Abstract
Hyperplastic synovial fibroblasts (SFs) serve a critical role in the pathogenesis of knee osteoarthritis (OA); however, the molecular mechanism involved in OA during synovial tissue hyperproliferation remains unclear. Growth arrest-specific gene 1 (GAS1), a cell growth repressor gene, was found to be downregulated in OASFs according to previous preliminary experiments. It was therefore hypothesized that reduced GAS1 expression may participate in the hyperproliferation of SFs in OA development, downstream of possible microRNA (miR) regulation, in hyperplastic OASFs. In the present study, GAS1 expression was indeed decreased in OASFs and interleukin-1β-induced SFs by reverse transcription-quantitative PCR and western blot analysis. Further cell viability assays, cell cycle and apoptosis analyses revealed that the overexpression of GAS1 can inhibited proliferation, induced cell cycle arrest and promoted apoptosis in SFs. In contrast, GAS1 knockdown in SFs accelerated cell proliferation, enhanced cell cycle progression and suppressed apoptosis. Notably, the suppressive effects of GAS1 were mediated through the inactivation of the PI3K-Akt pathway. Finally, miR-34a-5p and miR-181a-5p were predicted and subsequently verified to directly target the 3′-untranslated region of the GAS1 gene, downregulating GAS1 levels in OASFs and IL-1β-induced SFs. In conclusion, the present study demonstrated that downregulation of GAS1 can lead to the hyperproliferation of SFs in OA pathogenesis through the PI3K-Akt pathway, and miR-34a-5p and miR-181a-5p are potential regulators of GAS1 expression in OA. Therefore, it may be promising to investigate the potential of GAS1 as a novel therapeutic target for preventing SF hyperplasia in OA.
Keywords: osteoarthritis, synovial fibroblast, growth arrest specific gene 1, proliferation, apoptosis, PI3K-Akt pathway, microRNA-34a-5p, microRNA-181a-5p
Introduction
Knee osteoarthritis (OA) is one of the predominant causes of knee disability, which is characterized by the gradual deterioration of the articular cartilage, hypertrophic inflammatory synovium and subchondral osteophyte formation (1). OA causes joint pain, limitation of joint function and impaired mobility. This disease is widely regarded as being multifactorial rather than a simple degradative disease. Accumulating evidence has shown that low-grade chronic synovial inflammation is involved in the entire pathological process of OA (2). Proinflammatory cytokines, including interleukin-1β (IL-1β), tumor necrosis factor α (TNF-α) and interleukin 6 (IL-6), have been consistently found to be elevated in the synovial fluid, synovial tissues and extracellular matrix around the cartilage (3). Hyperactive and hyperplastic synovial tissues can lead to the synthesis and release of abundant cytokines, chemokines, matrix metalloproteinases and collagenases, which can in turn trigger macrophage infiltration, cartilage degradation and chondrocyte apoptosis (2,4). However, the molecular mechanism underlying the hyperproliferation of osteoarthritis synovial fibroblasts (OASFs) remains unclear.
A large number of genes have been found to be aberrantly expressed in OASFs compared with non-OASFs (5). According to a previous gene expression profiling analysis performed in our laboratory, it was found from microarray assays that growth arrest-specific gene 1 (GAS1) was downregulated in OASFs (Jin et al, unpublished). Growth arrest-specific gene 1 (GAS1) is a 37 kDa glycosyl-phosphatidylinositol-anchored protein (6). It has been shown to serve as a suppressive regulator of cell growth and as a mediator of apoptosis in a variety of normal or neoplastic of cell types, including mouse fibroblasts, human glioma, thyroid and breast cancer cells (7–9). The expression of GAS1 has been found to be reduced in, and negatively associated with the progression and metastasis of, various malignancies, including gastric, breast and colorectal cancer, and glioblastoma (9–13). Previous studies have revealed that GAS1 can directly inactivate RET, thereby inhibiting the RET-mediated PI3K-Akt/Bad tumorigenesis pathway (14,15). However, the biological role of GAS1 in SFs during OA pathogenesis is poorly understood. Therefore, it may be hypothesized that the downregulation of GAS1 can contribute to the proliferative phenotype of OASFs to aggravate inflammation in OA joints.
MicroRNAs (miRNAs or miRs) belong to a family of single-stranded non-coding RNA molecules (~20–26 nucleotides in length) that regulate mRNA translation and stability in multiple physiological and pathological settings (16). Accumulating evidence demonstrates that miRNAs can alter the epigenetic profiles of multiple cell types, in turn affecting their downstream biological and pathological processes, including cell proliferation, differentiation, invasion and angiogenesis (17). Indeed, numerous studies have reported that miR-146a, miR-155, miR-124a and miR-126 are widely implicated in inflammation or hyperplasia in synovial tissues and OA pathogenesis (18–20). Considering miRNA sequencing data (21–23) and Targetscan 7.2 prediction results, it was hypothesized that potential miRNA candidates that can directly regulate GAS1 expression may exist in OASFs.
The present study sought to elucidate the role of GAS1 in hyperplastic OASFs in addition to the molecular mechanisms involved. Subsequent experiments were applied to verify and investigate potential miRNA candidates that can regulate GAS1 expression in OASFs.
Materials and methods
Synovial tissue specimens
All human synovial specimens were acquired in the Department of Orthopedics, Tangdu Hospital of the Fourth Military Medical University (Xi'an, China) between May 2016 and September 2017. A total of 10 OA synovial tissue specimens were collected from end-stage OA patients (sex, 4 males and 6 females; age range, 55–78 years) with total knee arthroplasty and from seven non-OA counterparts (sex, 5 males and 2 females; age range, 34–67 years) following traumatic amputation, respectively. Patients with rheumatoid arthritis, potential bacterial infection of the knee joint and other inflammatory arthritis, including Lupus arthritis, ankylosing arthritis and seronegative arthritis were excluded from the study. Informed consent was obtained from all the participants concerned in the present study, and the study was approved by the Institutional Review Board of Tangdu Hospital, Fourth Military Medical University (Xi'an, China).
Primary culture of synovial fibroblasts and IL-1β stimulation
Synovial tissues freshly obtained from OA and non-OA patients (average tissue size, 1×1×0.5 cm) were washed with phosphate-buffered saline (PBS), minced thoroughly using a scalpel, and subjected to 2 h enzymatic digestion in DMEM/F12 (HyClone; GE Healthcare Life Sciences) supplemented with 1 mg/ml type I collagenase (Worthington Biochemical Corporation) on a shaking incubator (speed, 100 rpm) at 37°C. Cell suspensions were filtered using a 100-µm cell strainer (BD Biosciences) and placed in 35 cm2 tissue culture flasks containing DMEM/F12 supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin. The primary medium was then switched after 48 h incubation (37°C, 5% CO2) to remove dead tissue and non-adherent cells. A total of 1–2 passages were performed after 5 days of culture when the adherent cells approached 70% confluence. Following three generations of passage, highly purified SFs were used for further assays. Luciferase assays were performed in 293T cell lines, which were maintained in DMEM with 10% FBS in a constant-temperature incubator (37°C, 5% CO2). For IL-1β stimulation, the sub-cultured SFs at passage 3 were seeded into 6-well dishes at a density of 2×105 cells/well. After 48 h incubation, when the cell confluence was ~80%, cells were exposed to conditioned medium (DMEM/F12 with 1% FBS) supplemented with various concentrations (5, 10 or 20 ng/ml) of IL-1β (R&D Systems, Inc.). A negative control group was established by using an equivalent volume of PBS instead of IL-1β.
RNA extraction, cDNA synthesis and reverse transcription-quantitative PCR (RT-qPCR)
Total cellular RNA from non-OASFs and OASFs was isolated and purified using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocols. Subsequent cDNA synthesis for miRNA detection was performed using miDETECT A Track™ miRNA qPCR Starter Kit (Guangzhou RiboBio Co., Ltd.) from 1 µg total RNA of each sample, according to manufacturer's protocol. The temperature protocol for cDNA was set as follows: Poly (A) tailing at 37°C for 1 h, reverse transcription reaction at 42°C for 1 h, followed by inactivation at 72°C for 10 min. The qPCR reaction was performed in a Rotor-Gene Q PCR cycler system (Qiagen GmbH). The temperature protocol for miRNA qPCR was set as follows: Initial enzyme activation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 2 sec, annealing at 60°C for 20 sec, extension and fluorescence detection at 70°C for 10 sec. Subsequent cDNA synthesis for mRNA detection was performed using 5X All-In-One RT MasterMix kit (Applied Biological Materials, Inc.) from 1 µg total RNA, according to the manufacturer's protocol. The temperature program was set as follows: Incubation at 25°C for 10 min, followed by 42°C for 15 min and, finally, inactivation at 85°C for 5 min. Subsequent qPCR was performed using the EvaGreen® 2X qPCR MasterMix kit (Applied Biological Materials, Inc.) also in the Rotor-Gene Q PCR cycler system, according to the manufacturer's protocol. The thermocycling conditions for qPCR were set as follows: Initial enzyme activation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 sec, annealing and extension at 60°C for 1 min.
For miRNA expression detection, U6 was also measured and served as the internal control, whilst GAPDH was used as the internal control for mRNA expression detection. The oligonucleotide primers for qPCR were purchased from Sangon Biotech Co., Ltd. miScript Primers for miR-34a-5p, miR-203a-3p, miR-181a-5p and U6 were purchased from Qiagen, Inc. The sequences of qPCR primers used for mRNA and miRNA detection are listed in Table I. Results were normalized to the respective internal controls using the 2−ΔΔCq method (24).
Table I.
Sequences for primers, siRNAs, miRNA mimics and inhibitors.
| Name | Sequence (5′ to 3′) |
|---|---|
| qPCR primers for mRNA detection | |
| GAPDH | |
| Forward | GGAGCGAGATCCCTCCAAAAT |
| Reverse | GGCTGTTGTCATACTTCTCATGG |
| GAS1 | |
| Forward | ATGCCGCACCGTCATTGAG |
| Reverse | TCATCGTAGTAGTCGTCCAGG |
| qPCR primers for miRNA detection | |
| Hsa-miR-34a_1 Forward | UGGCAGUGUCUUGGUUGU |
| Hsa-miR-181a_2 Forward | AACAUUCAACGCUGUCGGUGAGU |
| Hsa-miR-203_1 Forward | GUGAAAUGUUUAGGACCACUAG |
| U6 Forward | CGCAAGGATGACACGCAAATTC |
| Universal Reverse | GACGAGGACTCGAGCTCAAGCT |
| Primers for GAS1 coding sequence subcloning | |
| Forward | ATCGAATTCCTTCCTGGTAATTCTTCACCTCTT |
| Reverse | ATCGCTCGAGAGTGGCCGATTGAAAGGTATATT |
| Primers for Luciferase assay | |
| GAS1-wt-3′UTR | |
| Forward | ATCGCTCGAGGTCCCACTTACCGATTCATTCT |
| Reverse | ATATGCGGCCGCTCACAATGGACTGTGGGTTT |
| GAS1-mt-3′UTR for miRNA-34a-5p | |
| Primer 1 | |
| Forward | TAAAAAAGCTCTGCTCTGCCATGTATGAAAGTCTC |
| Reverse | GAGACTTTCATACATGGCAGAGCAGAGCTTTTTTA |
| Primer 2 | |
| Forward | AAAAAAGCTCTGCTGTGCCATGTATGAAAGTCTCT |
| Reverse | AGAGACTTTCATACATGGCACAGCAGAGCTTTTTT |
| Primer 3 | |
| Forward | AAAAGCTCTGCTGAGCCATGTATGAAAGTCTC |
| Reverse | GAGACTTTCATACATGGCTCAGCAGAGCTTTT |
| Primer 4 | |
| Forward | AAAAGCTCTGCTGACCCATGTATGAAAGTCTCTTT |
| Reverse | AAAGAGACTTTCATACATGGGTCAGCAGAGCTTTT |
| Primer 5 | |
| Forward | TAAAAAAGCTCTGCTGACGCATGTATGAAAGTCTC |
| Reverse | GAGACTTTCATACATGCGTCAGCAGAGCTTTTTTA |
| GAS1-mt-3′UTR for miRNA-181a-5p | |
| Primer 1 | |
| Forward | GTTTAAATATGCGGAGTTTGTATATTGCCTCTGCTCC |
| Reverse | GGAGCAGAGGCAATATACAAACTCCGCATATTTAAAC |
| Primer 2 | |
| Forward | GTTTAAATATGCGGAGTTACTATATTGCCTCTGCTCC |
| Reverse | GGAGCAGAGGCAATATAGTAACTCCGCATATTTAAAC |
| siRNAs | |
| SiRNA-GAS1 | |
| Sense | CUACUACGACGAAGAAUAUTT |
| Anti-sense | AUAUUCUUCGUCGUAGUAGTT |
| SiRNA-NC | |
| Sense | UUCUCCGACGUGUCACGUTT |
| Anti-sense | ACGUGACACGUUCGGAGAATT |
| miRNA mimics and inhibitors | |
| miR-34a-5p mimics | UGGCAGUGUCUUAGCUGGUUGUAACCAGCUAAGACACUGCCAUU |
| miR-181a-5p mimics | AACAUUCAACGCUGUCGGUGAGUUCACCGACAGCGUUGAAUGUUUU |
| miR-NC | CAGUACUUUUGUGUAGUACAA |
| miR-34a-5p inhibitor | ACAACCAGCUAAGACACUGCCA |
| miR-181a-5p inhibitor | ACUCACCGACAGCGUUGAAUGUU |
GAS1, growth arrest specific-1; miR, microRNA; siRNA, small interfering RNA; NC, negative control; mt, mutant.
GAS1 plasmid construction
pcDNA3.1(+) plasmid (Invitrogen; Thermo Fisher Scientific, Inc.) was used for GAS1 overexpression vector construction. To obtain a GAS1 coding sequence (CDS) for subcloning, a PCR procedure was performed to generate a 1600-bp DNA product using a pair of primers containing EcoRI and XhoI restricting sites, using Taq DNA polymerase (Invitrogen; Thermo Fisher Scientific, Inc.) and template cDNA produced by from reverse transcription from non-OASF mRNA. The thermocycling protocol for this PCR reaction was set as follows: Initial denaturation at 95°C for 3 min, followed by 35 cycles of denaturation at 95°C for 30 sec, annealing at 56°C for 45 sec, and extension at 72°C for 1 min; The final extension is at 72°C for 10 min. primers for the PCR sub-cloning of GAS1 CDS are listed in Table I. The PCR products and pcDNA3.1(+) vector were first digested with EcoRI and XhoI and subsequently ligated. The assembled pcDNA3.1(+)-GAS1 (pcDNA-GAS1) was then ready for transfection, and pcDNA3.1(+) (pcDNA-vector) was also used as the negative control.
Nucleotide transfection into cells
Cells used for nucleotide transfection were distributed into the following groups: siRNA-GAS1, siRNA-NC, pcDNA-GAS1, pcDNA-vector and Blank control group. SFs in the siRNA-GAS1 and siRNA-NC groups were transfected with GAS1-specific siRNA and siRNA negative control, respectively. SFs in pcDNA-GAS1 and pcDNA-vector groups were transfected with reconstructed pcDNA3.1(+)-GAS1 plasmid and pcDNA3.1(+) plasmid, respectively. For miRNA transfection, miR-34a-5p and miR-181a-5p mimics and corresponding inhibitors were transfected into non-OASFs. miRNA mimics NC were used as negative control for miRNA mimics. siRNAs, miRNA mimics and inhibitors were purchased from Sangon Biotech Co., Ltd., and the sequences of these nucleotide products are listed in Table I. For the transfection procedure, SFs at passage 3 were trypsinized, distributed equally (2×105 per well) into 6-well plates and incubated under the condition of 37°C, 5% CO2. When 60% cell confluence was achieved, 2.5 µg plasmids, 75 pmol siRNA, miRNA mimics or inhibitors were added by mixing with 5 µl Lipofectamine® 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and 250 µl Opti-MEM (Gibco; Thermo Fisher Scientific, Inc.) per well, according to manufacturer's protocol. A blank group was set up by using the same volume of PBS in place of the nucleotides and Lipofectamine 2000. After 6 h incubation under 37°C and 5% CO2, the medium was changed to normal DMEM/F12. After another 48 h incubation under the same condition, cells were ready for subsequent experiments. The GAS1 mRNA expression and miRNAs expression were assessed using RT-qPCR 48 h following transfection according to standard protocols. To examine further if PI3K-Akt pathway activity is involved after GAS1 knockdown in non-OASFs, LY294002, an inhibitor of PI3K (50 µM; MedChemExpress) was added to the wells 2 h prior to siRNA-GAS1 transfection of non-OASFs, which is set as the siRNA-GAS1-LY group,
Immunofluorescence
Immunofluorescence detection was performed to verify siRNA transfection efficiency. At 48 h after transfection, cells from each group were inoculated into 8-well chamber slides (Thermo Fisher Scientific, Inc) at a density of 1×104 cells/well. Cells were subsequently cultured (37°C, 5% CO2) until 70% confluence was reached, following which they were washed with PBS and fixed with 4% paraformaldehyde at room temperature for 10 min, followed by permeabilization with 0.5% Triton X-100 (Sigma-Aldrich; Merck KGaA) for 30 min at room temperature. For subsequent blocking, 5% bovine serum albumin (BSA) solution (Sigma-Aldrich; Merck KGaA) was added to each chamber at room temperature for 1 h. The cells were then incubated with rabbit polyclonal anti-GAS1 antibody (1:200; cat. no. 17903-1-AP; ProteinTech Group, Inc.) at 4°C overnight, followed by washing with PBS and incubation with goat Cy3-conjugated anti-rabbit IgG secondary antibody (1:100; cat.no. SA00009-2; ProteinTech Group, Inc.) in the dark at room temperature for 1 h. Subsequently, the cells were incubated with DAPI Staining Solution (1:1,000; cat. no. 28718-90-3; Beyotime Institute of Biotechnology) in the dark at room temperature for 10 min and washed with PBS. Images of the cells were captured using Olympus IX71 fluorescent microscope (magnification, ×400; Olympus Corporation).
Protein extraction and western blotting assay
Cells at 80% confluence were lysed on ice using RIPA lysis buffer (Sigma-Aldrich; Merck KGaA) supplemented with proteinase inhibitor cocktail (100X; Beijing ComWin Biotech Co., Ltd.) and phosphatase inhibitor cocktail (Beijing Solarbio Science & Technology Co., Ltd.). After 30 min incubation on ice, cell protein lysates were collected and centrifuged under 15,000 × g for 10 min at 4°C. Protein concentrations were quantified using a bicinchoninic acid assay (Beijing ComWin Biotech Co., Ltd.). A total of 40 µg protein samples were separated by 10% SDS-PAGE (Bio-Rad Laboratories, Inc.) and transferred onto PVDF membranes (Thermo Fisher Scientific, Inc.). Membranes were first blocked in 5% (w/v) non-fat milk solution for 1 h at room temperature before being incubated in specific primary antibody (Ab) solutions at 4°C overnight. The primary Abs used were as follows: GAS1 polyclonal rabbit Ab (1:1,000; cat. no. 17903-1-AP; ProteinTech Group, Inc.); PI3K (p85) polyclonal rabbit Ab (1:1,000; cat. no. 4292; Cell Signaling Technology, Inc.); phosphorylated (p)-Akt (Ser473) polyclonal rabbit Ab (1:1,000; cat. no. 9271; Cell Signaling Technology, Inc.); total Akt polyclonal rabbit Ab (1:1,000; cat. no. 9272; Cell Signaling Technology, Inc.); cyclin-dependent kinase 2 (Cdk2) polyclonal rabbit Ab (1:1,000; cat. no. 2546; Cell Signaling Technology, Inc.); Bax polyclonal rabbit Ab (1:750; cat. no. ab199677; Abcam); and GAPDH polyclonal rabbit Ab (1:2,000; cat. no. 2118; Cell Signaling Technology, Inc.). Subsequently, membranes were incubated in room temperature with goat horseradish peroxidase-conjugated anti-rabbit IgG secondary Ab (1:5,000; cat. no. 7074; Cell Signaling Technology, Inc.) for 2 h, followed by extensive washing for 15 min three times. Protein bands were visualized using Immobilon Western Chemiluminescent HRP Substrate (EMD Millipore), and band intensities were measured using a ChemiDoc XRS chemiluminescence imaging system (Bio-Rad Laboratories, Inc.), according to the manufacturer's protocols. Densitometric analysis and quantification were performed using ImageJ (version 1.52a; National Institutes of Health) and were normalized to GAPDH. The Akt phosphorylation ratios were calculated using the following formula: (p-Akt/GAPDH)/(total Akt/GAPDH).
Cell viability assay
A Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) assay was applied to assess the cell viability of SFs. Transfected SFs were trypsinized and seeded in 96 well plates (1×104 cells/well) in triplicate followed by incubation at 37°C and 5% CO2. A total of 10 µl CCK-8 solution was then added to each well 0, 24, 48, 72 and 96 h after culture for 1 h, following which optical density (OD) was measured at 450 nm using an Infinite M200 Pro multifunctional microplate reader (Tecan Group, Ltd.).
Apoptosis and cell cycle detection by flow-cytometry
For apoptosis assays, SFs in suspension were collected, rinsed with ice-cold PBS and kept on ice at a concentration of 1×106 cells/ml in a volume of 500 µl 48 h after transfection. In total, 3 µl Annexin V-FITC (Nanjing Keygen Biotech. Co. Ltd.) was added to each sample and incubated in the dark room for 15 min at 4°C. Subsequently, 5 µl propidium iodide (PI; Nanjing Keygen Biotech. Co. Ltd.) was added and incubated for 5 min at 4°C. Following staining, samples were then subjected to flow cytometry analysis (NovoCyte®; ACEA Bioscience, Inc.).
For cell cycle analysis, transfected SFs were washed three times and fixed with 70% ethanol at 4°C overnight to a final concentration of 1×106 cells/ml. The cells were then rinsed with PBS and centrifuged (500 × g and 5 min at room temperature) to eliminate the fixation buffer, before they were mixed with 500 µl PI/RNase solution and incubated for 60 min in the dark at room temperature Finally, cell samples were measured using flow cytometry (NovoCyte) at a wavelength of 450 nm using the NovoExpress™ software (version 1.2.1; ACEA Biosciences, Inc.).
miRNA prediction
Candidate miRNAs and corresponding binding sites were predicted with the help of Targetscan website (version 7.2; http://www.targetscan.org/vert_72/). By entering GAS1 in the ‘Gene symbol’, a list of miRNA families whose seed regions match the 3′untranslated region (3′UTR) of GAS1 were obtained. In total, 18 miRNA families and corresponding binding sites were listed as ‘broadly conserved among vertebrates.’ Following a literature search, two previous studies were found showing differentially expressed miRNAs between OA and non-OA synovial tissues, synovial fluids or synovial fibroblasts (21,23). By pooling the data obtained from the predicted miRNA list and these two studies, three miRNA candidates potentially regulating GAS1 expression in OASFs were shortlisted.
Luciferase assay
For the luciferase assay, the psi-CHECK2 vector (Promega Corporation) was used to construct luciferase reporter plasmids. The wild-type fragment of GAS1 3′untranslated region (GAS1-wt-3′UTR) containing the binding sites for candidate miRNAs, was sub-cloned by PCR using specific primers containing restriction enzyme cutting sites for XhoI and NotI. The template cDNA used for cloning was acquired following reverse transcription from non-OASF mRNA. Mutant 3′UTR fragments of GAS1 (GAS1-mut-3′UTR), mutated at the predicted miR-34a-5p or miR-181a-5p binding sites, were created by site-directed mutagenesis in which consecutive bases were replaced to disrupt miR-34a-5p or miR-181a-5p binding. The sequences of GAS1-wt-3′UTR and GAS1-mt-3′UTR primers used were listed in Table I. PCR products and the psi-CHECK2 reporter vector were digested with XhoI and NotI for 16 h at 37°C and ligated overnight at 16°C. The assembled luciferase reporter vectors were amplified and purified, then 2.5 µg reconstructed reporter vector was co-transfected alongside 75 pmol miRNA mimics into 6-well plates of 293T cells using Lipofectamine 2000 reagent and Opti-MEM, according to manufacturers' protocols. The cells were then seeded into 96 well plates at a concentration of 1×104 cells/well. After 48 h incubation, luciferase activity was tested in each well using the Luc-Pair™ Duo-Luciferase HS Assay Kit (GeneCopoeia, Inc.) on an Infinite M200 Pro Multifunctional microplate reader (Tecan Group, Ltd.). All firefly luciferase activity was normalized to Renilla luciferase activity.
Statistical analysis
For RT-qPCR, CCK-8 cell viability and luciferase assays, data were normalized to corresponding controls and are presented as the mean ± SEM from ≥3 separate replicates. One-way ANOVA was performed for multiple comparisons, followed by Tukey's honest significant difference test for pairwise comparisons. Statistical analysis was performed using GraphPad prism 7.00 software (GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
GAS1 expression is downregulated in OASFs
The expression of GAS1 was first assessed using RT-qPCR and western blotting in non-OASFs and OASFs. In total, seven non-OASF and 10 OASFs samples were examined for RT-qPCR. For western blotting, three individual protein samples were randomly selected from each group for protein measurements. The expression of GAS1 was found to be downregulated in OASFs compared with non-OASFs on mRNA and protein levels (Fig. 1A and B), consistent with the findings of our previous gene profiling (Jin et al, unpublished), which led to further study into the function of GAS1 in OASFs.
Figure 1.
GAS1 expression is downregulated in OASFs and IL-1β-stimulated non-OASFs. (A) The relative expression of GAS1 mRNA and (B) protein in non-OASFs and OASFs. (C) The relative expression of GAS1 mRNA and (D) protein in non-OASFs treated with four ascending concentrations of IL-1β (0, 5, 10 and 20 ng/ml). *P<0.05 and **P<0.01. OASFs, osteoarthritis synovial fibroblasts; IL-1β, interleukin-1β; GAS1, growth arrest specific-1; NO, non-OASF; OA, OASF.
IL-1β treatment downregulates GAS1 expression in non-OASFs
IL-1β is a well-known pro-inflammatory cytokine which has been reported to exhibit growth-promoting properties in synovial tissue of OA patients (25). Therefore, multiple concentrations of IL-1β (5, 10 and 20 ng/ml) were used to stimulate non-OASFs. The expression of GAS1 in the IL-1β-stimulated groups was reduced at the mRNA and protein levels compared with corresponding the PBS control group, in a dose-dependent manner (Fig. 1C and D). This suggested that IL-1β stimulation may suppress GAS1 expression in a dose-dependent manner.
GAS1 overexpression inhibits SF proliferation and promotes apoptosis
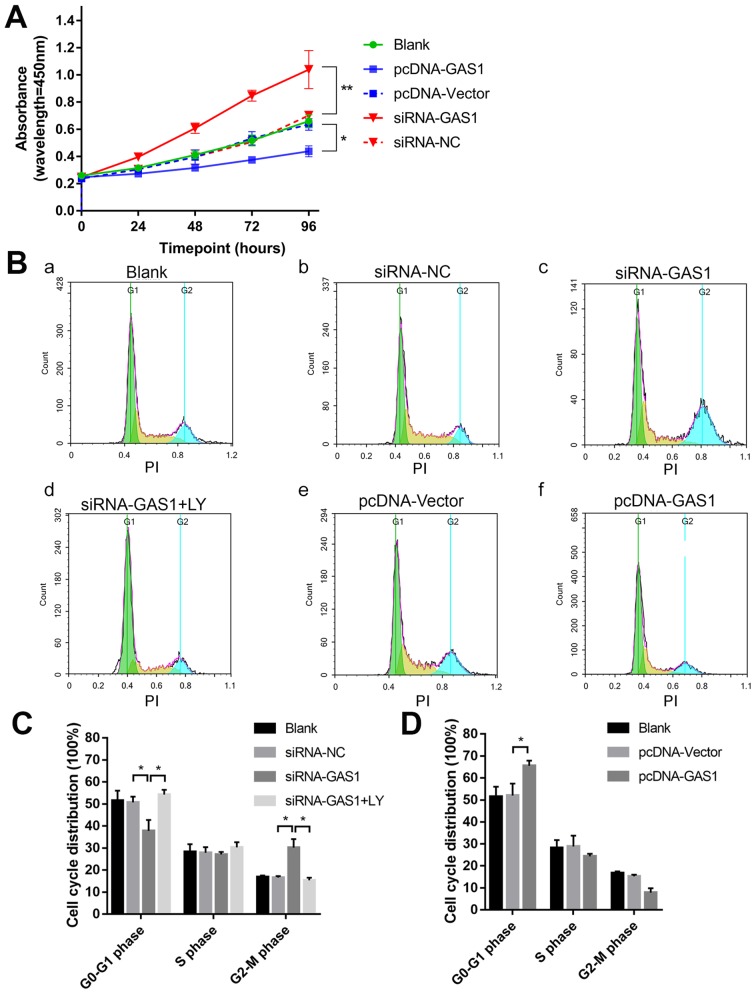
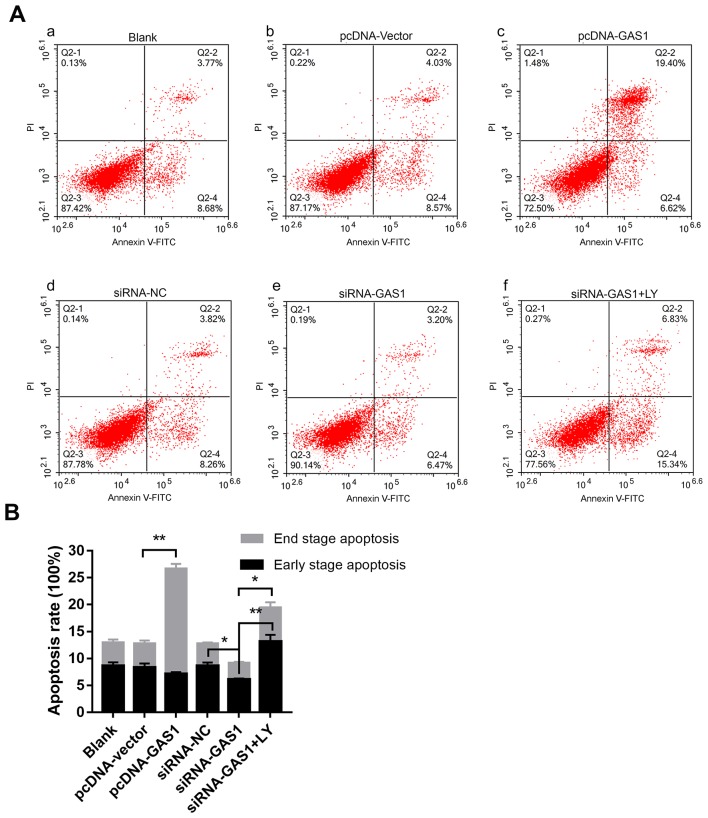
To evaluate the function of GAS1 in SFs, GAS1 expression was either overexpressed or knocked down in non-OASFs. Transfection efficiency was subsequently verified using qPCR and immunofluorescence (Fig. S1). The cell viability assay was assessed at different indicated time points using CCK-8 assay in each group (Fig. 2A). At 96 h timepoint, the OD values of the siRNA-GAS1 group were significantly higher compared with those in the siRNA-NC group (P=0.0013). In contrast, the OD value for the pcDNA-GAS1 group at 96 h timepoint was significantly lower compared with that in the pcDNA-vector group (P=0.0439). By observing the cell viability curves for each group, it was noticed that the siRNA-GAS1 group grew faster compared with other groups, whereas those in the pcDNA-GAS1 group grew at a less rapid rate. For the cell cycle analysis of each group were subsequently evaluated using flow cytometry at 48 h after transfection (Fig. 2B-D). siRNA-GAS1 transfection significantly increased the frequency of non-OASFs in the G2-M phase compared with cells transfected with siRNA-NC (P=0.038; Fig. 2B and C); while the frequency of cells in the G0-G1 phase was increased in the pcDNA-GAS1 group compared with those in the pcDNA-vector group (P=0.039; Fig. 2B and D). In terms of apoptosis (Fig. 3A and B), cells from the pcDNA-GAS1 group exhibited a significantly higher frequency of end-stage apoptosis compared with those in the pcDNA-vector group (P<0.001; Fig. 3A and B); cells in the siRNA-GAS1 group exhibited a slight but significant reduction in the frequency of early-stage apoptosis (P=0.004; Fig. 3A and B) Taken together, these findings suggested that GAS1 overexpression inhibit SF cell viability whilst promoting apoptosis, whereas GAS1 knockdown increases SF proliferation and inhibit apoptosis.
Figure 2.
GAS1 inhibits SF cell viability. (A) Cell viability curves of non-OASFs transfected with pcDNA-GAS1, pcDNA-vector, siRNA-GAS1 or siRNA-NC. *P<0.05 and **P<0.01 at 96 h. (B) Representative cell cycle plots of (Ba) the blank control group, and non-OASFs transfected with (Bb) siRNA-NC, (Bc) siRNA-GAS1, (Bd) siRNA-GAS1 (plus treatment with LY), (Be) pcDNA-vector and (Bf) pcDNA-GAS1, as measured using flow cytometry. (C) Quantified data of non-OASFs transfected with siRNA-GAS1, siRNA-NC and cells transfected with siRNA-GAS1 and treated with LY. (D) Quantified data of non-OASFs transfected with pcDNA-GAS1 or pcDNA-vector. *P<0.05. PI, propidium iodide; SF, synovial fibroblasts; OASFs, osteoarthritis synovial fibroblasts; GAS1, growth arrest specific-1; NC, negative control; siRNA, small interfering RNA; LY, LY294002.
Figure 3.
GAS1 promotes SF apoptosis. (A) Representative flow cytometry dot plots of (Aa) the blank control group, and non-OASFs transfected with (Ab) pcDNA-vector, (Ac) pcDNA-GAS1, (Ad) siRNA-NC, (Ae) siRNA-GAS1 and (Af) siRNA-GAS1 + LY treatment. (B) Quantified data and corresponding statistical analysis of early- and end-stage apoptotic cells from each of the aforementioned groups. *P<0.01 and **P<0.001. PE, phycoerythrin; APC, allophycocyanin; SF, synovial fibroblasts; OASFs, osteoarthritis synovial fibroblasts; GAS1, growth arrest specific-1; NC, negative control; siRNA, small interfering RNA; LY, LY294002.
GAS1 regulates SF proliferation and apoptosis through PI3K-Akt signaling
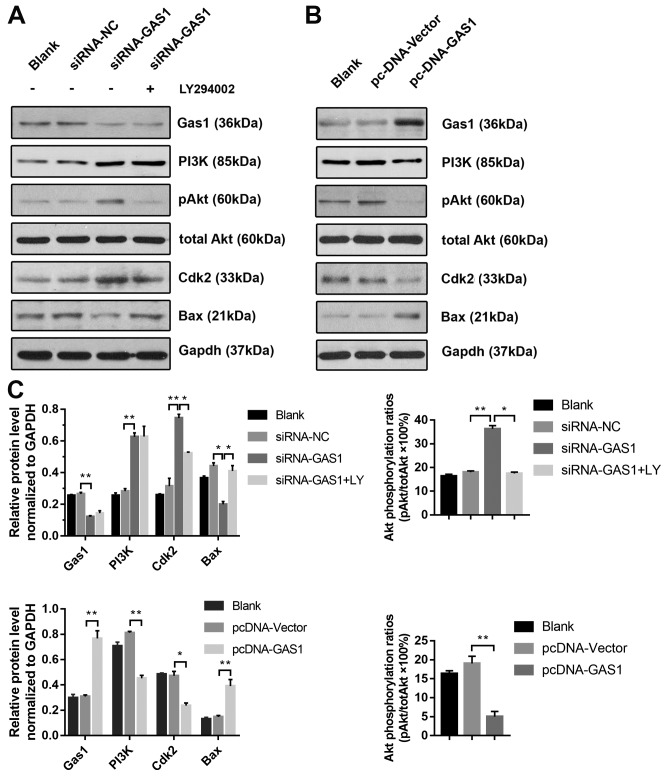
To further investigate the anti-proliferative effects of GAS1 in non-OASFs, the activity of the PI3K-Akt pathway, previously demonstrated to be crucial for cell proliferation as in prostate, lung and colorectal cancer cells (26–28) and previously reported to lie downstream of GAS1 (15), was next examined by applying LY294002, a pharmacological PI3K inhibitor. Transfection with siRNA-GAS1 increased the levels of PI3K (p85), p-Akt (Ser473) and Cdk2 expression whilst reducing the levels of Bax, a protein associated with apoptosis, compared with siRNA-NC group (Fig. 4A and C). The level of Akt phosphorylation was significantly reduced by treatment with LY294002 prior to siRNA-GAS1 transfection (referred to as the siRNA-GAS1 + LY group). Additionally, LY294002 treatment reduced Cdk2 expression whilst increasing Bax expression compared with untreated cells (Fig. 4A). In contrast, transfection with pcDNA-GAS1 reduced the level of PI3K (p85), p-Akt (Ser473) and Cdk2 whilst increasing the expression of Bax compared with those transfected with cells transfected with the pcDNA-vector (Fig. 4B and C). The role of PI3K/Akt signaling in the anti-proliferative effects of GAS1 in SFs was functionally assessed by analyzing the cell cycle and apoptosis in the presence of LY294002. Compared the siRNA-GAS1 group, SFs in the siRNA-GAS1 + LY group exhibited lower frequencies in G2-M phase (P=0.007) and higher frequencies of early- (P<0.001) and end-stage (P=0.009) apoptosis (Figs. 2B and C, and 3). The results of the present study suggested that GAS1 may function as a SF growth suppressor by inhibiting the PI3K-Akt pathway.
Figure 4.
GAS1 regulates SF proliferation and apoptosis through the PI3K-Akt pathway. (A) Representative blots demonstrating the expression of GAS1 and proteins associated with the PI3K/Akt pathway and apoptosis (PI3K, p-Akt, total Akt, Cdk2 and Bax) from non-OASFs transfected with siRNA-GAS1, siRNA-NC or cells transfected with siRNA-GAS1 + LY (50 µM) treatment. (B) Representative blots demonstrating the expression of GAS1 and proteins associated with the PI3K/Akt pathway and apoptosis (PI3K, p-Akt, total Akt, Cdk2 and Bax) from non-OASFs transfected with pcDNA-vector or pcDNA-GAS. (C) Quantification of densitometric data and Akt phosphorylation. *P<0.01 and **P<0.001. SF, synovial fibroblasts; OASFs, osteoarthritis synovial fibroblasts; p-Akt, phosphorylated Akt; Cdk2, cyclin-dependent kinase 2; GAS1, growth arrest specific-1; NC, negative control; siRNA, small interfering RNA; LY, LY294002.
miR-34a-5p and miR-181a-5p directly downregulate the expression of GAS1 in SFs
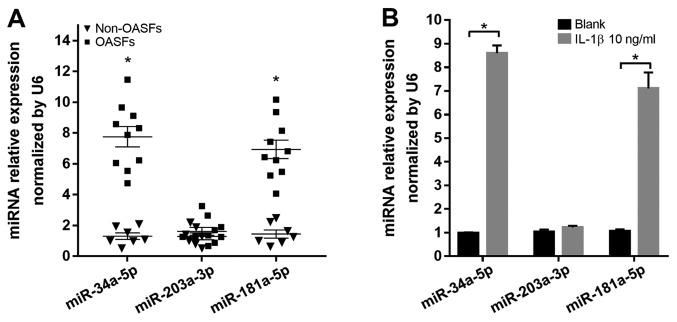
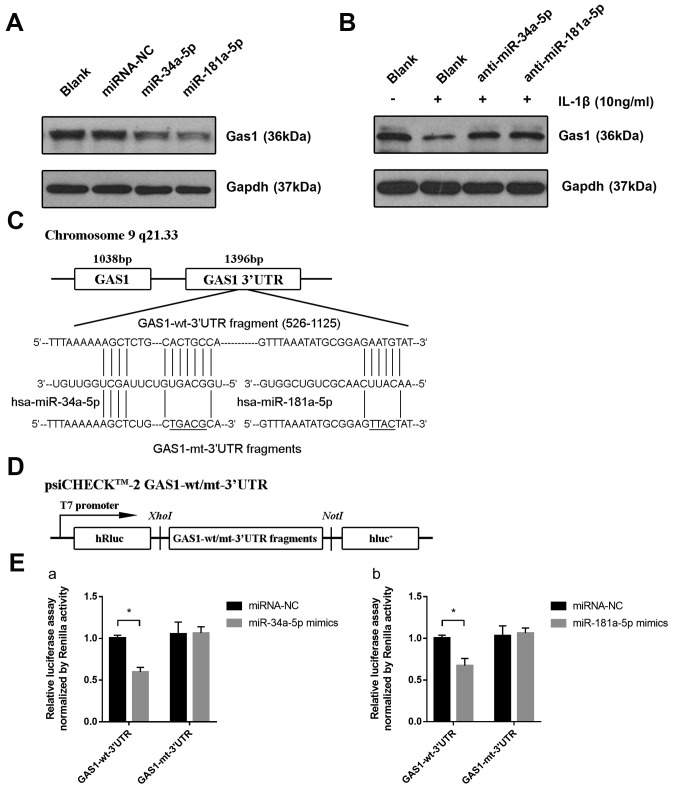
The mechanism by which IL-1β induces the downregulation of GAS1 remains unclear; therefore, it was hypothesized that endogenous small miRs, induced by pro-inflammatory factors, may modulate GAS1 expression by directly targeting the 3′UTR of GAS1 mRNA. In particular, three miRs, miR-34a-5p, miR-203a-3p and miR-181a-5p, were found to be overexpressed in OASFs according to a number of previously published reports (21,23) and were capable of binding to the 3′UTR of GAS1 mRNA, according to the Targetscan 7.2 predictions. Therefore, the expression of these three miRs in OASFs and non-OASFs were measured using RT-qPCR. OASFs exhibited higher levels of miR-34a-5p and miR-181a-5p expression, but not miR-203a-3p, compared with non-OASFs (Fig. 5A). In addition, miR-34a-5p and miR-181a-5p expression were significantly higher in non-OASFs stimulated with IL-1β (10 ng/ml) compared with the corresponding blank controls (Fig. 5B). To examine the relationship between the miRNA candidates and GAS1 expression in SFs, non-OASFs were transfected with miRNA mimics or inhibitors. Transfection efficiency was assessed by RT-qPCR (Fig. S2). GAS1 protein expression was downregulated when non-OASFs were transfected with miR-34a-5p or miR-181a-5p mimics compared with the negative control and blank groups (Fig. 6A). Consistently, pre-transfection with miR-34a-5p or miR-181a-5p inhibitors rescued the reduction in GAS1 expression induced by IL-1β, suggesting that IL-1β-induced GAS1 downregulation is potentially mediated by miR-34a-5p and miR-181a-5p (Fig. 6B). To further examine whether miR-34a-5p and miR-181a-5p could directly regulate GAS1 expression, psi-CHECK-2 reporter vectors containing the GAS1-wt-3′UTR and GAS1-mt-3′UTR were reconstructed (Fig. 6C and D), which were then transfected in 293T cells alongside miR-34a-5p or miR-181a-5p mimics. Luciferase assays confirmed that the reporter activity was significantly reduced following co-transfection with psiCHECK2-GAS1-wt-3′UTR and miR-34a-5p (P<0.001) or miR-181a-5p (P=0.002) mimics, compared with cells co-transfected with psiCHECK2-GAS1-mut-3′UTR and miR-34a-5p or miR-181a-5p mimics (Fig. 6E), suggesting that GAS1 is a direct target gene of miR-34a-5p and miR-181a-5p. Taken together, these observations suggested that IL-1β-induced GAS1 downregulation may be mediated by miR-34a-5p and miR-181a-5p.
Figure 5.
miR-34a-5p and miR-181a-5p expression are upregulated in OASFs and IL-1β-stimulated non-OASFs. (A) Relative expression levels of miR-34a-5p, miR-203a-3p and miR-181a-5p were measured in non-OASFs and OASFs. *P<0.001 vs. non-OASFs. (B) Relative expression levels of miR-34a-5p, miR-203a-3p and miR-181a-5p in non-OASFs and IL-1β (10 ng/ml)-treated non-OASFs. *P<0.001. SF, synovial fibroblasts; OASFs, osteoarthritis synovial fibroblasts; miR, microRNA; IL-1β, interleukin 1β.
Figure 6.
miR-34a-5p and miR-181a-5p directly downregulate the expression of GAS1 by targeting the GAS1 3′UTR. (A) Expression of GAS1 proteins in non-OASFs transfected with miRNA-NC, miR-34a-5p or miR-181a-5p mimics, determined by western blotting. (B) Expression of GAS1 protein in IL-1β (10 ng/ml)-stimulated non-OASFs transfected with miR-34a-5p or miR-181a-5p inhibitors. (C) Schematic diagram showing the sequences of potential binding sites for miR-34a-5p and miR-181a-5p in the 3′UTR of GAS1, as predicted using Targetscan 7.2 software. (D) Schematic diagram of the luciferase reporter vector constructs containing wild-type or mutated GAS1 3′UTR sequences. (E) Luciferase activities in 293T cells co-transfected with psiCHECK2-wt/mut-3′UTR and (Ea) miR-34a-5p or (Eb) miR-181a-5p mimics. *P<0.01. OASFs, osteoarthritis synovial fibroblasts; miR, microRNA; IL-1β, interleukin 1β; GAS1, growth arrest specific-1; 3′UTR, 3′untranslated region; NC, negative control; wt, wild-type; mut, mutant; luc, luciferase.
Discussion
OA is the most prevalent joint disease worldwide and is characterized by progressive degradation of the articular cartilage, sequentially leading to a loss of joint function and disability (1). However, the molecular mechanism of OA pathogenesis remains unclear. Oehler et al (29) reported that hyperplastic and inflammatory SFs are widely found in early- and late-stage OA by arthroscopic biopsy histology. According to a retrospective trial by Ayral et al (30), adjacent synovitis of the OA knee was found to be associated with the severity of cartilage lesions or chondropathy. Based on this evidence, it is reasonable to conclude that aberrant hyperplasia of SFs is involved in the progression of OA. In the local microenvironment of the OA joint, inflamed SFs proliferate and cumulatively release proinflammatory cytokines and matrix metalloproteinases, further aggravating inflammation and cartilage damage (31,32). Notably, in the present study it was observed that GAS1 was downregulated in OASFs and in IL-1β-stimulated non-OASFs. The level of GAS1 expression was negatively associated with the concentration of IL-1β exposure, which has not been previously reported.
GAS1 was first identified as a growth suppressive gene in 1988 (7). Schneider et al reported that in the mouse-derived fibroblast cell line NIH3T3, cell cycle arrest occurred in the G0-S phase when GAS1 was overexpressed (7). Del Sal et al (33) found that overexpression of GAS1 induced cell growth arrest in a p53-dependent manner. The inhibitory effect of GAS1 on tumorigenesis was subsequently investigated in a number of malignant neoplasms. According to gene expression profiling analyzes, GAS1 was found to be notably downregulated in various cancer cell lines compared with their non-cancerous counterparts (34), whilst several studies have also reported that the chromosomal site of the GAS1 gene is deleted in bladder and colorectal cancers (35,36). Lee et al (37) found that GAS1 expression can be repressed by proliferative factors, including c-Myc and v-Src, and Zhao et al (38) found that overexpression of GAS1 can retard cell growth during the pathogenesis of gastric cancer. Experiments from the present study also demonstrated the growth-inhibitory effects on SFs, in that knockdown of GAS1 expression significantly increased non-OASF proliferation as measured using CCK-8 assay, whilst overexpression of GAS1 resulted in the opposite effect, with cells arrested at the G0/G1 cell cycle phase.
In the present study, the overexpression of GAS1 induced late-stage apoptosis, whereas the silencing of GAS1 could slightly, but significantly, reduce early-stages of apoptosis. Indeed, another previous study had also reported the pro-apoptotic effects of GAS1 by activating caspase 3 in glioma cells (13), and Wang et al (11) found that GAS1 can promote chemotherapy-induced cell apoptosis in gastric cancer cell lines. GAS1 has also been tested using xenografts in vivo. Nude mice injected with lung cancer cells overexpressing GAS1 exhibited reductions in tumor growth (10). Interestingly, an opposite effect has also been reported by another study, which showed that in vascular endothelial cells, upregulation of GAS1 which is mediated by VE-cadherin and vascular endothelial growth factor signaling, can prevent cell apoptosis (39). The effect of GAS1 on apoptosis being dependent on the cellular context could be a reasonable explanation for the apparent discrepancy between these aforementioned studies. The results of the present study, in-keeping with the general pattern of other studies, confirmed the pro-apoptotic effects of GAS1 in SFs. However, there was only a slightly reduced apoptosis rate in response to GAS1 downregulation. In conclusion, the suppressive effects of GAS1 may maintain the balance of non-OASF growth, whereas its downregulation could induce hyperplasia in SFs during OA pathogenesis, suggesting GAS1 to be a potential target for the treatment of synovial hyperplasia in OA management.
Research from Cabrera et al (14) detailed the downstream signaling pathway of GAS1 in neural tumors. According to sequence alignments and secondary structure predictions, similar to glial cell-derived neurotrophic factor family receptor α (GFRα), GAS1 has a RET-binding domain (40). In the N2a neuroblastoma cell line, GAS1 can inactivate the PI3K-Akt pathway by directly inhibiting RET phosphorylation (14). The overactivation of the PI3K-Akt pathway and downstream signaling have been linked to SF survival and proliferation in abnormal synovial states, including rheumatoid arthritis and synovitis (41). Based on these findings, it was speculated in the present study that the apoptotic and cell cycle arrest effects of GAS1 may be dependent on the PI3K-Akt pathway (14,42). The expression levels of PI3K, p-Akt (Ser473) and Cdk2, a cell cycle regulator, were upregulated after GAS1 silencing, but were all conversely downregulated after GAS1 overexpression. In contrast, Bax, a gene associated with apoptosis involved in the PI3K-Akt downstream pathway, was downregulated after GAS1 overexpression. Treatment of SFs with LY294002, a PI3K-Akt inhibitor, following GAS1 knockdown, recovered the expression of Cdk2 and Bax to normal levels comparable with the negative control group, suggesting that the silencing of GAS1 promoted the hyperproliferation of SFs in a PI3K-Akt-dependent manner. Further cell cycle and apoptosis detection experiments on the siRNA-GAS1-LY group also provided support to this conclusion. As to why RET was not detected in the present study, it was suspected that inactivation of the PI3K-Akt pathway by GAS1 was not mediated by RET. Therefore, the molecular mechanisms underlying the functional role of GAS1 in cell growth suppression and in promoting apoptosis in SFs require further investigation.
The mechanism of IL-1β-induced GAS1 downregulation remains poorly understood. It was speculated that miRs could be important mediators of GAS1 expression due to their reported regulatory functions by post-transcriptionally binding to specific sequences of target gene 3′UTRs. Following investigations into miRNA-gene matching using a combination of miRNA expression profile data of OA and non-OASFs from other studies (21,23), three miRNAs, miR-34a-5p, miR-203a-3p and miR-181a-5p, which are found to be upregulated in OASFs and have potential binding sites in the GAS1 3′UTR as predicted by Targetscan 7.2, were selected for further analysis in the present study. miR-34a-5p and miR-181a-5p, but not miR-203a-3p, were found to be significantly overexpressed with a >7-fold change, in OASFs and IL-1β-treated non-OASFs, by RT-qPCR. Their potential contributions to regulating GAS1 expression in non-OASFs during the OA process were subsequently functionally validated. GAS1 expression was demonstrated to be downregulated in SFs transfected with miR-34a-5p or miR-181a-5p mimics, whilst IL-1β-induced GAS1 downregulation was reversed by transfection with miR-34a-5p and miR-181a-5p inhibitors. Luciferase assays verified further that miR-34a-5p and miR-181a-5p directly target the GAS1 3′UTR. miR-34a-5p has been reported to induce divergent effects on cell growth and apoptosis among different cell types (15,43–46). In some studies, miR-34a-5p was regarded as a tumor growth suppressor and apoptotic factor in multiple cancer cell types (43–46). By contrast, in papillary thyroid carcinoma, miR-34a-5p was demonstrated to be a tumorigenic and anti-apoptotic factor by directly downregulating GAS1, sequentially hindering cancer cell apoptosis through the PI3K-Akt/Bad pathway (15). Li et al (21) found that miR-34a-5p can be detected at relatively higher levels in the synovial fluid of late stage OA patients compared with early stage OA patients. Several researchers have confirmed that miR-34a can induce chondrocyte apoptosis during OA progression by targeting genes, including sirtuin 1, cellular communication network factor 1 and δ-like canonical Notch ligand 1 (47–49). The present study strongly suggested that OASFs, in addition to IL-1β-stimulated SFs, overexpressed miR-34a-5p, which is consistent with previous data regarding OA chondrocytes and the present observation that miR-34a-5p directly targets the GAS1 3′UTR. However, additional experiments are required to prove that miR-34a-5p can promote the hyperproliferation of SFs by directly targeting GAS1. miR-181a-5p, a member of the miR-181 family, has not been well studied in OA and other arthritic disease models. Elevated expression of miR-181a-5p was observed by miRNA expression sequencing profiling acquired in a post-traumatic OA mouse model (23). In cancer research, another study have previously proved that miR-181a-5p is a proliferative factor which can facilitate cell cycle progression in melanoma (50). In the present study, miR-181a-5p was found to be upregulated in OASFs or IL-1β-stimulated SFs, which negatively regulated GAS1 expression by directly targeting the GAS1 3′UTR, in a similar mechanism to that of miR34a-5p. To the best of our knowledge, the present study was the first to report the potential proliferative effects of miR-181a-5p in OASFs. As such, miR-34a-5p and miR-181a-5p could serve as novel therapeutic targets in OA treatment, though further comprehensive experimental analyzes would need to be performed.
In conclusion, the present study confirmed the downregulation of GAS1 in SFs of OA joints or in cytokine-induced SFs. By overexpressing or silencing GAS1 expression in non-OASFs, the suppressive role of GAS1 on SF hyperproliferation was observed, which was mechanistically found to involve inhibiting PI3K-Akt signaling. Finally, two miRNA candidates, miR-34a-5p and miR-181a-5p, were found to downregulate GAS1 expression in OASFs or in IL-1β-stimulated SFs, by directly targeting the GAS1 3′UTR. According to the present study, GAS1 could be regarded as a key mediator in regulating SF proliferation. In addition, a unique miR-34a-5p/miR-181a-5p-GAS1-PI3K-Akt axis has been potentially uncovered in the regulation of SF hyperplasia in OA joints, which could serve as a potential therapeutic target for preventing synovial hyperplasia in OA progression.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Hua Long and Dr Jun Chen, The Second Affiliated Hospital of Air Force Medical University (Tangdu Hospital of Fourth Military Medical University), Xi'an, China for collecting the synovial tissue specimens. The authors would also like acknowledge the technical assistance of Ms Shun Guo, The Second Affiliated Hospital of Air Force Medical University (Tangdu Hospital of Fourth Military Medical University) and Dr Weigang Zhang (Xijing Hospital, Xi'an, China) for technical assistance and manuscript revision. The authors are sincerely grateful to Professor Shizhen Emily Wang (University of California, San Diego, CA, USA) for laboratory technique training.
Funding
The present study was supported by grant from the Natural Science Foundation of China (grant no. 81072194).
Availability of data and materials
The data used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
CD and BAM designed the study. CD, XLW, NL and XYW performed the primary cultures of SFs. CD, XLW and NL performed the mRNA quantification and protein detection. CD, KLZ and XYW performed cell viability, apoptosis, and cell cycle experiments. CD and XLW performed luciferase assays. CD and KLZ prepared the figures. CD, HMZ, HPW, BW and MA collected and analyzed the data. CD, KLZ and HMZ wrote the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
All procedures in this study were in accordance with ethical standards, and were approved by the Institutional Review Board of Tangdu Hospital, Fourth Military Medical University (Xi'an, China) with informed consent obtained from all the participants concerned.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Glyn-Jones S, Palmer AJ, Agricola R, Price AJ, Vincent TL, Weinans H, Carr AJ. Osteoarthritis. Lancet. 2015;386:376–387. doi: 10.1016/S0140-6736(14)60802-3. [DOI] [PubMed] [Google Scholar]
- 2.Scanzello CR, Goldring SR. The role of synovitis in osteoarthritis pathogenesis. Bone. 2012;51:249–257. doi: 10.1016/j.bone.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kapoor M, Martel-Pelletier J, Lajeunesse D, Pelletier JP, Fahmi H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat Rev Rheumatol. 2011;7:33–42. doi: 10.1038/nrrheum.2010.196. [DOI] [PubMed] [Google Scholar]
- 4.Robinson WH, Lepus CM, Wang Q, Raghu H, Mao R, Lindstrom TM, Sokolove J. Low-grade inflammation as a key mediator of the pathogenesis of osteoarthritis. Nat Rev Rheumatol. 2016;12:580–592. doi: 10.1038/nrrheum.2016.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Del Rey MJ, Usategui A, Izquierdo E, Cañete JD, Blanco FJ, Criado G, Pablos JL. Transcriptome analysis reveals specific changes in osteoarthritis synovial fibroblasts. Ann Rheum Dis. 2012;71:275–280. doi: 10.1136/annrheumdis-2011-200281. [DOI] [PubMed] [Google Scholar]
- 6.Lee CS, Buttitta L, Fan CM. Evidence that the WNT-inducible growth arrest-specific gene 1 encodes an antagonist of sonic hedgehog signaling in the somite. Proc Natl Acad Sci USA. 2001;98:11347–11352. doi: 10.1073/pnas.201418298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54:787–793. doi: 10.1016/S0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- 8.Benitez JA, Arregui L, Vergara P, Segovia J. Targeted-simultaneous expression of Gas1 and p53 using a bicistronic adenoviral vector in gliomas. Cancer Gene Ther. 2007;14:836–846. doi: 10.1038/sj.cgt.7701076. [DOI] [PubMed] [Google Scholar]
- 9.Jiménez A, López-Ornelas A, Estudillo E, González-Mariscal L, González RO, Segovia J. A soluble form of GAS1 inhibits tumor growth and angiogenesis in a triple negative breast cancer model. Exp Cell Res. 2014;327:307–317. doi: 10.1016/j.yexcr.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 10.Evdokiou A, Cowled PA. Tumor-suppressive activity of the growth arrest-specific gene GAS1 in human tumor cell lines. Int J Cancer. 1998;75:568–577. doi: 10.1002/(SICI)1097-0215(19980209)75:4<568::AID-IJC13>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, Zhou X, Zhang Y, Zhu H, Zhao L, Fan L, Wang Y, Gang Y, Wu K, Liu Z, Fan D. Growth arrest-specific gene 1 is downregulated and inhibits tumor growth in gastric cancer. FEBS J. 2012;279:3652–3664. doi: 10.1111/j.1742-4658.2012.08726.x. [DOI] [PubMed] [Google Scholar]
- 12.Jiang Z, Xu Y, Cai S. Down-regulated GAS1 expression correlates with recurrence in stage II and III colorectal cancer. Hum Pathol. 2011;42:361–368. doi: 10.1016/j.humpath.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 13.Zamorano A, Lamas M, Vergara P, Naranjo JR, Segovia J. Transcriptionally mediated gene targeting of gas1 to glioma cells elicits growth arrest and apoptosis. J Neurosci Res. 2003;71:256–263. doi: 10.1002/jnr.10461. [DOI] [PubMed] [Google Scholar]
- 14.Cabrera JR, Sanchez-Pulido L, Rojas AM, Valencia A, Mañes S, Naranjo JR, Mellström B. Gas1 is related to the glial cell-derived neurotrophic factor family receptors alpha and regulates Ret signaling. J Biol Chem. 2006;281:14330–14339. doi: 10.1074/jbc.M509572200. [DOI] [PubMed] [Google Scholar]
- 15.Ma Y, Qin H, Cui Y. MiR-34a targets GAS1 to promote cell proliferation and inhibit apoptosis in papillary thyroid carcinoma via PI3K/Akt/Bad pathway. Biochem Biophys Res Commun. 2013;441:958–963. doi: 10.1016/j.bbrc.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 17.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79:581–588. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 18.Stanczyk J, Pedrioli DM, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE, Detmar M, Gay S, Kyburz D. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58:1001–1009. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- 19.Nakamachi Y, Kawano S, Takenokuchi M, Nishimura K, Sakai Y, Chin T, Saura R, Kurosaka M, Kumagai S. MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum. 2009;60:1294–1304. doi: 10.1002/art.24475. [DOI] [PubMed] [Google Scholar]
- 20.Gao J, Zhou XL, Kong RN, Ji LM, He LL, Zhao DB. microRNA-126 targeting PIK3R2 promotes rheumatoid arthritis synovial fibro-blasts proliferation and resistance to apoptosis by regulating PI3K/AKT pathway. Exp Mol Pathol. 2016;100:192–198. doi: 10.1016/j.yexmp.2015.12.015. [DOI] [PubMed] [Google Scholar]
- 21.Li YH, Tavallaee G, Tokar T, Nakamura A, Sundararajan K, Weston A, Sharma A, Mahomed NN, Gandhi R, Jurisica I, Kapoor M. Identification of synovial fluid microRNA signature in knee osteoarthritis: Differentiating early- and late-stage knee osteoarthritis. Osteoarthritis Cartilage. 2016;24:1577–1586. doi: 10.1016/j.joca.2016.01.625. [DOI] [PubMed] [Google Scholar]
- 22.Murata K, Yoshitomi H, Tanida S, Ishikawa M, Nishitani K, Ito H, Nakamura T. Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res Ther. 2010;12:R86. doi: 10.1186/ar3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kung LHW, Ravi V, Rowley L, Bell KM, Little CB, Bateman JF. Comprehensive expression analysis of microRNAs and mRNAs in synovial tissue from a mouse model of early post-traumatic osteoarthritis. Sci Rep. 2017;7:17701. doi: 10.1038/s41598-017-17545-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Inoue H, Takamori M, Nagata N, Nishikawa T, Oda H, Yamamoto S, Koshihara Y. An investigation of cell proliferation and soluble mediators induced by interleukin 1beta in human synovial fibroblasts: Comparative response in osteoarthritis and rheumatoid arthritis. Inflamm Res. 2001;50:65–72. doi: 10.1007/s000110050726. [DOI] [PubMed] [Google Scholar]
- 26.Li L, Ittmann MM, Ayala G, Tsai MJ, Amato RJ, Wheeler TM, Miles BJ, Kadmon D, Thompson TC. The emerging role of the PI3-K-Akt pathway in prostate cancer progression. Prostate Cancer Prostatic Dis. 2005;8:108–118. doi: 10.1038/sj.pcan.4500776. [DOI] [PubMed] [Google Scholar]
- 27.Sarris EG, Saif MW, Syrigos KN. The biological role of PI3K pathway in lung cancer. Pharmaceuticals. 2012;5:1236–1264. doi: 10.3390/ph5111236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng Y, Qian W, Zhang Y, Peng W, Li J, Gu Q, Ji D, Zhang Z, Wang Q, Zhang D, Sun Y. CDCA2 promotes the proliferation of colorectal cancer cells by activating the AKT/CCND1 pathway in vitro and in vivo. BMC Cancer. 2019;19:576. doi: 10.1186/s12885-019-5793-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oehler S, Neureiter D, Meyer-Scholten C, Aigner T. Subtyping of osteoarthritic synoviopathy. Clin Exp Rheumatol. 2002;20:633–640. [PubMed] [Google Scholar]
- 30.Ayral X, Pickering EH, Woodworth TG, Mackillop N, Dougados M. Synovitis: A potential predictive factor of structural progression of medial tibiofemoral knee osteoarthritis-results of a 1 year longitudinal arthroscopic study in 422 patients. Osteoarthritis Cartilage. 2005;13:361–367. doi: 10.1016/j.joca.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Sieghart D, Liszt M, Wanivenhaus A, Bröll H, Kiener H, Klösch B, Steiner G. Hydrogen sulphide decreases IL-1β-induced activation of fibroblast-like synoviocytes from patients with osteoarthritis. J Cell Mol Med. 2015;19:187–197. doi: 10.1111/jcmm.12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eymard F, Pigenet A, Citadelle D, Flouzat-Lachaniette CH, Poignard A, Benelli C, Berenbaum F, Chevalier X, Houard X. Induction of an inflammatory and prodegradative phenotype in autologous fibroblast-like synoviocytes by the infrapatellar fat pad from patients with knee osteoarthritis. Arthritis Rheumatol. 2014;66:2165–2174. doi: 10.1002/art.38657. [DOI] [PubMed] [Google Scholar]
- 33.Del Sal G, Ruaro EM, Utrera R, Cole CN, Levine AJ, Schneider C. Gas1-induced growth suppression requires a transactivation-independent p53 function. Mol Cell Biol. 1995;15:7152–7160. doi: 10.1128/MCB.15.12.7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Y, Prasad M, Lemon WJ, Hampel H, Wright FA, Kornacker K, LiVolsi V, Frankel W, Kloos RT, Eng C, et al. Gene expression in papillary thyroid carcinoma reveals highly consistent profiles. Proc Natl Acad Sci USA. 2001;98:15044–15049. doi: 10.1073/pnas.251547398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moriarty HT, Webster LR. Fragile sites and bladder cancer. Cancer Genet Cytogenet. 2003;140:89–98. doi: 10.1016/S0165-4608(02)00650-7. [DOI] [PubMed] [Google Scholar]
- 36.Konstantinova LN, Fleischman EW, Knisch VI, Perevozchikov AG, Kopnin BP. Karyotype peculiarities of human colorectal adenocarcinomas. Hum Genet. 1991;86:491–496. doi: 10.1007/BF00194640. [DOI] [PubMed] [Google Scholar]
- 37.Lee TC, Li L, Philipson L, Ziff EB. Myc represses transcription of the growth arrest gene gas1. Proc Natl Acad Sci USA. 1997;94:12886–12891. doi: 10.1073/pnas.94.24.12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao L, Pan Y, Gang Y, Wang H, Jin H, Tie J, Xia L, Zhang Y, He L, Yao L, et al. Identification of GAS1 as an epirubicin resistance-related gene in human gastric cancer cells with a partially randomized small interfering RNA library. J Biol Chem. 2009;284:26273–26285. doi: 10.1074/jbc.M109.028068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spagnuolo R, Corada M, Orsenigo F, Zanetta L, Deuschle U, Sandy P, Schneider C, Drake CJ, Breviario F, Dejana E. Gas1 is induced by VE-cadherin and vascular endothelial growth factor and inhibits endothelial cell apoptosis. Blood. 2004;103:3005–3012. doi: 10.1182/blood-2003-07-2459. [DOI] [PubMed] [Google Scholar]
- 40.Airaksinen MS, Holm L, Hätinen T. Evolution of the GDNF family ligands and receptors. Brain Behav Evol. 2006;68:181–190. doi: 10.1159/000094087. [DOI] [PubMed] [Google Scholar]
- 41.Hayer S, Pundt N, Peters MA, Wunrau C, Kühnel I, Neugebauer K, Strietholt S, Zwerina J, Korb A, Penninger J, et al. PI3Kgamma regulates cartilage damage in chronic inflammatory arthritis. FASEB J. 2009;23:4288–4298. doi: 10.1096/fj.09-135160. [DOI] [PubMed] [Google Scholar]
- 42.Mograbi B, Bocciardi R, Bourget I, Busca R, Rochet N, Farahi-Far D, Juhel T, Rossi B. Glial cell line-derived neurotrophic factor-stimulated phosphatidylinositol 3-kinase and Akt activities exert opposing effects on the ERK pathway: Importance for the rescue of neuroectodermic cells. J Biol Chem. 2001;276:45307–45319. doi: 10.1074/jbc.M101220200. [DOI] [PubMed] [Google Scholar]
- 43.Zhou Y, Ding BZ, Lin YP, Wang HB. MiR-34a, as a suppressor, enhance the susceptibility of gastric cancer cell to luteolin by directly targeting HK1. Gene. 2018;644:56–65. doi: 10.1016/j.gene.2017.10.046. [DOI] [PubMed] [Google Scholar]
- 44.Li T, Li L, Li D, Wang S, Sun J. MiR-34a inhibits oral cancer progression partially by repression of interleukin-6-receptor. Int J Clin Exp Pathol. 2015;8:1364–1373. [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao J, Guerrero A, Kelnar K, Peltier HJ, Bader AG. Synergy between next generation EGFR tyrosine kinase inhibitors and miR-34a in the inhibition of non-small cell lung cancer. Lung Cancer. 2017;108:96–102. doi: 10.1016/j.lungcan.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 46.Shi H, Zhou S, Liu J, Zhu J, Xue J, Gu L, Chen Y. miR-34a inhibits the in vitro cell proliferation and migration in human esophageal cancer. Pathol Res Pract. 2016;212:444–449. doi: 10.1016/j.prp.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 47.Yan S, Wang M, Zhao J, Zhang H, Zhou C, Jin L, Zhang Y, Qiu X, Ma B, Fan Q. MicroRNA-34a affects chondrocyte apoptosis and proliferation by targeting the SIRT1/p53 signaling pathway during the pathogenesis of osteoarthritis. Int J Mol Med. 2016;38:201–209. doi: 10.3892/ijmm.2016.2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang B, Ni J, Long H, Huang J, Yang C, Huang X. IL-1β-induced miR-34a up-regulation inhibits Cyr61 to modulate osteoarthritis chondrocyte proliferation through ADAMTS-4. J Cell Biochem. 2018;119:7959–7970. doi: 10.1002/jcb.26600. [DOI] [PubMed] [Google Scholar]
- 49.Zhang W, Hsu P, Zhong B, Guo S, Zhang C, Wang Y, Luo C, Zhan Y, Zhang C. MiR-34a enhances chondrocyte apoptosis, senescence and facilitates development of osteoarthritis by targeting DLL1 and regulating PI3K/AKT pathway. Cell Physiol Biochem. 2018;48:1304–1316. doi: 10.1159/000492090. [DOI] [PubMed] [Google Scholar]
- 50.Zhang L, He X, Li F, Pan H, Huang X, Wen X, Zhang H, Li B, Ge S, Xu X, et al. The miR-181 family promotes cell cycle by targeting CTDSPL, a phosphatase-like tumor suppressor in uveal melanoma. J Exp Clin Cancer Res. 2018;37:15. doi: 10.1186/s13046-018-0679-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used and/or analyzed during the present study are available from the corresponding author on reasonable request.