Abstract
Inflammasomes are multiprotein complexes formed in response to pathogens. NLRP1 and CARD8 are related proteins that form inflammasomes, but the pathogen-associated signal(s) and the molecular mechanisms controlling their activation have not been established. Inhibitors of the serine dipeptidyl peptidases DPP8 and DPP9 (DPP8/9) activate both NLRP1 and CARD8. Interestingly, DPP9 binds directly to NLRP1 and CARD8, and this interaction may contribute to the inhibition of NLRP1. Here, we use activity-based probes, reconstituted inflammasome assays, and mass spectrometry-based proteomics to further investigate the DPP9–CARD8 interaction. We show that the DPP9–CARD8 interaction, unlike the DPP9–NLRP1 interaction, is not disrupted by DPP9 inhibitors or CARD8 mutations that block autoproteolysis. Moreover, wild-type, but not catalytically inactive mutant, DPP9 rescues CARD8-mediated cell death in DPP9 knockout cells. Together, this work reveals that DPP9’s catalytic activity and not its binding to CARD8 restrains the CARD8 inflammasome and thus suggests the binding interaction likely serves some other biological purpose.
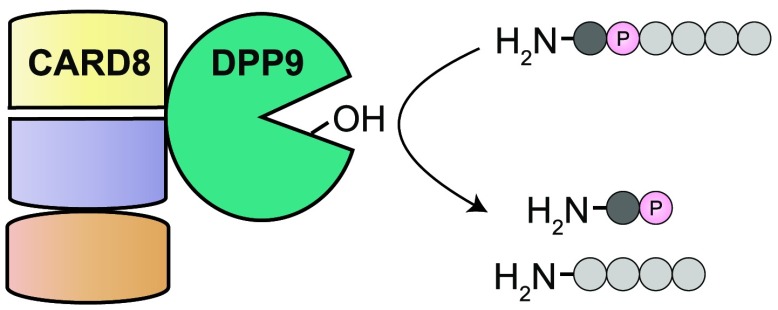
The innate immune system uses a wide array of germline-encoded receptors to detect pathogens.1 Several of these receptors recognize intracellular microbial structures and activities and form multiprotein complexes called inflammasomes.2,3 Inflammasomes recruit and activate the cysteine protease caspase-1, which in turn cleaves and activates the inflammatory cytokines IL-1β and IL-18 as well as GSDMD. The N-terminal fragment of GSDMD oligomerizes and forms pores in the plasma membrane, inducing an inflammatory form of cell death called pyroptosis (Figure S1).
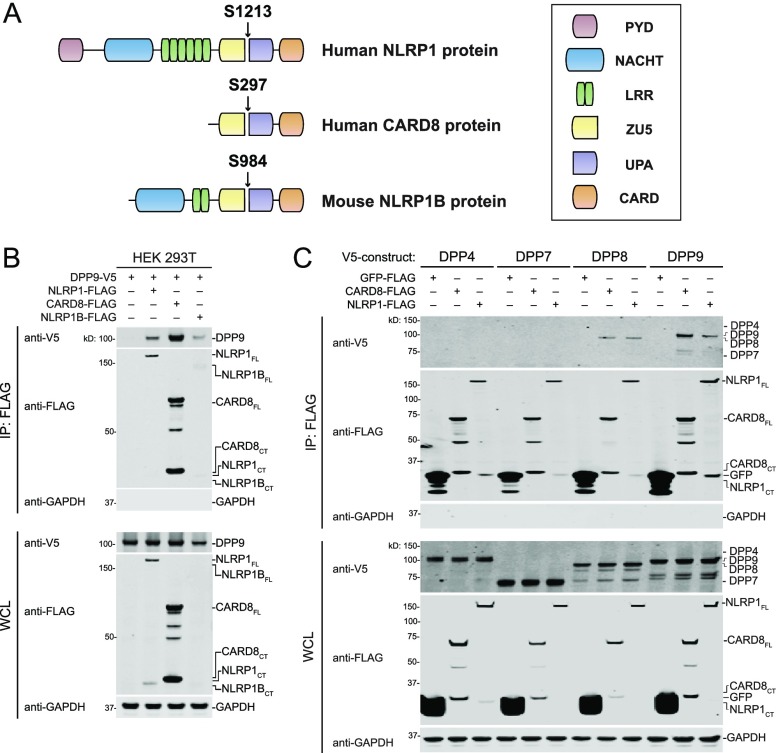
Several members of the nucleotide-binding domain leucine rich repeat (NLR) family of proteins form inflammasomes. Human NLRP1, the first protein discovered to form an inflammasome,4 consists of an N-terminal pyrin domain (PYD), followed by nucleotide-binding (NACHT), leucine-rich repeat (LRR), found in ZO-1 and UNC5 (ZU5), conserved in UNC5, PIDD, and Ankyrin (UPA), and caspase activation and recruitment (CARD) domains (Figure 1A). NLRP1 undergoes autoproteolysis near the C-terminus of its ZU5 domain, generating two fragments that remain associated in an inactive state.5−7 Humans also express CARD8, an inflammasome-forming protein similar to the C-terminal region of NLRP1 that also undergoes autoproteolysis within its ZU5 domain (Figure 1A).5 Rodents have homologues of NLRP1 lacking N-terminal PYDs (Figure 1A) but do not have CARD8 homologues.
Figure 1.
DPP8/9 association with ZU5-UPA-containing inflammasome proteins. (A) Graphical representation of NLRP1 and CARD8 proteins. The site of autoproteolysis for each protein is indicated. Allele 1 of mouse NLRP1B is shown. (B) Anti-FLAG IPs of lysates from HEK 293T cells transiently transfected with constructs encoding DPP9-V5 (1 μg) and the indicated FLAG-tagged inflammasome protein (1 μg). (C) Anti-FLAG IPs of lysates from HEK 293T cells transiently transfected with constructs encoding the indicated V5-tagged DPP (1 μg) and the indicated FLAG-tagged protein (1 μg). Immunoblots depict input whole cell lysate (WCL) and captured proteins (IP: FLAG).
The pathogen-associated signal that NLRP1 detects, if a single one exists, is not yet known. Bacillus anthracis lethal factor (LF), a zinc metalloprotease, activates a subset of rodent NLRP1 proteins,8 but not human NLRP1 or CARD8. LF directly cleaves the sensitive NLRP1 alleles near their N-termini,9−11 inducing the proteasomal degradation of their N-terminal fragments by the N-end rule pathway and releasing their C-terminal fragments to activate caspase-1.12,13 Several additional pathogens, including Shigella flexneri, Listeria monocytogenes, and Toxoplasma gondii, have also been reported to activate rodent NLRP1 inflammasomes.14−16 The Shigella flexneri IpaH7.8 ubiquitin ligase was recently shown to directly ubiquitinate and activate mouse NLRP1B,12 but the mechanism by which the other pathogens activate NLRP1, and whether they also activate human NLRP1 and CARD8, is not yet known.14−16
Several small molecules have been discovered to modulate inflammasome activity.17−19 These compounds have not only been used as chemical probes to study inflammasome regulation but have also attracted attention as potential pharmaceuticals. In particular, inhibitors of DPP8 and DPP9 (DPP8/9) were the first agents identified to activate all functional rodent NLRP1 proteins, human NLRP1, and human CARD8.18−22 DPP8/9 are highly related intracellular serine dipeptidyl peptidases that cleave N-terminal dipeptides from polypeptide substrates.23,24 DPP8/9 inhibitors, like LF, induce the proteasome-mediated degradation of the inflammasome N-terminal fragments, but the mechanistic basis of this response remains unknown.13,20 Zhong and co-workers recently reported that DPP9 directly binds to both human NLRP1 and CARD8 and suggested that the DPP9 protein itself, in addition to its catalytic activity, may play a role in inhibiting NLRP1.21 Here, we show that both NLRP1 and CARD8 do indeed associate with DPP9. However, unlike the DPP9-NLRP1 interaction, the DPP9–CARD8 interaction is not disrupted by DPP8/9 inhibitors, or even by large activity-based probes, and does not require CARD8 autoproteolysis. Moreover, the catalytic activity of DPP9 is required to rescue CARD8-mediated cell death in DPP9 knockout cells. Overall, this work reveals DPP9 activity, and not direct protein binding, inhibits the CARD8 inflammasome.
We first wanted to confirm that DPP9 interacts with human NLRP1 and CARD8. We therefore ectopically expressed V5-tagged DPP9 and FLAG-tagged NLRP1 or CARD8 in HEK 293T cells, harvested lysates, and performed anti-FLAG immunoprecipitations (IPs; Figure 1B). As previously reported,21 we observed that DPP9 bound to both CARD8 and NLRP1. We performed a similar experiment with mouse NLRP1B (allele 1) and found that it also bound to DPP9 (Figure 1B).
Although DPP9 is the primary enzyme that restrains the inflammasome, DPP8 plays a role in the absence of DPP9.19 DPP4 and DPP7 are highly similar to DPP8/9 but are not involved in inflammasome regulation.19,23 We thus tested whether these DPPs could also bind CARD8 and NLRP1. We found that DPP8, but not DPP4 or DPP7, was captured by both CARD8 and NLRP1 (Figure 1C). These results indicate that DPP9 is not the principal regulator of inflammasome activation (over DPP8) due to a unique ability to bind NLRP1 and CARD8.
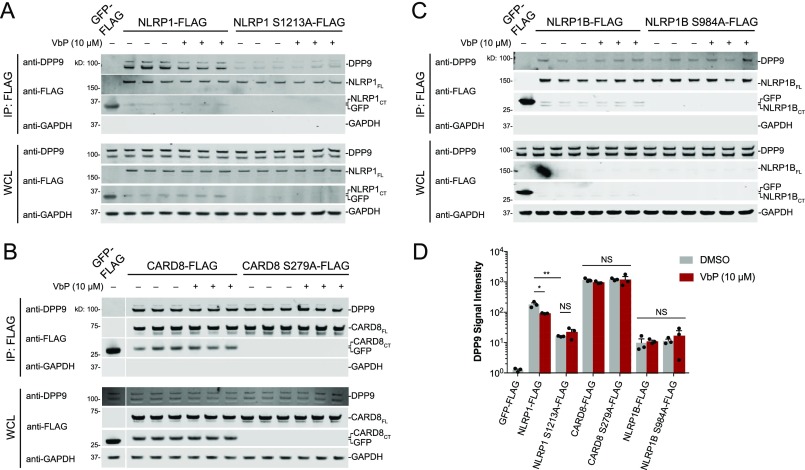
The NLRP1 FIIND domain, which is comprised of ZU5 and UPA subdomains, was reported to mediate the interaction with DPP9.21 Moreover, the autoproteolysis-deficient mutant NLRP1 F1212A protein exhibited impaired binding to DPP9, suggesting that autoproteolysis was required to create the binding interface. It remains unknown whether autoproteolysis is similarly required for NLRP1B and CARD8 binding to DPP9. We therefore expressed autoproteolysis-deficient mutant NLRP1 S1213A, CARD8 S279A, and NLRP1B S984A proteins and performed anti-FLAG IP experiments with either overexpressed DPP9 (Figure S2, Figure S3) or endogenous DPP9 (Figure 2). The latter experiment was performed in technical triplicate to enable quantification of the immunoprecipitated endogenous DPP9 (Figure 2D). NLRP1 S1213A was defective in binding to overexpressed (Figure S3) and endogenous DPP9 (Figure 2A,D), although binding was not completely eliminated. Surprisingly, the CARD8 and NLRP1B wild-type (WT) and mutant proteins bound similarly to DPP9 (Figure 2B–D, Figure S2, Figure S3). Thus, DPP9 does not bind to the neo-N-terminus of the autoproteolytic C-terminal fragment of CARD8 or NLRP1B. These data are consistent with our recent report that the NH2–Ser-Pro neo-N-terminal sequence of the NLRP1B C-terminal fragment is not a DPP9 substrate.24 Although CARD8 autoproteolysis is not required for DPP9 binding, only full-length CARD8, and not the isolated N- or C-terminal fragments, binds to DPP9 (Figure S2), as was previously observed with NLRP1.21
Figure 2.
CARD8 autoproteolysis not required for the DPP9–CARD8 interaction. (A–C) Anti-FLAG IPs of lysates from HEK 293T cells transiently transfected with constructs encoding the indicated FLAG-tagged protein (2 μg). Lysates were treated with either DMSO or VbP (10 μM) for 1 h prior to IP. Immunoblots depict input whole cell lysate (WCL) and captured proteins (IP: FLAG). (D) Quantitation of DPP9 immunoblots from A–C. Data are means ± SEM of three independent replicates. *p < 0.05; **p < 0.01; NS, not significant by two-sided Student’s t test.
Treatment of cells with small molecule DPP8/9 inhibitors, including Val-boroPro (VbP), was reported to disrupt the DPP9–NLRP1 interaction.21 On the basis of these data, the authors suggested that VbP might induce inflammasome activation, in part, by direct displacement of the DPP9–NLRP1 interaction. However, it was not established whether the DPP9 inhibitors directly disrupted the binding interface or whether the inhibition of DPP9 proteolytic activity in cells indirectly disrupted the interaction. To test if VbP directly displaced the protein–protein interaction, we treated NLRP1- and DPP9-containing lysates, rather than cells, with VbP before performing an anti-FLAG IP. We found that VbP only slightly reduced (∼2-fold) the binding of DPP9 to NLRP1. However, VbP did not impact the binding of DPP9 to NLRP1 S1213A, CARD8 (WT or S279A), or NLRP1B (WT or S984A; Figure 2).
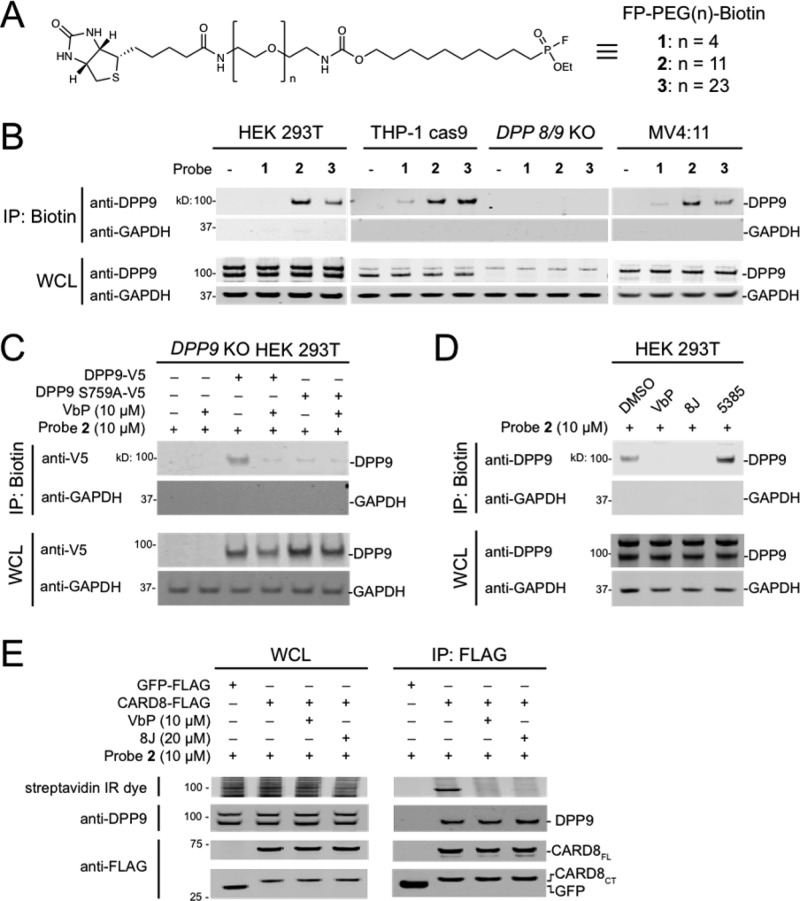
VbP is a relatively small compound (MW = 310). We hypothesized a larger inhibitor might more dramatically affect the protein–protein interactions, particularly if the interaction was proximal to DPP9’s active site. Activity-based probes (ABPs) are nonselective chemical probes that covalently modify the active sites of mechanistically related enzymes.25,26 Fluorophosphonate (FP)-based ABPs have been extensively used to profile the activity of serine hydrolases,27−29 including DPP8/9. We next synthesized a series of FP-biotin probes with various length (n = 4, 11, or 23) polyethene glycol (PEG) linkers that could be used to determine if larger inhibitors could disrupt the DPP9–CARD8 interaction (Figure 3A). We then treated lysates from HEK 293T, THP-1 Cas9 (stably expressing the Cas9 protein control), THP-1 DPP8/9 double knockout, and MV4;11 cells with these ABPs and enriched biotinylated proteins without protein denaturation (Figure 3B). The probes with the longer PEG chains, 2 and 3, captured DPP9 from the wild-type but not the DPP8/9 knockout cells (Figure 3B). The shorter probe 1 was less effective in pulling down DPP9, likely because the biotin failed to extend out of the DPP9 active-site channel. Consistent with this premise, previous applications of probe 1 required a denaturing step to capture biotinylated proteins.28 To confirm that capture of DPP9 was due to covalent modification of the catalytic serine, we treated lysates from cells ectopically expressing DPP9 or catalytically inactive DPP9 S759A with DMSO or VbP prior to labeling with 2 and enriching biotinylated proteins (Figure 3C). As expected, only DMSO-treated DPP9 was captured. Similarly, FP enrichment of endogenous DPP9 in HEK 293T lysates was blocked by VbP and the chemically distinct DPP8/9 inhibitor compound 8J, but not the DPP7 inhibitor 5385 (Figure 3D).19 To test whether 2 disrupted the DPP9–CARD8 interaction, we incubated lysates containing FLAG-tagged CARD8 with DMSO, VbP, or 8J, before treating with 2 (MW = 1087) and performing anti-FLAG IPs (Figure 3E). Consistent with our previous results, VbP- and 8J-treated DPP9 bound CARD8 but was not labeled by 2. In addition, we found that 2-labeled DPP9 equally bound CARD8, unequivocally demonstrating that inhibited DPP9 binds to CARD8. Moreover, as the long FP-biotin probe did not impact DPP9–CARD8 binding, the binding surface likely does not include the DPP9 active-site channel. Overall, these results demonstrate that DPP8/9 inhibitors modestly interfere with the direct binding of DPP9 to human NLRP1, but not CARD8.
Figure 3.
An extended activity-based probe does not disrupt the DPP9–CARD8 interaction. (A) Structure of FP-PEG(n)-Biotin probes. (B–D) Streptavidin enrichment of the indicated cell lysates with the indicated FP-biotin probes. In C, lysates from DPP9 KO HEK 293T cells transiently transfected with constructs encoding the indicated DPP9 protein (1 μg) were treated with VbP or DMSO for 1 h. In D, lysates from HEK 293T cells were treated with VbP (10 μM), 8J (20 μM), or 5385 (20 μM) for 1 h. (E) Lysates from HEK 293T cells transiently transfected with constructs encoding the indicated FLAG proteins were treated with VbP or 8J for 1 h, followed by probe 2 for 1 h, and subjected to anti-FLAG IP. Immunoblots depict input whole cell lysate (WCL) and captured proteins from either FP-biotin enrichment (IP: Biotin) or anti-FLAG IPs (IP: FLAG).
It remained possible that DPP8/9 inhibitors indirectly disrupt the CARD8–DPP9 interaction in cells, and that this disruption contributes to CARD8 inflammasome formation. To determine if VbP indirectly disrupts the CARD8–DPP9 interaction, we performed IP experiments from VbP-treated cells (Figure S4). Although VbP-induced degradation of the inflammasomes13,20 confounded analysis of the DPP9–inflammasome interactions, it appeared that CARD8 still bound approximately the same amount of DPP9 with and without VbP treatment. We should note Zhong and co-workers performed a similar experiment and observed that VbP treatment slightly reduced CARD8–DPP9 binding.21 We are unsure why our results differ, but our data collectively show that VbP, at most, only slightly disrupts the CARD8–DPP9 interaction in cells.
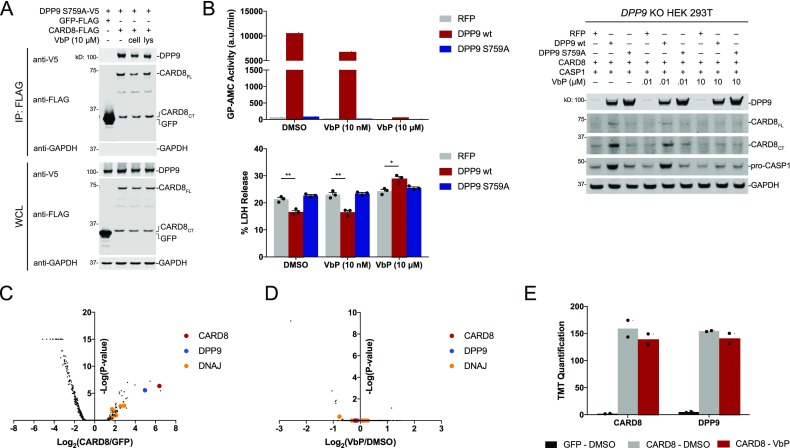
We next wanted to determine if DPP9 binding contributes to the inhibition of CARD8. If DPP9 does indeed inhibit CARD8 by direct binding, we hypothesized overexpression of the catalytically inactive DPP9 S759A protein would rescue spontaneous inflammasome activation in DPP9 deficient cells. Indeed, previous work showed that DPP9 S759A partially rescued spontaneous inflammasome formation in HEK 293T DPP8/9 double knockout cells expressing NLRP1 and ASC-GFP.21 We confirmed that DPP9 S759A binds to CARD8 (Figure S2, Figure 4A). Next, we confirmed that our DPP9 knockout HEK 293T cells were markedly deficient (∼84%) in dipeptidyl peptidase (i.e., GP-AMC cleavage) activity (Figure S5). These data are consistent with previous observations in HeLa cells,30 and we speculate that DPP9 plays a more important role than DPP8 in restraining the NLRP1 and CARD8 inflammasomes because DPP9 is responsible for the majority of intracellular postproline dipeptidase activity.19,30 We then stably reintroduced RFP, DPP9 WT, or DPP9 S759A in these knockout cells. As expected, DPP9 S759A expression did not alter GP-AMC activity in lysates, whereas DPP9 WT expression dramatically increased dipeptidase activity (Figure 4B). We then transiently transfected constructs encoding CARD8 and CASP1 into these cells and found that reintroduction of WT, but not S759A, DPP9 reduced LDH release (Figure 4B). Furthermore, inhibition of the reconstituted DPP9 activity with VbP restored cell death. We observed reduced levels of both CARD8 and CASP1 protein in cells without DPP9 activity (Figure 4B), likely due to the toxicity of their expression in these cells. Overall, these data show that DPP9’s catalytic activity, and not its CARD8 binding capacity, inhibits the CARD8 inflammasome.
Figure 4.
DPP9 proteolytic activity is necessary to rescue CARD8 mediated cell death. (A) Anti-FLAG IPs of lysates from HEK 293T cells transiently transfected with constructs encoding DPP9 S759A-V5 (1 μg) and the indicated FLAG-tagged protein (1 μg). VbP was added to the cells (“cell”) for 6 h before harvesting or to the cell lysates (“lys”) for 1 h after harvesting. Immunoblots depict input whole cell lysate (WCL) and captured proteins (IP: FLAG). (B) DPP9 KO HEK 293T cells stably expressing the indicated protein were transiently transfected with constructs encoding CARD8 (0.05 μg) and CASP1 (0.025 μg). After 36 h, cells were treated with DMSO or VbP (10 nM or 10 μM) for 6 h, before cell viability was evaluated by lactate dehydrogenase (LDH) release. Data are means ± SEM of three biological replicates. *p < 0.05, **p < 0.01 by two-sided Student’s t test. Lysates from these cells were harvested and protein levels evaluated by immunobloting and DPP activity assessed with GP-AMC assay. (C–E) HEK 293T cells were transiently transfected with constructs encoding GFP-FLAG or CARD8-FLAG. After 48 h, DMSO or VbP (10 μM) was added for an additional 24 h prior to anti-FLAG IP and MS analysis. Volcano plots depict the relative amounts of proteins in the CARD8–DMSO samples compared to the GFP-DMSO samples (C) and CARD8–VbP samples compared to the CARD8–DMSO samples (D). TMT quantification for CARD8 and DPP9 is shown (E).
Although DPP9 inhibition did not significantly affect the DPP9–CARD8 interaction, it remained possible that other CARD8 interactors were more dramatically influenced by DPP9 inhibition. To unbiasedly investigate CARD8 interactions, we ectopically expressed FLAG-tagged CARD8 or GFP in HEK 293T cells, treated cells with DMSO or VbP, and performed anti-FLAG IPs. The captured proteins were then eluted, digested with trypsin, labeled with tandem mass tags (TMT), and analyzed by mass spectrometry (Table S1). Consistent with previous results,21 DPP9 was strongly enriched in the CARD8 IP relative to the GFP IP (Figure 4C). Interestingly, a number of DNAJ chaperone proteins, including DNAJA2, were also enriched. We recently found that DNAJA2 is required for the activation of the NLRP1B inflammasome in RAW 264.7 cells,13 and these results suggest that CARD8 and NLRP1 inflammasomes are likely direct clients for DNAJA2 and related chaperones. We did not find any major differences, including in the amount of DPP9 binding, between VbP- and DMSO-treated samples (Figure 4D,E). These data further support that VbP does not directly disrupt the CARD8–DPP9 interaction and that protein binding is unlikely to directly regulate CARD8 activation.
In summary, here we have shown that NLRP1 and CARD8 interact with both DPP8 and DPP9. Interestingly, we found that the binding properties of CARD8 and NLRP1 to DPP9 are different—the NLRP1, but not the CARD8, interaction requires autoproteolysis and is modestly disrupted by chemical inhibition. Moreover, while DPP9 may inhibit NLRP1, in part, by direct protein binding,21 DPP9 binding does not appear to directly restrain the CARD8 inflammasome. The biological significance of these differences warrants future study. Overall, as binding seems to play a small, if any, role in direct inflammasome inhibition, we speculate that DPP9 instead binds to NLRP1 and CARD8 in order to localize dipeptidyl peptidase activity near the inflammasome in cells. Future investigations are needed to identify the key substrate(s) of DPP9 and to clarify the biological function of DPP9’s association with the inflammasome proteins.
Methods
General Experimental Procedures
A complete description of methods is given in Supporting Information.
Acknowledgments
We thank Z. Li, M. Miele, and R. Hendrickson for mass spectrometry assistance. This work was supported by the Josie Robertson Foundation (D.A.B.), a Stand Up to Cancer-Innovative Research Grant (Grant Number SU2C-AACR-IRG11-17 to D.A.B.; Stand Up to Cancer is a program of the Entertainment Industry Foundation; research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C), the Pew Charitable Trusts (D.A.B. is a Pew-Stewart Scholar in Cancer Research), the Pershing Square Sohn Cancer Research Alliance (D.A.B.), the NIH (R01 AI137168 to D.A.B.; F30 CA243444 to A.R.G.; T32 GM007739-Andersen to A.R.G; the MSKCC Core Grant P30 CA008748), an Alfred P. Sloan Foundation Research Fellowship (D.A.B.), Gabrielle’s Angel Foundation (D.A.B.), and the American Cancer Society (Postdoctoral Fellowship PF-17-224-01-CCG to C.Y.T.).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.9b00462.
Supplemental figures, biological methods, synthetic methods, compound characterization, and mass spectrometry data tables (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Janeway C. A. Jr.; Medzhitov R. (2002) Innate immune recognition. Annu. Rev. Immunol. 20, 197–216. 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Broz P.; Dixit V. M. (2016) Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16, 407–420. 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M.; Dixit V. M. (2014) Mechanisms and functions of inflammasomes. Cell 157, 1013–1022. 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Martinon F.; Burns K.; Tschopp J. (2002) The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 10, 417–426. 10.1016/S1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- D’Osualdo A.; Weichenberger C. X.; Wagner R. N.; Godzik A.; Wooley J.; Reed J. C. (2011) CARD8 and NLRP1 undergo autoproteolytic processing through a ZU5-like domain. PLoS One 6, e27396 10.1371/journal.pone.0027396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger J. N.; Lich J. D.; Dare L. C.; Cook M. N.; Brown K. K.; Duraiswami C.; Bertin J.; Gough P. J. (2012) Autolytic proteolysis within the function to find domain (FIIND) is required for NLRP1 inflammasome activity. J. Biol. Chem. 287, 25030–25037. 10.1074/jbc.M112.378323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frew B. C.; Joag V. R.; Mogridge J. (2012) Proteolytic processing of Nlrp1b is required for inflammasome activity. PLoS Pathog. 8, e1002659 10.1371/journal.ppat.1002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden E. D.; Dietrich W. F. (2006) Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat. Genet. 38, 240–244. 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- Levinsohn J. L.; Newman Z. L.; Hellmich K. A.; Fattah R.; Getz M. A.; Liu S.; Sastalla I.; Leppla S. H.; Moayeri M. (2012) Anthrax lethal factor cleavage of Nlrp1 is required for activation of the inflammasome. PLoS Pathog. 8, e1002638 10.1371/journal.ppat.1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmich K. A.; Levinsohn J. L.; Fattah R.; Newman Z. L.; Maier N.; Sastalla I.; Liu S.; Leppla S. H.; Moayeri M. (2012) Anthrax lethal factor cleaves mouse nlrp1b in both toxin-sensitive and toxin-resistant macrophages. PLoS One 7, e49741 10.1371/journal.pone.0049741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavarria-Smith J.; Vance R. E. (2013) Direct proteolytic cleavage of NLRP1B is necessary and sufficient for inflammasome activation by anthrax lethal factor. PLoS Pathog. 9, e1003452 10.1371/journal.ppat.1003452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandstrom A.; Mitchell P. S.; Goers L.; Mu E. W.; Lesser C. F.; Vance R. E. (2019) Functional degradation: A mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science 364, eaau1330. 10.1126/science.aau1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chui A. J.; Okondo M. C.; Rao S. D.; Gai K.; Griswold A. R.; Johnson D. C.; Ball D. P.; Taabazuing C. Y.; Orth E. L.; Vittimberga B. A.; Bachovchin D. A. (2019) N-terminal degradation activates the NLRP1B inflammasome. Science 364, 82–85. 10.1126/science.aau1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald S. E.; Chavarria-Smith J.; Boothroyd J. C. (2014) NLRP1 is an inflammasome sensor for Toxoplasma gondii. Infect. Immun. 82, 460–468. 10.1128/IAI.01170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli K. M.; Gorfu G.; Hassan M. A.; Printz M.; Crown D.; Leppla S. H.; Grigg M. E.; Saeij J. P.; Moayeri M. (2014) Inflammasome sensor NLRP1 controls rat macrophage susceptibility to Toxoplasma gondii. PLoS Pathog. 10, e1003927 10.1371/journal.ppat.1003927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman-Zenevich J.; Stuart S.; Abdel-Nour M.; Girardin S. E.; Mogridge J. (2017) Listeria monocytogenes and Shigella flexneri Activate the NLRP1B Inflammasome. Infect. Immun. 10.1128/IAI.00338-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll R. C.; Robertson A. A.; Chae J. J.; Higgins S. C.; Munoz-Planillo R.; Inserra M. C.; Vetter I.; Dungan L. S.; Monks B. G.; Stutz A.; Croker D. E.; Butler M. S.; Haneklaus M.; Sutton C. E.; Nunez G.; Latz E.; Kastner D. L.; Mills K. H.; Masters S. L.; Schroder K.; Cooper M. A.; O’Neill L. A. (2015) A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat. Med. 21, 248–255. 10.1038/nm.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okondo M. C.; Rao S. D.; Taabazuing C. Y.; Chui A. J.; Poplawski S. E.; Johnson D. C.; Bachovchin D. A. (2018) Inhibition of Dpp8/9 Activates the Nlrp1b Inflammasome. Cell Chem. Biol. 25, 262–267.e5. 10.1016/j.chembiol.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okondo M. C.; Johnson D. C.; Sridharan R.; Go E. B.; Chui A. J.; Wang M. S.; Poplawski S. E.; Wu W.; Liu Y.; Lai J. H.; Sanford D. G.; Arciprete M. O.; Golub T. R.; Bachovchin W. W.; Bachovchin D. A. (2017) DPP8 and DPP9 inhibition induces pro-caspase-1-dependent monocyte and macrophage pyroptosis. Nat. Chem. Biol. 13, 46–53. 10.1038/nchembio.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. C.; Taabazuing C. Y.; Okondo M. C.; Chui A. J.; Rao S. D.; Brown F. C.; Reed C.; Peguero E.; de Stanchina E.; Kentsis A.; Bachovchin D. A. (2018) DPP8/DPP9 inhibitor-induced pyroptosis for treatment of acute myeloid leukemia. Nat. Med. 24, 1151–1156. 10.1038/s41591-018-0082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong F. L.; Robinson K.; Teo D. E. T.; Tan K. Y.; Lim C.; Harapas C. R.; Yu C. H.; Xie W. H.; Sobota R. M.; Au V. B.; Hopkins R.; D’Osualdo A.; Reed J. C.; Connolly J. E.; Masters S. L.; Reversade B. (2018) Human DPP9 represses NLRP1 inflammasome and protects against autoinflammatory diseases via both peptidase activity and FIIND domain binding. J. Biol. Chem. 293, 18864–18878. 10.1074/jbc.RA118.004350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai K.; Okondo M. C.; Rao S. D.; Chui A. J.; Ball D. P.; Johnson D. C.; Bachovchin D. A. (2019) DPP8/9 inhibitors are universal activators of functional NLRP1 alleles. Cell Death Dis. 10, 587. 10.1038/s41419-019-1817-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum J. S.; Kozarich J. W. (2003) Prolyl peptidases: a serine protease subfamily with high potential for drug discovery. Curr. Opin. Chem. Biol. 7, 496–504. 10.1016/S1367-5931(03)00084-X. [DOI] [PubMed] [Google Scholar]
- Griswold A. R.; Cifani P.; Rao S. D.; Axelrod A. J.; Miele M. M.; Hendrickson R. C.; Kentsis A.; Bachovchin D. A. (2019) A Chemical Strategy for Protease Substrate Profiling. Cell Chem. Biol. 26, 901–907.e6. 10.1016/j.chembiol.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M. J.; Cravatt B. F. (2006) Mechanism-based profiling of enzyme families. Chem. Rev. 106, 3279–3301. 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- Cravatt B. F.; Wright A. T.; Kozarich J. W. (2008) Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 77, 383–414. 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Patricelli M. P.; Cravatt B. F. (1999) Activity-based protein profiling: the serine hydrolases. Proc. Natl. Acad. Sci. U. S. A. 96, 14694–14699. 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachovchin D. A.; Koblan L. W.; Wu W.; Liu Y.; Li Y.; Zhao P.; Woznica I.; Shu Y.; Lai J. H.; Poplawski S. E.; Kiritsy C. P.; Healey S. E.; DiMare M.; Sanford D. G.; Munford R. S.; Bachovchin W. W.; Golub T. R. (2014) A high-throughput, multiplexed assay for superfamily-wide profiling of enzyme activity. Nat. Chem. Biol. 10, 656–663. 10.1038/nchembio.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachovchin D. A.; Ji T.; Li W.; Simon G. M.; Blankman J. L.; Adibekian A.; Hoover H.; Niessen S.; Cravatt B. F. (2010) Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proc. Natl. Acad. Sci. U. S. A. 107, 20941–20946. 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R.; Parmentier N.; Moller U.; Urlaub H.; Van den Eynde B. J.; Melchior F. (2009) The cytoplasmic peptidase DPP9 is rate-limiting for degradation of proline-containing peptides. J. Biol. Chem. 284, 27211–27219. 10.1074/jbc.M109.041871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.