Abstract
Purpose:
To propose a rapid multi-slice T1 measurement method using time-resolved imaging of contrast kinetics (TRICKS) and a semi-automated image processing algorithm for comprehensive assessment murine kidney function using dynamic contrast-enhanced MRI (DCE-MRI).
Methods:
A multi-slice TRICKS sampling scheme was implemented in an established rapid T1 measurement method. A semi-automated image-processing scheme employing basic image processing techniques and machine learning was developed to facilitate image analysis. Reliability of the multi-slice technique in measuring renal perfusion and glomerular filtration rate (GFR) was tested in normal mice (n=7 for both techniques) by comparing to the validated single-slice technique. Utility of this method was demonstrated on mice after either sham surgery (n=7) or induction of unilateral renal artery stenosis (RAS, n=8). Renal functional parameters were extracted using a validated bi-compartment model.
Results:
The TRICKS sampling scheme achieved an acceleration factor of 2.7, allowing imaging of eight axial slices at 1.23 sec/scan. With the aid of the semi-automated scheme, image analysis required under 15-min for both kidneys per mouse. The multi-slice technique yielded renal perfusion and GFR values comparable to the single-slice technique. Model-fitted renal parameters successfully differentiated control and stenotic mouse kidneys, including renal perfusion (706.5±164.0 vs. 375.9±277.9mL/100g/min, P=0.002), blood flow (1.6±0.4 vs. 0.7±0.7mL/min, P<0.001), and GFR (142.9±17.9 vs. 58.0±42.8μL/min, P<0.001).
Conclusion:
The multi-slice TRICKS-based DCE-MRI technique, with a semi-automated image processing scheme, allows rapid and comprehensive measurement of murine kidney function.
Keywords: Dynamic contrast enhanced MRI, Time-resolved imaging of contrast kinetics, Kidney volume, Renal functional biomarkers, Machine learning
1. Introduction
Kidney disease a major public health issue [1] and may eventually progress to end-stage renal failure, which requires dialysis or kidney transplantation, imposing enormous socioeconomic burdens to affected individuals [2]. Furthermore, some forms of renal diseases may involve patchy lesions or heterogeneous involvement, such as polycystic kidney disease. Animal models of kidney diseases play an increasingly important role in investigating the pathophysiology underpinning the onset and progression of various renal diseases [3, 4]. For assessment of renal function, most researchers have been relying on relevant urine and serum biomarkers [5]. Nevertheless, these biomarkers are insensitive to early functional alterations [5, 6] and cannot evaluate single-kidney function.
Gadolinium-based dynamic contrast enhanced MRI (DCE-MRI) has emerged as a powerful tool for simultaneous characterization of single-kidney anatomy and function [7–9]. By following the passage of gadolinium contrast from input arteries through the renal parenchyma where renal perfusion, filtration, and tubular flow occur, renal functional parameters can be quantified with the aid of mathematical models [7, 10–12]. In addition, DCE-MRI also allows delineation of the dynamics of other paramagnetic metal chelates [13] and the dioxygen molecule [14, 15]. For reliable estimation of renal parameters, accurate delineation of contrast dynamics in the abdominal aorta and renal parenchyma is necessary. T1-weighted imaging is typically used to capture contrast transits in kidney, but suffers from inaccuracy due to the presence of B1 homogeneities [16, 17]. Alternatively, rapid T1 measurement insensitive to B1-inhomogeniety offers more reliable delineation of contrast dynamics, given the linear relationship between R1 (1/T1) and contrast concentration.
Recently, we developed a single-slice DCE-MRI technique and a modified two-compartment model for reliable evaluation of glomerular filtration rate (GFR) and renal perfusion in mice [18]. The rapid T1 measurement method saturation recovery snapshot-fast low angle shot (Snapshot-FLASH) was used to capture contrast dynamics in one axial slice across the renal hilum. The validity of this technique in measuring single-kidney GFR and perfusion has been demonstrated by comparing to reference standard inulin clearance method and to arterial spin labeling techniques, respectively. However, a single slice may not be representative of the entire kidney, especially in kidneys with focal disruptions or lesions. Moreover, an additional three-dimensional scan and labor-intensive image segmentation is needed for measurement of kidney volume in order to calculate whole kidney renal blood flow (RBF) and GFR. Therefore, development of a multi-slice DCE-MRI technique using rapid T1 measurement would be useful for simultaneous measurement of renal volume and hemodynamics in mice.
Undersampling in k-space is required to balance the tradeoff between high spatial and temporal resolution for kidney DCE-MRI. A number of fast imaging techniques [19–23] derived from keyhole imaging [24], including the time-resolved imaging for contrast kinetics (TRICKS) [25] have been developed to facilitate image acquisition. In TRICKS, the 3D Cartesian k-space is divided into four regions based on distance from the center k-space line in the phase encoding direction. The central region with lower spatial frequencies dominates the image contrast and is sampled more frequently than the three peripheral regions. Then view sharing is implemented during image reconstruction [25]. Such k-space undersampling techniques were originally developed to capture rapid acquisition of contrast passage in MR angiography and the TRICKS technique has recently been shown useful in renal DCE-MRI [26]. However, the TRICKS scheme has been applied in T1-weighted DCE-MRI, where the MR signal is typically at steady state. Its applicability in facilitating image acquisition during saturation recovery-based T1 measurement and the accuracy of the fitted renal parameters from the undersampled DCE-MRI images remain to be investigated.
In this study, we aimed to adapt the TRICKS scheme in a 2D T1 measurement method, i.e., multi-slice saturation recovery Snapshot-FLASH, for renal DCE-MRI in mice. Furthermore, because manual image segmentation is labor intensive and time consuming for the multi-slice dataset, a semi-automated scheme employing basic image processing techniques and machine learning was developed to facilitate DCE-MRI image analysis. The validity of multi-slice technique in measuring renal function was tested on normal mice by comparing to the validated single-slice technique. The utility of this multi-slice technique was demonstrated on mice with unilateral renal artery stenosis (RAS), which induces a fall in GFR and RBF in one kidney.
2. Materials and methods
2.1. Imaging Method.
During DCE-MRI, contrast dynamics were measured using the previously developed saturation recovery Snapshot-FLASH method [18]. A TRICKS-based k-space data acquisition scheme was developed to facilitate multi-slice imaging (Fig. 1). A total of 128 phase encoding lines were sampled during image acquisition. These lines were segmented into low (L, 28 center lines) and high (H, 100 peripheral lines) frequency lines (Fig. 1a). The H lines were further divided into five groups (noted by different colors), with each interleaving with lines from other groups in a sequential manner. During data acquisition, while the L lines were constantly sampled, only one of the H groups was acquired per repetition (Fig. 1b). As such, an acceleration factor of 2.7 was achieved by using this acquisition scheme, allowing imaging of eight slices at a temporal step size of 1.23 sec/scan. During each repetition, the eight slices were sampled during saturation recovery with delays from 0.25 to 1.05 sec (Fig. 1c).
Figure 1. The TRICKS sampling scheme used in the multi-slice saturation recovery Snapshot-FLASH technique.
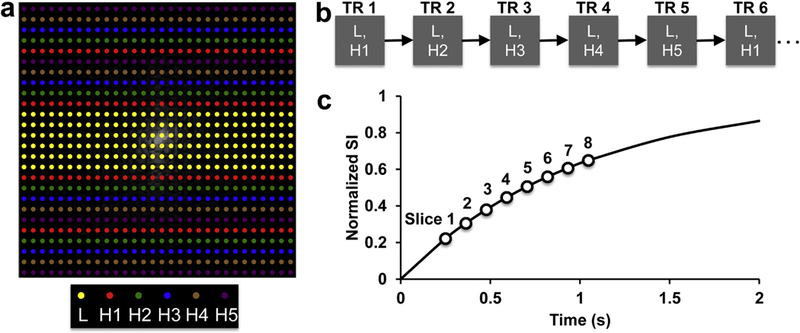
(a) The 128 phase encoding lines were segmented into low (L, 28 center lines) and high (H, 100 peripheral lines) frequency lines. The H lines were further divided into 5 groups (noted by different colors), with each interleaving with lines from other groups in a sequential manner. (b) During data acquisition, while the L lines were constantly sampled, only one of the H groups was acquired per repetition. (c) A total of eight slices were sampled during saturation recovery with delays from 0.25 to 1.05 sec. A T1 of 1 sec is assumed in the recovery curve.
2.2. Animals.
This study was approved by the local Institutional Animal Care and Use Committee. To test the feasibility and utility of the proposed DCE-MRI technique in both normal and diseased mouse kidneys, sixteen 12-week old male C57/BL6 mice underwent either sham or unilateral RAS surgeries. The surgical procedures have been described previously [27–29]. Briefly, anesthetized mice were placed prone on a warm heating pad at approximately 37°C. Animals were given Buprenorphine SR (0.5mg/kg) for 72 hours subcutaneously before surgery for analgesic treatment. Following a flank incision of approximately 1 cm proximal to the right kidney, the right renal artery was dissected from the renal vein. To induce RAS, a polytetrafluoroethylene cuff with 0.20 mm inner diameter and 0.5 mm length was placed around the renal artery and tied with 10–0 nylon sutures. Sham surgery was similarly performed but without cuff placement. Then the incisions were sutured and mice returned to their cages. Possible adverse effects include bleeding during surgery and skin infection after surgery, and mice were therefore treated with antibiotic topical ointment right after surgery. One mouse died during the RAS surgery due to bleeding. The remaining fifteen mice (sham, n=7 and RAS, n=8) all successfully underwent MRI two weeks after surgery and provided analyzable data.
In order to test the validity of the multi-slice technique, another cohort of seven age-matched male C57/BL6 mice with sham surgery underwent renal function measurement using the validated single-slice DCE-MRI technique, as described previously [18]. Only one slice across the kidney hilum was scanned, from which renal functional parameters, including renal perfusion and normalized GFR, were quantified. For the seven sham mice scanned by the multi-slice DCE-MRI technique, the same renal parameters were measured from one single slice across the renal hilum, and compared against those derived by the single-slice technique.
2.3. MRI.
All MRI studies were performed on a vertical 16.4 T scanner equipped with a single-element 38-mm inner diameter birdcage coil (Bruker, Billerica, MA). Upon initial anesthesia by 3% isoflurane, all mice underwent tail-vein catheterization by a home-made fine needle (30G), which was connected to a syringe pump (Braintree Scientific, Braintree, MA) by a polytetrafluoroethylene tubing (0.15 mm inner diameter) for injection of contrast agent during DCE-MRI. Animals were secured in a mouse cradle and placed vertically in the MR scanner. Maintenance of anesthesia during MRI was achieved by 1–2.5% isoflurane mixed with pure oxygen. Body temperature was kept between 36°C and 37°C by blowing warm air on the mice. Real-time visualization of animal respiration and body temperature were achieved using a physiological monitoring system (SA Instruments, Stony Brook, NY).
For kidney localization, an IntraGateFLASH sequence was used to image two axial slices across the hilum of the two kidneys and one coronal slice showing the largest cross-sectional area of both kidneys. Other imaging parameters were: TR 20 ms; TE 1.24 ms; flip angle 15°; slice thickness 1.0 mm; field of view 2.56×2.56 cm2; matrix size 128×128. The axial images were used to prescribe the 3D kidney volume scan and the coronal image for prescription of the eight axial slices of the DCE-MRI scans.
In order to test the accuracy of the multi-slice TRICKS technique in estimating mouse kidney volume, reference volumetric measurement was performed using a three-dimensional fast imaging with steady precession (3D-FISP) sequence. Imaging parameters were: TR 14 ms; TE 2.7 ms; flip-angle 20°; field of view 5.12×2.56×1.28 cm3; matrix size 256×128×64; number of averages 2.
In DCE-MRI, contrast dynamics were captured using the saturation recovery Snapshot-FLASH sequence combined with the multi-slice TRICKS technique for image acquisition. In order to achieve full magnetization saturation, three 0.1-ms nonselective 90° radiofrequency pulses each followed by spoil gradients in two directions were applied [17, 30]. Rapid T1 measurement was by acquisition of one proton density (M0) and one T1-weighted (Mt) image with certain delay time after saturation recovery. The eight axial slices were prescribed with slice distances from 1.5 to 2.0 mm, in order to cover both kidneys in the longitudinal direction. For each slice, ten proton density (M0) images were first acquired with a repetition time of 18 sec, which allowed full recovery of the longitudinal magnetization of the mouse kidney with a maximal T1 of ~3.1 s in inner medulla at 16.4 T [31]. Then following the acquisition of ten baseline T1-weighted (Mt) images, 20 μL of 37.5mM gadodiamide was injected through the tail-vein within 2 sec, after which the Mt images were acquired repetitively. Other imaging parameters were: encoding scheme, central; TR 17.9 ms; TE 0.8 ms; flip angle 15°; slice thickness 1.5 mm; field of view 2.56×2.56 cm2; matrix size 128×128; number of repetitions 100.
2.4. Image Analysis.
Reference kidney volume was measured from the 3D-FISP images using Analyze™ (version 12.0, Biomedical Imaging Resource, Mayo Clinic, MN). Renal volume was measured by manual segmentation of kidney on all frames where kidney was observed. The DCE-MRI dataset was analyzed using a MatLab® (Mathworks, Natick, MA) module developed in-house. For image reconstruction, view sharing of the non-sampled high frequency lines from four most adjacent frames was implemented. Images with noticeable motion artifacts were manually excluded. The remaining M0 images were averaged to increase signal-to-noise ratio. As described previously [9, 18], T1 maps were generated pixel-wise by a simple exponential fitting with T1 as the only unknown parameter, which was justified by the superior signal-to-noise ratio at 16.4 T (>100 in the M0 image) and nearly-perfect (>98%) saturation efficiency by our saturation module [18]. The accuracy of such two-point T1 measurement has been previously verified in both phantom and in-vivo studies [18]. Then changes in T1 relaxation rate ΔR1 from baseline, which is linearly proportional to the gadodiamide concentration, was used to measure the relative concentration of gadodiamide in tissue. This method assumes the relaxivity of gadodiamide at 16.4T is identical in all relevant compartments. To facilitate image analysis, a semi-automated scheme (Fig. 2) was developed to extract the arterial input function (AIF), kidney dynamics, and output.
Figure 2. Schematic illustrations of the semi-automated scheme for image analysis.
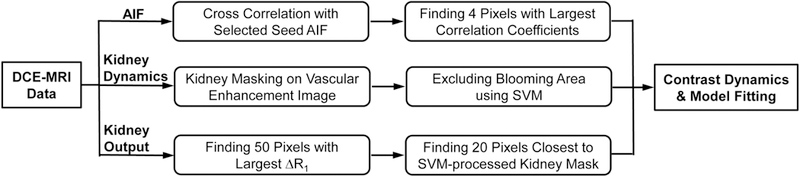
The AIF is measured from the abdominal aorta by using cross correlation. Kidney dynamics are measured from the renal parenchyma. First, kidney segmentation was performed by interactive image thresholding, opening, and edge smoothing on vascular enhancement images. Second, a supervised machine learning algorithm support vector machine (SVM) was trained and implemented, in order to exclude renal pelvic regions with gadolinium blooming effect. Kidney output was automatically measured from the slice containing the kidney hilum with a thresholding technique. Then extracted gadolinium dynamics were fitted by a bi-compartment model to measure renal parameters.
The AIF was measured from the abdominal aorta by using cross correlation. One seed pixel with representative AIF was manually selected. Cross correlation between this seed AIF and dynamic curves from all adjacent pixels in a 9×9 matrix was performed. For each slice, four pixels with the largest correlation coefficients were identified and used to calculate the AIF [18]. Then the averaged AIF curve from different slices was obtained for model input.
Kidney segmentation was performed by interactive image thresholding, opening, and edge smoothing on vascular enhancement images, which were obtained by subtracting images acquired at vascular phases of contrast enhancement from the baseline images. Optional manual segmentation was available for dysfunctional kidneys with little contrast enhancement. Then the kidney volume was estimated from these generated kidney masks and the IntraGateFLASH-acquired coronal image, assuming a linear change in kidney size between axial slices and cone-shaped kidney poles. Supposing mask area for one kidney is denoted as Si (i=1,2,…n, where n is the number of slices covering this kidney), slice gap as d, and distances between kidney poles to the closest imaging slices as du and di, then the kidney volume (V) is calculated as
In order to exclude renal pelvic regions with gadolinium blooming effect (signal voids due to significant T2* shortening induced by high gadolinium concentration), a supervised machine learning algorithm support vector machine (SVM) was trained and applied (Appendix). The contrast dynamic ΔR1 curve was used for SVM training and subsequent classification. This SVM algorithm provided a classification accuracy of 95%. This yielded effective kidney volume (Veff), from which the kidney dynamics were quantified by averaging the dynamic curves from all pixels. Kidney output curve was measured from the slice containing the kidney hilum. Fifty pixels with the largest ΔR1 were first identified, from which twenty pixels closest to the SVM-processed kidney mask of the same slice were selected to obtain the averaged kidney output curve. Manual segmentation of renal pelvis was performed for stenotic kidneys with modest pelvic enhancement.
The DCE-MRI dataset from the seven mice scanned using the single-slice DCE-MRI technique was analyzed by manually selecting image ROIs, as described previously [18]. Then contrast dynamics, including the AIF, kidney dynamics, and output curves, were measured for model fitting of renal parameters.
The measured contrast dynamics were fitted using a previously developed bi-compartmental model [18], which was modified by adding a transit delay of blood from abdominal aorta to kidney. In this model, blood flow from abdominal aorta enters the kidney, which is simplified as a combination of the vascular (plasma) and tubular (filtrates) compartments, gets filtered by glomeruli, and flows out of the kidney through the renal pelvis. Notably, the plasma AIF was calculated as AIF/(1-Hematocrit) and used as the input to the model fitting. This model contains a total of five unknown parameters, including the blood transit delay Td, perfusion rate constant kp, renal plasma volume fraction fp, normalized GFR kGFR, and contrast efflux rate constant kout. Parameter fitting was performed by minimizing residual differences between the measured and fitted data using the Levenberg-Marquardt algorithm. Then renal perfusion (mL/100g/min) was calculated as kp·fp·6000/(1-Hematocrit)·λ, where λ is blood/tissue partition coefficient for water (0.9ml/g) [32], RBF as Perfusion·Veff, and GFR as kGFR·Veff·60. Additionally, renal perfusion maps were fitted from the DCE-MRI images acquired within the first 20 seconds after contrast injection [33], which contains the first-pass the contrast bolus, using the model to demonstrate the feasibility of the proposed multi-slice technique in perfusion mapping.
2.5. Statistical Analysis.
All statistical analysis was performed using JMP 10.0 (SAS Institute, Cary, NC). Results were expressed as means±standard deviations. One-way analysis of variance was followed by Student’s unpaired or paired t-test for statistical comparisons between groups, as appropriate. Pearson correlation and Bland-Altman analysis were used for comparison of estimated kidney volumes by the multi-slice TRICKS technique and 3D-FISP. A P value smaller than 0.05 was considered statistically significant.
3. Results
Representative Mt images of four slices of one control kidney imaged using the proposed multi-slice TRICKS technique are shown in Fig 3. Full dynamic image series for control and RAS mice is shown in Supplemental Animations 1 and 2, respectively. Despite k-space under-sampling by TRICKS, view-sharing of k-space data from adjacent frames offered satisfactory image quality. The varying image contrast of different slices resulted from different T1 weighting due to their different saturation recovery times. Following acquisition of the baseline Mt images, strong signal enhancement in the abdominal aorta is observed after injection of the contrast agent. Then the renal cortex and medulla are sequentially intensified, reflecting renal perfusion, filtration, and tubular flow [34]. Notably, the high concentration of gadolinium in the renal pelvic region causes blooming effect, or signal dropout instead of increase, at later tubular phases. These regions need to be excluded to guarantee reliable curve fitting and parameter estimation.
Figure 3. Representative T-weighted images acquired using the proposed DCE-MRI technique.
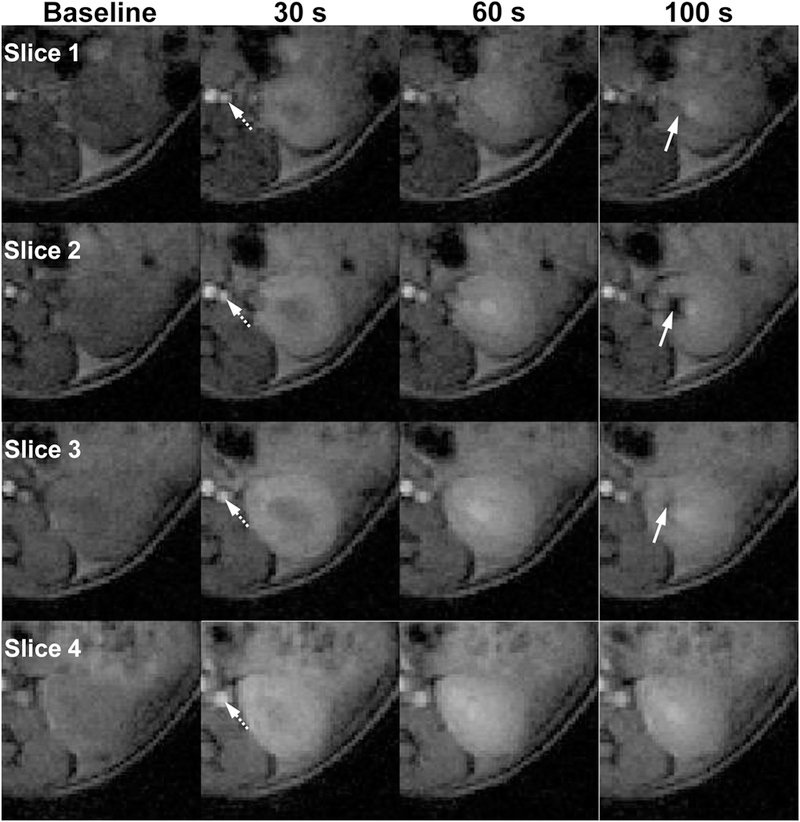
Images of four adjacent slices of one normal kidney at baseline, 30, 60, and 100 sec are shown. Sequential enhancement of different renal regions at different time points reflects renal perfusion, filtration, and tubular flow. Notably, gadolinium blooming effect causes signal dropout at late tubular phase, as indicated by white arrows.
With the aid of these semi-automated methods, image analysis including model fitting required under 15 min for both kidneys per mouse. Representative kidney masks semi-automatically selected using the proposed segmentation technique are shown in Fig. 4a. These masks reliably outlined both kidneys in all eight imaging slices. The estimated renal volume from these masks showed a good correlation with the reference kidney volume by 3D-FISP (Fig. 4b, R2=0.967, P<0.001). A Bland-Altman plot also demonstrated a good agreement between kidney volumes obtained by these two methods (Fig. 4c, mean difference, 0.98±20.34μL).
Figure 4. Measurement of kidney volume from the multi-slice DCE-MRI dataset.
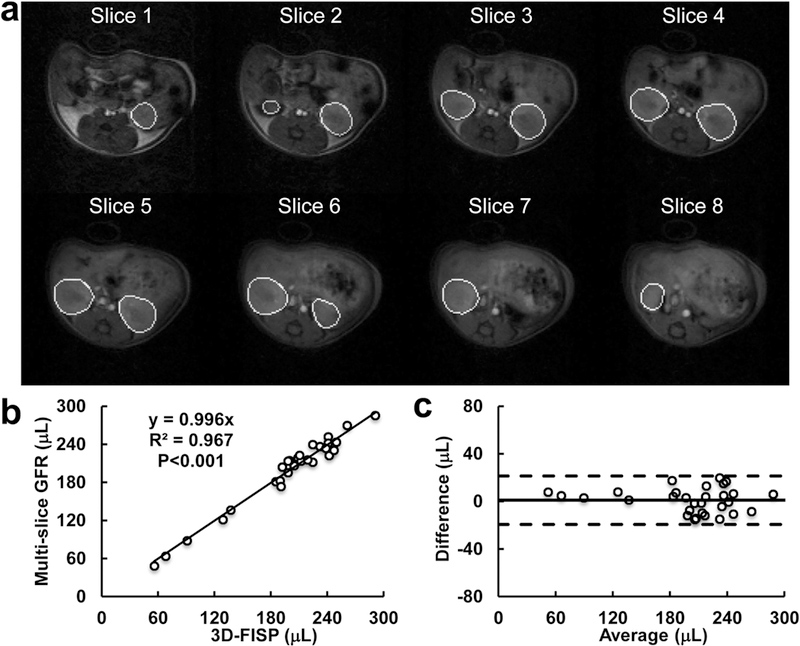
(a) Representative kidney masks semi-automatically selected using the proposed segmentation technique. The background images were obtained at early vascular phase. (b-c) Linear correlation (b) and Bland-Altman analysis (c) of kidney volume estimated from the DCE-MRI and 3D-FISP datasets.
The corresponding SVM-processed kidney masks overlaid on Mt images at late tubular phase are shown in Fig. 5a. The generated linear SVM reliably detected and excluded pixels with gadolinium blooming effect. The automatically selected pixels for measurement of contrast output curves in the left and right kidneys are also highlighted in slice 3 and 5, respectively. Representative AIF, kidney dynamics and output curves are shown in Fig. 5b, which are similar to our previous observations [18]. The AIF is characterized by a transient peak and gradual decrease. An initial dramatic signal increase occurs in the renal parenchyma due to perfusion, followed by a second rise as a result of tubular accumulation of contrast, and a subsequent decrease caused by contrast outflow. The kidney output curve also sees a similar but smaller increase indicating low medullary perfusion, followed by a plateau before another drastic increase due to arrival of contrast following tubular flow, and eventually a gradual decrease due to contrast clearance from kidney.
Figure 5. Exclusion of blooming areas and model fitting of contrast dynamics.
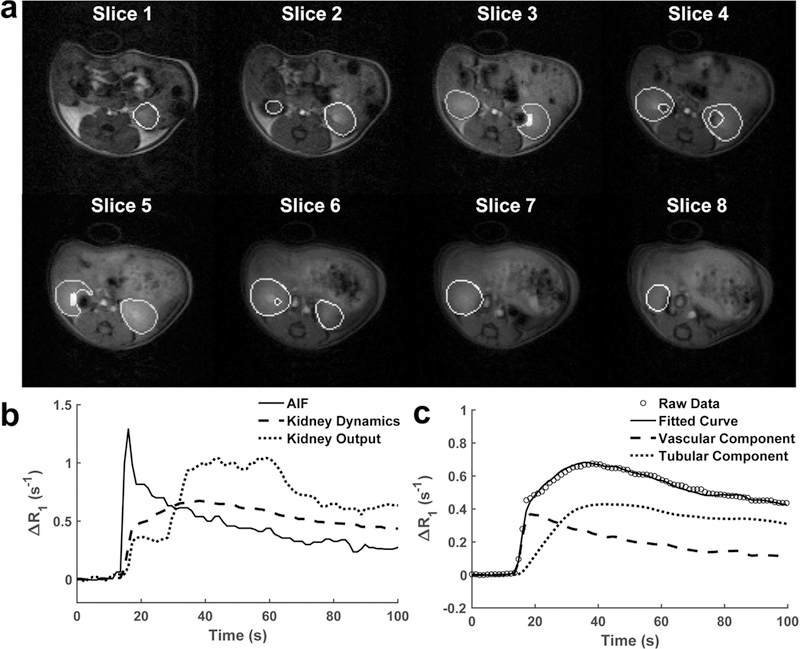
(a) SVM-processed kidney masks overlaid on Mt images at late tubular phase. Pixels automatically selected for measurement of kidney output curves are also highlighted on slice 3 and 5 for the left and right kidneys, respectively. (b) Representative extracted AIF, kidney dynamics and output curves. (c) The representative raw and model-fitted gadolinium dynamics as well as the vascular and tubular components in the renal parenchyma.
The representative raw and model-fitted gadolinium dynamics as well as the vascular and tubular components in the renal parenchyma are shown in Fig. 5c. The bi-compartment model provided a good fitting to the experimentally-acquired data at both vascular and tubular phases. Model-fitted parameters are shown in Table 1. No difference was observed in the blood transit delay (Td) between the control, stenotic and contralateral kidneys. The perfusion rate constant kp in the stenotic kidney was significantly decreased compared to controls (P=0.009). Lower renal plasma volume (fP) was also found in the stenotic kidneys, as compared to both control (p=0.002) and contralateral (p=0.020) kidneys. Similarly, the normalized GFR (kGFR) dropped in the stenotic kidneys (P<0.001 vs. control and contralateral kidneys). The contrast efflux rate constant (kout) were similar in the three groups of kidneys.
Table 1.
Model-fitted parameters in control, stenotic, and contralateral kidneys
| Control Kidney | Stenotic Kidney | Contralateral Kidney | |
|---|---|---|---|
| Td (s) | 0.17±0.13 | 0.26±0.11 | 0.21±0.10 |
| kp (s−1) | 0.46±0.12 | 0.30±0.11* | 0.37±0.16 |
| fp | 0.18±0.04 | 0.11±0.06* | 0.17±0.03† |
| kGFR (s−1) | 0.011±0.001 | 0.006±0.002* | 0.012±0.001† |
| kout (s−1) | 0.014±0.003 | 0.011±0.007 | 0.013±0.004 |
Data expressed as mean±standard deviation.
P<0.05 compared to control kidney.
P<0.05 compared to stenotic kidney.
As shown in Fig. 6a, comparable renal perfusion maps in control mouse kidneys were obtained by the single- (left) and multi-slice (right) techniques. The multi-slice method adequately captured the heterogeneity of intra-renal perfusion, showing that the renal cortex has the highest perfusion, followed by the outer medulla, inner medulla, and papilla. This perfusion heterogeneity showed a good spatial concordance with an anatomical mouse kidney section (right) obtained ex-vivo. The multi-slice technique yielded comparable renal perfusion (Fig. 6b, 706.5±164.0 vs. 719.0±228.8mL/100g/min) and normalized GFR kGFR (Fig. 6c, 0.011±0.001 vs. 0.011±0.001 s−1) values compared to the single-slice technique, supporting the high reliability of the multi-slice technique in measuring renal function despite the implementation of TRICKS undersampling and view sharing in k-space. For the multi-slice technique, the renal perfusion and kGFR derived from the slice across the renal hilum were also similar to those obtained from all imaged slices (Table 1), justifying the measurement of renal function in murine RAS kidneys using the single-slice DCE-MRI technique, as described previously [18].
Figure 6. Validation of multi-slice DCE-MRI technique by the validated single-slice technique.
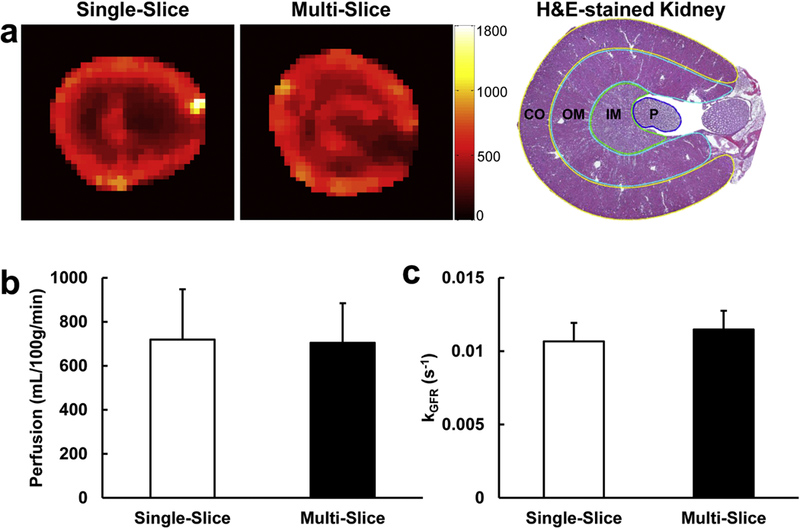
(a) Representative renal perfusion maps acquired using the single-slice (left) and multi-slice (middle) techniques. The unit of the color bar is mL/100g/min. A representative H&E-stained mouse kidney section (right) is also shown with cortex (CO), outer medulla (OM), inner medulla (IM), and papilla (P) segmented by yellow, cyan, green, and blue lines, respectively. The multi-slice DCE-MRI technique yielded comparable renal perfusion (b) and normalized GFR koFR (c), as compared to the validated single-slice technique.
Renal perfusion maps of the control, stenotic, and contralateral kidneys (Fig. 7a.) show differential perfusion in the renal cortex and medulla in all kidneys. The measured renal perfusion, volume, blood flow and GFR by the proposed multi-slice TRICKS-based DCE-MRI technique are shown in Fig. 7b–e. Significant drops in the stenotic (375.9±277.9mL/100g/min, P=0.002) and contralateral (532.3±148.0 mL/100g/min, P=0.022) kidney perfusion were also observed (Fig. 7b), as compared to the control kidneys (706.5±164.0mL/100g/min). The stenotic kidneys (137.8±65.8μL) in RAS mice were smaller than the control (213.3±24.0μL, P<0.001) and contralateral (228.2±34.3μL, P=0.004) kidneys (Fig. 7c). As a result of fall in both volume and perfusion, the stenotic kidney RBF (0.7±0.7mL/min) was lower than in the control (1.6±0.4mL/min, P<0.001) and contralateral (1.5±0.6mL/min, P=0.031) kidneys (Fig. 7d). Similarly, significant decrease in GFR was observed in the stenotic kidneys (58.0±42.8μL/min), as compared to the control (142.9±17.9μL/min, P<0.001) and contralateral (147.7±19.3μL/min, P<0.001) kidneys (Fig. 7e).
Figure 7. Utility of the proposed multi-slice DCE-MRI technique in differentiating renal functional differences in control and RAS mice.
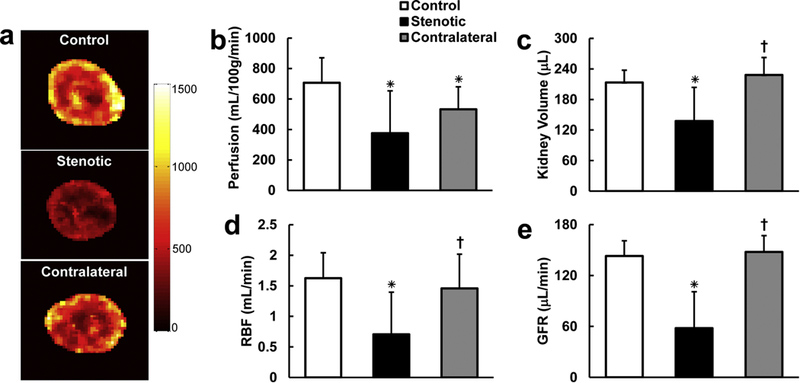
Representative renal perfusion maps (a) and measured renal perfusion (b), volume (c), RBF (d), and GFR (e) in control, stenotic, and contralateral kidneys. The unit of the color bar is mL/100g/min. Data expressed as mean±standard deviation. *P<0.05 compared to control kidney. †P<0.05 compared to stenotic kidney. RAS, renal artery stenosis; RBF, renal blood flow; GFR, glomerular filtration rate.
4. Discussion
We have proposed a DCE-MRI technique for comprehensive measurement of murine kidney size and function. A multi-slice T1 measurement method Snapshot-FLASH with TRICKS sampling has been developed for tracing contrast dynamics in eight axial slices with a temporal step size of 1.23 sec/scan. A semi-automated image-processing scheme employing basic image segmentation algorithms and machine learning has also been proposed to facilitate image analysis. The validity of the multi-slice technique was also demonstrated by comparing to the validated single-slice technique in normal mice. The kidney volume estimated from DCE-MRI agreed well with that measured using a three-dimensional volume scan. The multi-slice DCE-MRI technique estimates of renal parameters, including renal perfusion, normalized GFR, RBF, and GFR, also successfully differentiated normal and RAS mouse kidneys.
The proposed DCE-MRI technique may provide a powerful tool for comprehensive renal functional assessment in mouse models of kidney disease. The last two decades saw substantial development of various spontaneous, genetic and induced mouse models for investigation of pathogenesis and mechanisms of human kidney diseases [4]. Commonly used serum or urine biomarkers are neither accurate nor sufficient for evaluation of renal function. By contrast, DCE-MRI provides a valuable tool for simultaneous assessment of single-kidney structure and function. Our recently developed single-slice DCE-MRI technique provided reliable estimation of renal perfusion and GFR, as compared to inulin-clearance and arterial spin labeling methods, respectively [18]. However, heterogeneous renal diseases may not lend themselves to accurate assessment of renal functional biomarkers using a single slice. In this study, this method underwent further development and gained multi-slice capability by utilizing the TRICKS sampling scheme, thus allowing simultaneous measurement of kidney volume, perfusion, RBF, and GFR, with the aid of the bi-compartmental model. For the first time, we have shown that the TRICKS scheme can be successfully implemented in saturation recovery-based rapid T1 measurement method for tracing contrast dynamics in murine kidneys.
The proposed DCE-MRI technique with TRICKS sampling offers several advantages for rapid and reliable measurement of contrast dynamics. First, saturation recovery does not require full magnetization recovery and thus enables rapid T1 measurement [17, 29]. Although only one single point along the recovery curve was sampled for each slice, the high saturation efficiency (>98%) allows reliable T1 measurement by assuming a simple mono-exponential recovery from zero, as demonstrated in previous studies [18, 29]. Second, normalization of T1-weighed image by the proton density image automatically corrects B1 field inhomogeneity, and thus avoids biased estimation of contrast concentration. Moreover, the TRICKS sampling scheme yields an acceleration factor of 2.7, allowing full kidney coverage with a temporal step size of 1.23 sec/scan, which has been shown to be sufficient for reliable estimation of renal function in mouse kidneys [18]. Additionally, for the first time we demonstrated the feasibility of implementing TRICKS in DCE-MRI employing saturation recovery-based rapid T1 measurement. With these, similar contrast dynamics were obtained, as compared to the previous single-slice technique [18]. Given the view sharing at relatively high temporal resolution and low acceleration factor, our multi-slice TRICKS technique, as compared to the single-slice technique, was still capable of differentiating perfusion at different renal regions, as characterized by the highest perfusion in the renal cortex, followed by the outer medulla, inner medulla, and papilla. Importantly, the measured renal perfusion and GFR agreed well with those measured using the single-slice technique, supporting the validity of the multi-slice technique in measuring renal function. The measured renal perfusion in control mice also agreed well with those assessed by non-DCE MR method arterial spin labeling [35, 36] and ultrasonography [37]. Our measured single-kidney GFR in control mice also closely agreed with those obtained by the FITC-Inulin clearance method [38, 39]. Therefore, the proposed DCE-MRI technique provides reliable measurement of mouse kidney function.
Distinctive spatiotemporal patterns of contrast dynamics were exploited in the proposed semi-automated scheme to facilitate image processing. While the particular dynamics in the AIF allowed its measurement using cross correlation, the strong signal enhancement at the early vascular and late tubular phases was utilized for delineation of kidney dynamics and output by the thresholding-based algorithms [40–42]. Notably, rapid renal clearance of contrast agents in mice compared to some other species [7, 8, 34], as well as its uni-papillary structure concentrating the filtered contrast in a small region, causes more pronounced gadolinium blooming effect in the renal pelvic region, which needs to be excluded for reliable measurement of renal function. To avoid time-consuming and user-dependent manual exclusion, the machine learning algorithm SVM, which has been successfully used for localization of tumors in DCE-MRI [43–47], was employed to take advantage of the distinctive features in contrast dynamics with the blooming effect, for automated and reliable exclusion. With the aid of such semi-automated scheme, image analysis on both kidneys could be accomplished within approximately 15 minutes, including time for operator-required exclusion of images with motion artifacts and manual segmentation of renal pelvis in stenotic kidneys with minimal enhancement.
Although the current DCE-MRI technique is proposed for measurement of murine kidney function, it may also be applicable in human kidneys after further technical development. Importantly, more slices are needed to cover larger kidneys, which may prolong the sampling time after saturation recovery. Fortunately, lower temporal resolution may suffice in human kidney DCE-MRI [48]. However, caution is needed to avoid sampling at recovery times near full magnetization recovery, which may confound estimation of contrast concentrations. For this, additional saturation modules may be needed during data acquisition.
This study faces limitations. First, the current DCE-MRI technique remains 2D rather than 3D. This is partly due to the T1 measurement method, i.e., saturation recovery Snapshot-FLASH, which requires accurate timing of signal recovery after saturation. Two-dimensional imaging provides faster image readout per slice and less motion and blurring artifacts. Second, given that the principles of the 2D measurements of GFR had been previously validated against a reference standard [18], additional validation of the measured renal functional parameters was not undertaken. However, similar values in both normal and RAS kidneys were obtained compared to those by the previously validated single-slice technique [18], supporting the validity of the current multi-slice technique. Third, future studies are needed to further develop the potential of this technique to identify local foci of decreased renal function. Additionally, other fast imaging techniques, such as compressed sensing or k-t acceleration methods, may be used to further expedite image acquisition. Nevertheless, their applicability in DCE-MRI employing saturation recovery-based rapid T1 measurement and accuracy in assessing renal function remain to be investigated.
In conclusion, the proposed multi-slice TRICKS-based DCE-MRI technique allows rapid and reliable tracking of contrast dynamics in whole mouse kidneys. Our developed semi-automated image processing scheme may be useful in facilitating extraction of contrast dynamics. With the aid of the validated bi-compartment model, the current DCE-MRI technique allows comprehensive measurement of kidney volume, renal perfusion, blood flow, and GFR in mouse kidneys.
Supplementary Material
Acknowledgements
This study was partly supported by National Institutes of Health Grants DK104273, DK102325, DK120292, and HL123160.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Jha V, Garcia-Garcia G, Iseki K, Li Z, Naicker S, Plattner B, et al. Chronic kidney disease: global dimension and perspectives. Lancet 2013;382:260–72. 10.1016/S0140-6736(13)60687-X. [DOI] [PubMed] [Google Scholar]
- [2].2017 USRDS Annual Data Report: Executive Summary. American Journal of Kidney Diseases 2018;71:S1–S8. 10.1053/j.ajkd.2018.01.003. [DOI] [Google Scholar]
- [3].Hewitson TD, Ono T, Becker GJ. Small animal models of kidney disease: a review. Methods Mol Biol 2009;466:41–57. 10.1007/978-1-59745-352-3_4. [DOI] [PubMed] [Google Scholar]
- [4].Yang HC, Zuo Y, Fogo AB. Models of chronic kidney disease. Drug Discov Today Dis Models 2010;7:13–9. / 10.1016/j.ddmod.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Urbschat A, Obermuller N, Haferkamp A. Biomarkers of kidney injury. Biomarkers 2011;16 Suppl 1:S22–30. 10.3109/1354750X.2011.587129. [DOI] [PubMed] [Google Scholar]
- [6].Perrone RD, Madias NE, Levey AS. Serum creatinine as an index of renal function: new insights into old concepts. Clin Chem 1992;38:1933–53. [PubMed] [Google Scholar]
- [7].Annet L, Hermoye L, Peeters F, Jamar F, Dehoux JP, Van Beers BE. Glomerular filtration rate: assessment with dynamic contrast-enhanced MRI and a cortical-compartment model in the rabbit kidney. J Magn Reson Imaging 2004;20:843–9. 10.1002/jmri.20173. [DOI] [PubMed] [Google Scholar]
- [8].Sourbron SP, Michaely HJ, Reiser MF, Schoenberg SO. MRI-measurement of perfusion and glomerular filtration in the human kidney with a separable compartment model. Invest Radiol 2008;43:40–8. 10.1097/RLI.0b013e31815597c5. [DOI] [PubMed] [Google Scholar]
- [9].Vivier PH, Storey P, Rusinek H, Zhang JL, Yamamoto A, Tantillo K, et al. Kidney function: glomerular filtration rate measurement with MR renography in patients with cirrhosis. Radiology 2011;259:462–70. 10.1148/radiol.11101338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Buckley DL, Shurrab AE, Cheung CM, Jones AP, Mamtora H, Kalra PA. Measurement of single kidney function using dynamic contrast-enhanced MRI: comparison of two models in human subjects. J Magn Reson Imaging 2006;24:1117–23. 10.1002/jmri.20699. [DOI] [PubMed] [Google Scholar]
- [11].Lee VS, Rusinek H, Bokacheva L, Huang AJ, Oesingmann N, Chen Q, et al. Renal function measurements from MR renography and a simplified multicompartmental model. Am J Physiol Renal Physiol 2007;292:F1548–59. 10.1152/ajprenal.00347.2006 [DOI] [PubMed] [Google Scholar]
- [12].Zhang JL, Rusinek H, Bokacheva L, Lerman LO, Chen Q, Prince C, et al. Functional assessment of the kidney from magnetic resonance and computed tomography renography: impulse retention approach to a multicompartment model. Magn Reson Med 2008;59:278–88. 10.1002/mrm.21489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hingorani DV, Bernstein AS, Pagel MD . A review of responsive MRI contrast agents: 2005–2014. Contrast Media Mol Imaging 2015;10:245–65. 10.1002/cmmi.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Linnik IV, Scott ML, Holliday KF, Woodhouse N, Waterton JC, O’Connor JP, et al. Noninvasive tumor hypoxia measurement using magnetic resonance imaging in murine U87 glioma xenografts and in patients with glioblastoma. Magn Reson Med 2014;71:1854–62. 10.1002/mrm.24826. [DOI] [PubMed] [Google Scholar]
- [15].O’Connor JP, Boult JK, Jamin Y, Babur M, Finegan KG, Williams KJ, et al. Oxygen-Enhanced MRI Accurately Identifies, Quantifies, and Maps Tumor Hypoxia in Preclinical Cancer Models. Cancer Res 2016;76:787–95. 10.1158/0008-5472.CAN-15-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Azlan CA, Di Giovanni P, Ahearn TS, Semple SI, Gilbert FJ, Redpath TW. B1 transmission-field inhomogeneity and enhancement ratio errors in dynamic contrast-enhanced MRI (DCE-MRI) of the breast at 3T. J Magn Reson Imaging 2010;31:234–9. 10.1002/jmri.22018. [DOI] [PubMed] [Google Scholar]
- [17].Jiang K, Li W, Jiao S, Castel L, Van Wagoner DR, Yu X. Rapid multislice T1 mapping of mouse myocardium: Application to quantification of manganese uptake in alpha-Dystrobrevin knockout mice. Magn Reson Med 2015;74:1370–9. 10.1002/mrm.25533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jiang K, Tang H, Mishra PK, Macura SI, Lerman LO. Measurement of Murine Single-Kidney Glomerular Filtration Rate Using Dynamic Contrast-Enhanced MRI. Magn Reson Med 2018;79:2935–43. 10.1002/mrm.26955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Doyle M, Walsh EG, Blackwell GG, Pohost GM. Block regional interpolation scheme for k-space (BRISK): a rapid cardiac imaging technique. Magn Reson Med 1995;33:163–70. 10.1002/mrm.1910330204. [DOI] [PubMed] [Google Scholar]
- [20].Parrish T, Hu X. Continuous update with random encoding (CURE): a new strategy for dynamic imaging. Magn Reson Med 1995;33:326–36. 10.1002/mrm.1910330307. [DOI] [PubMed] [Google Scholar]
- [21].Hennig J, Scheffler K, Laubenberger J, Strecker R. Time-resolved projection angiography after bolus injection of contrast agent. Magn Reson Med 1997;37:341–5. 10.1002/mrm.1910370306. [DOI] [PubMed] [Google Scholar]
- [22].Tsao J, Boesiger P, Pruessmann KP. k-t BLAST and k-t SENSE: dynamic MRI with high frame rate exploiting spatiotemporal correlations. Magn Reson Med 2003;50:1031–42. 10.1002/mrm.10611. [DOI] [PubMed] [Google Scholar]
- [23].Saranathan M, Rettmann DW, Hargreaves BA, Clarke SE, Vasanawala SS. Differential Subsampling with Cartesian Ordering (DISCO): a high spatio-temporal resolution Dixon imaging sequence for multiphasic contrast enhanced abdominal imaging. J Magn Reson Imaging 2012;35:1484–92. 10.1002/jmri.23602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].van Vaals JJ, Brummer ME, Dixon WT, Tuithof HH, Engels H, Nelson RC, et al. “Keyhole” method for accelerating imaging of contrast agent uptake. J Magn Reson Imaging 1993;3:671–5. 10.1002/jmri.1880030419. [DOI] [PubMed] [Google Scholar]
- [25].Korosec FR, Frayne R, Grist TM, Mistretta CA. Time-resolved contrast-enhanced 3D MR angiography. Magn Reson Med 1996;36:345–51. 10.1002/mrm.1910360304. [DOI] [PubMed] [Google Scholar]
- [26].Song T, Laine AF, Chen Q, Rusinek H, Bokacheva L, Lim RP, et al. Optimal k-space sampling for dynamic contrast-enhanced MRI with an application to MR renography. Magn Reson Med 2009;61:1242–8. 10.1002/mrm.21901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ebrahimi B, Crane JA, Knudsen BE, Macura SI, Grande JP, Lerman LO. Evolution of cardiac and renal impairment detected by high-field cardiovascular magnetic resonance in mice with renal artery stenosis. J Cardiovasc Magn Reson 2013;15:98 10.1186/1532-429X-15-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jiang K, Ferguson CM, Ebrahimi B, Tang H, Kline TL, Burningham TA, et al. Noninvasive Assessment of Renal Fibrosis with Magnetization Transfer MR Imaging: Validation and Evaluation in Murine Renal Artery Stenosis. Radiology 2017;283:77–86. 10.1148/radiol.2016160566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jiang K, Tang H, Mishra PK, Macura SI, Lerman LO. A rapid T1 mapping method for assessment of murine kidney viability using dynamic manganese-enhanced magnetic resonance imaging. Magn Reson Med 2018;80:190–9. 10.1002/mrm.27025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Li W, Griswold M, Yu X. Rapid T1 mapping of mouse myocardium with saturation recovery Look-Locker method. Magn Reson Med 2010;64:1296–303. 10.1002/mrm.22544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hedayat AF, Park KH, Kwon TG, Woollard JR, Jiang K, Carlson DF, et al. Peripheral vascular atherosclerosis in a novel PCSK9 gain-of-function mutant Ossabaw miniature pig model. Transl Res 2018;192:30–45. 10.1016/j.trsl.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rajendran R, Lew SK, Yong CX, Tan J, Wang DJ, Chuang KH. Quantitative mouse renal perfusion using arterial spin labeling. NMR Biomed 2013;26:1225–32. 10.1002/nbm.2939. [DOI] [PubMed] [Google Scholar]
- [33].Zollner FG, Zimmer F, Klotz S, Hoeger S, Schad LR. Renal perfusion in acute kidney injury with DCE-MRI: deconvolution analysis versus two-compartment filtration model. Magn Reson Imaging 2014;32:781–5. 10.1016/j.mri.2014.02.014. [DOI] [PubMed] [Google Scholar]
- [34].Krier JD, Ritman EL, Bajzer Z, Romero JC, Lerman A, Lerman LO. Noninvasive measurement of concurrent single-kidney perfusion, glomerular filtration, and tubular function. Am J Physiol Renal Physiol 2001;281:F630–8. 10.1152/ajprenal.2001.281.4.F630. [DOI] [PubMed] [Google Scholar]
- [35].Gao Y, Goodnough CL, Erokwu BO, Farr GW, Darrah R, Lu L, et al. Arterial spin labeling-fast imaging with steady-state free precession (ASL-FISP): a rapid and quantitative perfusion technique for high-field MRI. NMR Biomed 2014;27:996–1004. 10.1002/nbm.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Prevost VH, Girard OM, Callot V, Cozzone PJ, Duhamel G. Fast imaging strategies for mouse kidney perfusion measurement with pseudocontinuous arterial spin labeling (pCASL) at ultra high magnetic field (11.75 tesla). J Magn Reson Imaging 2015;42:999–1008. 10.1002/jmri.24874. [DOI] [PubMed] [Google Scholar]
- [37].Grifoni SC, Chiposi R, McKey SE, Ryan MJ, Drummond HA. Altered whole kidney blood flow autoregulation in a mouse model of reduced beta-ENaC. Am J Physiol Renal Physiol 2010;298:F285–92. 10.1152/ajprenal.00496.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Qi Z, Whitt I, Mehta A, Jin J, Zhao M, Harris RC, et al. Serial determination of glomerular filtration rate in conscious mice using FITC-inulin clearance. Am J Physiol Renal Physiol 2004;286:F590–6. 10.1152/ajprenal.00324.2003. [DOI] [PubMed] [Google Scholar]
- [39].Thibodeau JF, Holterman CE, Burger D, Read NC, Reudelhuber TL, Kennedy CR. A novel mouse model of advanced diabetic kidney disease. PLoS One 2014;9:e113459. 10.1371/journal.pone.0113459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].de Priester JA, Kessels AG, Giele EL, den Boer JA, Christiaans MH, Hasman A, et al. MR renography by semiautomated image analysis: performance in renal transplant recipients. J Magn Reson Imaging 2001;14:134–40. 10.1002/jmri.1163. [DOI] [PubMed] [Google Scholar]
- [41].Rusinek H, Boykov Y, Kaur M, Wong S, Bokacheva L, Sajous JB, et al. Performance of an automated segmentation algorithm for 3D MR renography. Magn Reson Med 2007;57:1159–67. 10.1002/mrm.21240. [DOI] [PubMed] [Google Scholar]
- [42].Zollner FG, Sance R, Rogelj P, Ledesma-Carbayo MJ, Rorvik J, Santos A, et al. Assessment of 3D DCE-MRI of the kidneys using non-rigid image registration and segmentation of voxel time courses. Comput Med Imaging Graph 2009;33:171–81. 10.1016/j.compmedimag.2008.11.004. [DOI] [PubMed] [Google Scholar]
- [43].Artan Y, Haider MA, Langer DL, van der Kwast TH, Evans AJ, Yang Y, et al. Prostate cancer localization with multispectral MRI using cost-sensitive support vector machines and conditional random fields. IEEE Trans Image Process 2010;19:2444–55. 10.1109/TIP.2010.2048612. [DOI] [PubMed] [Google Scholar]
- [44].Levman J, Leung T, Causer P, Plewes D, Martel AL. Classification of dynamic contrast-enhanced magnetic resonance breast lesions by support vector machines. IEEE Trans Med Imaging 2008;27:688–96. 10.1109/TMI.2008.916959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Torheim T, Malinen E, Kvaal K, Lyng H, Indahl UG, Andersen EK, et al. Classification of dynamic contrast enhanced MR images of cervical cancers using texture analysis and support vector machines. IEEE Trans Med Imaging 2014;33:1648–56. 10.1109/TMI.2014.2321024. [DOI] [PubMed] [Google Scholar]
- [46].Artzi M, Liberman G, Nadav G, Blumenthal DT, Bokstein F, Aizenstein O, et al. Differentiation between treatment-related changes and progressive disease in patients with high grade brain tumors using support vector machine classification based on DCE MRI. J Neurooncol 2016;127:515–24. 10.1007/s11060-016-2055-7. [DOI] [PubMed] [Google Scholar]
- [47].Deng W, Luo L, Lin X, Fang T, Liu D, Dan G, et al. Head and Neck Cancer Tumor Segmentation Using Support Vector Machine in Dynamic Contrast-Enhanced MRI. Contrast Media Mol Imaging 2017;2017:8612519. 10.1155/2017/8612519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Michaely HJ, Sourbron SP, Buettner C, Lodemann KP, Reiser MF, Schoenberg SO. Temporal constraints in renal perfusion imaging with a 2-compartment model. Invest Radiol 2008;43:120–8. 10.1097/RLI.0b013e3181583b0c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


