Abstract
Background
The present study aimed to use the targeted capture and sequencing technique to diagnose adult hereditary spherocytosis (HS). These results were compared with clinical features and laboratory examinations to explore the diagnosis of HS.
Methods
Whole blood and clinical data from ten patients with HS were collected. Genomic DNA was extracted, and a library was prepared. Exomes of patients with ten HS-related genes encoding red cell membrane skeleton protein were captured and sequenced. Bioinformatics analyses were carried out throughout the 1000 Genomes Project, ExAC, dbSNP147, and 1000 Normal Han Population databases.
Results
Gene mutations were found in 9 out of 10 cases of HS. Our data validation showed 90% specificity. Three types of gene mutations were found, including 6 cases of SPTB, 3 cases of ANK1, and 2 cases of SLC4A1. There were 4 mutation forms, including nonsense mutation, missense mutation, shear mutation, and code shift mutation, all of which were new, heterozygous mutations. These variations were predicted to be pathogenic in four databases.
Conclusions
Our data demonstrate that targeted gene enrichment and sequencing methods were an efficient tool for determining genetic etiologies of red blood cell (RBC) membrane disorders and can facilitate accurate diagnosis and genetic counseling. They are also in good agreement with the clinical results.
Keywords: Hereditary spherocytosis (HS), ANK1, SPTB, SLC4A1, targeted capture
Introduction
Hereditary spherocytosis (HS) is the most frequent form of inherited hemolytic anemia and is characterized by the presence of spherical-shaped erythrocytes on the peripheral blood smear, anemia, jaundice, hemolysis, splenomegaly, and gallstones. The clinical manifestations of HS vary widely, ranging from asymptomatic patients to severely anemic, transfusion-dependent patients (1). HS is diagnosed on the basis of positive family history, in addition to laboratory tests which include red blood cell (RBC) morphology, the acidified glycerol hemolysis test (AGLT50), osmotic fragility (OF) test, and Eosin-5'-maleimide (EMA) binding test with a negative direct antiglobulin test. The OF test has been considered the gold standard screening test for HS but provides false-negative findings in about 25% of patients (2). EMA measurement and SDS-polyacrylamide gel electrophoresis (SDS-PAGE) of erythrocyte membrane proteins are also useful for screening HS, but standardization of these methods are currently lacking, and the experimental steps are complex (1,3). None of the screening tests can accurately detect all cases of HS because the clinical phenotypes are widely variable, ranging from asymptomatic to severely affected (4).
HS is most commonly associated with autosomal dominant (AD) inheritance, although non-dominant and autosomal recessive (AR) inheritance has also been described. Its prevalence is 1 in 2,000 in the Northern European population (5), and the prevalence in China is about 1 in 100,000 people (6). RBC membranes from HS patients show qualitative and/or quantitative abnormalities of RBC membrane structure-related proteins. The defects in membrane components increase membrane fragility and induce vesiculation, all which result in continuous membrane loss, decreased surface area, and take the shape of spherocytes with decreased deformability.
To date, mutations in at least five genes (ANK1, EPB4.2, SLC4A1, SPTA1, and SPTB), encoding ankyrin, protein 4.2, band 3, α-spectrin, and β-spectrin protein respectively, have been associated with HS (4). As a consequence of unclear morphological features and unreliable screening tests, the diagnosis of HS can be difficult. Molecular testing is useful for primary differential or confirming a diagnosis of HS.
Sanger sequencing is primarily performed in order to identify the causative mutations in single-gene disorders. The method for sequencing is low flux, time-consuming, and laborious. Recent advances in molecular technologies have helped to identify unexpected candidate genes in numerous inherited disorders. Various next-generation sequencing (NGS)-based methods have been developed, including whole-genome sequencing, exome sequencing, and gene panels (7). NGS allows us to overcome the limitations of the current diagnostic methods and provides additional information to assess the pathogenicity of identified genetic variants by comprehensive genotype-phenotype analyses.
The present study used the targeted capture and sequencing technique to diagnose adult HS. These results were compared with clinical features and laboratory examinations to explore the diagnosis of HS.
Methods
Sample collection
Ten cases of HS patient data from 7 men and 3 women, were collected from January 2008 to December 2017 in Nanjing First Hospital. The median age at diagnosis was 48.5 years (ranged from 30 to 67 years). All patients were asked in detail about medical and family history. The positive results (yellow staining of sclera and skin, splenomegaly, cholecystolithiasis) of physical examination were recorded. All patients underwent the following examinations: blood and urine routine, total bilirubin, direct and indirect bilirubin, gallbladder and spleen ultrasound, oxidative-fermentative (OF) test, AGLT50 test, peripheral blood smear, and scanning electron microscopy. The diagnosis of HS is usually based on a combination of clinical and family histories, physical examination (e.g., jaundice, splenomegaly) and laboratory findings. The EDTA anticoagulant peripheral blood 5 ml was extracted from the patients and was put in −20 °C refrigeration and prepared for testing.
EMA labeling test using flow cytometry
The EMA-binding test was performed as described by King et al. (3) with a few adjustments. Fifteen thousand RBCs were collected to calculate the average fluorescence intensity and detect the channel FL1. Median fluorescent intensities (at least 4 mice/strains) were measured with a FACSVantageSE/DiVA (BD BioSciences) flow cytometer. A small amount of EMA dye reserve solution was stored in a refrigerator at −800 °C for no more than 6 months.
Observation of RBC morphology by scanning electron microscope
Heparinized RBCs were rapidly fixed in 4% glutaraldehyde prepared in 0.2 mol/L phosphate buffer (pH7.2), centrifuged at low speed, and washed with pH7.2, 0.1 M phosphate buffer. Dehydration was carried out with the following ethanol gradient: 30%, 50%, 70%, 90%, 100%; 15 minutes at a time. After drying at a critical point and spraying gold (current: 15 mA, time: 60 seconds), the sliding glass was observed under FEI QUANTA 200 scanning electron microscope.
Molecular analysis for HS (MyGenostics Inc., Beijing, China)
DNA library preparation
The whole genome DNA was extracted from 5 mL of peripheral blood of patients. After dilution of 3–5 µg DNA, 150 BP fragments were prepared by Covariss2 ultrasound. The whole genome library was prepared by using the NEBNext DNA library preparation kit (Illumina NEB Company). The size selected product was PCR amplified (each sample was tagged with a unique index during this procedure), purified by magnetic beads (Beckman Company), and the final product was validated using the Agilent Bioanalyzer.
Targeted gene enrichment and sequencing
In brief, by using GenCap gene sequence capture technology (Beijing MyGenostics Company), the capture probes of RBC-related pathogenic genes were hybridized with the whole genome library of patients, and bound and adsorbed with magnetic beads encapsulating biotin and streptavidin. The DNA fragments of non-target regions were eluted, and high throughput sequencing analysis was carried out with the new generation sequencer, Illumina hiseq2000.
Bioinformatics analysis
After sequencing the target area, the readout was filtered and processed to remove the connection and low-quality data (quality value >20) and low coverage data (depth >10) from the sequencing data. Using SOAPaligner software to compare the reference genome, the data obtained were analyzed by standard process information, and the results of single nucleotide polymorphisms (SNPs) and insertion-deletion mutation (InDels) were obtained. At the same time, effective data such as sequencing depth, coverage, and homogeneity were counted. To determine pathogenicity, nonsynonymous variants were evaluated by four algorithms, Ployphen, SIFT, PANTHER, and Pmut, as described previously.
Pathogenicity prediction
The structural changes caused by the mutations were predicted and analyzed by SWISS-MODEL (http://swissmodel.expasy.org). By inputting the wild-type and mutated protein sequence on the website, we were able to acquire the 3D structure. The relevant parameters were analyzed by with reference to the related literature.
Results
Characteristics of clinical and laboratory examination
Clinical characteristics and laboratory findings in 10 genetically identified Chinese HS are shown in Table 1. Seven patients had a distinct familial inheritance history (except No. 8, 9, 10). Their clinical manifestations varied widely. Some of them had yellow sclera and skin from childhood. Other patients had obvious clinical symptoms in adulthood. Laboratory examination showed that there were 8 cases of anemia and 9 cases of reticulocytosis in the 10 cases of HS. AGLT50 tests were positive in all patients. Among the 10 cases, 2 individuals showed increased OF test scores, but 8 cases were normal, 2 of which were positive after being incubated. Three abnormal types of RBCs were observed in the peripheral blood smear including, spherical/acanthocyte/stomatocyte/ erythrocytes, small concentrated red cells, and irregular red cells (with mushroom, helmet, spine, or oval shape). The B-ultrasound examination showed that 9 out of 10 cases had splenomegaly (except for No. 10) and all patients had gallstones (No. 6 still has a urinary stone).
Table 1. Clinical and laboratory characteristics of 10 patients with HS.
| Patient | Sex | Age | Family history | Hb (g/L) | Ret (%) | Spherocytes (%) | Hyperdense cells (%) | AGLT50 (s) | OF (g/L) (start-complete hemolysis) | EMA | Gallstone | Length × thickness of spleen (mm) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 60 | y | 107 | 15.6 | 18 | 36 | 76 | 4.4–3.4; 6.0–4.6 (incubated) | no | + | 168×60 |
| 2 | F | 32 | Y | 55 | 19 | 16 | 46 | 69 | 4.4–3.4; 6.0–4.6 (incubated) | 4.5 | + | 135×46 |
| 3 | M | 30 | Y | 90 | 8.6 | 28 | 52 | 78 | 4.4–3.4; 6.0–4.6 (incubated) | no | + | 154×68 |
| 4 | F | 67 | Y | 94 | 5.6 | 30 | 48 | 200 | 4.2–3.4; 4.6–3.4 (incubated) | 4.5 | + | 150×45 |
| 5 | M | 40 | Y | 136 | 3,2 | 38 | 50 | 220 | 4.2–3.4; 4.6–3.4 (incubated) | 6.6 | + | 145×52 |
| 6 | M | 61 | Y | 68 | 8.6 | 46 | 60 | 98 | 4.4–3.4; 6.0–4.6 (incubated) | 5.84 | + | 210×88 |
| 7 | M | 67 | y | 143 | 9.2 | 32 | 40 | 120 | 4.4–3.4; 6.0–4.6 (incubated) | 7.52 | + | 176×65 |
| 8 | F | 63 | N | 100 | 3.2 | 17 | 64 | 90 | 5.0–3.8 | 6.69 | + | 188×69 |
| 9 | M | 46 | N | 90 | 8.9 | 68 | 80 | 160 | 4.4–3.4; 6.0–4.6 (incubated) | no | + | 172×68 |
| 10 | M | 48 | N | 85 | 1.1 | 18 | 38 | 200 | 4.2–3.4; 4.6–3.4 (incubated) | 5.86 | + | 138×35 |
Hb, hemoglobin; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; Ret, reticulocyte; IBIL, indirect bilirubin.
Targeted gene enrichment and sequencing results
Gene mutations were found in 9 cases of HS with Illumina hiseq-2000 high throughput target gene capture sequencing. The detection rate of gene mutation was 90%. Three types of gene mutations were found in 9 patients of HS, including 6 cases of SPTB, 3 cases of ANK1 (the same family), and 2 cases of SLC4A1 (the same family). The mutation in EPB41 and EPB42 were not detected. There were 4 mutation forms: nonsense mutation, missense mutation, shear mutation, and code shift mutation. These were all heterozygous mutations and can be considered new gene mutations, as none of them have been reported in the HGMD database (see Table 2).
Table 2. Mutation gene sequencing results with 10 cases of HS.
| Patient | Chromosome | Gene | Transcript | Exon | Nucleotide | Amino acid | Het/Hom | Mutation type | HGMD databases |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Chr8-41581138 | ANK1 | NM001142446 | 8 | c.824C>G | p.P275R | het | Missense mutation | No report |
| 2 | Chr8-41581138 | ANK1 | NM001142446 | 8 | c.824C>G | p.P275R | het | Missense mutation | No report |
| 3 | Chr8-41581138 | ANK1 | NM001142446 | 8 | c.824C>G | p.P275R | het | Missense mutation | No report |
| 4 | Chr17-42338094-42338096 | SLC4A1 | NM000342 | 5 | c.256-258GAG>TTCTC | p.E86Ffs | het | frameshift mutation | No report |
| 5 | Chr17-42338094-42338096 | SLC4A1 | NM000342 | 5 | c.256-258GAG>TTCTC | p.E86Ffs | het | frameshift mutation | No report |
| 6 | Chr14-65241110 | SPTB | NM001024858 | 23 | c.4973+5G>A | splicing | het | Splice mutation | No report |
| 7 | Chr14-65241110 | SPTB | NM001024858 | 23 | c.4973+5G>A | splicing | het | Splice mutation | No report |
| 8 | Chr4-106156811 | TET2 | NM001127208 | 3 | c.1712G>A | p.R571H | - | Missense mutation | Report |
| 9 | Chr14-65249093 | SPTB | NM01024858 | 19 | c.4181G>A | p.W1394X | het | Nonsense mutation | No report |
| 10 | Chr14-65271746 | SPTB | NM001024858 | 2 | c.211G>A | p.V71M | het | Missense mutation | No report |
Het, heterozygous; hom, homozygous; HS, hereditary spherocytosis.
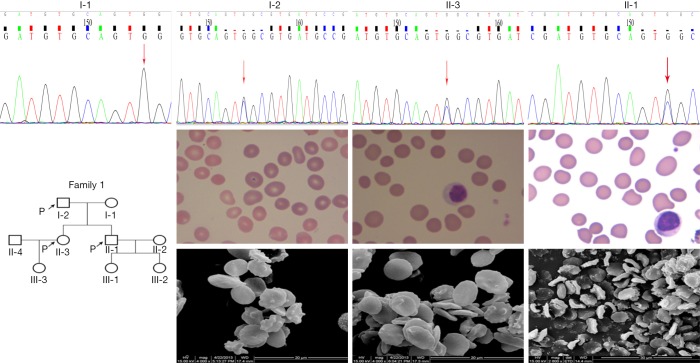
A de novo heterozygous ANK1 variant (c.824C>G, the 824th nucleotides of the coding region are mutated from cytosine to guanine) was identified in No.1 (father), 2 (daughter), and 3 (son) (see Figure 1). This mutation belongs to the missense mutation that results in amino acid change (p.P275R, the 275th amino acids were changed from proline to arginine). The inherited form of the disease is an AD inheritance. No cases of this mutation were retrieved in the 1000 Genome Project, ExAC, dbSNP147, or 1000 normal Han Population databases. It was predicted to be a pathogenic variation in SIFT software, a possible pathogenic variation in PolyPhen_2 software, and the cause of the disease in the MutationTaster software. Three patients showed typical clinical or laboratory evidence of hemolytic anemia. The MCF units of EMA binding tests for No.2 were below normal levels (4.5, normal MCF value 11.8±2.6).
Figure 1.
Family tree, ANK1 mutation, light microscope (Wright-Giemsa stain, ×1,000) and scanning electron images (4,000×; scale bar =10 µm) of RBC morphology in No.1 (father, I-2), 2 (daughter, II-3), 3 (son, II-1).
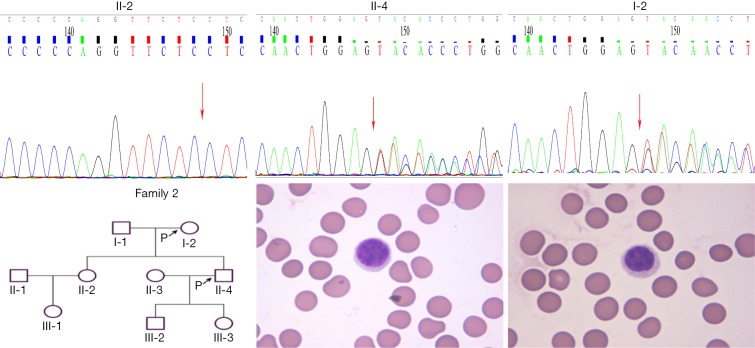
A new heterozygous SLC4A1 mutation (c.256_258GAG>TTCTC, amino acid change p.E86Ffsm, frameshift mutation) was confirmed in the second family, No. 5 (son) and No.4 (mother) (see Figure 2); this mutation also has not been reported in the above four databases. There was no variation found in the sister. The father’s specimen has not been received. The inherited form was also AD inheritance.
Figure 2.
Family tree, SLC4A1 mutation, light microscope (Wright-Giemsa stain, ×1,000), and scanning electron images (4,000×; scale bar =10 µm) of RBC morphology in No.4 (mother, I-2), and 5 (son, II-4).
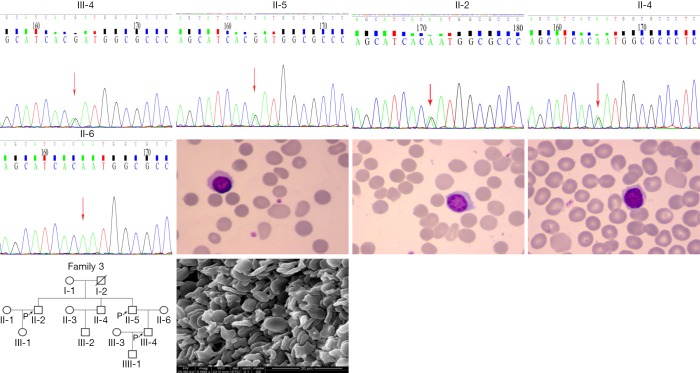
The splicing mutation in exon 23 of the SPTB gene was found in No. 6 (second brother) and No. 7 (first brother) (see Figure 3), specifically, c.4973+5G>A (the 5th nucleotide of intron changed from guanine to adenine); this mutation also has not been reported in the above four databases. The variation may affect normal exon splicing and result in deletion of intron or exon sequences in mature RNA. Thus, the structure and function of spectrin were affected in the observed cases. The daughter of patient No. 6 was also identified with the same mutation. However, no hemolytic anemia was found, and blood routine examination was also normal.
Figure 3.
Family tree, SPTB mutation, light microscope (Wright-Giemsa Stain,×1,000) and scanning electron images (4,000×; scale bar =10 µm) of RBC morphology in No.6 (second brother, II-5) and 7(first brother, II-2).
The heterozygous SPTB variant (c.4181G>A, the 4181st nucleotide of the coding region from guanine to adenine, resulting in an amino acid change of p.W1394X and a nonsense mutation) was detected in patient No. 9, but a non-genetic family history was not found; this mutation also has not been reported in the above four databases. MutationTaster software predicted a pathogenic variation, which led to the early termination of the SPTB gene transcription and complete protein product could not be synthesized.
Patient No. 10 showed c.211G>A (the 71st amino acid mutated from valine to methionine, the 211th nucleotide-nucleotide changed from guanine to adenine, resulting in an amino acid change of p.V71M and a missense mutation). The mutation had a minimum allele frequency of 0.0002 in the 1000 Genomes Project database. This SPTB variant was predicted as deleterious, probably damaging, and with a high score by SIFT, PolyPhen_2 software. In addition to this, the MutationTaster software directly predicted it as the cause of the disease. The only clinical inconsistency is the lack of splenomegaly.
Although the gene mutation-related erythrocyte membrane skeleton protein was not detected in patient No.8 or her three children, there was a strong suspicion of HS based on the clinical history and identification of spherocytes on the smear. Moreover, her hemoglobin and bilirubin returned to normal after splenectomy. This could be attributed to the technical limitation of the NGS sequencing assay or novel gene mutations.
Discussion
The reasons for the difficulty or misdiagnosis of HS are also related to the factors outlined below. HS is a rare disease, and the age of onset and severity of the disease also vary considerably. Undoubtedly, clinicians of the past did not know enough about HS. However, the most important problem could be the lack of a specific index in many laboratory tests for HS. Over the years, RBC membranopathy was screened and diagnosed by using conventional methods including the patient’s clinical/family history, RBC morphology, membrane protein analysis, and chemistry results (OF, EMA, etc.). In regards to the diagnostic pathway of the red cell membrane disorders, blood smear microscopy is fundamental and can indicate important information; it should thus be performed in all individuals with a positive family history of hemolytic anemia and a negative direct antiglobulin test. Our results support this view. The spherical and concentrated RBC counts of peripheral blood smears in all patients increased significantly, exceeding 10%. Furthermore, the electron microscopy scanning was applied to erythrocyte morphological observation. Erythrocyte morphology manifested as single or multiple acanthocytes or was speculated on red cell surfaces. The red cell body shape was an irregular dish-shape or flower-shape. The biconcave discoid shape disappeared in HS patients. However, these morphological features were not found in other hemolytic anemia cases, such as autoimmune hemolytic anemia (AIHA) (data not published). These findings enrich the diagnostic means available to clinicians.
It has been recommended that the first step of screening tests for red cell membrane defects should include the panel of OF and EMA-binding tests. Previously, the OF test was considered to be the gold standard for HS diagnosis; however, we found that the positive detection rate was not high in our experiments. Even after incubation, the positive detection rate was not much improved. In the EMA test, the results of HS overlapped with other anemia and hemolytic diseases. Therefore, it can be only used as a screening test.
The 2011 Commonwealth HS guidelines for diagnosis recommend (8) that those who have a family history of HS, the typical clinical features of HS, and abnormal blood parameters (MCHC, reticulocytosis, spherocytes), should be diagnosed with HS without further examination. A combination of high predictive values was recommended for analysis. EMA plus AGLT was the first choice. The EMA test was directly aimed at the defect of HS membrane structure, and AGLT reflects the change of the surface/volume ratio of the membrane. The combination of the two methods achieved a sensitivity of almost 100%. The sensitivities of other experimental combinations including OF plus AGLT and EMA plus OF were 97% and 95% respectively (8,9). The improvement of hemolytic symptoms after splenectomy can also help support HS diagnosis. Nevertheless, there are still a few patients, especially those with mild HS, who cannot be diagnosed early.
The development of the new generation of sequencing technology has opened new avenues in the research of modern genomics. Sequence capture sequencing of target gene hybridized with genomic DNA (solid or liquid phase) can customize specific probes in the genomic region of interest. The DNA fragment of the target genome area is enriched and then sequenced using NGS technology (7). Compared with whole-genome sequencing and under the same cost, the coverage is deeper, and the accuracy of the data is higher while being more convenient, economical, and efficient. Compared with the first generation sequencing method (Sanger method), it has the advantages of high throughput, low cost, and time-saving (10). Target area capture technology combined with current high-throughput sequencing technology can be applied to repeat sequences in exon regions and candidate genome regions, and it can also be used to discover and analyze SNP loci to help find many complex disease-related genes and loci (11). Our study shows that 9 out of 10 patients were identified with related gene mutations of erythrocyte membrane protein. Our NGS data validation detected gene mutations in 9 out of the 10 patients. According to statistics, about 85% of the monogenic disease mutation loci are located in the exons. Therefore, the clinical application of this technique has greatly improved the diagnostic rate of HS (12).
HS is a rare disease in China. Between 2006 and 2017, we confirmed another 15 cases of HS (a total of 25 cases). Only in 2017 did we apply gene sequencing technology to HS diagnosis. In the past two years, systematic research reports have been seen in succession, and there have been more than 30 cases of HS. The results of two reports show that ANK1 and SPTB are the most common mutant genes in Chinese HS patients, mainly with missense mutations and nonsense mutations. There was no significant correlation between the mutation of HS-related genes and the severity of HS (13). In the second study, all of the ANK1 or SPTB mutations, scattered throughout the entire genes, were non-recurrent; and most of them were null mutations, which might cause HS via a haploinsufficiency mechanism. De novo mutations in ANK1 or SPTB often occur with an unexpectedly high frequency (87.5% and 64.2%, respectively) (14). A South Korean study also yielded similar results (15).
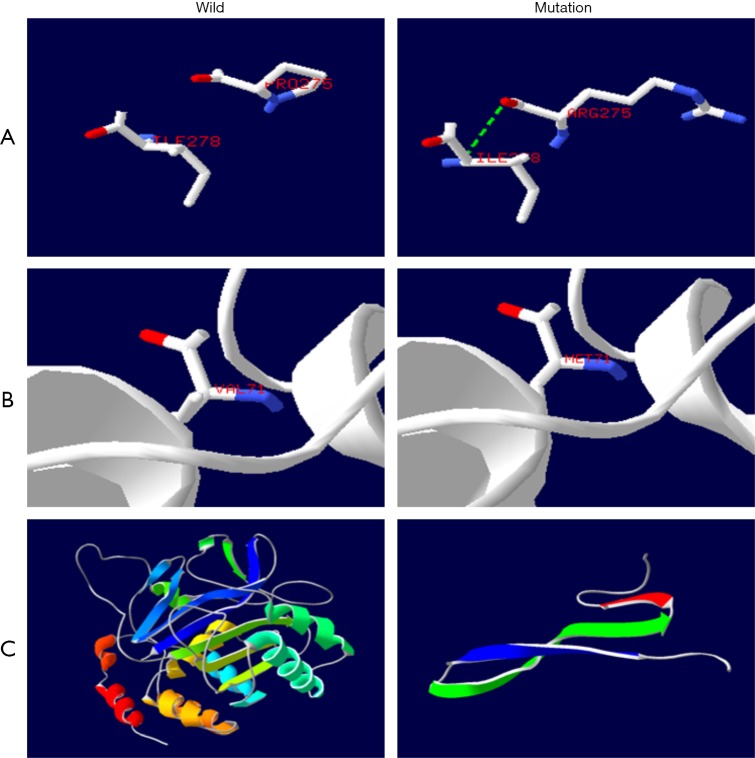
Apart from this, we can analyze the possible defects of protein function induced by a gene mutation. The ANK1 gene encodes the anchor protein of 206-kD, which contains three domains: the anchor protein repeat sequence domain, the ZU5 domain, and the death domain (16). One end is connected to the junction point of the tail of the beta chain of the contractile protein; the other end is connected to the band 3 protein, and the membrane skeleton is fixed in the lipid bilayer, which plays a key role in the stability of the membrane (17). The mutation occurs on the repeated sequence of the ANK1 protein. The protein’s three-dimensional structure prediction model showed that the mutation of the ANK1 gene in the patients No. 1, 2, 3 resulted in 275 proline mutation to arginine and side chain changes. This produces a hydrogen bond with 278 isoleucine (see Figure 4A) (18). This change may cause defects in the mass of the ANK1 protein, resulting in a weakening of the interaction between the membrane skeleton and the lipid bilayer. Thus, the lipid bilayer becomes unstable. Part of the lipid vesicles formed by budding form was lost, the erythrocyte membrane surface area decreased, and the biconcave changed into a sphere with erythrocyte morphology (19).
Figure 4.
Prediction map of the three-dimensional structure of protein (protein structure prediction Website-SWISS-MODEL). (A) (ANK1): there is an additional hydrogen bond between 275 P → R and 278 amino acids, and the side chain of amino acids has also changed. (B) (SPTB): the side chain of amino acid changed when the amino acid V → M was in the 71st position. (C) (SLC4A1): the side chain of amino acid changed when the amino acid V → M was in the 71st position.
The SPTB gene expresses the 246 kD spectrin beta1 subunit. Spectrin belongs to the fibrous protein of the red cell membrane stretch, consisting of two similar beta and alpha subunits which form heterodimers in reverse parallel arrangement. It contains three domains: the calponin homology domain of the CH domain (calponin-homology), the ankyrin repeat domain (spectrin repeat), and the C terminal domain (20). The terminal end of polymers formed by multiple heterodimers are combined with actin and transmembrane proteins to form a “junction complex”, which forms a grid structure deformation in the cell membrane, supports lipid bilayers and maintains erythrocytes’ biconcave disc shape. The mutation of the SPTB gene may lead to the non-formation of the normal polymer structure, which results in weakened protein function or the loss of the protein altogether. The erythrocyte membrane loses its skeleton protein support, thus deforming and triggering the hemolytic anemia (21). Spectrin has 2 actin-binding regions and 4.1R binding regions which are located in the 2 CH domain. Spectrin beta subunits combine with actin and 4.1R by the CH domain (22). The three-dimensional structure prediction model of the protein showed that the mutation of No. 10 resulted in 71 valine changing to methionine while there was also a side chain change (see Figure 4B). The normal combination of spectrin with actin and 4.1R may be disturbed by this. It leads to hereditary elliptocytary anemia (HE) and the instability of the mechanical properties of the erythrocyte membrane.
The 102 kD band3 protein of the SLC4A1 gene encoding, also known as the anion exchanger, is the most abundant protein on the human erythrocyte membrane, accounting for 25% of the total membrane protein. It contains 3 domains: the hydrophilic N terminal cytoplasmic domain, which mainly acts as the anchor location of other membrane-related proteins; cytoskeleton, a hydrophobic transmembrane domain with an anion exchange function containing 14 segments of the transmembrane region; the C terminal domain, which is reinserted into the cytoskeleton (17). The variation in No. 4 and 5 occurred on the cytoplasmic domain of band3 (see Figure 4C). The protein prediction model of the three-dimensional structure shows that the frameshift mutation leads to the premature termination of transcription, resulting in incomplete protein structure and inability to exercise the normal function of the band3 protein. The connection between the band3 protein and the ANK1 protein weakens, thus also weakening the vertical interaction between the membrane skeleton and the lipid bilayer, resulting in the instability of the lipid bilayer and the subsequent spheroidization and hemolysis of erythrocytes (23).
In summary, our study demonstrated the usefulness of the targeted capture and sequencing technique in the diagnosis of HS. Our NGS data validation shows 90% sensitivity. Additional studies involving larger cohorts must be conducted to investigate the distribution of causative gene mutations in patients with HS in China.
Acknowledgments
The authors would like to thank all patients for their selfless cooperation and for referring other patients to us or helping us in the diagnosis in one way or another. MyGenostics Inc. helped us finish the work of genetic sequence capture sequencing. All authors reviewed the manuscript and approved the final version.
Funding: This work was supported by grants from the research of Key Projects of Nanjing Municipal Committee for Health and Family Planning (project number: ZKX17025).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.King MJ, Garçon L, Hoyer JD, et al. ICSH guidelines for the laboratory diagnosis of nonimmune hereditary red cell membrane disorders. Int J Lab Hematol 2015;37:304-25. 10.1111/ijlh.12335 [DOI] [PubMed] [Google Scholar]
- 2.Bianchi P, Fermo E, Vercellati C, et al. Diagnostic power of laboratory tests for hereditary spherocytosis: a comparison study in 150 patients grouped according to molecular and clinical characteristics. Haematologica 2012;97:516-23. 10.3324/haematol.2011.052845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.King MJ, Behrens J, Rogers C, et al. Rapid flow cytometric test for the diagnosis of membrane cytoskeleton-associated haemolytic anaemia. Br J Haematol 2000;111:924-33. [PubMed] [Google Scholar]
- 4.Farias MG. Advances in laboratory diagnosis of hereditary spherocytosis. Clin Chem Lab Med 2017;55:944-8. 10.1515/cclm-2016-0738 [DOI] [PubMed] [Google Scholar]
- 5.Perrotta S, Gallagher PG, Mohandas N. Hereditary spherocytosis. Lancet 2008;372:1411-26. 10.1016/S0140-6736(08)61588-3 [DOI] [PubMed] [Google Scholar]
- 6.Wang C, Cui Y, Li Y, et al. A systematic review of hereditary spherocytosis reported in Chinese biomedical journals from 1978 to 2013 and estimation of the prevalence of the disease using a disease model. Intractable Rare Dis Res 2015;4:76-81. 10.5582/irdr.2015.01002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xue Y, Ankala A, Wilcox WR, et al. Solving the molecular diagnostic testing conundrum for Mendelian disorders in the era of next-generation sequencing: single-gene, gene panel, or exome/genome sequencing. Genet Med 2015;17:444-51. 10.1038/gim.2014.122 [DOI] [PubMed] [Google Scholar]
- 8.Bolton-Maggs PH, Langer JC, Iolascon A, et al. General Haematology Task Force of the British Committee for Standards in Haematology. Guidelines for the diagnosis and management of hereditary spherocytosis-2011 update. Br J Haematol 2012;156:37-49. 10.1111/j.1365-2141.2011.08921.x [DOI] [PubMed] [Google Scholar]
- 9.Crisp RL, Solari L, Vota D, et al. A prospective study to assess the predictive value for hereditary spherocytosis using five laboratory tests(cryohemolysis test, eosin-5'-maleimide flow cytometry, osmoti fragility test, autohemolysis test, and SDS-PAGE on 50 hereditary spherocytosis families in Argentina. Ann Hematol 2011, 90:625-634. 10.1007/s00277-010-1112-0 [DOI] [PubMed] [Google Scholar]
- 10.Agarwal AM, Nussenzveig RH, Reading NS, et al. Clinical utility of next-generation sequencing in the diagnosis of hereditary haemolytic anaemias. Br J Haematol 2016;174:806-14. 10.1111/bjh.14131 [DOI] [PubMed] [Google Scholar]
- 11.Russo R, Andolfo I, Manna F, et al. Multi-gene panel testing improves diagnosis and management of patients with hereditary anemias. Am J Hematol 2018;93:672-82. 10.1002/ajh.25058 [DOI] [PubMed] [Google Scholar]
- 12.Roy NB, Wilson EA, Henderson S, et al. A novel 33-Gene targeted resequencing panel provides accurate, clinical-grade diagnosis and improves patient management for rare inherited anaemias. Br J Haematol 2016;175:318-30. 10.1111/bjh.14221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng GX, Yang WR, Zhao X, et al. The characteristic of hereditary spherocytosis related gene mutation in 37 Chinese hereditary spherocytisis patients. Zhonghua Xue Ye Xue Za Zhi 2018;39:898-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang R, Yang S, Xu M, et al. Exome sequencing confirms molecular diagnoses in 38 Chinese families with hereditary spherocytosis. Sci China Life Sci 2018;61:947-53. 10.1007/s11427-017-9232-6 [DOI] [PubMed] [Google Scholar]
- 15.Choi HS, Choi Q, Kim JA, et al. Molecular diagnosis of hereditary spherocytosis by multi-gene target sequencing in Korea: matching with osmotic fragility test and presence of Spherocyte. Orphanet J Rare Dis 2019;14:114. 10.1186/s13023-019-1070-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lux SE, John KM, Bennett V. Analysis of cDNA for human erythrocyte ankyrin indicates a repeated structure with homology to tissue-differentiation and cell-cycle control proteins. Nature 1990;344:36-42. 10.1038/344036a0 [DOI] [PubMed] [Google Scholar]
- 17.Ipsaro JJ, Huang L, Mondondragon A. Structures of the spectrin-ankyrin interaction binding domains. Blood 2009;113:5385-93. 10.1182/blood-2008-10-184358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biasini M, Bienert S, Waterhouse A, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Research 2014;42:W252-8. 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barcellini W, Bianchi P, Fermo E, et al. Hereditary red cell membrane defects: diagnostic and clinical aspects. Blood Transfus 2011;9:274-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winkelmann JC, Chang JG, Tse WT, et al. Full-length sequence of the cDNA for human erythroid beta-spectrin. J Biol Chem 1990;265:11827-32. [PubMed] [Google Scholar]
- 21.Hughes MR, Anderson N, Maltby S, et al. A novel ENU-generated truncation mutation lacking the spectrin-binding and C-terminal regulatory domains of Ank1 models severe hemolytic hereditary spherocytosis. Exp Hematol 2011;39:305-20. 10.1016/j.exphem.2010.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang DQ, Wang YP, Wang WH, et al. Interaction between protein 4.1R and spectrin heterodimers. Mol Med Rep 2011;4:651-4. [DOI] [PubMed] [Google Scholar]
- 23.He Y, Jia S, Dewan RK, et al. Novel mutations in patients with hereditary red blood cell membrane disorders using next-generation sequencing. Gene 2017;627:556-62. 10.1016/j.gene.2017.07.009 [DOI] [PubMed] [Google Scholar]