15.25 | 9.00 | Specific for Gen | 20.36 | 91.63 |
| GO:0008203 | cholesterol metabolic process | 2.0E-6 | 1.3E-6 | 19.6 | 11 | 20.59 | 7.00 | Specific for Gen | 25.30 | 88.56 |
| GO:1902652 | secondary alcohol metabolic process | 3.2E-6 | 1.3E-6 | 19.6 | 11 | 18.92 | 7.00 | Specific for Gen | 25.30 | 88.56 |
| GO:0006695 | cholesterol biosynthetic process | 8.7E-6 | 1.3E-6 | 19.6 | 11 | 31.25 | 5.00 | Specific for Gen | 17.96 | 89.82 |
| GO:1902653 | secondary alcohol biosynthetic process | 13.0E-6 | 1.3E-6 | 19.6 | 11 | 27.78 | 5.00 | Specific for Gen | 17.96 | 89.82 |
| GO:1901615 | organic hydroxy compound metabolic process | 33.0E-6 | 1.3E-6 | 19.6 | 11 | 7.69 | 10.00 | Specific for Gen | 18.53 | 92.66 |
| GO:0044283 | small molecule biosynthetic process | 60.0E-6 | 1.3E-6 | 19.6 | 11 | 5.85 | 12.00 | Specific for Gen | 29.87 | 89.62 |
| GO:0046165 | alcohol biosynthetic process | 100.0E-6 | 1.3E-6 | 19.6 | 11 | 17.86 | 5.00 | Specific for Gen | 17.96 | 89.82 |
| GO:0008610 | lipid biosynthetic process | 310.0E-6 | 1.3E-6 | 19.6 | 11 | 4.86 | 12.00 | Specific for Gen | 15.69 | 94.16 |
| GO:0006066 | alcohol metabolic process | 340.0E-6 | 1.3E-6 | 19.6 | 11 | 8.33 | 7.00 | Specific for Gen | 25.30 | 88.56 |
| GO:0016053 | organic acid biosynthetic process | 1.0E-3 | 1.3E-6 | 19.6 | 11 | 5.97 | 8.00 | Specific for Gen | 32.52 | 86.72 |
| GO:0046394 | carboxylic acid biosynthetic process | 1.0E-3 | 1.3E-6 | 19.6 | 11 | 5.97 | 8.00 | Specific for Gen | 32.52 | 86.72 |
| GO:1901607 | alpha-amino acid biosynthetic process | 1.0E-3 | 1.3E-6 | 19.6 | 11 | 10.42 | 5.00 | Specific for Gen | 33.08 | 82.71 |
| GO:0008652 | cellular amino acid biosynthetic process | 1.2E-3 | 1.3E-6 | 19.6 | 11 | 9.80 | 5.00 | Specific for Gen | 33.08 | 82.71 |
| GO:0031099 | Regeneration | 2.2E-3 | 1.3E-6 | 19.6 | 11 | 4.94 | 8.00 | Specific for Gen | 32.52 | 86.72 |
| GO:0006520 | cellular amino acid metabolic process | 5.6E-3 | 1.3E-6 | 19.6 | 11 | 4.15 | 8.00 | Specific for Gen | 22.58 | 90.31 |
| GO:0022600 | digestive system process | 6.0E-3 | 1.3E-6 | 19.6 | 11 | 12.00 | 3.00 | Specific for Gen | 26.42 | 79.25 |
| GO:1901605 | alpha-amino acid metabolic process | 6.3E-3 | 1.3E-6 | 19.6 | 11 | 5.08 | 6.00 | Specific for Gen | 28.72 | 86.17 |
| GO:0007586 | Digestion | 10.0E-3 | 1.3E-6 | 19.6 | 11 | 9.38 | 3.00 | Specific for Gen | 26.42 | 79.25 |
| GO:0072330 | monocarboxylic acid biosynthetic process | 14.0E-3 | 1.3E-6 | 19.6 | 11 | 5.97 | 4.00 | Specific for Gen | 38.69 | 77.37 |
| GO:0031331 | positive regulation of cellular catabolic process | 15.0E-3 | 1.3E-6 | 19.6 | 11 | 5.71 | 4.00 | Both treatments | 58.03 | 58.03 |
| GO:0009896 | positive regulation of catabolic process | 17.0E-3 | 1.3E-6 | 19.6 | 11 | 5.48 | 4.00 | Both treatments | 58.03 | 58.03 |
| GO:0097164 | ammonium ion metabolic process | 27.0E-3 | 1.3E-6 | 19.6 | 11 | 6.25 | 3.00 | Specific for Gen | 26.42 | 79.25 |
| GO:0015748 | organophosphate ester transport | 33.0E-3 | 1.3E-6 | 19.6 | 11 | 5.77 | 3.00 | Specific for Gen | 26.42 | 79.25 |
| GO:0006633 | fatty acid biosynthetic process | 33.0E-3 | 1.3E-6 | 19.6 | 11 | 5.77 | 3.00 | Specific for Gen | 26.42 | 79.25 |
| GO:0001878 | Response to yeast | 3.4E-6 | 4.8E-6 | 17.7 | 4 | 25.00 | 6.00 | Specific for Gen | 43.08 | 71.80 |
| GO:0009620 | response to fungus | 9.5E-6 | 4.8E-6 | 17.7 | 4 | 20.69 | 6.00 | Specific for Gen | 43.08 | 71.80 |
| GO:0031099 | Regeneration | 2.2E-3 | 350.0E-6 | 11.5 | 8 | 4.94 | 8.00 | Specific for Gen | 32.52 | 86.72 |
| GO:0042060 | wound healing | 2.8E-3 | 350.0E-6 | 11.5 | 8 | 4.68 | 8.00 | Specific for Gen | 52.30 | 73.22 |
| GO:0022600 | digestive system process | 6.0E-3 | 350.0E-6 | 11.5 | 8 | 12.00 | 3.00 | Specific for Gen | 26.42 | 79.25 |
| GO:0007586 | Digestion | 10.0E-3 | 350.0E-6 | 11.5 | 8 | 9.38 | 3.00 | Specific for Gen | 26.42 | 79.25 |
| GO:0042246 | tissue regeneration | 41.0E-3 | 350.0E-6 | 11.5 | 8 | 4.08 | 4.00 | Specific for Gen | 38.69 | 77.37 |
| GO:0007599 | Hemostasis | 43.0E-3 | 350.0E-6 | 11.5 | 8 | 5.08 | 3.00 | Specific for E2 | 68.26 | 45.51 |
| GO:0007596 | blood coagulation | 43.0E-3 | 350.0E-6 | 11.5 | 8 | 5.08 | 3.00 | Specific for E2 | 68.26 | 45.51 |
| GO:0050817 | Coagulation | 48.0E-3 | 350.0E-6 | 11.5 | 8 | 4.84 | 3.00 | Specific for E2 | 68.26 | 45.51 |
| GO:0006979 | Response to oxidative stress | 3.1E-3 | 3.9E-3 | 8.0 | 1 | 7.25 | 5.00 | Both treatments | 53.89 | 53.89 |
| GO:0031589 | cell-substrate adhesion | 8.6E-3 | 10.0E-3 | 6.6 | 5 | 7.02 | 4.00 | Specific for E2 | 64.60 | 43.07 |
| GO:0007160 | cell-matrix adhesion | 30.0E-3 | 10.0E-3 | 6.6 | 5 | 6.00 | 3.00 | Specific for E2 | 79.25 | 26.42 |
| GO:0006730 | One-carbon metabolic process | 13.0E-3 | 13.0E-3 | 6.3 | 0 | 8.57 | 3.00 | Specific for Gen | 0.00 | 100.00 |
| GO:0051241 | negative regulation of multicellular organismal process | 12.0E-3 | 14.0E-3 | 6.2 | 3 | 4.32 | 6.00 | Specific for Gen | 51.88 | 77.82 |
| GO:0001945 | Lymph vessel development | 35.0E-3 | 35.0E-3 | 4.8 | 2 | 5.56 | 3.00 | Specific for E2 | 68.26 | 45.51 |
| GO:0000188 | Inactivation of MAPK activity | 260.0E-6 | 39.0E-3 | 4.7 | 10 | 22.22 | 4.00 | Specific for Gen | 53.43 | 71.24 |
| GO:0043407 | negative regulation of MAP kinase activity | 420.0E-6 | 39.0E-3 | 4.7 | 10 | 19.05 | 4.00 | Specific for Gen | 53.43 | 71.24 |
| GO:0043409 | negative regulation of MAPK cascade | 1.1E-3 | 39.0E-3 | 4.7 | 10 | 14.29 | 4.00 | Specific for Gen | 53.43 | 71.24 |
| GO:0043405 | regulation of MAP kinase activity | 1.5E-3 | 39.0E-3 | 4.7 | 10 | 7.32 | 6.00 | Both treatments | 64.85 | 64.85 |
| GO:0071901 | negative regulation of protein serine/threonine kinase activity | 1.5E-3 | 39.0E-3 | 4.7 | 10 | 12.50 | 4.00 | Specific for Gen | 53.43 | 71.24 |
| GO:0033673 | negative regulation of kinase activity | 2.1E-3 | 39.0E-3 | 4.7 | 10 | 6.67 | 6.00 | Both treatments | 64.85 | 64.85 |
| GO:0006469 | negative regulation of protein kinase activity | 2.1E-3 | 39.0E-3 | 4.7 | 10 | 6.74 | 6.00 | Both treatments | 64.85 | 64.85 |
| GO:0051348 | negative regulation of transferase activity | 2.6E-3 | 39.0E-3 | 4.7 | 10 | 6.25 | 6.00 | Both treatments | 64.85 | 64.85 |
| GO:0001933 | negative regulation of protein phosphorylation | 2.9E-3 | 39.0E-3 | 4.7 | 10 | 6.12 | 6.00 | Both treatments | 64.85 | 64.85 |
| GO:0042326 | negative regulation of phosphorylation | 3.0E-3 | 39.0E-3 | 4.7 | 10 | 6.00 | 6.00 | Both treatments | 64.85 | 64.85 |
| GO:1902532 | negative regulation of intracellular signal transduction | 7.0E-3 | 39.0E-3 | 4.7 | 10 | 4.96 | 6.00 | Both treatments | 64.85 | 64.85 |
| GO:0071900 | regulation of protein serine/threonine kinase activity | 7.1E-3 | 39.0E-3 | 4.7 | 10 | 4.92 | 6.00 | Both treatments | 64.85 | 64.85 |
| GO:0001706 | endoderm formation | 14.0E-3 | 39.0E-3 | 4.7 | 10 | 8.11 | 3.00 | Specific for Gen | 45.51 | 68.26 |
| GO:0001704 | formation of primary germ layer | 17.0E-3 | 39.0E-3 | 4.7 | 10 | 5.48 | 4.00 | Specific for Gen | 53.43 | 71.24 |
| GO:0007492 | endoderm development | 49.0E-3 | 39.0E-3 | 4.7 | 10 | 4.76 | 3.00 | Specific for Gen | 45.51 | 68.26 |
| GO:0042541 | Hemoglobin biosynthetic process | 1.3E-3 | 62.0E-3 | 4.0 | 6 | 23.08 | 3.00 | Specific for Gen | 26.42 | 79.25 |
| GO:0020027 | hemoglobin metabolic process | 1.5E-3 | 62.0E-3 | 4.0 | 6 | 21.43 | 3.00 | Specific for Gen | 26.42 | 79.25 |
| GO:0055076 | transition metal ion homeostasis | 48.0E-3 | 62.0E-3 | 4.0 | 6 | 4.84 | 3.00 | Specific for Gen | 26.42 | 79.25 |
Table 8.
Enrichment of KEGG pathways by the genes differentially expressed in response to E2 or to Gen (Analyses “E2” vs “Gen”). Significantly enriched pathways (FDR < 0.05), identified by their KEGG identifier, are grouped according to 6 functionally related groups obtained by ClueGO analysis. Each group is named after its most significant term (lowest FDR) and is highlighted in bold. Functionally related groups are sorted by highest enrichment score, calculated as [- Log2 (group FDR)].
| KEGG Identifier | KEGG ID FDR | Group FDR | Enrichment Score | Groups | % Associated Genes | Nr. Genes | Treatment | %Genes Specific for E2 | %Genes Specific for Gen |
|---|---|---|---|---|---|---|---|---|---|
| Steroid biosynthesis | 210.0E-9 | 180.0E-9 | 22.4 | 0 | 33.33 | 7.00 | Specific for Gen | 0.00 | 100.00 |
| ECM-receptor interaction | 1.3E-3 | 1.1E-3 | 9.8 | 4 | 8.64 | 7.00 | Specific for E2 | 66.84 | 40.11 |
| Alanine, aspartate and glutamate metabolism | 7.6E-3 | 2.6E-3 | 8.6 | 5 | 10.00 | 4.00 | Specific for Gen | 21.53 | 86.14 |
| Arginine biosynthesis | 9.5E-3 | 2.6E-3 | 8.6 | 5 | 12.00 | 3.00 | Specific for Gen | 0.00 | 100.00 |
| DNA replication | 8.4E-3 | 5.4E-3 | 7.5 | 1 | 10.53 | 4.00 | Specific for Gen | 25.00 | 75.00 |
| p53 signaling pathway | 11.0E-3 | 9.5E-3 | 6.7 | 3 | 6.76 | 5.00 | Both treatments | 53.89 | 53.89 |
| Proteasome | 14.0E-3 | 14.0E-3 | 6.2 | 2 | 7.14 | 4.00 | Specific for E2 | 86.14 | 21.53 |
Table 9.
Enrichment of GO Biological Processes by the genes differentially expressed after 1 day or 5 days (Analyses “1d” vs “5d”). Significantly enriched biological processes (FDR < 0.05), identified by their ID and term description, are grouped according to 11 functionally related networks (GO group) obtained by ClueGO analysis. Each group is named after its most significant term (lowest FDR), highlighted in bold, which was chosen for GOTerm representation in Fig. 3 of the associated MS in JSBMB [1]. Functionally related groups are sorted by highest enrichment score, calculated as [- Log2 (group FDR)]. The classification of each term as specifically enriched in the responses after 1 day or 5 days (or both), obtained by ClueGO cluster analysis, is also shown. For groups including terms enriched in more than one list the classification of the leading term was adopted for the whole group.
| GOID | GOTerm | Term FDR | Group FDR | Enrichment Score | GOGroups | % Associated Genes | Nr. Genes | Time | %Genes Specific for 1d | %Genes Specific for 5d |
|---|---|---|---|---|---|---|---|---|---|---|
| GO:0043207 | Response to external biotic stimulus | 490.0E-12 | 100.0E-12 | 33.2 | 7 | 8.54 | 21.00 | Specific for 5 days | 36.49 | 72.99 |
| GO:0051707 | response to other organism | 490.0E-12 | 100.0E-12 | 33.2 | 7 | 8.54 | 21.00 | Specific for 5 days | 36.49 | 72.99 |
| GO:0009615 | response to virus | 26.0E-6 | 100.0E-12 | 33.2 | 7 | 12.96 | 7.00 | Specific for 5 days | 14.29 | 85.71 |
| GO:0009617 | response to bacterium | 1.0E-3 | 100.0E-12 | 33.2 | 7 | 5.20 | 9.00 | Specific for 5 days | 39.30 | 78.60 |
| GO:0043207 | Response to external biotic stimulus | 490.0E-12 | 8.9E-9 | 26.7 | 9 | 8.54 | 21.00 | Specific for 5 days | 36.49 | 72.99 |
| GO:0051707 | response to other organism | 490.0E-12 | 8.9E-9 | 26.7 | 9 | 8.54 | 21.00 | Specific for 5 days | 36.49 | 72.99 |
| GO:0051591 | response to cAMP | 33.0E-6 | 8.9E-9 | 26.7 | 9 | 75.00 | 3.00 | Specific for 5 days | 45.51 | 68.26 |
| GO:0009617 | response to bacterium | 1.0E-3 | 8.9E-9 | 26.7 | 9 | 5.20 | 9.00 | Specific for 5 days | 39.30 | 78.60 |
| GO:0046683 | response to organophosphorus | 1.7E-3 | 8.9E-9 | 26.7 | 9 | 20.00 | 3.00 | Specific for 5 days | 45.51 | 68.26 |
| GO:0014074 | response to purine-containing compound | 1.7E-3 | 8.9E-9 | 26.7 | 9 | 20.00 | 3.00 | Specific for 5 days | 45.51 | 68.26 |
| GO:0032496 | response to lipopolysaccharide | 1.9E-3 | 8.9E-9 | 26.7 | 9 | 8.33 | 5.00 | Specific for 5 days | 33.08 | 82.71 |
| GO:0002237 | response to molecule of bacterial origin | 2.2E-3 | 8.9E-9 | 26.7 | 9 | 7.94 | 5.00 | Specific for 5 days | 33.08 | 82.71 |
| GO:0042493 | response to drug | 10.0E-3 | 8.9E-9 | 26.7 | 9 | 9.38 | 3.00 | Specific for 5 days | 45.51 | 68.26 |
| GO:0009612 | response to mechanical stimulus | 23.0E-3 | 8.9E-9 | 26.7 | 9 | 6.67 | 3.00 | Specific for 5 days | 45.51 | 68.26 |
| GO:0016126 | Sterol biosynthetic process | 2.0E-9 | 76.0E-9 | 23.6 | 11 | 31.03 | 9.00 | Specific for 1 day | 84.82 | 21.21 |
| GO:0006694 | steroid biosynthetic process | 2.8E-9 | 76.0E-9 | 23.6 | 11 | 19.30 | 11.00 | Specific for 1 day | 78.95 | 26.32 |
| GO:0016125 | sterol metabolic process | 6.6E-9 | 76.0E-9 | 23.6 | 11 | 20.83 | 10.00 | Specific for 1 day | 86.42 | 19.21 |
| GO:0008202 | steroid metabolic process | 7.0E-9 | 76.0E-9 | 23.6 | 11 | 14.63 | 12.00 | Specific for 1 day | 80.73 | 24.22 |
| GO:1901617 | organic hydroxy compound biosynthetic process | 600.0E-9 | 76.0E-9 | 23.6 | 11 | 15.25 | 9.00 | Specific for 1 day | 84.82 | 21.21 |
| GO:0008203 | cholesterol metabolic process | 1.8E-6 | 76.0E-9 | 23.6 | 11 | 20.59 | 7.00 | Specific for 1 day | 80.21 | 26.74 |
| GO:1902652 | secondary alcohol metabolic process | 2.9E-6 | 76.0E-9 | 23.6 | 11 | 18.92 | 7.00 | Specific for 1 day | 80.21 | 26.74 |
| GO:0006695 | cholesterol biosynthetic process | 7.9E-6 | 76.0E-9 | 23.6 | 11 | 31.25 | 5.00 | Specific for 1 day | 89.82 | 17.96 |
| GO:1902653 | secondary alcohol biosynthetic process | 12.0E-6 | 76.0E-9 | 23.6 | 11 | 27.78 | 5.00 | Specific for 1 day | 89.82 | 17.96 |
| GO:1901615 | organic hydroxy compound metabolic process | 30.0E-6 | 76.0E-9 | 23.6 | 11 | 7.69 | 10.00 | Specific for 1 day | 86.42 | 19.21 |
| GO:0044283 | small molecule biosynthetic process | 54.0E-6 | 76.0E-9 | 23.6 | 11 | 5.85 | 12.00 | Specific for 1 day | 68.82 | 45.88 |
| GO:0046165 | alcohol biosynthetic process | 92.0E-6 | 76.0E-9 | 23.6 | 11 | 17.86 | 5.00 | Specific for 1 day | 89.82 | 17.96 |
| GO:0006066 | alcohol metabolic process | 330.0E-6 | 76.0E-9 | 23.6 | 11 | 8.33 | 7.00 | Specific for 1 day | 80.21 | 26.74 |
| GO:0016053 | organic acid biosynthetic process | 990.0E-6 | 76.0E-9 | 23.6 | 11 | 5.97 | 8.00 | Specific for 5 days | 54.20 | 65.04 |
| GO:0046394 | carboxylic acid biosynthetic process | 990.0E-6 | 76.0E-9 | 23.6 | 11 | 5.97 | 8.00 | Specific for 5 days | 54.20 | 65.04 |
| GO:1901607 | alpha-amino acid biosynthetic process | 1.0E-3 | 76.0E-9 | 23.6 | 11 | 10.42 | 5.00 | Specific for 5 days | 49.63 | 66.17 |
| GO:0008652 | cellular amino acid biosynthetic process | 1.1E-3 | 76.0E-9 | 23.6 | 11 | 9.80 | 5.00 | Specific for 5 days | 49.63 | 66.17 |
| GO:0072330 | monocarboxylic acid biosynthetic process | 14.0E-3 | 76.0E-9 | 23.6 | 11 | 5.97 | 4.00 | Both times | 58.03 | 58.03 |
| GO:0031331 | positive regulation of cellular catabolic process | 16.0E-3 | 76.0E-9 | 23.6 | 11 | 5.71 | 4.00 | Specific for 5 days | 43.07 | 64.60 |
| GO:0009896 | positive regulation of catabolic process | 17.0E-3 | 76.0E-9 | 23.6 | 11 | 5.48 | 4.00 | Specific for 5 days | 43.07 | 64.60 |
| GO:0097164 | ammonium ion metabolic process | 27.0E-3 | 76.0E-9 | 23.6 | 11 | 6.25 | 3.00 | Specific for 1 day | 79.25 | 26.42 |
| GO:0001878 | Response to yeast | 3.1E-6 | 4.8E-6 | 17.7 | 5 | 25.00 | 6.00 | Both times | 50.00 | 50.00 |
| GO:0009620 | response to fungus | 8.6E-6 | 4.8E-6 | 17.7 | 5 | 20.69 | 6.00 | Both times | 50.00 | 50.00 |
| GO:0031099 | Regeneration | 2.1E-3 | 150.0E-6 | 12.7 | 8 | 4.94 | 8.00 | Specific for 1 day | 75.88 | 43.36 |
| GO:0042060 | wound healing | 2.7E-3 | 150.0E-6 | 12.7 | 8 | 4.68 | 8.00 | Specific for 1 day | 73.22 | 52.30 |
| GO:0042246 | tissue regeneration | 42.0E-3 | 150.0E-6 | 12.7 | 8 | 4.08 | 4.00 | Specific for 1 day | 77.37 | 38.69 |
| GO:0007599 | hemostasis | 44.0E-3 | 150.0E-6 | 12.7 | 8 | 5.08 | 3.00 | Specific for 5 days | 45.51 | 68.26 |
| GO:0007596 | blood coagulation | 44.0E-3 | 150.0E-6 | 12.7 | 8 | 5.08 | 3.00 | Specific for 5 days | 45.51 | 68.26 |
| GO:0050817 | coagulation | 49.0E-3 | 150.0E-6 | 12.7 | 8 | 4.84 | 3.00 | Specific for 5 days | 45.51 | 68.26 |
| GO:0006979 | Response to oxidative stress | 2.9E-3 | 3.9E-3 | 8.0 | 0 | 7.25 | 5.00 | Specific for 1 day | 60.00 | 40.00 |
| GO:0031589 | Cell-substrate adhesion | 8.6E-3 | 10.0E-3 | 6.6 | 3 | 7.02 | 4.00 | Specific for 5 days | 43.07 | 64.60 |
| GO:0009266 | Response to temperature stimulus | 15.0E-3 | 19.0E-3 | 5.7 | 2 | 7.89 | 3.00 | Specific for 1 day | 100.00 | 0.00 |
| GO:0000188 | Inactivation of MAPK activity | 230.0E-6 | 29.0E-3 | 5.1 | 10 | 22.22 | 4.00 | Specific for 5 days | 38.69 | 77.37 |
| GO:0043407 | negative regulation of MAP kinase activity | 400.0E-6 | 29.0E-3 | 5.1 | 10 | 19.05 | 4.00 | Specific for 5 days | 38.69 | 77.37 |
| GO:0043409 | negative regulation of MAPK cascade | 1.0E-3 | 29.0E-3 | 5.1 | 10 | 14.29 | 4.00 | Specific for 5 days | 38.69 | 77.37 |
| GO:0043405 | regulation of MAP kinase activity | 1.4E-3 | 29.0E-3 | 5.1 | 10 | 7.32 | 6.00 | Specific for 5 days | 28.72 | 86.17 |
| GO:0071901 | negative regulation of protein serine/threonine kinase activity | 1.4E-3 | 29.0E-3 | 5.1 | 10 | 12.50 | 4.00 | Specific for 5 days | 38.69 | 77.37 |
| GO:0033673 | negative regulation of kinase activity | 1.9E-3 | 29.0E-3 | 5.1 | 10 | 6.67 | 6.00 | Specific for 5 days | 54.36 | 67.96 |
| GO:0006469 | negative regulation of protein kinase activity | 1.9E-3 | 29.0E-3 | 5.1 | 10 | 6.74 | 6.00 | Specific for 5 days | 54.36 | 67.96 |
| GO:0051348 | negative regulation of transferase activity | 2.5E-3 | 29.0E-3 | 5.1 | 10 | 6.25 | 6.00 | Specific for 5 days | 54.36 | 67.96 |
| GO:0001933 | negative regulation of protein phosphorylation | 2.7E-3 | 29.0E-3 | 5.1 | 10 | 6.12 | 6.00 | Specific for 5 days | 54.36 | 67.96 |
| GO:0042326 | negative regulation of phosphorylation | 2.8E-3 | 29.0E-3 | 5.1 | 10 | 6.00 | 6.00 | Specific for 5 days | 54.36 | 67.96 |
| GO:1902532 | negative regulation of intracellular signal transduction | 7.0E-3 | 29.0E-3 | 5.1 | 10 | 4.96 | 6.00 | Specific for 5 days | 54.36 | 67.96 |
| GO:0071900 | regulation of protein serine/threonine kinase activity | 7.1E-3 | 29.0E-3 | 5.1 | 10 | 4.92 | 6.00 | Specific for 5 days | 28.72 | 86.17 |
| GO:0001706 | endoderm formation | 15.0E-3 | 29.0E-3 | 5.1 | 10 | 8.11 | 3.00 | Specific for 5 days | 26.42 | 79.25 |
| GO:0001704 | formation of primary germ layer | 17.0E-3 | 29.0E-3 | 5.1 | 10 | 5.48 | 4.00 | Specific for 5 days | 21.53 | 86.14 |
| GO:0001945 | Lymph vessel development | 37.0E-3 | 39.0E-3 | 4.7 | 4 | 5.56 | 3.00 | Specific for 1 day | 68.26 | 45.51 |
| GO:0009142 | Nucleoside triphosphate biosynthetic process | 44.0E-3 | 44.0E-3 | 4.5 | 1 | 5.08 | 3.00 | Specific for 5 days | 0.00 | 100.00 |
| GO:0042541 | Hemoglobin biosynthetic process | 1.2E-3 | 62.0E-3 | 4.0 | 6 | 23.08 | 3.00 | Specific for 5 days | 26.42 | 79.25 |
| GO:0020027 | hemoglobin metabolic process | 1.4E-3 | 62.0E-3 | 4.0 | 6 | 21.43 | 3.00 | Specific for 5 days | 26.42 | 79.25 |
| GO:0055076 | transition metal ion homeostasis | 49.0E-3 | 62.0E-3 | 4.0 | 6 | 4.84 | 3.00 | Specific for 5 days | 26.42 | 79.25 |
Table 10.
Enrichment of KEGG pathways by the genes differentially expressed after 1 day or 5 days (analyses “1d” vs “5d”). Significantly enriched pathways (FDR < 0.05), identified by their KEGG identifier, are grouped according to 7 functionally related groups obtained by ClueGO analysis.
| KEGG Identifier | KEGG ID FDR | Group FDR | Enrichment Score | Groups | % Associated Genes | Nr. Genes | Time | %Genes Specific for 1d | %Genes Specific for 5d |
|---|---|---|---|---|---|---|---|---|---|
| p53 signaling pathway | 11.0E-3 | 11.0E-3 | 6.5 | 5 | 6.76 | 5.00 | Both times | 53.89 | 53.89 |
| Steroid biosynthesis | 210.0E-9 | 210.0E-9 | 22.2 | 0 | 33.33 | 7.00 | Specific for 1d | 100.00 | 0.00 |
| ECM-receptor interaction | 1.3E-3 | 1.3E-3 | 9.6 | 6 | 8.64 | 7.00 | Specific for 1d | 66.84 | 40.11 |
| Alanine, aspartate and glutamate metabolism | 7.6E-3 | 7.6E-3 | 7.0 | 1 | 10.00 | 4.00 | Specific for 1d | 64.60 | 43.07 |
| DNA replication | 8.4E-3 | 8.4E-3 | 6.9 | 3 | 10.53 | 4.00 | Specific for 5 d | 25.00 | 75.00 |
| Arachidonic acid metabolism | 12.0E-3 | 12.0E-3 | 6.4 | 2 | 7.84 | 4.00 | Specific for 5 d | 25.00 | 75.00 |
| Proteasome | 14.0E-3 | 14.0E-3 | 6.2 | 4 | 7.14 | 4.00 | Specific for 5 d | 21.53 | 86.14 |
2. Experimental design, materials, and methods
2.1. Experimental set-up and sampling
The experimental set-up generating the analysed RNAs has been previously described by Pinto et al. [1,2]. Immature sea bass (n = 10/experimental group) received intraperitoneal injections of 5 mg/kg E2 or 5 mg/kg Gen in coconut oil or injection of coconut oil alone (control groups). Individual scales were plucked with forceps from the same region of the skin (below the dorsal fin) in each fish, frozen in liquid nitrogen and stored at −80 °C until total RNA extraction.
2.2. Total RNA extraction
An automated Maxwell 16 Instrument and the SEV (standard elution volume) total RNA purification kit (Promega, Madison, Wisconsin, USA) were used for the extraction of total RNA from n = 15 scales/individual sea bass. Scales in lysis buffer were mechanically disrupted with an Ultra Turrax homogenizer (IKA, Staufen, Germany) and a dispersing element specialized for fibrous tissues. The extracted total RNA was concentrated by precipitation with 2 vol (V) of 100% ethanol and 1/10 V of 3 M sodium acetate pH 5.2 and resuspended in 20–30 μl of Milli-Q filtered water.
RNA quantity and quality were measured in a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Between 3 and 10 μg of RNA were digested with DNase, using a rigorous treatment regime by carrying out two sequential digestions of 30 min with 2U of TURBO DNAse, as recommended in the instructions of the DNA-free kit (Ambion, Thermo Fisher Scientific, Waltham, Massachusetts, USA).
2.3. RNA-seq library preparation
The prepared DNase treated RNAs were precipitated with 5 vol of 100% ethanol and shipped in refrigerated conditions to the Shanghai Ocean University Sequencing Service, Shanghai, China. Their quality was assessed using a Bioanalyser 2100 (Agilent Technologies, Santa Clara, California, USA), to confirm that all RNAs had an RNA integrity number (RIN) greater than 8. Library preparation was conducted using a TruSeq mRNA library prep kit (Illumina, San Diego, California, USA), following the suppliers' instructions. For each library, 0.5 μg of each of the 30 individual RNAs were used (n = 4–6 individual libraries per treatment, see Table 1). Sequencing of paired-end (100 bp) reads was carried out using an Illumina Hi-Seq 1500, at the Shanghai Ocean University Sequencing Service.
2.4. Sequencing and data analysis
Quality control and trimming of the produced reads was carried out with FastQC and Cutadapt [3,4], using a Phred quality score cut-off of 20.
Good quality (filtered) reads (Table 1) were then mapped and assembled using the sea bass reference genome assembly from June 2012 (dicLab v1.0c) and the annotation from July 2013 [5,6], by running TopHat and Cufflinks packages with the data and using the default parameters [7,8]. Relative expression levels were obtained in fpkm (fragments per kilobase of transcript per million fragments mapped) for each sea bass gene and differential expression between experimental conditions was evaluated using Cuffdiff.
Differential expression was evaluated using pairwise comparisons between a) the scales of E2-or Gen-treated fish compared to control fish at the same sampling time (E1d vs C1d, Gen1d vs C1d, E5d vs C5d or Gen5d vs C5d) or b) between the control groups over time (C1d vs C5d). Two different stringency conditions were used to identify differentially expressed genes: those passing the condition FDR <0.05 and those satisfying both conditions FDR <0.05 and a ≥ 2-fold change in expression between the two compared groups.
The common or specific genes identified between different lists of E2 or Gen DE genes were represented by area proportional Venn diagrams generated with BioVenn [9,10], using their Cufflinks XLOC identifiers (see Supplementary Table 1). Similarities between global transcriptome changes identified for the six experimental conditions (C1d, E1d, Gen1d, C5d, E5d and Gen5d) were evaluated by hierarchical clustering with Cluster 3.0 [11,12], using median centered expression data after Log2 transformation of normalized gene expression (fpkm) levels. The uncentered correlation option and complete linkage options were used to cluster group arrays (experimental conditions).
2.5. Gene and functional annotations
The 749 genes for which differential expression was detected with FDR <0.05 were subjected to a multistep automatic gene annotation strategy. Stand-alone Blastx analyses of the corresponding Cufflinks transcripts (individual sequences extracted from the sea bass annotated genes coding sequences, between the mapping positions) were run against the Swiss-Prot curated protein database [13,14] and the unreviewed GenBank protein database [15,16], with expect values (E) set at a maximum of 10−10. These Blastx results were then combined and compared with the Cufflinks mappings to the annotated sea bass genome [5,6]. For genome mapping, the annotation of the closest gene was used as long as a maximum distance of 1000 bp existed between this gene and the mapping position of the Cufflinks transcript.
A hierarchical preference order was defined to assign annotation matches to the differentially expressed genes, which was Swiss-Prot hits > sea bass genome hits > GenBank hits, as represented in Fig. 1. Genes were annotated using Swiss-Prot hits for all genes that gave significant hits with this curated database; when no Swiss-Prot hits were found genes were annotated using genome mapping and genes with no significant annotation using the two previous databases were annotated using the non-curated Genbank protein database.
In addition, 54% of the 332 DE genes with ≥ 2-fold change were manually curated to verify the accuracy of the annotations. For this process, individual Cufflinks transcript sequences extracted from the mapping positions were re-annotated using individual BLAT versus the sea bass genome [5,6], compared with the sea bass annotations of these and close by genes. Their predicted coding sequences were blasted against the Swiss-Prot and GenBank protein databases, followed by a careful verification by multisequence alignments carried out with MultAlin [17,18].
Gene ontology (GO) and pathway (KEGG) enrichment analyses were carried out using Cytoscape v3.5.1 [19] and ClueGO plug-in v2.3.2 [20], with Cluepedia v1.3.2. The zebrafish (Danio rerio) orthologues for the 371 genes found to be DE with E2 or Gen treatments at FDR<0.05 were identified using stand-alone BlastX (with E value < 10−10) against the Ensembl zebrafish protein predictions (GRC Zebrafish Build 10, INSDC Assembly GCA_000002035.3 from Sep 2014, downloaded in Feb 2017 at https://www.ensembl.org/ [21]). D. rerio Ensembl protein IDs corresponding to each list of DE genes (“All” = all genes regulated by E2 and/or Gen; “E2” or “Gen” = genes regulated by each treatment irrespective of the sampling time and “1d” or “5d” = genes regulated after 1 or 5 days by either E2 or Gen) were then submitted to the Cytoscape/ClueGO plug-in. This was run using the following settings: enrichment analysis (right-sided hypergeometric test) using GO Biological Process (GO BP, levels 3–8) terms for D. rerio updated on 13/05/2017; Benjamini–Hochberg false discovery rate (FDR) correction with only terms with FDR ≤ 0.05 and a minimum of three genes/4% being considered significant; Initial group size (for grouping into functionally related networks of enriched terms) set as 1 and group merging at 50%, with a Kappa-statistics score threshold set at 0.4. The enrichment score for each functionally related network group was calculated as -Log2 (group FDR). The leading terms for each group were selected based on their highest enrichment score (lowest term FDR) and were used for group naming in tables and condensed bar plot representations and for evaluation of treatment specific enrichment. Parallel enrichment analyses were carried out for the Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways for each DE list, using the same parameters and strategy as described for GO BP.
Acknowledgements
The research leading to these results was funded by the Foundation for Science and Technology of Portugal, through projects PTDC/AAG-GLO/4003/2012 and CCMAR/Multi/04326/2019. PISP was funded by an FCT post-doctoral fellowship SFRH/BPD/84033/2012. The authors thank Dr Manuel Manchado (IFAPA El Toruno, Cadiz, Spain) for advice on gene enrichment analyses and Rui S.R. Machado (SysBioLab, CBMR, University of Algarve, Portugal) for heat map representation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jsbmb.2019.105448.
Contributor Information
Patricia I.S. Pinto, Email: ppinto@ualg.pt.
Deborah M. Power, Email: dpower@ualg.pt.
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Pinto P.I.S., Andrade A., Moreira C., Zapater C., Thorne M., Santos S., Estêvão M.D., Gomez A., Canario A.V.M., Power D.M. Genistein and estradiol have common and specific impacts on the sea bass (Dicentrarchus labrax) skin-scale barrier. J. Steroid Biochem. Mol. Biol. 2019;195:105448. doi: 10.1016/j.jsbmb.2019.105448. [DOI] [PubMed] [Google Scholar]
- 2.Pinto P.I., Estevao M.D., Andrade A., Santos S., Power D.M. Tissue responsiveness to estradiol and genistein in the sea bass liver and scale. J. Steroid Biochem. Mol. Biol. 2016;158:127–137. doi: 10.1016/j.jsbmb.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 3.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17(1):10–12. [Google Scholar]
- 4.Andrews S. 2010. Fastqc, a Quality Control Tool for High Throughput Sequence Data.http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- 5.Tine M., Kuhl H., Gagnaire P.A., Louro B., Desmarais E., Martins R.S., Hecht J., Knaust F., Belkhir K., Klages S., Dieterich R., Stueber K., Piferrer F., Guinand B., Bierne N., Volckaert F.A., Bargelloni L., Power D.M., Bonhomme F., Canario A.V., Reinhardt R. European sea bass genome and its variation provide insights into adaptation to euryhalinity and speciation. Nat. Commun. 2014;5:5770. doi: 10.1038/ncomms6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuhl H., Reinhardts R. 2012. European Seabass (Dicentrarchus labrax) Genome.http://seabass.mpipz.mpg.de/ [Google Scholar]
- 7.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trapnell C. 2012. Cufflinks.https://cole-trapnell-lab.github.io/cufflinks/ [Google Scholar]
- 9.Hulsen T. 2007-2019. BioVenn Website.http://www.biovenn.nl/ [Google Scholar]
- 10.Hulsen T., de Vlieg J., Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Hoon M.J., Imoto S., Nolan J., Miyano S. Open source clustering software. Bioinformatics. 2004;20(9):1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 12.de Hoon M.J. 2004-2019. Cluster 3.0 Software.http://bonsai.hgc.jp/∼mdehoon/software/cluster/software.htm#ctv [Google Scholar]
- 13.Uniprot Consortium, the Uniprot Database. 2002–2019. https://www.uniprot.org/uniprot/ [Google Scholar]
- 14.Uniprot consortium, UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45(D1):D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2013;41(Database Issue):D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.NCBI . 2004-2019. The National Center for Biotechnology Information (NCBI) Downloads.ftp://ftp.ncbi.nlm.nih.gov/ [Google Scholar]
- 17.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corpet F. 2000–2019. MultAlin Multiple Sequence Alignment Website.http://multalin.toulouse.inra.fr/multalin/ [Google Scholar]
- 19.Isserlin R., Merico D., Voisin V., Bader G.D. Enrichment Map - a Cytoscape app to visualize and explore OMICs pathway enrichment results. F1000Research. 2014;3:141. doi: 10.12688/f1000research.4536.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pages F., Trajanoski Z., Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curwen V., Eyras E., Andrews T.D., Clarke L., Mongin E., Searle S.M., Clamp M. The Ensembl automatic gene annotation system. Genome Res. 2004;14(5):942–950. doi: 10.1101/gr.1858004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.