Highlights
-
•
Lesion segmentation is an important tool in multiple sclerosis (MS) research.
-
•
Several automated methods exist, e.g., supervised, trained or deep learning-based.
-
•
Convolutional neural networks (CNN) show promising results for lesion segmentation.
-
•
CNN method nicMSlesions can be trained with input from one single-subject.
-
•
With minimal input, nicMSlesions outperforms e.g. LST-LPA and LesionTOADS.
Keywords: MRI, Multiple sclerosis, Automatic lesion segmentation, Convolutional neural networks
Abbreviations: BIANCA, brain intensity abnormality classification algorithm; CNN, convolutional neural network; EDSS, expanded disease disability status scale; ICC, intra-class correlation coefficient; IQR, interquartile range; LST-LPA, lesion segmentation toolbox with lesion probability algorithm; LesionTOADS, lesion-topology preserving anatomical segmentation; MICCAI, medical image computing and computer assisted intervention; MS, multiple sclerosis; SI, Dice's similarity index
Abstract
Purpose
Accurate lesion segmentation is important for measurements of lesion load and atrophy in subjects with multiple sclerosis (MS). International MS lesion challenges show a preference of convolutional neural networks (CNN) strategies, such as nicMSlesions. However, since the software is trained on fairly homogenous training data, we aimed to test the performance of nicMSlesions in an independent dataset with manual and other automatic lesion segmentations to determine whether this method is suitable for larger, multi-center studies.
Methods
Manual lesion segmentation was performed in fourteen subjects with MS on sagittal 3D FLAIR images from a 3T GE whole-body scanner with 8-channel head coil. We compared five different categories of automated lesion segmentation methods for their volumetric and spatial agreement with manual segmentation: (i) unsupervised, untrained (LesionTOADS); (ii) supervised, untrained (LST-LPA and nicMSlesions with default settings); (iii) supervised, untrained with threshold adjustment (LST-LPA optimized for current data); (iv) supervised, trained with leave-one-out cross-validation on fourteen subjects with MS (nicMSlesions and BIANCA); and (v) supervised, trained on a single subject with MS (nicMSlesions). Volumetric accuracy was determined by the intra-class correlation coefficient (ICC) and spatial accuracy by Dice's similarity index (SI). Volumes and SI were compared between methods using repeated measures ANOVA or Friedman tests with post-hoc pairwise comparison.
Results
The best volumetric and spatial agreement with manual was obtained with the supervised and trained methods nicMSlesions and BIANCA (ICC absolute agreement > 0.968 and median SI > 0.643) and the worst with the unsupervised, untrained method LesionTOADS (ICC absolute agreement = 0.140 and median SI = 0.444). Agreement with manual in the single-subject network training of nicMSlesions was poor for input with low lesion volumes (i.e. two subjects with lesion volumes ≤ 3.0 ml). For the other twelve subjects, ICC varied from 0.593 to 0.973 and median SI varied from 0.535 to 0.606. In all cases, the single-subject trained nicMSlesions segmentations outperformed LesionTOADS, and in almost all cases it also outperformed LST-LPA.
Conclusion
Input from only one subject to re-train the deep learning CNN nicMSlesions is sufficient for adequate lesion segmentation, with on average higher volumetric and spatial agreement with manual than obtained with the untrained methods LesionTOADS and LST-LPA.
1. Introduction
Multiple sclerosis (MS) is an autoimmune disorder of the central nervous system, characterized by neurodegeneration and demyelination. To enable both atrophy and lesion load measurements in subjects with MS, accurate lesion segmentation is necessary. However, manual lesion segmentation is labor intensive and highly time consuming. Therefore, several automatic lesion segmentation methods have been developed (Garcia-Lorenzo et al., 2013; Danelakis et al., 2018) with varying amounts of manual input and output optimization possibilities. In general, methods can be supervised and/or trained, i.e. based on a previous set of training images that comes with the algorithm (supervised) or based on a set of training images specific to the dataset in which the method is to be applied (trained).
International MS lesion challenges have shown that especially convolutional neural networks (CNN) deep learning strategies perform well for MS lesion segmentation (Commowick et al., 2018). However, CNN methods have the disadvantage that they still require a lot of manual reference input data in order to construct the network in the MR domain of choice. Recently, a method was published showing that input from only one single subject could be sufficient for the cascaded CNN method nicMSlesions to outperform manual delineation (Valverde et al., 2019). The nicMSlesions software consists of an 11 layer CNN source model trained using the public MS databases of the Medical Image Computing and Computer Assisted Intervention (MICCAI) society (Valverde et al., 2017). Because that original training data was fairly homogeneous, with most data acquired in 3T Siemens systems, we aimed to test the performance of nicMSlesions in an independent dataset with manual lesion segmentations and to compare it with other segmentation methods. In this way, we will be able to determine whether this deep learning method is a reliable lesion segmentation method when compared to other existing lesion segmentation methods, with special attention for the use of minimal input data (i.e., few manual delineations available) for its applicability in real-world data.
In this study, we will focus on four different segmentation methods, with different configurations splitting them in (i) unsupervised, untrained methods (Lesion-Topology preserving Anatomical Segmentation (LesionTOADS) (Shiee et al., 2010)); (ii) supervised, untrained methods (Lesion Segmentation Toolbox with Lesion Probability Algorithm (LST-LPA) (Schmidt, 2017) and the default network of nicMSlesions (Valverde et al., 2019)); (iii) supervised, untrained methods with threshold adjustment (LST-LPA); (iv) supervised, trained methods (FMRIB Software Library (FSL) Brain Intensity AbNormality Classification Algorithm (BIANCA) (Griffanti et al., 2016) and the trained network of nicMSlesions); and (v) supervised, trained method with minimal input (single-subject trained network of nicMSlesions). We quantified volumetric and spatial agreement with manual for all methods and also tested their performance in lesion-negative, healthy control images, in order to determine which type of method is this most suitable (i.e., best performance with least manual labor) for larger, multi-center studies.
2. Methods
2.1. Subjects
From a larger cohort of subjects with RRMS, a total of fourteen subjects scanned between December 2016 and June 2017 were included in this study. Subjects included were over 18 years of age, diagnosed with RRMS for a maximum of five years with a maximum expanded disease disability status scale (EDSS) score of 5.0, and received either first line disease modifying therapy or no therapy at all. Patients were not eligible for participation if they had switched medication in the 6 months prior to their visit, if they had received second-line treatment in the past, or if they had received steroid treatment in the 3 months prior to the MRI examination. Further exclusion criteria were past or current neurological or immunological syndromes other than MS, and inability to undergo MRI examination. From the same cohort, data of five healthy controls was also included in the present study.
This study was approved by the local institutional medical ethics committee and written informed consent was obtained from all individuals, according to the Declaration of Helsinki.
2.2. MRI examination
Subjects underwent extensive MRI examination on a 3T whole-body MR scanner (GE Discovery MR750) with an 8-channel phased-array head coil. The protocol included a sagittal 3D T1-weighted fast spoiled gradient echo sequence (FSPGR with TR/TE/TI = 8.2/3.2/450 ms and voxel size 1.0 × 1.0 × 1.0 mm) and a sagittal 3D T2-weighted fluid attenuated inversion recovery sequence (FLAIR with TR/TE/TI = 8000/130/2338 ms with voxel size 1.0 × 1.0 × 1.2 mm).
2.3. MR imaging data analysis
For this research, we investigated the performance of four different lesion segmentation methods in comparison to manual segmentation, all with different levels of optimization and training possibilities, allowing us to study the following five categories of lesion segmentation methods: (i) unsupervised, untrained segmentation, without optimization; (ii) supervised, untrained segmentation, without optimization; (iii) supervised, untrained segmentation, with threshold adjustment; (iv) supervised, fully trained segmentation, with optimization; and (v) supervised, trained segmentation with minimal input. Details on the different lesion segmentation methods are described below and an overview of these methods is shown in Table 1 and Fig. 1.
Table 1.
Overview of the different lesion segmentation methods investigated in this study for their performance to manual segmentation.
| Supervision | Training | Optimization | Method |
|---|---|---|---|
| no | No | no | (1) LesionTOADS |
| yes | No | no | (2) LST-LPA default (3) nicMSlesions default |
| yes | No | yes | (4) LST-LPA adjusted-threshold |
| yes | yes (n = 14) | yes | (5) nicMSlesions optimized (6) BIANCA |
| yes | yes (n = 1) | No | (7) nicMSlesions single-subject |
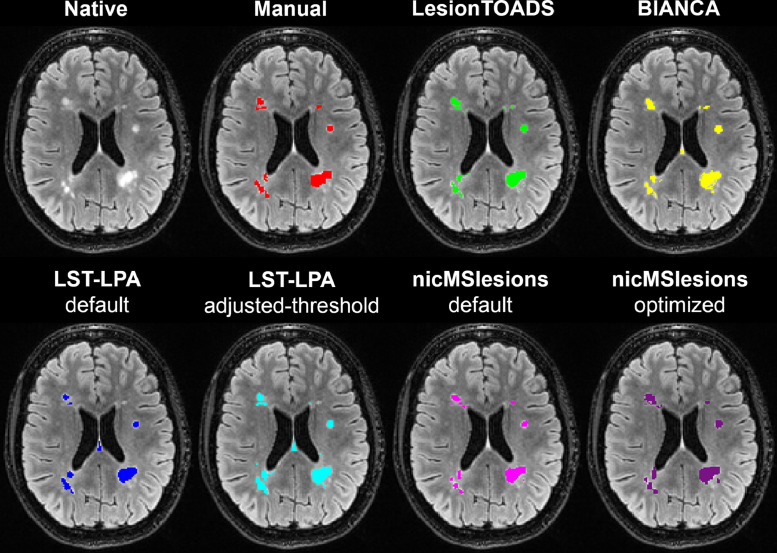
Fig. 1.
Example lesion segmentation shown for the different lesion segmentation methods on the original 3D FLAIR image of a 36 year old female with RRMS: manual (red); LesionTOADS (green), BIANCA (yellow), LST-LPA default (blue), LST-LPA adjusted-threshold (turquoise), nicMSlesions default (pink), and nicMSlesions optimized (purple). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.3.1. Manual segmentation
An expert rater (MLV; experience > 10 years) manually delineated the lesions on the 3D FLAIR images. For this, lesions were defined as hyper-intense regions compared to the surrounding tissue with a size of at least three voxels. The rater had access to the 3D T1 images for reference. Three subjects were rated twice to calculate intra-rater agreement.
2.3.2. Unsupervised, untrained segmentation without optimization: LesionTOADS
Lesion-Topology preserving Anatomical Segmentation (LesionTOADS)(Shiee et al., 2010) uses a statistical lesion atlas based on a topology preserving anatomical atlas. Preprocessing consisted of bias field correction and brain extraction from both T1 and FLAIR images, as well as affine linear registration of the brain extracted images from T1 to FLAIR. The method results in a binary lesion segmentation map.
2.3.3. Supervised, untrained segmentation without optimization: lst-LPA and nicMSlesions default
LST-LPA default: Lesion Segmentation Toolbox with Lesion Prediction Algorithm (LST-LPA) (Schmidt 2017) uses voxel-wise binary regression with spatially varying intercepts. No preprocessing is applied, because the algorithm performs the necessary bias field correction and affine registration of T1 to FLAIR images as part of the pipeline. Because the output is a probabilistic map, a threshold has to be defined to obtain binary segmentation files, with the default probability threshold set to 0.5.
nicMSlesionsnic default: MSlesions is a deep learning method based on cascaded convolutional neural networks that, in contrast to most trained or deep learning methods, can be used when limited amounts of manual input data are available. For the supervised, untrained segmentation, we segmented our data using the default parameters of the network (called baseline 2ch) from nicMSlesions with the outcome probability map thresholded at 0.5. No preprocessing was required.
2.3.4. Supervised, untrained segmentation with threshold adjustment: lst-LPA adjusted-threshold
Threshold adjustment of LST-LPA output was performed by varying the probability threshold from 0.10 to 0.80 with step-size 0.05 and selecting the threshold that resulted in the highest mean similarity index (SI) to manual across all fourteen subjects. Note that this configuration of LST-LPA is dependent on the manual delineations, since they are required for the probability threshold optimization.
2.3.5. Supervised, fully trained segmentation, with optimization: nicMSlesions optimized and BIANCA
nicMSlesions optimized: Next to the default baseline 2ch trained segmentation of nicMSlesions, the method allows full re-training of the neural network (all 11 layers) with our own dataset, evaluating performance by using a leave-one-out cross-validation approach (i.e. train on thirteen subjects and apply on the fourteenth subject). No preprocessing was required. From this output, we determined the optimal probability threshold in the same way as for LST-LPA. Although the minimal lesion size can be optimized for nicMSlesions as well, we chose to keep this at the default value of five voxels, since optimization of this parameter only has very limited effect on the performance of the segmentation (Valverde et al., 2017).
BIANCA: BIANCA (Griffanti et al., 2016) is a trained segmentation method based on the k-nearest neighbor algorithm. Preprocessing of the data consisted of bias field correction and brain extraction from both T1 and FLAIR images, as well as affine registration of the brain extracted images from T1 to FLAIR. Since BIANCA allows for many parameters to be set, we first optimized BIANCA for our full (n = 14) dataset with leave-one-out cross-validation. First, we optimized the optimal probability and cluster-size thresholds. Then, we optimized: (1) the number of lesion and non-lesion training points; (2) the location of the non-lesion training points; (3) use of patch and patch size; and (4) spatial weighting.
2.3.6. Supervised, trained segmentation with minimal input: nicMSlesions single-subject
Last, we tested a single-subject configuration of nicMSlesions, in which the last 1 or 2 layers of the 11-layer neural network were re-trained from single-subject input only, again using leave-one-out cross-validation. In our data, nicMSlesions re-trains one layer for input lesion volumes below 5 ml, and two layers for higher input lesion volume. Here, the default probability threshold of 0.5 was used as well.
2.4. Statistics
Per segmentation method, true and false positives and negatives were extracted from the native FLAIR images, and corresponding sensitivity and 1-specificity were calculated. Furthermore, two-way random intra-class correlation coefficient (ICC) for absolute agreement and for consistency were calculated to determine volumetric accuracy. Dice's similarity index (SI) compared to manual was calculated to quantify spatial accuracy.
Statistical analysis was performed in IBM SPSS Statistics for Windows, version 22.0 (IBM Corp., Armonk, N.Y., USA). Repeated measures ANOVA was used for volumetric agreement with manual per segmentation method and Friedman Tests for spatial agreement with manual per segmentation method. For the single-subject analyses of nicMSlesions, Wilcoxon Signed Ranks tests were used.
For the repeated measures ANOVA, Mauchly's test of sphericity was performed to assess equal variances of the differences between all within-subject factors. When the assumption of sphericity was violated, degrees of freedom were corrected using Huyn–Feldt estimates of sphericity. When appropriate, post-hoc analyses were conducted using Mann–Whitney U tests (unpaired) or Wilcoxon Signed Ranks tests (paired).
Inter quartile range was determined by the 25th and 75th percentile. Results were considered statistically significant upon p-value < 0.05.
3. Results
3.1. Demographics
Five of the fourteen subjects with RRMS were male (36%). Mean age was 37.1 ± 5.3 years (range 26.4–47.7) and mean disease duration 3.1 ± 1.4 years (range 0.6–4.7 years). Disease modifying treatment was used by ten subjects (dimethyl fumarate n = 3; glatiramer acetate n = 2; interferon-β n = 3; teriflunomide n = 2) and median EDSS was 3.5 (range 1.0–4.0).
From the healthy controls, one of five subjects was male (20%) and mean age was 34.3 ± 8.3 years (range 22.9–45.7).
3.2. Optimization of lesion segmentation methods
Optimization was performed on LST-LPA, nicMSlesions and BIANCA based on the highest SI to manual segmentation volume. Results are depicted in Supplemental Table 1 showing an optimal probability threshold of 0.25 for LST-LPA and 0.40 for nicMSlesions. Optimization of BIANCA (Supplemental Table 2) in our dataset resulted in the following settings: probability threshold 0.99; cluster size threshold 3; 2000 lesion points and equal number of non-lesion points in the training set; any location of the non-lesion training points; 3D patch with size 5; and spatial weighting 2.
3.3. Performance of lesion segmentation methods: untrained and trained on n = 14 subjects with MS
Volumetric and spatial reliability of the manual segmentations (i.e., intra-rater agreement) were good: for the three images that were segmented twice, ICC for absolute agreement was 0.867 and mean SI was 0.76 ± 0.04.
The median lesion volume according to manual delineation was 7.91 ml (interquartile range [IQR]: 4.26–10.15 ml). The performance of the different segmentation methods in subjects with RRMS is shown in Table 2. Note that the single-subject trained nicMSlesions is added to the table as an average over all fourteen configurations, but results from these variants separately are described in Section 3.4. A scatter-plot showing the lesion segmentation volumes compared to manual is depicted in Fig. 2.
Table 2.
Volumetric and spatial accuracy of the various grouped lesion segmentation methods compared to manual in the fourteen subjects with RRMS.
| RRMS n = 14 | Lesion volume | ICC absolute agreement | ICC consistency | True positive volume | True negative volume | False positive volume | False negative volume | Sensitivity | 1–Specificity (10E-3) | SI to manual |
|---|---|---|---|---|---|---|---|---|---|---|
| Manual | 7.91 (4.26–10.15) | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| LesionToads | 3.67 ⁎⁎⁎ (2.57–4.23) | 0.140 (−0.119–0.503) | 0.300 (−0.253–0.705) | 2.78 (1.18–3.28) | 11,092 (11,090–11,095) | 0.72 (0.48–1.11) | 5.07 (3.20–6.20) | 0.312 (0.230–0.405) | 0.065 (0.043–0.100) | 0.444 (0.336–0.542) |
| LST-LPA default | 4.40 ⁎⁎⁎ (1.86–7.19) | 0.73 (−0.059–0.939) | 0.947 (0.845–0.983) | 3.21 (1.48–5.81) | 11,091 (11,089–11,096) | 1.06 (0.44–2.20) | 4.02 (2.94–5.80) | 0.414 (0.298–0.520) | 0.096 (0.039–0.198) | 0.528 (0.425–0.581) |
| LST-LPA adjusted-threshold | 7.64 (3.82–11.15) | 0.917 (0.769–0.972) | 0.917 (0.764–0.973) | 4.46 (2.28–7.16) | 11,089 (11,086–11,095) | 2.95 (1.62–5.48) | 2.72 (2.07–4.40) | 0.592 (0.431–0.637) | 0.266 (0.145–0.494) | 0.568 (0.481–0.607) |
| NicMSlesions default | 8.91 * (5.03–12.51) | 0.872 (0.506–0.962) | 0.911 (0.747–0.971) | 4.26 (2.23–6.18) | 11,089 (11,085–11,094) | 4.75 (2.91–6.40) | 3.13 (1.81–4.85) | 0.553 (0.429–0.641) | 0.428 (0.262–0.577) | 0.490 (0.424–0.586) |
| NicMSlesions optimized | 7.39 (3.25–9.93) | 0.975 (0.928–0.992) | 0.975 (0.925–0.992) | 5.41 (2.22–7.11) | 11,091 (11,088–11,096) | 2.00 (1.27–3.05) | 2.32 (2.02–3.61) | 0.698 (0.536–0.736) | 0.180 (0.114–0.275) | 0.660 (0.613–0.716) |
| NicMSlesions single-subject # | 5.33 ⁎⁎⁎ (2.87–7.97) | 0.746 (0.189–0.893) | 0.854 (0.809–0.889) | 3.57 (1.77–5.29) | 11,091 (11,089–11,096) | 1.54 (0.95–2.60) | 3.59 (2.55–4.93) | 0.501 (0.401–0.591) | 0.139 (0.06–0.235) | 0.568 (0.490–0.638) |
| BIANCA | 7.54 (4.37–10.25) | 0.968 (0.905–0.990) | 0.966 (0.898–0.989) | 5.11 (2.67–6.79) | 11,090 (11,087–11,095) | 2.93 (1.92–3.78) | 2.67 (2.08–3.75) | 0.639 (0.521–0.686) | 0.264 (0.173–0.341) | 0.643 (0.514–0.675) |
Volumes are shown as median with interquartile range with the first and last quartile; intraclass correlation coefficients (ICC) are shown with 95% confidence interval. Note that the positive and negative volumes are extracted from the native FLAIR image.
nicMSlesions single-subject output is an average over all fourteen variants (also see Section 3.4).Statistics from repeated measures ANOVA with post-hoc pairwise Wilcoxon Signed Ranks test; * p < 0.05; ⁎⁎p < 0.01; ⁎⁎⁎p < 0.001.
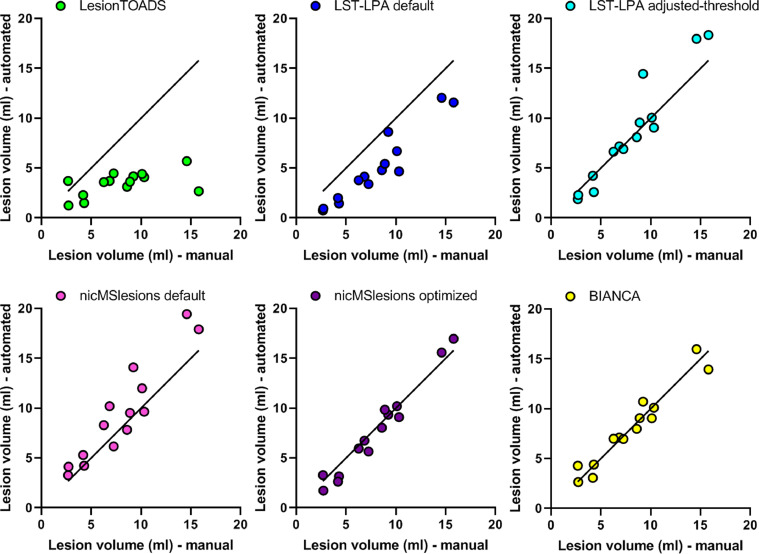
Fig. 2.
Scatter plots of the automated segmentation lesion volumes versus manual lesion volumes obtained from LesionTOADS (green), LST-LPA default (blue), LST-LPA adjusted-threshold (turquoise), nicMSlesions default (pink), nicMSlesions optimized (purple), and BIANCA (yellow). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The two supervised and trained methods nicMSlesions optimized and BIANCA showed the highest volumetric agreement (ICC absolute agreement = 0.975 and 0.968, respectively), as well as the highest sensitivity to lesions (0.698 and 0.639, respectively) and the best spatial agreement with manual (SI = 0.660 and 0.643, respectively). From the two supervised, untrained methods (i.e., LST-LPA default and adjusted-threshold, and nicMSlesions default), LST-LPA showed the best volumetric and spatial agreement, as well as best sensitivity to lesions, both for the default and for the probability threshold optimized configuration. The unsupervised, untrained method LesionTOADS performed worst on all volumetric and spatial measures, except for lesion specificity, which was higher than for the other methods.
Statistical analysis showed a significant effect of segmentation method on lesion volumes (F(6,78)=35.435, p < 0.001), with post-hoc testing showing this difference between manual and the methods LesionTOADS (p = 0.001), LST-LPA default (p = 0.001), nicMSlesions default (p = 0.022), but not for LST-LPA adjusted-threshold (p = 0.778), nicMSlesions optimized (p = 0.300) or BIANCA (p = 0.925). Friedman test for SI showed a main effect of method as well (χ2(5) = 56.082, p < 0.001), with post-hoc testing showing that these differences were significant between all combinations of methods except between: (a) LesionTOADS and nicMSlesions default; and (b) LST-LPA default and nicMSlesions default.
3.4. Performance of lesion segmentation methods: trained on n = 1 subject with MS
Next, we looked at the results from nicMSlesions with single-subject input. In this case, we excluded the subject that was used for training from the volumetric outcomes, leading to fourteen different mean lesion volumes for manual and for nicMSlesions single-subject (Table 3). Here, upon training with the single-subject with the lowest lesion volume (2.68 ml), nicMSlesions failed in one of the thirteen remaining subjects to segment the lesions; this was the only failure.
Table 3.
Volumetric and spatial accuracy of the different single-subjects configuration of nicMSlesions, each evaluated on the remaining thirteen subjects.
| Manual volume (ml) of single-subject used for training i.e., subject excluded from analysis | Median manual lesion volume (ml) with IQR | Median automated lesion volume (ml) with IQR | ICC absolute agreement with 95% CI | ICC consistency with 95% CI | Median Dice's similarity index to manual labels with IQR |
|---|---|---|---|---|---|
| 2.68 # | 8.73 (6.40–10.25) | 2.94 ** (2.45–4.24) | 0.131 (−0.088–0.499) | 0.396 (−0.201–0.779) | 0.363 (0.311–0.417) |
| 2.72 | 8.58 (5.27–10.20) | 8.18 (5.17–9.71) | 0.760 (0.397–0.919) | 0.758 (0.379–0.919) | 0.521 (0.462–0.581) |
| 4.19 | 8.58 (5.27–10.20) | 7.88 (4.63–10.79) | 0.941 (0.819–0.982) | 0.937 (0.806–0.980) | 0.574 (0.500–0.640) |
| 4.29 | 8.58 (5.22–10.20) | 8.02 (4.94–10.24 | 0.973 (0.914–0.992) | 0.971 (0.908–0.991) | 0.595 0.564–0.663) |
| 6.26 | 8.58 (4.34–10.20) | 5.44 ** (2.44–7.20) | 0.753 (−0.071–0.944) | 0.927 (0.779–0.977) | 0.596 (0.517–0.620) |
| 6.85 | 8.58 (4.34–10.20) | 5.48 ** (2.87–7.59) | 0.856 (−0.014–0.969) | 0.955 (0.861–0.986) | 0.562 (0.520–0.635) |
| 7.24 | 8.58 (4.34–10.20) | 6.09 ** 2.78–7.74) | 0.844 (−0.029–0.967) | 0.953 (0.853–0.985) | 0.599 (0.557–0.668) |
| 8.58 | 7.24 (4.24–10.20) | 6.15 ** (3.07–8.15) | 0.930 (0.284–0.985) | 0.972 (0.910–0.991) | 0.595 (0.529–0.663) |
| 8.88 | 7.24 (4.24–10.20) | 4.68 ** (2.31–650) | 0.727 (−0.072–0.938) | 0.930 (0.787–0.978) | 0.575 (0.439–0.630) |
| 9.22 | 7.24 (4.24–10.20) | 5.50 ** (2.19–7.34) | 0.819 (−0.052–0.963) | 0.963 (0.883–0.989) | 0.535 (0.479–0.653) |
| 10.09 | 7.24 (4.24–9.76) | 5.45 ** (2.39–7.23) | 0.855 (−0.034–0.970) | 0.962 (0.881–0.988) | 0.590 (0.513–0.648) |
| 10.31 | 7.24 (4.24–9.66) | 6.34 ** (2.58–8.09) | 0.885 (0.073–0.975) | 0.959 (0.870–0.987) | 0.606 (0.524–0.676) |
| 14.59 | 7.24 (4.24–9.66) | 4.81 ** (2.37–6.68) | 0.696 (−0.073–0.929) | 0.923 (0.769–0.976) | 0.571 (0.473–0.623) |
| 15.79 | 7.24 (4.24–9.66) | 3.86 ** (2.01–4.94) | 0.593 (−0.066–0.896) | 0.907 (0.723–0.971) | 0.540 (0.427–0.561) |
Abbreviations: IQR gives the interquartile range with the first and last quartile. Statistics from Wilcoxon Signed Ranks test; * p < 0.05; ⁎⁎p < 0.01; ⁎⁎⁎p < 0.001.
ICC for absolute agreement was lowest upon training single-subjects with the two lowest and the two highest lesion volumes (ICC absolute agreement ≤ 0.760). ICC consistency was also lower upon training single-subjects with low lesion volume (ICC consistency ≤ 0.758), but not upon input from subjects with higher lesion volumes. The best volumetric agreement was seen upon training using data of the single-subjects with lesion volumes of 4.19 or 4.29 ml (ICC absolute agreement = 0.941 and 0.973, respectively, and ICC consistency = 0.937 and 0.971, respectively).
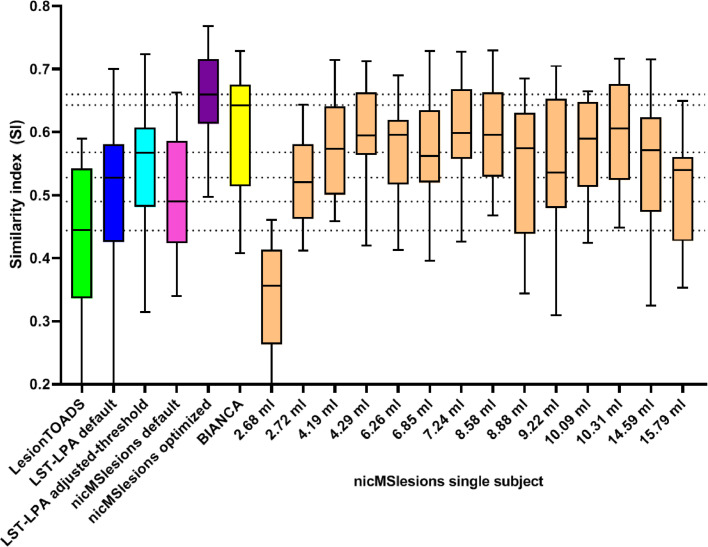
Spatial agreement for the single-subject configuration of nicMSlesions was good, with median SI varying from 0.363 (input lesion volume: 2.68 ml) to 0.606 (input lesion volume: 10.31 ml) and an average median SI of 0.559 (Table 3). Excluding the two subjects with lesion volumes ≤ 3.0 ml (i.e., the minimum lesion volume recommended by nicMSlesions for single-subject training (Valverde et al., 2019)), lowest median SI was 0.535 (input lesion volume: 9.22 ml) and the average median SI over all remaining twelve subjects was 0.578.
# One subject's images could not be segmented by this single-subject variant of nicMSlesions; therefore volume and Dice's similarity index are determined over twelve instead of thirteen subjects.
Wilcoxon Signed Ranks tests showed that in eleven out of fourteen cases, nicMSlesions single-subject volumetric output differed significantly from manual volumes.
Compared to the other supervised, trained methods BIANCA and nicMSlesions optimized, the single-subject trained nicMSlesions showed worse volumetric and spatial (Fig. 3) agreement, although this was highly dependent on the subject that was used for the training of the network. However, the single-subject training of nicMSlesions showed better volumetric and spatial agreement than the unsupervised, untrained method LesionTOADS in all thirteen successfully segmented cases; and also better than the supervised, untrained method LST-LPA default in ten (volumetric) or nine (spatial) of thirteen cases. The other two supervised, untrained methods (LST-LPA adjusted-threshold and nicMSlesions default) generally also performed worse than the single-subject nicMSlesions configuration.
Fig. 3.
Box-and-whiskers plot (min-to-max, line at median) showing Dice's similarity index (SI) in comparison to the manual lesion segmentation for LesionTOADS (green), LST-LPA default (blue), LST-LPA adjusted-threshold (turquoise), nicMSlesions default (pink), nicMSlesions optimized (purple), BIANCA (yellow), and the fourteen different nicMSlesions single-subject variants (orange). Horizontal dotted lines indicate the medians of the other automated lesion segmentation methods. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5. Performance of lesion segmentation methods: healthy controls
Last, we tested the various methods in healthy controls. The mean lesion volume measured in the five subjects, as well as the range from min-to-max is shown in Table 4. Since the manual lesion volume was 0 in all subjects, no volumetric or spatial reliability could be calculated.
Table 4.
Volumetric accuracy of the various grouped lesion segmentation methods compared to manual in the five healthy control subjects.
| HC n = 5 | Mean lesion volume ± SD (range min-to-max) |
|---|---|
| LesionToads | 0.81 ± 0.23 (0.45–1.08) |
| LST-LPA default | 0.40 ± 0.22 (0.07–0.67) |
| LST-LPA adjusted-threshold | 1.36 ± 0.11 (0.40–2.49) |
| NicMSlesions default | 2.23 ± 1.77 (0.00–5.18) |
| NicMSlesions optimized | 0.32 ± 0.43 (0.00–1.53) |
| NicMSlesions single-subject | 0.27 ± 0.34 (0.00–0.94) |
| BIANCA | 1.81 ± 0.83 (0.86–3.38) |
Results show that the measured lesion volumes in the healthy controls were generally low in all segmentation methods. However, the supervised, untrained method LST-LPA with threshold adjustment, the supervised, untrained version of nicMSlesions and the supervised and trained method BIANCA showed relatively higher false positive lesion volumes in these healthy controls, which was also seen in the subjects with MS (Table 2). The lowest false positive rate was seen when the single-subject trained network of nicMSlesions was used to segment the healthy control subjects.
4. Discussion
In this research, we compared four different lesion segmentation methods in different configurations to investigate the suitability of the deep learning cascaded CNN method nicMSlesions with input from only one manually delineated subject for multi-center trials. Although the supervised and trained methods nicMSlesions (re-train of full 11-layer cascaded CNN with own data) and BIANCA (optimized for own data) show the best volumetric and spatial agreement with manual, the single-subject configuration of nicMSlesions outperformed the unsupervised, untrained method LesionTOADS and the supervised, untrained method LST-LPA for both volumetric and spatial agreement with manual. Furthermore, the single-subject trained network of nicMSlesions showed the least false positives when tested on healthy controls.
There is a need for automatic lesion segmentation in MS, not only to assess lesion accrual itself, but also to assess structural brain changes such as atrophy, because the presence of MS lesions significantly hampers accurate brain segmentation (Gonzalez-Villa et al., 2017). Several reviews have been published showing an abundance of available MS lesion segmentation methods(Mortazavi et al., 2012; Garcia-Lorenzo et al., 2013) and since the emergence of machine learning even more methods have surfaced, as becomes evident in the various international MS lesion segmentation challenges (Carass et al., 2017; Commowick et al., 2018; Scully et al., 2008). An important feature of automated and accurate lesion segmentation, is robustness to new data from “unseen” centers, with potentially different MR vendors and acquisition protocols than those used during development (de Sitter et al., 2017). In this study, we show that re-training of the nicMSlesions network with data from an unseen center is possible with as little as one manual delineated subject as input data, showing the potential of this method for multi-center studies in MS. Important to note is that a cluster size threshold can be customized for this single-subject trained network of nicMSlesions as well, which may even further decrease the false positive rate of the segmentation.
One limitation of nicMSlesions is that for input lesion volumes below a certain threshold (in our dataset 5.0 ml), nicMSlesions has not enough data points to re-train the last two layers of the network, and therefore it only re-trains the last layer. Segmentation problems for low input lesion volumes are present in other automated lesion segmentation methods as well (Khayati et al., 2008; Schmidt et al., 2012; Steenwijk et al., 2013), although none of these methods use single-subject input. In our data, we see that training on subjects with lesion volumes below 3.0 ml yields less accurate segmentations than training on subjects with higher lesion volumes. However, training on subjects with lesion volumes between 3.0 and 5.0 ml shows good performance, even though only the last layer of the network was re-trained in these cases. Valverde et al. (2019) propose that this may be due to the importance of higher lesion numbers over higher lesion load, suggesting that input for re-training of the method should be chosen carefully in order to obtain the best results with the cascaded CNN method. The other configurations of nicMSlesions tested here (i.e., the default 11-layer CNN and the 11-layer re-trained CNN with probability threshold optimization) show no problems with the segmentation when four out of the fourteen subjects had lesion volumes below 5.0 ml, but we have no data when the network is re-trained with input from subjects with lesion volumes below 5.0 ml only, which should be tested in future work to assess the suitability of the method for cohorts with low lesion load or few lesions.
Our results as shown in Fig. 3 also seem to suggest that choosing training subjects that are most similar to a large part of the study population may be advantageous: while performance is roughly stable across the central part of the lesion volume distribution, partial re-training with subjects with an extreme lesion volume (i.e., very low or very high) appears to lead to reduced SI compared to manual.
We have shown that the default configuration of nicMSlesions can be optimized for protocols from an individual scanner. Even in a harmonized multi-center study with scanners from the same vendor, considerable differences in lesion volume in the same subject have been described (Shinohara et al., 2017). Hardware differences such as coil configuration, gradient and radio frequency amplifiers, and other differences such as acquisition parameters, spatial resolution, and filters used in image reconstruction, can have substantial effects on the appearance of lesion in FLAIR images and on their quantification using automated software. Therefore it is not unreasonable to suspect that training on one's own specific type of images could improve performance of the nicMSlesions segmentation, and our results suggest that this is indeed the case.
As in comparable papers on image analysis methods, while the main message can be appreciated visually from inspecting the graphs and tables reflecting measured data, we did include some statistical analyses. It should be noted that such statistical testing results are reported in this paper as auxiliary information. Given the relatively small number of subjects and the fact that quite a number of comparisons are of interest, as well as the more general debate on null hypothesis testing and the legitimacy of p-values (McShane et al., 2019), the resulting p-values should be interpreted with caution and are by no means intended to suggest any definitive answers to the questions posed.
This study has some limitations. First, manual delineation was performed by one single rater, thereby the gold standard presented here may be biased towards the individual rater. Furthermore, the data presented here is from one MR vendor, thereby no information on multi-center data could be included. However, the main objective of this study was to investigate the performance of the single-subject configuration of the deep learning cascaded CNN of nicMSlesions in comparison to other methods, while varying the amount of manual input and optimization possibilities, since this has not yet been reported. Of course, although all methods have already shown their multi-vendor validity in their separate proof-of-concept papers, these data should be replicated in other datasets to further prove that single-subject nicMSlesions outperforms the untrained methods LesionTOADS and LST-LPA. Because this paper focused on the nicMSlesions software, and because retraining is not as readily available for LST-LPA as for nicMSlesions, we did not also retrain LST-LPA using our own data. Such retraining could improve performance of LST-LPA which may affect the reported comparisons. However, it should be noted that the retraining of LST-LPA would require a group of patients rather than a single subject. Last, longitudinal performance of the single-subject configuration of nicMSlesions has not yet been shown, which should be further investigated.
In conclusion, the cascaded CNN lesion segmentation method nicMSlesions can be successfully re-trained with a limited amount of manual input, e.g., with only one manual delineation from new data, showing higher volumetric and spatial agreement with manual lesion segmentation than obtained with the commonly used untrained lesion segmentation methods LST-LPA and LesionTOADS. The method should be further optimized for cohorts with low lesion loads and in longitudinal, multi-center studies.
Funding
This work was supported by the Dutch Multiple Sclerosis Research Foundation (grant number 14-876 MS).
Declaration of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.nicl.2019.102074.
Appendix. Supplementary materials
References
- Carass A., Roy S., Jog A., Cuzzocreo J.L., Magrath E., Gherman A., Button J., Nguyen J., Prados F., Sudre C.H., Jorge Cardoso M., Cawley N., Ciccarelli O., Wheeler-Kingshott C.A.M., Ourselin S., Catanese L., Deshpande H., Maurel P., Commowick O., Barillot C., Tomas-Fernandez X., Warfield S.K., Vaidya S., Chunduru A., Muthuganapathy R., Krishnamurthi G., Jesson A., Arbel T., Maier O., Handels H., Iheme L.O., Unay D., Jain S., Sima D.M., Smeets D., Ghafoorian M., Platel B., Birenbaum A., Greenspan H., Bazin P.L., Calabresi P.A., Crainiceanu C.M., Ellingsen L.M., Reich D.S., Prince J.L., Pham D.L. Longitudinal multiple sclerosis lesion segmentation: resource and challenge. Neuroimage. 2017;148:77–102. doi: 10.1016/j.neuroimage.2016.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commowick O., Istace A., Kain M., Laurent B., Leray F., Simon M., Pop S.C., Girard P., Ameli R., Ferre J.C., Kerbrat A., Tourdias T., Cervenansky F., Glatard T., Beaumont J., Doyle S., Forbes F., Knight J., Khademi A., Mahbod A., Wang C.L., McKinley R., Wagner F., Muschelli J., Sweeney E., Roura E., Llado X., Santos M.M., Santos W.P., Silva A.G., Tomas-Fernandez X., Urien H., Bloch I., Valverde S., Cabezas M., Vera-Olmos F.J., Malpica N., Guttmann C., Vukusic S., Edan G., Dojat M., Styner M., Warfield S.K., Cotton F., Barillot C. Objective evaluation of multiple sclerosis lesion segmentation using a data management and processing infrastructure. Sci. Rep. 2018;8:13650. doi: 10.1038/s41598-018-31911-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danelakis A., Theoharis T., Verganelakis D.A. Survey of automated multiple sclerosis lesion segmentation techniques on magnetic resonance imaging. Comput. Med. Imaging Graph. 2018;70:83–100. doi: 10.1016/j.compmedimag.2018.10.002. [DOI] [PubMed] [Google Scholar]
- de Sitter A., Steenwijk M.D., Ruet A., Versteeg A., Liu Y., Schijndel R.A.van, Pouwels P.J.W., Kilsdonk I.D., Cover K.S., van Dijk B.W., Ropele S., Rocca M.A., Yiannakas M., Wattjes M.P., Damangir S., Frisoni G.B., Sastre-Garriga J., Rovira A., Enzinger C., Filippi M., Frederiksen J., Ciccarelli O., Kappos L., Barkhof F., Vrenken H., M. S. group and G. for NEU Performance of five research-domain automated WM lesion segmentation methods in a multi-center MS study. Neuroimage. 2017;163:106–114. doi: 10.1016/j.neuroimage.2017.09.011. [DOI] [PubMed] [Google Scholar]
- Garcia-Lorenzo D., Francis S., Narayanan S., Arnold D.L., Collins D.L. Review of automatic segmentation methods of multiple sclerosis white matter lesions on conventional magnetic resonance imaging. Med. Image Anal. 2013;17(1):1–18. doi: 10.1016/j.media.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Villa S., Valverde S., Cabezas M., Pareto D., Vilanova J.C., Ramio-Torrenta L., Rovira A., Oliver A., Llado X. Evaluating the effect of multiple sclerosis lesions on automatic brain structure segmentation. Neuroimage Clin. 2017;15:228–238. doi: 10.1016/j.nicl.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffanti L., Zamboni G., Khan A., Li L., Bonifacio G., Sundaresan V., Schulz U.G., Kuker W., Battaglini M., Rothwell P.M., Jenkinson M. BIANCA (Brain intensity abnormality classification algorithm): a new tool for automated segmentation of white matter hyperintensities. Neuroimage. 2016;141:191–205. doi: 10.1016/j.neuroimage.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khayati R., Vafadust M., Towhidkhah F., Nabavi M. Fully automatic segmentation of multiple sclerosis lesions in brain MR FLAIR images using adaptive mixtures method and Markov random field model. Comput. Biol. Med. 2008;38(3):379–390. doi: 10.1016/j.compbiomed.2007.12.005. [DOI] [PubMed] [Google Scholar]
- McShane B.B., Gal D., Gelman A., Robert C., Tackett J.L. Abandon statistical significance. Am. Stat. 2019;73:235–245. (sup1) [Google Scholar]
- Mortazavi D., Kouzani A.Z., Soltanian-Zadeh H. Segmentation of multiple sclerosis lesions in MR images: a review. Neuroradiology. 2012;54(4):299–320. doi: 10.1007/s00234-011-0886-7. [DOI] [PubMed] [Google Scholar]
- Schmidt P. LMU München; 2017. Bayesian Inference for Structured Additive Regression Models for Large-Scale Problems with Applications to Medical Imaging. [Google Scholar]
- Schmidt P., Gaser C., Arsic M., Buck D., Forschler A., Berthele A., Hoshi M., Ilg R., Schmid V.J., Zimmer C., Hemmer B., Muhlau M. An automated tool for detection of FLAIR-hyperintense white-matter lesions in multiple sclerosis. Neuroimage. 2012;59(4):3774–3783. doi: 10.1016/j.neuroimage.2011.11.032. [DOI] [PubMed] [Google Scholar]
- Shiee N., Bazin P.L., Ozturk A., Reich D.S., Calabresi P.A., Pham D.L. A topology-preserving approach to the segmentation of brain images with multiple sclerosis lesions. Neuroimage. 2010;49(2):1524–1535. doi: 10.1016/j.neuroimage.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara R.T., Oh J., Nair G., Calabresi P.A., Davatzikos C., Doshi J., Henry R.G., Kim G., Linn K.A., Papinutto N., Pelletier D., Pham D.L., Reich D.S., Rooney W., Roy S., Stern W., Tummala S., Yousuf F., Zhu A., Sicotte N.L., Bakshi R., Cooperative N. Volumetric analysis from a harmonized multisite brain MRI study of a single subject with multiple sclerosis. AJNR Am. J. Neuroradiol. 2017;38(8):1501–1509. doi: 10.3174/ajnr.A5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwijk M.D., Pouwels P.J., Daams M., van Dalen J.W., Caan M.W., Richard E., Barkhof F., Vrenken H. Accurate white matter lesion segmentation by k nearest neighbor classification with tissue type priors (kNN-TTPs) Neuroimage Clin. 2013;3:462–469. doi: 10.1016/j.nicl.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully M., Magnotta V., Gasparovic C., Pelligrimo P., Feis D.-L., Bockholt H.J. 3D segmentation in the clinic: A grand challenge II at MICCAI 2008 - MS lesion segmentation. Grand Challenge Work. Mult. Scler. Lesion Segm. Challenge. 2008 [Google Scholar]; https://www.researchgate.net/publication/28359621_3D_segmentation_in_the_clinic_A_grand_challenge_II_at_MICCAI_2008_-_MS_lesion_segmentation
- Valverde S., Cabezas M., Roura E., Gonzalez-Villa S., Pareto D., Vilanova J.C., Ramio-Torrenta L., Rovira A., Oliver A., Llado X. "Improving automated multiple sclerosis lesion segmentation with a cascaded 3D convolutional neural network approach. Neuroimage. 2017;155:159–168. doi: 10.1016/j.neuroimage.2017.04.034. [DOI] [PubMed] [Google Scholar]
- Valverde S., Salem M., Cabezas M., Pareto D., Vilanova J.C., Ramio-Torrenta L., Rovira A., Salvi J., Oliver A., Llado X. "One-shot domain adaptation in multiple sclerosis lesion segmentation using convolutional neural networks. Neuroimage Clin. 2019;21 doi: 10.1016/j.nicl.2018.101638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.