Abstract
Background
Time-restricted feeding (TRF) has been proved to improve general health in adults. However, according to our previous study, this regimen failed to show similar protective effect in pediatric population. Gut microbiota has been proved to play a vital part in the whole process. Although previous studies have defined the commensal flora as a real-time indicator of health conditions in adults, our study aimed to investigate whether the unfavorable TRF feeding schedule during childhood would cause long-term variations in murine model.
Methods
We randomly assigned 120 4-week-old Kunming mice (half male and half female) to four feeding regimens: TRF.AD, time-restricted feeding during the childhood phase before switched to ad libitum feeding pattern as adults; TRF.TRF, continuously treated with time-restricted feeding; AD.TRF, went through time-restricted feeding only in adulthood; AD.AD, always had ad libitum access to food. After 8 weeks of dietary intervention, faeces were harvested from 12-week-old adult mice (one subject per cage), and gut microbiota was subsequently analysed via DNA extraction and 16s rRNA sequencing.
Results
Mice on identical diet for four weeks but went through different feeding patterns during childhood showed distinctive traits in gut microbiota. Differences existed in both the α diversity and specific groups of bacteria under different taxonomical levels.
Conclusions
Feeding pattern in the childhood had long-term impact on mice gut flora that cannot be wiped out in adulthood.
Keywords: Time-restricted feeding (TRF), gut microbiota, juvenile mice
Introduction
Time-restricted feeding (TRF) with no food intake for 14–16 hours during the day, has been profoundly proved to play protective role against multiple disease states in adults. Our previous study has demonstrated that the gut flora shift caused by TRF bridged the feeding pattern and its health-related outcomes (1). However, our study group also proved that improper feeding schedule during the childhood causes pleiotropic adverse effects in later life in murine model, also largely attributed to the change in the gut microorganisms (2). With the improvement of sequencing technologies, researchers are equipped with more precise methodologies to expand the microflora related discovery to another magnitude (3). Subsequently, emerging evidence has demonstrated that microbial communities are closely associated with health condition (4). As the interactions between gut flora and the digestive tract contribute to the gut homeostasis, a major role of dysbiosis of resident micro-organisms in colorectal cancer (CRC), inflammatory bowel disease (IBD), and other digestive related conditions were revealed (5,6). Compelling evidence have illustrated that microbiome have a direct impact on host energy yielding pathways, which makes it capable of influencing metabolic condition phenotypes, including obesity, dyslipidemia, type 2 diabetes, etc. (7,8). And regarding those diseases that are not directly linked to gut flora, the breaks of microbiota-host mutualism have been shown to accompany the disease progression (9), associated with infectious diseases (10), and disturbed host immune system (11).
As accumulated evidence associate microbiota with a large entity of diseases, researchers are enthusiastic in seeking effective methods to manipulate the gut flora, making it favorable for better health condition. Dietary preferences, including the diet content as well as the feeding schedule, are inevitably one of the most important environmental factors impacting gut microbial composition. A body of knowledge is accumulating that point to the gut microbiota as a mediator of dietary impact on the host status (12,13). It is known that malnutrition in earlier life has long-term effect on microbiota (14), but no existing study has ever demonstrated the effect of different feeding patterns on microbiota, in the setting that identical total caloric intake and nutritional structure were available. As it’s challenging to gather evidence from large-scale controlled diet study in human, we designed this controlled murine experiment to indicate whether feeding pattern during childhood has a permanent impact on mice commensal microbiota.
Methods
Animals
All animal procedures were performed in accordance with the Institutional Animal Care and Use Committee (IACUC) guidelines of the Salk Institute, and permitted by Animal Welfare Committee of Peking Union Medical College Hospital. A total of 120 Kunming mice (half male and half female) (Beijing Vital River Laboratory Animal Technology Co., Ltd) at 3 weeks of age were entrained to a 12:12 light-dark cycle.
Feeding schedule and diets
After one week of accommodation, all subjects were randomly assigned to 4 feeding patterns. Under TRF pattern, all subjects were allowed to eat between zeitgeber time (ZT) 13 (ZT0: lights on; ZT13: 1 h after lights off) and ZT21 (3 h before lights on), namely 8 hours per day. Within this 8-hour period, total caloric intake was unrestricted. Both groups were treated with normal chow (GB14924.3-2010 Standard: 29% protein, 13% fat, 58% carbohydrate) by reference: LabDiet-5010. The experimental intervention lasted for 8 weeks, crossing the childhood period (first 4 weeks, when mice were 5–8 weeks of age) and part of the adulthood (latter 4 weeks, when mice were 9–12 weeks of age). The above mentioned 4 feeding regimens were: TRF.AD, time-restricted feeding during the childhood phase before switched to ad libitum feeding pattern as adults; TRF.TRF, continuously treated with time-restricted feeding; AD.TRF, went through time-restricted feeding only in adulthood; AD.AD, always had ad libitum access to food. The last alphabet (M/F) in the group name indicated the gender of subjects. M, male mice; F, female mice.
Gut microbiota analysis
Samples were collected at the end of 8 weeks. Three mice from each group (one mouse per cage) were sacrificed at ZT 21. Lethal dosage of anesthetics was applied before feces from rectum were harvested through laparotomy. Samples were flash froze and resuspended and digested before lysis. DNA from the lysate was extracted, precipitated, washed, and resuspended. 16S rRNA gene sequence tags, corresponding to the hypervariable V1–V3 region, were generated using the 454-pyrosequencing platform. Operational taxonomic unit (OTU)-based classification was used to generate data profiles. Alpha diversity, beta diversity, and structural analyses of the gut microbiota were performed, to indicate diversity between and within groups, as well as the composition of the flora.
Statistical analysis
Data were analyzed by Stata/MP 14.0. Comparisons between two groups were performed by the Wilcox test. P value was indicated when necessary.
Results
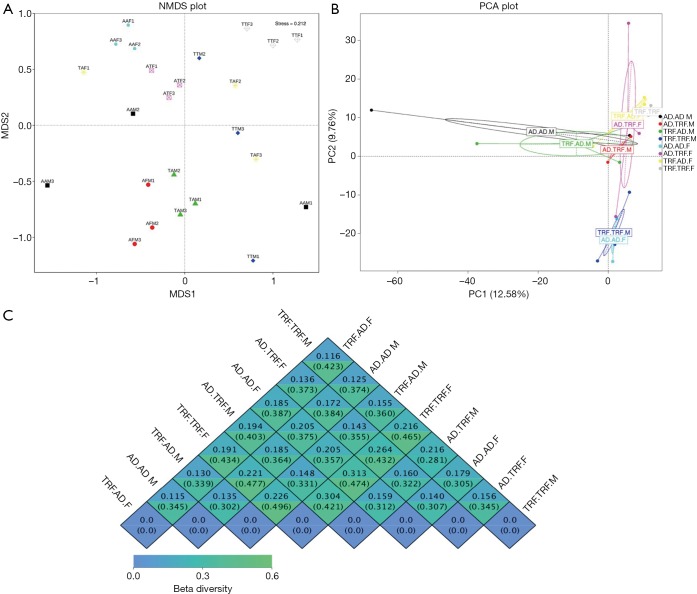
NMDS and PCA are both analyses reflecting β diversity of different experimental groups. In our study, they both suggested that subjects under the same feeding pattern, but raised in different cages had very similar microbial structural traits (Figure 1A,B). To note, if samples in a group share very few common features, regions encircling homogeneous individuals in PCA analysis might fail to be automatically generated by analytic software. Figure 1C displayed detailed β diversity data between different groups.
Figure 1.
β diversity analysis at 12 weeks of age. (A) Nonmetric multidimensional scaling (NMDS) and (B) principal component analysis (PCA) figures showed that individuals fitted well in their groups. (C) β diversity heat map showed by groups. TRF.AD, time-restricted feeding during the childhood phase before switched to ad libitum feeding pattern as adults; TRF.TRF, continuously treated with time-restricted feeding; AD.TRF, went through time-restricted feeding only in adulthood; AD.AD, always had ad libitum access to food. The last alphabet (M/F) in the group name indicated the gender of subjects. M, male mice; F, female mice (n=3).
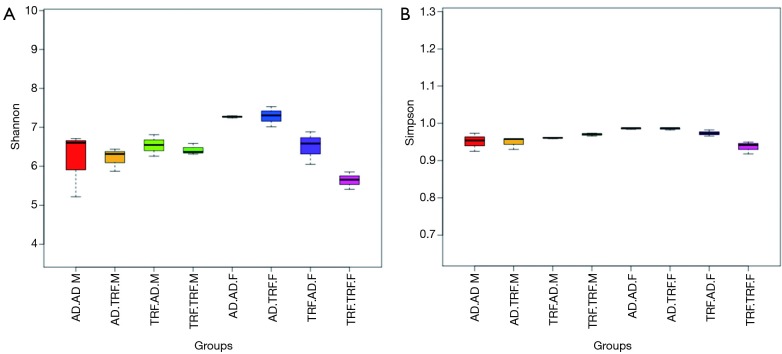
Regarding α diversity, Shannon and Simpson analysis were applied to evaluate the overall abundance and equipollency of the commensal flora respectively (Figure 2). Mice treated with TRF during earlier age were associated with less diversified gut microbiota when compared to subjects fed ad libitum (detailed data were shown in Table 1). In contrary, the difference regarding diversity was not so significant between different dietary patterns in adulthood. Variations existed between different genders. Female mice were found to have more diversified and evenly distributed bacteria ecosystem.
Figure 2.
α diversity analysis at 12 weeks of age. Shannon analysis (A) indicated the total number of bacteria species of each group. Simpson analysis (B) showed the equality of different groups. TRF.TRF, continuously treated with time-restricted feeding; AD.TRF, went through time-restricted feeding only in adulthood; AD.AD, always had ad libitum access to food. The last alphabet (M/F) in the group name indicated the gender of subjects. M, male mice, F, female mice (n=3).
Table 1. Wilcox test of α diversity between groups.
| Group | Shannon analysis | Simpson analysis | |||||
|---|---|---|---|---|---|---|---|
| Difference | P value | Significance | Difference | P value | Significance | ||
| Different diet during childhood | |||||||
| AD.AD.F-TRF.AD.F | 8.667 | 0.030 | * | 18.000 | 0.000 | *** | |
| AD.TRF.F-TRF.TRF.F | 19.000 | 0.000 | *** | 18.167 | 0.000 | *** | |
| AD.AD.M-TRF.AD.M | −1.333 | 0.719 | −2.000 | 0.464 | |||
| AD.TRF.M-TRF.TRF.M | −2.333 | 0.531 | −8.167 | 0.007 | ** | ||
| Different diet during adulthood | |||||||
| AD.AD.F-AD.TRF.F | −1.000 | 0.787 | −0.167 | 0.951 | |||
| TRF.AD.F-TRF.TRF.F | 9.333 | 0.021 | * | 12.333 | 0.000 | *** | |
| AD.AD.M-AD.TRF.M | 2.333 | 0.531 | 2.000 | 0.464 | |||
| TRF.AD.M-TRF.TRF.M | 1.333 | 0.719 | −4.167 | 0.138 | |||
| Same diet but different gender | |||||||
| AD.AD.F-AD.AD.M | 10.333 | 0.011 | * | 13.333 | 0.000 | *** | |
| AD.TRF.F-AD.TRF.M | 13.667 | 0.002 | ** | 15.500 | 0.000 | *** | |
| TRF.AD.F-TRF.AD.M | 0.333 | 0.928 | 5.667 | 0.049 | * | ||
| TRF.TRF.F-TRF.TRF.M | −7.667 | 0.052 | −10.833 | 0.001 | ** | ||
Significance: *, P<0.05; **, P<0.01; ***, P<0.001. TRF.AD, time-restricted feeding during the childhood phase before switched to ad libitum feeding pattern as adults; TRF.TRF, continuously treated with time-restricted feeding; AD.TRF, went through time-restricted feeding only in adulthood; AD.AD, always had ad libitum access to food; M, male mice; F, female mice.
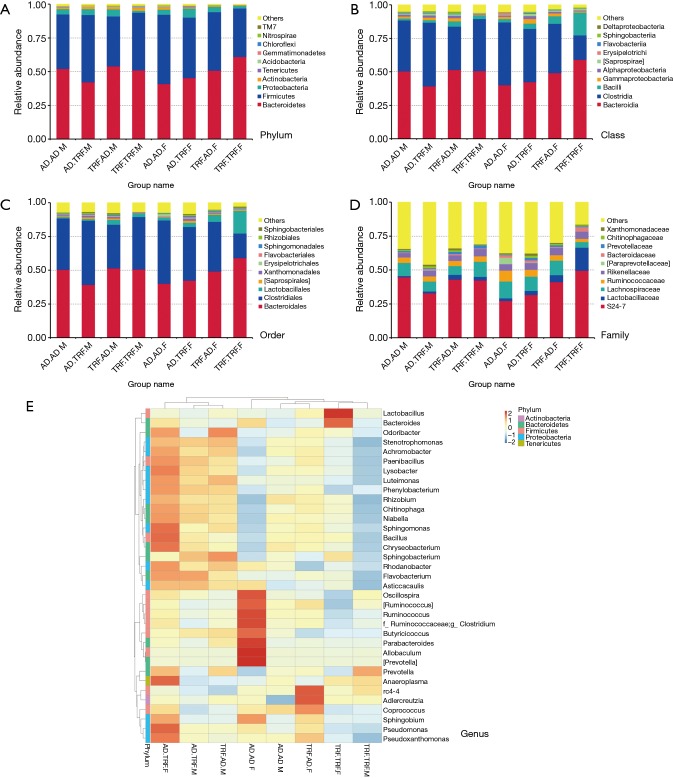
Analysis was executed under five different levels (phylum, class, order, family, and species) (Figure 3). As the largest entity, proportion of Bacteroidetes accounted for approximately 50% among all phylums, and Firmicutes came after it under this level. Mice went through restricted feeding as children were characterized with fewer proportions of Firmicutes. Typically, adding up the rest phylums together took up no more than 10%. Compared to TRF.AD groups, abundance of Clostridiales order and Ruminococcaceae family were greater in AD.AD mice in regardless of gender. Other differences of bacteria distribution among experimental subjects were available in our database.
Figure 3.
Component analysis under different taxonomical level. (A) Top 10 phylums of each group; (B) top 10 classes of each group; (C) top 10 orders of each group; (D) top 10 families of each group; (E) taxa-heatmap showing the relative abundance of the most common genus of each group. TRF.TRF, continuously treated with time-restricted feeding; AD.TRF, went through time-restricted feeding only in adulthood; AD.AD, always had ad libitum access to food. The last alphabet (M/F) in the group name indicated the gender of subjects. M, male mice, F, female mice (n=3).
Discussion
TRF is a distinct feeding schedule that does not restrict total caloric intake or nutritional structure, making it an ideal model to independently investigate the effect of feeding pattern on gut microbiota. TRF pattern allows 8–10 hours of feeding every day, but without any limitations of food access during this period. In adult model, even though TRF and ad libitum (AD) fed on similar amount of food, TRF attenuates fattening traits and relieves metabolic disorders (15). The mechanism of TRF predominantly attributes to its role as an oscillator in the communication of diet and innate circadian clock (16). It is also demonstrated in our previous investigation that commensal bacteria work as the intermediary agent both in juvenile and adult mice (1,2,17).
According to our data, although TRF during childhood failed to present similar protective effect against metabolic disorders as observed in adults (2), the changes of microbiota were undoubted. Previously, the plasticity of the gut ecosystem to environmental factors such as diet was highlighted via sequencing approach to show the composition and functional changes (18). But what we found through this experiment denied the extrapolation of this plasticity to pediatric mice. Instead, both α diversity and taxa analysis provided strong evidence that TRF brought evident alterations of intestinal microbial community, and which legacy effect persisted until adulthood. To specify, decreased overall abundance and equipollency were shown in TRF.AD mice groups comparing to AD.AD groups, which is unfavorable upon general healthy conditions. In addition, TRF.AD groups had low percentage of protective bacteria groups against overweight, IBD, and CRC. Surprisingly, compared to the difference caused by childhood feeding, the difference regarding diversity was not so significant between different dietary patterns in adulthood. As the samples were harvested when all mice were 12 weeks of age, those who were treated with different feeding interventions but switched to same feeding schedule were provided with 4 weeks of accommodation to the new diet. If the mice subjects provided identical intestinal environment to their commensal flora, a period of 3 to 4 days is adequate to develop very similar microbial communities according to the diet, in regardless of what feeding schedule they went through previously. However, the apparent gaps presented between TRF.AD group and AD.AD group, as well as TRF.TRF group and AD.TRF group demonstrated that feeding pattern caused irreversible structural alterations to the intestines. The above finding suggested that eating schedule in youth should be emphasized more for their close correlation to the entraining of gut ecosystem.
A healthy gut ecosystem is essential for the growth and development of pediatric mice, and feeding pattern is among the factors impacting the profile of microorganisms. Previous researches reported the role of microbiota in the maturation of the host immune system and its legacy effect on future health outcomes (19). Now that we have demonstrated that TRF caused alteration of microbial structure during childhood, these changes might further contribute to future disease susceptibility. This mechanism potentially explained the reason why childhood TRF causes irreversible negative health outcomes reported in our previous study (2). It was hypothesized that healthy growth is partially dependent on normal postnatal development of the intestinal microbial community and that perturbations are related to feeding (20). Through gut flora, feeding pattern during childhood phase can independently facilitate tolerance to environmental exposures, and irreversibly define disease progression in later life.
Conclusions
TRF in childhood has remarkable long-term influence on juvenile mice gut microbiota. This inappropriate eating schedule during earlier ages leads to a disturbed microbiota-host relationship, and the impairments can hardly be adequately repaired in later life.
Acknowledgments
We sincerely thank Huayu Yang and Haihong Zhang for the guidance of experimental techniques.
Funding: This study was supported by CAMS Innovation Fund for Medical Sciences (2016-I2M-1-001).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Our study was conducted with the permission of the Animal Welfare Committee of Peking Union Medical College Hospital (permission no. XHDW-2015-1098).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Hu D, Mao Y, Xu G, et al. Gut flora shift caused by time-restricted feeding might protect the host from metabolic syndrome, inflammatory bowel disease and colorectal cancer. Transl Cancer Res 2018;7:1282-9. 10.21037/tcr.2018.10.18 [DOI] [Google Scholar]
- 2.Hu D, Mao Y, Xu G, et al. Time-restricted feeding causes irreversible metabolic disorders and gut microbiota shift in pediatric mice. Pediatr Res 2019;85:518-26. 10.1038/s41390-018-0156-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Metzker ML. Sequencing technologies-the next generation. Nat Rev Genet 2010;11:31-46. 10.1038/nrg2626 [DOI] [PubMed] [Google Scholar]
- 4.Ericsson AC, Davis JW, Spollen W, et al. Effects of vendor and genetic background on the composition of the fecal microbiota of inbred mice. PLoS One 2015;10:e0116704. 10.1371/journal.pone.0116704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen P. Enteric dysbiosis, gut barrier and liver disease. Hepatobiliary Surg Nutr 2018;7:38-40. 10.21037/hbsn.2017.12.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zackular JP, Rogers MA, Ruffin MT, 4th, et al. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila) 2014;7:1112-21. 10.1158/1940-6207.CAPR-14-0129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012;490:55-60. 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- 8.Xie G, Wang X, Liu P, et al. Distinctly altered gut microbiota in the progression of liver disease. Oncotarget 2016;7:19355-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan MLY, El-Nezami H. Targeting gut microbiota in hepatocellular carcinoma: probiotics as a novel therapy. Hepatobiliary Surg Nutr 2018;7:11-20. 10.21037/hbsn.2017.12.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schieber AM, Lee YM, Chang MW, et al. Disease tolerance mediated by microbiome E. coli involves inflammasome and IGF-1 signaling. Science 2015;350:558-63. 10.1126/science.aac6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams WB, Liao HX, Moody MA, et al. HIV-1 VACCINES. Diversion of HIV-1 vaccine-induced immunity by gp41-microbiota cross-reactive antibodies. Science 2015;349:aab1253. 10.1126/science.aab1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sonnenburg JL, Backhed F. Diet-microbiota interactions as moderators of human metabolism. Nature 2016;535:56-64. 10.1038/nature18846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sonnenburg ED, Smits SA, Tikhonov M, et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature 2016;529:212-5. 10.1038/nature16504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanton LV, Charbonneau MR, Salih T, et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 2016. doi: . 10.1126/science.aad3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaix A, Zarrinpar A, Miu P, et al. Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab 2014;20:991-1005. 10.1016/j.cmet.2014.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thaiss CA, Levy M, Korem T, et al. Microbiota Diurnal Rhythmicity Programs Host Transcriptome Oscillations. Cell 2016;167:1495-510.e12. 10.1016/j.cell.2016.11.003 [DOI] [PubMed] [Google Scholar]
- 17.Mukherji A, Kobiita A, Ye T, et al. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell 2013;153:812-27. 10.1016/j.cell.2013.04.020 [DOI] [PubMed] [Google Scholar]
- 18.Faith JJ, Guruge JL, Charbonneau M, et al. The long-term stability of the human gut microbiota. Science 2013;341:1237439. 10.1126/science.1237439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohnmacht C, Marques R, Presley L, et al. Intestinal microbiota, evolution of the immune system and the bad reputation of pro-inflammatory immunity. Cell Microbiol 2011;13:653-9. 10.1111/j.1462-5822.2011.01577.x [DOI] [PubMed] [Google Scholar]
- 20.Blanton LV, Barratt MJ, Charbonneau MR, et al. Childhood undernutrition, the gut microbiota, and microbiota-directed therapeutics. Science 2016;352:1533. 10.1126/science.aad9359 [DOI] [PubMed] [Google Scholar]