Abstract
The genus Yersinia comprises species that differ widely in their pathogenic potential and public-health significance. Yersinia pestis is responsible for plague, while Yersinia enterocolitica is a prominent enteropathogen. Strains within some species, including Y. enterocolitica, also vary in their pathogenic properties. Phenotypic identification of Yersinia species is time-consuming, labour-intensive and may lead to incorrect identifications. Here, we developed a method to automatically identify and subtype all Yersinia isolates from their genomic sequence. A phylogenetic analysis of Yersinia isolates based on a core subset of 500 shared genes clearly demarcated all existing Yersinia species and uncovered novel, yet undefined Yersinia taxa. An automated taxonomic assignment procedure was developed using species-specific thresholds based on core-genome multilocus sequence typing (cgMLST). The performance of this method was assessed on 1843 isolates prospectively collected by the French National Surveillance System and analysed in parallel using phenotypic reference methods, leading to nearly complete (1814; 98.4 %) agreement at species and infra-specific (biotype and serotype) levels. For 29 isolates, incorrect phenotypic assignments resulted from atypical biochemical characteristics or lack of phenotypic resolution. To provide an identification tool, a database of cgMLST profiles and reference taxonomic information has been made publicly accessible (https://bigsdb.pasteur.fr/yersinia). Genomic sequencing-based identification and subtyping of any Yersinia is a powerful and reliable novel approach to define the pathogenic potential of isolates of this medically important genus.
Keywords: Yersinia, phylogenetics, core-genome multilocus sequence typing, species, identification, genotyping
Data Summary
This whole-genome shotgun project was deposited at GenBank/ENA/DDBJ under the accession numbers CABHPT00000000 to CABIIH00000000, and CABIIP00000000 to CABIJR00000000 (BioProject number PRJEB33414). Core-genome multilocus sequence typing profiles have been made available through the BIGSdb – Yersinia database at https://bigsdb.pasteur.fr/yersinia.
Impact Statement.
High-quality genomic sequences of bacterial isolates can now be obtained rapidly and affordably, opening up new perspectives for isolate identification and epidemiological investigation. In our study, we developed a genomic sequence analysis method applicable to all members of the genus Yersinia for identification of clinical, veterinary, food or environmental isolates at species and strain levels. We have made this bioinformatics procedure accessible online to enable the reliable identification of Yersinia isolates and to evaluate their pathogenic potential from their assembled genome sequences. The sharing of reference genomic sequence data and using a unified genotyping scheme will advance the discovery of novel Yersinia species, as illustrated herein, and will facilitate the genetic relatedness studies that are required when there is suspicion of an outbreak.
Introduction
The genus Yersinia , a member of the family Enterobacteriaceae , is currently composed of 19 species and includes 3 prominent human pathogens: the agent of plague, Yersinia pestis, and the enteropathogens Yersinia enterocolitica and Yersinia pseudotuberculosis [1]. Whereas the isolation of Y. pseudotuberculosis is rare, Y. enterocolitica represents the third main cause of diarrhoea of bacterial origin in temperate and cold countries [2]. Other Yersinia species are non-pathogenic for humans; Yersinia ruckeri is a fish pathogen [3] and Yersinia entomophaga is an insect pathogen [4]. Given the heterogeneous pathogenic potential of Yersinia members, identification at species and sometimes infra-species levels is essential for patient follow-up, and to guide the deployment of public-health measures. In addition, the taxonomy of the genus Yersinia is evolving dynamically, with eight novel species since 2005 [4–11]. Among these, Yersinia wautersii is the only one with a pathogenic potential for humans [11]. Therefore, a generally applicable identification strategy that would encompass all Yersinia species, including yet undescribed ones, would be useful.
Currently, the complete characterization of a Yersinia isolate is performed in reference laboratories and relies on phenotypic tests, such as the ability to grow on particular carbon sources, sero-agglutination, enzymatic tests and motility [1, 12–14]. This characterization allows all isolates to be assigned to species and serotypes, and to biotypes within the species Y. enterocolitica and Yersinia intermedia [15, 16]. The complete characterization of a Y. enterocolitica isolate is essential because it distinguishes the highly pathogenic biotype 1B, the non-pathogenic biotype 1A, and the low-pathogenic biotypes 2, 3, 4 and 5. Phenotypic characterization is labour-intensive, requires multiple assays and is time-consuming (7 days). Moreover, misidentifications may occur given that the distinction between some taxa relies on only a single metabolic trait, and that some isolates exhibit atypical metabolic profiles. Although MALDI-TOF MS is increasingly used for Yersinia species identification, this technique has limitations. For example, it does not distinguish the four species within the Y. pseudotuberculosis complex, including non-pathogenic Yersinia similis [11] and highly pathogenic Y. pestis [17–19]. Moreover, MALDI-TOF MS cannot discriminate the highly pathogenic biotype 1B of Y. enterocolitica from the non-pathogenic biotype 1A [20].
Identification of Yersinia isolates at the species and infra-specific levels has benefited from the development of sequence-based phylogenetic methods, including the highly reproducible and portable multilocus sequence typing (MLST) approach. A genotyping strategy based on five genes was first developed in 2005, but it did not have sufficient resolution to distinguish the various biotypes of Y. enterocolitica [21]. Later on, a seven-gene MLST scheme was set up to differentiate the three human pathogenic species ( Y. enterocolitica , Y. pseudotuberculosis and Y. pestis ), but this scheme was not applicable at the genus level [22]. Finally, a genus-wide seven-gene MLST scheme was developed [23], and allowed both the identification of Yersinia species and the differentiation of Y. enterocolitica biotypes; thus, representing a relevant alternative to the reference phenotypic method, especially for metabolically atypical isolates. However, classical MLST records variation at approximately 1/1000 of the Yersinia full genome sequence length, which limits its discriminatory power and restricts phylogenetic information. Whole-genome sequences can now be obtained readily using high-throughput sequencing technology and can be leveraged for core-genome MLST (cgMLST) genotyping [24], which provides much improved resolution and phylogenetic precision, as shown for some other bacterial pathogens [25–28].
In this work, we aimed to develop a Yersinia genus-wide strain identification and characterization method based on cgMLST. A phylogenetic analysis using genomes from public sequence repositories combined with genomes of isolates received routinely at the French National Reference Laboratory for Plague and other Yersinioses was performed to delineate current Yersinia species, as well as yet undefined taxa. Next, a genus-wide cgMLST scheme was developed to record nucleotide variation at 500 shared gene loci. Finally, an automated process for strain identification and characterization was set up, validated and implemented in the daily surveillance of Yersinia isolates in France. To disseminate the method, a database of cgMLST profiles and their corresponding identification was made publicly accessible using the web-based tool BIGSdb (Bacterial Isolate Genome Sequence Database) [29, 30], allowing external users to identify, type and subtype their Yersinia isolates using their own genomic sequences.
Methods
Isolate collection
All Yersinia isolates came from the strain collection of the French Yersinia National Reference Laboratory (YNRL), which comprises more than 41 000 strains, including members of all known Yersinia species (except Y. pestis ). Most strains (~26 000; 63 %) were isolated in France, whereas ~15 000 strains came from other countries throughout the world. Additionally, the Yersinia Research Unit (Institut Pasteur, Paris) maintains a collection of ~1800 Y. pestis strains isolated worldwide.
Phenotypic characterization
Yersinia isolates were identified at the YRNL using API20E and API50CH strips, tween-esterase activity [13], pyrazinamidase activity [12] and mannitol-mobility at 28 °C. Y. enterocolitica strains were biotyped according to Wauters biogrouping scheme [15]. Y. enterocolitica and Y. pseudotuberculosis strains were serotyped with a set of 47 and 5 O-antigen-specific rabbit antisera, respectively [31]. Identification of Y. pestis isolates included a specific phage lysis test [32].
Genomic sequencing
A total of 802 isolates from the YNRL were sequenced using an Illumina NextSeq 500 instrument. DNA extraction was performed using a PureLink genomic DNA mini kit (Invitrogen) following the manufacturer’s instructions, except that DNA was eluted in 150 µl H2O (Ambion). Sequencing libraries were prepared using a Nextera XT DNA library preparation kit (Illumina). Paired-end reads of 150 nt were obtained using the Mid Output or High Output kits (Illumina). Trimming and clipping were performed using AlienTrimmer v0.4.0 [33]. Redundant or over-represented reads were reduced using the khmer software package v1.3 [34]. Finally, sequencing errors were corrected using Musket v1.1 [35]. A de novo assembly was performed for each strain using SPAdes v3.12.0 [36] with the pre-processed reads. A minimum sequencing depth of 50× was obtained for each genome. Contigs with significantly low coverage compared to the others were discarded as putative contaminants. On average, genomes were assembled into 208 contigs (min=20; max=2051) with a total size of 4.6 Mb (min=3.6 Mb; max=5.4 Mb) and with an N50 value of 86 065 (min=18 825; max=490 559).
Constitution of a genome dataset
A total of 544 publicly available assembled genomes were downloaded in February 2016 (Tables 1 and S1, available with the online version of this article) from the GenBank repository, representing all Yersinia species according to the recognized taxonomy at that time, except Y. entomophaga . In addition, the genome sequences of 802 Yersinia isolates received at the YNRL (Tables 1 and S1), and previously characterized phenotypically, were determined and used in this work. Together, this constituted a reference dataset of 1346 assembled genomes (Tables 1 and S1). Finally, the cgMLST scheme was validated on 1843 additional isolates received at the YNRL between 2016 and mid-2017 that were sequenced and phenotyped in parallel.
Table 1.
Species and bioserotype composition of the genomic dataset
|
Species |
Bioserotype |
Origin |
Total |
|
|---|---|---|---|---|
|
Public genomes |
YNRL |
|||
|
6 |
14 |
20 |
||
|
2 |
12 |
14 |
||
|
4 |
17 |
21 |
||
|
1A |
35 |
42 |
77 |
|
|
1B |
13 |
3 |
16 |
|
|
2/O:9 |
28 |
8 |
36 |
|
|
2–3/O:5,27 |
15 |
7 |
22 |
|
|
3/O:3 |
0 |
19 |
19 |
|
|
4 |
26 |
115 |
141 |
|
|
5 |
7 |
4 |
11 |
|
|
Unknown |
8 |
0 |
8 |
|
|
0 |
2 |
2 |
||
|
22 |
19 |
41 |
||
|
16 |
7 |
23 |
||
|
13 |
11 |
24 |
||
|
2 |
5 |
7 |
||
|
10 |
11 |
21 |
||
|
1 |
0 |
1 |
||
|
2 |
0 |
2 |
||
|
270 |
20 |
290 |
||
|
43 |
442 |
485 |
||
|
6 |
12 |
18 |
||
|
8 |
11 |
19 |
||
|
5 |
13 |
18 |
||
|
2 |
5 |
7 |
||
|
Undefined |
0 |
3 |
3 |
|
|
Total |
544 |
802 |
1346 |
|
Design of theYersinia cgMLST scheme
From the 1346 reference genomes, 200 genomes representative of genus Yersinia diversity were selected in the following way. Genome assemblies made up of more than 500 contigs with N50 values below 10 000 nt were discarded. From the remaining genomes, we calculated pairwise genome distances using the program andi [37]. From the resulting matrix, we defined 200 clusters based on an UPGMA (unweighted pair group method with arithmetic mean) hierarchical clustering. For each cluster, one representative with the largest N50 value was selected, leading to a subset of 200 phylogenetically representative genomes (Table S1). A total of 1672 genes present in more than 95 % of these 200 representative genomes was defined as the core genes. For each of these core genes, a file containing all alleles (typically between 50 and 100) was generated. Each file was parsed, and genes were removed if (i) a character other than A, T, G or C was present in any of the sequences, (ii) the gene had a paralogue or (iii) there was a gap of more than 6 nt in the multiple sequence alignment. These filtering criteria led to the selection of 500 core genes deemed suitable for cgMLST analysis.
Phylogenetic analyses
In order to perform a phylogenetic analysis of the genus Yersinia , a representative subset was built from the 3878 Yersinia genomes available in our database. First, this set of isolates was partitioned into 30 clusters based on a UPGMA clustering from the pairwise dissimilarities between allelic profiles. Second, as these 30 clusters were of varying size n (n=1 to 1849 isolates), each was partitioned into 7 log10 n UPGMA sub-clusters, and the isolate with the smallest number of missing alleles was selected within each sub-cluster, leading to a final set of 236 isolates representative of the genus Yersinia . For each of the 500 genes of the cgMLST scheme, the corresponding allele sequences were translated, and a multiple sequence alignment was performed using mafft v7.407 [38]. The 500 multiple sequence alignments were concatenated into a unique supermatrix of 139 050 amino acid characters that was used to infer a maximum-likelihood phylogenetic tree using iq-tree v1.6.3 [39] with the evolutionary model JTT+F+R5 selected by minimizing the BIC criterion [40]. Following a similar approach, datasets within Y. enterocolitica and Y. pseudotuberculosis species led to two supermatrices of characters for 246 (+28 outgroup) and 294 (+20 outgroup) representative isolates, respectively, that were phylogenetically analysed with the evolutionary models JTT+F+R3 and JTT+F+R2, respectively.
Average nucleotide identity (ANI) estimation
Using the 236 representative Yersinia genomes (see above), the ANI was estimated for each pair of isolates using fastANI v1.1 [41].
BIGSdb
A database was created for the genus Yersinia in the Institut Pasteur MLST and whole-genome MLST resource (https://bigsdb.pasteur.fr), which uses the BIGSdb software tool, designed to store and analyse sequence data for bacterial isolates [29, 30]. All de novo assembled genomes were uploaded into the isolates database, and the reference alleles of the 500 cgMLST loci were defined in the linked database of reference sequences and profiles (‘seqdef’). Using BIGSdb functionalities, a scan of the genome sequence was performed for each isolate using parameters (min 80 % identity, min 80 % alignment, blastn word size of 20 nt) to check for the presence of each core gene and to determine its allele number. Allelic profiles identifiers (cgST) were defined for genomes with 50 or fewer missing alleles. The BIGSdb – Yersinia database of cgMLST profiles is accessible at https://bigsdb.pasteur.fr/yersinia/.
Taxonomic assignment
To assign Yersinia isolates to taxonomic categories based on their cgMLST allelic profiles, a set of allelic difference proportion cut-offs was determined at different levels of the Yersinia phylogenetic structure. For this, a minimum spanning tree was built from the pairwise dissimilarities between allelic profiles containing fewer than 50 missing alleles. For a given group of isolates (e.g. species), the corresponding subtree was extracted from the minimum spanning tree and the length of its largest edge was defined as the cut-off associated with this group of isolates.
Validation of the cgMLST method
For 1843 isolates received prospectively between 2016 and mid-2017 at the YNRL, taxonomic assignment obtained using the cgMLST method was compared to the phenotypic characterization performed in parallel. The above-defined stringent thresholds used to define species or lineages were re-evaluated based on isolates with no taxonomic assignment (Fig. S1). For isolates presenting discrepancies between the two characterization methods, phenotyping was repeated.
Seven-gene MLST
Genomes were scanned for the seven loci of the genus-wide MLST scheme of McNally and colleagues [23]. From February 15th 2019, for each of the seven MLST loci, the alleles were downloaded from the PubMLST website (https://pubmlst.org/yersinia/), translated and aligned at the amino acid level using mafft v7.407 [38]. Each of the seven multiple amino acid sequence alignments was converted into a position-specific score matrix (PSSM) [42] using blast+ v2.6.0 [43]. For each of the 236 representative Yersinia genomes (see above), the seven MLST alleles were searched with tblastn by using the corresponding PSSM as query, and extracted. The seven allele files were aligned using mafft, concatenated and phylogenetically analysed using iq-tree v1.6.3 [39] with the evolutionary model TIMe+I+G4 selected by minimizing the BIC criterion [40].
Time-to-results
The time-to-results of the cgMLST and phenotypic approaches were computed based on 1440 and 1113 isolates, respectively. cgMLST timing started with the date of receipt, and included strain isolation and growth, DNA extraction/purification, library preparation and sequencing on the sequencing core facility of Institut Pasteur, Paris (France), as well as demultiplexing, pre-processing, assembly and cgMLST allele calling. Phenotypic time-to-result was calculated from isolate receipt to taxonomic and bioserotype assignment as described above. To compare the time-to-results of both methods, a Student's t-test was performed.
Results
Phylogenetic structure of the genus Yersinia
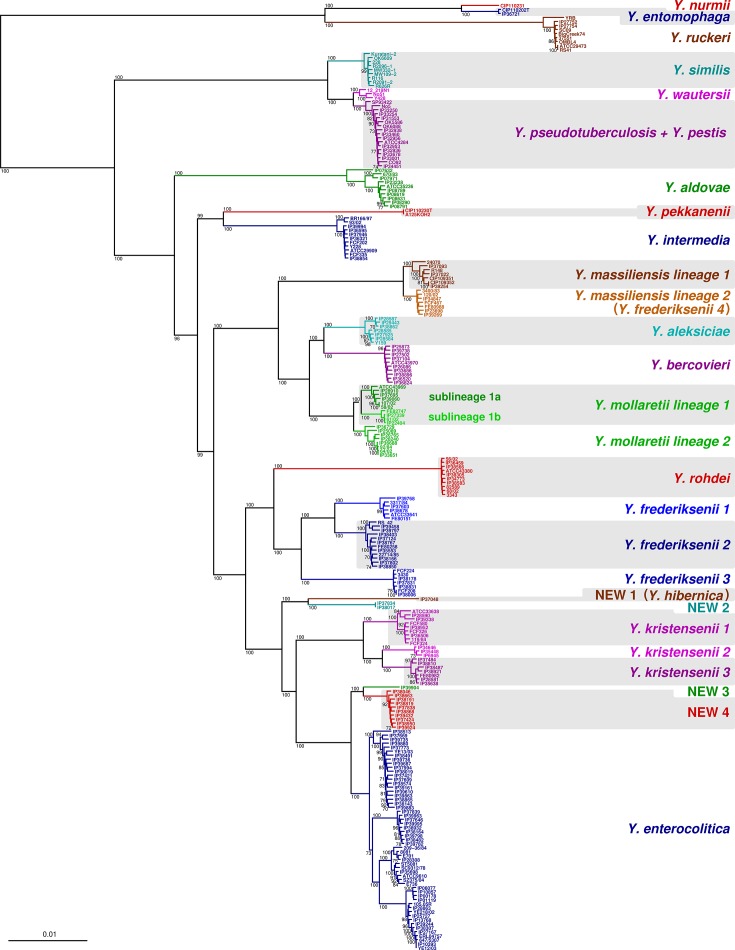
A phylogenetic analysis of 236 genomes representing the diversity of the genus Yersinia was performed based on the concatenation of 500 multiple sequence alignments, revealing a neat structuration of Yersinia into strongly demarcated clades (Fig. 1). Although most lineages fitted with phenotypic species assignments, a number of taxonomic inconsistencies and yet undescribed Yersinia groups were uncovered. The potential taxonomic rank of each demarcated clade was, therefore, evaluated based on ANI (Table S2) in the light of the proposed bacterial species delineation cut-off of 95–96 % ANI [44, 45]. As a first step, the use of a stringent delineation cut-off value of 96 % ANI led to 26 groups being defined, labelled as follows: Yersinia aldovae; Yersinia aleksiciae; Yersinia bercovieri; Y. enterocolitica ; Y. entomophaga ; Yersinia frederiksenii 1, 2 and 3; Y. intermedia ; Yersinia kristensenii 1, 2 and 3; Yersinia massiliensis lineages 1 and 2; Yersinia mollaretii lineages 1 and 2; Yersinia nurmii; Yersinia pekkanenii; Y. pseudotuberculosis (also containing Y. wautersii and Y. pestis isolates); Yersinia rohdei; Y. ruckeri ; Y. similis ; and NEW1 to NEW4 (Fig. 1, Table S2). Second, taxa with current standing in Yersinia taxonomy, but belonging to the same ANI group as other taxa ( Y. pestis and Y. wautersii ), were individualized as distinct groups. Therefore, genomes of our dataset were classified into 28 groups in total. Note that this classification is not a taxonomic proposal, but is used instead as an operational classification. The correspondence between phenotypic and genome-based phylogenetic classifications is summarized in Table 2 and provided for individual isolates in Table S1. We hereafter summarize these observations.
Fig. 1.
Maximum-likelihood phylogenetic tree of the genus Yersinia based on 500 concatenated multiple sequence alignments. Only bootstrap-based branch support values >70 % are shown. Bar, 0.01 amino acid substitutions per character.
Table 2.
Correspondence between genotypic and phenotypic characterization
|
Genotypic characterization |
Phenotypic characterization |
||
|---|---|---|---|
|
Species |
Lineage |
Species |
Infra-specific category |
|
1Aa |
Biotype 1A |
||
|
|
1Ab |
|
|
|
1B |
Biotype 1B |
||
|
2/3-9a |
Biotype 2/O:9 |
||
|
2/3-9b |
|||
|
|
2/3-5a |
Bioserotype 2–3/O5,27 |
|
|
|
2/3-5b |
|
|
|
3-3a |
Bioserotype 3/O:3 |
||
|
3-3b |
|||
|
3–3 c |
|||
|
3-3d |
|||
|
4 |
Bioserotype 4/O:3 |
||
|
5 |
Biotype 5 |
||
|
1 to 32 |
21 O-serotypes |
||
|
|
|
||
|
1 (sublineage 1a and 1b) |
|
||
|
2 |
|||
|
|
Diverse serotypes |
||
|
|
Diverse serotypes |
||
|
|
|||
|
|
|||
|
1 |
|
||
|
2 |
|||
|
|
|
||
|
|
|
||
|
|
|
||
|
|
|
||
|
|
|
||
|
|
|
||
|
|
|
||
|
|
|
||
|
|
|
||
|
|
|
||
|
|
|
||
|
NEW 1 ( Y. hibernica ) |
|
Yersinia sp. |
|
|
NEW 2 |
|
Yersinia sp. |
|
|
NEW 3 |
|
Biotype 1A |
|
|
NEW 4 |
|
Biotype 1A |
|
Non-human-pathogenic species Y. nurmii , Y. entomophaga , Y. ruckeri , Y. aldovae , Y. pekkanenii , Y. intermedia , Y. aleksiciae , Y. bercovieri and Y. rohdei each corresponded to a single clade (Fig. 1). In general, they were represented by a few isolates or showed little phylogenetic substructure. ANI values obtained within each group were all >96.6 %. In contrast, the maximum ANI value observed between the members of these species and their closest relatives was 94.7 % (Table S1). These observations are concordant with the species status of these nine Yersinia members.
Isolates identified as Y. mollaretii based on phenotypic characterization fell into two main lineages, one of them being further subdivided into two sublineages (Fig. 1). We defined lineage 1 as the one containing the type strain ATCC43969T. ANI values between the three lineages or sublineages were 95.6–96.6 % on average, whereas values within each of them were >98 %. The results show that Y. mollaretii represents a single clade, which could possibly be subdivided into three species and/or subspecies [46].
Isolates identified as Y. frederiksenii based on phenotypic characterization fell into four lineages (Fig. 1). Y. frederiksenii lineage 1 was defined as comprising the type strain ATCC33641T. Whereas lineages 2 and 3 were closely related to lineage 1, lineage 4 was in fact associated with Y. massiliensis (Fig. 1). ANI values among lineages 1 to 3 were 89.1 % on average, whereas ANI was >98.4 % within each lineage (Table S2). Lineage 4 (=Y. massiliensis lineage 2 in Fig. 1) displayed ANI values of 95.8 % on average with isolates phenotypically characterized as Y. massiliensis , including the type strain CIP109351T, whereas each of these two groups was homogeneous (ANI >98.6 %). While isolates of Y. frederiksenii lineage 4 are positive for rhamnose fermentation, Y. massiliensis isolates are negative. This result shows that Y. frederiksenii lineage 4 may be defined as a rhamnose-positive subspecies of Y. massiliensis, which we therefore labelled as Y. massiliensis lineage 2 (Fig. 1). Altogether, isolates that are currently identified phenotypically as Y. frederiksenii correspond to four separate clades that could be regarded as distinct species.
Isolates that were identified phenotypically as Y. kristensenii were monophyletic and subdivided into three main lineages (Fig. 1). Lineage 1 contained the type strain ATCC33638T; ANI values (Table S2) were 93.5 % among lineages but >98.9 % within lineage, showing that Y. kristensenii isolates actually correspond to three separate clades derived from a single ancestor and that may be considered as three species.
The phylogenetic analysis revealed the existence of additional novel lineages. First, two unique lineages, which we call Yersinia new species 1 (NEW 1) and Yersinia new species 2 (NEW 2; Fig. 1), corresponded to isolates that could not be identified based on phenotypic characterization due to atypical metabolic characteristics. Lineages NEW 1 and NEW 2 were phylogenetic sister groups (Fig. 1) separated by 90.1 % ANI (Table S2). They comprised only one and two isolates, respectively (ANI within lineage NEW 2 : 99.9%). Second, two additional lineages, which we call NEW 3 and NEW 4 (Fig. 1), were made up by isolates phenotypically characterized as Y. enterocolitica biotype 1A. These lineages comprised one and ten isolates, respectively, and were closely related to Y. enterocolitica . ANI values between the two lineages were 94.7 % on average (>99.3 % within NEW 4), and were 93.9 and 94.2 %, respectively, with Y. enterocolitica (Table S2). Altogether, these results show that lineages NEW 1 to NEW 4 may represent four additional Yersinia species. Of note, NEW 1 was recently described as the new species Yersinia hibernica [10]: the ANI between its type strain (accession numbers of its chromosome and plasmid are CP032487 and CP032488) and strain IP37048 (NEW 1) is 99.9 %.
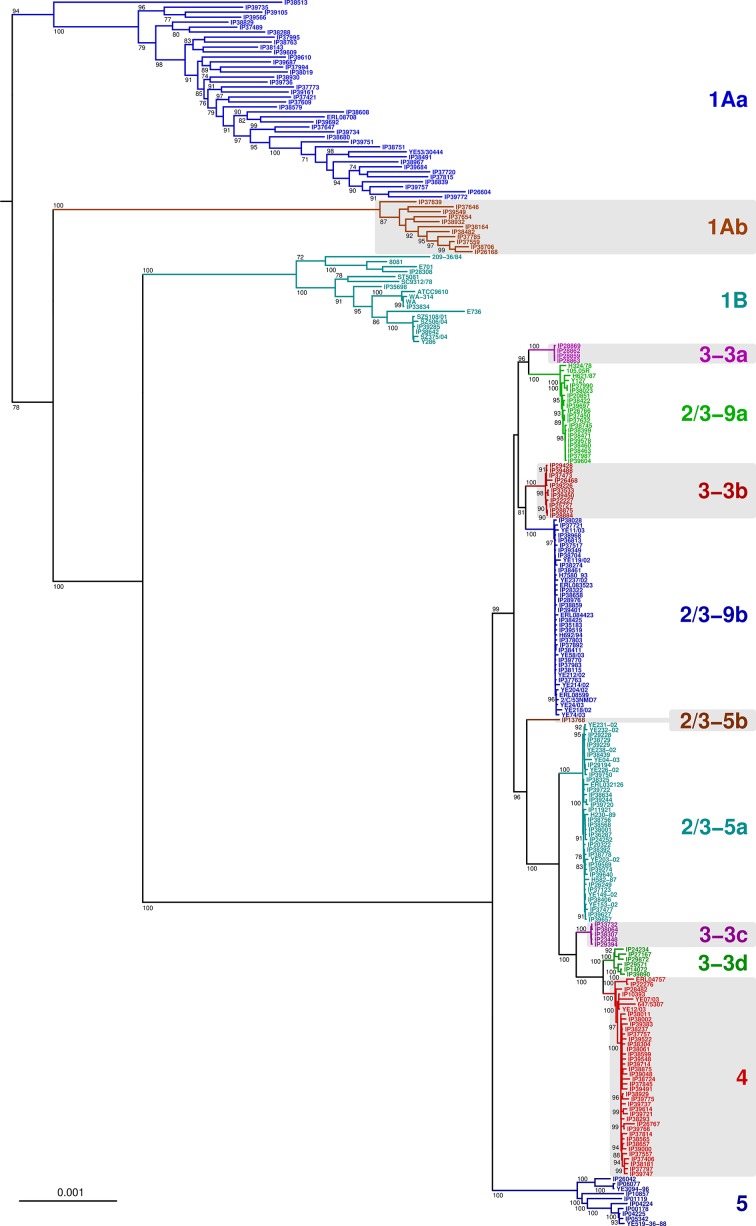
Population structure of Y. enterocolitica
Y. enterocolitica isolates clustered in a single clade (Fig. 1), and ANI values among them (ranging from 95.7 to 100 %) (Table S2) confirmed that they may be regarded as a single species. To provide a detailed view of the population structure of this species, a phylogenetic analysis was performed with 246 Y. enterocolitica isolates (Fig. 2). Non-pathogenic isolates identified as biotype 1A by phenotypic characterization fell into two separate clades, which we call 1Aa and 1Ab. These two lineages represented the earliest diverging branches of Y. enterocolitica , consistent with the hypothesis of evolution of pathogenic members from a non-pathogenic ancestor [47]. Y. enterocolitica isolates of biotypes 1B, 4 and 5 clustered into three different clades that were concordant with biotype assignments (Fig. 2). In contrast, other bioserotypes were in fact subdivided into two or more unrelated sublineages. First, isolates identified as bioserotype 2–3/O:9 fell into two unrelated sublineages for which we propose the names 2/3-9a and 2/3-9b. Likewise, isolates identified as bioserotype 2–3/O:5 fell into two different sublineages for which the names 2/3-5a and 2/3-5b are proposed; sublineage 2/3-5b comprised a single strain, but was clearly distinct from sublineage 2/3-5a isolates (ANI 99.5 to 99.6 %, data not shown). Finally, isolates identified as bioserotype 3/O:3 fell into four different and clearly distinct sublineages for which the names 3-3a, 3-3b, 3–3 c and 3-3d are proposed. Whereas 3–3 c and 3-3d were associated with the biotype 4 clade, sublineages 3-3a and 3-3b were associated with sublineages 2/3-9a and 2/3-9b, respectively.
Fig. 2.
Maximum-likelihood phylogenetic tree of Y. enterocolitica species based on 500 concatenated multiple sequence alignments. The tree was rooted with isolates from the groups NEW 3 and NEW 4 (not shown). Only bootstrap-based branch support values >70 % are shown. Bar, 0.001 amino acid substitutions per character.
Population structure of the Y. pseudotuberculosis complex
The Y. pseudotuberculosis species complex comprises Y. similis , Y. pseudotuberculosis , Y. pestis and Y. wautersii [11]. These four taxa clustered in a single clade in the Yersinia phylogenetic tree (Fig. 1). Non-pathogenic Y. similis isolates fell into a lineage that emerged first, with the three other species being tightly associated, consistent with previous works [11, 48]. ANI values (Table S2) among Y. similis isolates were >99.4 %, and they differed by 95.0 % on average with isolates from the three other species. These results support Y. similis as a separate species [7]. To provide details on the phylogenetic relationships among Y. wautersii , Y. pseudotuberculosis and Y. pestis isolates, an analysis was conducted with 294 strains from the Y. pseudotuberculosis complex (Fig. 3). Y. wautersii isolates fell into an early branching lineage clearly separated from the Y. pseudotuberculosis+Y. pestis clade. ANI values (Table S2) between these two lineages were 97.5 % on average (>99.2 % within each lineage), illustrating that Y. wautersii was defined at the species rank despite being more related to Y. pseudotuberculosis than the 95–96 % cut-off value. Isolates phenotypically characterized as Y. pseudotuberculosis or Y. pestis fell into 33 different sublineages, one of which comprised all Y. pestis isolates (Fig. 3). Considering its extreme pathogenicity and its peculiar ecological niche, the Y. pestis sublineage remains defined as a separate species, even though it emerged from within Y. pseudotuberculosis as recognized previously [47, 49, 50]. The 32 other sublineages making up the Y. pseudotuberculosis population structure were analysed with respect to the distribution of O-serotypes. Several individual O-serotypes were observed in different sublineages. The most conspicuous case is serotype O:1, which was widely distributed across sublineages; but this was also the case for serotypes O:2 to O:5. Besides, isolates of a single sublineage (e.g. 5, 8, 11 and 14) could differ in their O-serotypes, cgMLST scheme, definition of thresholds and taxonomic assignment.
Fig. 3.
Maximum-likelihood phylogenetic tree of Y. pseudotuberculosis , Y. wautersii and Y. pestis based on 500 concatenated multiple sequence alignments. The tree was rooted with Y. similis (not shown). Only bootstrap-based branch support values >70 % are shown. Bar, 0.001 amino acid substitutions per character. AAG, Auto-agglutinable; NAG, non-agglutinable; NT, non-typable as Y. pestis does not harbour the O-antigen; O, O antigen serotype; Unk, unknown serotype.
To capture the phylogenetic structure of Yersinia using cgMLST, an intuitive and highly reproducible bacterial strain genotyping method was implemented [24, 26], for which species- and sublineage-specific thresholds were defined (see Methods and Fig. S1). This set of cut-off values allowed all reference isolates to be grouped in agreement with their phylogenetic classification. To evaluate this method for strain identification, 1843 Yersinia isolates received prospectively at the YNRL between 2016 and mid-2017 were analysed by the automatic cgMLST taxonomic assignment procedure. Details of the isolates used for this evaluation are presented in Tables 3 and 4. For 1814 (98.4 %) isolates, genotypic assignment was consistent with phenotypic characterization (Table 3). Taxonomic assignments showed that the validation dataset comprised 669 non-pathogenic Yersinia isolates belonging to nine different species, 1116 pathogenic Y. enterocolitica isolates and 27 Y. pseudotuberculosis isolates.
Table 3.
Validation of the genotypic characterization
Consistent characterization with both methods was found for 1814 strains (out of 1843).
|
Phenotypic characterization |
Genotypic characterization |
||||
|---|---|---|---|---|---|
|
Species |
Biotype or serotype |
No. |
Species |
Lineage |
No. |
|
2 |
2 |
||||
|
|
10 |
|
10 |
||
|
|
45 |
|
3 |
||
|
|
34 |
||||
|
|
8 |
||||
|
|
18 |
|
18 |
||
|
|
7 |
|
3 |
||
|
|
4 |
||||
|
|
1 |
1 |
1 |
||
|
|
4 |
|
4 |
||
|
|
9 |
|
9 |
||
|
1A |
573 |
1Aa |
565 |
||
|
|
1Ab |
8 |
|||
|
2/O:5–27 |
16 |
2/3-5a |
16 |
||
|
2/O:9 |
147 |
2/3-9a |
11 |
||
|
|
|
|
2/3-9b |
136 |
|
|
3/O:3 |
3 |
3-3b |
2 |
||
|
|
|
|
|
3–3 c |
1 |
|
4/O:3 |
950 |
4 |
950 |
||
|
O:3 |
1 |
14 |
1 |
||
|
O:1 |
26 |
2 |
3 |
||
|
4 |
2 |
||||
|
7 |
4 |
||||
|
10 |
5 |
||||
|
12 |
2 |
||||
|
15 |
6 |
||||
|
16 |
3 |
||||
|
17 |
1 |
||||
|
Yersinia sp. |
|
2 |
NEW 2 |
|
2 |
|
Total |
|
1814 |
|
|
1814 |
Table 4.
Correspondence between the genotypic and phenotypic characterizations for the 29 non-concordant strains (out of 1843)
|
Phenotypic characterization |
Genotypic characterization |
||||
|---|---|---|---|---|---|
|
Species |
Biotype or serotype |
No. |
Species |
Lineage |
No. |
|
1A |
21 |
NEW4 |
|
21 |
|
|
|
4 |
2 |
4 |
||
|
1A/O:5 |
1 |
|
1 |
||
|
2/O:27 |
1 |
3–3 c |
1 |
||
|
|
2/O:5–27 |
1 |
1Ab |
1 |
|
|
|
3/O:3 |
1 |
4 |
1 |
|
|
Total |
|
29 |
|
|
29 |
Only 29 (1.6 %) isolates received a genotypic assignment that was discordant with phenotypic identification. Based on phenotypic characterization, these isolates belong mainly to two species: Y. enterocolitica and Y. frederiksenii (Table 4). First, 21 isolates were phenotypically characterized as Y. enterocolitica biotype 1A, whereas they actually belong to the new species temporarily called NEW 4. Second, four isolates phenotypically characterized as Y. frederiksenii actually belong to the above-described Y. massiliensis lineage 2. Third, one non-pathogenic Y. enterocolitica 1A/O:5 was identified as a Y. frederiksenii isolate by cgMLST. New phenotypic and genotypic characterizations were performed and confirmed the initial discrepancy. Fourth, one isolate (IP38164) identified as a pathogenic Y. enterocolitica 2/O5-27 was actually characterized by cgMLST as a Y. enterocolitica lineage 1Ab (non-pathogenic) isolate. This isolate exhibited a positive reaction for pyrazinamidase activity, which is a hallmark of the non-pathogenic Yersinia strains. Thus, we can consider that genotypic characterization allowed a correct taxonomic assignment of a non-pathogenic isolate. Fifth, isolate IP38493 phenotypically characterized as Y. enterocolitica 3/O:3 was classified by cgMLST as Y. enterocolitica lineage 4 (corresponding to bioserotype 4/O:3 based on phenotypic characterization). After investigation, it turned out that this isolate belongs to the phage type VIII, which is restricted to biotype 4 strains. We can conclude that isolate IP38493 was phenotypically misidentified due to an atypical positive xylose reaction, which is the only difference between biotypes 3 and 4. Finally, the last inconsistency was represented by isolate IP38064 identified as Y. enterocolitica 2/O:27 but belonging in fact to lineage 3–3 c (corresponding to bioserotype 3/O:3). This isolate presented an atypical phenotype for this lineage, as all biotype 2 strains are either of serotype O:5 or O:9. After further analysis, it turned out that this isolate also had a positive O:3 antiserum reaction, and had an atypical positive reaction for indole production, which is the only difference between biotypes 2 and 3. We concluded that IP38064 belongs to bioserotype 3/O:3, consistent with its Y. enterocolitica lineage 3–3 c genotypic assignment.
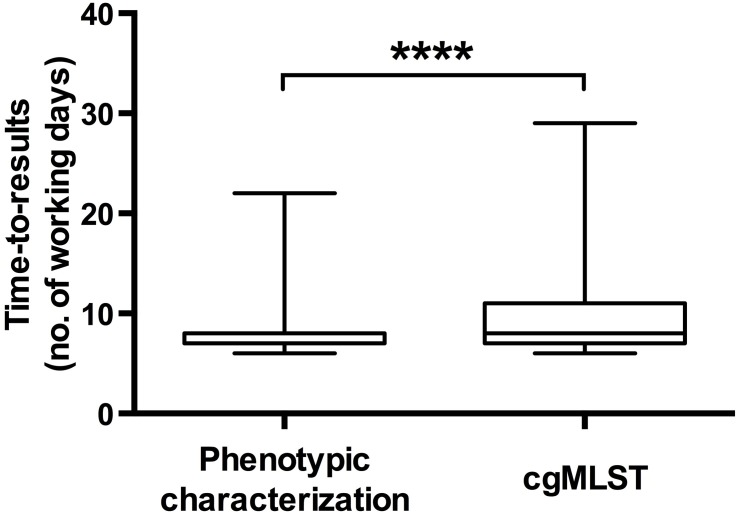
The time-to-results values for the two approaches were compared based on 1113 isolates phenotypically characterized in 2017 and on 1440 isolates characterized by cgMLST in 2018. Based on our current sequencing pipeline organization, cgMLST was slower by 1.3 days compared to phenotypic characterization (Fig. 4): 8.8 versus 7.5 days, respectively (P<0.0001). Whereas the time-to-results of the phenotypic characterization cannot be optimized, it would be possible to shorten the cgMLST process, as currently only two sequencing runs per week are performed on the core facility.
Fig. 4.
Time-to-results comparison between cgMLST and phenotypic methods. Calculations are based on 1113 isolates received in 2017 at the YNRL for the phenotypic characterization (mean=7.5 days) and 1440 isolates received in 2018 for the cgMLST (mean=8.8 days). Statistical analysis was performed with Student's t-test. ****, Difference highly significant, with P value <0.0001.
Phylogenetic tree of the genus Yersinia based on seven-gene MLST
Classical seven-gene MLST can be used without access to high-throughput sequencing. Therefore, we evaluated the genus-wide seven-gene MLST scheme developed by McNally and colleagues [23] on the 236 Yersinia genomes used to construct the 500-gene-based phylogenetic tree. The results showed that most clades, including potential novel species (see above), are neatly distinguished. Of note, Y. wautersii isolates did not belong to a single clade based on seven-gene MLST data, in contrast to cgMLST results (Fig. 1). Moreover, Y. pekkanenii and NEW 1+NEW 2 clades were not positioned at the same locations in the two trees (Figs 1 and S2), with higher bootstrap support values observed based on the 500 genes. These results show that seven-gene MLST is a reliable approach for Yersinia identification, although it is expected to be less reliable for phylogenetic classification given that a much lower number of genes are used.
Discussion
By analysing the largest set of whole-genome sequences representing the diversity of Yersinia to date, we have provided an updated view of the phylogenetic structure of this important bacterial genus. Our phylogenetic analysis confirms the neat demarcation of Yersinia into clearly distinct clades, most of which correspond to previously defined species. However, this work also uncovered a number of clades that may represent entirely novel species, or novel subdivisions within existing taxa. Overall, we identified eight novel clades that appear to represent new species based on the current ANI-based bacterial species definition: two novel clades provisionally labelled as NEW 1 and NEW 2, which are currently unidentified using classical approaches, as well as: (i) two clades currently identified as Y. frederiksenii and labelled Y. frederiksenii 2 and 3 ( Y. frederiksenii 1 is considered as Y. frederiksenii sensu stricto, because this lineage includes the type strain ATCC33641T; (ii) two clades currently identified as Y. kristensenii, labelled Y. kristensenii 2 and 3 ( Y. kristensenii 1 is considered as Y. kristensenii sensu stricto, because this lineage includes the type strain ATCC33638T); and (iii) two clades currently identified as Y. enterocolitica (NEW 3 and NEW 4). Furthermore, we uncovered clear subdivisions into separate lineages within the species Y. massiliensis , Y. mollaretii , Y. enterocolitica and Y. pseudotuberculosis . The clinical or epidemiological significance of these subdivisions largely remains to be defined. The results of our large genomic survey underline the need for a taxonomic update of the genus Yersinia . Although the ANI metric is a useful guide for deciding on the taxonomic status of phylogenetic lineages, the rigid application of a single threshold for a given taxonomic rank (e.g. species) would lead to inconsistencies with current taxonomy (e.g. Y. pestis and Y. wautersii ). Besides ANI values, it is highly relevant to consider the biology of organisms, their phenotypic characteristics including pathogenesis, and their phylogenetic breadth and internal structure.
Some of the clades or sublineages distinguished by cgMLST concur with new subgroups previously recognized using other methods. For instance, Reuter et al. [47] described that Y. frederiksenii strains were separated into two species clusters, SC8 and 9, which appear to correspond to Y. frederiksenii lineage 3 and lineage 1, respectively (Fig. 1). Moreover, strains previously classified as species cluster 14 [47] and Y. massiliensis genomospecies 2 [51] correspond to our Y. frederiksenii lineage 4/Y. massiliensis lineage 2. Finally, the recently described novel species Y. hibernica [10] corresponds to clade NEW 1.
Reference phenotyping approaches failed to recognize most novel clades and lineages. Occasional evolution of biochemical traits within species can result in some strains presenting atypical phenotypes, sometimes leading to misidentification, and isolates with the same phenotype may actually belong to different species on the genome level (Table S1). This clearly illustrates the limitations of current phenotypic characterizations of some of the Yersinia species. As phenotypic characterization, which is currently the most widely used approach, is labour-intensive and time-consuming (full characterization requires 7 to 8 days), alternative reference identification methods are clearly needed, rather than developing novel discriminant biochemical traits.
The phenotypic Vitek GN card or MS (MALDI-TOF) represent alternative identification methods, but are not fully reliable for Yersinia species [52, 53]. In addition, phenotypic identification at the infra-specific level and, thus, prediction of a strain’s pathogenic potential, is not possible. For instance, neither species of the Y. pseudotuberculosis complex nor biotype 1A nor 1B strains of Y. enterocolitica can be distinguished using MALDI-TOF MS [11, 17, 20]. Moreover, reference database updates would be needed for these methods to allow identification of novel taxa.
We implemented a genus-wide cgMLST genotyping system based on 500 highly conserved protein-encoding genes that were assembled with no nucleotide ambiguities, had no paralogue and presented limited size variation. These stringent criteria were used to maximize genotyping reliability and, therefore, led to a subset of genes much smaller than the actual core genome of Yersinia . Whereas cgMLST schemes with larger number of core genes are typically used to maximize discrimination for epidemiology and surveillance of single species [26, 54], the purpose of our cgMLST scheme is species-level and bioserotype-level identifications. Although most Y. enterocolitica cases are sporadic, some outbreaks have been described and subtyping methods were operational in these cases to identify contamination sources [55–57]. Whether our 500-gene cgMLST scheme could also be a useful tool in epidemiological investigations of Yersinia outbreaks remains to be evaluated.
The cgMLST approach presents the advantages of automation, standardization, reproducibility and simplicity of interpretation, which are important criteria for the use of genome sequence data in clinical microbiology [24, 58]. Using cgMLST genotypes (allelic profiles), we defined species- and lineage-specific thresholds for identification of Yersinia strains. While the initial definition of thresholds was based on the maximum cgMLST distance observed within groups of reference isolates, these thresholds were relaxed based on observed maximal distances (Table S3) when analysing prospective isolates. This allowed us to increase the identification rate and could be further adapted in the future if more distant genotypes are encountered within some lineages (Table S4). Given that Yersinia is so strongly structured phylogenetically, it is possible to increase the identification thresholds with no negative impact on reliability. Our evaluation using a prospective set of 1843 genomes produced in the framework of a national surveillance programme demonstrated the power of this approach for Yersinia identification at species- and infra-species bio-serotype levels. Furthermore, our cgMLST approach allows the clear distinction of all the species of the Y. pseudotuberculosis complex ( Y. pestis , Y. similis , Y. wautersii and Y. pseudotuberculosis ). The correspondence established in our system between phenotypic and genotypic characterization (Tables 2 and S4) enables backward compatibility [58–60] of the cgMLST strategy with classical Yersinia characterization, which is still extensively used worldwide. Furthermore, the cgMLST approach enables unambiguous classification of phenotypically atypical isolates. To make the cgMLST approach broadly available, the Yersinia BIGSdb was set up and made available at https://bigsdb.pasteur.fr/yersinia/. Genomic sequences can be analysed directly from the BIGSdb interface (Fig. S3). Alternately, cgMLST profiles and their attached identification information can be downloaded locally and used internally for confidential characterization of Yersinia genomic sequences. A recent survey revealed that national reference laboratories are increasingly using whole-genome sequence typing methods for surveillance of communicable diseases [61]. The method developed here has the potential to become a universally shared reference method for the identification of Yersinia isolates worldwide.
Data bibliography
1. Savin C, Martin L, Bouchier C, Filali S, Chenau J et al. GenBank BioProjects PRJEB3982, PRJEB3984, PRJEB3985, PRJEB3986, PRJEB3987, PRJEB3988, PRJEB3989, PRJEB3990, PRJEB3991 and PRJEB3992 (2014).
2. Reuter S, Connor TR, Barquist L, Walker D, Feltwell T et al. ENA studies PRJEB2116 and PRJEB2117 (2014).
3. Nguyen SV, Muthappa DM, Hurley D, Donoghue O, McCabe E et al. GenBank accession numbers CP032487 and CP032488 (2019).
Supplementary Data
Funding information
This project received funding from Santé Publique France (C.S., A.-S.L.G., E.C.) and from the Institut Pasteur (all authors).
Acknowledgements
We thank Sylvie Brémont for performing phenotypic characterization of isolates, and Andreea Alexandru, Maud Vanpeene and Vincent Enouf (sequencing core facility, Institut Pasteur) for set-up of the DNA preparation conditions and for genomic sequencing. This work used the computational and storage services (TARS cluster) provided by the IT department at the Institut Pasteur, Paris. We acknowledge the continuous support of Keith Jolley (Oxford University) for the development of the BIGSdb web application.
Author contributions
Conceptualization: C.S., A.C., E.C., S.B. Data curation: C.S., A.C., J.G., A.-S.L.G. Formal analysis: C.S., A.C., J.G. Funding acquisition: C.S., A.-S.L.G., E.C., S.B. Investigation: C.S., A.C., J.G. Methodology: A.C., J.G. Project administration: C.S. Resources: C.S., A.C., J.G. Software: A.C., J.G. Supervision: E.C., J.P.-C., S.B. Validation: C.S., A.C., J.G., A.-S.L.G. Visualization: C.S., A.C. Writing – original draft: C.S., S.B. Writing – review & editing: all authors.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: ANI, average nucleotide identity; cgMLST, core-genome multilocus sequence typing; MLST, multilocus sequence typing; UPGMA, unweighted pair group method with arithmetic mean; YNRL, Yersinia National Reference Laboratory.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Four supplementary tables and three supplementary figures are available with the online version of this article.
References
- 1.Carniel E, Autenrieth I, Cornelis G, Fukushima H, Guinet F, et al. Y. enterocolitica and Y. pseudotuberculosis. In: Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. The Prokaryotes. New York: Dworkin; 2006. pp. 270–398. [Google Scholar]
- 2.EFSA. ECDC The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. Efsa J. 2018;16:5500. doi: 10.2903/j.efsa.2017.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross AJ, Rucker RR, Ewing WH. Description of a bacterium associated with redmouth disease of rainbow trout (Salmo gairdneri) Can J Microbiol. 1966;12:763–770. doi: 10.1139/m66-103. [DOI] [PubMed] [Google Scholar]
- 4.Hurst MRH, Becher SA, Young SD, Nelson TL, Glare TR. Yersinia entomophaga sp. nov., isolated from the New Zealand grass grub Costelytra zealandica . Int J Syst Evol Microbiol. 2011;61:844–849. doi: 10.1099/ijs.0.024406-0. [DOI] [PubMed] [Google Scholar]
- 5.Sprague LD, Neubauer H. Yersinia aleksiciae sp. nov. Int J Syst Evol Microbiol. 2005;55:831–835. doi: 10.1099/ijs.0.63220-0. [DOI] [PubMed] [Google Scholar]
- 6.Merhej V, Adékambi T, Pagnier I, Raoult D, Drancourt M. Yersinia massiliensis sp. nov., isolated from fresh water. Int J Syst Evol Microbiol. 2008;58:779–784. doi: 10.1099/ijs.0.65219-0. [DOI] [PubMed] [Google Scholar]
- 7.Sprague LD, Scholz HC, Amann S, Busse HJ, Neubauer H. Yersinia similis sp. nov. Int J Syst Evol Microbiol. 2008;58:952–958. doi: 10.1099/ijs.0.65417-0. [DOI] [PubMed] [Google Scholar]
- 8.Murros-Kontiainen A, Johansson P, Niskanen T, Fredriksson-Ahomaa M, Korkeala H, et al. Yersinia pekkanenii sp. nov. Int J Syst Evol Microbiol. 2011;61:2363–2367. doi: 10.1099/ijs.0.019984-0. [DOI] [PubMed] [Google Scholar]
- 9.Murros-Kontiainen A, Fredriksson-Ahomaa M, Korkeala H, Johansson P, Rahkila R, et al. Yersinia nurmii sp. nov. Int J Syst Evol Microbiol. 2011;61:2368–2372. doi: 10.1099/ijs.0.024836-0. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen SV, Muthappa DM, Hurley D, Donoghue O, McCabe E, et al. Yersinia hibernica sp. nov., isolated from pig-production environments. Int J Syst Evol Microbiol. 2019;69:2023–2027. doi: 10.1099/ijsem.0.003422. [DOI] [PubMed] [Google Scholar]
- 11.Savin C, Martin L, Bouchier C, Filali S, Chenau J, et al. The Yersinia pseudotuberculosis complex: characterization and delineation of a new species, Yersinia wautersii . Int J Med Microbiol. 2014;304:452–463. doi: 10.1016/j.ijmm.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Kandolo K, Wauters G. Pyrazinamidase activity in Yersinia enterocolitica and related organisms. J Clin Microbiol. 1985;21:980–982. doi: 10.1128/jcm.21.6.980-982.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee WH. Two plating media modified with Tween 80 for isolating Yersinia enterocolitica . Appl Env Microbiol. 1977;33:215–216. doi: 10.1128/aem.33.1.215-216.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wauters G, Aleksić S, Charlier J, Schulze G. Somatic and flagellar antigens of Yersinia enterocolitica and related species. Contrib Microbiol Immunol. 1991;12:239–243. [PubMed] [Google Scholar]
- 15.Wauters G, Kandolo K, Janssens M. Revised biogrouping scheme of Yersinia enterocolitica . Contrib Microbiol Immunol. 1987;9:14–21. [PubMed] [Google Scholar]
- 16.Martin L, Leclercq A, Savin C, Carniel E. Characterization of atypical isolates of Yersinia intermedia and definition of two new biotypes. J Clin Microbiol. 2009;47:2377–2380. doi: 10.1128/JCM.02512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gérôme P, Le Flèche P, Blouin Y, Scholz HC, Thibault FM, et al. Yersinia pseudotuberculosis ST42 (O:1) strain misidentified as Yersinia pestis by mass spectrometry analysis. Genome Announc. 2014;2:e00435-14. doi: 10.1128/genomeA.00435-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ayyadurai S, Flaudrops C, Raoult D, Drancourt M. Rapid identification and typing of Yersinia pestis and other Yersinia species by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. BMC Microbiol. 2010;10:285. doi: 10.1186/1471-2180-10-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harch SAJ, Jennison AV, Bastian I. Yersinia pseudotuberculosis bacteraemia: a diagnostic dilemma in the era of MALDI-TOF mass spectrometry. Pathology. 2019;51:434–436. doi: 10.1016/j.pathol.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Rizzardi K, Wahab T, Jernberg C. Rapid subtyping of Yersinia enterocolitica by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) for diagnostics and surveillance. J Clin Microbiol. 2013;51:4200–4203. doi: 10.1128/JCM.01416-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kotetishvili M, Kreger A, Wauters G, Morris JG, Sulakvelidze A, et al. Multilocus sequence typing for studying genetic relationships among Yersinia species. J Clin Microbiol. 2005;43:2674–2684. doi: 10.1128/JCM.43.6.2674-2684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duan R, Liang J, Shi G, Cui Z, Hai R, et al. Homology analysis of pathogenic Yersinia species Yersinia enterocolitica, Yersinia pseudotuberculosis, and Yersinia pestis based on multilocus sequence typing. J Clin Microbiol. 2014;52:20–29. doi: 10.1128/JCM.02185-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall M, Chattaway MA, Reuter S, Savin C, Strauch E, et al. Use of whole-genus genome sequence data to develop a multilocus sequence typing tool that accurately identifies Yersinia isolates to the species and subspecies levels. J Clin Microbiol. 2015;53:35–42. doi: 10.1128/JCM.02395-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maiden MCJ, Jansen van Rensburg MJ, Bray JE, Earle SG, Ford SA, et al. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol. 2013;11:728–736. doi: 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis. 2014;20:1812–1820. doi: 10.3201/eid2011.140206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moura A, Criscuolo A, Pouseele H, Maury MM, Leclercq A, et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes . Nat Microbiol. 2016;2:16185. doi: 10.1038/nmicrobiol.2016.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jolley KA, Hill DMC, Bratcher HB, Harrison OB, Feavers IM, et al. Resolution of a meningococcal disease outbreak from whole-genome sequence data with rapid web-based analysis methods. J Clin Microbiol. 2012;50:3046–3053. doi: 10.1128/JCM.01312-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cody AJ, Bray JE, Jolley KA, McCarthy ND, Maiden MCJ. Core genome multilocus sequence typing scheme for stable, comparative analyses of Campylobacter jejuni and C. coli human disease isolates. J Clin Microbiol. 2017;55:2086–2097. doi: 10.1128/JCM.00080-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jolley KA, Maiden MCJ. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Guern A-S, Martin L, Savin C, Carniel E. Yersiniosis in France: overview and potential sources of infection. Int J Infect Dis. 2016;46:1–7. doi: 10.1016/j.ijid.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 32.CDC. WHO . Laboratory Manual of Plague Diagnostic Tests. Atlanta, GA; Geneva: Centers for Disease Control and Prevention; World Health Organization; 2000. [Google Scholar]
- 33.Criscuolo A, Brisse S. AlienTrimmer: a tool to quickly and accurately TRIM off multiple short contaminant sequences from high-throughput sequencing reads. Genomics. 2013;102:500–506. doi: 10.1016/j.ygeno.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 34.Crusoe MR, Alameldin HF, Awad S, Boucher E, Caldwell A, et al. The khmer software package: enabling efficient nucleotide sequence analysis. F1000Res. 2015;4:900. doi: 10.12688/f1000research.6924.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Schröder J, Schmidt B. Musket: a multistage k-mer spectrum-based error corrector for Illumina sequence data. Bioinformatics. 2013;29:308–315. doi: 10.1093/bioinformatics/bts690. [DOI] [PubMed] [Google Scholar]
- 36.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haubold B, Klötzl F, Pfaffelhuber P. andi: fast and accurate estimation of evolutionary distances between closely related genomes. Bioinformatics. 2015;31:1169–1175. doi: 10.1093/bioinformatics/btu815. [DOI] [PubMed] [Google Scholar]
- 38.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Konstantinidis KT, Tiedje JM. Towards a genome-based taxonomy for prokaryotes. J Bacteriol. 2005;187:6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2016;32:929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reuter S, Connor TR, Barquist L, Walker D, Feltwell T, et al. Parallel independent evolution of pathogenicity within the genus Yersinia . Proc Natl Acad Sci USA. 2014;111:6768–6773. doi: 10.1073/pnas.1317161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laukkanen-Ninios R, Didelot X, Jolley KA, Morelli G, Sangal V, et al. Population structure of the Yersinia pseudotuberculosis complex according to multilocus sequence typing. Environ Microbiol. 2011;13:3114–3127. doi: 10.1111/j.1462-2920.2011.02588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, et al. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis . Proc Natl Acad Sci USA. 1999;96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bercovier H, Mollaret HH, Alonso JM, Brault J, Fanning GR, et al. Intra- and interspecies relatedness of Yersinia pestis by DNA hybridization and its relationship to Yersinia pseudotuberculosis . Curr Microbiol. 1980;4:225–229. doi: 10.1007/BF02605861. [DOI] [Google Scholar]
- 51.Souza RA, Falcão DP, Falcão JP. Emended description of Yersinia massiliensis . Int J Syst Evol Microbiol. 2011;61:1094–1097. doi: 10.1099/ijs.0.021840-0. [DOI] [PubMed] [Google Scholar]
- 52.Linde HJ, Neubauer H, Meyer H, Aleksic S, Lehn N. Identification of Yersinia species by the Vitek GNI card. J Clin Microbiol. 1999;37:211–214. doi: 10.1128/jcm.37.1.211-214.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morka K, Bystroń J, Bania J, Korzeniowska-Kowal A, Korzekwa K, et al. Identification of Yersinia enterocolitica isolates from humans, pigs and wild boars by MALDI TOF MS. BMC Microbiol. 2018;18:86. doi: 10.1186/s12866-018-1228-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bouchez V, Guglielmini J, Dazas M, Landier A, Toubiana J, et al. Genomic sequencing of Bordetella pertussis for epidemiology and global surveillance of whooping cough. Emerg Infect Dis. 2018;24:988–994. doi: 10.3201/eid2406.171464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin L, Cabanel N, Lesoille C, Ménard T, Carniel E. Investigation of an unusual increase in human yersinioses in Creuse, France. Int J Infect Dis. 2015;34:76–78. doi: 10.1016/j.ijid.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 56.Saraka D, Savin C, Kouassi S, Cissé B, Koffi E, et al. Yersinia enterocolitica, a neglected cause of human enteric infections in Côte d'Ivoire. PLoS Negl Trop Dis. 2017;11:e0005216. doi: 10.1371/journal.pntd.0005216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Virtanen S, Laukkanen-Ninios R, Ortiz Martínez P, Siitonen A, Fredriksson-Ahomaa M, et al. Multiple-locus variable-number tandem-repeat analysis in genotyping Yersinia enterocolitica strains from human and porcine origins. J Clin Microbiol. 2013;51:2154–2159. doi: 10.1128/JCM.00710-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rossen JWA, Friedrich AW, Moran-Gilad J, Genomic ESG, Molecular D. Practical issues in implementing whole-genome-sequencing in routine diagnostic microbiology. Clin Microbiol Infect. 2018;24:355–360. doi: 10.1016/j.cmi.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 59.Bletz S, Mellmann A, Rothgänger J, Harmsen D. Ensuring backwards compatibility: traditional genotyping efforts in the era of whole genome sequencing. Clin Microbiol Infect. 2015;21::347.e1–347.e4. doi: 10.1016/j.cmi.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 60.van Belkum A, Tassios PT, Dijkshoorn L, Haeggman S, Cookson B, et al. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infect. 2007;13 (Suppl. 3):1–46. doi: 10.1111/j.1469-0691.2007.01786.x. [DOI] [PubMed] [Google Scholar]
- 61.Revez J, Espinosa L, Albiger B, Leitmeyer KC, Struelens MJ, et al. Survey on the use of whole-genome sequencing for infectious diseases surveillance: rapid expansion of European national capacities, 2015–2016. Front Public Health. 2017;5:347. doi: 10.3389/fpubh.2017.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.