Abstract
The present study aimed to investigate differentially expressed genes (DEGs) in whole blood (WB) obtained from patients with lumbar disc prolapse (LDP) and healthy volunteers. A total of 8 patients with LDP and 8 healthy volunteers were recruited. An Agilent SurePrint G3 human gene expression microarray 8×60 K was used to perform the microarray analyses. R was employed to identify DEGs, which were then subjected to bioinformatics analysis, including a Gene Ontology (GO) analysis, Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway analysis and protein-protein interaction (PPI) network analysis. DEGs in the degenerative annulus fibrosis (AF) and nucleus pulposus (NP) compared with non-degenerative tissues were also identified based on microarray data and the intersections of the three were assessed. Furthermore, reverse transcription-quantitative (RT-q)PCR was performed to confirm the aberrant expression levels of selected DEGs in the WB of all subjects. A total of 161 DEGs between LDP patients and the healthy controls were identified (128 upregulated and 33 downregulated). These DEGs were enriched in 293 biological process, 36 cellular component and 21 molecular function GO terms, as well as in 24 KEGG pathways. The PPI network contained 4 submodules, and Toll-like receptor 4 had the highest degree centrality. A total of 22 DEGs were common to the three groups of DEGs. The RT-qPCR assay confirmed that the expression levels of cytochrome P450 family 27 subfamily A member 1, superoxide dismutase 2, protein disulfide isomerase family A member 4, FKBP prolyl isomerase 11 and ectonucleotide pyrophosphatase/phosphodiesterase 4 were significantly different between the patient group and the volunteer group. In conclusion, several genes were identified as potential biomarkers in WB that should be further explored in future studies to determine their potential application in the clinical treatment and diagnosis of LDP, and the present bioinformatics analysis revealed several GO terms, KEGG pathways and submodules of the PPI network that may be involved in LDP, although the exact mechanisms remain elusive.
Keywords: lumbar disc prolapse, differentially expressed genes, whole blood, enrichment analysis, protein-protein interaction network, reverse transcription-quantitative PCR
Introduction
Lumbar disc prolapse (LDP) is a common type of intervertebral disk degeneration disease (DDD) caused by intervertebral disc degeneration (IDD). LDP results in serious lower back pain and sciatia, causes short-term disability and affects ~5% of the public population (1).
The molecular biological mechanism of DDD has been studied for numerous years, with the primary focus on IDD (2–7), and substantial progress has been made in the pathophysiology of IDD (8–15). However, the specific mechanisms underlying LDP, a specific type of DDD, have rarely been studied individually, and almost all studies on IDD are based on the analysis of intervertebral disc (IVD) tissues. The IVD is a complex structure containing annulus fibrosis (AF) and nucleus pulpous (NP), and the pathophysiological processes that occur in these two components during degeneration are distinctly different. Therefore, completely distinguishing between the two is difficult when collecting IVD tissue. Thus, the etiopathogeneses of LDP and IDD remain incompletely understood. The current limitation in the understanding of the pathogenesis has restricted the clinical treatment of this condition and obtaining a more comprehensive interpretation of the mechanisms underlying LDP is therefore critical for the development of successful therapeutic strategies.
To date, only one study has explored the gene expression profile of peripheral blood mononuclear cells obtained from patients with IDD and limited data are available on the transcriptome characteristics of whole blood (WB) in either IDD or LDP. Thus, in the present study, microarray analysis was used to investigate the mRNA transcriptome characteristics of WB obtained from LDP patients. Differentially expressed genes (DEGs) in WB (WB-DEGs) were identified and subjected to functional analyses. DEGs in the degenerative AF (AF-DEGs) and NP (NP-DEGs) compared with non-degenerative tissues were also identified based on microarray data and the intersections of the WB-DEGs, AF-DEGs and NP-DEGs were evaluated. Reverse transcription-quantitative (RT-q)PCR assays were also performed using WB from the same 8 IDD patients and 8 healthy volunteers to measure the expression levels of selected WB-DEGs and DEGs common to WB-DEGs, AF-DEGs and NP-DEGs (common DEGs).
Materials and methods
Human WB collection and ethics statement
A total of 8 patients, including 4 males and 4 females aged between 33 and 60 years, as well as 8 volunteers, including 4 males and 4 females aged between 19 and 23 years, were recruited for the present study. For the patient group, the inclusion criteria were as follows: Age of 35–60 years; severe lower back pain and sciatica within 4 weeks; LDP confirmed by MRI; and no drug use within 3 months. The development of LDP is based on IDD, which is an age-related process, with the degree of histopathologic degeneration increasing with age. All the patients in the present study were diagnosed with LDP following MRI examination. Previous studies have not described the degree of histopathologic degeneration that may be observed in IDD via MRI. LDP is a common disease in middle-aged and elderly people and therefore, all patients recruited into the present study were aged between 35–60 years. However, control group patients were not age matched, as older individuals may have exhibited more degeneration, making it harder to compare those with the disease to those that were healthy. Younger controls were therefore selected for the current study. Thus, the expression of certain genes associated with LDP may not be statistically different between the two groups. Young people (<24 years) are therefore more representative of those without IDD or low IDD. Tsai et al (16) compared specimens obtained from young and elderly individuals to investigate gene expression in IDD. The current study therefore recruited young people as controls, with all volunteers attending college. The inclusion criteria were as follows: Age between 18 and 25 years; never experienced lower back pain or sciatica; and no drug use within 3 months. For the two groups, the exclusion criteria were as follows: Other types of spinal disease, deformities, trauma, surgery, basic metabolic disease, rheumatism, congenital disease or congenital disability, tuberculosis and tumors. A total of 10 ml of fasting WB was collected from the left medial cubital vein of each participant between 7:00 and 7:30 a.m. All WB samples were immediately incubated in a PAX gene Blood RNA tube (BD Biosciences) for 36–72 h at −20°C and then sent to Shanghai Bohao Biotechnology Co., Ltd. for gene chip hybridization screening.
All procedures described in the present study were authorized by the Ethics Committee of the Sichuan Provincial Orthopedic Hospital (Chengdu), and all patients and donors enrolled in the study provided written informed consent to participate in the study. The WB samples were collected between April 2018 and August 2018 at Sichuan Provincial Orthopedic Hospital (Chengdu).
Microarray analysis
Chip scanning was accomplished on an Agilent Microarray Scanner platform (Agilent Technologies, Inc.). Gene chip hybridization screening was performed between September 2018 and October 2018 using an Agilent SurePrint G3 human gene expression microarray 8×60 K at Shanghai Biotechnology Co., Ltd. following the standard protocol of Agilent Technologies, Inc. The complete data sets containing the gene expression profiles of the WB samples were uploaded to the gene expression omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) and are accessible under the accession no. GSE124272.
Identification of WB-DEGs
The raw data obtained in the chip scan were normalized using the limma package in R. To identify WB-DEGs, the Quantile algorithm was used, and the signal value was log2-normalized. Student's t-test was performed with a threshold P-value of 0.05 and absolute fold change (FC) of 2 to identify WB-DEGs. For genes with multiple probes, the average FC value was used.
Enrichment analysis
The enrichment analysis included Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. The method adopted in the present study was Fisher's exact test and the clusterProfiler package from R/bioconductor was used to perform the enrichment analysis. The criteria selected were the number of genes that were enriched in a certain term/GO term ≥2 and a P-value <0.05. The most significant terms/pathways obtained by the enrichment analysis were ranked according to the enrichment factor calculated as follows: Enrichment factor = (number of DEGs in a term/total number of DEGs)/(total number of genes in the database in a term/total number of genes in the database).
Protein-protein interaction (PPI) network and module analysis
DEGs were mapped using the Search Tool for the Retrieval of Interacting Genes (STRING; http://string-db.org) using the default instructions. Disconnected nodes in the network were excluded. A PPI network of nodes with a combined score >0.4 was then constructed using Cytoscape software (V3.6.1; http://www.cytoscape.org/). Degree centrality was calculated with the plug-in CentiScaPe and the plug-in MCODE was applied to filter out significant submodules according to the following criteria: ‘degree cutoff = 2’, ‘node score cutoff = 0.2’, ‘k-core = 2’ and ‘max depth = 100’.
IVD microarray data
The microarray dataset GSE70362 based on the Affymetrix GPL17810 platform [HG-U133_Plus_2] was downloaded from the GEO database. This dataset contains gene expression data for 24 AF and 24 NP samples from the post-mortem tissues from 19 donors aged between 21 and 82 years. All samples were collected from T12/L1 to L4/L5 at <12 h after declaring brain death. The original signal intensity data in CEL-format files and classification based on the Thompson grading system (17) were downloaded for analysis for the present study. To annotate the data, the original probe IDs were transformed into gene symbols. Any unidentified probes were discarded. For genes with multiple probes, the average FC value was used. Within each set of 24 samples (AF or NP), 8 samples classified as Thompson grade I or I–II were considered non-degenerative, and 16 samples classified as Thompson grades II to V were considered degenerative.
Identification of common DEGs
GeneSpring GX 11.5 software (Agilent Technologies, Inc.) was employed to identify the AF-DEGs and NP-DEGs. The initial data were pre-processed by using the Robust Multi-array Average procedure. Paired t-tests were performed with a threshold P-value of 0.05 and FC of 1.5 to identify AF-DEGs and NP-DEGs. When taking the intersection of the three, WB-DEGs with FC>1.5 were filtered.
Total RNA extraction, complementary (c)DNA synthesis and RT-qPCR
Total RNA was extracted from blood cells and purified using a PX Blood RNA Kit (Omega Bio-Tek, Inc.) following the manufacturer's protocol. Each RNA sample was reverse-transcribed into cDNA using a ReverTra Ace qPCR Kit (Toyobo) and the cDNA was subsequently amplified using real-time qPCR. Gene expression levels were quantified using a 7500 HT Sequence Detection System with Power SYBR Green PCR Master Mix (both from Applied Biosystems; Thermo Fisher Scientific, Inc.). Primers for specific genes were designed by Primer Express (V3.0.1 http://www.thermofisher.com) and synthesized by Sangon Biotech Co. All forward and reverse primers used for qPCR are listed in Table SI. The thermocycling conditions consisted of an initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 10 sec, 60°C for 1 min, and a final extension at 60°C for 1 min. The β-actin gene was used as an internal control, and gene expression levels were normalized to those of β-actin according to the 2−ΔΔCq method (17).
Statistical analysis
The RT-qPCR data are expressed as the mean ± standard deviation. Statistical analyses were performed using Student's t-test in GraphPad Prism 6.0 (GraphPad Software Inc.). P<0.05 was considered to indicate statistical significance.
Results
WB-DEGs
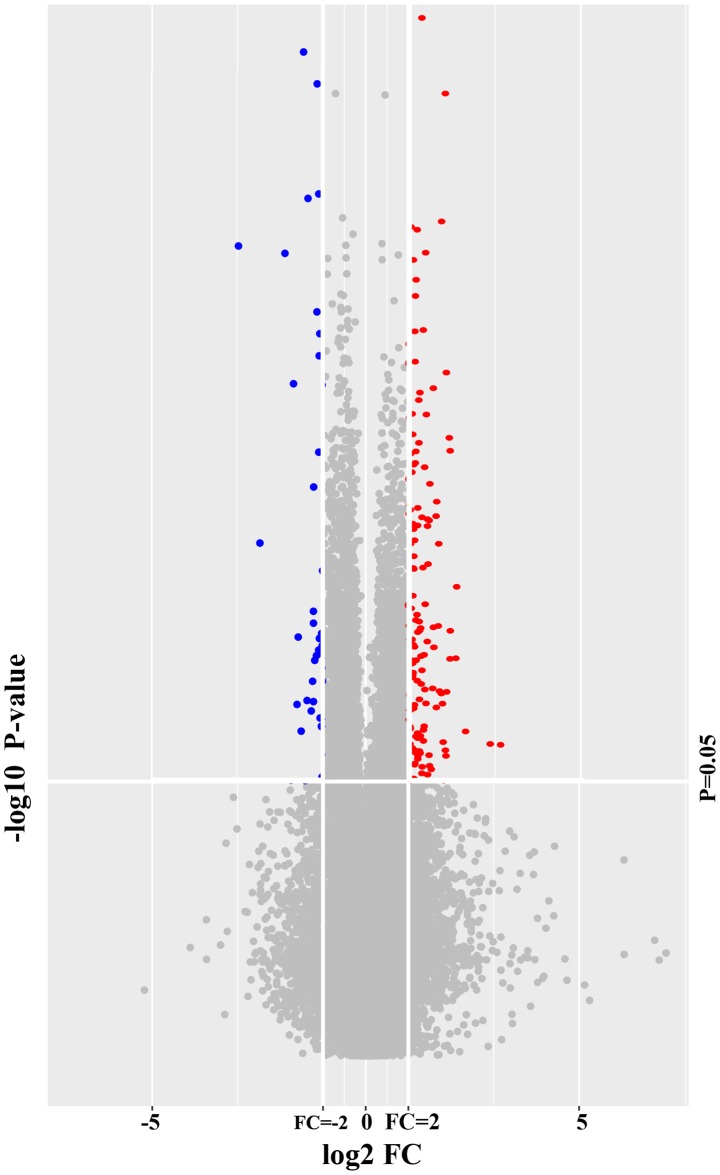
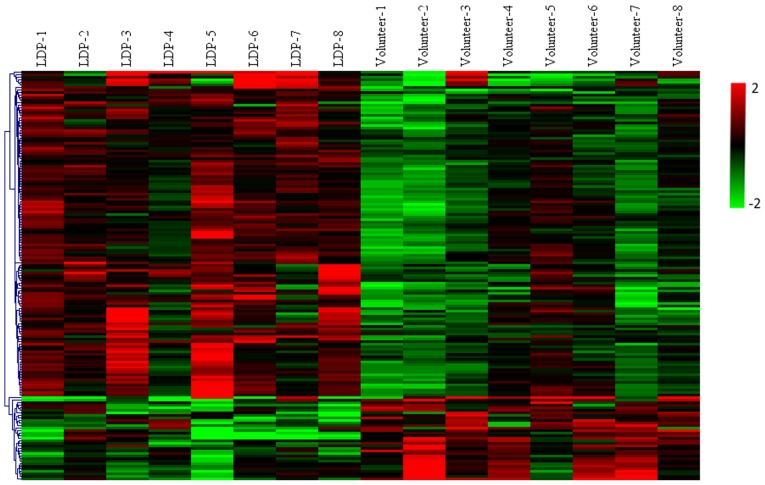
A total of 58,341 probe sets corresponding to 34,758 genes were obtained using the Agilent SurePrint G3 human gene expression microarray 8×60 K. A total of 161 genes in WB were identified to be differentially expressed between LDP patients and healthy volunteers with P<0.05 and FC>2. All of these DEGs are listed in Table SII. These DEGs represented 0.28% of the total transcriptome. A total of 128 WB-DEGs were upregulated and 33 were downregulated. A volcano plot (Fig. 1) and an expression heat map (Fig. 2) were constructed for the DEGs identified.
Figure 1.
Volcano plot indicating that a total of 161 genes in the whole blood were differentially expressed between the patients with lumbar disk prolapse and healthy volunteers (P<0.05 and FC>2), including 128 upregulated genes and 33 downregulated genes (red, upregulated; blue, downregulated). FC, fold change.
Figure 2.
Heatmap displaying hierarchical clustering of the expression of the 161 differentially expressed genes in whole blood (P<0.05; absolute fold change >2) between the LDP patient group and the healthy volunteer group, including 128 upregulated and 33 downregulated genes. The original data were normalized using the z-score to indexes between −2 and 2. Red, upregulated; green, downregulated. LDP, lumbar disk prolapse.
Enrichment analysis
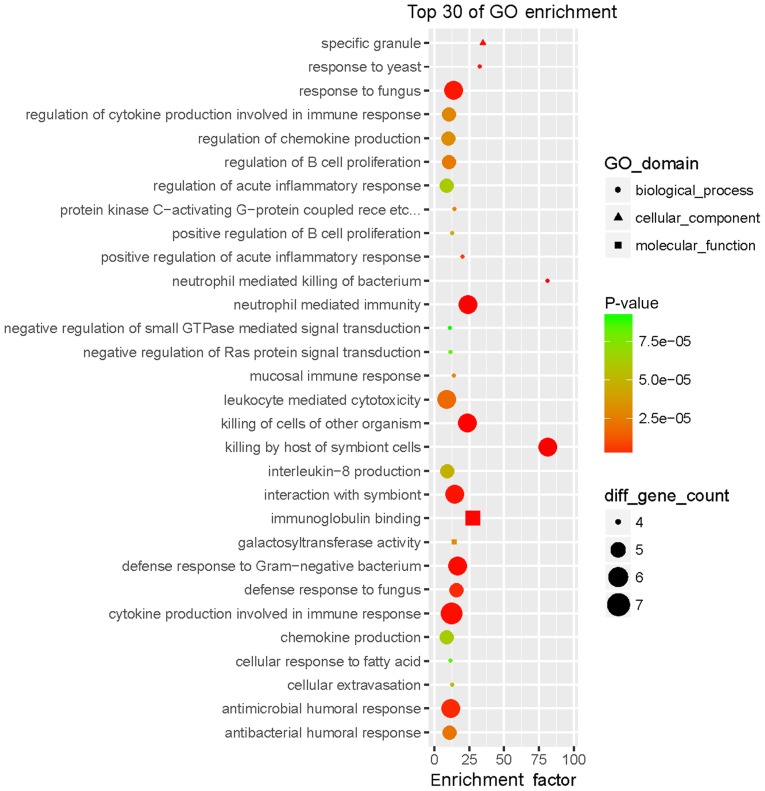
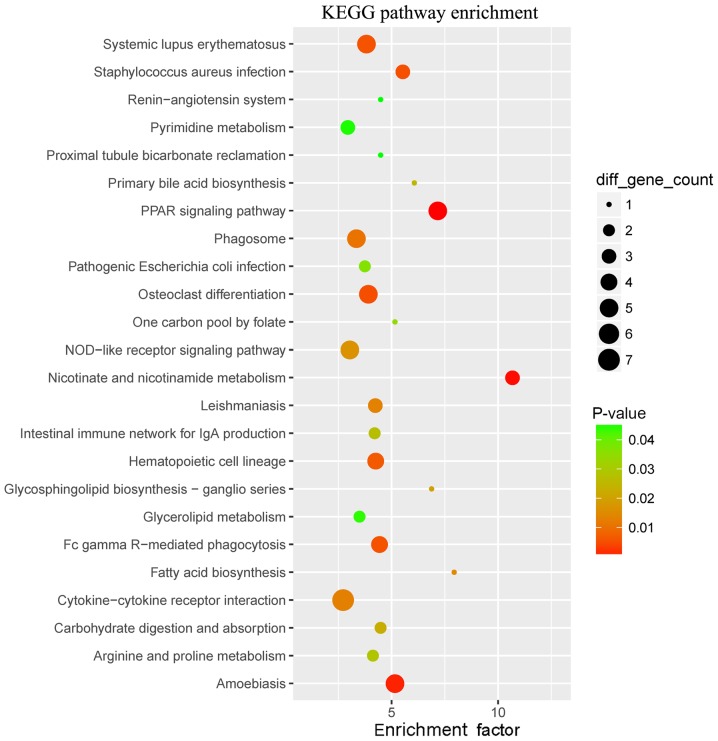
The GO enrichment analysis revealed that the WB-DEGs were enriched in 293 biological process (BP), 36 cellular component (CC) and 21 molecular function (MF) terms. Fig. 3 presents the top 30 GO terms of the 161 DEGs ranked according to their enrichment factor and Table I presents data regarding the top 10 GO terms. Of the GO terms in the category BP, ‘neutrophil-mediated killing of bacterium’ and ‘killing by host of symbiont cells’ had the highest enrichment factors, ‘specific granule’ had the highest enrichment factor in the category CC and ‘immunoglobulin binding’ had the highest enrichment factor in the category MF. The WB-DEGs were enriched in a total of 24 KEGG pathways, as presented in Fig. 4 and Table II, of which ‘primary bile acid biosynthesis’ and ‘one-carbon pool by folate’ had the highest enrichment factors.
Figure 3.
Top 30 GO terms of the 161 DEGs in whole blood ranked according to the enrichment factor. The enrichment factor was defined as follows: (Number of DEGs in a term/total number of DEGs)/(total number of genes in the database in a term/total number of genes in the database). The term ‘diff gene count’ indicates the number of DEGs enriched in a GO term. GO, gene ontology; DEG, differentially expressed gene.
Table I.
Top 10 GO terms ranked according to enrichment factors.
| GO ID | GO term | Description | Gene ratio (%) | P-value | Q-value | Count | Genes | Enrichment factor |
|---|---|---|---|---|---|---|---|---|
| GO:0070944 | BP | Neutrophil-mediated killing of bacterium | 2.61 | 3.15 ×10−8 | 1.46×10−5 | 4 | CTSG, TREM1, AZU1, NCF1 | 81.34 |
| GO:0051873 | BP | Killing by host of symbiont cells | 3.92 | 5.03×10−11 | 5.43×10−8 | 6 | CTSG, TREM1, NCF1, ELANE, CAMP, AZU1 | 81.34 |
| GO:0042581 | CC | Specific granule | 2.61 | 7.51×10−7 | 1.28×10−4 | 4 | ANXA3, OLFM4, CAMP, ADAM8 | 34.86 |
| GO:0001878 | BP | Response to yeast | 2.61 | 9.94×10−7 | 1.53×10−4 | 4 | NCF1, CAMP, ELANE, ADM | 32.54 |
| GO:0019865 | MF | Immunoglobulin binding | 3.27 | 1.87×10−7 | 4.65×10−5 | 5 | FCGR2B, IGJ, FCAR, FCGR1A, FCGR1B | 27.73 |
| GO:0002446 | BP | Neutrophil-mediated immunity | 3.92 | 3.90×10−8 | 1.58×10−5 | 6 | CTSG, ANXA3, ELANE, AZU1, TREM1, NCF1 | 24.4 |
| GO:0031640 | BP | Killing of cells of other organism | 3.92 | 4.75×10−8 | 1.54×10−5 | 6 | CTSG, CAMP, ELANE, AZU1, TREM1, NCF1 | 23.62 |
| GO:0002675 | BP | Positive regulation of acute inflammatory response | 2.61 | 7.17×10−6 | 7.03×10−4 | 4 | FFAR2, FFAR3, ADAM8, OSM | 20.34 |
| GO:0050829 | BP | Defense response to Gram-negative bacterium | 3.92 | 3.97×10−7 | 9.18×10−5 | 6 | AZU1, TREM1, TLR4, SLC11A1, CAMP, ADM | 16.64 |
| GO:0050832 | BP | Defense response to fungus | 3.27 | 3.13×10−6 | 3.49×10−4 | 5 | NCF1, CTSG, ELANE, CAMP, ADM | 16.05 |
‘Gene ratio’ is the ratio of the count obtained to the count expected. ‘Count’ is the number of DEGs enriched in a GO term. The ‘Genes’ column lists the gene symbols of the DEGs enriched in the corresponding GO term. BP, biological process; CC, cellular component; MF, molecular function; GO, gene ontology. Gene names are defined in Table SII.
Figure 4.
KEGG pathway enrichment of 161 WB-DEGs. The WB-DEGs were enriched in a total of 24 KEGG pathways. The enrichment factor was defined as follows: (Number of DEGs in a term/total number of DEGs)/(total number of genes in the database in a term/total number of genes in the database). The term ‘diff gene count’ refers to the number of DEGs enriched in a KEGG pathway. WB-DEGs, differentially expressed genes in whole blood; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Table II.
Top 10 KEGG pathways ranked according to enrichment factors.
| Pathway ID | Description | Gene ratio (%) | P-value | Q-value | Count | Genes | Enrichment factor |
|---|---|---|---|---|---|---|---|
| hsa04614 | Renin-angiotensin system | 1.59 | 0.04 | 0.16 | 1 | CTSG | 4.49 |
| hsa04964 | Proximal tubule bicarbonate reclamation | 1.59 | 0.04 | 0.16 | 1 | SLC4A4 | 4.49 |
| hsa00240 | Pyrimidine metabolism | 4.76 | 0.04 | 0.17 | 3 | ENTPD1, TYMS, NT5C2 | 2.95 |
| hsa00561 | Glycerolipid metabolism | 3.17 | 0.04 | 0.17 | 2 | DGAT2, GK | 3.50 |
| hsa05130 | Pathogenic Escherichia coli infection | 3.17 | 0.04 | 0.15 | 2 | TLR5, TLR4 | 3.75 |
| hsa00670 | One carbon pool by folate | 1.59 | 0.03 | 0.15 | 1 | TYMS | 5.16 |
| hsa00330 | Arginine and proline metabolism | 3.17 | 0.03 | 0.13 | 2 | ARG1, SAT1 | 4.13 |
| hsa04672 | Intestinal immune network for IgA production | 3.17 | 0.03 | 0.13 | 2 | TNFSF13B, CCR9 | 4.21 |
| hsa00120 | Primary bile acid biosynthesis | 1.59 | 0.03 | 0.13 | 1 | CYP27A1 | 6.07 |
| hsa04973 | Carbohydrate digestion and absorption | 3.17 | 0.02 | 0.13 | 2 | MGAM, SLC2A5 | 4.49 |
‘Gene ratio’ is the ratio of the count obtained to the count expected. ‘Count’ is the number of DEGs enriched in a KEGG pathway. The ‘Genes’ column lists gene symbols of the DEGs enriched in the corresponding KEGG pathway. DEG, differentially expressed gene; KEGG, Kyoto Encyclopedia of Genes and Genomes; hsa, Homo sapiens. Gene names are defined in Table SII.
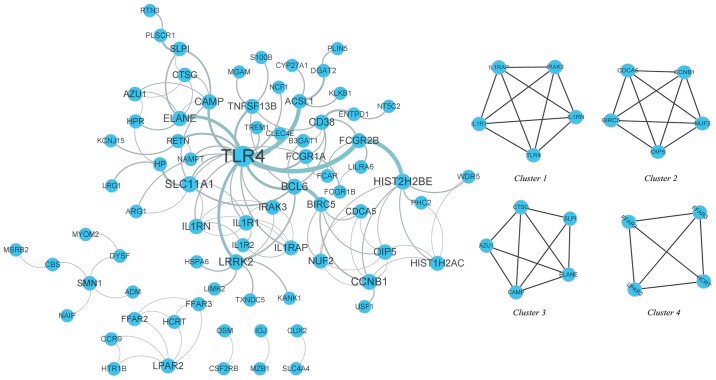
PPI network and submodules
The DEGs were mapped using the STRING database, and a PPI network was then constructed using Cytoscape software. The PPI network included 146 connected nodes and 119 edges. The MCODE analysis revealed that 19 WB-DEGs were clustered in 4 submodules in the PPI network (Fig. 5). To identify the hub nodes, the degree centrality of each node was calculated with the plug-in CentiScaPe, indicating that Toll-like receptor 4 (TLR4) had the highest degree centrality (Table III).
Figure 5.
PPI network of the WB-DEGs and submodules visualized in Cytoscape software. All nodes with a combined score >0.4 are provided, and there were 146 connected nodes and 119 edges in the PPI network. The size of the nodes represents the degree centrality and TLR4 was identified as the hub gene in the PPI network with the highest degree centrality. MCODE analysis revealed that 19 WB-DEGs were clustered in 4 submodules in the PPI network. PPI, protein-protein interaction; WB-DEGs, differentially expressed genes in whole blood; TLR4, Toll-like receptor 4.
Table III.
Degree centrality of the 19 DEGs calculated by CentiScaPe.
| Node/gene | Degree centrality | MCODE cluster | MCODE score |
|---|---|---|---|
| TLR4 | 19 | 1 | 4.0 |
| IL1R1 | 6 | 1 | 4.0 |
| IL1RN | 6 | 1 | 4.0 |
| IRAK3 | 5 | 1 | 4.0 |
| IL1RAP | 5 | 1 | 4.0 |
| CCNB1 | 7 | 2 | 4.0 |
| OIP5 | 6 | 2 | 4.0 |
| NUF2 | 5 | 2 | 4.0 |
| BIRC5 | 5 | 2 | 4.0 |
| CDCA5 | 4 | 2 | 4.0 |
| CAMP | 6 | 3 | 2.7 |
| ELANE | 6 | 3 | 2.7 |
| AZU1 | 4 | 3 | 3.0 |
| CTSG | 4 | 3 | 2.7 |
| SLPI | 4 | 3 | 3.0 |
| LPAR2 | 5 | 4 | 3.0 |
| HCRT | 3 | 4 | 3.0 |
| FFAR2 | 3 | 4 | 3.0 |
| FFAR3 | 3 | 4 | 3.0 |
MCODE analysis revealed that 19 DEGs were clustered in 4 submodules. DEG, differentially expressed gene. Gene names are defined in Table SII.
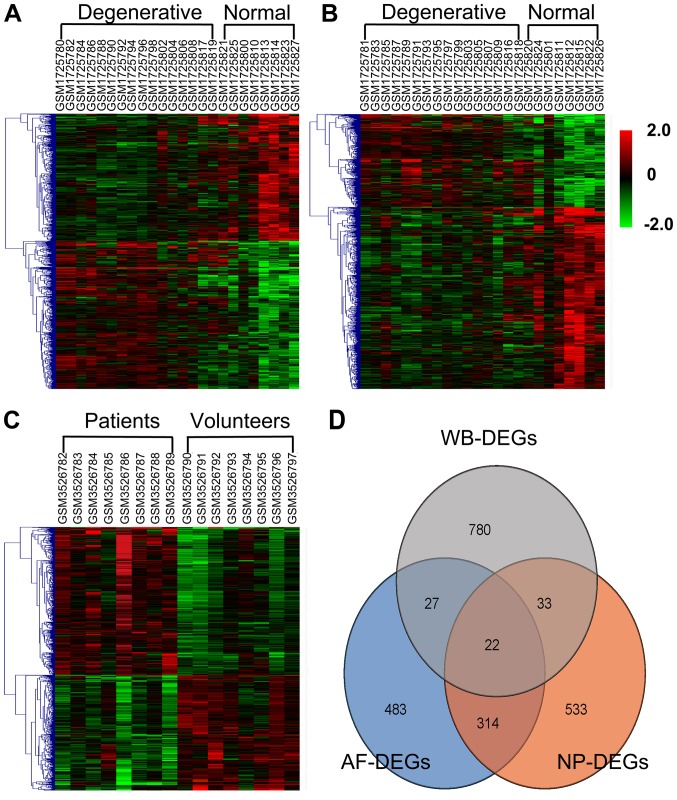
Common DEGs
WB-DEGs, AF-DEGs and NP-DEGs with a threshold P-value of 0.05 and FC of 1.5 were identified, and statistical data regarding these DEGs are presented in Table IV. Hierarchical heat maps of WB-DEGs, AF-DEGs and NP-DEGs are provided in Fig. 6A-C, respectively. As illustrated in a Venn diagram (Fig. 6D), 22 DEGs that overlapped among the three groups and were identified as common DEGs. Data regarding the dysregulation of these 22 common DEGs are provided in Table V.
Table IV.
Statistical data regarding the DEGs in WB, AF and NP.
| Type of sample | Total DEGs | Proportion of transcriptome (%) | Upregulated (n) | Downregulated (n) |
|---|---|---|---|---|
| WB | 862 | 1.48 | 484 | 378 |
| AF | 846 | 1.58 | 455 | 391 |
| NP | 902 | 1.65 | 305 | 597 |
DEGs, differentially expressed genes; WB, whole blood; AF, annulus fibrosis; NP, nucleus pulposus.
Figure 6.
Heatmap and Venn diagram of AF-DEGs, NP-DEGs and WB-DEGs. Heatmaps of (A) AF-DEGs - the gene sample (GSM) number for each column is 1725780, 1725782, 1725784, 1725786, 1725788, 1725790, 1725792, 1725794, 1725796, 1725798, 1725802, 1725804, 1725806, 1725808, 1725817, 1725819, 1725821, 1725825, 1725800, 1725810, 1725813, 1725814, 1725823, 1725827 from left to right; (B) NP-DEGs - the GSM number for each column is 1725781, 1725783, 1725785, 1725787, 1725789, 1725791, 1725793, 1725795, 1725797, 1725799, 1725803, 1725805, 1725807, 1725809, 1725816, 1725818, 1725820, 1725824, 1725801, 1725811, 1725812, 1725815, 1725822, 17258276 from left to right; (C) WB-DEGs - the GSM number for each column is 3526782, 3526783, 3526784, 3526785, 3526786, 3526787, 3526788, 3526789, 3526790, 3526791, 3526792, 3526793, 3526794, 3526795, 3526796, 3526797 from left to right. (D) Venn diagram illustrating the numbers of exclusive and 22 DEGs overlapping among the WB-DEGs, AF-DEGs and NP-DEGs. DEGs, differentially expressed genes; WB-DEGs, DEGs in whole blood; AF-DEGs, DEGs in annulus fibrosis; NP-DEGs, DEGs in nucleus pulposus.
Table V.
Dysregulation of 22 common DEGs in WB, AF and NP.
| Fold change | |||
|---|---|---|---|
| Gene symbol | WB-DEGs | AF-DEGs | NP-DEGs |
| ABLIM1 | −1.59 | −1.73 | −2.97 |
| ACKR3 | −1.53 | −2.77 | −2.21 |
| ARG1 | 7.36 | −1.90 | −2.95 |
| AVIL | 2.211 | −2.44 | −2.13 |
| BACH2 | −1.78 | −2.38 | −2.20 |
| C1orf21 | −1.57 | −1.69 | −1.69 |
| ENPP4 | −1.55 | 1.80 | 2.05 |
| FKBP11 | −1.60 | 1.76 | 1.79 |
| FRY | 1.61 | −1.68 | 1.83 |
| HSPA13 | −1.59 | 1.91 | 1.66 |
| KLF3-AS1 | −1.86 | −2.07 | −1.63 |
| MRVI1 | 2.43 | −1.70 | −1.61 |
| PDIA4 | −1.74 | 1.99 | 1.68 |
| SLC19A3 | 1.54 | −1.67 | −1.66 |
| SLC25A37 | 1.59 | −1.50 | 1.53 |
| SOD2 | 1.59 | 2.80 | 1.54 |
| SSH2 | 1.76 | −1.90 | −2.56 |
| THEM4 | −1.53 | −1.63 | −1.66 |
| TNFAIP6 | 2.65 | 2.22 | 3.00 |
| TREM1 | 2.12 | 3.02 | 2.87 |
| ZBTB41 | −1.53 | 1.52 | 1.58 |
| ZNF185 | 1.52 | −1.66 | −2.19 |
DEGs, differentially expressed genes; WB, whole blood; AF, annulus fibrosis; NP, nucleus pulposus. Gene names are defined in Table SII.
Expression of DEGs
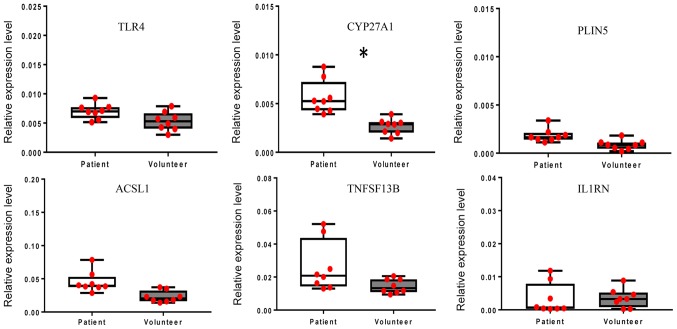
RT-qPCR was performed using WB samples to verify the gene chip hybridization results for selected DEGs. Of the WB-DEGs, the expression of TLR4, cytochrome P450 family 27 subfamily A member 1 (CYP27A1), perilipin 5 (PLIN5), acyl-CoA synthetase long-chain family member 1 (ACSL1), tumor necrosis factor superfamily member 13b (TNFSF13B) and interleukin 1 receptor antagonist (IL1RN) was validated. As presented in Fig. 7, the expression levels of TLR4, CYP27A1, PLIN5, ACSL1 and TNFSF13B were higher in WB obtained from LDP patients compared with those in WB obtained from healthy volunteers, while the expression level of IL1RN was lower in WB obtained from LDP patients compared with that in WB obtained from healthy volunteers. Furthermore, the results suggested that the gene expression levels of CYP27A1 were significantly different between the patient group and the control group (P<0.05).
Figure 7.
Confirmation of the aberrant expression of the differentially expressed genes in whole blood TLR4, CYP27A1, PLIN5, ACSL1, TNFSF13B and IL1RN. All reverse transcription-quantitative PCR experiments were performed as three independent replicates and the average Cq value was used to calculate the ΔCq and ΔΔCq value regarding the internal reference. The relative expression levels are provided as 2−ΔΔCq (n=8). The gene expression levels of CYP27A1 were significantly different between the patient group and the control group. *P<0.05. TLR4, Toll-like receptor 4; CYP27A1, cytochrome P450 family 27 subfamily A member 1; PLIN5, perilipin 5; ACSL1, acyl-CoA synthetase long-chain family member 1; TNFSF13B, tumor necrosis factor superfamily member 13b; IL1RN, interleukin 1 receptor antagonist; Cq, quantification cycle.
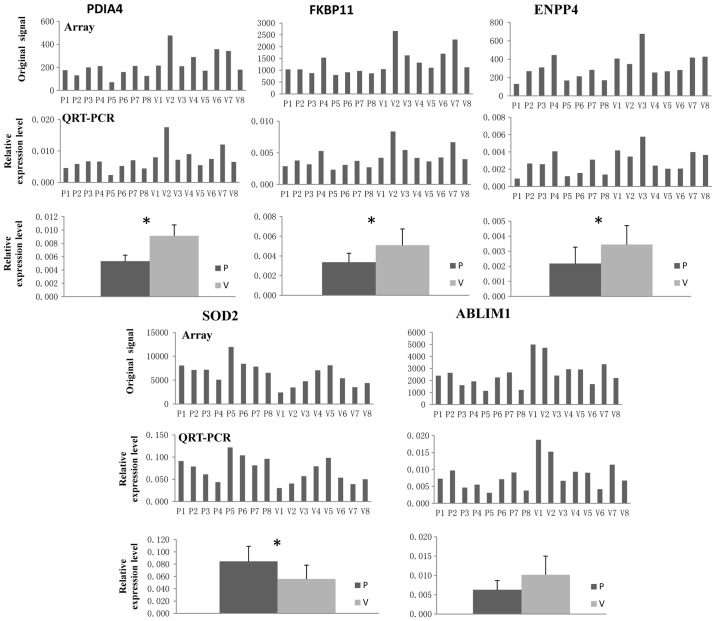
Of the common DEGs, the expression of protein disulfide isomerase family A member 4 (PDIA4), FKBP prolyl isomerase 11 (FKBP11), ectonucleotide pyrophosphatase/phosphodiesterase 4 (ENPP4), superoxide dismutase 2 (SOD2) and actin-binding LIM protein 1 (ABLIM1) were validated. As presented in Fig. 8, the expression of PDIA4, FKBP11, ENPP4 and ABLIM1 was downregulated in WB from IDD patients compared with that in healthy volunteers, whereas the expression of SOD2 in WB from IDD patients was significantly higher than that in healthy volunteers. Furthermore, the results suggested that the gene expression levels of PDIA4, FKBP11, ENPP4 and SOD2 were significantly different between the patient group and volunteer group (P<0.05). The results obtained by RT-qPCR analysis of WB were consistent with those of the gene chip hybridization, suggesting that the microarray data correlated well with the RT-qPCR data, demonstrating the reliability of the microarray results.
Figure 8.
Experimental confirmation of DEGs common to WB-DEGs, AF-DEGs and NP-DEGs, PDIA4, FKBP11, ENPP4, SOD2 and ABLIM1 in whole blood. All RT-qPCR experiments were performed as three independent replicates and the average Cq value was used to calculate the ΔCq and ΔΔCq value regarding the internal reference. The relative expression levels are provided as 2−ΔΔCq (n=8). The gene expression levels of PDIA4, FKBP11, ENPP4 and SOD2 were significantly different between the patient group and the control group. *P<0.05. Cq, quantification cycle; SOD2, superoxide dismutase 2; PDIA4, protein disulfide isomerase family A member 4; FKBP11, FKBP prolyl isomerase 11; ENPP4, ectonucleotide pyrophosphatase/phosphodiesterase 4; ABLIM1, actin-binding LIM protein 1; RT-qPCR, reverse transcription-quantitative; P, patient; V, healthy volunteer.
Discussion
Lower back pain and sciatica caused by LDP are substantial sources of pain in patients and seriously affect their quality of life. As the prevalence of IDD increases with age, with up to 90% of asymptomatic individuals aged >60 years presenting with IDD (18), lower back pain observed in IDD has been extensively investigated (19). However, the mechanisms underlying LDP and IDD are not exactly the same. Previous studies have mostly focused on AF and NP (8–15), which may only be evaluated during surgery. However, to the best of our knowledge, they have not investigated the gene expression in WB obtained from patients with LDP.
In the present study, a total of 161 DEGs were identified in WB obtained from patients with LDP vs. healthy volunteers, including 128 up- and 33 downregulated genes. GO enrichment indicated that ‘neutrophil-mediated killing of bacterium’ and ‘killing by host of symbiont cells’ had the highest enrichment factor in the category BP, ‘specific granule’ had the highest enrichment factor in the category CC and ‘immunoglobulin binding’ had the highest enrichment factor in MF. Pathway enrichment analysis indicated that ‘primary bile acid biosynthesis’ and ‘one-carbon pool by folate’ had the highest enrichment factors. To the best of our knowledge, no previous study has demonstrated the role of these GO terms and pathways in LDP; thus, they may represent new directions for studying LDP. In the PPI network, 4 submodules were identified and TLR4 with the highest degree centrality was identified as the hub gene in the PPI network, indicating that these submodules and TLR4 may have an important role in LDP. The results of the bioinformatics analysis require further experimental verification. Furthermore, 22 common DEGs between WB-DEGs, AF-DEGs and NP-DEGs were identified. The RT-qPCR results confirmed that the expression levels of CYP27A1, SOD2, PDIA4, FKBP11 and ENPP4 were significantly different between the patient group and volunteer group (P<0.05); thus, these genes may represent potential biomarkers in WB for LDP.
In a genome-wide expression analysis, Schubert et al (13) identified 5 AF markers and 6 NP markers. None of these 11 markers was dysregulated in the WB obtained from the patients of the present study. Guo et al (9) compared the gene expression profiles of degenerative and non-degenerative IVDs and identified 35 DEGs with a FC>2 that overlapped between AF and NP. None of these 35 common DEGs overlapped with the DEGs identified in WB in the present study. Using a microarray analysis, Kazezian et al (10) identified 17 molecular markers of AF degeneration. However, none of those molecular markers was among the WB-DEGs determined in the present study. Zhang et al (15) compared the results of an expression array analysis of peripheral blood mononuclear cells obtained from IDD patients and those obtained from non-IDD individuals and identified 62 DEGs, including 33 upregulated and 24 downregulated genes. Between their and the present study, only the upregulation of Annexin A3 was consistent.
TLR4 is a member of the TLR family and was reported to be expressed in IVD cells in AF and NP (20). TLR4 has been demonstrated to substantially contribute to neuropathic pain (21), and the activation of TLR4 has been indicated to result in the initiation of the inflammatory cascade and the inhibition of extracellular matrix anabolism in IVD (20). Inflammation has an important role in IDD (22,23), and IDD has been characterized as a disease involving extracellular matrix degradation (24). In the present study, TLR4 was identified as the hub gene in the PPI network, indicating that it may be crucial to the process of LDP. CYP27A1 has been previously demonstrated to promote cholesterol efflux (25,26), and patients with higher serum lipid levels of total cholesterol are at a higher risk of lumbar disc herniation (27). On the other hand, elevated blood lipid levels of cholesterol contribute to atherosclerosis, and aortic atherosclerosis-induced damage to the lumbar nutrition supply has been associated with IDD (28). IDD is an age-associated process that is accompanied and accelerated by the accumulation of oxidative damage and inflammation (29). Blood vessels grow into the IVD during the process of IDD (30), and the resulting neovascularization exposes the avascular tissue of the IVD to high oxygen tension and subsequent oxidative injury (31). SOD2 has an anti-oxidative role by converting superoxide radicals into H2O2, which may be broken down into harmless H2O and O2 by anti-oxidative enzymes (32) and is critical for the regulation of oxidative stress resistance (33). Excessive reactive oxygen species may cause protein, DNA and membrane damage and are associated with cellular inflammatory responses by inducing the expression of cytokines and chemokines (34). The inflammatory response has a critical role in the initiation and progression of IDD (22,35–38). Thus, the elimination of superoxide radicals by SOD2 may be considered an anti-inflammatory process. SOD2 may be protective against IVD through anti-oxidant and anti-inflammatory mechanisms in the process of IDD. Data regarding the biological actions of PDIA4, FKBP11 and ENPP4 in IDD are limited, and their roles in IDD require further investigation.
In conclusion, in the present study, CYP27A1, SOD2, PDIA4, FKBP11 and ENPP4 were identified as potential WB biomarkers of LDP, and TLR4 was identified as a hub gene in the PPI network. Furthermore, several GO terms, KEGG pathways and submodules of the PPI network may be involved in LDP through unknown mechanisms. LDP is based on IDD and the degree of histopathologic degeneration of LDD increases with age (39). All of the patients of the present study were confirmed to have LDP by MRI. To the best of our knowledge, previous studies have not described the degree of histopathologic degeneration at which IDD may be observed by MRI. Young individuals are more representative of subjects without IDD or low IDD; thus, young individuals were recruited as controls, as previously described (16). In the case of such groupings, WB-DEGs may be linked to other aging-associated processes and not just IDD. Thus, the intersection of the WB-DEGs, AF-DEGs and NP-DEGs was focused on to obtain molecular markers in WB. However, age may be a confounding factor that affects gene expression. The present results still require further experimental verification with large sample sizes and mechanistic study.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- LDP
lumbar disc prolapse
- DDD
disc degeneration disease
- IDD
intervertebral disc degeneration
- IVD
intervertebral disc
- AF
annulus fibrosis
- NP
nucleus pulpous
- WB
whole blood
- RT-qPCR
reverse transcription-quantitative PCR
- DEGs
differentially expressed genes
- FC
fold change
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PPI
protein-protein interaction
- BP
biological process
- CC
cellular component
- MF
molecular function
- TLR4
Toll-like receptor 4
- CYP27A1
cytochrome P450 family 27 subfamily A member 1
- PLIN5
perilipin 5
- ACSL1
acyl-CoA synthetase long-chain family member 1
- TNFSF13B
tumor necrosis factor superfamily member 13b
- IL1RN
interleukin 1 receptor antagonist
Funding
The present study was supported by the Sichuan Science and Technology Program, China (grant no. 2018SZ0075).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the GEO database (accession no. GSE124272).
Authors' contributions
YW, GD and LTL designed the current study, and together with LL, LJ, SWL, SL, FW, WD and YL, performed the data analysis and wrote the manuscript. GD and YW obtained funding and together with LTL, LL, LJ and SWL, advised on the study design and writing. SL, FW, WD and YL contributed to writing and English proofreading. All authors made substantial contributions to the conception and design of the current study, and gave final approval of the version to be published.
Ethics approval and consent to participate
The design of the present study adhered to the tenets of the Declaration of Helsinki and the protocol was reviewed and approved by the institutional review board and ethics committee of Sichuan Provincial Orthopedic Hospital (approval no. 2018SZ0075). All patients and donors enrolled in the study provided written informed consent to participate in this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ahsan K, Najmus-Sakeb, Hossain A, Khan SI, Awwal MA. Discectomy for primary and recurrent prolapse of lumbar intervertebral discs. J Orthop Surg (Hong Kong) 2012;20:7–10. doi: 10.1177/230949901202000102. [DOI] [PubMed] [Google Scholar]
- 2.Kepler CK, Ponnappan RK, Tannoury CA, Risbud MV, Anderson DG. The molecular basis of intervertebral disc degeneration. Spine J. 2013;13:318–330. doi: 10.1016/j.spinee.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Adams MA, Roughley PJ. What is intervertebral disc degeneration, and what causes it? Spine. 2006;31:2151–2161. doi: 10.1097/01.brs.0000231761.73859.2c. [DOI] [PubMed] [Google Scholar]
- 4.Daly C, Ghosh P, Jenkin G, Oehme D, Goldschlager T. A review of animal models of intervertebral disc degeneration: Pathophysiology, regeneration, and translation to the clinic. BioMed Res Int. 2016;2016:5952165. doi: 10.1155/2016/5952165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beattie PF. Current understanding of lumbar intervertebral disc degeneration: A review with emphasis upon etiology, pathophysiology, and lumbar magnetic resonance imaging findings. J Orthop Sports Phys Ther. 2008;38:329–340. doi: 10.2519/jospt.2008.2768. [DOI] [PubMed] [Google Scholar]
- 6.Weber KT, Jacobsen TD, Maidhof R, Virojanapa J, Overby C, Bloom O, Quraishi S, Levine M, Chahine NO. Developments in intervertebral disc disease research: Pathophysiology, mechanobiology, and therapeutics. Curr Rev Musculoskelet Med. 2015;8:18–31. doi: 10.1007/s12178-014-9253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colombini A, Lombardi G, Corsi MM, Banfi G. Pathophysiology of the human intervertebral disc. Int J Biochem Cell Biol. 2008;40:837–842. doi: 10.1016/j.biocel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 8.Chen K, Wu D, Zhu X, Ni H, Wei X, Mao N, Xie Y, Niu Y, Li M. Gene expression profile analysis of human intervertebral disc degeneration. Genet Mol Biol. 2013;36:448–454. doi: 10.1590/S1415-47572013000300021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo W, Zhang B, Li Y, Duan HQ, Sun C, Xu YQ, Feng SQ. Gene expression profile identifies potential biomarkers for human intervertebral disc degeneration. Mol Med Rep. 2017;16:8665–8672. doi: 10.3892/mmr.2017.7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kazezian Z, Gawri R, Haglund L, Ouellet J, Mwale F, Tarrant F, O'Gaora P, Pandit A, Alini M, Grad S. Gene expression profiling identifies interferon signalling molecules and IGFBP3 in human degenerative annulus fibrosus. Sci Rep. 2015;5:15662. doi: 10.1038/srep15662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu C, Fei HD, Sun ZY, Tian JW. Bioinformatic analysis of the microarray gene expression profile in degenerative intervertebral disc cells exposed to TNF-α. Eur Rev Med Pharmacol Sci. 2015;19:3332–3339. [PubMed] [Google Scholar]
- 12.Markova DZ, Kepler CK, Addya S, Murray HB, Vaccaro AR, Shapiro IM, Anderson DG, Albert TJ, Risbud MV. An organ culture system to model early degenerative changes of the intervertebral disc II: Profiling global gene expression changes. Arthritis Res Ther. 2013;15:R121. doi: 10.1186/ar4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schubert AK, Smink JJ, Arp M, Ringe J, Hegewald AA, Sittinger M. Quality assessment of surgical disc samples discriminates human annulus fibrosus and nucleus pulposus on tissue and molecular level. Int J Mol Sci. 2018;19:E1761. doi: 10.3390/ijms19061761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang Y, Wang S, Liu Y, Wang X. Microarray analysis of genes and gene functions in disc degeneration. Exp Ther Med. 2014;7:343–348. doi: 10.3892/etm.2013.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang YG, Guo X, Sun Z, Jia G, Xu P, Wang S. Gene expression profiles of disc tissues and peripheral blood mononuclear cells from patients with degenerative discs. J Bone Miner Metab. 2010;28:209–219. doi: 10.1007/s00774-009-0120-4. [DOI] [PubMed] [Google Scholar]
- 16.Tsai TT, Lai PL, Liao JC, Fu TS, Niu CC, Chen LH, Lee MS, Chen WJ, Fang HC, Ho NY, et al. Increased periostin gene expression in degenerative intervertebral disc cells. Spine J. 2013;13:289–298. doi: 10.1016/j.spinee.2013.01.040. [DOI] [PubMed] [Google Scholar]
- 17.Thompson JP, Pearce RH, Schechter MT, Adams ME, Tsang IK, Bishop PB. Preliminary evaluation of a scheme for grading the gross morphology of the human intervertebral disc. Spine. 1990;15:411–415. doi: 10.1097/00007632-199005000-00012. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Brinjikji W, Luetmer PH, Comstock B, Bresnahan BW, Chen LE, Deyo RA, Halabi S, Turner JA, Avins AL, James K, et al. Systematic literature review of imaging features of spinal degeneration in asymptomatic populations. AJNR Am J Neuroradiol. 2015;36:811–816. doi: 10.3174/ajnr.A4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luoma K, Riihimäki H, Luukkonen R, Raininko R, Viikari-Juntura E, Lamminen A. Low back pain in relation to lumbar disc degeneration. Spine. 2000;25:487–492. doi: 10.1097/00007632-200002150-00016. [DOI] [PubMed] [Google Scholar]
- 21.Rajan NE, Bloom O, Maidhof R, Stetson N, Sherry B, Levine M, Chahine NO. Toll-Like Receptor 4 (TLR4) expression and stimulation in a model of intervertebral disc inflammation and degeneration. Spine. 2013;38:1343–1351. doi: 10.1097/BRS.0b013e31826b71f4. [DOI] [PubMed] [Google Scholar]
- 22.Nicotra L, Loram LC, Watkins LR, Hutchinson MR. Toll-like receptors in chronic pain. Exp Neurol. 2012;234:316–329. doi: 10.1016/j.expneurol.2011.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Risbud MV, Shapiro IM. Role of cytokines in intervertebral disc degeneration: Pain and disc content. Nat Rev Rheumatol. 2014;10:44–56. doi: 10.1038/nrrheum.2013.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shamji MF, Setton LA, Jarvis W, So S, Chen J, Jing L, Bullock R, Isaacs RE, Brown C, Richardson WJ. Proinflammatory cytokine expression profile in degenerated and herniated human intervertebral disc tissues. Arthritis Rheum. 2010;62:1974–1982. doi: 10.1002/art.27444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vo NV, Hartman RA, Yurube T, Jacobs LJ, Sowa GA, Kang JD. Expression and regulation of metalloproteinases and their inhibitors in intervertebral disc aging and degeneration. Spine J13. 2013:331–341. doi: 10.1016/j.spinee.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Bahr S, Movin T, Papadogiannakis N, Pikuleva I, Rönnow P, Diczfalusy U, Björkhem I. Mechanism of accumulation of cholesterol and cholestanol in tendons and the role of sterol 27-hydroxylase (CYP27A1) Arterioscler Thromb Vasc Biol. 2002;22:1129–1135. doi: 10.1161/01.ATV.0000022600.61391.A5. [DOI] [PubMed] [Google Scholar]
- 27.Escher G, Krozowski Z, Croft KD, Sviridov D. Expression of sterol 27-hydroxylase (CYP27A1) enhances cholesterol efflux. J Biol Chem. 2003;278:11015–11019. doi: 10.1074/jbc.M212780200. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Zhao Y, Wang M, Si M, Li J, Hou Y, Jia J, Nie L. Serum lipid levels are positively correlated with lumbar disc herniation - a retrospective study of 790 Chinese patients. Lipids Health Dis. 2016;15:80. doi: 10.1186/s12944-016-0248-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kauppila LI. Atherosclerosis and disc degeneration/low-back pain--a systematic review. Eur J Vasc Endovasc Surg. 2009;37:661–670. doi: 10.1016/j.ejvs.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Bank RA, Bayliss MT, Lafeber FP, Maroudas A, Tekoppele JM. Ageing and zonal variation in post-translational modification of collagen in normal human articular cartilage. The age-related increase in non-enzymatic glycation affects biomechanical properties of cartilage. Biochem J. 1998;330:345–351. doi: 10.1042/bj3300345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ali R, Le Maitre CL, Richardson SM, Hoyland JA, Freemont AJ. Connective tissue growth factor expression in human intervertebral disc: Implications for angiogenesis in intervertebral disc degeneration. Biotech Histochem. 2008;83:239–245. doi: 10.1080/10520290802539186. [DOI] [PubMed] [Google Scholar]
- 32.Nasto LA, Robinson AR, Ngo K, Clauson CL, Dong Q, St Croix C, Sowa G, Pola E, Robbins PD, Kang J, et al. Mitochondrial-derived reactive oxygen species (ROS) play a causal role in aging-related intervertebral disc degeneration. J Orthop Res. 2013;31:1150–1157. doi: 10.1002/jor.22320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flekac M, Skrha J, Hilgertova J, Lacinova Z, Jarolimkova M. Gene polymorphisms of superoxide dismutases and catalase in diabetes mellitus. BMC Med Genet. 2008;9:30. doi: 10.1186/1471-2350-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landis GN, Tower J. Superoxide dismutase evolution and life span regulation. Mech Ageing Dev. 2005;126:365–379. doi: 10.1016/j.mad.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 35.Nel A, Xia T, Mädler L, Li N. Toxic potential of materials at the nanolevel. Science. 2006;311:622–627. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- 36.Tian Y, Yuan W, Fujita N, Wang J, Wang H, Shapiro IM, Risbud MV. Inflammatory cytokines associated with degenerative disc disease control aggrecanase-1 (ADAMTS-4) expression in nucleus pulposus cells through MAPK and NF-κB. Am J Pathol. 2013;182:2310–2321. doi: 10.1016/j.ajpath.2013.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H, Tian Y, Wang J, Phillips KL, Binch AL, Dunn S, Cross A, Chiverton N, Zheng Z, Shapiro IM, et al. Inflammatory cytokines induce NOTCH signaling in nucleus pulposus cells: Implications in intervertebral disc degeneration. J Biol Chem. 2013;288:16761–16774. doi: 10.1074/jbc.M112.446633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Molinos M, Almeida CR, Caldeira J, Cunha C, Gonçalves RM, Barbosa MA. Inflammation in intervertebral disc degeneration and regeneration. J R Soc Interface. 2015;12:20150429. doi: 10.1098/rsif.2015.0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boos N, Weissbach S, Rohrbach H, Weiler C, Spratt KF, Nerlich AG. Classification of age-related changes in lumbar intervertebral discs: 2002 Volvo Award in basic science. Spine. 2002;27:2631–2644. doi: 10.1097/00007632-200212010-00002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the GEO database (accession no. GSE124272).