Abstract
Liver cancer exhibits the fourth most common cause of cancer-associated mortality worldwide. Due to the rapid growth, solid tumors undergo severe hypoxia and produce high levels of extracellular adenosine to maintain homeostasis. A previous study indicated that the hypoxic condition in liver cancer increased hepatic adenosine, which is known to facilitate cancer survival and proliferation. Extracellular adenosine has been revealed to regulate pathological and physiological processes in cells and tissues. However, its pathophysiological role in liver cancer remains undetermined. Emerging evidence has indicated that the adenosine A2B receptor promotes the progression of liver cancer. Therefore, it was hypothesized that HIF-1α is a transcriptional regulator of A2B in human liver cancer. The current study determined A2B expression of a number of liver cancer cell lines and performed functional studies of HIF-1α as a master transcriptional regulator of hepatic A2B signaling during hypoxic conditions. The current study aimed to identify the promoter region of A2B, which has a hypoxia response element, by performing luciferase assays. The present study demonstrated that reduced HIF-1α expression is associated with low expression of A2B, and HIF-1α overexpression is associated with A2B induction. Furthermore, the siRNA-mediated downregulation of A2B inhibited the growth and proliferation of HepG2, which is a liver cancer cell line. The relationship between HIF-1α and A2B expression was also identified in human liver cancer specimens. In conclusion, the current study indicated that A2B is induced by the HIF-1α transcriptional regulator during hypoxia, and it may be a potential pharmacologic and therapeutic target for the treatment of patients with liver cancer.
Keywords: liver cancer, adenosine A2B receptor, hypoxia inducible factor-1α, adenosine, hypoxia
Introduction
Liver cancer is the fourth leading cause of cancer mortality worldwide (1,2). Despite significant progress over the past decades, the clinical outcomes of patients with liver cancer remain poor and an in-depth understanding of the molecular pathogenesis of liver cancer could facilitate the development of novel diagnostic and therapeutic techniques.
Adenosine is an endogenous nucleoside that controls many physiological processes through interactions with adenosine receptors, such as A1, A2A, A2B, and A3 (3). Adenosine A2 receptors have two segmented isoforms; high-affinity A2A receptor which is highly concentrated in the striatum, and relatively low-affinity A2B receptors which present throughout the brain (4). Generally, adenosines can be found eluted into the extracellular matrix of ordinary solid organs under low-oxygen conditions (5). However, in cancerous organs, the signaling pathway generated between adenosine and its receptor has been shown to coordinate adenosine accumulation and the increased levels of adenosine in the extracellular fluid of solid tumors stimulate cancer cell proliferation and tumor angiogenesis (6). The relationship between the tumor progression and adenosine A2B receptor expression has been investigated in many types of tumors including bladder (7), breast (8), colon (9), and prostate (10) cancers. While some researchers have shown that A2B is highly expressed in cancerous tissues in patients with HCC (11), the regulatory pathways and the function of A2B in liver cancer remain undiscovered.
The adenosine A2B receptor is reported to be transcriptionally induced by tumor necrosis factor-alpha (TNF-α) (12), interferon-gamma (IFN-γ) (13), and hypoxia-inducible factor-1α (HIF-1α) (14,15). HIF-1α is an oxygen-regulated subunit of HIF-1, a heterodimeric transcription factor consisting of α- and β-subunits. Intra-tumoral hypoxia is the major cause of increased HIF-1 activity in human HCC (16), and HIF-1α protein stabilization in cancer cells leading to the upregulation of multiple HIF-1 target genes that are required for HCC tumor growth, as demonstrated by both genetic and pharmacologic loss-of-function studies (17). HIF-1α mediated expression of adenosine A2B receptor activations in hypoxia has reported in endothelial cells (15), acute lung injury (18), and breast cancer (19). In addition to this, many researchers suggested that hypoxia is one factor for inducing A2B expressions, however, direct HIF-1α mediated A2B increment during tumor progression, especially in liver cancer has not been investigated yet. Therefore, we designed experiments to verify whether transcriptional regulation pathways control the endogenous hepatic adenosine signaling during a low-oxygen condition of liver cancer cells. Furthermore, we also investigated the potential HIF-1α binding regions in the A2B promoter genes. We believe our study will contribute to the development of new therapeutic targets to treat liver cancers.
Materials and methods
Cell culture and hypoxic conditions
We purchased human liver cancer cell lines, including HepG2 (KCLB no. 88065; passage 21), the hepatoblastoma cell line (20), and the hepatocellular carcinoma (HCC) cell lines SK-Hep1 (KCLB no. 30052; passage 49) and SNU-449 (KCLB no. 00449; passage 28) from the Korean Cell Line Bank (KCLB; Seoul, Republic of Korea). HepG2 and SK-Hep1 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) high-glucose (Hyclone; Thermo Fisher Scientific, Inc.), supplemented with 10% fetal bovine serum (FBS; Mediatech) and 100 U/ml penicillin/streptomycin (HyClone). The HCC SNU-449 cells were maintained in RPMI-1640 (Invitrogen; Thermo Fisher Scientific, Inc.), supplemented with 10% FBS and 100 U/ml penicillin/streptomycin. We cultured all the cells in a standard humidified incubator at 37°C in an atmosphere of 5% CO2. To induce hypoxic conditions, we placed cells in a hypoxia incubator (MCO-18M; Sanyo) filled with a mixture of 5% CO2, 94.5% N2, and 0.5% O2 gas. Authentication of the cell lines was done using short tandem repeat (STR) profiling by the Korean Cell Line Bank (Seoul National University College of Medicine, Seoul, Republic of Korea; http://cellbank.snu.ac.kr/) with proper STR references. The cell lines were tested for mycoplasma contamination before use and were negative for mycoplasma.
Establishment of A2B luciferase constructs
We used a pGL3 basic luciferase vector as a control plasmid and performed luciferase assays with previously designed constructs (15). The full-length construct (1095 bp) had a hypoxia-response element (HRE). The other truncated construct (477 bp) lacked an HRE. Additionally, we used a mutant form of the 1095 bp plasmid with a modified sequence (ACGTG was altered to AATCG).
A2B reporter assay
To confirm the transcriptional activity of the adenosine A2B receptor, we used HepG2 cells as an easily transfectable cellular model. First, to investigate the A2B promoter region, we used HepG2 cells to measure the reporter gene activity. We co-transfected cells with 2 µg of A2B-Luc promoter-reporter and 0.02 µg Renilla reporter vector for 24 h using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. Next, we exposed the cells to hypoxic conditions for up to 12 h. To prepare whole protein lysates, we used 1× passive lysis buffer (cat no. E1910; Promega). To perform the luciferase assay, we followed the protocol of the Dual-Luciferase Reporter Assay (Promega).
RNA interference
Small interfering RNAs (siRNA) specific to either HIF-1α (HIF-1α-siRNA), A2B (A2B-siRNA), or scrambled sequences (scr-siRNA) were prepared by Bioneer Corporation. We used 0.5 µg of siRNA for transfections using Lipofectamine® 2000 reagent (Invitrogen). The sequences of siRNAs were: Scrambled-siRNA (scr-siRNA), sense, 5′-CCUACGCCACCAAUUUCGU(dTdT)-3′, antisense, 5′-ACGAAAUUGGUGGCGUAGG(dTdT)-3′; HIF-1α-siRNA, sense, 5′-GUGGUUGGAUCUAACACUA(dTdT)-3′, antisense, 5′-UAGUGUUAGAUCCAACCAC(dTdT)-3′; A2B-siRNA, sense, 5′-GAGACUUCCGCUACACUUU(dTdT)-3′, antisense, 5′-AAAGUGUAGCGGAAGUCUC(dTdT)-3′.
HIF-1α plasmid overexpression
We seeded HepG2 cells into 6-well plates at a density of 1.0×106 cells per well and incubated them for 24 h at 37°C. The cells were transfected with a HIF-1-pcDNA3.1-expressing plasmid using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). We confirmed HIF-1α overexpression by western blot. The negative control plasmid pcDNA3.1(+) and the HIF-1-pcDNA3.1-expressing plasmids were purchased from Addgene. We treated cells with range from 3 to 300 nM of echinomycin (cat no. 5520; Tocris Bioscience), a specific inhibitor of HIF-1 DNA binding activity, to achieve chemical interference of HIF activity.
BrdU assay
Exponentially growing HepG2 cells were digested, centrifuged, collected, and inoculated into 96-well culture plates at a density of 5.0×103 cells/well. After culturing for 24 h in DMEM containing 10% FBS, the cells were transfected with scrambled-siRNA (scr-siRNA) and A2B-siRNA. HepG2 cell proliferation activity was analyzed using BrdU cell proliferation assay kits (cat no. 2750; Millipore), 24, 48, 72, and 96 h after siRNA transfection. The BrdU reagent was added to the culture wells at each time point, and the plates were incubated at 37°C for an additional 4 h. The cells were then fixed, and the BrdU levels were detected colorimetrically following the manufacturer's instructions. To plot the growth curve, a microplate reader (Sunrise TECAN, Labx, Canada) was used to measure absorbance at 450 nm. Experiments were repeated four times.
Cell growth and viability test
Cell growth and viability were determined by performing a 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay in 12 well cell culture plates (NEST Biotechnology). HepG2 cells were seeded at a density of 5.0×104 per well for 24 h and cultured in DMEM containing 10% FBS. The cells were then transfected with scr-siRNA or A2B-siRNA. After incubation for 24, 48, and 72 h, 20 µl of MTT solution (5 mg/ml in phosphate buffer saline) was added to each well, and the plates were incubated in the dark for 4 h at 37°C. Cell growth and viability were expressed as the percentage of absorbance of MTT-treated cells relative to that of untreated cells at a wavelength of 540 nm. Measurements for all treatment groups were taken four times.
Patient tissue samples
We collected five individual human HCC specimens from patients with HCC, who visited the Asan Medical Center (Division of Liver Transplantation and Hepatobiliary Surgery; Seoul, Republic of Korea) between 2014 and 2015 (Table I). None of the patients received any form of treatment before surgical procedures. We separately collected small pieces of the tumor and adjacent normal tissues and snap-froze them into liquid nitrogen. Frozen samples were stored at −80°C until use. The Institutional Review Board of Asan Medical Center reviewed and approved the collection and use of patient specimens (approval no. 2016-0582). All patients who provided tissue samples agreed to donate their specimens and provided written informed consents.
Table I.
Clinical characteristic of five patients with HCC in the Asan Medical Center.
| Patients | P1 | P2 | P3 | P4 | P5 |
|---|---|---|---|---|---|
| Grading | Grade III | Grade III | Grade III | Grade III | Grade III |
| Gender | F | M | M | M | F |
| Age (years) | 38 | 54 | 62 | 65 | 68 |
| Risk Factors | HBV | HBV | HBV | HBV | HBV |
| AST (U/l) | 174 | 82 | 133 | 191 | 190 |
| ALT (U/l) | 128 | 72 | 87 | 147 | 137 |
| Albumin (g/dl) | 3.5 | 3.6 | 3.3 | 3.0 | 2.8 |
| Tumor size (cm) | 3.7×3.3×3.3 | 2.4×1.5×1.0 | 4.5×4.0×3.3 | 11.6×10.8×9.6 | 14.2×10.8×7.5 |
P, patient; AST, aspartate aminotransferase; ALT, alanine aminotransferase; HBV, hepatitis B virus; HCC, hepatocellular carcinoma.
RNA extraction and RT-PCR
We isolated total RNA from human livers and liver cancer cells using QIAzol reagent (Qiagen). We homogenized the frozen tissues and liver cancer cell suspensions in QIAzol reagent. The homogenates were mixed thoroughly after adding chloroform and were centrifuged at 16,000 × g for 15 min. The aqueous phase was removed, and the RNA was precipitated with isopropyl alcohol. RNA was pelleted, washed with 70% ethanol, dried, and eluted using DEPC-treated water. We used a spectrophotometer Nanodrop 2000 (Nanodrop) to quantify the final concentration.
We quantified transcription of relevant genes using real-time reverse transcription-polymerase chain reaction (RT-PCR) in a CFX Connect Real-Time PCR Detection System (Bio-Rad) with 5× HOT FIREPol EvaGreen qPCR Supermix (Solis BioDyne), according to the manufacturer's instructions. In brief, the samples were first denatured at 95°C for 15 min, followed by 40 cycles of denaturation at 95°C for 15 sec, annealing at 55°C-60°C for 15 sec, and elongation at 72°C for 20 sec. The data are expressed as the fold changes in the treatment groups relative to the control level and were normalized to GAPDH levels using the delta-delta Ct methods (21). Table II lists the sense and antisense primer sequences.
Table II.
Sequences of Primers used in the study.
| Gene | Primer sequence |
|---|---|
| Adora1 | Forward 5′-TGCACTGGCCTGTTCTGTAG-3′ |
| Reverse 5′-CTGCCTCTCCCACGTACAAT-3′ | |
| Adora2a | Forward 5′-GGAGTTTGCCCCTTCCTAAG-3′ |
| Reverse 5′-CTGCTTCCTCAGAACCCAAG-3′ | |
| Adora2b | Forward 5′-ATCTCCAGGTATCTTCTC-3′ |
| Reverse 5′-GTTGGCATAATCCACACAG-3′ | |
| Adora3 | Forward 5′-CCTGGGCATCACAATCCACT-3′ |
| Reverse 5′-ACCCTCTTGTATCTGACGGTA-3′ | |
| Gapdh | Forward 5′-GAGTCAACGGATTTGGTCGT-3′ |
| Reverse 5′-TTGATTTTGGAGGGATCTCG-3′ |
Protein isolation and western blotting
We extracted protein samples from ~2.0×106 cells or 70 mg of frozen liver tissues. The samples were homogenized in lysis buffer (RIPA; Biosesang), quantified with BCA protein assay kit (Thermo Fisher Scientific, Inc.), and immunoblotted with anti-adenosine A2B receptor antibody (1:1,000 dilution; ab40002, Abcam) followed by rabbit anti-goat IgG HRP (1:20,000 dilution; A5420, Sigma-Aldrich); or anti-HIF-1α antibody (1:1,000 dilution; cat no. 610959, BD Biosciences) followed by goat anti-mouse IgG HRP (1;5,000; sc-2005, Santa Cruz Biotechnology). We also probed the nitrocellulose membranes with actin-peroxidase conjugate (1:20,000 dilution; A3854, Sigma-Aldrich). The protein signals were detected using an enhanced chemiluminescence solution (SuperSignal™ West Femto; Thermo Fisher Scientific, Inc.), and images were obtained using the ImageQuant LAS 4000 system (GE Healthcare Biosciences).
Statistical analysis
We performed all statistical analyses using the GraphPad Prism 6.0 software (GraphPad Software). All other data are presented as the mean ± SD. For western blot analyses, we repeated each experiment three times. We used a one-way analysis of variance (ANOVA) followed by Bonferroni's multiple comparison test. P<0.05 was considered to indicate a statistically significant difference.
Results
Hypoxia triggered A2B induction is time-dependent in liver cancer cells
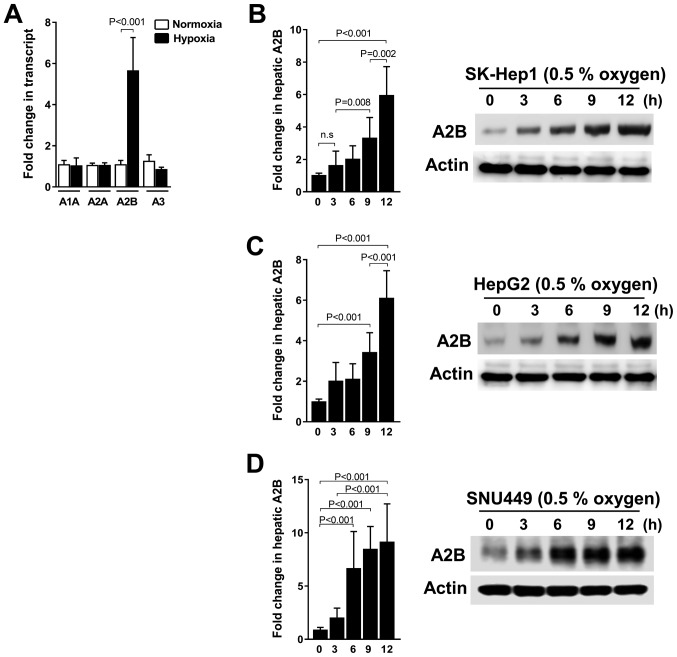
In this study, we investigated whether A2B is upregulated under hypoxic conditions or not. Thus, we grew three liver cancer cell lines, SK-Hep1, HepG2, and SNU449, under hypoxic conditions. We consistently detected the significant expression of only A2B mRNA levels compared to the expression of other receptor genes (A1, A2A, and A3) under low-oxygen conditions (Fig. 1A). As shown, the A2B mRNA level and protein expressions were expressed in a time-dependent manner during hypoxia in SK-Hep1 cells (Fig. 1B). Additionally, A2B is upregulated under hypoxic conditions in HepG2 (Fig. 1C) and SNU449 cells (Fig. 1D). These data reveal a selective induction of A2B during hypoxic conditions.
Figure 1.
Adenosine receptor isoform expression profiles in liver cancer cell lines and A2B induction following hypoxic growth of representative liver cancer cell lines. (A) Transcript expression profiles of adenosine receptor isoforms, including A1, A2A, A2B and A3, in liver cancer cell lines (SK-Hep1, HepG2 and SNU-449). (B) Transcriptional and protein expressions of A2B in liver cancer cell lines (B) SK-HEP1, (C) HepG2 the hepatoblastoma cell line and (D) SNU-449 grown under hypoxic (0.5% O2) or normal conditions. Transcriptional expression patterns and levels were detected using reverse transcription-quantitative PCR. The results were analyzed relative to GAPDH and according to fold changes relative to the control. For western blot analysis, actin was used as a loading control. Data are presented as the mean ± standard deviation.
HIF-1α directly regulated the expression of A2B by binding to the HRE in the A2B promoter region
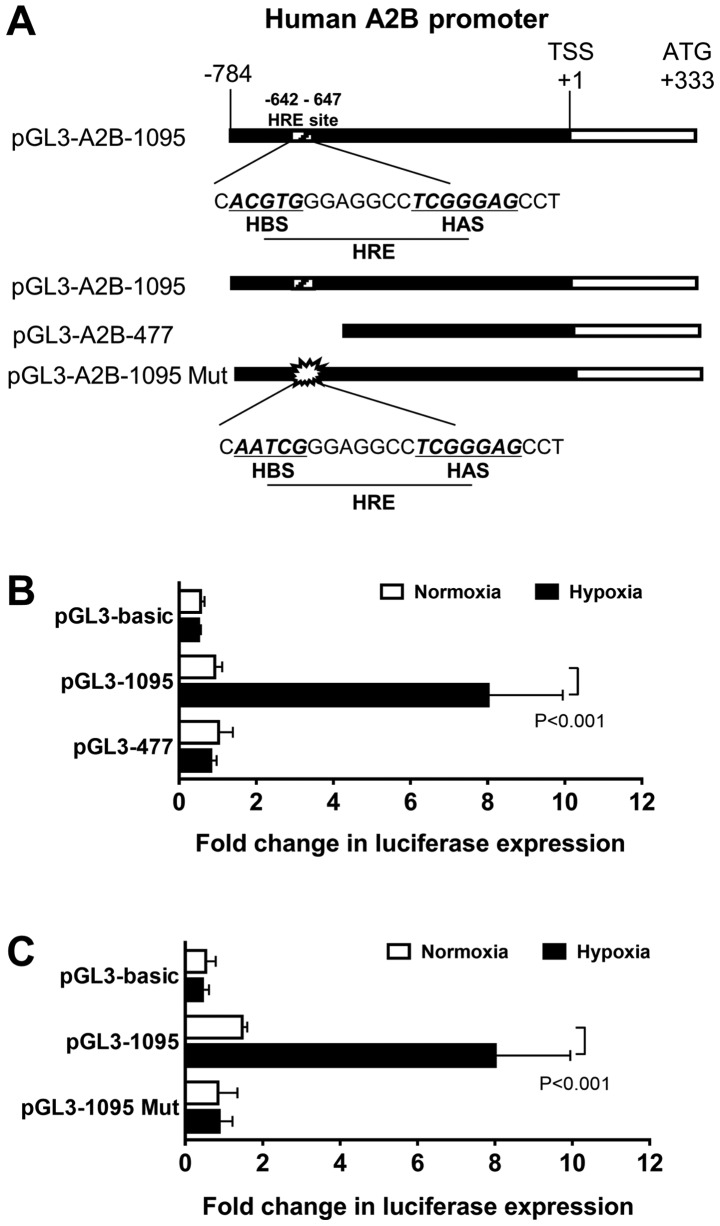
To verify if A2B is transcriptionally induced by HIF-1α, we first searched for the human A2B promoter region, which has an HRE, containing −642 to −647 bp, at the 5′-CACGTGG-3′ sequence and its typical HIF-1α ancillary HAS site (5′-CGGGGAG-3′) at −546 to −541 (Fig. 2A). To investigate the function of the HIF-1α binding site, we used two modified constructs, a full-length promoter construct (1095 bp) and a truncated construct (477 bp) of HIF-1α (Fig. 2B). As shown in the results, the full-length HRE construct-transfected cells had significantly higher activity than the truncated construct. Next, to define whether the full-length construct was acted upon by HIF-1α, we performed luciferase assays with site-directed mutagenesis constructs. The mutant form of this construct had an HRE AATCG change from ACGTG (Fig. 2C). The mutant construct (1095 Mut) exhibited low luciferase assay activity levels, as hypothesized. These results confirmed the A2B promoter region has an HRE site in it.
Figure 2.
Luciferase reporter assays were performed to analyze A2B transcriptional activity under hypoxic conditions. (A) Map of luciferase reporter constructs indicating the position of HRE-related A2B promoter (−642 to −647). Relative positions of the HBS and HAS. (B) HepG2 cells were transfected with a full-length construct pGL3-1095 or a truncation construct pGL3-477 plasmid. The empty pGL3-basic vector was transfected as a control vector. All transfected cells were grown under 0.5% O2 hypoxia conditions for 12 h (P<0.05; n=5). Renilla expression was used to normalize all luciferase expressions. (C) For site-directed mutagenesis, cells were transfected with a pGL3-1095 Mut construct and cultured under hypoxic conditions for 12 h. The luciferase assay activity was measured and normalized with Renilla (P<0.05; n=4). Data are expressed as the mean ± standard deviation. A2B, adenosine A2B receptor; HBS, HIF binding site; HAS, HIF ancillary site; Mut, mutant.
HIF-1α upregulates A2B expression during hypoxia
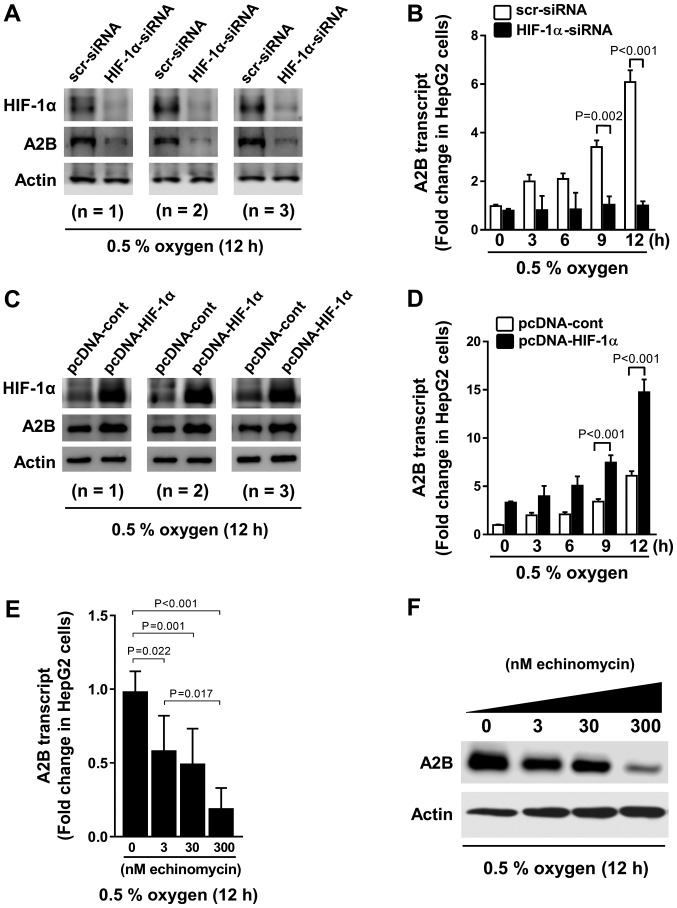
To confirm the HRE function within the promoter region of A2B, we performed experiments with HIF-1α siRNA and HIF-1α pcDNA plasmids. We transfected HIF-1α siRNA for 12 h (Fig. 3A) and then exposed the cells to hypoxia for different durations of time (Fig. 3B). HIF-1α siRNA-transfected cells displayed low A2B expression levels while HIF-1α is being absent. In contrast, the cells overexpressing HIF-1α exhibited high A2B expression levels (Fig. 3C) and hypoxia time dependency (Fig. 3D). Moreover, we treated cells with echinomycin (a cell-permeable inhibitor of HIF-1α-mediated gene transcription) and observed a dose-dependent decrease in the A2B mRNA and protein expression levels during low-oxygen conditions (Fig. 3E and F). Our results suggest HIF-1α is a transcriptional regulator of A2B.
Figure 3.
A2B expression profiles in liver cancer cell lines with gain- or loss- of HIF-1α function under hypoxic conditions. Western blot analyses of the expression of (A) HIF-1α and A2B and A2B transcript levels under hypoxic (0.5% oxygen) conditions for (B) up to 12 h in HepG2 cell lines. Cells were transfected with either scrambled-siRNA or HIF-1α-siRNA. Western blot analyses of (C) HIF-1α and A2B expression and A2B transcript levels under hypoxic (0.5% oxygen) conditions for (D) up to 12 h in HepG2 cell lines. Cells were transfected with either empty vector pcDNA3.1 (pcDNA-cont) or HIF-1α-pcDNA3.1 (pcDNA-HIF-1α) plasmid. (E) Cells were treated with different doses of echinomycin, a HIF-1α chemical inhibitor, for (F) up to 12 h. The expression of A2B transcript and protein levels were analyzed using RT-qPCR and western blot analysis, respectively. Data are presented as the mean ± standard deviation (n=4). Actin was used as a loading control housekeeping gene for use in western blot analysis. GAPDH was used as the housekeeping gene for RT-qPCR. A2B, adenosine A2B receptor; HIF-1α, hypoxia inducible factor-1α; siRNA, small-interfering RNA; RT-q, reverse transcription-quantitative.
Silencing of A2B in liver cancer cell suppressed cell growth and proliferation
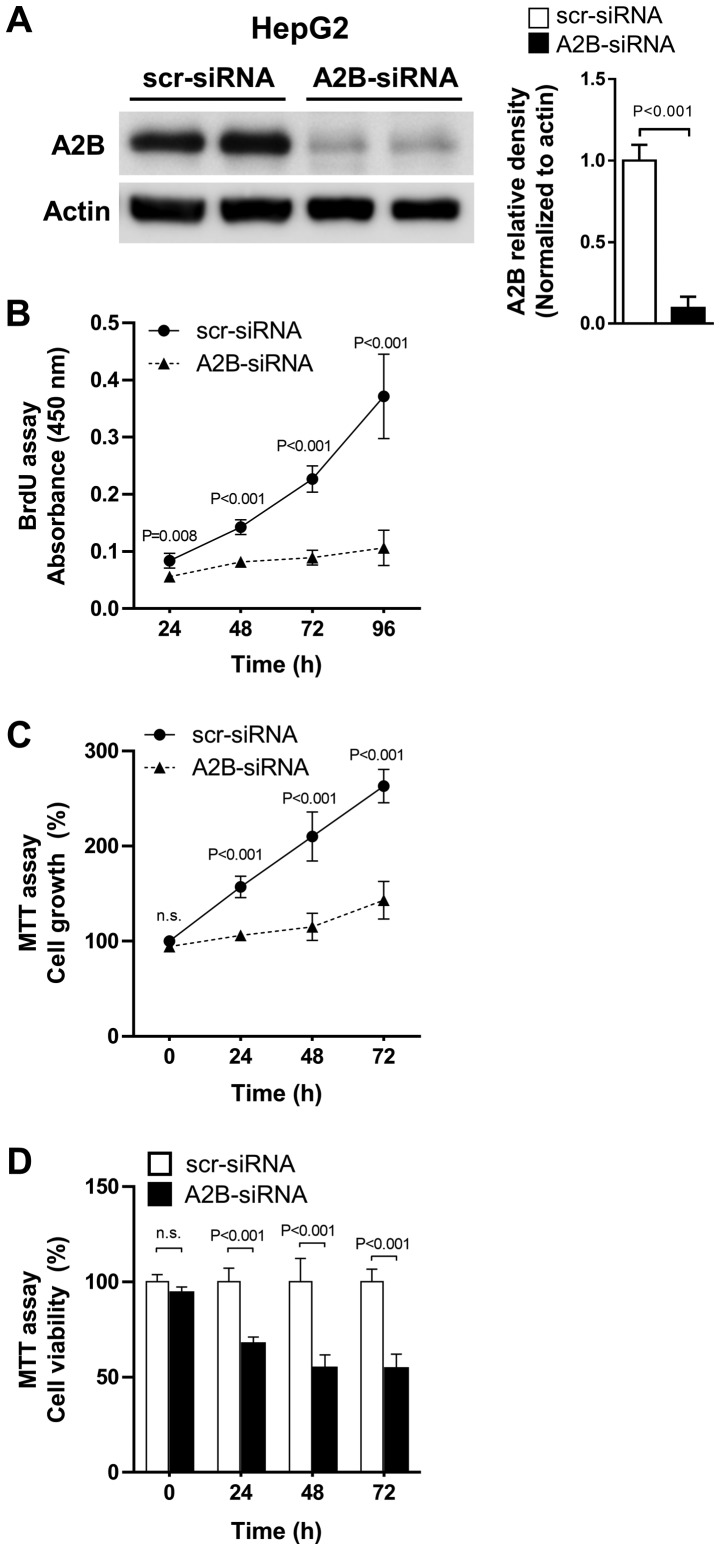
As increased expression of adenosine A2B was observed in liver cancer cells, we tested whether the direct knock-down of A2B expression could affect cell growth and proliferation in liver cancer cells. We transfected HepG2 (the hepatoblastoma cell line) cells with A2B-siRNA and maintained them for up to 96 h. The A2B-siRNA effectively inhibited A2B expressions compared to scr-siRNA (Fig. 4A). Treatment with A2B-siRNA for 24 to 96 h significantly (P<0.001) decreased cell proliferation, as revealed by the BrdU assay (Fig. 4B). Similarly, the MTT assay also showed that cell growth was greatly decreased when cells were treated with A2B-siRNA (Fig. 4C). Additionally, the relative cell viabilities were significantly (P<0.001) lowered after transfection of cells with A2B-siRNA (Fig. 4D). As A2B deficient liver cancer cells show consistent decrease in cell growth and proliferation, we could conclude that A2B takes an important role in liver cancer cell proliferation.
Figure 4.
siRNA-mediated A2B silencing inhibits HepG2 cell proliferation. (A) HepG2 cells were transfected with either scr-siRNA or A2B-siRNA. Transfection efficiency of A2B-siRNA was measured using western blot analysis. Actin was used as a loading control. (B) Cell proliferation was determined by measuring BrdU incorporation for up to 96 h (n=4). (C) Relative cell growth rates were examined by the MTT assay. (D) Cell viability of cells at each time-point was measured by MTT assay. Data are presented as the mean ± standard deviation (n=4). Cells transfected with scr-siRNA was used as control and P<0.05 was considered to indicate a statistically significant result. siRNA, small-interfering; A2B, adenosine A2B receptor; scr, scrambled.
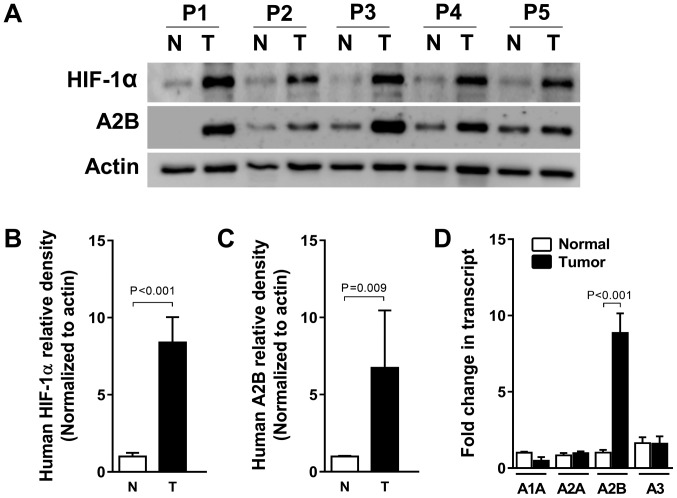
HIF-1α levels are associated with A2B transcript and protein levels in human liver cancer specimens
Solid tumors, including liver cancer, usually experience hypoxic conditions due to the fast growth (22). To confirm the relationship between adenosine A2B receptor expression and human liver cancer, we tested A2B induction from both tumor tissues and the adjacent normal tissues came from identical individuals diagnosed as a liver cancer. As expected, HIF-1α protein expression was high in the five human liver cancer specimens compared to its expression in normal tissues. A2B protein expression levels were also increased, suggesting that HIF-1α correlates with A2B in tumors, according to the fold changes of HIF-1α and A2B densitometry results (Fig. 5A-C). Collectively, HIF-1α, as a master transcription factor, can bind to the A2B promoter region and induce expression of A2B, thereby promoting tumor growth and survival. Moreover, we also confirmed the higher level of A2B gene expression in the HCC specimens by qRT-PCR. By contrast, we found no associations between the expression levels of mRNAs of A1, A2A, or A3 between normal and HCC specimens (Fig. 5D).
Figure 5.
Protein levels of A2B and HIF-1α measured in human HCC specimens and in non-cancerous tissue specimens, and mRNA levels of A2B isoforms. (A) HIF-1α and A2B protein expression were detected using western blot analysis. Specimens from five patients (P1-P5) who underwent HCC hepatectomy and non-cancerous tissue specimens around the tumors. Actin was used as a loading control. Relative expression of (B) HIF-1α and (C) A2B by densitometric quantification of western blots. (D) mRNA levels of A2B and other adenosine isoform receptors from five patients. The expression levels are presented relative to those of the control housekeeping gene GAPDH. The results are presented as the mean ± standard deviation; P<0.05; Student's t-test. A2B, adenosine A2B receptor; HIF-1α, hypoxia inducible factor-1α; HCC, hepatocellular carcinoma; T, HCC hepatectomy; N, non-cancerous tissue specimens.
Discussion
In this study, we defined the various expression patterns of adenosine receptor isoforms from three different liver cancer cell lines and human liver cancer specimens. We found that the A2B expression patterns were persistently higher than those of A1, A2A, and A3 receptor subtypes in liver cancer cell lines when maintained under low-oxygen conditions. Those patterns were also observed in human liver cancers. Under the hypothesis that hypoxia-mediated HIF-1α activation leads to liver cancer cell proliferation by activating adenosine A2B receptors, we followed transcription factor binding assays utilizing A2B promoter constructs and constructs with site-directed mutagenesis and have identified HIF-1α as the key regulator of A2B-induction during hypoxic conditions. We also investigated loss- and gain-of-function using HIF-1α siRNA and overexpressing vectors. We found that the expression levels of A2B were modulated HIF-1α-dependently. In addition to this, echinomycin treatment dose-dependently inhibited A2B expressions under hypoxia, which indicate transcriptional induction of A2B by HIF-1α. We also showed that active deprivation of A2B expression by siRNA suppresses cell growth and proliferation of the hepatoblastoma cell line, HepG2. Those results indicate the activation of hepatic A2B signals during low oxygen condition is related to liver cancer cell proliferation. Consistent with our in vitro results, human liver cancer specimens showed elevated levels of HIF-1α along with A2B in both transcript and protein levels. These data implicate HIF-1α in the regulation of adenosine-elicited cancer growth in liver.
We have previously studied the HIF-1α stabilization during hypoxic conditions in liver cell lines (23) and identified the protective role of HIF-1α stabilization mediated by hypoxia-induced inhibition of succinate dehydrogenase, concomitant increases in glycolytic capacity, and improved tricarboxylic acid (TCA) cycle function in alveolar epithelium (24). Also the pharmacologic studies with HIF activator or inhibitor treatment implicated HIF-1α-stabilization increases in liver cancer proliferation (25).
On the other hand, studies have shown the key roles of adenosine in HCC cell proliferation (26). Adenosine also upregulates endothelial cell proliferation by associating with A2B in porcine and rat arterial endothelial cells (27). In retinal endothelial cells, activated A2B can initiate neovascularization through increased angiogenic growth factor expression (28). In contrast, adenosine inhibits the growth of cardiac fibroblasts and aortic and vascular smooth muscle cells through the activation of A2B, in addition to increasing the proliferation of peripheral micro-vessels, but exerts the opposite effect on cardiac tissue and capillaries (29). Thus, A2B can promote cancer proliferation and expansion by initiating neovascularization around cancer masses (30).
We showed that A2B is induced under hypoxic conditions. In general, solid cancers, such as breast, lung, colon, and prostate cancers undergo similar low-oxygen growth conditions (31); moreover, to maintain homeostasis under low-oxygen supply conditions, specific genes need to be systematically activated and expressed or inactivated.
It was previously reported that bleomycin, a chemical irritant which induce HIF-1α, can mediate A2B receptor expression while inducing adenosine accumulation in acute lung injury (32). In addition, HIF-1α mediated expression of A2B in endothelial cells (15), lung injury (18), breast cancer (19), and dendritic cells (33) has been reported. Nevertheless, the hypoxia mediated A2B signaling mechanism promoting liver cancer cell proliferation is yet to be elucidated.
Other researchers have suggested that adenosine receptors are highly controlled and, thus, adenosine responses might be regulated by the surface expression of adenosine receptors. Consistent with our study, Eltzschig et al reported that microarray analyses of cDNA derived from endothelial cells subjected to various periods of hypoxia revealed significant changes in the adenosine receptor's profiles, wherein the prominent phenotypic change favored A2B expression with concomitant downregulation of A1 and A2 subtypes (34). In another study, Feoktistov and Biaggioni suggested that hypoxia increases A2B expression levels releasing vascular endothelial growth factor through decreasing adenosine (35). Human endothelial and smooth muscle cells have also been shown to express A2B (36). Moreover, hypoxia reduces matrix metalloproteinase-9 (MMP-9) production by human monocyte-derived dendritic cells and requires the activation of adenosine A2B via the cAMP/protein kinase (PKA) signaling pathway (37,38). Collectively, these data suggest that A2B increases HIF-1α dependently under hypoxic conditions and might promote liver cancer cell proliferation.
In summary, we investigated the expressions of A2B and HIF-1α in liver cancer cells to find an association suggesting that under hypoxic conditions, HIF-1α induces A2B expression to maintain tumor proliferation. Our results show that the upregulated A2B plays a critical role in liver cancer cell growth, suggesting A2B as a potential target for drug design; thus, an effective A2B inhibitor might be useful for treating liver cancers.
Acknowledgements
The authors would like to thank Dr Holger K. Eltzschig (University of Texas Health Science Center at Houston, TX, USA) for providing the human A2B-Luc reporter construct.
Glossary
Abbreviations
- HCC
hepatocellular carcinoma
- HIF-1α
hypoxia inducible factor-1α
- HRE
hypoxia response element
- A2B
adenosine A2B receptor
Funding
The present study was supported by Asan Institute for Life Sciences (grant no. 15-662 and 18-IT0622), the National Research Foundation of Korea (grant no. NRF-2015K1A4A3046807), the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (grant no. NRF-2017R1D1A1B04032429) and the Yuhan Corporation (grant no. 2015-0908) and a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant no. HI15C0972).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
JHK and ET designed the study. JL and JK conducted the experiments. JHK, GWS, YIY, SH, GCP and SGL performed human specimen handling. YHJ, VAK, BJK and NK contributed to the interpretation of the results. GCP and ET were involved in manuscript writing and analyzing the data. SGL, GCP and ET supervised the work. All authors provided critical feedback and conducted the research, analyzed the data and prepared the manuscript.
Ethics approval and consent to participate
The Institutional Review Board (IRB) of Asan Medical Center (Seoul, Republic of Korea) reviewed and approved the collection and use of patient specimens (approval no. 2016-0582). All patients who provided tissue samples agreed to donate their specimens and provided written informed consents.
Patient consent for publication
No applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380:1450–1462. doi: 10.1056/NEJMra1713263. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends-an update. Cancer Epidemiol Biomarkers Prev. 2016;25:16–27. doi: 10.1158/1055-9965.EPI-15-0578. [DOI] [PubMed] [Google Scholar]
- 3.Collis MG, Hourani SM. Adenosine receptor subtypes. Trends Pharmacol Sci. 1993;14:360–366. doi: 10.1016/0165-6147(93)90094-Z. [DOI] [PubMed] [Google Scholar]
- 4.Haskó G, Pacher P, Sylvester Vizi E, Illes P. Adenosine receptor signaling in the brain immune system. Trends Pharmacol Sci. 2005;26:511–516. doi: 10.1016/j.tips.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haskó G, Linden J, Cronstein B, Pacher P. Adenosine receptors: Therapeutic aspects for inflammatory and immune diseases. Nat Rev Drug Discov. 2008;7:759–770. doi: 10.1038/nrd2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blay J, White TD, Hoskin DW. The extracellular fluid of solid carcinomas contains immunosuppressive concentrations of adenosine. Cancer Res. 1997;57:2602–2605. [PubMed] [Google Scholar]
- 7.Cekic C, Sag D, Li Y, Theodorescu D, Strieter RM, Linden J. Adenosine A2B receptor blockade slows growth of bladder and breast tumors. J Immunol. 2012;188:198–205. doi: 10.4049/jimmunol.1101845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panjehpour M, Castro M, Klotz KN. Human breast cancer cell line MDA-MB-231 expresses endogenous A2B adenosine receptors mediating a Ca2+ signal. Br J Pharmacol. 2005;145:211–218. doi: 10.1038/sj.bjp.0706180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma DF, Kondo T, Nakazawa T, Niu DF, Mochizuki K, Kawasaki T, Yamane T, Katoh R. Hypoxia-inducible adenosine A2B receptor modulates proliferation of colon carcinoma cells. Hum Pathol. 2010;41:1550–1557. doi: 10.1016/j.humpath.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 10.Vecchio EA, Tan CY, Gregory KJ, Christopoulos A, White PJ, May LT. Ligand-independent adenosine A2B receptor constitutive activity as a promoter of prostate cancer cell proliferation. J Pharmacol Exp Ther. 2016;357:36–44. doi: 10.1124/jpet.115.230003. [DOI] [PubMed] [Google Scholar]
- 11.Xiang HJ, Liu ZC, Wang DS, Chen Y, Yang YL, Dou KF. Adenosine A(2b) receptor is highly expressed in human hepatocellular carcinoma. Hepatol Res. 2006;36:56–60. doi: 10.1016/j.hepres.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Kolachala V, Asamoah V, Wang L, Obertone TS, Ziegler TR, Merlin D, Sitaraman SV. TNF-alpha upregulates adenosine 2b (A2b) receptor expression and signaling in intestinal epithelial cells: A basis for A2bR overexpression in colitis. Cell Mol Life Sci. 2005;62:2647–2657. doi: 10.1007/s00018-005-5328-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xaus J, Mirabet M, Lloberas J, Soler C, Lluis C, Franco R, Celada A. IFN-gamma up-regulates the A2B adenosine receptor expression in macrophages: A mechanism of macrophage deactivation. J Immunol. 1999;162:3607–3614. [PubMed] [Google Scholar]
- 14.Eltzschig HK, Ibla JC, Furuta GT, Leonard MO, Jacobson KA, Enjyoji K, Robson SC, Colgan SP. Coordinated adenine nucleotide phosphohydrolysis and nucleoside signaling in posthypoxic endothelium: Role of ectonucleotidases and adenosine A2B receptors. J Exp Med. 2003;198:783–796. doi: 10.1084/jem.20030891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong T, Westerman KA, Faigle M, Eltzschig HK, Colgan SP. HIF-dependent induction of adenosine A2B receptor in hypoxia. FASEB J. 2006;20:2242–2250. doi: 10.1096/fj.06-6419com. [DOI] [PubMed] [Google Scholar]
- 16.Stolze IP, Mole DR, Ratcliffe PJ. Regulation of HIF: Prolyl hydroxylases. Novartis Found Symp. 2006;272:15–36. [PubMed] [Google Scholar]
- 17.Semenza GL. HIF-1: Upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20:51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eckle T, Kewley EM, Brodsky KS, Tak E, Bonney S, Gobel M, Anderson D, Glover LE, Riegel AK, Colgan SP, Eltzschig HK. Identification of hypoxia-inducible factor HIF-1A as transcriptional regulator of the A2B adenosine receptor during acute lung injury. J Immunol. 2014;192:1249–1256. doi: 10.4049/jimmunol.1100593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan J, Lu H, Samanta D, Salman S, Lu Y, Semenza GL. Hypoxia-inducible factor 1-dependent expression of adenosine receptor 2B promotes breast cancer stem cell enrichment. Proc Natl Acad Sci USA. 2018;115:E9640–E9648. doi: 10.1073/pnas.1809695115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.López-Terrada D, Cheung SW, Finegold MJ, Knowles BB. Hep G2 is a hepatoblastoma-derived cell line. Hum Pathol. 2009;40:1512–1515. doi: 10.1016/j.humpath.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 21.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C (T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 22.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tak E, Lee S, Lee J, Rashid MA, Kim YW, Park JH, Park WS, Shokat KM, Ha J, Kim SS. Human carbonyl reductase 1 upregulated by hypoxia renders resistance to apoptosis in hepatocellular carcinoma cells. J Hepatol. 2011;54:328–339. doi: 10.1016/j.jhep.2010.06.045. [DOI] [PubMed] [Google Scholar]
- 24.Eckle T, Brodsky K, Bonney M, Packard T, Han J, Borchers CH, Mariani TJ, Kominsky DJ, Mittelbronn M, Eltzschig HK. HIF1A reduces acute lung injury by optimizing carbohydrate metabolism in the alveolar epithelium. PLoS Biol. 2013;11:e1001665. doi: 10.1371/journal.pbio.1001665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 26.Tak E, Jun DY, Kim SH, Park GC, Lee J, Hwang S, Song GW, Lee SG. Upregulation of P2Y2 nucleotide receptor in human hepatocellular carcinoma cells. J Int Med Res. 2016;44:1234–1247. doi: 10.1177/0300060516662135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubey RK, Gillespie DG, Jackson EK. A(2B) adenosine receptors stimulate growth of porcine and rat arterial endothelial cells. Hypertension. 2002;39:530–535. doi: 10.1161/hy0202.103075. [DOI] [PubMed] [Google Scholar]
- 28.Grant MB, Tarnuzzer RW, Caballero S, Ozeck MJ, Davis MI, Spoerri PE, Feoktistov I, Biaggioni I, Shryock JC, Belardinelli L. Adenosine receptor activation induces vascular endothelial growth factor in human retinal endothelial cells. Circ Res. 1999;85:699–706. doi: 10.1161/01.RES.85.8.699. [DOI] [PubMed] [Google Scholar]
- 29.Mustafa SJ, Morrison RR, Teng B, Pelleg A. Adenosine Receptors in Health and Disease. Springer; 2009. Adenosine receptors and the heart: Role in regulation of coronary blood flow and cardiac electrophysiology; pp. 161–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merighi S, Mirandola P, Varani K, Gessi S, Leung E, Baraldi PG, Tabrizi MA, Borea PA. A glance at adenosine receptors: Novel target for antitumor therapy. Pharmacol Ther. 2003;100:31–48. doi: 10.1016/S0163-7258(03)00084-6. [DOI] [PubMed] [Google Scholar]
- 31.Hockel M, Vaupel P. Tumor hypoxia: Definitions and current clinical, biologic, and molecular aspects. J Natl Cancer Inst. 2001;93:266–276. doi: 10.1093/jnci/93.4.266. [DOI] [PubMed] [Google Scholar]
- 32.Volmer JB, Thompson LF, Blackburn MR. Ecto-5′-nucleotidase (CD73)-mediated adenosine production is tissue protective in a model of bleomycin-induced lung injury. J Immunol. 2006;176:4449–4458. doi: 10.4049/jimmunol.176.7.4449. [DOI] [PubMed] [Google Scholar]
- 33.Yang M, Ma C, Liu S, Shao Q, Gao W, Song B, Sun J, Xie Q, Zhang Y, Feng A, et al. HIF-dependent induction of adenosine receptor A2b skews human dendritic cells to a Th2-stimulating phenotype under hypoxia. Immunol Cell Biol. 2010;88:165–171. doi: 10.1038/icb.2009.77. [DOI] [PubMed] [Google Scholar]
- 34.Eltzschig HK, Eckle T, Mager A, Küper N, Karcher C, Weissmüller T, Boengler K, Schulz R, Robson SC, Colgan SP. ATP release from activated neutrophils occurs via connexin 43 and modulates adenosine-dependent endothelial cell function. Circ Res. 2006;99:1100–1108. doi: 10.1161/01.RES.0000250174.31269.70. [DOI] [PubMed] [Google Scholar]
- 35.Feoktistov I, Biaggioni I. Pharmacological characterization of adenosine A2B receptors: Studies in human mast cells co-expressing A2A and A2B adenosine receptor subtypes. Biochem Pharmacol. 1998;55:627–633. doi: 10.1016/S0006-2952(97)00512-1. [DOI] [PubMed] [Google Scholar]
- 36.Igata M, Motoshima H, Tsuruzoe K, Kojima K, Matsumura T, Kondo T, Taguchi T, Nakamaru K, Yano M, Kukidome D, et al. Adenosine monophosphate-activated protein kinase suppresses vascular smooth muscle cell proliferation through the inhibition of cell cycle progression. Circ Res. 2005;97:837–844. doi: 10.1161/01.RES.0000185823.73556.06. [DOI] [PubMed] [Google Scholar]
- 37.Zhao P, Li XG, Yang M, Shao Q, Wang D, Liu S, Song H, Song B, Zhang Y, Qu X. Hypoxia suppresses the production of MMP-9 by human monocyte-derived dendritic cells and requires activation of adenosine receptor A2b via cAMP/PKA signaling pathway. Mol Immunol. 2008;45:2187–2195. doi: 10.1016/j.molimm.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Chen H, Koupenova M, Yang D, Sume SS, Trackman PC, Ravid K. Regulation of MMP-9 expression by the A2b adenosine receptor and its dependency on TNF-α signaling. Exp Hematol. 2011;39:525–530. doi: 10.1016/j.exphem.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.