Abstract
Non-small cell lung cancer (NSCLC) is the leading cause of lung cancer-associated mortality. Recent studies revealed that long non-coding (lnc)RNAs have crucial roles in human cancers. The present study was the first, to the best of our knowledge, to indicate that the lncRNA transducer of ERBB2, 1-antisense 1 (TOB1-AS1) acts as a tumor suppressor in NSCLC. Knockdown of TOB1-AS1 significantly induced NSCLC cell migration, invasion and proliferation. It was also demonstrated that the higher expression of TOB1-AS1 in NSCLC samples was associated with longer overall survival time. Furthermore, a TOB1-AS1-mediated competing endogenous RNA network in NSCLC was constructed, including Homo sapiens (hsa)-microRNA (miR)-27a-3p, hsa-miR-23a-3p, hsa-miR-23b-3p, hsa-miR-27b-3p, hsa-miR-23c, dynein cytoplasmic 2 light intermediate chain 1, E4F transcription factor 1, TSPY-like 4, component of oligomeric Golgi complex 7, inositol hexakisphosphate kinase 2 and deltex E3 ubiquitin ligase 3. Of note, dysregulation of targets of TOB1-AS1 was associated with the prognosis of NSCLC patients. The present study suggested that TOB1-AS1 may serve as a novel biomarker for NSCLC.
Keywords: non-small cell lung cancer, long non-coding RNA, transducer of ERBB2 1 antisense 1, metastasis, competing endogenous RNA network
Introduction
In the past decade, one of the most noteworthy discoveries in the field of biology is that non-protein-coding RNAs (ncRNAs) have crucial roles in human diseases (1). MicroRNAs (miRNAs or miRs) (2), long non-coding RNAs (lncRNAs) (3), transfer RNA-derived small RNA (4) and circular RNAs (5) have been indicated to be up- or downregulated in multiple types of cancer and to be associated with the prognosis of affected patients. Among these ncRNAs, lncRNAs have attracted an increased amount of attention. lncRNAs have important roles in regulating cancer cell apoptosis (6), metastasis (7), autophagy (8), angiogenesis (9) and epithelial-to-mesenchymal transition (EMT) (10). lncRNAs are able to interact with proteins. For instance, lncRNA EPIC1 binds to MYC to promote the expression of cell cycle regulators (11). lncRNAs have been demonstrated to function as miRNA sponges and to be involved in post-transcriptional regulation of gene expression. For instance, lncRNA activated by transforming growth factor-β competitively sponges miR-200 to activate the expression of zinc finger E-box binding homeobox 1 (ZEB1) and ZEB2 during EMT (12). lncRNAs have also been indicated to interact with chromatin. For instance, nuclear enriched abundant transcript 1 (NEAT1) is a regulator of nuclear paraspeckles, which has been reported to bind to the promoters of a series of targets in prostate cancer (13). However, the functions of the majority of lncRNAs in cancer have remained elusive.
Non-small cell lung cancer (NSCLC) constitutes ~80% of all lung cancer cases and is the most common type of human lung cancer. There is evidence to indicate that ncRNAs have key roles in the tumorigenesis and progression of lung cancer. lncRNAs may function as either oncogenes or tumor suppressors in lung cancer. For instance, lncRNA MIR4435-2 host gene (HG) has been reported to promote lung cancer progression by activating β-catenin signaling (14). lncRNA myocardial infarction associated transcript has been indicated to promote NSCLC proliferation and metastasis by inducing matrix metalloproteinase 9 expression (15). MIR22HG has a tumor-suppressive role in lung cancer by regulating Y-box binding protein 1, MET and p21 (16). However, lncRNA-low expression in tumor (LET) inhibits NSCLC cell migration and invasion (17). Growth arrest specific 5-antisense 1 (AS1) has also been indicated to suppress NSCLC metastasis by reducing ZEB1, N-cadherin, vimentin and/or Snail1 (18). Furthermore, certain lncRNAs have been reported to be potential biomarkers for NSCLC. For instance, LET is downregulated in NSCLC and low expression of LET is associated with a more favorable prognosis (19). The exploration of the functions of lncRNAs in NSCLC may provide potential diagnostic and prognostic markers for NSCLC.
Transducer of ERBB2, 1 (TOB1) is a well-known tumor suppressor, which exerts its roles by negatively regulating the activity of receptor tyrosine-kinase ERBB2 (20). TOB1 was indicated to be downregulated in multiple human cancer types, including liver cancer (21), breast cancer (22) and lung cancer (23). For instance, a previous study reported that overexpression of TOB1 significantly suppresses lung cancer cell proliferation and metastasis (23). TOB1 was demonstrated to suppress cancer cell proliferation and metastasis via a series of downstream regulators, including cyclin D1, AKT signaling, BCL-2, BCL-XL and SMAD4 (22,24–26). LncRNA TOB1-AS1 originates from the TOB1 gene cluster. A previous study suggested that TOB1-AS1 acts as a tumor suppressor in cervical cancer (27). TOB1-AS1 inhibits cervical cancer cell proliferation, cell cycle and metastasis through interacting with miR-27b (27). However, the roles of TOB1-AS1 in lung cancer have remained largely elusive.
In the present study, lncRNA TOB1-AS1 was identified to be downregulated in NSCLC samples by analyzing public datasets. Bioinformatics analysis and experimental validation were performed to reveal the potential functions of TOB1-AS1 in NSCLC cells. The results of the present study demonstrate that TOB1-AS1 may be a potential biomarker for NSCLC.
Materials and methods
Cell lines and transfection
A549 and H1299 cells were obtained from the American Type Culture Collection. The A549 and H1299 cells were cultured in RPMI-1640 medium (HyClone; GE Healthcare) containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.). The NSCLC cells were cultured at 37°C in a humidified incubator with 5% CO2. Small interfering RNA (siRNA) targeting TOB1-AS1 (si-TOB1-AS1) and non-specific control siRNA (si-NC) were synthesized by GenePharma Co., Ltd. and transfected into cells by using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). The sequences of si-TOB1-AS1 were as follows: si-RNA1, 5′-GCACCCGATTAATTGAATA-3′; si-RNA2, 5′-GCGACTCGGATCCGTTTAT-3′; si-NC, 5′-TTCTCCGAACGTGTCACGT-3′. The knockdown efficacy was determined by reverse transcription-quantitative (RT-q) PCR. Cells were harvested 24 h following transfection for use in the subsequent experiments.
RT-qPCR
For detection of the subcellular localization of TOB1-AS1 in NSCLC cells, cytoplasmic and nuclear extracts were prepared using NE-PER™ nuclear and cytoplasmic extraction reagents (Pierce; Thermo Fisher Scientific, Inc.). Then, total cytoplasmic and nuclear RNA were extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, complementary (c)DNA was synthesized using the RevertAid First Strand cDNA Synthesis kit (Promega Corporation) in accordance with the manufacturer's protocol. qPCR was performed using iQ™ SYBR-Green Supermix (Bio-Rad Laboratories, Inc.). The following primers were used for qPCR: TOB1-AS1 forward, 5′-GCCAGGCCTAGAAGCTTTTG-3′ and reverse, 5′-TCTTCCCACCCCTTCTCCTA-3′; GAPDH forward, 5′-TGACTTCAACAGCGACACCCA-3′ and reverse, 5′-CACCCTGTTGCTGTAGCCAAA-3′, dynein cytoplasmic 2 light intermediate chain 1 (DYNC2LI1) forward, 5′-TGAACCACCAAAACCAACCT-3′ and reverse, 5′-TGGTGTGTTGTGCCCTTTTG-3′; E4F transcription factor 1 (E4F1) forward, 5′-GTTGCTGGGCCAGGAGG-3′ and reverse, 5′-GATGTCTGCTGCCAGAGAGG-3′; TSPY-like 4 (TSPYL4) forward, 5′-CTGGCTGTGTCCTGATACCC-3′ and reverse, 5′-CCAATTTGGGTTGATGGCCG-3′; component of oligomeric Golgi complex 7 (COG7) forward, 5′-GAATGCAACTTGCTGCCGAA-3′ and reverse, 5′-AGCTTGGCAGAAATCACAGC-3′, inositol hexakisphosphate kinase 2 (IP6K2) forward, 5′-AGACCCCTAAGGACTGGGTG-3′ and reverse, 5′-ACAAGACTTCAGATTTCTTTAGCCA-3′; Deltex E3 Ubiquitin Ligase 3 (DTX3) forward, 5′-TTCCATGGGCTTTCCAGGTC-3′ and reverse, 5′-GCCATTCTGGACAGGACGAA-3′. The 2−ΔΔCq method was used to calculate the relative expression levels (28).
Cell migration and invasion assay
The cell migration and invasion assays were performed as previously described (29).
Cell proliferation assay
The Cell Counting kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) was used to examine cell proliferation. All procedures were performed according to the manufacturer's protocols. The absorbance at 450 nm was measured and the absorbance at 615 nm was used as a reference using a microplate reader (BioTek Instruments, Inc.).
Public database analysis
In the present study, the expression levels of TOB1-AS1 in lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC) and normal lung tissues were evaluated by using Gene Expression Profiling Interactive Analysis (GEPIA) databases (http://gepia.cancer-pku.cn/index.html) (30). Informed consent had been obtained from all lung cancer patients (31). LncATLAS dataset (http://lncatlas.crg.eu/) was used to evaluate the subcellular localization of TOB1-AS1 in A549, HeLa and K562 cells (32).
Kaplan-Meier plotter database analysis
The Kaplan-Meier plotter online database (http://kmplot.com/analysis/index.php?p=service&cancer=lung) (33) was used to assess the prognostic significance of TOB1-AS1 expression in lung cancer. In order to evaluate the prognostic value of TOB1-AS1, the patient samples were divided into two groups: High expression and low expression, on the basis of the median expression of TOB1-AS1 (high expression, top 50%; low expression, bottom 50%). The Kaplan-Meier survival plots were obtained by entering the survival time [overall survival (OS) and post-progression survival (PPS; PPS was recorded as the time from tumor progression until death or was censored on the date of the last follow-up consultation)] (34) of patients with NSCLC. The P-values of the log-rank test and hazard ratio with 95% confidence intervals were also calculated.
Construction of ceRNA networks
First, interactions between miRNA and mRNA, and between miRNA and lncRNA were obtained from StarBase v2.0 (http://starbase.sysu.edu.cn/) (35) and Targetscan v7.1 database (http://www.targetscan.org/vert_71/) (36), which were used to comprehensively analyze miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale crosslinking immunoprecipitation-sequencing data. Subsequently, the ceRNA networks were generated using Cytoscape Software (version 3.6.0; http://www.cytoscape.org) (37).
Statistical analysis
Data analysis was performed using SPSS 24.0 software (IBM Corp.) and figures were created using GraphPad Prism 7 software (GraphPad Software, Inc.). Statistical comparisons between groups of normalized data were performed using the Student's t-test according to the test conditions. One-way analysis of variance followed by the Newman-Keuls post-hoc test was used to identify the differences among multiple groups. P<0.05 was considered to indicate a statistically significant difference with a 95% confidence level.
Results
TOB1-AS1 is downregulated in NSCLC
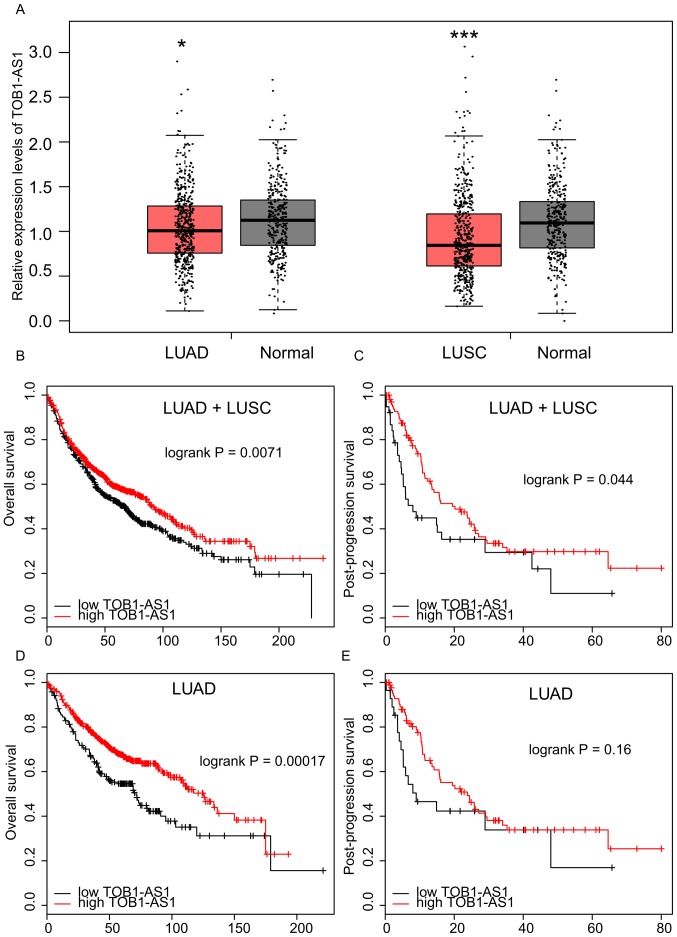
The expression levels of TOB1-AS1 in NSCLC and normal tissues have so far remained elusive. The public dataset GEPIA was used in the present study. As presented in Fig. 1A, the results revealed that TOB1-AS1 was significantly downregulated in LUAD and lung squamous cell carcinoma (LUSC) compared with the matched normal samples.
Figure 1.
Long non-coding RNA TOB1-AS1 is downregulated in NSCLC. (A) TOB1-AS1 expression levels were downregulated in LUAD and LUSC samples compared to normal tissues. (B and C) Higher expression of TOB1-AS1 in NSCLC samples was associated with longer (B) OS and (C) PPS time. (D and E) The association between expression levels of TOB1-AS1 and (D) OS or (E) PPS time in LUAD were also analyzed. *P<0.05 and ***P<0.01 vs. normal lung tissues. NSCLC, non-small cell lung cancer; OS, overall survival; PPS, post-progression survival; TOB1-AS1, transducer of ERBB2, 1-antisense 1; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Higher expression of TOB1-AS1 is associated with a better prognosis in NSCLC
The association between TOB1-AS1 expression and OS time or PPS time in NSCLC was analyzed using the Kaplan-Meier Plotter database. As presented in Fig. 1B and C, it was observed that NSCLC patients with a higher expression of TOB1-AS1 had a longer OS and PPS time than those with low expression. Furthermore, a higher expression of TOB1-AS1 in LUAD was associated with a longer OS time (Fig. 1D). However, no significant associations were observed between TOB1-AS1 expression and PPS time in the LUAD patients (Fig. 1E) or between TOB1-AS1 expression and OS or PPS time in the LUSC samples (data not shown).
TOB1-AS1 is localized in the cytoplasm and nucleus
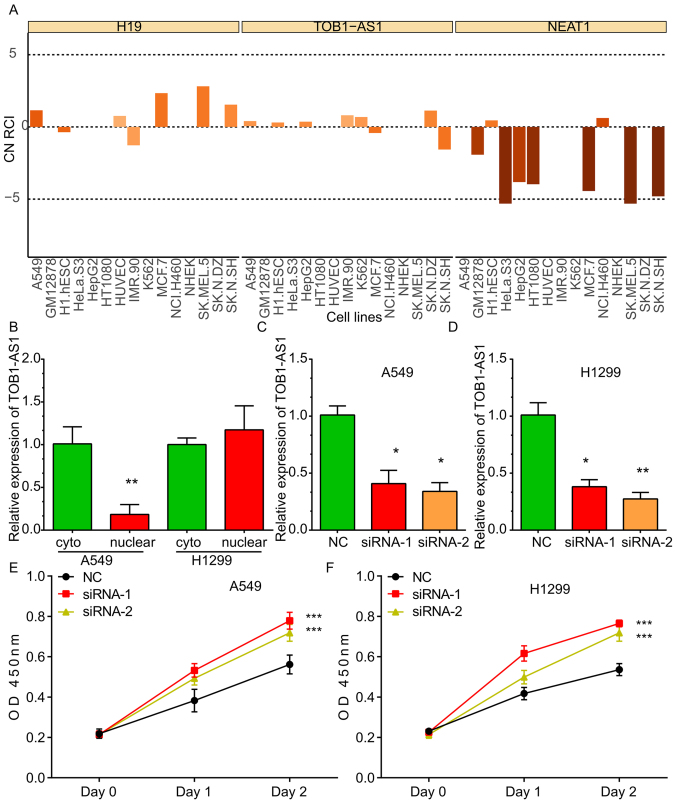
By analyzing the LncATLAS dataset, it was indicated that TOB1-AS1 was localized in the cytoplasm and nucleus of the majority of the human cancer cell lines, including A549, HeLa and K562 cells. The expression pattern of H19 located in the cytoplasm and that of NEAT1 located in the nuclei of cells were used as controls (Fig. 2A). Furthermore, RT-qPCR was used to detect the subcellular location of TOB1-AS1. As presented in Fig. 2, the TOB1-AS1 levels in the cytoplasm were higher compared with the nucleus in A549 cells, whereas TOB1-AS1 levels in the cytoplasm were comparable with that in the nucleus in H1299 cell lines (Fig. 2B).
Figure 2.
Knockdown of TOB1-AS1 induces proliferation of non-small cell lung cancer cells. (A) Analysis of the LncATLAS dataset indicated that TOB1-AS1 localized in the cytoplasm and nucleus in most of the cell lines. NEAT1 was used as a positive control of nuclear genes and H19 was used as a positive control of cytoplasmic genes. (B) Reverse transcription-quantitative PCR analysis indicated that TOB1-AS1 was located in the cytoplasm and nucleus of the A549 and H1299 cell lines. (C and D) Knockdown efficiency of TOB1-AS1 by using siRNA-1 and siRNA-2 in (C) A549 and (D) H1299 cells compared with NC. *P<0.05 and **P<0.01, vs. NC. (E and F) Knockdown of TOB1-AS1 induced cell proliferation in (E) A549 and (F) H1299 cells. ***P<0.001 vs. NC. TOB1-AS1, transducer of ERBB2, 1-antisense 1; NEAT1, nuclear paraspeckle assembly transcript 1; siRNA, small interfering RNA; NC, negative control; OD, optical density; Cyto, cytosol; CN RCI, Cytoplasmic/Nuclear concentration index (RCI).
In order to explore the molecular roles of TOB1-AS1 in NSCLC, loss-of-function assays were performed in the A549 and H1299 cells. For this, two siRNAs against TOB1-AS1 were designed, which effectively reduced the expression of TOB1-AS1 in the A549 (Fig. 2C) and H1299 (Fig. 2D) cells.
TOB1-AS1 knockdown promotes NSCLC cell proliferation
To investigate the effects of TOB1-AS1 knockdown on the proliferation of NSCLC cells, CCK-8 assays were performed. The results revealed that the proliferation rate of the cells transfected with si-TOB1-AS1 was significantly upregulated compared with that in the control group of A549 (Fig. 2E) and H1299 cells (Fig. 2F).
TOB1-AS1 knockdown promotes NSCLC cell migration and invasion
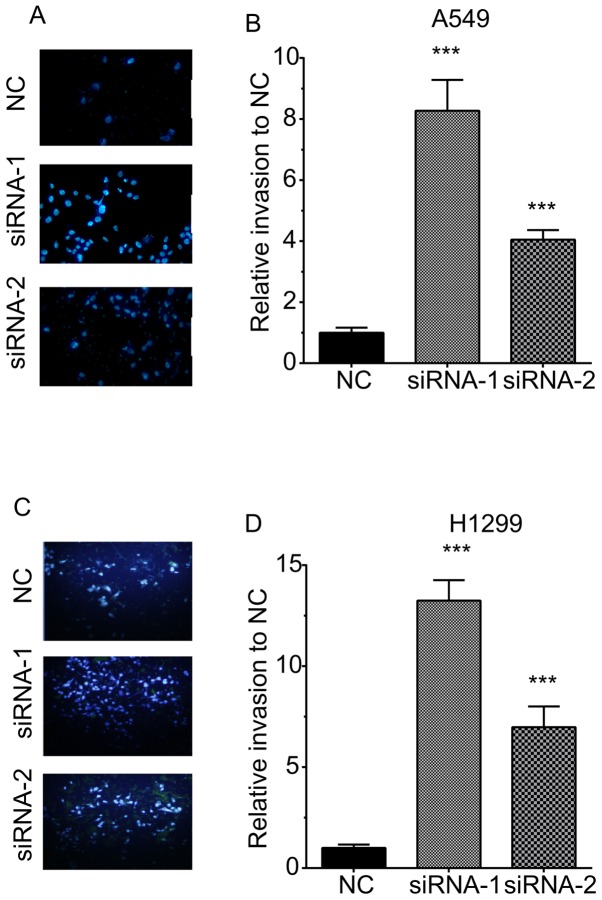
A Transwell assay was then performed to determine the effects of TOB1-AS1 knockdown on cell migration (Fig. 3). The results revealed that the numbers of migrating cells in the TOB1-AS1 knockdown group were significantly increased by 3.67- and 2.3-fold of those in the control group of A549 cells (Fig. 3A,B). Furthermore, in H1299 cells, TOB1-AS1 knockdown promoted cell migration by 5.3- and 3.3-fold of that in the control group (Fig. 3C,D).
Figure 3.
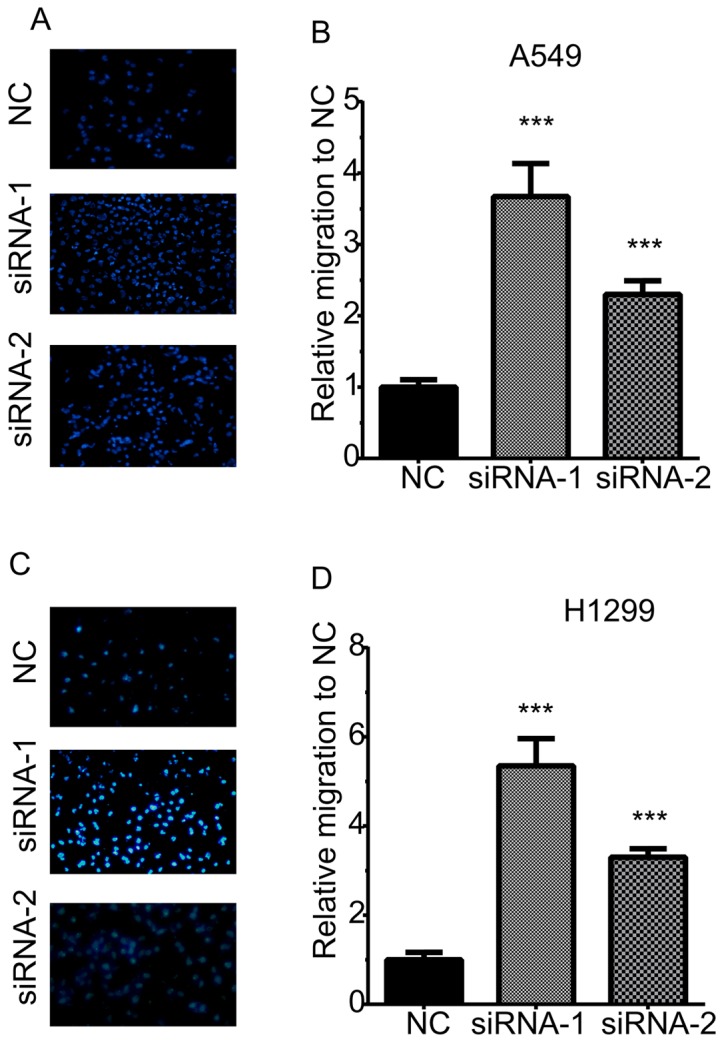
Knockdown of TOB1-AS1 induces migration of non-small cell lung cancer cells. (A and B) Knockdown of TOB1-AS1 potentiated the cell migration compared to control group in A549 cells. (A) Representative images of migratory A549 cells for each of the three conditions. (B) Quantified data of (A). (C and D) Knockdown of TOB1-AS1 induced migration of H1299 cells. (C) Representative images of migratory H1299 cells for each of the three conditions. (D) Quantified data of (C). ***P<0.001 vs. NC. Magnification, ×200; TOB1-AS1, transducer of ERBB2, 1-antisense 1; siRNA, small interfering RNA; NC, negative control.
The invasive ability of H1299 cells was also detected by quantifying the cells transgressing through Matrigel in a Transwell chamber. As illustrated in Fig. 3, TOB1-AS1 knockdown promoted cell invasion. The numbers of invasive cells were increased by 8.3- and 4.1-fold in A549 cells transfected with si-TOB1-AS1 compared with those in the negative control group (Fig. 4A,B). The numbers of invasive cells were increased by 13.25- and 6.98-fold in H1299 cells transfected with si-TOB1-AS1-1 and si-TOB1-AS1-2, respectively, in comparison with those in the negative control group (Fig. 4C,D).
Figure 4.
Knockdown of TOB1-AS1 induces invasion of non-small cell lung cancer cells. (A and B) Knockdown of TOB1-AS1 potentiated the cell invasion compared to control group in A549 cells. (A) Representative images of invasive A549 cells for each of the three conditions. (B) Quantified data of (A). (C and D) Knockdown of TOB1-AS1 induced invasion of H1299 cells. (C) Representative images of invasive H1299 cells for each of the three conditions. (D) Quantified data of (C). ***P<0.001 vs. NC. Magnification, ×200; TOB1-AS1, transducer of ERBB2, 1-antisense 1; siRNA, small interfering RNA; NC, negative control.
Construction of TOB1-AS1-mediated ceRNA network in NSCLC
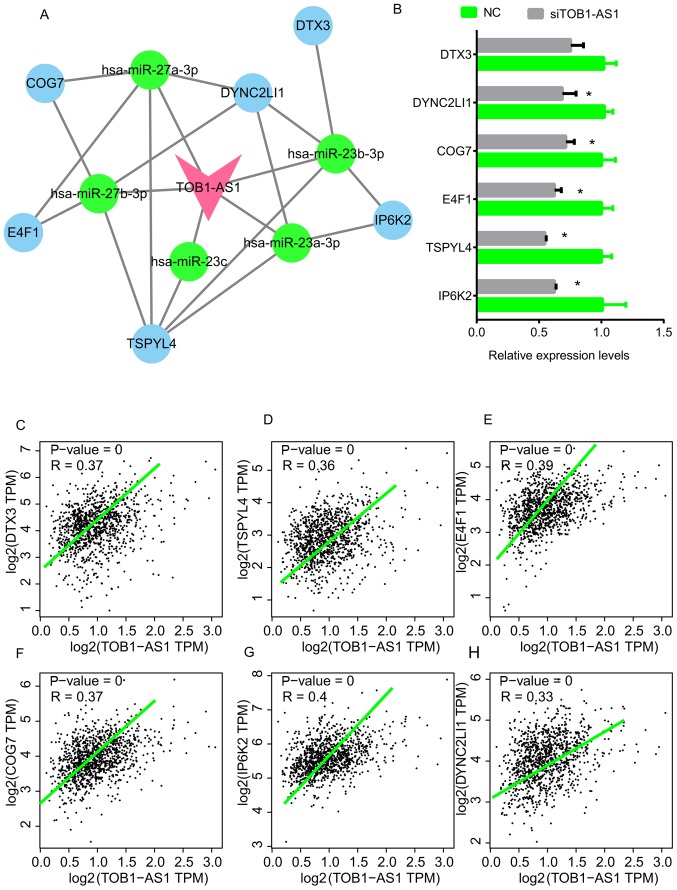
In order to elucidate the mechanisms of action of TOB1-AS1 regarding NSCLC progression, a TOB1-AS1-mediated ceRNA network in NSCLC was constructed. The co-expressed genes with a Pearson's score of >0.4 were selected as targets of TOB1-AS1. The Starbase and Targetscan databases were used to predict the potential miRNAs mediating the regulatory interaction between TOB1-AS1 and its targets. As presented in Fig. 5, a total of 5 miRNAs [Homo sapiens (hsa)-miR-27a-3p, hsa-miR-23a-3p, hsa-miR-23b-3p, hsa-miR-27b-3p and hsa-miR-23c] and 6 mRNAs (DYNC2LI1, E4F1, TSPYL4, COG7, IP6K2 and DTX3) were included in this network (Fig. 5A). In addition, the expression levels of these 6 mRNAs were measured after the silencing of TOB1-AS1 in A549 cells. The results indicated that knockdown of TOB1-AS1 using siRNA-1 significantly suppressed the expression of DYNC2LI1, E4F1, TSPYL4, COG7 and IP6K2 (Fig. 5B). By analyzing the GEPIA dataset, the expression of TOB1-AS1 in NSCLC was found to be positively correlated with DTX3, TSPYL4, E4F1, IP6K2, COG7 and DYNC2LI1 (Fig. 5C-H).
Figure 5.
TOB1-AS1-mediated ceRNA network in NSCLC and key genes. (A) A TOB1-AS1 mediated ceRNA network in NSCLC was constructed, pink node represents TOB1-AS1; blue nodes represent mRNAs; green nodes represent miRNAs. (B) The expression levels of DTX3, TSPYL4, E4F1, IP6K2, COG7 and DYNC2LI1 in A549 cells were detected after the knockdown of TOB1-AS1. *P<0.05 vs. NC. (C-H) The expression of TOB1-AS1 in NSCLC was positively correlated with (C) DTX3, (D) TSPYL4, (E) E4F1, (F) IP6K2, (G) COG7 and (H) DYNC2LI1 by using the GEPIA dataset. NSCLC, non-small cell lung cancer; TOB1-AS1, transducer of ERBB2, 1-antisense 1; siRNA, small interfering RNA; NC, negative control; ceRNA, competing endogenous RNA; hsa, Homo sapiens; miR, microRNA; DYNC2LI1, dynein cytoplasmic 2 light intermediate chain 1; E4F1, E4F transcription factor 1; TSPYL4, TSPY-like 4; COG7, component of oligomeric Golgi complex 7; IP6K2, inositol hexakisphosphate kinase 2; DTX3, deltex E3 ubiquitin ligase 3; TPM, transcript per million.
Dysregulation of TOB1-AS1 targets in NSCLC is associated with prognosis
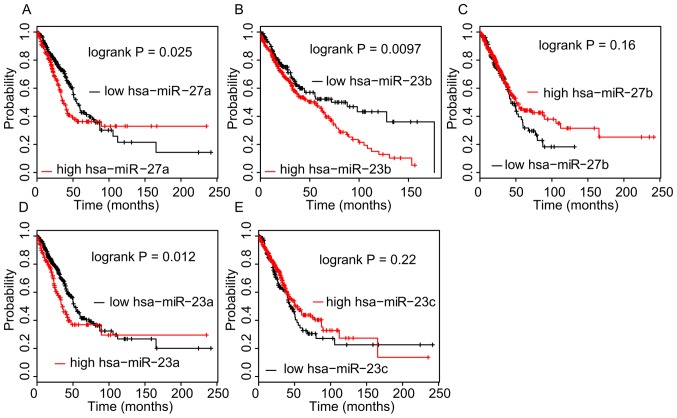
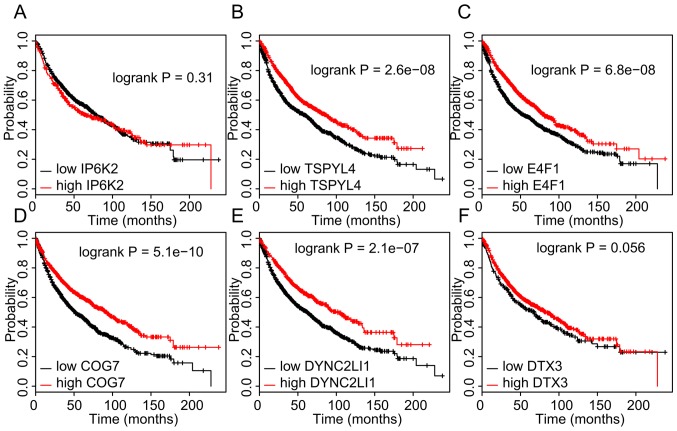
The association between the expression levels of ceRNAs and OS time in NSCLC was then analyzed in the TCGA dataset using the Kaplan-Meier method. As presented in Fig. 6, a higher expression of hsa-miR-27a-3p (P=0.025), hsa-miR-23a-3p (P=0.012) and hsa-miR-23b-3p (P=0.0097) in NSCLC tissues was significantly associated with a shorter OS time. However, analysis of the Kaplan-Meier Plotter database indicated that a higher expression of DYNC2LI1, E4F1, TSPYL4, and COG7 in NSCLC was associated with a longer OS time (Fig. 7).
Figure 6.
Higher expression of hsa-miR-27a-3p, hsa-miR-23a-3p, hsa-miR-23b-3p in non-small cell lung cancer samples was associated with shorter OS time. (A-E) Kaplan-Meier analysis of the dataset from The Cancer Genome Atlas was performed to assess the influence of high/low expression of (A) hsa-miR-27a-3p, (B) hsa-miR-23b-3p, (C) hsa-miR-27b-3p, (D) hsa-miR-23a-3p and (E) hsa-miR-23c on OS. P<0.05 was considered to indicate a statistically significant difference with a 95% confidence level. hsa, Homo sapiens; miR, microRNA; OS, overall survival.
Figure 7.
Higher expression of DYNC2LI1, E4F1, TSPYL4, and COG7 in non-small cell lung cancer samples is associated with longer OS time. (A-F) Kaplan-Meier analysis of the dataset from The Cancer Genome Atlas was performed to assess the influence of high/low expression of (A) IP6K2, (B) TSPYL4, (C) E4F1, (D) COG7, (E) DYNC2LI1 and (F) DTX3 on OS. P<0.05 was considered to indicate a statistically significant difference with a 95% confidence level. OS, overall survival; DYNC2LI1, dynein cytoplasmic 2 light intermediate chain 1; E4F1, E4F transcription factor 1; TSPYL4, TSPY-like 4; COG7, component of oligomeric Golgi complex 7; IP6K2, inositol hexakisphosphate kinase 2; DTX3, deltex E3 ubiquitin ligase 3.
Discussion
The mechanisms underlying NSCLC have remained to be fully elucidated. Recent studies have demonstrated that lncRNAs have crucial roles in regulating NSCLC-associated pathways. For instance, p53 inducible cancer associated RNA transcript 1 suppresses NSCLC proliferation and invasion by targeting the AKT1 signaling pathway (38). Small nucleolar RNA host gene 1 promotes NSCLC progression by activating Wnt/β-catenin signaling (39). However, the functions of the majority of lncRNAs in NSCLC remain elusive. To the best of our knowledge, the present study was the first to demonstrate that lncRNA TOB1-AS1 expression was reduced in NSCLC samples compared with that in normal tissues. Higher expression levels of TOB1-AS1 were associated with a better prognosis in NSCLC. These analyses indicated that TOB1-AS1 may serve as a novel biomarker for NSCLC.
TOB1-AS1 is a recently discovered lncRNA. A study on cervical cancer demonstrated that TOB1-AS1 functions as a tumor suppressor (27). TOB1-AS1 inhibits cervical cancer cell proliferation, the cell cycle and metastasis. Regarding possible mechanisms, TOB1-AS1 was reported to interact with miR-27b to degrade its expression (27). The results of the present study were consistent with those of previous studies. The present results indicated that TOB1-AS1 was localized in the cytoplasm and nucleus. Knockdown of TOB1-AS1 induced cell proliferation. Furthermore, silencing of TOB1-AS1 promoted A549 and H1299 cell migration and invasion. These results suggest that TOB1-AS1 functions as a tumor suppressor in NSCLC. Of note, siRNA-1 and siRNA-2 appeared to carry the same transfection efficiency despite siRNA-1 being more effective in inhibiting migration/invasion. This may have been due to off-target effects of the siRNAs. In spite of this, the present results indicate that knockdown of TOB1-AS1 promoted NSCLC proliferation and migration. With the development of the clustered regularly interspaced short palindromic repeat and associated nuclease Cas9 method (40), knockout assays may be a more powerful tool to explore the potential roles of lncRNAs in human cancers and to overcome off-target effects.
The mechanisms of action of TOB1-AS1 in NSCLC remain to be fully elucidated. To gain further insight, a TOB1-AS1-mediated ceRNA network in NSCLC was constructed in the present study, including hsa-miR-27a-3p, hsa-miR-23a-3p, hsa-miR-23b-3p, hsa-miR-27b-3p, hsa-miR-23c, DYNC2LI1, E4F1, TSPYL4, COG7, IP6K2 and DTX3. Further analysis revealed that these TOB1-AS1 targets were also dysregulated and associated with the prognosis of NSCLC. miR-27a has been reported to regulate cell proliferation, migration and invasion in NSCLC (41–43), while miR-23a was indicated to promote NSCLC metastasis (44). miR-27b was demonstrated to inhibit NSCLC growth and invasion by targeting LIM domain kinase 1 (45). E4F1 is an ubiquitin E3 ligase for p53 and regulates p53 transcription functions (46). These studies, together with the present results, suggest that TOB1-AS1 and its targets have crucial roles in NSCLC.
Of note, the present study had several limitations. First, gain-of-function assays should be further performed to validate the roles of TOB1-AS1 in NSCLC. TOB1-AS1 is a novel lncRNA. Despite the RNA sequence provided in the NCBI database, the accurate 5'sequence and 3'sequence still require to be validated using 5′race and 3′race assays. Furthermore, in vivo assays should be performed to further validate the suppressive roles of TOB1-AS1 in mouse models (47). Such a study may provide useful information to understand the molecular functions of TOB1-AS1 in NSCLC.
In conclusion, the present study demonstrated for the first time, at least to the best of our knowledge, that TOB1-AS1 has a tumor-suppressive role in NSCLC. Knockdown of TOB1-AS1 significantly induced NSCLC migration, invasion and proliferation. Furthermore, microarray and bioinformatics analyses were performed to explore the underlying mechanisms of the regulation of NSCLC metastasis by TOB1-AS1. It was also revealed that TOB1-AS1 in NSCLC samples was associated with a longer OS time. Despite the loss-of-functions assays, the roles of TOB1-AS1 on NSCLC still require further validation. However, the present study clearly demonstrated that TOB1-AS1 may serve as a novel biomarker for NSCLC.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (grant nos. 81373621 and 81774166), the Excellent Academic Leaders of Health and Family Planning System in Shanghai (grant no. 2017BR044), Clinical Science and Technology Innovation Project of the Science and Technology Commission of Shanghai Municipality (grant no. 16401970800) and was sponsored by the Shanghai Sailing Program (grant no. 17YF1419700).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JHT and LSL designed the study. WJS and HL performed experiments and wrote the manuscript. ZJQ and FFQ performed bioinformatics analysis. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: Background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 3.Kopp F, Mendell JT. Functional classification and experimental dissection of long Noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pekarsky Y, Croce CM. Noncoding RNA genes in cancer pathogenesis. Adv Biol Regul. 2019;71:219–223. doi: 10.1016/j.jbior.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18:6. doi: 10.1186/s12943-018-0934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao C, Wang X, Liu Y, Wang M, Yan B, Jiang Y, Shi Y, Shen Y, Liu X, Lai W, et al. A G3BP1-interacting lncRNA promotes ferroptosis and apoptosis in cancer via nuclear sequestration of p53. Cancer Res. 2018;78:3484–3496. doi: 10.1158/0008-5472.CAN-17-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng SJ, Chen HY, Ye Z, Deng SC, Zhu S, Zeng Z, He C, Liu ML, Huang K, Zhong JX, et al. Hypoxia-induced LncRNA-BX111 promotes metastasis and progression of pancreatic cancer through regulating ZEB1 transcription. Oncogene. 2018;37:5811–5828. doi: 10.1038/s41388-018-0382-1. [DOI] [PubMed] [Google Scholar]
- 8.Liu CY, Zhang YH, Li RB, Zhou LY, An T, Zhang RC, Zhai M, Huang Y, Yan KW, Dong YH, et al. LncRNA CAIF inhibits autophagy and attenuates myocardial infarction by blocking p53-mediated myocardin transcription. Nat Commun. 2018;9:29. doi: 10.1038/s41467-017-02280-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao J, Du P, Cui P, Qin Y, Hu C, Wu J, Zhou Z, Zhang W, Qin L, Huang G. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene. 2018;37:4094–4109. doi: 10.1038/s41388-018-0250-z. [DOI] [PubMed] [Google Scholar]
- 10.Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF, Waye MM. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–22525. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang Y, Jia L, Li S, Cancer Genome Atlas Research Network. Xie W, Yang D. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that Interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell. 2018;33:706–720.e9. doi: 10.1016/j.ccell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 13.Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K, Kossai M, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian H, Chen L, Huang J, Wang X, Ma S, Cui F, Luo L, Ling L, Luo K, Zheng G. The lncRNA MIR4435-2HG promotes lung cancer progression by activating β-catenin signalling. J Mol Med (Berl) 2018;96:753–764. doi: 10.1007/s00109-018-1654-5. [DOI] [PubMed] [Google Scholar]
- 15.Lai IL, Yang CA, Lin PC, Chan WL, Lee YT, Yen JC, Chang YS, Chang JG. Long noncoding RNA MIAT promotes non-small cell lung cancer proliferation and metastasis through MMP9 activation. Oncotarget. 2017;8:98148–98162. doi: 10.18632/oncotarget.21465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su W, Feng S, Chen X, Yang X, Mao R, Guo C, Wang Z, Thomas DG, Lin J, Reddy RM, et al. Silencing of long noncoding RNA MIR22HG triggers cell Survival/Death signaling via oncogenes YBX1, MET, and p21 in lung cancer. Cancer Res. 2018;78:3207–3219. doi: 10.1158/0008-5472.CAN-18-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S, Zhao H, Li J, Zhang A, Wang H. Downregulation of long non-coding RNA LET predicts poor prognosis and increases Notch signaling in non-small cell lung cancer. Oncotarget. 2017;9:1156–1168. doi: 10.18632/oncotarget.23452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Y, Lyu H, Liu H, Shi X, Song Y, Liu B. Downregulation of the long noncoding RNA GAS5-AS1 contributes to tumor metastasis in non-small cell lung cancer. Sci Rep. 2016;6:31093. doi: 10.1038/srep31093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun GG, Lu YF, Cheng YJ, Hu WN. The expression of BTG1 is downregulated in NSCLC and possibly associated with tumor metastasis. Tumour Biol. 2014;35:2949–2957. doi: 10.1007/s13277-013-1379-6. [DOI] [PubMed] [Google Scholar]
- 20.Lee HS, Kundu J, Kim RN, Shin YK. Transducer of ERBB2.1 (TOB1) as a Tumor suppressor: A mechanistic perspective. Int J Mol Sci. 2015;16:29815–29828. doi: 10.3390/ijms161226203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho KJ, Do NL, Otu HH, Dib MJ, Ren X, Enjyoji K, Robson SC, Terwilliger EF, Karp SJ. Tob1 is a constitutively expressed repressor of liver regeneration. J Exp Med. 2010;207:1197–1208. doi: 10.1084/jem.20092434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang YW, Nasto RE, Varghese R, Jablonski SA, Serebriiskii IG, Surana R, Calvert VS, Bebu I, Murray J, Jin L, et al. Acquisition of estrogen independence induces TOB1-related mechanisms supporting breast cancer cell proliferation. Oncogene. 2016;35:1643–1656. doi: 10.1038/onc.2015.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiao Y, Sun KK, Zhao L, Xu JY, Wang LL, Fan SJ. Suppression of human lung cancer cell proliferation and metastasis in vitro by the transducer of ErbB-2.1 (TOB1) Acta Pharmacol Sin. 2012;33:250–260. doi: 10.1038/aps.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu D, Zhou W, Wang S, Zhou Z, Wang S, Chen L. Tob1 enhances radiosensitivity of breast cancer cells involving the JNK and p38 pathways. Cell Biol Int. 2015;39:1425–1430. doi: 10.1002/cbin.10545. [DOI] [PubMed] [Google Scholar]
- 25.Schulze-Topphoff U, Casazza S, Varrin-Doyer M, Pekarek K, Sobel RA, Hauser SL, Oksenberg JR, Zamvil SS, Baranzini SE. Tob1 plays a critical role in the activation of encephalitogenic T cells in CNS autoimmunity. J Exp Med. 2013;210:1301–1309. doi: 10.1084/jem.20121611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun KK, Zhong N, Yang Y, Zhao L, Jiao Y. Enhanced radiosensitivity of NSCLC cells by transducer of erbB2.1 (TOB1) through modulation of the MAPK/ERK pathway. Oncol Rep. 2013;29:2385–2391. doi: 10.3892/or.2013.2403. [DOI] [PubMed] [Google Scholar]
- 27.Yao J, Li Z, Yang Z, Xue H, Chang H, Zhang X, Li T, Guo K. Long noncoding RNA TOB1-AS1, an epigenetically silenced gene, functioned as a novel tumor suppressor by sponging miR-27b in cervical cancer. Am J Cancer Res. 2018;8:1483–1498. [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Yi C, Wan X, Zhang Y, Fu F, Zhao C, Qin R, Wu H, Li Y, Huang Y. SNORA42 enhances prostate cancer cell viability, migration and EMT and is correlated with prostate cancer poor prognosis. Int J Biochem Cell Biol. 2018;102:138–150. doi: 10.1016/j.biocel.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 30.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research Network, corp-author. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mas-Ponte D, Carlevaro-Fita J, Palumbo E, Hermoso Pulido T, Guigo R, Johnson R. LncATLAS database for subcellular localisation of long noncoding RNAs. RNA. 2017;23:1080–1087. doi: 10.1261/rna.060814.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imai H, Mori K, Wakuda K, Ono A, Akamatsu H, Shukuya T, Taira T, Kenmotsu H, Naito T, Kaira K, et al. Progression-free survival, post-progression survival, and tumor response as surrogate markers for overall survival in patients with extensive small cell lung cancer. Ann Thoracic Med. 2015;10:61–66. doi: 10.4103/1817-1737.146885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou KR, Liu S, Cai L, Bin L, et al. ENCORI: The encyclopedia of RNA interactomes [Google Scholar]
- 36.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang C, Su C, Song Q, Dong F, Yu S, Huo J. LncRNA PICART1 suppressed non-small cell lung cancer cells proliferation and invasion by targeting AKT1 signaling pathway. Am J Transl Res. 2018;10:4193–4201. [PMC free article] [PubMed] [Google Scholar]
- 39.Cui Y, Zhang F, Zhu C, Geng L, Tian T, Liu H. Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/β-catenin signaling pathway. Oncotarget. 2017;8:17785–17794. doi: 10.18632/oncotarget.14854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Revi Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chae DK, Ban E, Yoo YS, Kim EE, Baik JH, Song EJ. MIR-27a regulates the TGF-β signaling pathway by targeting SMAD2 and SMAD4 in lung cancer. Mol Carcinog. 2017;56:1992–1998. doi: 10.1002/mc.22655. [DOI] [PubMed] [Google Scholar]
- 42.Yang Y, Zang A, Jia Y, Shang Y, Zhang Z, Ge K, Zhang J, Fan W, Wang B. Genistein inhibits A549 human lung cancer cell proliferation via miR-27a and MET signaling. Oncol Lett. 2016;12:2189–2193. doi: 10.3892/ol.2016.4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miao Y, Li J, Qiu X, Li Y, Wang Z, Luan Y. miR-27a regulates the self renewal of the H446 small cell lung cancer cell line in vitro. Oncol Rep. 2013;29:161–168. doi: 10.3892/or.2012.2095. [DOI] [PubMed] [Google Scholar]
- 44.Cao M, Li Y, Lu H, Meng Q, Wang L, Cai L, Dong X. MiR-23a-mediated migration/invasion is rescued by its target, IRS-1, in non-small cell lung cancer cells. J Cancer Res Clin Oncol. 2014;140:1661–1670. doi: 10.1007/s00432-014-1725-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun Y, Xu T, Cao YW, Ding XQ. Antitumor effect of miR-27b-3p on lung cancer cells via targeting Fzd7. Eur Rev Med Pharmacol Sci. 2017;21:4113–4123. [PubMed] [Google Scholar]
- 46.Caramel J, Lacroix M, Le Cam L, Sardet C. E4F1 connects the Bmi1-ARF-p53 pathway to epidermal stem cell-dependent skin homeostasis. Cell Cycle. 2011;10:866–867. doi: 10.4161/cc.10.6.14974. [DOI] [PubMed] [Google Scholar]
- 47.Yang Liu, Liu G. lncRNA BANCR suppresses cell viability and invasion and promotes apoptosis in non-small-cell lung cancer cells in vitro and in vivo. Cancer Manag Res. 2019;11:3565–3574. doi: 10.2147/CMAR.S194848. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.