Abstract
Naked mole-rats are long-lived animals that show unusual resistance to hypoxia, cancer and ageing. Protein deimination is an irreversible post-translational modification caused by the peptidylarginine deiminase (PAD) family of enzymes, which convert arginine into citrulline in target proteins. Protein deimination can cause structural and functional protein changes, facilitating protein moonlighting, but also leading to neo-epitope generation and effects on gene regulation. Furthermore, PADs have been found to regulate cellular release of extracellular vesicles (EVs), which are lipid-vesicles released from cells as part of cellular communication. EVs carry protein and genetic cargo and are indicative biomarkers that can be isolated from most body fluids. This study was aimed at profiling deiminated proteins in plasma and EVs of naked mole-rat. Key immune and metabolic proteins were identified to be post-translationally deiminated, with 65 proteins specific for plasma, while 42 proteins were identified to be deiminated in EVs only. Using protein-protein interaction network analysis, deiminated plasma proteins were found to belong to KEEG (Kyoto Encyclopedia of Genes and Genomes) pathways of immunity, infection, cholesterol and drug metabolism, while deiminated proteins in EVs were also linked to KEEG pathways of HIF-1 signalling and glycolysis. The mole-rat EV profiles showed a poly-dispersed population of 50–300 nm, similar to observations of human plasma. Furthermore, the EVs were assessed for three key microRNAs involved in cancer, inflammation and hypoxia. The identification of post-translational deimination of critical immunological and metabolic markers contributes to the current understanding of protein moonlighting functions, via post-translational changes, in the longevity and cancer resistance of naked mole-rats.
Keywords: peptidylarginine deiminases (PADs), protein deimination, naked mole-rat (Heterocephalus glaber), extracellular vesicles (EVs), immunity, metabolism, microRNA (miR21; miR155; miR210)
1. Introduction
Peptidylarginine deiminases (PADs) are phylogenetically conserved calcium-dependent enzymes that post-translationally convert arginine into citrulline in target proteins in an irreversible manner. This can cause structural changes in target proteins and affect protein function, gene regulation and generation of neoepitopes [1,2,3,4,5,6]. Such post-translational changes in proteins may also allow for protein moonlighting, an evolutionary acquired phenomenon facilitating proteins to exhibit several physiologically relevant functions from within one polypeptide chain [7,8].
PADs and associated protein deimination are crucial players in cancer, autoimmune and neurodegenerative diseases [4,5,6,9], with recent indications also for PAD-mediated mechanisms in ageing [10]. Furthermore, critical roles have been identified for PADs and PAD-mediated protein deimination in response to hypoxia and in CNS regeneration [11,12,13,14,15] as well as in tissue remodelling and immunity [16,17,18]. Importantly, PADs have been found to have key roles in the regulation of extracellular vesicle (EV) release [19,20,21,22]. EVs are found in most body fluids and participate in cellular communication via transfer of cargo proteins and genetic material [6,23,24,25,26]. EVs isolated from serum, plasma and other body fluids can therefore be useful health biomarkers [27,28]. Hitherto, work on EVs has mainly been in the context of human pathologies and recent comparative immunology studies on EVs and EV cargo have been performed [29], including the assessment of deiminated EV protein cargo [30,31,32].
PADs have been identified throughout phylogeny from bacteria to mammals, with five tissue specific PAD isozymes in mammals, three in chicken, one in bony and cartilaginous fish [1,16,17,31,33] and PAD homologues in parasites [34], fungi [35] and bacteria [22]. While five PAD isozymes have been described in the naked mole-rat (PADI1, Gene ID: 101722077; PADI2, Gene ID: 101721485; PADI3, Gene ID: 101722435; PADI4, Gene ID: 101722785; PADI6, Gene ID: 101723122), no studies have been carried out on their deiminated protein products or the putative physiological relevance of such post-translational deimination in the physiology of the naked mole-rat.
The naked mole-rat is a burrowing rodent and eusocial mammal, and the only species in the genus Heterochephalus of the family Heterochepalidae [36,37]. They have a set of highly unusual physical traits, many of which are thought to derive from their highly-social and putatively hypoxic and hypercapnic subterranean lifestyle. For example, naked mole-rats are among the most hypoxia-tolerant mammal presently identified and tolerate minutes of anoxia, hours at 3% O2, and days to weeks at 8% O2 [38,39,40,41]. The key to tolerating prolonged hypoxia is to match metabolic demand to reduced energy (O2) supply [42,43,44,45], and in acute severe hypoxia (3% O2), the metabolic rate of adult naked mole-rats decreases up to 85% [40]. However, naked mole-rats remain conscious and active, albeit to a reduced degree [46,47,48]. These findings indicate that naked mole-rats are capable of significant metabolic plasticity within their natural environment. Conversely, naked mole-rats are largely non-responsive to hypercapnia and associated acidity-related pain responses are largely absent [49,50]. Naked mole-rats also have numerous adaptations that are not as obviously linked to their natural habitat, including a remarkable resistance to cancer [51,52], they are the only mammalian thermo-conformer and almost entirely ectothermic for regulation of body temperature [53,54] and they have remarkable longevity [55,56,57,58]. These traits make the naked mole-rat an important animal model for a range of human diseases and for furthering understanding of pathways underlying cancer resistance and longevity [59,60,61]. However, little is known about the immune system of naked mole-rats. As PAD-mediated pathways and EVs are increasingly recognized as key players in immune responses and metabolism, and related to a range of human inflammatory pathologies and cancer, a study on these parameters in mole-rat is warranted.
In the current study, plasma and plasma-derived EVs were profiled in naked mole-rats and assessed for deiminated protein profiles as well as three key microRNAs (miRs) related to inflammation and hypoxic resistance. For the first time we report on post-translational deimination of key immune and metabolic proteins in naked mole-rat and species-specific EV profiles.
2. Results
2.1. PAD Homologues in Naked Mole-Rat Plasma
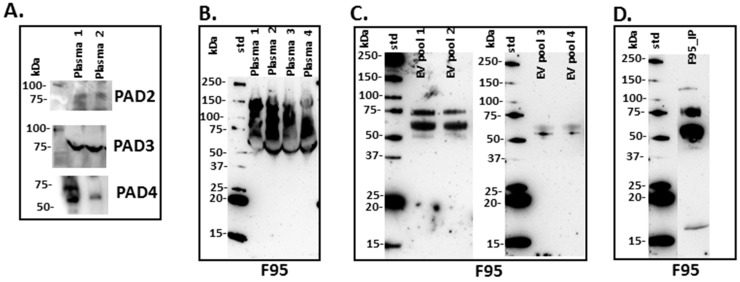
Using PAD-isozyme specific antibodies, generated against human PADs, positive bands were observed by Western blotting and indicated PAD homologue proteins in mole-rat plasma at the expected size of approximately 70–75 kDa for PAD2, PAD3 and PAD4 (Figure 1A).
Figure 1.
Peptidylarginine deiminases (PADs) and deiminated proteins in naked mole-rat plasma and plasma-extracellular vesicles (EVs). (A) PAD positive bands were identified at the expected size of approximately 70–75 kDa using the human PAD2, PAD3 and PAD4 specific antibodies in naked mole-rat plasma. (B) Total deiminated proteins were identified in naked mole-rat plasma (n = 4) using the F95 pan-deimination specific antibody. (C) Total deiminated proteins were identified in naked mole-rat plasma-EVs using the F95 pan-deimination specific antibody (EV pools from plasma of 4 individuals are shown, respectively). (D) The F95-enriched IP fraction from mole-rat plasma (from a pool of 5 individual mole-rat plasma; F95_IP) is shown. The molecular weight marker is indicated next to each blot.
2.2. Deiminated Protein Profiles of Naked Mole-Rat Plasma and Plasma-Derived EVs
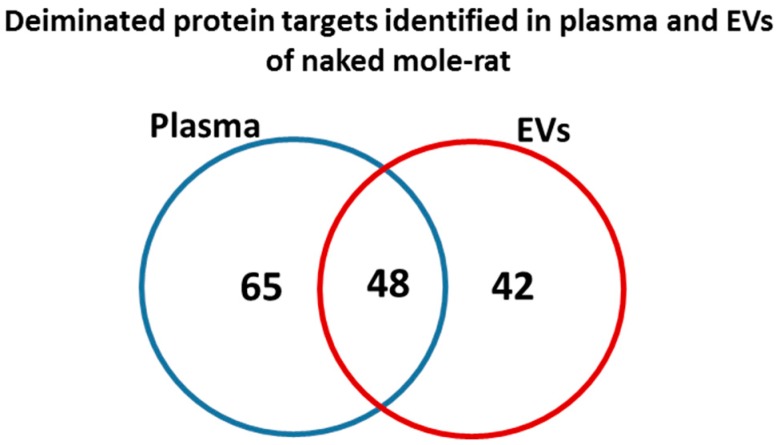
Total deiminated proteins were detected by Western blotting with the pan-deimination F95 antibody in mole-rat plasma and plasma-derived EVs, revealing a range of proteins mainly between 50–150 kDa (Figure 1B). The mono-specific F95 antibody was used in this study for the identification of deiminated proteins, as it has been developed against a deca-citrullinated peptide and is predicted to react with all deiminated/citrullinated proteins based on 100% sequence homology (MABN328 Merck), and it has been used to identify deiminated proteins in human and animals from diverse taxa [11,12,16,18,19,21,22,31]. Deiminated proteins in mole-rat were also detected in the plasma-derived EVs, mainly in the size range of 20–100 kDa (Figure 1C). Deiminated protein candidates in plasma and EVs were further identified by F95 enrichment (see F95 enriched fraction from plasma assessed by Western blotting, Figure 1D) and LC-MS/MS analysis (Table 1 and Table 2; Supplementary Tables S1 and S2). In plasma, 112 species-specific protein hits were identified (Table 1 and Supplementary Table S1) while in EVs, 80 protein hits were identified (Table 2 and Supplementary Table S2). Overall, 48 proteins overlapped between plasma and plasma-derived EVs, while 65 proteins were specific for whole plasma only and 42 proteins for EVs only (Figure 2). The protein lists for deiminated proteins identified in naked mole-rat plasma and plasma-EVs were submitted to STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) analysis (https://string-db.org/) to predict putative protein-protein interaction networks (Figure 3 and Figure 4).
Table 1.
Deiminated proteins identified by F95 enrichment in total plasma of naked mole-rat (Heterocephalus glaber). Deiminated proteins were isolated by immunoprecipitation using the pan-deimination F95 antibody. The F95 enriched eluate was analysed by LC-MS/MS and peak list files were submitted to mascot. Species-specific peptide sequence hits scoring with H. glaber are included and total score is shown. Protein hits identified in whole plasma only (but not in plasma EVs) are highlighted in pink. For full LC-MS/MS data analysis see Supplementary Table S1.
| Protein Name | Symbol | Total Score (p < 0.05) # |
|---|---|---|
| Apolipoprotein B-100 | G5AZB7_HETGA | 6077 |
| Alpha-2-macroglobulin | G5BPM1_HETGA | 4974 |
| Complement C3 | G5C0N5_HETGA | 4300 |
| Serotransferrin | G5BQA9_HETGA | 4058 |
| Serum albumin | G5B5P2_HETGA | 3394 |
| Fibronectin | G5BHR4_HETGA | 3333 |
| Fibrinogen beta chain | G5BML3_HETGA | 2638 |
| Kininogen-1 | G5BT86_HETGA | 2276 |
| Fibrinogen alpha chain | G5BML2_HETGA | 2094 |
| Ceruloplasmin | G5BSL1_HETGA | 2022 |
| Histidine-rich glycoprotein | G5BT87_HETGA | 1851 |
| Complement C4-A | G5C3H6_HETGA | 1759 |
| Complement factor H | G5BM71_HETGA | 1646 |
| Plasminogen | G5BC53_HETGA | 1548 |
| Inter-alpha-trypsin inhibitor heavy chain H4 | G5BUN4_HETGA | 1455 |
| Fibrinogen gamma chain | G5BML1_HETGA | 1338 |
| Complement factor B | G5C3H1_HETGA | 1313 |
| Plasma kallikrein | G5BNV2_HETGA | 1167 |
| Antithrombin-III | G5ARS6_HETGA | 1093 |
| Coagulation factor V | G5CB46_HETGA | 1092 |
| Complement C5 | G5AXS5_HETGA | 1002 |
| Hemoglobin subunit beta | G5BS33_HETGA | 941 |
| Transcobalamin-2 isoform 1 | G5AVP0_HETGA | 913 |
| Hemopexin | G5BBR0_HETGA | 899 |
| Coagulation factor XIII B chain | G5BM72_HETGA | 821 |
| Protein AMBP | G5B1Y4_HETGA | 817 |
| N-acetylmuramoyl-L-alanine amidase | G5BYP3_HETGA | 812 |
| Coagulation factor XII | G5BQ09_HETGA | 802 |
| Apolipoprotein A-I | APOA1_HETGA | 798 |
| Hemoglobin subunit alpha | G5BXY1_HETGA | 784 |
| Coagulation factor XIII A chain | G5BAS8_HETGA | 779 |
| Hemoglobin subunit beta | G5BYJ8_HETGA | 759 |
| Alpha-1-antiproteinase S | G5B496_HETGA | 724 |
| Inter-alpha-trypsin inhibitor heavy chain H3 | G5BUN3_HETGA | 682 |
| Vitronectin | G5BVN8_HETGA | 652 |
| Complement factor I | G5AQM1_HETGA | 647 |
| Hemoglobin subunit epsilon-1 | G5BS35_HETGA | 642 |
| Inter-alpha-trypsin inhibitor heavy chain H2 | G5AXV8_HETGA | 584 |
| Apolipoprotein E | G5CBM7_HETGA | 581 |
| Inhibitor of carbonic anhydrase | G5BQB0_HETGA | 537 |
| Inter-alpha-trypsin inhibitor heavy chain H1 | G5BUN2_HETGA | 482 |
| Four and a half LIM domains protein 1 | G5CA61_HETGA | 469 |
| Haptoglobin | G5B5U6_HETGA | 464 |
| C4b-binding protein | G5BP10_HETGA | 438 |
| L-lactate dehydrogenase | G5AKA3_HETGA | 437 |
| Insulin-like growth factor-binding protein complex acid labile chain | G5BY64_HETGA | 403 |
| Catalase | G5AXV0_HETGA | 382 |
| Fetuin-B | G5BT88_HETGA | 347 |
| Alpha-2-HS-glycoprotein | G5BT89_HETGA | 337 |
| Selenoprotein P | G5APA7_HETGA | 328 |
| Vitamin D-binding protein | G5BE53_HETGA | 317 |
| Adiponectin | G5BT83_HETGA | 283 |
| von Willebrand factor | G5CAN6_HETGA | 274 |
| Beta-2-glycoprotein 1 | G5BGY7_HETGA | 268 |
| Basement membrane-specific heparan sulfate proteoglycan core protein | G5BI06_HETGA | 267 |
| Gelsolin | G5AXS0_HETGA | 259 |
| Aspartyl aminopeptidase | G5AKJ4_HETGA | 256 |
| Complement C1q subcomponent subunit A | G5BHZ8_HETGA | 246 |
| Ficolin-3 | G5AUT5_HETGA | 241 |
| Mannan-binding lectin serine protease 1 | G5BTD5_HETGA | 228 |
| Alpha-1-antichymotrypsin | G5B491_HETGA | 228 |
| Carboxypeptidase N catalytic chain | G5AYP3_HETGA | 210 |
| Complement C1q subcomponent subunit C | G5BHZ7_HETGA | 191 |
| Mannan-binding lectin serine protease 2 | G5C521_HETGA | 188 |
| Prothrombin | G5ATC4_HETGA | 177 |
| Complement C1q tumour necrosis factor-related protein 3 | G5BQ97_HETGA0 | 177 |
| Nucleoside diphosphate kinase | G5C4L3_HETGA | 168 |
| L-lactate dehydrogenase | G5BEG2_HETGA | 167 |
| Alpha-2-antiplasmin | G5BCV1_HETGA | 164 |
| Keratin, type II cytoskeletal 6B | G5ALS1_HETGA | 152 |
| Carbonic anhydrase 2 | G5ATW7_HETGA | 143 |
| Proprotein convertase subtilisin/kexin type 9 | G5APQ4_HETGA | 137 |
| Angiotensinogen | G5BQI5_HETGA | 121 |
| 6-phosphogluconate dehydrogenase, decarboxylating | G5C530_HETGA | 91 |
| Nesprin-1 | G5C0E1_HETGA | 87 |
| Complement C1q subcomponent subunit B | G5BHZ6_HETGA | 87 |
| Angiopoietin-related protein 6 | G5B341_HETGA | 85 |
| Hepatocyte growth factor activator | G5BZF5_HETGA | 84 |
| Calpain-1 catalytic subunit | G5B6L3_HETGA | 84 |
| Sulfhydryl oxidase | G5AYL7_HETGA | 80 |
| Keratin, type II cytoskeletal 5 | G5ALS3_HETGA | 76 |
| Profilin | G5BH50_HETGA | 75 |
| 40S ribosomal protein S27a | G5B8W4_HETGA | 68 |
| Xanthine dehydrogenase/oxidase | G5B3Z0_HETGA | 68 |
| Plasma protease C1 inhibitor | G5BLJ5_HETGA | 67 |
| Collectin-11 | G5C7L1_HETGA | 66 |
| Extracellular matrix protein 1 | G5BH40_HETGA | 64 |
| Actin, gamma-enteric smooth muscle | G5AXH0_HETGA | 61 |
| Collectin-10 | G5C9R8_HETGA | 61 |
| Keratin, type I cytoskeletal 17 | G5B0M4_HETGA | 58 |
| Contactin-1 | G5BGJ6_HETGA | 55 |
| Dynein heavy chain 1, axonemal | G5BUL8_HETGA | 50 |
| Leucine-rich repeat-containing protein 15 | G5ALE6_HETGA | 43 |
| Transcriptional regulator ATRX | G5C0I5_HETGA | 42 |
| Transmembrane protein 127 | G5BMW1_HETGA | 41 |
| TRAF3-interacting protein 1 | G5BHH8_HETGA | 41 |
| Mannose-binding protein A | G5C4H7_HETGA | 40 |
| Hyaluronan-binding protein 2 | G5BKD5_HETGA | 39 |
| Fer3-like protein | G5BZJ1_HETGA | 38 |
| Complement component C7 | G5C4R4_HETGA | 38 |
| PITH domain-containing protein | G5BHY3_HETGA | 38 |
| 1,4-alpha-glucan-branching enzyme | G5BA56_HETGA | 37 |
| tRNA pseudouridine synthase | G5BB54_HETGA | 37 |
| SRRM2-like protein | G5BG61_HETGA | 34 |
| Putative carboxypeptidase PM20D1 | G5BX50_HETGA | 34 |
| Olfactory receptor | G5C741_HETGA | 33 |
| Protein CASC5 | G5BLZ4_HETGA | 33 |
| Conserved oligomeric Golgi complex subunit 1 | G5B5M3_HETGA | 32 |
| Small G protein signalling modulator 1 | G5BGN5_HETGA | 32 |
| Eukaryotic translation initiation factor 3 subunit E | G5BD61_HETGA | 31 |
| HEAT repeat-containing protein 2 | G5AZ15_HETGA | 31 |
| Proline-rich protein 11 | G5AXY5_HETGA | 31 |
| Peroxiredoxin-1 | G5ARW1_HETGA | 30 |
| Pre-mRNA-processing factor 17 | G5B0Y2_HETGA | 30 |
| Ribonucleoside-diphosphate reductase | G5B9C2_HETGA | 30 |
# Ions score is −10 * Log(p), where p is the probability that the observed match is a random event. Individual ions scores > 30 indicate identity or extensive homology (p < 0.05). Protein scores are derived from ions scores as a non-probabilistic basis for ranking protein hits. Cut-off was set at Ions score 30. Protein hits identified in whole plasma only (but not in plasma EVs) are highlighted in pink.
Table 2.
Deiminated proteins identified by F95 enrichment in plasma-derived EVs of naked mole-rat (Heterocephalus glaber). Deiminated proteins were isolated by immunoprecipitation using the pan-deimination F95 antibody. The F95 enriched eluate was analysed by LC-MS/MS and peak list files were submitted to mascot. Species-specific peptide sequence hits scoring with H. glaber are included and total score is shown. Protein hits identified in plasma-EVs only are highlighted in blue. For full LC-MS/MS data analysis see Supplementary Table S2.
| Protein Name | Symbol | Total Score (p < 0.05) # |
|---|---|---|
| Alpha-2-macroglobulin | G5BPM1_HETGA | 2978 |
| Serum albumin | G5B5P2_HETGA | 2570 |
| Serotransferrin | G5BQA9_HETGA | 1976 |
| Keratin, type II cytoskeletal 6B | G5ALS1_HETGA | 1459 |
| Complement C3 | G5C0N5_HETGA | 1440 |
| Keratin, type II cytoskeletal 5 | G5ALS3_HETGA | 1211 |
| Kininogen-1 | G5BT86_HETGA | 1196 |
| Histidine-rich glycoprotein | G5BT87_HETGA | 1186 |
| Keratin, type I cytoskeletal 10 | G5AX66_HETGA | 1130 |
| Keratin, type I cytoskeletal 14 | G5B0M6_HETGA | 1038 |
| Keratin, type II cytoskeletal 1 | G5ALS8_HETGA | 1011 |
| Fibrinogen beta chain | G5BML3_HETGA | 974 |
| Fibrinogen alpha chain | G5BML2_HETGA | 956 |
| Apolipoprotein B-100 | G5AZB7_HETGA | 834 |
| Keratin, type I cytoskeletal 13 | G5B0N0_HETGA | 701 |
| Keratin, type I cytoskeletal 17 | G5B0M4_HETGA | 673 |
| Desmoplakin | G5BAT4_HETGA | 670 |
| Hemoglobin subunit alpha | G5BXY1_HETGA | 591 |
| Hemoglobin subunit beta | G5BS33_HETGA | 581 |
| Complement C4-A | G5C3H6_HETGA | 571 |
| Hemoglobin subunit beta | G5BYJ8_HETGA | 568 |
| Apolipoprotein A-I | APOA1_HETGA | 499 |
| Keratin, type II cytoskeletal 79 | G5BJ37_HETGA | 468 |
| Actin, cytoplasmic 1 | G5BI78_HETGA | 444 |
| Keratin, type II cytoskeletal 4 | G5BJ36_HETGA | 443 |
| Fibrinogen gamma chain | G5BML1_HETGA | 382 |
| Ceruloplasmin | G5BSL1_HETGA | 350 |
| Apolipoprotein E | G5CBM7_HETGA | 338 |
| Inter-alpha-trypsin inhibitor heavy chain H4 | G5BUN4_HETGA | 332 |
| Protein AMBP | G5B1Y4_HETGA | 280 |
| Inhibitor of carbonic anhydrase | G5BQB0_HETGA | 244 |
| Hemopexin | G5BBR0_HETGA | 239 |
| Histone H4 | G5BKL3_HETGA | 238 |
| Plasminogen | G5BC53_HETGA | 232 |
| Alpha-1-antiproteinase S | G5B496_HETGA | 217 |
| Hemoglobin subunit epsilon-1 | G5BS35_HETGA | 213 |
| Junction plakoglobin | G5B0M0_HETGA | 202 |
| N-acetylmuramoyl-L-alanine amidase | G5BYP3_HETGA | 190 |
| Inter-alpha-trypsin inhibitor heavy chain H3 | G5BUN3_HETGA | 190 |
| Keratin, type II cytoskeletal 7 | G5BL96_HETGA | 182 |
| Fibronectin | G5BHR4_HETGA | 164 |
| Glyceraldehyde-3-phosphate dehydrogenase | G5CAP7_HETGA | 154 |
| Coagulation factor XII | G5BQ09_HETGA | 152 |
| Antithrombin-III | G5ARS6_HETGA | 150 |
| Catenin beta-1 | G5ALX2_HETGA | 126 |
| Plakophilin-1 | G5B3A4_HETGA | 120 |
| Annexin | G5AWC0_HETGA | 119 |
| Keratin, type II cytoskeletal 3 | G5ALT2_HETGA | 118 |
| 40S ribosomal protein S27a | G5B8W4_HETGA | 116 |
| Inter-alpha-trypsin inhibitor heavy chain H2 | G5AXV8_HETGA | 106 |
| Alpha-2-antiplasmin | G5BCV1_HETGA | 86 |
| Complement factor B | G5C3H1_HETGA | 81 |
| Elongation factor 1-alpha | G5ALK7_HETGA | 78 |
| Angiotensinogen | G5BQI5_HETGA | 77 |
| Histone H2B | G5BH20_HETGA | 77 |
| Plasma kallikrein | G5BNV2_HETGA | 72 |
| Beta-enolase | G5BW96_HETGA | 70 |
| Vitamin D-binding protein | G5BE53_HETGA | 57 |
| Alpha-1-antichymotrypsin | G5B491_HETGA | 55 |
| Keratin, type II cytoskeletal 80 | G5BL95_HETGA | 53 |
| Heat shock cognate 71 kDa protein | G5B170_HETGA | 51 |
| Leucine-rich repeat-containing protein KIAA1731 | G5C3Y1_HETGA | 50 |
| Cathepsin D | G5C2G1_HETGA | 49 |
| Basement membrane-specific heparan sulfate proteoglycan core protein | G5BI06_HETGA | 47 |
| Skin-specific protein 32 | G5BUY9_HETGA | 46 |
| Desmocollin-1 | G5C312_HETGA | 46 |
| Protein-arginine deiminase type-6 | G5BZN1_HETGA | 46 |
| Cryptochrome-1 | G5C454_HETGA | 45 |
| Complement C1q subcomponent subunit C | G5BHZ7_HETGA | 45 |
| p2Y purinoceptor 12 | G5C102_HETGA | 44 |
| Alpha-1B-glycoprotein | G5B7K8_HETGA | 44 |
| Exportin-1 | G5C1Y9_HETGA | 44 |
| Putative hexokinase HKDC1 | G5B183_HETGA | 43 |
| Transcriptional regulator ATRX | G5C0I5_HETGA | 42 |
| Coiled-coil domain-containing protein 57 | G5BJ98_HETGA | 42 |
| Microtubule plus-end tracking protein TIP150 | G5ATU1_HETGA | 40 |
| Tubulin alpha-1C chain | G5AQ00_HETGA | 38 |
| Fer3-like protein | G5BZJ1_HETGA | 38 |
| Amino acid transporter | G5BYQ9_HETGA | 36 |
| Ventricular zone-expressed PH domain-containing protein-like protein 1 | G5B253_HETGA | 35 |
| SRRM2-like protein | G5BG61_HETGA | 35 |
| N6-adenosine-methyltransferase 70 kDa subunit | G5BFU9_HETGA | 33 |
| Proline-rich protein 11 | G5AXY5_HETGA | 33 |
| Carbonic anhydrase 2 | G5ATW7_HETGA | 32 |
| 26S proteasome non-ATPase regulatory subunit 3 | G5BRY1_HETGA | 31 |
| Serine/threonine-protein kinase LATS1 | G5BMD1_HETGA | 31 |
| Transmembrane gamma-carboxyglutamic acid protein 1 | G5C418_HETGA | 31 |
| Collagen alpha-2(I) chain | G5ANK8_HETGA | 31 |
| Bcl-2-associated transcription factor 1 | G5C1A7_HETGA | 30 |
# Ions score is −10 * Log(p), where p is the probability that the observed match is a random event. Individual ions scores > 30 indicated identity or extensive homology (p < 0.05). Protein scores were derived from ions scores as a non-probabilistic basis for ranking protein hits. Cut-off was set at Ions score 30. Protein hits identified in whole plasma only (but not in plasma EVs) are highlighted in blue.
Figure 2.
Deiminated proteins identified in naked mole-rat plasma and plasma-EVs. Species specific hits identified for deiminated proteins in naked mole-rat plasma and EVs showed 112 total proteins identified in plasma and 80 in EVs, respectively. Of these, 48 protein hits were overlapping, while 64 proteins were specific for whole plasma and 32 for plasma-EVs only, respectively.
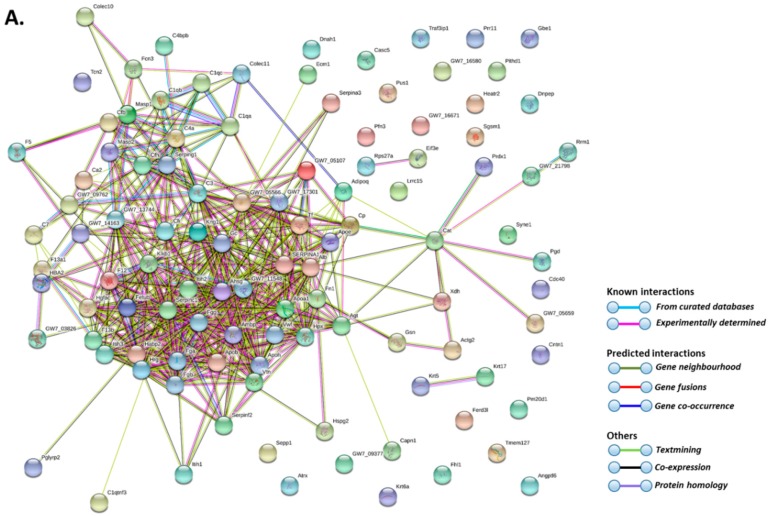
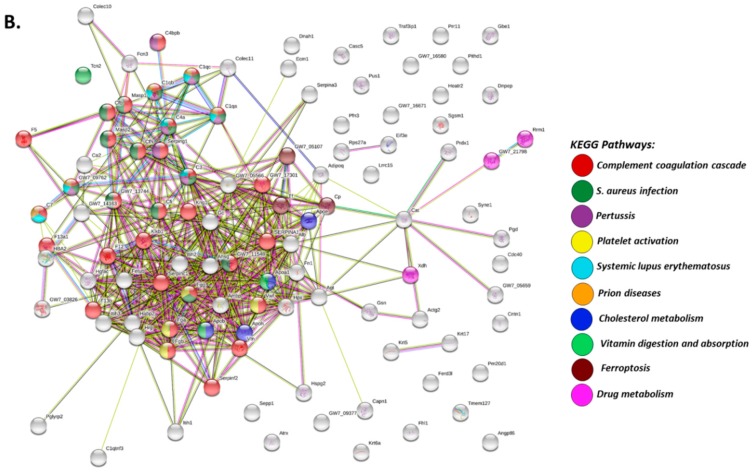
Figure 3.
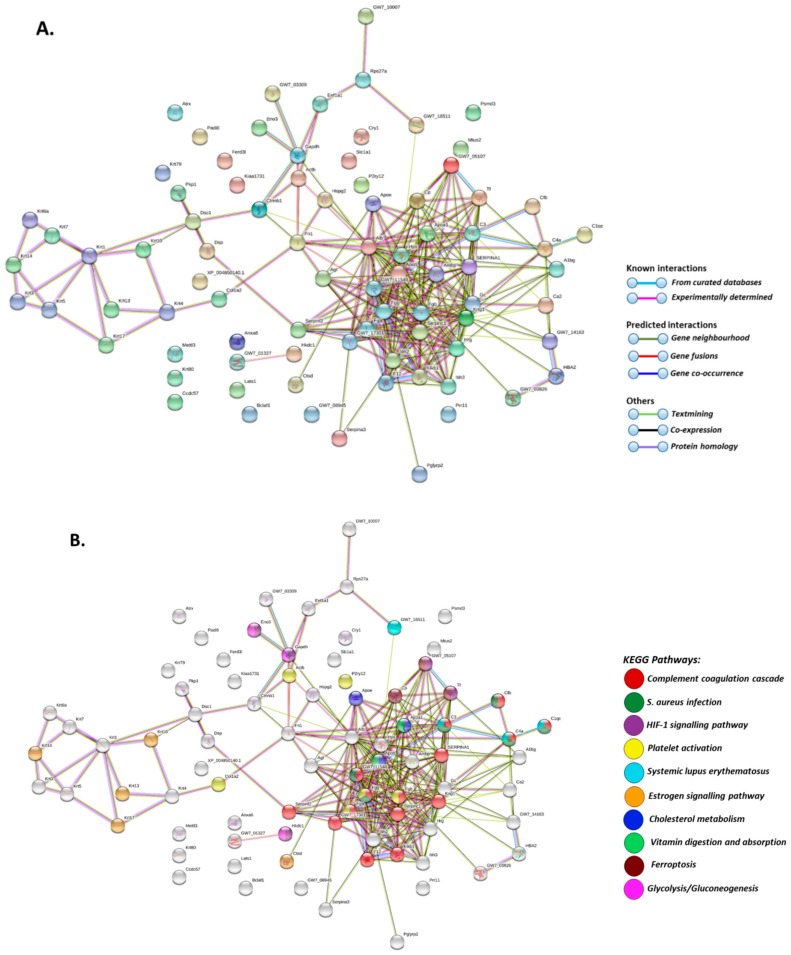
Protein-protein interaction networks of deiminated proteins identified in naked mole-rat plasma. Reconstruction of protein-protein interactions based on known and predicted interactions using STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) analysis. (A) Coloured nodes represent query proteins and first shell of interactors. (B) KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways relating to the identified proteins and reported in STRING are highlighted as follows: red = complement and coagulation cascade; dark green = Staphylococcus aureus infection; purple = pertussis; yellow = platelet activation; light blue = systemic lupus erythematosus (SLE); orange = prion diseases; dark blue = cholesterol metabolism; light green = vitamin digestion and absorption; dark red = ferroptosis; pink = drug metabolism. Coloured nodes represent query proteins and first shell of interactors; white nodes are second shell of interactors. Coloured lines indicate whether protein interactions are identified via known interactions (curated databases, experimentally determined), predicted interactions (gene neighbourhood, gene fusion, gene co-occurrence) or via text mining, co-expression or protein homology (see the colour key for connective lines included in the figure).
Figure 4.
Protein-protein interaction networks of deiminated proteins identified in plasma-EVs of naked mole-rat. Reconstruction of protein-protein interactions based on known and predicted interactions using STRING analysis. (A) Coloured nodes represent query proteins and first shell of interactors. (B) KEGG pathways relating to the identified proteins and reported in STRING are highlighted as follows: red = complement and coagulation cascade; dark green = Staphylococcus aureus infection; purple = HIF-signalling pathway; yellow = platelet activation; light blue = systemic lupus erythematosus (SLE); orange = oestrogen signalling pathway; dark blue = cholesterol metabolism; light green = vitamin digestion and absorption; pink = glycolysis/gluconeogenesis. Coloured nodes represent query proteins and first shell of interactors, white nodes are second shell of interactors. Coloured lines indicate whether protein interactions are identified via known interactions (curated databases, experimentally determined), predicted interactions (gene neighbourhood, gene fusion, gene co-occurrence) or via text mining, co-expression or protein homology (see the colour key for connective lines included in the figure).
2.3. Characterisation of Extracellular Vesicles in Naked Mole-Rat Plasma
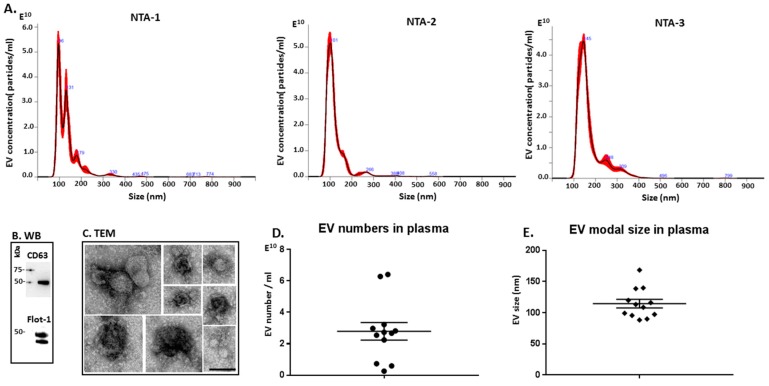
EVs isolated from naked mole-rat plasma were characterised for size distribution using nanoparticle tracking analysis (NTA; Figure 5A), by Western blotting using EV-specific markers (Figure 5B) and by morphological analysis using transmission electron microscopy (TEM; Figure 5C). A poly-dispersed population of EVs mainly in the size range of 50 to 300 nm was observed, with some variations between individual animals (Figure 5A), and also with respect to EV yield from plasma (Figure 5D) and modal EV size (Figure 5E). Overall, the main EV peaks were detected at approximately 100–140 nm (Figure 5A,E), with the modal size of EVs falling mainly in the range of 90–115 nm, although this varied somewhat between animals and some outliers were detected (Figure 5E). Western blotting confirmed that the naked mole-rat EVs were positive for the EV-specific markers CD63 and Flot-1 (Figure 5B). Typical EV morphology was confirmed by TEM (Figure 5C).
Figure 5.
Extracellular vesicle profiling in naked mole-rat plasma. (A) Nanoparticle tracking analysis (NTA) shows a size distribution of EVs from naked mole-rat in the range of mainly 50 to 300 nm, with representative NTA profiles of EVs from 3 different animals (NTA-1, NTA-2, NTA-3). (B) Western blotting analysis confirms that naked mole-rat EVs are positive for the phylogenetically conserved EV-specific markers CD63 and Flot-1. (C) Transmission electron microscopy (TEM) analysis of naked mole-rat plasma-derived EVs shows typical EV morphology; a composite figure is shown and the scale bar (100 nm) applies for all images in the panel. (D) EV yield in plasma of 12 individual naked mole-rats is shown. (E) EV modal size in plasma of 12 individual naked mole-rats is presented.
2.4. MicroRNA Analysis of Naked Mole-Rat EVs
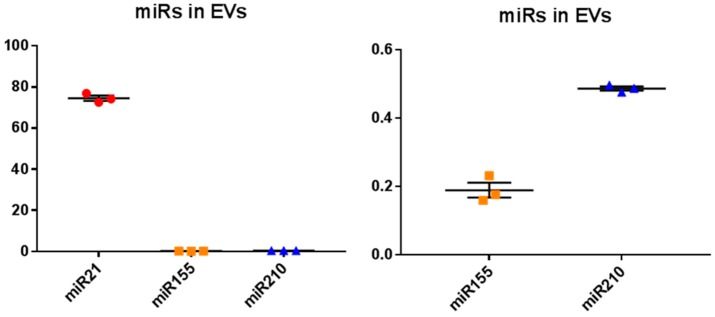
EVs isolated from naked mole-rat plasma were assessed for the relative expression of the immune- and cancer-related miR21, the inflammatory-related miR155 and the metabolic- and hypoxia-related miR210. The highest relative miR levels of the three miRs tested, were observed for miR21 in naked mole-rat plasma-EVs, 394-fold higher than for miR155 and 153-fold higher than for miR210, respectively (Figure 6A). The relative expression of the hypoxia- and metabolic-related miR210 was 2.6–fold higher than the inflammatory miR155 (Figure 6B), which overall showed comparatively the lowest relative levels of expression of the three miRs tested (Figure 6A,B).
Figure 6.
MicroRNA (miR) analysis of three key miRs related to cancer, inflammation, metabolism and hypoxia, in naked mole-rat plasma-derived EVs. (A). The onco-related miR21 showed the highest relative expression of the three miRs tested, being 394-fold higher than miR155 and 153-fold higher than miR210, respectively (p < 0.0001; n = 3). (B). The relative expression of the hypoxia and metabolic related miR210 was 2.6–fold higher than the inflammatory miR155 (p = 0.0002; n = 3); (miR21, miR155 and miR210 expression is represented as red circles, orange squares and blue triangles, respectively).
3. Discussion
For the first time, post-translationally deiminated proteins are described in naked mole-rat (Heterocephalus glaber) plasma, unravelling novel aspects of post-translational deimination of key proteins involved in immune defences and metabolism. PAD homologues were identified in naked mole-rat plasma by Western blotting via cross reaction with human PAD2, which is the phylogenetically most conserved form of PAD [1,16,31]. PAD-positive protein bands were also observed for PAD3 and PAD4, via cross-reaction with PAD isozyme-specific human antibodies, at an expected size of approximately 70 kDa, similar to that reported for other mammalian PADs. Such protein detection corresponds with PAD isozymes that have been identified in the naked mole-rat genome (PADI1, Gene ID: 101722077; PADI2, Gene ID: 101721485; PADI3, Gene ID: 101722435; PADI4, Gene ID: 101722785; PADI6, Gene ID: 101723122). Deiminated proteins were detected and identified both in whole plasma and in plasma-derived EVs. The identity of specific deimination protein candidates in plasma and plasma-EVs was assessed using F95-enrichment in tandem with LC-MS/MS analysis. The F95 mono-specific antibody used in the current study was developed against a deca-citrullinated peptide and has been verified to detect deiminated/citrullinated proteins in diverse taxa [11,12,16,18,19,21,22,31]. In the naked mole-rat, species-specific protein hits revealed 48 common deiminated proteins in plasma and EVs by F95-enrichment, which included some key immune and metabolic related proteins, while 65 deiminated protein hits were specific for plasma and 42 deiminated protein hits specific for EVs only.
As assessed by STRING analysis, the PPI enrichment p-value for deiminated proteins identified in naked mole-rat plasma and as cargo in plasma-derived EVs was found to be <1.0 × 10−16 for both. Such an enrichment value indicates that the identified network of proteins has significantly more interactions than expected. Therefore, these deiminated proteins have more interactions among themselves than would be expected for a random set of proteins of similar size, drawn from the genome. Such an enrichment indicates that the proteins, as a group, are at least partially biologically connected. Deiminated target proteins identified in whole naked mole-rat plasma belonged to the following KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways: complement coagulation cascade, platelet activation, cholesterol metabolism, vitamin digestion and adsorption, drug metabolism as well as bacterial infection (Staphylococcus aureus and pertussis), autoimmunity (systemic lupus erythematosus) and prion diseases (Figure 3B). Deiminated target proteins identified in EVs of naked mole-rat plasma belonged to KEGG pathways of the complement coagulation cascade, platelet activation, HIF- (hypoxia-inducible factor) signalling pathway, glycolysis/gluconeogenesis, cholesterol metabolism, vitamin digestion and adsorption, oestrogen signalling pathway, as well as bacterial infection (Staphylococcus aureus) and autoimmunity (systemic lupus erythematosus) (Figure 4B).
Of interest is that deiminated protein candidates involved in KEGG-pathways for HIF-1-signalling, the master regulator of oxygen homeostasis, seem enriched in the plasma-EVs, indicating a role for EV-mediated transport of such proteins in hypoxic signalling. It is worth noting that naked mole-rats have a high endogenous expression of HIF due to mutation in the VHL (Von Hippel-Lindau disease tumor suppressor) domain [62], which could possibly explain the elevated HIF-1 signalling related targets identified here. Furthermore, deiminated proteins identified in EVs were also enriched for glycolysis and gluconeogenesis KEGG pathways. Our findings indicate that protein deimination may play hitherto unidentified roles in the unusual hypoxia resistance and metabolism of the naked mole-rat, including via EV-transport in cellular communication, also under normal physiological conditions. In addition, the presence of deiminated histone H2B and H4 in EVs may be of some interest as histone deimination is well known to contribute to epigenetic regulation including in cancer [4,6], and the naked mole-rat has been found to have a particularly stable epigenome, which may contribute to the cancer resistance and longevity observed in these animals [63]. Furthermore, an abundance of deiminated complement components identified in both plasma and plasma-EVs may indicate roles for functional diversity of the complement system via post-translational deimination in the naked mole-rat. This may play various roles in naked mole-rat immune responses as recent studies have identified some unusual characteristics, including atypical immune surveillance and a greater reliance on myeloid-biased innate immunity [64]. Also noteworthy is the identification of deiminated adiponectin in naked mole-rat plasma identified here, as adiponectin in humans is the most abundant secreted adipokine with pleiotropic roles in metabolism [65,66], glucose regulation [67,68,69], longevity [70], regeneration and cancer [71,72,73]. Such deimination of adiponectin has not been studied and may add to some of its protein moonlighting function and be of relevance in the context of the unusual metabolism of the naked mole-rat.
As part of EV-mediated cellular communication in physiology and pathologies, the transport of microRNAs (miRs) is well acknowledged. There is increasing interest, reflected in a range of studies, in furthering our understanding of how such EV-mediated transport may play a part in physiological and pathophysiological processes. MiRs are highly conserved small non-coding RNAs that control gene expression and regulate biological processes by targeting messenger RNAs (mRNAs). MiRs can, for example, inhibit post-transcriptional translation of mRNA as well as enhance mRNA degradation [74]. Some expression profiling has been carried out in naked mole-rats, mainly at the transcriptome level [75,76], although no studies have assessed miRs in EVs of naked mole-rat plasma. This study focused on assessing three key miRs known to be involved in cancer, inflammation and hypoxia due to the unusual resistance of naked mole-rats to cancer, ageing and hypoxia. MiR21 is a main immunoregulatory and onco-related miR and is also associated with chronic diseases [77,78,79]. MiR21 is strongly conserved throughout evolution and while many experimentally verified targets of miR21 are tumour suppressors, miR21 is also linked to cardiac disease and oxidative stress [80]. Less is known about the physiological roles of miR21. In the current study, miR21 was found to be by far the highest miR expressed in EVs of naked mole-rat out of the three miRs tested. Roles for miR21 in immune responses of naked mole-rat have not been reported in detail and the expression of miR21 in EVs has not been assessed in naked mole-rat before.
In mammals, miR155 is known to be a major inflammatory related miR, linked to inflammatory and stress responses [81]. Here, miR155 was found to be the least expressed of the three miRs tested in naked mole-rat plasma-EVs, possibly indicating that this miR may be a contributing factor to the “anti-inflammatory” state of mole-rats, which may have some relation to their longevity and cancer resistance. MiR210 is known to be a major miR induced under hypoxia and has an important role in mitochondrial metabolism, DNA damage response, cell proliferation and apoptosis [74]. MiR210 has an important role in regulating mitochondrial metabolism [82] and cell glycolytic activity, as well as being linked to inflammation [83]. MiR210 has been identified as a regulator of the hypoxia pathway and was found to have pro-apoptotic functions under normoxic conditions, but anti-apoptotic effects under hypoxic conditions [84,85]. In the current study, miR210 was found to be more highly expressed in mole-rat plasma than the inflammatory miR155. As naked mole-rats are known to be hypoxia tolerant animals and to exhibit marked changes in their metabolic substrate use and metabolic demand in hypoxia [41,86], miR210 may have functional roles in metabolic control, possibly contributing to the well-known longevity of these animals. As this is the first study to assess the expression of these three onco-, inflammatory- and metabolic-related miRs in naked mole-rat plasma-EVs, it remains to be fully understood what specific functions the EV-mediated transport of these miRs play in the unusual physiology of naked mole-rats.
Here, for the first time, we report the protein deimination profiles of plasma and plasma-derived EVs in naked mole-rats. Post-translational deimination of major key immune and metabolic factors in naked mole-rats was identified and related to key KEGG pathways of inflammation, metabolism and oxygen transport. Our findings highlight novel aspects of protein moonlighting via post-translational deimination, including via EV-mediated transport. Research on EVs is a relatively new field in comparative animal models, and to our knowledge this is the first characterisation of EVs and associated protein and selected miR cargo markers in naked mole-rats. Furthermore, as PADs have been found to play major roles in the regulation of EV release [19,20,21,22,34], their contribution to EV-mediated cell communication in response to physiological and pathophysiological changes in naked mole-rats remains to be further investigated. Findings in long-lived mammals that display cancer resistance, including naked mole-rats, may be of considerable translational value for furthering our understanding of the mechanisms underlying cancer resistance for improved development of human cancer therapies [59].
In continuation of the current study, the assessment of changes in deiminated proteins and EV profiles, including protein and genetic EV-cargo, may be of great interest in studies using this unique animal model to further understanding of the hitherto novel and understudied mechanisms involved in cancer and ageing.
4. Materials and Methods
4.1. Sampling of Naked Mole-Rat Plasma
Naked mole-rats were group-housed in interconnected multi-cage systems at 30 °C and 21% O2 in 50% humidity with a 12L:12D light cycle. Animals were fed fresh tubers, vegetables, fruit and Pronutro cereal supplement ad libitum. Animals were not fasted prior to experimental trials. All experimental procedures were approved by the University of Ottawa Animal Care Committee in accordance with the Animals for Research Act and by the Canadian Council on Animal Care (protocol # 2535). Non-breeding (subordinate) naked mole-rats do not undergo sexual development or express sexual hormones and thus we did not take sex into consideration when evaluating our results [87]. Blood was collected from 12 adult (~ 1–2 years old) subordinate naked mole-rats following live cervical dislocation and rapid decapitation. Blood was collected in Eppendorf tubes pre-coated with a 10% EDTA (ethylenediaminetetraacetic acid) solution. Plasma was isolated by centrifugation at 5000× g for 5 min. The isolated plasma was aliquoted and immediately frozen at −80 °C until further use.
4.2. Extracellular Vesicle Isolation and Nanoparticle Tracking Analysis (NTA)
EVs were isolated by step-wise centrifugation according to our established protocols using ultracentrifugation and the recommendations of MISEV2018 (the minimal information for studies of extracellular vesicles 2018; [88]). Mole-rat plasma were diluted 1:4 in ultra-filtered (using a 0.22 μm filter) Dulbecco’s PBS (100 μL plasma added to 400 μL DPBS) and then centrifuged at 4000× g for 30 min at 4 °C for removal of aggregates and apoptotic bodies. The supernatants were collected and centrifuged further at 100,000× g for 1 h at 4 °C. The EV-enriched pellets were washed in 1 mL DPBS and ultra-centrifuged at 100,000× g for 1 h at 4 °C. The final EV pellets were resuspended in 100 µL DPBS and frozen at −80 °C until further use. For NTA, based on Brownian motion of particles in suspension, the EV pellets were diluted 1/100 in DPBS and applied to the NanoSight NS300 system (Malvern Panalytical Ltd., Malvern, UK) in conjunction with a syringe pump to ensure continuous flow of the sample. Five 60 sec videos were recorded for each sample, with approximately 40–60 particles per frame, and the replicate histograms generated were averaged.
4.3. Transmission Electron Microscopy (TEM)
EVs were isolated from individual plasma as described above. For TEM, the EV pellets were fixed with 2.5% glutaraldehyde in 100 mM sodium cacodylate buffer (pH 7.0) for 1 h at 4 °C. The EVs were then resuspended in 100 mM sodium cacodylate buffer (pH 7.0), placed on to a grid with a glow discharged carbon support film and stained with 2% aqueous Uranyl Acetate (Sigma-Aldrich, Gillingham, UK). Individual EVs were imaged by TEM using a Morada CCD camera (EMSIS GmbH, Münster, Germany) and processed via iTEM (EMSIS).
4.4. Western Blotting Analysis
Mole-rat plasma and plasma-EVs (an EV pellet derived from 100 µL plasma, reconstituted in 100 µL PBS) were diluted 1:1 in 2 × Laemmli sample buffer, boiled for 5 min at 100 °C and separated by SDS-PAGE on 4–20% TGX gels (BioRad, Watford, UK). Approximately 5 μg protein was loaded per lane. Following SDS-PAGE, proteins were transferred to nitrocellulose membranes using semi-dry Western blotting; even transfer was assessed by PonceauS (Sigma-Aldrich, Gillingham, UK) staining. The membranes were blocked in 5% BSA in TBS-T for 1 h at room temperature (RT) and thereafter incubated with the following primary antibodies diluted in TBS-T: F95 pan-deimination antibody (MABN328, Merck, Watford, UK, 1/1000); PAD2 (ab50257, Abcam, Cambridge, UK, 1/1000); PAD3 (ab50246, Abcam, 1/1000); PAD4 (ab50247, Abcam, 1/1000); and two EV-specific markers: CD63 (ab216130, Abcam, 1/1000); Flot-1 (ab41927, Abcam, 1/2000). After primary antibody incubation overnight at 4 °C on a shaking platform, the membranes were washed for 3 × 10 min in TBS-T at RT and thereafter incubated with the appropriate HRP-conjugated secondary antibodies (anti-rabbit IgG BioRad or anti-mouse IgM BioRad, diluted 1/4000 in TBS-T) for 1 h, at RT. Membranes were washed for 5 × 10 min in TBS-T, followed by 1 × 10 min in TBS proteins bands were visualised using enhanced chemiluminescence (ECL, Amersham, Buckinghamshire, UK) and digital images were obtained using the UVP BioDoc-ITTM System (Thermo Fisher Scientific, Hemel Hempstead, UK).
4.5. Immunoprecipitation and Protein Identification
Deiminated proteins in plasma and in plasma-derived EVs were immunoprecipitated by enrichment with the F95 pan-deimination antibody (MABN328, Merck, Watford, UK), which has been developed against a deca-citrullinated peptide and specifically detects proteins modified by citrullination [89]. The mono-specific F95 antibody is predicted to react with all deiminated/citrullinated proteins based on 100% sequence homology and has for example been used to identify deiminated proteins in human, mouse, rat, chicken and teleost fish tissue [11,12,16,18,19,21,22,31,89]. The Catch and Release immunoprecipitation kit (Merck, Watford, UK) was used according to the manufacturer’s instructions. For F95 enrichment, plasma was pooled from 5 individual animals (5 × 20 μL), while for EVs, total protein was first extracted from the EV pellets derived from 100 μL plasma per animal, using 100 μL RIPA + buffer on ice for 2 h followed by centrifugation at 16,000× g for 30 min to collect the supernatant containing the proteins. The immunoprecipitation was carried out on a rotating platform overnight at 4 °C, and the F95 bound proteins were eluted using denaturing elution buffer according to the manufacturer’s instructions (Merck). The F95 enriched eluates were then either analysed by Western blotting or by liquid chromatography with tandem mass spectrometry (LC-MS/MS; Cambridge Proteomics, Cambridge, UK). Peak files obtained were submitted to Mascot (Matrix Science). An in-house database (Cambridge proteomics) for naked mole-rat was used for the identification of species-specific protein hits (CCP_Heterocephalus_glaber Heterocephalus_glaber_20190911; 21449 sequences; 10466552 residues).
4.6. MicroRNA Analysis
EV isolates from individual naked mole-rat plasma (from 100 μL plasma as before) were assessed for relative expression of 3 key microRNAs (miRs) related to oncogenic, inflammatory and metabolic activity. These selected miRs included two cancer and immune-related miRs, miR21 and miR155, and miR210 for hypoxia and metabolic activity. Total RNA was extracted from mole-rat plasma EVs (prepared as before) using Trizol (Sigma-Aldrich, Gillingham, UK). The purity and concentration of the isolated RNA were measured using the NanoDrop Spectrophotometer at 260 nm and 280 nm absorbance. The cDNA was produced using the qScript microRNA cDNA Synthesis Kit (Quantabio, Beverly, MA, USA) according to the manufacturer’s instructions and used to assess the expression of miR21, miR155 and miR210. Reference RNAs used for the normalization of miR expression levels were U6-snRNA and has-let-7a-5p. The PerfeCTa SYBR Green SuperMix (Quantabio, Beverly, MA, USA) was used together with MystiCq microRNA qPCR primers for the miR21 (hsa-miR-21-5p), mir155 (hsa-miR-155-5p) and miR210 (hsa-miR-210-5p). All miR primers were obtained from Sigma-Aldrich (UK). Thermocycling conditions were used as follows: denaturation at 95 °C for 2 min, followed by 40 cycles of 95 °C for 2 s, 60 °C for 15 s, and extension at 72 °C for 15 s. The 2ΔΔCt method [90] was used for calculating relative miR expression levels and for normalisation. Each experiment was performed in 3 individuals, in triplicate.
4.7. Statistical Analysis
The histograms and graphs were prepared using the Nanosigh NS300 software (Malvern Panalytical Ltd., Malvern, UK) and GraphPad Prism version 7 (GraphPad Software, San Diego, CA, USA). Experiments were repeated in triplicate, histograms represent mean of data and standard error of mean (SEM) is indicated by the error bars. Significant differences were considered as p ≤ 0.05, following one-way ANOVA or Student’s t-test.
5. Conclusions
Here, for the first time, we report the protein deimination profiles of plasma and plasma-derived EVs in naked mole-rats. Post-translational deimination of major key immune and metabolic proteins in naked mole-rats was identified and related to key KEGG pathways of inflammation, metabolism and oxygen transport. Our findings highlight novel aspects of protein moonlighting via post-translational deimination, including via EV-mediated transport of such proteins in cellular communication. Three key microRNAs for oncogenic, inflammatory and metabolic/hypoxia function were also assessed in mole-rat plasma EVs. In continuation of the current study, the assessment of changes in deiminated proteins and EV profiles, including protein and microRNA EV-cargo may be of great interest in studies using this unique animal model to further understanding of hitherto novel and understudied mechanisms involved in cancer, inflammatory diseases and ageing.
Acknowledgments
The authors would like to thank Yagnesh Umrania and Michael Deery at the Cambridge Centre for Proteomics, UK, for the LC-MS/MS analysis. Thanks are due to The Guy Foundation for funding the purchase of equipment utilised in this work.
Abbreviations
| BSA | Bovine Serum Albumin |
| CD63 | CD63 antigen; granulophysin; lysosomal-associated membrane protein 3 |
| CNS | Central Nervous System |
| ECL | Enhanced Chemiluminescence |
| EVs | Extracellular Vesicles |
| F95 | Pan-deimination/citrullination antibody |
| FBS | Foetal Bovine Serum |
| Flot-1 | Flotillin-1 |
| HIF | Hypoxia-inducible Factor |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| kDa | Kilodalton |
| LC-MS/MS | Liquid Chromatography Mass Spectrometry |
| miR | microRNA |
| NTA | Nanoparticle Tracking Analysis |
| PAD | Peptidylarginine Deiminase |
| SDS-PAGE | Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis |
| TBS | Tris Buffered Saline |
| TEM | Transmission Electron Microscopy |
| WB | Western Blotting |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/21/5378/s1. Supplementary Table S1. Deiminated proteins identified by F95 enrichment in total plasma of naked mole-rat (Heterocephalus glaber). Deiminated proteins were isolated by immunoprecipitation using the pan-deimination F95 antibody. The F95 enriched eluate was analysed by LC-MS/MS and peak list files were submitted to mascot. Peptide sequences for the protein hits, their m/z values and individual scores are listed. Supplementary Table S2. Deiminated proteins identified by F95 enrichment in plasma-EVs of naked mole-rat (Heterocephalus glaber). Deiminated proteins were isolated by immunoprecipitation using the pan-deimination F95 antibody. The F95 enriched eluate was analysed by LC-MS/MS and peak list files were submitted to mascot. Peptide sequences for the protein hits, their m/z values and individual scores are listed.
Author Contributions
Data curation, S.L.; Formal analysis, P.U.-O. and S.L.; Funding acquisition, M.E.P. and S.L.; Investigation, P.U.-O. and S.L.; Methodology, M.E.P.; P.U.-O.; K.W.H.; I.K. and S.L.; Project administration, S.L.; Resources, M.E.P.; I.K., P.U.-O. and S.L.; Supervision, S.L.
Funding
This work was supported in parts by a University of Westminster Start-up Grant to SL and a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant and the Canada Research Chairs Program to MEP.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Vossenaar E.R., Zendman A.J., van Venrooij W.J., Pruijn G.J. PAD, a growing family of citrullinating enzymes: Genes, features and involvement in disease. Bioessays. 2003;25:1106–1118. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- 2.György B., Toth E., Tarcsa E., Falus A., Buzas E.I. Citrullination: A posttranslational modification in health and disease. Int. J. Biochem. Cell Biol. 2006;38:1662–1677. doi: 10.1016/j.biocel.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Bicker K.L., Thompson P.R. The protein arginine deiminases: Structure, function, inhibition, and disease. Biopolymers. 2013;99:155–163. doi: 10.1002/bip.22127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang S., Wang Y. Peptidylarginine deiminases in citrullination, gene regulation, health and pathogenesis. Biochim. Biophys. Acta Gene Regul. Mech. 2013;1829:1126–1135. doi: 10.1016/j.bbagrm.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Witalison E.E., Thompson P.R., Hofseth L.J. Protein Arginine Deiminases and Associated Citrullination: Physiological Functions and Diseases Associated with Dysregulation. Curr. Drug Targets. 2015;16:700–710. doi: 10.2174/1389450116666150202160954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lange S., Gallagher M., Kholia S., Kosgodage U.S., Hristova M., Hardy J., Inal J.M. Peptidylarginine Deiminases-Roles in Cancer and Neurodegeneration and Possible Avenues for Therapeutic Intervention via Modulation of Exosome and Microvesicle (EMV) Release? Int. J. Mol. Sci. 2017;18:1196. doi: 10.3390/ijms18061196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henderson B., Martin A.C. Protein moonlighting: A new factor in biology and medicine. Biochem. Soc. Trans. 2014;42:1671–1678. doi: 10.1042/BST20140273. [DOI] [PubMed] [Google Scholar]
- 8.Jeffrey C.J. Protein moonlighting: What is it, and why is it important? Philos. Trans. R. Soc. B Biol. Sci. 2018;373:20160523. doi: 10.1098/rstb.2016.0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohanan S., Cherrington B.D., Horibata S., McElwee J.L., Thompson P.R., Coonrod S.A. Potential role of peptidylarginine deiminase enzymes and protein citrullination in cancer pathogenesis. Biochem. Res. Int. 2012 doi: 10.1155/2012/895343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong S.L., Wagner D.D. Peptidylarginine deiminase 4: A nuclear button triggering neutrophil extracellular traps in inflammatory diseases and aging. FASEB J. 2018;32:6358–6370. doi: 10.1096/fj.201800691R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lange S., Gögel S., Leung K.Y., Vernay B., Nicholas A.P., Causey C.P., Thompson P.R., Greene N.D., Ferretti P. Protein deiminases: New players in the developmentally regulated loss of neural regenerative ability. Dev. Biol. 2011;355:205–214. doi: 10.1016/j.ydbio.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lange S., Rocha-Ferreira E., Thei L., Mawjee P., Bennett K., Thompson P.R., Subramanian V., Nicholas A.P., Peebles D., Hristova M., et al. Peptidylarginine deiminases: Novel drug targets for prevention of neuronal damage following hypoxic ischemic insult (HI) in neonates. J. Neurochem. 2014;130:555–562. doi: 10.1111/jnc.12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lange S. Peptidylarginine Deiminases as Drug Targets in Neonatal Hypoxic-Ischemic Encephalopathy. Front. Neurol. 2016;7:22. doi: 10.3389/fneur.2016.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sase T., Arito M., Onodera H., Omoteyama K., Kurokawa M.S., Kagami Y., Ishigami A., Tanaka Y., Kato T. Hypoxia-induced production of peptidylarginine deiminases and citrullinated proteins in malignant glioma cells. Biochem. Biophys. Res. Commun. 2017;482:50–56. doi: 10.1016/j.bbrc.2016.10.154. [DOI] [PubMed] [Google Scholar]
- 15.Yu R., Li C., Sun L., Jian L., Ma Z., Zhao J., Liu X. Hypoxia induces production of citrullinated proteins in human fibroblast-like synoviocytes through regulating HIF1α. Scand. J. Immunol. 2018;87:e12654. doi: 10.1111/sji.12654. [DOI] [PubMed] [Google Scholar]
- 16.Magnadottir B., Hayes P., Hristova M., Bragason B.Þ., Nicholas A.P., Dodds A.W., Gudmundsdottir S., Lange S. Post-translational Protein Deimination in Cod (Gadus morhua L.) Ontogeny—Novel Roles in Tissue Remodelling and Mucosal Immune Defences? Dev. Comp. Immunol. 2018;87:157–170. doi: 10.1016/j.dci.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Magnadottir B., Hayes P., Gísladóttir B., Bragason B.Þ., Hristova M., Nicholas A.P., Guðmundsdóttir S., Lange S. Pentraxins CRP-I and CRP-II are post-translationally deiminated and differ in tissue specificity in cod (Gadus morhua L.) ontogeny. Dev. Comp. Immunol. 2018;87:1–11. doi: 10.1016/j.dci.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 18.Magnadottir B., Bragason B.T., Bricknell I.R., Bowden T., Nicholas A.P., Hristova M., Gudmundsdottir S., Dodds A.W., Lange S. Peptidylarginine Deiminase and Deiminated Proteins are detected throughout Early Halibut Ontogeny—Complement Components C3 and C4 are post-translationally Deiminated in Halibut (Hippoglossus hippoglossus L.) Dev. Comp Immunol. 2019;92:1–19. doi: 10.1016/j.dci.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 19.Kholia S., Jorfi S., Thompson P.R., Causey C.P., Nicholas A.P., Inal J., Lange S. A Novel Role for Peptidylarginine Deiminases (PADs) in Microvesicle Release: A Therapeutic Potential for PAD Inhibitors to Sensitize Prostate Cancer Cells to Chemotherapy. J. Extracell. Vesicles. 2015;4:26192. doi: 10.3402/jev.v4.26192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kosgodage U.S., Trindade R.P., Thompson P.T., Inal J.M., Lange S. Chloramidine/Bisindolylmaleimide-I-Mediated Inhibition of Exosome and Microvesicle Release and Enhanced Efficacy of Cancer Chemotherapy. Int. J. Mol. Sci. 2017;18:1007. doi: 10.3390/ijms18051007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kosgodage U.S., Onganer P.U., Maclatchy A., Nicholas A.P., Inal J.M., Lange S. Peptidylarginine Deiminases Post-translationally Deiminate Prohibitin and Modulate Extracellular Vesicle Release and miRNAs 21 and 126 in Glioblastoma Multiforme. Int. J. Mol. Sci. 2018;20:103. doi: 10.3390/ijms20010103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosgodage U.S., Matewele P., Mastroianni G., Kraev I., Brotherton D., Awamaria B., Nicholas A.P., Lange S., Inal J.M. Peptidylarginine Deiminase Inhibitors Reduce Bacterial Membrane Vesicle Release and Sensitize Bacteria to Antibiotic Treatment. Front. Cell. Infect. Microbiol. 2019;9:227. doi: 10.3389/fcimb.2019.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inal J.M., Ansa-Addo E.A., Lange S. Interplay of host-pathogen microvesicles and their role in infectious disease. Biochem. Soc. Trans. 2013;41:258–262. doi: 10.1042/BST20120257. [DOI] [PubMed] [Google Scholar]
- 24.Colombo M., Raposo G., Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 25.Turchinovich A., Drapkina O., Tonevitsky A. Transcriptome of Extracellular Vesicles: State-of-the-Art. Front. Immunol. 2019;10:202. doi: 10.3389/fimmu.2019.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vagner T., Chin A., Mariscal J., Bannykh S., Engman D.M., Di Vizio D. Protein Composition Reflects Extracellular Vesicle Heterogeneity. Proteomics. 2019;19:1800167. doi: 10.1002/pmic.201800167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hessvik N.P., Llorente A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018;75:193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramirez S.H., Andrews A.M., Paul D., Pachter J.S. Extracellular vesicles: Mediators and biomarkers of pathology along CNS barriers. Fluids Barriers CNS. 2018;15:19. doi: 10.1186/s12987-018-0104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iliev D., Strandskog G., Nepal A., Aspar A., Olsen R., Jørgensen J., Wolfson D., Ahluwalia B.S., Handzhiyski J., Mironova R. Stimulation of exosome release by extracellular DNA is conserved across multiple cell types. FEBS J. 2018;285:3114–3133. doi: 10.1111/febs.14601. [DOI] [PubMed] [Google Scholar]
- 30.Magnadóttir B., Kraev I., Guðmundsdóttir S., Dodds A.W., Lange S. Extracellular vesicles from cod (Gadus morhua L.) mucus contain innate immune factors and deiminated protein cargo. Dev. Comp. Immunol. 2019;99:103397. doi: 10.1016/j.dci.2019.103397. [DOI] [PubMed] [Google Scholar]
- 31.Criscitiello M.F., Kraev I., Lange S. Deiminated proteins in extracellular vesicles and plasma of nurse shark (Ginglymostoma cirratum)—Novel insights into shark immunity. Fish Shellfish Immunol. 2019;92:249–255. doi: 10.1016/j.fsi.2019.06.012. [DOI] [PubMed] [Google Scholar]
- 32.Lange S., Kraev I., Magnadóttir B., Dodds A.W. Complement component C4-like protein in Atlantic cod (Gadus morhua L.)—Detection in ontogeny and identification of post-translational deimination in serum and extracellular vesicles. Dev. Comp. Immunol. 2019;101:103437. doi: 10.1016/j.dci.2019.103437. [DOI] [PubMed] [Google Scholar]
- 33.Rebl A., Köllner B., Anders E., Wimmers K., Goldammer T. Peptidylarginine deiminase gene is differentially expressed in freshwater and brackish water rainbow trout. Mol. Biol. Rep. 2010;37:2333–2339. doi: 10.1007/s11033-009-9738-5. [DOI] [PubMed] [Google Scholar]
- 34.Gavinho B., Rossi I.V., Evans-Osses I., Lange S., Ramirez M.I. Peptidylarginine deiminase inhibition abolishes the production of large extracellular vesicles from Giardia intestinalis, affecting host-pathogen interactions by hindering adhesion to host cells. bioRxiv. 2019 doi: 10.1101/586438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El-Sayed A.S.A., Shindia A.A., AbouZaid A.A., Yassin A.M., Ali G.S., Sitohy M.Z. Biochemical characterization of peptidylarginine deiminase-like orthologs from thermotolerant Emericella dentata and Aspergillus nidulans. Enzyme Microb. Technol. 2019;124:41–53. doi: 10.1016/j.enzmictec.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Woods C.A., Kilpatrick C.W. Infraorder Hystricognathi. In: Wilson D.E., Reeder D.M., editors. Mammal Species of the World: A Taxonomic and Geographic Reference. 3rd ed. Johns Hopkins University Press; Baltimore, MD, USA: 2005. p. 1542. [Google Scholar]
- 37.O’Riain M.J., Faulkes C.G. African mole rats: Eusociality, relatedness and ecological constraints. In: Korb. J., Heinze. J., editors. Ecology of Social Evolution. Springer; Berlin/Heidelberg, Germany: 2008. pp. 207–223. [Google Scholar]
- 38.Chung D., Dzal Y.A., Seow A., Milsom W.K., Pamenter M.E. Naked mole rats exhibit metabolic but not ventilatory plasticity following chronic sustained hypoxia. Proc. Biol. Sci. 2016;283:20160216. doi: 10.1098/rspb.2016.0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pamenter M.E., Dzal Y.A., Milsom W.K. Adenosine receptors mediate the hypoxic ventilatory response but not the hypoxic metabolic response in the naked mole rat during acute hypoxia. Proc. Biol. Sci. 2015;282:20141722. doi: 10.1098/rspb.2014.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pamenter M.E., Lau G.Y., Richards J.G., Milsom W.K. Naked mole rat brain mitochondria electron transport system flux and H+ leak are reduced during acute hypoxia. J. Exp. Biol. 2018;221:jeb171397. doi: 10.1242/jeb.171397. [DOI] [PubMed] [Google Scholar]
- 41.Park T.J., Reznick J., Peterson B.L., Blass G., Omerbasic D., Bennett N.C., Kuich P.H.J.L., Zasada C., Browe B.M., Hamann W., et al. Fructose-driven glycolysis supports anoxia resistance in the naked mole-rat. Science. 2017;356:305–308. doi: 10.1126/science.aab3896. [DOI] [PubMed] [Google Scholar]
- 42.Hochachka P.W. Defense strategies against hypoxia and hypothermia. Science. 1986;231:234–241. doi: 10.1126/science.2417316. [DOI] [PubMed] [Google Scholar]
- 43.Hochachka P.W., Buck L.T., Doll C.J., Land S.C. Unifying theory of hypoxia tolerance: Molecular/metabolic defense and rescue mechanisms for surviving oxygen lack. Proc. Natl. Acad. Sci. USA. 1996;93:9493–9498. doi: 10.1073/pnas.93.18.9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buck L.T., Pamenter M.E. Adaptive responses of vertebrate neurons to anoxia—Matching supply to demand. Respir. Physiol. Neurobiol. 2006;154:226–240. doi: 10.1016/j.resp.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 45.Buck L.T., Pamenter M.E. The hypoxia-tolerant vertebrate brain: Arresting synaptic activity. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2018;224:61–70. doi: 10.1016/j.cbpb.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 46.Ilacqua A.N., Kirby A.M., Pamenter M.E. Behavioural responses of naked mole rats to acute hypoxia and anoxia. Biol. Lett. 2017;13:20170545. doi: 10.1098/rsbl.2017.0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Houlahan C.R., Kirby A.M., Dzal Y.A., Fairman G.D., Pamenter M.E. Divergent behavioural responses to acute hypoxia between individuals and groups of naked mole rats. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2018;224:38–44. doi: 10.1016/j.cbpb.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 48.Kirby A.M., Fairman G.D., Pamenter M.E. Atypical behavioural, metabolic and thermoregulatory responses to hypoxia in the naked mole rat (Heterocephalus glaber) J. Zool. 2018;305:106–115. doi: 10.1111/jzo.12542. [DOI] [Google Scholar]
- 49.Park T.J., Lu Y., Juttner R., Smith E.S., Hu J., Brand A., Wetzel C., Milenkovic N., Erdmann B., Heppenstall P.A., et al. Selective inflammatory pain insensitivity in the African naked mole-rat (Heterocephalus glaber) PLoS Biol. 2008;6:e13. doi: 10.1371/journal.pbio.0060013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Branigan T., Elkhalifa S., Pamenter M.E. Behavioural responses to environmental hypercapnia in two eusocial species of African mole rats. J. Comp. Physiol. A Neuroethol. Sens. Neural. Behav. Physiol. 2018;204:811–819. doi: 10.1007/s00359-018-1283-z. [DOI] [PubMed] [Google Scholar]
- 51.Seluanov A., Hine C., Azpurua J., Feigenson M., Bozzella M., Mao Z., Catania K.C., Gorbunova V. Hypersensitivity to contact inhibition provides a clue to cancer resistance of naked mole-rat. Proc. Natl. Acad. Sci. USA. 2009;106:19352–19357. doi: 10.1073/pnas.0905252106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian X., Azpurua J., Hine C., Vaidya A., Myakishev-Rempel M., Ablaeva J., Mao Z., Nevo E., Gorbunova V., Seluanov A. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature. 2013;499:346–349. doi: 10.1038/nature12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woodley R., Buffenstein R. Thermogenic changes with chronic cold exposure in the naked mole-rat (Heterocephalus glaber) Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2002;133:827–834. doi: 10.1016/S1095-6433(02)00199-X. [DOI] [PubMed] [Google Scholar]
- 54.Vandewint A.L., Zhu-Pawlowsky A.J., Kirby A., Tattersall G.J., Pamenter M.E. Evaporative cooling and vasodilation mediate thermoregulation in naked mole-rats during normoxia but not hypoxia. J. Therm. Biol. 2019;84:228–235. doi: 10.1016/j.jtherbio.2019.07.011. [DOI] [PubMed] [Google Scholar]
- 55.Lagunas-Rangel F.A., Chávez-Valencia V. Learning of nature: The curious case of the naked mole rat. Mech. Ageing Dev. 2017;164:76–81. doi: 10.1016/j.mad.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 56.Munro D., Baldy C., Pamenter M.E., Treberg J.R. The exceptional longevity of the naked mole-rat may be explained by mitochondrial antioxidant defenses. Aging Cell. 2019;18:e12916. doi: 10.1111/acel.12916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Munro D., Pamenter M.E. Comparative studies of mitochondrial reactive oxygen species in animal longevity: Technical pitfalls and possibilities. Aging Cell. 2019;18:e13009. doi: 10.1111/acel.13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Viltard M., Durand S., Pérez-Lanzón M., Aprahamian F., Lefevre D., Leroy C., Madeo F., Kroemer G., Friedlander G. The metabolomic signature of extreme longevity: Naked mole rats versus mice. Aging. 2019;11:4783–4800. doi: 10.18632/aging.102116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seluanov A., Gladyshev V.N., Vijg J., Gorbunova V. Mechanisms of cancer resistance in long-lived mammals. Nat. Rev. Cancer. 2018;18:433–441. doi: 10.1038/s41568-018-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ruby J.G., Smith M., Buffenstein R. Naked Mole-Rat mortality rates defy gompertzian laws by not increasing with age. Elife. 2018;7:e31157. doi: 10.7554/eLife.31157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stenvinkel P., Shiels P.G. Long-lived animals with negligible senescence: Clues for ageing research. Biochem. Soc. Trans. 2019;47:1157–1164. doi: 10.1042/BST20190105. [DOI] [PubMed] [Google Scholar]
- 62.Kim E.B., Fang X., Fushan A.A., Huang Z., Lobanov A.V., Han L., Marino S.M., Sun X., Turanov A.A., Yang P., et al. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature. 2011;479:223–227. doi: 10.1038/nature10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tan L., Ke Z., Tombline G., Macoretta N., Hayes K., Tian X., Lv R., Ablaeva J., Gilbert M., Bhanu N.V., et al. Naked Mole Rat Cells Have a Stable Epigenome that Resists iPSC Reprogramming. Stem Cell Rep. 2017;9:1721–1734. doi: 10.1016/j.stemcr.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hilton H.G., Rubinstein H.G., Janki N.D., Ireland P., Bernstein A.T., Wright N., Finkle K.M., Martin-McNulty D., Roy M., et al. Single-cell transcriptomics of the naked mole-rat reveals unexpected features of mammalian immunity. bioRxiv. 2019:597195. doi: 10.1101/597195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frankenberg A.D.V., Reis A.F., Gerchman F. Relationships between adiponectin levels, the metabolic syndrome, and type 2 diabetes: A literature review. Arch. Endocrinol. Metab. 2017;61:614–622. doi: 10.1590/2359-3997000000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spracklen C.N., Karaderi T., Yaghootkar H., Schurmann C., Fine R.S., Kutalik Z., Preuss M.H., Lu Y., Wittemans L.B.L., Adair L.S., et al. Exome-Derived Adiponectin-Associated Variants Implicate Obesity and Lipid Biology. Am. J. Hum. Genet. 2019;105:15–28. doi: 10.1016/j.ajhg.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamauchi T., Kamon J., Minokoshi Y., Ito Y., Waki H., Uchida S., Yamashita S., Noda M., Kita S., Ueki K., et al. Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nat. Med. 2002;8:1288–1295. doi: 10.1038/nm788. [DOI] [PubMed] [Google Scholar]
- 68.Yamauchi T., Kamon J., Ito Y., Tsuchida A., Yokomizo T., Kita S., Sugiyama T., Miyagishi M., Hara K., Tsunoda M., et al. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature. 2003;423:762–769. doi: 10.1038/nature01705. [DOI] [PubMed] [Google Scholar]
- 69.Kadowaki T., Yamauchi T. Adiponectin and adiponectin receptors. Endocr. Rev. 2005;26:439–451. doi: 10.1210/er.2005-0005. [DOI] [PubMed] [Google Scholar]
- 70.Chen Y.L., Tao J., Zhao P.J., Tang W., Xu J.P., Zhang K.Q., Zou C.G. Adiponectin receptor PAQR-2 signaling senses low temperature to promote C. elegans longevity by regulating autophagy. Nat. Commun. 2019;10:2602. doi: 10.1038/s41467-019-10475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fiaschi T. Mechanisms of Adiponectin Action. Int. J. Mol. Sci. 2019;20:2894. doi: 10.3390/ijms20122894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fiaschi T., Magherini F., Gamberi T., Modesti P.A., Modesti A. Adiponectin as a tissue regenerating hormone: More than a metabolic function. Cell. Mol. Life Sci. 2014;71:1917–1925. doi: 10.1007/s00018-013-1537-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parida S., Siddharth S., Sharma D. Adiponectin, obesity, and cancer: Clash of the bigwigs in health and disease. Int. J. Mol. Sci. 2019;20:2519. doi: 10.3390/ijms20102519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bavelloni A., Ramazzotti G., Poli A., Piazzi M., Focaccia E., Blalock W., Faenza I. MiRNA-210: A Current Overview. Anticancer Res. 2017;37:6511–6521. doi: 10.21873/anticanres.12107. [DOI] [PubMed] [Google Scholar]
- 75.Tang J., Ning R., Zeng B., Li Y. Molecular Evolution of PTEN Pseudogenes in Mammals. PLoS ONE. 2016;11:e0167851. doi: 10.1371/journal.pone.0167851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Altwasser R., Paz A., Korol A., Manov I., Avivi A., Shams I. The transcriptome landscape of the carcinogenic treatment response in the blind mole rat: Insights into cancer resistance mechanisms. BMC Genom. 2019;20:17. doi: 10.1186/s12864-018-5417-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Musso G., Cassader M., Cohney S., De Michieli F., Pinach S., Saba F., Gambino R. Fatty Liver and Chronic Kidney Disease: Novel Mechanistic Insights and Therapeutic Opportunities. Diabetes Care. 2016;39:1830–1845. doi: 10.2337/dc15-1182. [DOI] [PubMed] [Google Scholar]
- 78.Juźwik C.A., Drake S.S., Zhang Y., Paradis-Isler N., Sylvester A., Amar-Zifkin A., Douglas C., Morquette B., Moore C.S., Fournier A. microRNA dysregulation in neurodegenerative diseases: A systematic review. Prog. Neurobiol. 2019;26:101664. doi: 10.1016/j.pneurobio.2019.101664. [DOI] [PubMed] [Google Scholar]
- 79.Li J., Li K., Chen X. Inflammation-regulatory microRNAs: Valuable targets for intracranial atherosclerosis. J. Neurosci. Res. 2019;97:1242–1252. doi: 10.1002/jnr.24487. [DOI] [PubMed] [Google Scholar]
- 80.Xu H.X., Pan W., Qian J.F., Liu F., Dong H.Q., Liu Q.J. MicroRNA-21 contributes to the puerarin-induced cardioprotection via suppression of apoptosis and oxidative stress in a cell model of ischemia/reperfusion injury. Mol. Med. Rep. 2019;20:719–727. doi: 10.3892/mmr.2019.10266. [DOI] [PubMed] [Google Scholar]
- 81.Xiaoyan W., Pais E.M., Lan L., Jingrui C., Lin M., Fordjour P.A., Guanwei F. MicroRNA-155: A Novel Armamentarium Against Inflammatory Diseases. Inflammation. 2017;40:708–716. doi: 10.1007/s10753-016-0488-y. [DOI] [PubMed] [Google Scholar]
- 82.Chen Z., Li Y., Zhang H., Huang P., Luthra R. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene. 2010;29:4362–4368. doi: 10.1038/onc.2010.193. [DOI] [PubMed] [Google Scholar]
- 83.Voloboueva L.A., Sun X., Xu L., Ouyang Y.-B., Giffard R.G. Distinct effects of miR-210 reduction on neurogenesis: Increased neuronal survival of inflammation but reduced proliferation associated with mitochondrial enhancement. J. Neurosci. 2017;37:3072–3084. doi: 10.1523/JNEUROSCI.1777-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Favaro E., Ramachandran A., McCormick R., Gee H., Blancher C., Crosby M., Devlin C., Blick C., Buffa F., Li J.-L., et al. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS ONE. 2010;5:e10345. doi: 10.1371/journal.pone.0010345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang X., Le Q.-T., Giaccia A.J. MiR-210—Micromanager of the hypoxia pathway. Trends Mol. Med. 2010;16:230–237. doi: 10.1016/j.molmed.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pamenter M.E., Dzal Y.A., Thompson W.A., Milsom W.K. Do naked mole rats accumulate a metabolic acidosis or an oxygen debt in severe hypoxia? J. Exp. Biol. 2019;222:jeb191197. doi: 10.1242/jeb.191197. [DOI] [PubMed] [Google Scholar]
- 87.Holmes M.M., Goldman B.D., Goldman S.L., Seney M.L., Forger N.G. Neuroendocrinology and sexual differentiation in eusocial mammals. Front. Neuroendocrinol. 2009;30:519–533. doi: 10.1016/j.yfrne.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Théry C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G.K., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nicholas A.P., Whitaker J.N. Preparation of a monoclonal antibody to citrullinated epitopes: Its characterization and some applications to immunohistochemistry in human brain. Glia. 2002;37:328–336. doi: 10.1002/glia.10039. [DOI] [PubMed] [Google Scholar]
- 90.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 −ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.