Abstract
SUMOylation is a reversible and highly dynamic post-translational modification of target proteins by small ubiquitin-like modifiers (SUMO). It is orchestrated by SUMO-activating, -conjugating, and -ligating enzymes in a sequential manner and is important in regulating a myriad of predominantly nuclear processes. DeSUMOylation is achieved by SUMO-specific proteases (SENPs). Deregulation of SUMOylation and deSUMOylation results in cellular dysfunction and is linked to various diseases, including cancer. In recent years, SENPs have emerged as potential therapeutic targets. In this review, we will describe the inhibitors and activity-based probes of SENPs. Furthermore, we will summarize the biochemical assays available for evaluating the activity of SENPs to identify inhibitors.
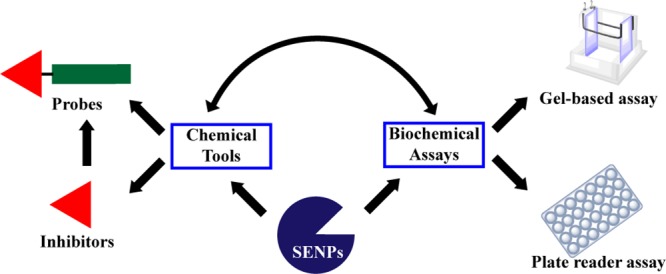
SUMOylation is a reversible and highly dynamic post-translational modification of lysine residues in target proteins by small ubiquitin-like modifiers (SUMOs). SUMOylation of proteins can affect their activity, stability, localization, and interaction with other proteins. SUMOylation is important in regulating a multitude of cellular processes, including cell cycle progression, genome stability, transcription, and DNA repair.1−3 SUMO is attached to target proteins (Figure 1) by a SUMO-activating enzyme (E1), a SUMO-conjugating enzyme (E2), and a SUMO ligase (E3)4 in a stepwise manner. First, a thioester bond is formed between the active site cysteine of the SUMO E1 and the C-terminus of SUMO, which requires ATP. Next, SUMO is transferred to the catalytic cysteine residue of the SUMO E2 UBC9. In the last step, a SUMO E3 stimulates the E2-mediated conjugation of SUMO to target proteins. This results in an isopeptide linkage between the C-terminus of SUMO and the ε-amino group of a lysine residue within the target protein. This lysine residue is frequently located in the SUMOylation consensus motif ψKxE, where ψ is an amino acid with a large hydrophobic side-chain. SUMO can be conjugated to a single lysine residue in proteins (mono-SUMOylation) or multiple lysine residues (multi-SUMOylation) or form SUMO chains (poly-SUMOylation).
Figure 1.
Schematic overview of the pathway of SUMOylation and deSUMOylation. Precursor SUMO (pro-SUMO) must be activated by a SENP into its mature form before entering the pathway of SUMOylation. Mature SUMO forms a thioester with SUMO-activating enzyme (E1) in an ATP-dependent manner. Subsequently, SUMO is transferred to the SUMO-conjugating enzyme (E2). Finally, a SUMO ligase (E3) stimulates the E2-mediated conjugation of SUMO to a lysine in target proteins. Deconjugation of SUMO from target proteins is catalyzed by SENPs.
Mammalian cells express at least three SUMO isoforms, SUMO-1, SUMO-2, and SUMO-3. SUMO is translated as an inactive precursor (pro-SUMO) and is activated by SUMO-specific proteases (SENPs), exposing a diglycine motif that is required for conjugation.5,6 SENPs are also required for deconjugation of SUMOs from target proteins, thereby tightly regulating the SUMOylation levels of individual target proteins required for normal cell physiology. Mammalian cells express six SENPs, designated SENP1–3 and SENP5–7. All are cysteine proteases and have catalytic triads (Cys–His–Asp) in a conserved protease domain. They differ in their subcellular localization, substrate specificity, and selectivity for SUMO precursor processing versus deconjugation (Table 1).5−7 Recently, three other SUMO proteases were identified with low sequence identity to SENPs: deSUMOylating isopeptidase 1 (DESI1), DESI2, and ubiquitin-specific protease-like 1 (USPL1).8,9
Table 1. Biological Properties of SENPs.
| SENPs | localization | substrate preference | precursor processing | deconjugation | refs |
|---|---|---|---|---|---|
| SENP1 | nuclear pore and nuclear foci | SUMO-1/2/3 | yes | yes | (19−21) |
| SENP2 | nuclear pore and nuclear foci | SUMO-2/3 > SUMO-1 | yes | yes | (22−24) |
| SENP3 | nucleolus | SUMO-2/3 | unknown | yes | (25) |
| SENP5 | nucleolus and mitochondria | SUMO-2/3 | yes | yes | (25, 26) |
| SENP6 | nucleoplasm | poly SUMO-2/3 | no | yes | (27−29) |
| SENP7 | nucleoplasm | poly SUMO-2/3 | no | yes | (27, 30, 31) |
Deregulation of SENPs leads to cellular dysfunction and is linked to various human diseases, including cancer.10−12 SENP1 increases the transcriptional activity of androgen receptor (AR) in AR signaling, which correlates with the development of prostate cancer.13 In addition, SENP1 and SENP3 deSUMOylate and stabilize hypoxia-inducible factor 1α (HIF-1α) during hypoxia, a key step in forming new blood vessels and supporting tumor growth.14,15 Furthermore, increased levels of SENP1 and SENP3 were found in colon, prostate, ovary, and lung cancers.12 Elevated levels of SENP5 associate with poor prognosis in breast cancer patients.16 Moreover, silencing of SENP5 resulted in the inhibition of cell proliferation in oral squamous cell carcinoma.17,18 Collectively, this makes SENPs attractive targets for cancer therapeutics.
Early research on SENP inhibitors and activity based probes focused on assembling an electrophilic reactive group at the C-terminal glycine residue of full length or truncated SUMOs. In recent years, some small molecule inhibitors and probes were also developed. In addition, some well-established biochemical assays are available to measure the activity of SENPs. In this review, we will discuss the inhibitors and activity-based probes of SENPs and the assays for investigating the activity of SENPs in vitro.
Inhibitors and Activity-Based Probes of SENPs
Peptide and Protein-Based Inhibitors and Activity-Based Probes of SENPs
In 2000, the first inhibitor of SUMO proteases 1 (Figure 2) was reported by Mossessova and Lima.32 The authors elegantly generated Smt3 aldehyde through the in situ reduction of a Ulp1 (a SUMO protease of budding yeast) and Smt3 (the yeast SUMO homologue), using sodium borohydride.
Figure 2.
Overview of inhibitors and probes of SENPs. Compounds 1–6 are peptide- and protein-based inhibitors and activity-based probes of SENPs, compounds 7–19 are small molecule inhibitors and activity based probes of SENPs. (Of these inhibitors, only compounds 12 and 13 have been demonstrated to be active in cells.)
In 2003, Hemelaar et al. reported332 and that SUMO-1-VS (Figure 2), containing a vinyl sulfone (VS) reactive functional group at the C-terminus of SUMO-1, could form an irreversible adduct with the catalytic domain of SENP2. The authors further confirmed that the active site cysteine of SENP2 is required for the catalysis, since the formation of an adduct was prevented after the incubation of alkylating agent NEM (N-ethylmaleimide) and SENP2. They also showed that SUMO-1-VS could react with SUMO E1 and SUMO E2 covalently, although to a lesser extent than with SENP2. The commercially available SUMO-VS probes34 have been developed with an HA tag at the N-terminus to detect SENPs by immunoblotting.
Since vinyl sulfones are able to react with the active site cysteine residues of SENPs, Borodovsky et al. developed35 a series of biotinylated VS-based probes harboring either 5, 9, or 13 amino acids of the SUMO-1 C-terminus to profile SENPs. In a cell lysate labeling experiment, all of them proved efficient at labeling SENP1 equally, but only the probe containing five-amino-acid residues, 3 (Figure 2), showed selective labeling of SENP1.
In 2012, Dobrota et al. reported a peptidyl activity based probe 4 (Figure 2) for SENP1 and SENP2.36 This probe contains the final seven amino acids of SUMO-2 with a reactive electrophile fluoromethylketone at its C-terminus, showing a preference for SENP1/2 above other cysteine proteases in labeling experiments. However, its inhibitory activity on SENP2 was twice as high compared to SENP1. Moreover, this probe could compete with SUMO-1 from a SUMO-1–SENP1 complex, suggesting that it has the same binding site on SENP1.
In 2018, our group37 reported the total chemical synthesis of all three isoforms of SUMO based probes 5 (Figure 2), with an alkyne and a rhodamine dye incorporated at the C-terminus and the N-terminus, respectively. Alkynes were discovered as covalent warheads for cysteine proteases by Ekkebus et al. and Sommer et al.(38,39) The labeling result was consistent with the substrate preference of SENPs. SENP1 and SENP2 could be labeled by all three isoforms of SUMO probes. SENP3 and SENP7 had a more clear preference for SUMO-2/3 probes than SENP6. In addition, in order to assess whether SENPs proteolytically cleave SUMO chains, a K11 linked diSUMO-2 probe 6 (Figure 2) was synthesized.
Small Molecule Inhibitors and Activity Based Probes
In 2011, Ponder et al. identified one compound, 7 (Figure 2), as an inhibitor of recombinant SENP1 of human parasite pathogen Plasmodium falciparum (pfSENP1) from a library of irreversible cysteine protease inhibitors.40 This lead compound, 7, has a non-natural peptide backbone and an epoxide reactive group, displaying inhibitory activity on pfSENP1 with an IC50 of 17.9 μM. Medicinal chemistry efforts led to compound 8 (Figure 2) with the same potency as 7. Both compound 7 and 8 could effectively inhibit human SENP1 and SENP2. Compound 7 showed an IC50 of 9.0 μM and 4.7 μM on SENP1 and SENP2, respectively, while compound 8 showed a higher potency with an IC50 of 7.1 μM and 3.7 μM on SENP1 and SENP2, respectively. Furthermore, the authors designed a series of compounds harboring an acyloxymethylketone (AOMK) reactive group to explore the inhibitory activity on human SENPs.41 Compound 9 (Figure 2) with a QTGG natural amino acid sequence was the most potent inhibitor for SENP1 (IC50 = 3.6 μM) and SENP2 (IC50 = 0.25 μM), while compound 10 (Figure 2) with a LRGG sequence was the most potent inhibitor for SENP6 (IC50 = 4.2 μM) and SENP7 (IC50 = 4.3 μM). The most potent epoxide and AOMK inhibitors 8 and 9 were converted into biotinylated and Cy5 fluorescently labeled probes. All of them showed activity toward SENP1 and SENP2, but only the AOMK based probe 11 (Figure 2) was highly specific in cell lysate labeling experiments.
In 2011, Zhou et al. developed a panel of benzodiazepine based compounds,42 aiming to change the poor pharmacokinetic property of peptide inhibitors. Two compounds, 12 and 13 (Figure 2), with the best inhibitory activity were discovered, showing an IC50 of 15.5 μM and 9.2 μM against SENP1 and an IC50 of 13.0 μM and 35.7 μM against prostate cancer cells PC3. The structure–activity relationship revealed that the C4 formyl group is required for the inhibitory activity by forming a covalent adduct with the catalytic site cysteine of SENP1.
At the same time, Uno et al. developed a series of 1-[4-(N-benzylamino) phenyl]-3-phenylurea derivatives as selective SENP1 inhibitors based on hypoxia inducible factor inhibitors.43 At first, the target protein SENP1 was found in a pulldown experiment when using a biotin conjugated probe based on HIF-1α inhibitor 14a (Figure 2). From there, SENP1 inhibitor 14b (Figure 2) was obtained with an IC50 value of 29.6 μM.
In 2012, Zhang et al. reported 2-(4-chlorophenyl)-2-oxoethyl 4-benzamidobenzoates as noncovalent SENP1 inhibitors.44 The lead compound, 15a (Figure 2), was identified by virtual screening of 180 000 compounds. According to the docking model of 15a-SENP1, the structure relationship was further investigated and two compounds with different substitutes at the meta position of the benzoate showed stronger inhibitory activity: compound 15b (Figure 2) contains a benzyl group with an IC50 of 1.1 μM, which is probably caused by a π–π interaction with the phenyl ring of Phe496 of SENP1, while compound 15c (Figure 2) contains an electron-withdrawing bromine atom with an IC50 of 1.2 μM.
In 2013, Madu and co-workers reported a new class of noncovalent inhibitors of SENPs containing 2-fold symmetry by virtual screening.45 The inhibition kinetics of these inhibitors are noncompetitive, confirmed by nuclear magnetic resonance. Among them, compound 16 (Figure 2) showed the highest potency against SENP1 and SENP2, showing different inhibitory activity for SUMO-1 (IC50 = 32.8 μM, IC50 = 1.42 μM) and SUMO-2 (IC50 = 1.88 μM, IC50 = 1.1 μM) precursors.
In 2014, Kumar et al. characterized a class of 1,2,5-oxadiazoles as SENP2 inhibitors using in silico screening.46 From the initial round of screening of 4 000 000 compounds, two compounds, 17a and 18a (Figure 2), were used for a second round of screening of analogs to improve the inhibitory potency. The two best compounds, 17b and 18b (Figure 2), of each scaffold, containing a Cl atom at the 4-position, showed an IC50 of 5.9 μM and 3.7 μM on SENP2, respectively. The structure–activity relationships further revealed that the electron withdrawing groups at the 4-position are of great importance in increasing SENP2 inhibitory activity. However, compound 17b also displayed moderate SENP1 inhibitory activity with an IC50 of 9.7 μM.
In 2016, Zhou et al. found 11 different scaffolds with SENP1 inhibitory activity by virtual screening of a 200 000 compound library.47 The most potent compound, 19 (Figure 3), with an IC50 of 3.5 μM was found by structural modifications of these scaffolds.
Figure 3.
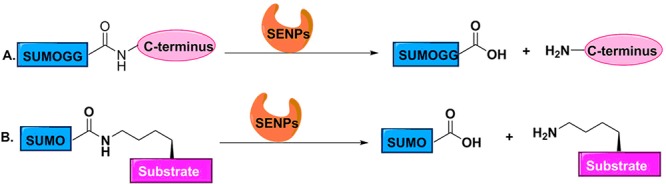
Gel-based assays. (A) Pro-SUMO substrate can used to measure the endopeptidase activity of SENPs. (B) Isopeptide-linked SUMOylated substrates can be used to measure the isopeptidase activity of SENPs).
Biochemical Assays for Measuring Activity of SENPs
Gel-Based Assays
Pro-SUMOs48 and isopeptide-linked SUMOylated substrates49 have been used in gel-based assays to measure the activity of SENPs (Figure 3). Pro-SUMOs contain short C-terminal sequences present in precursor SUMOs, while isopeptide linked SUMOylated substrates contain an isopeptide bond between the C-terminal glycine of SUMO and the lysine residue of substrates. Both of them can be recognized and hydrolyzed by SENPs. The fraction of remaining pro-SUMO or isopeptide-linked SUMOylated substrates can be visualized by Coomassie Blue staining followed by SDS-PAGE, which has a linear relationship with endopeptidase or isopeptidase activity of SENPs, respectively. Mencía and Lorenzo50 have established a method to obtain large quantities of physiological SUMOylated proteins in E. coli. RanGAP1-SUMO is the substrate primarily utilized to study the deconjugation activity of SENPs, but not the only substrate. This method has the advantage of using the physiological SUMO but is difficult to quantitate and laborious.51 Ponder et al. have used this assay to identify inhibitors of pfSENP.40
Fluorescence Based Assays
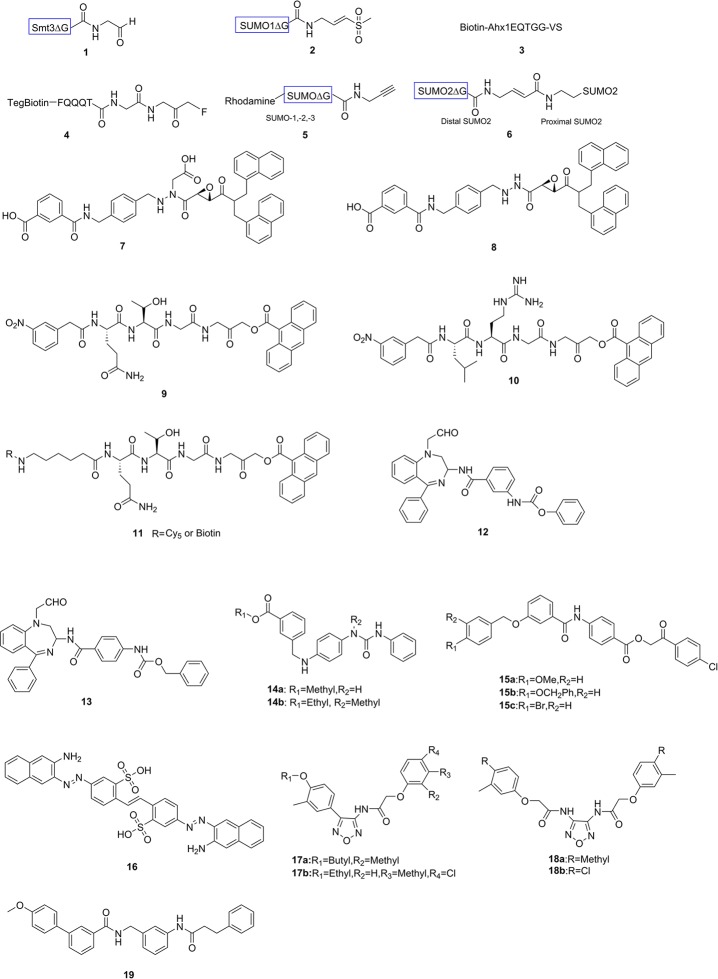
The substrate of a fluorescence resonance energy transfer (FRET)-based assay (Figure 4) consists of two fluorescent proteins, donor cyan fluorescent protein (CFP) and acceptor yellow fluorescent protein (YFP), at the N-terminus and C-terminus of SUMO, respectively.52 When SUMO is not processed, these two fluorescent proteins are in proximity, and the substrate has a strong FRET signal at the YFP emission wavelength. Once SUMO is processed by SENPs, the distance between them will increase. YFP will no longer be excited, and therefore, the only light that will be emitted comes from CFP. The FRET signal of YFP emission versus CFP emission can be used to monitor the activity of SENPs. The assay is quantitative and real-time but lacks sensitivity and therefore requires a high concentration of substrate.53 This leads to measurement problems due to background noise.
Figure 4.
Fluorescence based assay. (A) A FRET-based assay. (B) An AMC-based fluorogenic assay. (C) An AFC-based fluorogenic assay. (D) SUMO-PLA2 based enzyme coupled assay. (E) Bioluminescence-based assay reagents.
Fluorogenic Assays
This assay is based on a fluorophore attaching to the substrate of SENPs via an amide bond (Figure 4). SENPs can recognize and hydrolyze the amide bond, releasing the fluorophore. The fluorescent signal can be easily read out by fluorometry to evaluate the activity of SENPs. Dang et al. reported54 the first SUMO based fluorogenic substrate of SENPs, SUMO-AMC (7-amido-4-methylcoumarin). In 2008, Drag et al.(55) started employing specific tetrapeptide fluorogenic substrates to monitor activities of SENPs. They found that SENP1, -2, and -5 have a preference for QTGG-AFC (7-amino-4-trifluoromethylcoumarin), which represents the C-terminus of mature SUMO-1/2/3, while SENP6 and -7 do not show activity on QTGG-AFC. Surprisingly, they found that SENP6 and -7 have a preference for LRGG-AFC, which resembles the C-terminus of ubiquitin and Nedd8. Ponder et al.(41) have used these two tetrapeptide substrates to identify inhibitors of SENPs. This assay is sensitive, but the detection of fluorophores requires wavelengths in the UV range, and many compounds absorb or emit in this range, which can result in false positives. It should be noted that these short fluorogenic peptide substrates do not mimic the full interaction of SUMO and SENPs that recognizes key elements of the entire Ub fold of SUMO. Consequently, inhibitors that target this interface would be missed when using these short peptide substrates.
An Enzyme Coupled Assay Based on a SUMO Phospholipase A2 Fusion Protein
An assay reagent called SUMO–CHOP (Figure 4) has been developed56 by Nicholson et al. using a SUMO-PLA2 (phospholipase A2) fusion protein as a substrate. Once the substrate is bound to a SENP, the amide linkage between SUMO and the PLA2 will be cleaved. PLA2 will be activated as it requires a free amino terminus for activity. Subsequently, PLA2 can cleave the fluorogenic substrate C6-Nbd-PC to generate a fluorescent signal. The fluorescent signal obtained upon PLA2 cleavage can be used to measure the activity of SENPs. However, false positives will occur if compounds can inhibit PLA2 directly. A commercially available SUMO–CHOP reporter has been used to evaluate the inhibitory activity of benzodiazepine-based compounds.42
Bioluminescence-Based Assay Reagents
RLRGG-luciferin is the first substrate reported to be used in bioluminescence-based assays to measure the activity of SENPs52,57 (Figure 4). It is a pentapeptide substrate containing the GG motif, which can be recognized by SENPs, ubiquitin, and Nedd8 proteases.57 It does not represent a physiological and specific substrate for SENPs, and it does not contain extended binding sites for SENPs. But it can provide useful information on whether a candidate compound inhibits the catalytic activity of a SENP. In 2012, Orcutt et al. developed57 SUMO-2-luciferin as a substrate which mimics the full length SUMO physiological substrate with enhanced sensitivity and specificity. Once the substrate is recognized and cleaved by SENPs, the amino-terminus of luciferin can be released and subsequently oxidized by luciferase to produce light. The relative light units can be used to monitor the activity of SENPs. The background noise in this assay is very low since luciferin cannot produce luminescence by itself, making this assay more sensitive. However, some test compounds may inhibit luciferase, thus resulting in false positives.
Conclusion
SENPs are potential drug targets due to their involvement in the development of various diseases, particularly cancer. Different types of inhibitors and probes are currently available to study SENPs. Although the probes are very potent reagents reporting SENP activity, small molecule inhibitors so far lack potency and selectivity. Good probe molecules are urgently needed to study the biology of SUMO proteases. More selective and potent chemical tools of SENPs will be helpful in shedding light on their possible mechanisms in tumorigenesis, and in exploring new strategies for cancer therapy. However, it is a major challenge to discover inhibitors and probes with isoform specificity, since all SENPs are cysteine proteases and process similar substrates. The second main challenge is to identify selective fluorescent activity based probes to report the activity of SENPs in cells and animals. The continuing development of more and better biochemical assays will accelerate the discovery of chemical tools.
Acknowledgments
Work in the Vertegaal and Ovaa laboratories is funded by NWO VICI grants and a joint ZonMW TOP grant. Y.J. is a recipient of a CSC scholarship. H.O. is a shareholder of the company UbiQ.
Glossary
Keywords
- SUMO
small ubiquitin-like modifier that can be covalently attached to target proteins
- SUMOylation
a reversible and highly dynamic post-translational modification by attaching SUMO to lysine residues of target proteins, which is orchestrated by SUMO-activating, -conjugating, and -ligating enzymes in a sequential manner
- SUMO-specific proteases (SENPs)
enzymes that can activate SUMO into its mature form before entering into SUMOylation and catalyze the deconjugation of SUMO from target proteins
- DeSUMOylation
deconjugation of SUMO from target proteins that is catalyzed by SENPs
- SUMO-activating enzyme (E1)
the enzyme that initiates SUMOylation by forming a thioester bond with SUMO in an ATP-dependent manner
- SUMO-conjugating enzyme (E2)
the enzyme that transfers SUMO to its substrate
- SUMO ligase (E3)
the enzyme that stimulates the E2-mediated conjugation of SUMO to target proteins
- Biochemical assays
assays that can be used to evaluate the activity of SENPs
The authors declare the following competing financial interest(s): H.O. is shareholder of the company UbiQ.
References
- Dasso M. (2008) Emerging roles of the SUMO pathway in mitosis. Cell Div. 3, 5. 10.1186/1747-1028-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S. P.; Durocher D. (2013) Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell 49, 795–807. 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Muller S.; Ledl A.; Schmidt D. (2004) SUMO: a regulator of gene expression and genome integrity. Oncogene 23, 1998–2008. 10.1038/sj.onc.1207415. [DOI] [PubMed] [Google Scholar]
- Gareau J. R.; Lima C. D. (2010) The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871. 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey C. M.; Wilson N. R.; Hochstrasser M. (2012) Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 13, 755–766. 10.1038/nrm3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drag M.; Salvesen G. S. (2008) DeSUMOylating enzymes--SENPs. IUBMB Life 60, 734–742. 10.1002/iub.113. [DOI] [PubMed] [Google Scholar]
- Kumar A.; Zhang K. Y. (2015) Advances in the development of SUMO specific protease (SENP) inhibitors. Comput. Struct. Biotechnol. J. 13, 204–211. 10.1016/j.csbj.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin E. J.; Shin H. M.; Nam E.; Kim W. S.; Kim J. H.; Oh B. H.; Yun Y. (2012) DeSUMOylating isopeptidase: a second class of SUMO protease. EMBO Rep. 13, 339–346. 10.1038/embor.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz S.; Chachami G.; Kozaczkiewicz L.; Winter U.; Stankovic-Valentin N.; Haas P.; Hofmann K.; Urlaub H.; Ovaa H.; Wittbrodt J.; Meulmeester E.; Melchior F. (2012) Ubiquitin-specific protease-like 1 (USPL1) is a SUMO isopeptidase with essential, non-catalytic functions. EMBO Rep. 13, 930–938. 10.1038/embor.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bawa-Khalfe T.; Yeh E. T. H. (2010) SUMO Losing Balance: SUMO Proteases Disrupt SUMO Homeostasis to Facilitate Cancer Development and Progression. Genes Cancer 1, 748–752. 10.1177/1947601910382555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge K. D.; Park-Sarge O. K. (2009) Sumoylation and human disease pathogenesis. Trends Biochem. Sci. 34, 200–205. 10.1016/j.tibs.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler J. S.; Dejean A. (2017) SUMO and the robustness of cancer. Nat. Rev. Cancer 17, 184–197. 10.1038/nrc.2016.143. [DOI] [PubMed] [Google Scholar]
- Kaikkonen S.; Jääskeläinen T.; Karvonen U.; Rytinki M. M.; Makkonen H.; Gioeli D.; Paschal B. M.; Palvimo J. J. (2009) SUMO-Specific Protease 1 (SENP1) Reverses the Hormone-Augmented SUMOylation of Androgen Receptor and Modulates Gene Responses in Prostate Cancer Cells. Mol. Endocrinol. 23, 292–307. 10.1210/me.2008-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.; Zuo Y.; Zhang H.; Kang X.; Yue F.; Yi Z.; Liu M.; Yeh E. T.; Chen G.; Cheng J. (2010) Induction of SENP1 in endothelial cells contributes to hypoxia-driven VEGF expression and angiogenesis. J. Biol. Chem. 285, 36682–36688. 10.1074/jbc.M110.164236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.; Han Y.; Wang Y.; Sun X.; Yan S.; Yeh E. T. H.; Chen Y.; Cang H.; Li H.; Shi G.; Cheng J.; Tang X.; Yi J. (2009) SENP3 is responsible for HIF-1 transactivation under mild oxidative stress via p300 de-SUMOylation. EMBO J. 28, 2748–2762. 10.1038/emboj.2009.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashman R.; Cohen H.; Ben-Hamo R.; Zilberberg A.; Efroni S. (2014) SENP5 mediates breast cancer invasion via a TGFbetaRI SUMOylation cascade. Oncotarget 5, 1071–1082. 10.18632/oncotarget.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. G.; Xie H. H.; Li N.; Wu K.; Qiu J. G.; Shen D. M.; Huang C. J. (2015) SUMO-specific protease 6 promotes gastric cancer cell growth via deSUMOylation of FoxM1. Tumor Biol. 36, 9865–9871. 10.1007/s13277-015-3737-z. [DOI] [PubMed] [Google Scholar]
- Stefanska B.; Cheishvili D.; Suderman M.; Arakelian A.; Huang J.; Hallett M.; Han Z. G.; Al-Mahtab M.; Akbar S. M. F.; Khan W. A.; Raqib R.; Tanvir I.; Khan H. A.; Rabbani S. A.; Szyf M. (2014) Genome-Wide Study of Hypomethylated and Induced Genes in Patients with Liver Cancer Unravels Novel Anticancer Targets. Clin. Cancer Res. 20, 3118–3132. 10.1158/1078-0432.CCR-13-0283. [DOI] [PubMed] [Google Scholar]
- Shen L. N.; Dong C.; Liu H.; Naismith J. H.; Hay R. T. (2006) The structure of SENP1-SUMO-2 complex suggests a structural basis for discrimination between SUMO paralogues during processing. Biochem. J. 397, 279–288. 10.1042/BJ20052030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L.; Tatham M. H.; Dong C.; Zagorska A.; Naismith J. H.; Hay R. T. (2006) SUMO protease SENP1 induces isomerization of the scissile peptide bond. Nat. Struct. Mol. Biol. 13, 1069–1077. 10.1038/nsmb1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XU Z.; AU S. W. N. (2005) Mapping residues of SUMO precursors essential in differential maturation by SUMO-specific protease, SENP1. Biochem. J. 386, 325–330. 10.1042/BJ20041210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Saitoh H.; Matunis M. J. (2002) Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell. Biol. 22, 6498–6508. 10.1128/MCB.22.18.6498-6508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverter D.; Lima C. D. (2004) A Basis for SUMO Protease Specificity Provided by Analysis of Human Senp2 and a Senp2-SUMO Complex. Structure 12, 1519–1531. 10.1016/j.str.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Hang J.; Dasso M. (2002) Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 277, 19961–19966. 10.1074/jbc.M201799200. [DOI] [PubMed] [Google Scholar]
- Gong L.; Yeh E. T. H. (2006) Characterization of a Family of Nucleolar SUMO-specific Proteases with Preference for SUMO-2 or SUMO-3. J. Biol. Chem. 281, 15869–15877. 10.1074/jbc.M511658200. [DOI] [PubMed] [Google Scholar]
- Zunino R.; Schauss A.; Rippstein P.; Andrade-Navarro M.; McBride H. M. (2007) The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. J. Cell Sci. 120, 1178–1188. 10.1242/jcs.03418. [DOI] [PubMed] [Google Scholar]
- Alegre K. O.; Reverter D. (2011) Swapping small ubiquitin-like modifier (SUMO) isoform specificity of SUMO proteases SENP6 and SENP7. J. Biol. Chem. 286, 36142–36151. 10.1074/jbc.M111.268847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D.; Ayaydin F.; Kolli N.; Tan S. H.; Anan T.; Kametaka A.; Azuma Y.; Wilkinson K. D.; Dasso M. (2006) SUSP1 antagonizes formation of highly SUMO2/3-conjugated species. J. Cell Biol. 174, 939–949. 10.1083/jcb.200510103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alegre K. O.; Reverter D. (2014) Structural insights into the SENP6 Loop1 structure in complex with SUMO2. Protein Sci. 23, 433–441. 10.1002/pro.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima C. D.; Reverter D. (2008) Structure of the human SENP7 catalytic domain and poly-SUMO deconjugation activities for SENP6 and SENP7. J. Biol. Chem. 283, 32045–32055. 10.1074/jbc.M805655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L. N.; Geoffroy M. C.; Jaffray E. G.; Hay R. T. (2009) Characterization of SENP7, a SUMO-2/3-specific isopeptidase. Biochem. J. 421, 223–230. 10.1042/BJ20090246. [DOI] [PubMed] [Google Scholar]
- Mossessova E.; Lima C. D. (2000) Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5, 865–876. 10.1016/S1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- Hemelaar J.; Borodovsky A.; Kessler B. M.; Reverter D.; Cook J.; Kolli N.; Gan-Erdene T.; Wilkinson K. D.; Gill G.; Lima C. D.; Ploegh H. L.; Ovaa H. (2004) Specific and Covalent Targeting of Conjugating and Deconjugating Enzymes of Ubiquitin-Like Proteins. Mol. Cell. Biol. 24, 84–95. 10.1128/MCB.24.1.84-95.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson K. D.; Gan-Erdene T.; Kolli N. (2005) Derivitization of the C-Terminus of Ubiquitin and Ubiquitin-like Proteins Using Intein Chemistry: Methods and Uses. Methods Enzymol. 399, 37–51. 10.1016/S0076-6879(05)99003-4. [DOI] [PubMed] [Google Scholar]
- Borodovsky A.; Ovaa H.; Meester W. J. N.; Venanzi E. S.; Bogyo M. S.; Hekking B. G.; Ploegh H. L.; Kessler B. M.; Overkleeft H. S. (2005) Small-Molecule Inhibitors and Probes for Ubiquitin- and Ubiquitin-Like-Specific Proteases. ChemBioChem 6, 287–291. 10.1002/cbic.200400236. [DOI] [PubMed] [Google Scholar]
- Dobrota C.; Fasci D.; Hadade N. D.; Roiban G. D.; Pop C.; Meier V. M.; Dumitru I.; Matache M.; Salvesen G. S.; Funeriu D. P. (2012) Glycine fluoromethylketones as SENP-specific activity based probes. ChemBioChem 13, 80–84. 10.1002/cbic.201100645. [DOI] [PubMed] [Google Scholar]
- Mulder M. P. C.; Merkx R.; Witting K. F.; Hameed D. S.; El Atmioui D.; Lelieveld L.; Liebelt F.; Neefjes J.; Berlin I.; Vertegaal A. C. O.; Ovaa H. (2018) Total Chemical Synthesis of SUMO and SUMO-Based Probes for Profiling the Activity of SUMO-Specific Proteases. Angew. Chem., Int. Ed. 57, 8958–8962. 10.1002/anie.201803483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekkebus R.; van Kasteren S. I.; Kulathu Y.; Scholten A.; Berlin I.; Geurink P. P.; de Jong A.; Goerdayal S.; Neefjes J.; Heck A. J.; Komander D.; Ovaa H. (2013) On terminal alkynes that can react with active-site cysteine nucleophiles in proteases. J. Am. Chem. Soc. 135, 2867–2870. 10.1021/ja309802n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S.; Weikart N. D.; Linne U.; Mootz H. D. (2013) Covalent inhibition of SUMO and ubiquitin-specific cysteine proteases by an in situ thiol-alkyne addition. Bioorg. Med. Chem. 21, 2511–2517. 10.1016/j.bmc.2013.02.039. [DOI] [PubMed] [Google Scholar]
- Ponder E. L.; Albrow V. E.; Leader B. A.; Bekes M.; Mikolajczyk J.; Fonovic U. P.; Shen A.; Drag M.; Xiao J.; Deu E.; Campbell A. J.; Powers J. C.; Salvesen G. S.; Bogyo M. (2011) Functional characterization of a SUMO deconjugating protease of Plasmodium falciparum using newly identified small molecule inhibitors. Chem. Biol. 18, 711–721. 10.1016/j.chembiol.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrow V. E.; Ponder E. L.; Fasci D.; Bekes M.; Deu E.; Salvesen G. S.; Bogyo M. (2011) Development of small molecule inhibitors and probes of human SUMO deconjugating proteases. Chem. Biol. 18, 722–732. 10.1016/j.chembiol.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Z.; Wang W.; Wang L.; Wen D.; Zhao Y.; Wang Q.; Meng Q.; Chen G.; Wu Y.; Zhou H. (2011) Design, synthesis, and biological evaluation of benzodiazepine-based SUMO-specific protease 1 inhibitors. Bioorg. Med. Chem. Lett. 21, 6389–6392. 10.1016/j.bmcl.2011.08.101. [DOI] [PubMed] [Google Scholar]
- Uno M.; Koma Y.; Ban H. S.; Nakamura H. (2012) Discovery of 1-[4-(N-benzylamino)phenyl]-3-phenylurea derivatives as non-peptidic selective SUMO-sentrin specific protease (SENP)1 inhibitors. Bioorg. Med. Chem. Lett. 22, 5169–5173. 10.1016/j.bmcl.2012.06.084. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Wen D.; Huang Z.; Huang M.; Luo Y.; Liu B.; Lu H.; Wu Y.; Peng Y.; Zhang J. (2012) 2-(4-Chlorophenyl)-2-oxoethyl 4-benzamidobenzoate derivatives, a novel class of SENP1 inhibitors: Virtual screening, synthesis and biological evaluation. Bioorg. Med. Chem. Lett. 22, 6867–6870. 10.1016/j.bmcl.2012.09.037. [DOI] [PubMed] [Google Scholar]
- Madu I. G.; Namanja A. T.; Su Y.; Wong S.; Li Y. J.; Chen Y. (2013) Identification and characterization of a new chemotype of noncovalent SENP inhibitors. ACS Chem. Biol. 8, 1435–1441. 10.1021/cb400177q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A.; Ito A.; Takemoto M.; Yoshida M.; Zhang K. Y. (2014) Identification of 1,2,5-oxadiazoles as a new class of SENP2 inhibitors using structure based virtual screening. J. Chem. Inf. Model. 54, 870–880. 10.1021/ci4007134. [DOI] [PubMed] [Google Scholar]
- Zhao Y.; Wang Z.; Zhang J.; Zhou H. (2016) Identification of SENP1 inhibitors through in silico screening and rational drug design. Eur. J. Med. Chem. 122, 178–184. 10.1016/j.ejmech.2016.06.018. [DOI] [PubMed] [Google Scholar]
- Mikolajczyk J.; Drag M.; Békés M.; Cao J. T.; Ronai Z. e.; Salvesen G. S. (2007) Small Ubiquitin-related Modifier (SUMO)-specific Proteases. J. Biol. Chem. 282, 26217–26224. 10.1074/jbc.M702444200. [DOI] [PubMed] [Google Scholar]
- Reverter D.; Lima C. D. (2009) Preparation of SUMO Proteases and Kinetic Analysis Using Endogenous Substrates. Methods Mol. Biol. 497, 225–239. 10.1007/978-1-59745-566-4_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencía M.; Lorenzo V. d. (2004) Functional transplantation of the sumoylation machinery into Escherichia coli. Protein Expression Purif. 37, 409–418. 10.1016/j.pep.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Tatham M. H.; Hay R. T. (2009) FRET-based in vitro assays for the analysis of SUMO protease activities. Methods Mol. Biol. 497, 253–268. 10.1007/978-1-59745-566-4_17. [DOI] [PubMed] [Google Scholar]
- Madu I. G., and Chen Y. (2012) Assays for investigating deSUMOylation enzymes, in Current Protocols in Molecular Biology, Chapter 10, Unit 10.30, Wiley. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Song Y.; Madahar V.; Liao J. (2012) Quantitative Forster resonance energy transfer analysis for kinetic determinations of SUMO-specific protease. Anal. Biochem. 422, 14–21. 10.1016/j.ab.2011.12.019. [DOI] [PubMed] [Google Scholar]
- Dang L. C.; Melandri F. D.; Stein R. L. (1998) Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry 37, 1868–1879. 10.1021/bi9723360. [DOI] [PubMed] [Google Scholar]
- Drag M.; Mikolajczyk J.; Krishnakumar I. M.; Huang Z.; Salvesen G. S. (2008) Activity profiling of human deSUMOylating enzymes (SENPs) with synthetic substrates suggests an unexpected specificity of two newly characterized members of the family. Biochem. J. 409, 461–469. 10.1042/BJ20070940. [DOI] [PubMed] [Google Scholar]
- Nicholson B.; Leach C. A.; Goldenberg S. J.; Francis D. M.; Kodrasov M. P.; Tian X.; Shanks J.; Sterner D. E.; Bernal A.; Mattern M. R.; Wilkinson K. D.; Butt T. R. (2008) Characterization of ubiquitin and ubiquitin-like-protein isopeptidase activities. Protein Sci. 17, 1035–1043. 10.1110/ps.083450408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orcutt S. J.; Wu J.; Eddins M. J.; Leach C. A.; Strickler J. E. (2012) Bioluminescence assay platform for selective and sensitive detection of Ub/Ubl proteases. Biochim. Biophys. Acta, Mol. Cell Res. 1823, 2079–2086. 10.1016/j.bbamcr.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]