Abstract
Stenotrophomonas maltophilia is emerging as an important cause of disease in nosocomial and community-acquired settings, including bloodstream, wound and catheter-associated infections. Cystic fibrosis (CF) airways also provide optimal growth conditions for various opportunistic pathogens with high antibiotic tolerance, including S. maltophilia . Currently, there is no rapid, cost-effective and accurate molecular method for detecting this potentially life-threatening pathogen, particularly in polymicrobial specimens, suggesting that its true prevalence is underestimated. Here, we used large-scale comparative genomics to identify a specific genetic target for S. maltophilia , with subsequent development and validation of a real-time PCR assay for its detection. Analysis of 167 Stenotrophomonas spp. genomes identified a conserved 4 kb region in S. maltophilia , which was targeted for Black Hole Quencher assay design. Our assay yielded the positive detection of 89 of 89 (100%) clinical S. maltophilia strains, and no amplification of 23 non- S. maltophilia clinical isolates. S. maltophilia was detected in 10 of 16 CF sputa, demonstrating the assay's utility for direct detection in respiratory specimens. The assay demonstrated good sensitivity, with limits of detection and quantitation on pure culture of ~10 and ~100 genome equivalents, respectively. Our assay provides a highly specific, sensitive and cost-effective method for the accurate identification of S. maltophilia , and will improve the diagnosis and treatment of this under-recognized pathogen by enabling its accurate and rapid detection from polymicrobial clinical and environmental samples.
Keywords: Stenotrophomonas maltophilia, diagnostics, comparative genomics
Data Summary
All 167 genomes used for reconstructing Stenotrophomonas spp. phylogeny are described in (available with the online version of this article), including accession numbers obtained from the National Center for Biotechnology Information Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra).
Impact Statement.
An over-reliance on antibiotics in recent decades has led to the unintended rise of antimicrobial-resistant opportunistic bacterial pathogens that are difficult to treat. Incorrect or under-diagnosis of pathogens, especially naturally antimicrobial-resistant species, has dire consequences for patient health and effective antibiotic stewardship measures. One example is S. maltophilia , a bacterium with naturally high antibiotic tolerance that is increasingly being recognized as a life-threatening pathogen, particularly in nosocomial settings. Current methods for identifying S. maltophilia rely on selective culture or mass spectrometry, neither of which can reliably detect S. maltophilia in polymicrobial or low-abundance specimens. Here, we use a large-scale comparative genomics approach to reconstruct the phylogeny of Stenotrophomonas spp. from 167 publicly available genomes, with subsequent identification of a conserved 4 kb region of S. maltophilia , which was targeted for real-time PCR assay development. We demonstrate that our new assay is sensitive and specific, and provides accurate, cost-effective and rapid detection of S. maltophilia from cultures and polymicrobial specimens. The real-time PCR format provides an accessible way to accurately assess the true prevalence of S. maltophilia , which is essential for guiding antibiotic treatment options and improving patient outcomes. Finally, our study highlights the utility of comparative genomics for robust taxonomic reassignment of speciation errors that are, unfortunately, common in public databases.
Introduction
Stenotrophomonas maltophilia is a Gram-negative, intrinsically multidrug-resistant bacterium that is ubiquitous in aqueous environments, such as soils, plant roots, and water treatment and distribution systems [1]. Whilst conventionally overlooked as a laboratory contaminant, or as a common commensal in hospitalized patients, S. maltophilia is increasingly being recognized as an important nosocomial pathogen in its own right, due to its ability to cause life-threatening disease in immunocompromised individuals [2]. This opportunistic pathogen has been isolated from a variety of hospital settings, including taps, sinks, central venous catheters, ice machines, and water fountains, reinforcing its nosocomial importance [1, 3, 4]. S. maltophilia most commonly infects people with meningitis, cancer, chronic obstructive pulmonary disease or cystic fibrosis (CF), with pneumonia, bacteraemia, and wound and urinary infections being the most frequent clinical manifestations [5, 6]. Risk factors for S. maltophilia infection include prolonged hospitalization, neutropenia, catheterization and previous use of broad-spectrum antibiotics [7]. The recommended antibiotic treatment for S. maltophilia infections is co-trimoxazole; however, resistance towards this antibiotic combination has been documented [2, 8, 9]. Indeed, treatment options are limited for S. maltophilia , with this pathogen also exhibiting resistance towards several antibiotic classes, including fluoroquinolones, macrolides, β-lactams, aminoglycosides, carbapenems, tetracyclines, polymyxins, chloramphenicol and cephalosporins [1, 2]. With a mortality rate approaching 70 %, the importance of timely identification and effective treatment of S. maltophilia infections is paramount [10].
S. maltophilia is a common pathogen in CF airways due to its ability to evade many antipseudomonal antibiotics, with chronic S. maltophilia infection associated with an increased risk of respiratory disease and mortality [11, 12]. CF is an autosomal recessive genetic disorder effecting multiple organs; however, its pathogenesis is most prominent in airways, with ~90 % of CF deaths associated with respiratory failure [13]. The excessive production of mucus in CF airways provides optimal growth conditions for opportunistic pathogens, which drives most CF morbidity and mortality. Molecular methods have confirmed that CF lower airways harbour diverse microbial communities, with Pseudomonas aeruginosa and Burkholderia cepacia complex species of greatest concern due to frequent rapid respiratory decline in people infected with these pathogens [14, 15]. However, other co-infecting opportunistic pathogens, such as Achromobacter spp., S. maltophilia , Staphylococcus aureus , Haemophilus influenzae and certain fungal species (e.g. Aspergillus), are also prominent in CF airways, and are known to contribute to pathogenesis [14]. Indeed, recent studies have shown a mutualistic relationship between S. maltophilia and P. aeruginosa in CF airways, with compounds produced by S. maltophilia under exposure to certain antibiotics, such as imipenem, supporting the survival of otherwise antibiotic-susceptible P. aeruginosa strains [16]. Furthermore, these S. maltophilia compounds can enhance P. aeruginosa stress tolerance, increasing polymyxin tolerance [17]. These studies highlight the importance of S. maltophilia in CF airway pathogenesis, particularly during antibiotic treatment, and emphasize the need for correct species identification in polymicrobial infections.
Although there are a variety of diagnostic methods available for S. maltophilia detection, such as PCR amplicon sequencing, VITEK MS identification or key morphological characteristics on growth media, these methods suffer from issues such as limited access to equipment with a large capital expenditure [e.g. ~US $200 000 (£162 586, £1=$1.23) for VITEK MS instrumentation], high per-assay cost, the need for highly trained personnel, laboriousness, slow turnaround time, the requirement for purified colonies, and misidentification issues [16, 18, 19]. For example, S. maltophilia and P. aeruginosa exhibit colony colour differences when grown on bromothymol blue-containing media, which reflects their different metabolic processes [16]. However, the use of media containing bromothymol blue is not routine and, thus, the retrieval of S. maltophilia from polymicrobial specimens requires clinical expertise in identifying appropriate culture media for differentiation of this bacterium from other pathogens. As a non-exhaustive list, S. maltophilia has been misidentified as several other organisms, including Bordetella bronchiseptica , Alcaligenes faecalis , Burkholderia cepacia and numerous Pseudomonas species, which are common in clinical settings, including CF sputa [1, 20]. Diagnostic inconsistencies in S. maltophilia detection from clinical specimens can lead to inappropriate or even detrimental treatment [16], particularly for those patients requiring urgent care. Therefore, there is a need to accurately identify this emerging pathogen to improve antibiotic-treatment regimens, stewardship and patient outcomes. Here, we report the development and validation of a black hole quencher (BHQ) probe-based real-time PCR assay for the specific detection of S. maltophilia . Our results indicate that our real-time PCR assay is more sensitive than routine culture for detecting S. maltophilia , particularly in polymicrobial respiratory specimens.
Methods
Genomes of Stenotrophomonas spp. and closely related species
All non-redundant Stenotrophomonas spp. genomes available in the National Center for Biotechnology Information (NCBI) that were generated using paired-end Illumina sequencing were downloaded from the Sequence Read Archive database (SRA; https://www.ncbi.nlm.nih.gov/sra) as of December 2018. Additional non-redundant Stenotrophomonas spp. genomes were downloaded from the NCBI GenBank database. In total, 167 publicly available genomes were accessible for this study (Table S1), represented by S. maltophilia (including all ' Stenotrophomonas pavanii '; n=132), Stenotrophomonas acidaminiphila (n=4), Stenotrophomonas bentonitica (n=3), Stenotrophomonas chelatiphaga (n=1), Stenotrophomonas daejeonensis (n=1), Stenotrophomonas ginsengisoli (n=1), Stenotrophomonas humi (n=1), Stenotrophomonas indicatrix (n=6), Stenotrophomonas koreensis (n=1), Stenotrophomonas lactitubi (n=1), Stenotrophomonas nitritireducens (n=2), ' Stenotrophomonas panacihumi ' (n=1), Stenotrophomonas pictorum (n=1), Stenotrophomonas rhizophila (n=2), Stenotrophomonas terrae (n=1) and unassigned Stenotrophomonas spp. (n=9). Three strains listed on the NCBI database as Pseudomonas geniculata (95, AM526 and N1) were included in the phylogenomic analysis to confirm that they were in fact S. maltophilia . Sequence data from assembled genomes were converted to simulated 100 bp paired-end Illumina reads at 85× coverage using art version MountRainier [21]. SRA data were quality-filtered using Trimmomatic v0.33 [22] using previously described filtering parameters [23] prior to analysis.
Bioinformatic analysis to identify a conserved S. maltophilia genetic locus not found in other Stenotrophomonas spp.
Default settings of the haploid comparative genomics pipeline SPANDx v3.2.1 [24] were used to phylogenetically delineate S. maltophilia from non- S. maltophilia species and, subsequently, to identify S. maltophilia -conserved loci. Illumina reads for the publicly available strains (Table S1) were mapped to the closed 4.85 Mbp S. maltophilia K279a genome (GenBank accession no. NC_010943.1) [25]. Phylogenomic reconstruction of the 167 taxa was performed using 31 246 core, biallelic SNPs using the maximum parsimony function of paup* v4.0a.164 [26], rooted with S. daejeonensis JCM 16244 (GenBank accession no. LDJP01000001.1), and with bootstrapping carried out using 1000 replicates. To identify genetic loci present in all S. maltophilia but absent in all other organisms, including other Stenotrophomonas spp., the BEDcov output [27] from SPANDx was examined. Candidate regions were assessed further for specificity and PCR assay design.
To identify the origin of the formate dehydrogenase loci in S. indicatrix RS1 and RS7 strains, alignment of the RS7 (GenBank accession no. RKSR01000015.1) contig encoding this genetic region (~117 kb) was compared with the K279a genome using progressiveMauve v20150226 build 10 [28].
S. maltophilia probe-based real-time PCR assay design
Upon identification of a putative S. maltophilia -specific genetic target, sequence alignments were used to locate conserved regions. Although some variation was allowed within the amplicon, primer- and probe-binding regions required 100 % sequence identity in all S. maltophilia strains to avoid false negatives. Oligo self-dimers and heterodimers were assessed in silico using NetPrimer (http://www.premierbiosoft.com/netprimer/) and Beacon Designer (http://www.premierbiosoft.com/qOligo/Oligo.jsp/), with configurations resulting in ΔG values of <−8.0 (NetPrimer) and <−4.0 (Beacon Designer) excluded. The following sequences and probe label were chosen for the assay: Smalto-For 5′-AAGGACAAGGCGATGACCATC-3′, Smalto-Rev 5′-CCCCACCACGAYTTCATCA-3′ and Smalto-Probe 5′-FAM-CAGAACGACATCTGGTTGGCG-BHQ1-3′, resulting in an amplicon length of 344 bp. NCBI microbial nucleotide Discontiguous MegaBLAST (http://blast.ncbi.nlm.nih.gov/) analysis of this amplicon was used to determine assay specificity for only S. maltophilia . No mismatches in the probe-binding site were tolerated in any S. maltophilia strain, whereas ≥2 mismatches in the probe-binding site were considered sufficient for conferring S. maltophilia specificity.
Microbiological cultures and CF sputum DNA extractions
A total of 89 S . maltophilia isolates were obtained for PCR testing: S. maltophilia control strain LMG 957, 16 sputum-derived isolates cultured from individuals with CF identified either by amplified rRNA gene restriction analysis (n=8 [29]) or in silico multilocus sequence typing (n=8 [30]), and 72 isolates from various clinical presentations that had previously been identified as S. maltophilia by the Pathology Queensland Central Laboratory, Brisbane, Australia, according to the VITEK2 GNI card (98 % of isolates) or VITEK MS (2 % of isolates). Strains were grown on Luria–Bertani (LB) agar for 24 h at 37 °C. DNA was extracted from a small (toothpick head) swatch of the primary culture by heat soaking in 80 µl of a 5 % Chelex solution at 95 °C for 10 min, with the resultant DNA diluted 1 : 10 with molecular grade H2O prior to real-time PCR testing.
Sixteen sputa collected from nine Australian adult CF patients underwent DNA extraction using the Zymo Quick-DNA miniprep plus kit (ZYMO Research) protocol, with the addition of an enzymatic lysis solution (20 mg lysozyme ml−1, 22 U lysostaphin ml−1 and 250 U mutanolysin ml−1) at 37 °C for 1–3 h prior to the addition of Proteinase K. Three patients had sputa collected over three time points (range: 13–46 days), and one patient had two sputa collected 320 days apart. ‘Day 1’ samples represent sputa collected on the day of intravenous antibiotic commencement.
S. maltophilia real-time PCR assay validation and specificity testing
Each PCR consisted of 1× SsoAdvanced universal probes supermix (Bio-Rad), optimized primer and probe concentrations of 0.30 and 0.35 µM, respectively (Macrogen), 1 µl DNA template, and RNase/DNase-free PCR grade water (Thermo Fisher Scientific), to a final per-reaction volume of 5 µl. Optimized thermocycling parameters included an initial hot start activation at 95 °C for 2 min, followed by 45 cycles of 95 °C for 3 s and 60 °C for 10 s, using the CFX96 Touch real-time PCR detection system (Bio-Rad). Results were analysed using CFX Maestro v4.1.2433.1219 software.
The 89 S . maltophilia isolates and 16 CF sputa were tested using the optimized PCR conditions to ensure accurate detection of S. maltophilia DNA in known positive isolates, and in sputum samples from which S. maltophilia was variably detected using selective culture (horse blood agar, McConkey agar, Burkholderia cepacia agar base and Bacitracin agar base). Relative S. maltophilia abundance in sputa was determined by performing a 16S rRNA gene real-time PCR as described elsewhere [31] and calculating the cycles-to-threshold difference (∆C t) between 16S rRNA gene and the S. maltophilia assays. Further specificity testing was performed on 28 non- S. maltophilia DNA: Burkholderia thailandensis (n=1), Burkholderia territorii (n=4), Burkholderia cenocepacia (n=1), Enterobacter aerogenes (n=1), Enterobacter cloacae (n=4), Klebsiella oxytoca (n=1), Klebsiella pneumoniae (n=2), P. aeruginosa (n=11), Staphylococcus aureus (n=1) and Staphylococcus epidermidis (n=2).
Limit of detection (LoD) and limit of quantification (LoQ) were determined for our newly designed S. maltophilia real-time PCR assay using S. maltophilia DNA serial dilutions of 50 ng µl−1 to 0.05 fg µl−1 across eight replicates per dilution. Twenty-four no-template controls were also included. The upper and lower LoD/LoQ limits were determined as described elsewhere [32].
Results
Comparative genomics of Stenotrophomonas spp. identifies a S. maltophilia -specific gene target
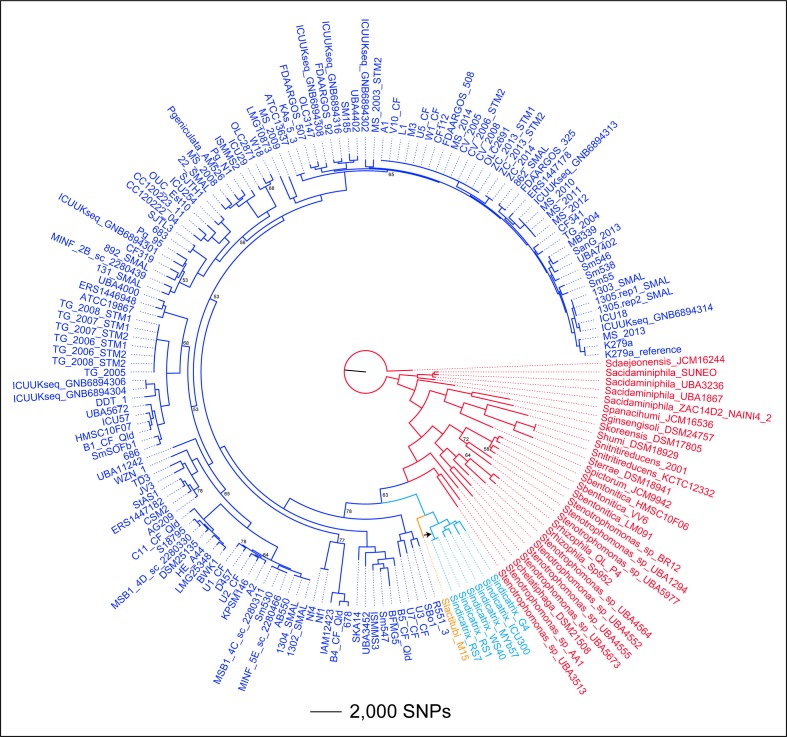
Phylogenomic reconstruction of 167 Stenotrophomonas spp. genomes using 31 306 core-genome, biallelic, orthologous SNPs demonstrated a close relationship between S. maltophilia and two recently described Stenotrophomonas species, S. indicatrix and S. lactitubi [33], and a clear distinction of these taxa from other Stenotrophomonas spp. (Fig. 1). This phylogenomic analysis was used to delineate S. maltophilia from other species, and to modify incorrect species designations for 43 taxa, including reclassification of all 4 ' S. pavanii ', 3 P. geniculata and 17 Stenotrophomonas sp. as S. maltophilia , and 9 S. maltophilia as Stenotrophomonas spp. (Table S1).
Fig. 1.
Phylogenomic analysis of Stenotrophomonas spp. Dark blue, S. maltophilia (target species); red, distantly related Stenotrophomonas spp.; light blue, S. indicatrix ; orange, S. lactitubi . The tree was rooted with S. daejeonensis JCM16244. Consistency index=0.20. Branches with bootstrap values with <80 % support are labelled. The black arrow indicates the phylogenetic position of a probable homologous recombination event from an S. maltophilia strain to a recent common ancestor of the RS1–RS7 clade. This recombination event involved genes encoding formate dehydrogenase.
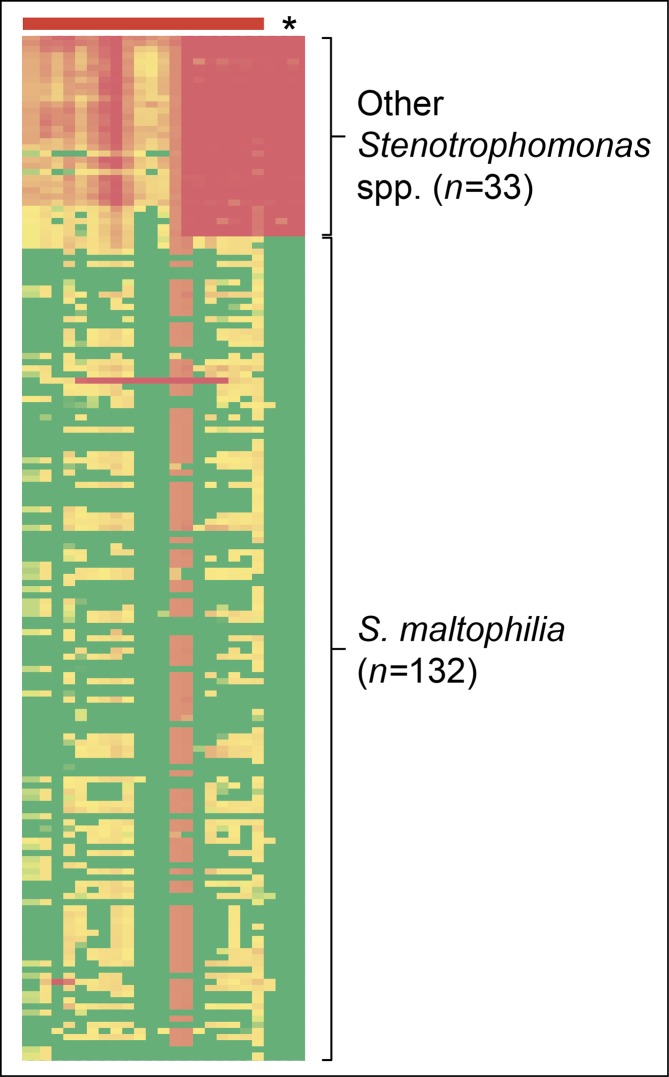
Following identification of the species boundary for S. maltophilia , we next identified loci specific for S. maltophilia . BEDcov outputs identified a conserved 4 kb region in all 132 S . maltophilia genomes that was either absent or highly divergent in the other Stenotrophomonas spp., including S. indicatrix and S. lactitubi (Fig. 2). The genetic coordinates for these loci are 3 935 000–3 939 000 in S. maltophilia K279a, spanning coding regions for formate dehydrogenase (α, β and γ subunits; encoded by fdnG, fdnH and fdnI, respectively). Formate dehydrogenase is also encoded by other pathogens, including P. aeruginosa [34] and Burkholderia spp. [35]. To account for this factor, blast analysis was next performed to identify genetic regions that had high conservation across all S. maltophilia strains but were divergent in all other species, including P. aeruginosa and Burkholderia spp. Using this approach, we targeted our S. maltophilia -specific assay to permit amplification of S. maltophilia , to the exclusion of all other species.
Fig. 2.
Heatmap of the SPANDx BEDcov output from Stenotrophomonas spp. used to identify Stenotrophomonas maltophilia -specific loci. A region of 6 kb in total (including a 4 kb region encoding formate dehydrogenase α, β and γ subunits) was found to be highly conserved in S. maltophilia but absent or highly divergent in other Stenotrophomonas spp., including S. indicatrix and S. lactitubi (asterisk). The 20 kb region immediately downstream of this 4 kb locus (red horizontal bar) is included for comparison. Green, >99 % coverage across the BEDcov window according to BWA-MEM read mapping; red, <20 % coverage; orange through pale green, 20–99 % coverage.
Upon identification of a suitable 344 bp S . maltophilia -specific amplicon (coordinates 3 935 993–3 936 336 in K279a; located within fdnG), a microbial discontiguous MegaBLAST analysis was conducted to confirm specificity (performed 15 March 2019). For all available S. maltophilia genomes, 100 % sequence identity for both primers and probe was attained. Unexpectedly, the genomes of two recently published S. indicatrix strains, which were isolated from soil in Lebanon (RS1 and RS7; GenBank accession numbers NZ_RKSQ00000000 and NZ_RKSR00000000, respectively), provided close blast hits to this amplicon, despite this locus being absent or highly divergent in the four S. indicatrix genomes that were used in the phylogenomic analysis. Fortunately, RS1 and RS7 contained two SNPs in the probe-binding region, including a mismatch at the very 3′ end, which would be expected to inhibit or substantially reduce fluorophore detection in these strains due to poor probe-binding kinetics.
Origin of formate dehydrogenase loci in S. indicatrix RS1 and RS7
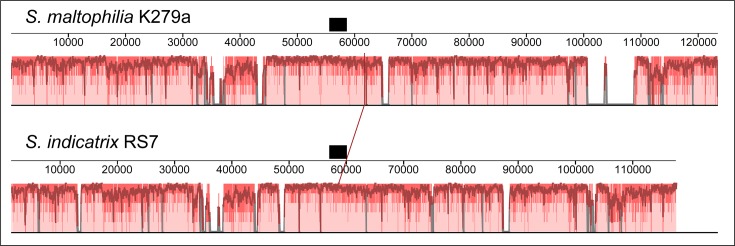
To investigate the origin for the formate dehydrogenase loci in two of the six S. indicatrix strains, we examined the synteny surrounding this set of genes in S. indicatrix RS7 compared with S. maltophilia K279a. This analysis revealed that there was good synteny in both upstream and downstream flanking regions of the formate dehydrogenase loci between S. maltophilia and S. indicatrix (Fig. 3). Comparison of RS7 with S. indicatrix WS40 revealed that the latter strain lacked a ~12.4 kb region encompassing coordinates 53 849–66 256 in the RS7 RKSR01000015.1 contig, which includes the fdnG locus. Given that RS1 and RS7 are genetically highly similar (Fig. 1), and no other S. indicatrix possess the formate dehydrogenase loci, it is likely that a recent common ancestor of RS1 and RS7 acquired these loci from a S. maltophilia strain via homologous recombination, with subsequent genetic divergence occurring between S. indicatrix and S. maltophilia .
Fig. 3.
ProgressiveMauve alignment of the ~118 kb Stenotrophomonas indicatrix RS7 contig, RKSR01000015.1, against Stenotrophomonas maltophilia K279a. The region encoding the formate dehydrogenase α subunit, fdnG, is represented by a black bar. This analysis reveals good synteny between S. maltophilia and S. indicatrix at genetic regions both uptream and downstream of the formate dehydrogenase locus. Other S. indicatrix strains (not shown) lack a ~12.4 kb region compared with RS7 and S. maltophilia that includes this formate dehydrogenase locus.
blast analysis identifies further S. maltophilia misclassification
In addition to RS1 and RS7, blast analysis revealed several non- S. maltophilia matches to this amplicon, including three P. geniculata strains, several misclassified Stenotrophomonas spp. and even misclassifications assigned to distant bacteria e.g. Acinetobacter baumannii (4300STDY7045681; GenBank accession no. UFGS00000000.1), P. aeruginosa (E15_London_28_01_14; GenBank accession no. CVWF00000000.1) and Pseudomonas spp. (UBA10046; GenBank accession DPXK00000000.1). In all cases, these isolates were confirmed to be S. maltophilia according to Microbes blast analysis of the whole genome, thereby representing species assignment errors in the NCBI database, and demonstrating the specificity of our S. maltophilia target.
S. maltophilia real-time PCR assay specificity
To further assess assay specificity, DNA from 89 S . maltophilia isolates collected from acute and chronic infections, including an abdominal wound (n=1), an abdomen aspirate (n=1), blood (n=12), a bronchial washing (n=1), CF sputum (n=26), endotracheal tubes (n=18), a joint swab (n=1), non-CF sputum (n=21), a portacath tip (n=1), an ulcer swab (n=1) and urine (n=6), were tested. The real-time PCR assay accurately detected 100 % of the S. maltophilia isolates but did not amplify across any of the tested non- S. maltophilia species [ Burkholderia thailandensis (n=1), P. aeruginosa (n=11), K. pneumoniae (n=2), K. oxytoca (n=1), E. cloacae (n=4), E. aerogenes (n=1), Staphylococcus epidermidis (n=2) and Staphylococcus aureus (n=1)]. As S. maltophilia is known to be a common bacterium in CF sputa, we next tested the assay across 16 CF sputum samples obtained from nine patients. Of these, 10 sputa demonstrated S. maltophilia presence at various abundances, with 16S rRNA gene to S. maltophilia assay ∆C t values ranging from 2.5 (SCHI0019 day 11; the same as pure S. maltophilia DNA control) to 17.0 (Table 1). In these CF sputum samples, S. maltophilia was frequently co-detected alongside mucoid and non-mucoid P. aeruginosa (Table 1). In two longitudinal samples (SCHI0020 and SCHI0021), S. maltophilia was only detected at very low levels, according to ∆C t values, at the day 1 time point, with the two subsequent sputa being PCR-negative for this organism. In contrast, S. maltophilia persisted in the other two longitudinal samples (SCHI0002 and SCHI0019). There was an 71.4 % congruence between the two methods, with two PCR-positive sputa (SCHI0020 day 1 and SCHI0021 day 1) being negative by culture (Table 1). However, these two specimens had the lowest S. maltophilia load (∆C t values of 13.8 and 17.0), indicating higher sensitivity with the real-time PCR.
Table 1.
Real-time PCR quantification of Stenotrophomonas maltophilia on 16 sputa obtained from CF airways, with concurrent culture diagnosis
|
Sample |
ΔC t* |
Culture result |
Other known culture growth |
|---|---|---|---|
|
S. maltophilia isolate control |
2.5 |
Positive |
Pure isolate |
|
SCHI0002 day 1 |
7.9 |
Positive |
Mucoid and non-mucoid P. aeruginosa |
|
SCHI0002 day 320 |
6.0 |
Positive |
Mucoid P. aeruginosa |
|
SCHI0008 |
na |
nd |
nd |
|
SCHI0010 |
na |
Negative |
Mucoid and non-mucoid P. aeruginosa |
|
SCHI0011 |
12.4 |
nd |
nd |
|
SCHI0013 |
9.7 |
nd |
nd |
|
SCHI0014 |
8.0 |
Positive |
No other bacterial growth |
|
SCHI0019 day 1 |
6.7 |
Positive |
|
|
SCHI0019 day 11 |
2.5 |
nd |
nd |
|
SCHI0019 day 46 |
6.5 |
nd |
nd |
|
SCHI0020 day 1 |
17.0 |
Negative |
Mucoid P. aeruginosa |
|
SCHI0020 day 6 |
na |
nd |
nd |
|
SCHI0020 day 31 |
na |
nd |
nd |
|
SCHI0021 day 1 |
13.8 |
Negative |
MRSA, mucoid and non-mucoid P. aeruginosa |
|
SCHI0021 day 5 |
na |
nd |
nd |
|
SCHI0021 day 13 |
na |
nd |
nd |
MRSA, Methicillin-resistant Staphylococcus aureus; na, no amplification; nd, not determined.
*Cycles-to-threshold difference.
S. maltophilia real-time PCR assay sensitivity
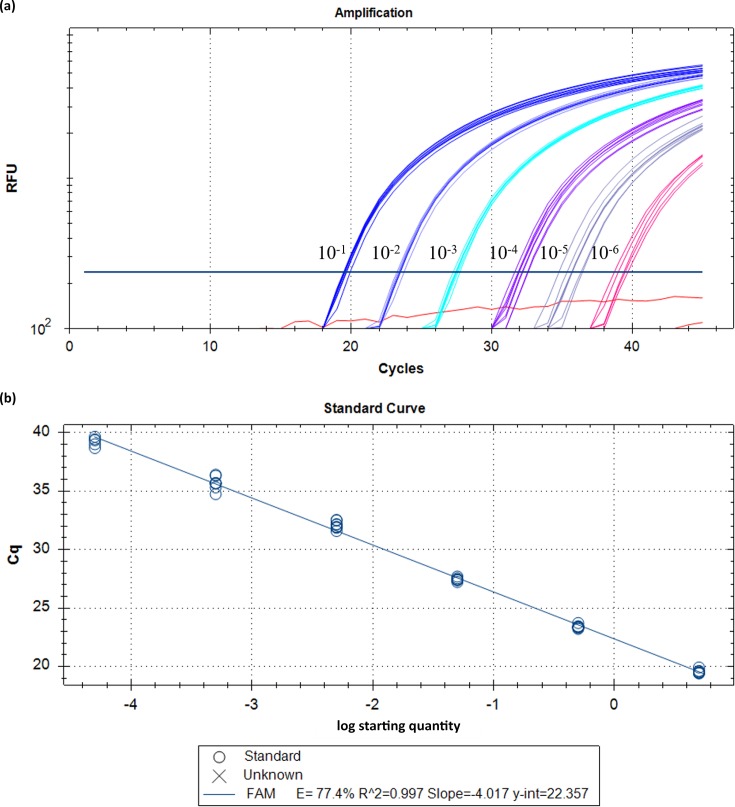
To assess assay sensitivity, LoD and LoQ values were determined (Fig. 4). Using a 10-fold DNA dilution series (50 ng µl−1 to 0.05 fg µl−1), the LoD for this assay was ~5 fg µl−1 or ~9 genome equivalents (GEs), and the LoQ was 0.5 pg µl−1 or ~94 GEs.
Fig. 4.
LoD and LoQ values for the S. maltophilia-specific real-time PCR assay. To determine the LoD and LoQ, serial dilutions of a S. maltophilia-positive control DNA sample were performed across eight replicates, ranging from 50 ng µl–1 to 0.05 fg µl−1 (a). To assess the correlation coefficient, a standard curve was also included for these dilutions, resulting in an R 2 of 0.997 (b). LoD and LoQ were identified as ~5 fg µl−1 and ~0.5 pg µl−1, respectively. All 24 negative controls (red) were negative. RFU, relative fluorescence units; Cq, quantification cycle (also known as C t [cycles-to-threshold]).
Discussion
S. maltophilia is emerging as an important multidrug-resistant nosocomial pathogen, being amongst the top three most common non-fermentative Gram-negative bacilli identified in hospitalized patients [5, 36]. Despite S. maltophilia being well-adapted to many environments, most infections occur in immunocompromised individuals in the nosocomial setting [10], although community-acquired infections are also on the rise [1]. Here, we describe what is to the best of our knowledge the first real-time PCR assay to detect S. maltophilia with 100 % accuracy in purified colonies, and demonstrate that this assay is superior to microbiological culture for detecting this multidrug-resistant bacterium in polymicrobial respiratory specimens collected from CF patients.
There is currently a lack of a rapid, cost-effective, accessible and accurate diagnostic method for S. maltophilia detection, particularly from polymicrobial clinical specimens such as CF sputa. As S. maltophilia is thought to be the only Stenotrophomonas species to cause human disease, mass spectrometry (MS)-based systems such as VITEK 2 and VITEK MS are a common diagnostic method in large, centralized pathology laboratories. However, the accuracy of species determination using MS is heavily dependent on the quality of the associated databases, and it is currently unknown whether other Stenotrophomonas spp. can be accurately differentiated from S. maltophilia on these systems. In addition, access to this instrument is limited to well-resourced laboratories owing to a large barrier-to-entry cost [~US $200 000 (£162 586)] [37–39]. Furthermore, as CF sputa are polymicrobial, overgrowth of other bacteria on selective culture plates is common. This is particularly problematic for VITEK diagnosis when S. maltophilia is present in low abundance or the patient is co-infected with mucoid P. aeruginosa . From a genotyping standpoint, 16S rRNA gene PCR has been used to identify S. maltophilia in blood samples for patients undergoing chemotherapy for leukemia [40], and a multiplex PCR targeting P. aeruginosa , S. maltophilia and Burkholderia cepacia successfully identified S. maltophilia in 85 % of cases [41]. However, these assays have either not been optimized to avoid non-specific amplification in other Stenotrophomonas spp. and members of the closely related genus Xanthomonas , or they require downstream processing (e.g. gel electrophoresis, Sanger sequencing) to confirm results, which is laborious, time-consuming and raises potential laboratory contamination issues.
Therefore, the purpose of this study was to use large-scale comparative genomics to identify a S. maltophilia-specific genetic target, and to subsequently design a highly specific and accurate real-time PCR-based assay for identifying S. maltophilia . Using this approach, we identified a genetic region with high specificity for S. maltophilia , which was subsequently targeted for assay development. We found that our newly developed assay correctly identified 89 S . maltophilia isolates with 100 % accuracy. The accuracy and specificity of this assay is both highly sensitive and selective for S. maltophilia , with an LoQ and LoD of ~94 and ~9 GEs, respectively. We chose the BHQ probe real-time PCR format due to its relatively inexpensive up-front cost [~US $25 000 (£30 750) for real-time instrumentation], low per-reaction cost [~US $0.80 (£0.98) per sample when performed in duplicate], high-throughput capacity, closed-tube format (which eliminates post-PCR contamination concerns), simple set-up and rapid turnaround-time (~1 h). This format also enables robust identification of target species in polymicrobial specimens. Although not examined in this study, the multi-fluorophore capacity of many real-time PCR instruments also enables multiplexing of probe-based assays for the simultaneous identification of multiple organisms in a single specimen, leading to further cost reductions.
Our in silico and laboratory results indicate that all non- S. maltophilia micro-organisms failed to amplify, with the possible exception of two S. indicatrix strains, RS1 and RS7, the genomes of which only became available subsequent to assay design. S. indicatrix is a newly identified Stenotrophomonas species [33] that has so far been isolated from dirty dishes in Germany (strain WS40 [33]), sewage in China (strain G4; unpublished), a rotting apple in Germany (Myb57; unpublished), soil in Lebanon (strains RS1 and RS7; unpublished) and a human respiratory infection in Germany (ICU300 [42]). Comparative analysis of RS7 and S. maltophilia indicated recent homologous recombination of the formate dehydrogenase locus targeted by our study; however, blastn analysis showed that there were two SNPs in the probe-binding region, including a SNP at the 5′ ultimate base of the BHQ probe, which would likely result in poor or no amplification. Taken together, we show that our assay is highly specific for S. maltophilia , particularly in clinical samples, but it also has applicability for testing environmental samples, such as hospital water supplies.
Although the quality of life and life expectancy for people with CF has markedly increased in recent decades due to improvements in antibiotic treatments and clinical management, persistent polymicrobial infections in CF airways remain the primary cause of morbidity and mortality [43]. A recent longitudinal study of a single CF patient’s airways using a cutting-edge metatranscriptomic approach, which measures only the ‘active’ microbial population through mRNA characterization, revealed that S. maltophilia was the second most prevalent bacterium behind P. aeruginosa in the 6 months prior to death [43]. Our results also revealed a high prevalence of S. maltophilia in adult CF sputa, with 10/16 samples positive for this bacterium according to our assay. As some of these sputa had concurrent culture results, we demonstrated an improvement in detection with 71.4 % congruence to real-time PCR results, with two culture-negative samples returning as positive by real-time PCR (SCHI0020 day 1 and SCHI0021 day 1). While our sample size is low, this finding demonstrates that our assay may have a higher sensitivity for detecting S. maltophilia in CF clinical specimens than culture methods, although it cannot be ruled out that S. maltophilia PCR positivity may be due to the presence of DNA in the CF sputa from a recently eradicated S. maltophilia infection. Of four longitudinally collected sputa, one patient (SCHI0019) had S. maltophilia at all time points (days 1, 11 and 46; Table 1) despite intravenous meropenem and tobramycin antibiotic therapy administered on day 2 of admission, and another patient, SCHI0002, was positive for S. maltophilia in samples that were collected nearly 12 months apart (days 1 and 320), indicating either long-term airway persistence or reinfection with this organism. In one sputum sample from SCHI0019 (day 11), the ∆C t value between the 16S rRNA gene and S. maltophilia PCRs was identical to that of pure S. maltophilia culture (Table 1), indicating that S. maltophilia had become the dominant, and potentially sole, bacterial species in this specimen. Although outside the scope of this study, this finding demonstrates the potential for S. maltophilia to persist and dominate in CF airways following antibiotic-driven microbiome perturbations, which may have implications for rapid re-infection with more formidable pathogens such as P. aeruginosa . A further two patients, SCHI0020 and SCHI0021, had S. maltophilia at day 1, but subsequent samples (up to day 31 and 13, respectively) were PCR-negative following intravenous ceftazidime and tobramycin antibiotic treatment phase administered on day 2 of admission, indicating successful eradication of S. maltophilia in these cases. Future work will entail testing our newly developed assay across larger CF sputum panels, including longitudinal samples, to further examine the potential mutualistic relationships between S. maltophilia and other pathogens such as P. aeruginosa , and to assess assay performance directly on clinical specimens to further reduce sample processing timeframes.
In conclusion, the ability to accurately, rapidly and cheaply detect S. maltophilia is critical for understanding the prevalence of this underappreciated opportunistic pathogen and for reducing its burden of disease. The implementation of this assay in the clinical setting will enable researchers, clinicians and pathologists to more accurately identify this multidrug-resistant bacterium, particularly in isolates that have been ruled out as other multidrug-resistant Gram-negative pathogens, such as P. aeruginosa or Burkholderia spp. Finally, the correct and rapid identification of S. maltophilia will improve antibiotic stewardship measures by enabling more targeted eradication of this pathogen, and in polymicrobial infections such as those commonly found in CF airways, S. maltophilia eradication may reduce the prevalence and persistence of more serious pathogens such as P. aeruginosa , leading to improved quality of life and lifespans for people with CF.
Data bibliography
1. Hagstrom A et al. BioProject PRJNA19369 (2008).
2. Lira F et al. BioProject PRJEA89665 (2012).
3. Lucas S et al. BioProject PRJNA17107 (2013).
4. DOE Joint Genome Institute. BioProject PRJNA185300 (2013).
5. Lucas S et al. BioProject PRJNA53943 (2013).
6. Roach D.J. et al. BioProject PRJNA267549 (2014).
7. Kanamori H et al. BioProject RJDB3841 (2015).
8. Sanger BioProject PRJEB6891 (2015).
9. Tang H et al. BioProject PRJNA156719 (2015).
10. Pak TR et al. BioProject PRJNA277366 (2015).
11. Saffarian A et al. BioProject PRJNA278502 (2015).
12. Patil PP et al. BioProject PRJNA284363, PRJNA284364, PRJNA284366(2015).
13. Ormerod K. BioProject PRJNA285410 (2015).
14. Vinuesa P et al. BioProject PRJNA296415 (2015).
15. Crossman LC et al. BioProject PRJNA30351 (2015).
16. Liu W. BioProject PRJEB15263 (2016).
17. Varghese N. BioProjectPRJEB16042, PRJEB17324 (2016).
18. Mitreva M et al. BioProject PRJNA269850, PRJNA269851, PRJNA272632 (2016).
19. Patil PP et al. BioProject PRJNA284369, PRJNA284375, PRJNA284376, PRJNA284378, PRJNA299446 (2016).
20. Pan X et al. BioProject PRJNA286061 (2016).
21. Park H. BioProject PRJNA310387 (2016).
22. Nazaret S, Bodilis J. BioProject PRJNA323844 (2016).
23. Wei Y et al. BioProject PRJNA326321 (2016).
24. Blow F, Darby AC. BioProject PRJNA326914 (2016).
25. Oh D-K, Kim KR. BioProject PRJNA330867 (2016).
26. Kozyreva V. BioProject PRJNA341407 (2016).
27. Richard D. BioProject RJNA344031 (2016).
28. Hattie C. BioProject PRJNA344912 (2016).
29. Niu B et al. BioProject PRJNA357031 (2016).
30. Varghese N. BioProject PRJEB21383, PRJEB22431 (2017).
31. Sanger BioProject PRJEB8666 (2017).
32. Patil PP et al. BioProject PRJNA299448 (2017).
33. Dong H et al. BioProject PRJNA321363 (2017).
34. Ge S. BioProject PRJNA347873 (2017).
35. Parks DH et al. BioProject PRJNA348753 (2017).
36. Esposito A et al. BioProject PRJNA350620 (2017).
37. Mohapatra B, Sar P. BioProject PRJNA352524 (2017).
38. Aslam F, Yasmin A. BioProject PRJNA358642 (2017).
39. Zhineng W. BioProject PRJNA388045 (2017).
40. Fouts D et al. BioProject PRJNA390523 (2017).
41. Weber M et al. BioProject PRJNA414363 (2017).
42. Castro-Jaimes S et al. BioProject PRJNA421960 (2017).
43. Sanger BioProject PRJEB20809 (2018).
44. Sichtig H et al. BioProject PRJEB20827, PRJNA231221 (2018).
45. Riera N et al. BioProject PRJNA369256 (2018).
46. Ho BC. BioProject PRJNA374779 (2018).
47. Glady-Croue J et al. BioProject PRJNA381518 (2018).
48. Yang W. BioProject PRJNA400855 (2018).
49. Garrido-Sanz D et al. BioProject PRJNA412168 (2018).
50. Parks DH et al. BioProject PRJNA417962 (2018).
51. Medina-Cordoba LK et al. BioProject PRJNA418312 (2018).
52. Chen M-X. BioProject PRJNA427609 (2018).
53. Kenzaka T, Tani K. BioProject PRJNA430144 (2018).
54. Xiong W. BioProject PRJNA437214 (2018).
55. Pelletier D et al. BioProject PRJNA440801 (2018).
56. Xu J. BioProject PRJNA445756 (2018).
57. Venturi V et al. BioProject PRJNA463055 (2018).
58. Xiong W. BioProject PRJNA474584 (2018).
59. Cao G. BioProject PRJNA486733 (2018).
60. Cooper A. BioProject PRJNA489399 (2018).
61. Li R. BioProject PRJNA505368 (2018).
62. Fouts DE et al. BioProject PRJNA508495 (2018).
63. Tokajian ST, Khnayzer RS. BioProject PRJNA496409, PRJNA496522 (2019).
Supplementary Data
Funding information
This study was funded by the University of the Sunshine Coast and Advance Queensland [awards AQRF13016-17RD2 (D. S. S.) and AQIRF0362018 (E. P. P.)]. T. J. K. is the recipient of a National Health and Medical Research Council Early Career Fellowship (1088448).
Acknowledgements
We wish to thank Kay Ramsay (formerly of the QIMR Berghofer Medical Research Institute; currently at the University of Otago) for assistance with Stenotrophomonas maltophilia identification from CF sputum.
Author contributions
Conceptualization, P.N.A.H., S.C.B., D.S.S. and E.P.P.; methodology and experimental design, T.A.F., M.G.B., D.S.S. and E.P.P.; validation, T.A.F., M.G.B. and E.P.P.; sample collection, S.C.B., H.B., T-K.N., T.J.K. and G.R.N.; writing – original draft preparation, T.A.F. and E.P.P.; manuscript review and editing, M.G.B., P.N.A.H., S.C.B., H.B., T-K.N., T.J.K., G.R.N., D.S.S. and E.P.P.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
This study was approved by the Prince Charles Hospital Human Research Ethics Committee (HREC/13/QPCH/127) and consent was obtained from all individuals.
Footnotes
Abbreviations: BHQ, black hole quencher; CF, cystic fibrosis; GE, genome equivalent; LoD, limit of detection; LoQ, limit of quantification; NCBI, National Center for Biotechnology Information.
All supporting data, code and protocols have been provided within the article or through supplementary data files. One supplementary table is available with the online version of this article.
References
- 1.Brooke JS. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev. 2012;25:2–41. doi: 10.1128/CMR.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adegoke AA, Stenström TA, Okoh AI. Stenotrophomonas maltophilia as an emerging ubiquitous pathogen: looking beyond contemporary antibiotic therapy. Front Microbiol. 2017;8:2276. doi: 10.3389/fmicb.2017.02276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trifonova A, Strateva T. Stenotrophomonas maltophilia – a low-grade pathogen with numerous virulence factors. Infect Dis. 2019;51:168–178. doi: 10.1080/23744235.2018.1531145. [DOI] [PubMed] [Google Scholar]
- 4.Waters V, Yau Y, Prasad S, Lu A, Atenafu E, et al. Stenotrophomonas maltophilia in cystic fibrosis: serologic response and effect on lung disease. Am J Respir Crit Care Med. 2011;183:635–640. doi: 10.1164/rccm.201009-1392OC. [DOI] [PubMed] [Google Scholar]
- 5.Looney WJ, Narita M, Mühlemann K. Stenotrophomonas maltophilia: an emerging opportunist human pathogen. Lancet Infect Dis. 2009;9:312–323. doi: 10.1016/S1473-3099(09)70083-0. [DOI] [PubMed] [Google Scholar]
- 6.Jeon YD, Jeong WY, Kim MH, Jung IY, Ahn MY, et al. Risk factors for mortality in patients with Stenotrophomonas maltophilia bacteremia. Medicine. 2016;95:e4375. doi: 10.1097/MD.0000000000004375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Senol E, DesJardin J, Stark PC, Barefoot L, Snydman DR. Attributable mortality of Stenotrophomonas maltophilia bacteremia. Clin Infect Dis. 2002;34:1653–1656. doi: 10.1086/340707. [DOI] [PubMed] [Google Scholar]
- 8.Gales AC, Jones RN, Forward KR, Sader JJ, et al. Emerging importance of multidrug-resistant Acinetobacter species and Stenotrophomonas maltophilia as pathogens in seriously ill patients: geographic patterns, epidemiological features, and trends in the SENTRY antimicrobial surveillance program (1997–1999) Clin Infect Dis. 2001;32:S104–S113. doi: 10.1086/320183. [DOI] [PubMed] [Google Scholar]
- 9.Al-Jasser AM. Stenotrophomonas maltophilia resistant to trimethoprim-sulfamethoxazole: an increasing problem. Ann Clin Microbiol Antimicrob. 2006;5:23. doi: 10.1186/1476-0711-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falagas ME, Kastoris AC, Vouloumanou EK, Dimopoulos G. Community-acquired Stenotrophomonas maltophilia infections: a systematic review. Eur J Clin Microbiol Infect Dis. 2009;28:719–730. doi: 10.1007/s10096-009-0709-5. [DOI] [PubMed] [Google Scholar]
- 11.Parkins MD, Floto RA. Emerging bacterial pathogens and changing concepts of bacterial pathogenesis in cystic fibrosis. J Cyst Fibros. 2015;14:293–304. doi: 10.1016/j.jcf.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Waters V, Atenafu EG, Lu A, Yau Y, Tullis E, et al. Chronic Stenotrophomonas maltophilia infection and mortality or lung transplantation in cystic fibrosis patients. J Cyst Fibros. 2013;12:482–486. doi: 10.1016/j.jcf.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Klimova B, Kuca K, Novotny M, Maresova P. Cystic fibrosis revisited - a review study. Med Chem. 2017;13:102–109. doi: 10.2174/1573406412666160608113235. [DOI] [PubMed] [Google Scholar]
- 14.Sherrard LJ, Tunney MM, Elborn JS. Antimicrobial resistance in the respiratory microbiota of people with cystic fibrosis. The Lancet. 2014;384:703–713. doi: 10.1016/S0140-6736(14)61137-5. [DOI] [PubMed] [Google Scholar]
- 15.Muhlebach MS, Hatch JE, Einarsson GG, McGrath SJ, Gilipin DF, et al. Anaerobic bacteria cultured from cystic fibrosis airways correlate to milder disease: a multisite study. Eur Respir J. 2018;52:1800242. doi: 10.1183/13993003.00242-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kataoka D, Fujiwara H, Kawakami T, Tanaka Y, Tanimoto A, et al. The indirect pathogenicity of Stenotrophomonas maltophilia . Int J Antimicrob Agents. 2003;22:601–606. doi: 10.1016/S0924-8579(03)00244-9. [DOI] [PubMed] [Google Scholar]
- 17.Ryan RP, Fouhy Y, Garcia BF, Watt SA, Niehaus K, et al. Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa . Mol Microbiol. 2008;68:75–86. doi: 10.1111/j.1365-2958.2008.06132.x. [DOI] [PubMed] [Google Scholar]
- 18.Gallo SW, Ramos PL, Ferreira CAS, Dias de Oliveira S. A specific polymerase chain reaction method to identify Stenotrophomonas maltophilia . Mem Inst Oswaldo Cruz. 2013;108:390–391. doi: 10.1590/S0074-02762013000300020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinot C, Deredjian A, Nazaret S, Brothier E, Cournoyer B, et al. Identification of Stenotrophomonas maltophilia strains isolated from environmental and clinical samples: a rapid and efficient procedure. J Appl Microbiol. 2011;111:1185–1193. doi: 10.1111/j.1365-2672.2011.05120.x. [DOI] [PubMed] [Google Scholar]
- 20.Denton M, Kerr KG. Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia . Clin Microbiol Rev. 1998;11:57–80. doi: 10.1128/CMR.11.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang W, Li L, Myers JR, Marth GT. ART: a next-generation sequencing read simulator. Bioinformatics. 2012;28:593–594. doi: 10.1093/bioinformatics/btr708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price EP, Viberg LT, Kidd TJ, Bell SC, Currie BJ, et al. Transcriptomic analysis of longitudinal Burkholderia pseudomallei infecting the cystic fibrosis lung. Microb Genom. 2018;4:mgen.0.000194. doi: 10.1099/mgen.0.000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarovich DS, Price EP. SPANDx: a genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets. BMC Res Notes. 2014;7:618. doi: 10.1186/1756-0500-7-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crossman LC, Gould VC, Dow JM, Vernikos GS, Okazaki A, et al. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 2008;9:R74. doi: 10.1186/gb-2008-9-4-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swofford DL, Sullivan J. Sunderland, Massachusetts: Sinauer Associates; 2003. [Google Scholar]
- 27.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kidd TJ, Ramsay KA, Hu H, Bye PTP, Elkins MR, et al. Low rates of Pseudomonas aeruginosa misidentification in isolates from cystic fibrosis patients. J Clin Microbiol. 2009;47:1503–1509. doi: 10.1128/JCM.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wood ME, Stockwell RE, Johnson GR, Ramsay KA, Sherrard LJ, et al. Cystic fibrosis pathogens survive for extended periods within cough-generated droplet nuclei. Thorax. 2019;74:87–90. doi: 10.1136/thoraxjnl-2018-211567. [DOI] [PubMed] [Google Scholar]
- 31.Yang S, Lin S, Kelen GD, Quinn TC, Dick JD, et al. Quantitative multiprobe PCR assay for simultaneous detection and identification to species level of bacterial pathogens. J Clin Microbiol. 2002;40:3449–3454. doi: 10.1128/JCM.40.9.3449-3454.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price EP, Dale JL, Cook JM, Sarovich DS, Seymour ML, et al. Development and validation of Burkholderia pseudomallei-specific real-time PCR assays for clinical, environmental or forensic detection applications. PLoS One. 2012;7:e37723. doi: 10.1371/journal.pone.0037723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weber M, Schünemann W, Fuß J, Kämpfer P, Lipski A. Stenotrophomonas lactitubi sp. nov. and Stenotrophomonas indicatrix sp. nov., isolated from surfaces with food contact. Int J Syst Evol Microbiol. 2018;68:1830. doi: 10.1099/ijsem.0.002732. [DOI] [PubMed] [Google Scholar]
- 34.Panmanee W, Su S, Schurr MJ, Lau GW, Zhu X, et al. The anti-sigma factor mucA of Pseudomonas aeruginosa: dramatic differences of a mucA22 vs. a ΔmucA mutant in anaerobic acidified nitrite sensitivity of planktonic and biofilm bacteria in vitro and during chronic murine lung infection. PLoS One. 2019;14:e0216401. doi: 10.1371/journal.pone.0216401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hatrongjit R, Packdibamrung K. A novel NADP+-dependent formate dehydrogenase from Burkholderia stabilis 15516: screening, purification and characterization. Enzyme Microb Technol. 2010;46:557–561. doi: 10.1016/j.enzmictec.2010.03.002. [DOI] [Google Scholar]
- 36.Zhao J, Xing Y, Liu W, Ni W, Wei C, et al. Surveillance of dihydropteroate synthase genes in Stenotrophomonas maltophilia by LAMP: implications for infection control and initial therapy. Front Microbiol. 2016;7:1723. doi: 10.3389/fmicb.2016.01723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.AbdulWahab A, Taj-Aldeen SJ, Ibrahim EB, Talaq E, Abu-Madi M, et al. Discrepancy in MALDI-TOF MS identification of uncommon gram-negative bacteria from lower respiratory secretions in patients with cystic fibrosis. Infect Drug Resist. 2015;8:83–88. doi: 10.2147/IDR.S80341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gautam V, Sharma M, Singhal L, Kumar S, Kaur P, et al. MALDI-TOF mass spectrometry: an emerging tool for unequivocal identification of non-fermenting gram-negative bacilli. Indian J Med Res. 2017;145:665–672. doi: 10.4103/ijmr.IJMR_1105_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nashid N, Yau Y. Evaluation of the matrix-assisted laser desorption ionization time-of-flight mass spectrometry for the identification of cystic fibrosis pathogens. Open Forum Infect Dis. 2016;3 (Suppl. 1):177. doi: 10.1093/ofid/ofw172.44. [DOI] [Google Scholar]
- 40.Nakamura A, Sugimoto Y, Ohishi K, Sugawara Y, Fujieda A, et al. Diagnostic value of PCR analysis of bacteria and fungi from blood in empiric-therapy-resistant febrile neutropenia. J Clin Microbiol. 2010;48:2030–2036. doi: 10.1128/JCM.01700-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.da Silva Filho LVF, Tateno AF, Velloso LdeF, Levi JE, Fernandes S, et al. Identification of Pseudomonas aeruginosa, Burkholderia cepacia complex, and Stenotrophomonas maltophilia in respiratory samples from cystic fibrosis patients using multiplex PCR. Pediatr Pulmonol. 2004;37:537–547. doi: 10.1002/ppul.20016. [DOI] [PubMed] [Google Scholar]
- 42.Steinmann J, Mamat U, Abda EM, Kirchhoff L, Streit WR, et al. Analysis of phylogenetic variation of Stenotrophomonas maltophilia reveals human-specific branches. Front Microbiol. 2018;9:806. doi: 10.3389/fmicb.2018.00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cobián Güemes AG, Lim YW, Quinn RA, Conrad DJ, Benler S, et al. Cystic fibrosis rapid response: translating multi-omics data into clinically relevant information. mBio. 2019;10:e00431-19. doi: 10.1128/mBio.00431-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.