Abstract
Genome-wide screening using CRISPR coupled with nuclease Cas9 (CRISPR/Cas9) is a powerful technology for the systematic evaluation of gene function. Statistically principled analysis is needed for the accurate identification of gene hits and associated pathways. Here, we describe how to perform computational analysis of CRISPR screens using the MAGeCKFlute pipeline. MAGeCKFlute combines the MAGeCK and MAGeCK-VISPR algorithms and incorporates additional downstream analysis functionalities. MAGeCKFlute is distinguished from other currently available tools by being a comprehensive pipeline that contains a series of functions for analyzing CRISPR screen data. This protocol explains how to use MAGeCKFlute to perform quality control, normalization, batch effect removal, copy number bias correction, gene hit identification, and downstream functional enrichment analysis for CRISPR screens. We also describe gene identification and data analysis in CRISPR screens involving drug treatment. Completing the entire MAGeCKFlute pipeline requires approximately two hours on a desktop computer running Linux or Mac OS and with R support. The MAGeCKFlute package is available at http://www.bioconductor.org/packages/release/bioc/html/MAGeCKFlute.html.
Introduction
CRISPR(Clustered Regularly Interspaced Short Palindromic Repeats)/Cas9 is a powerful technology to target desired genomic sites for gene editing or activity modulation via specific single-guide RNAs (sgRNAs)1. CRISPR screening is a high-throughput technology to investigate the functions of many genes in a single experiment. In the screening experiment, sgRNAs are designed, synthesized and cloned into a lentivirus library, which is subsequently transduced into cells at a low multiplicity of infection (MOI) to ensure only one sgRNA copy is integrated per cell. A sgRNA usually contains 18–20 nucleotides complementary to its target and guides the Cas9 enzyme to a specific DNA location where Cas9 induces a double-strand break. The repair of such a break by the cell often leads to a knockout of the targeted gene. Cells are cultured under different experimental settings, and the sgRNAs incorporated into the host genome are replicated with the host cell division.
Genome-wide CRISPR screens2,3 allow for a systematic investigation of gene functions in various contexts4. The screening procedure can be categorized into knockout screens5,6,7 and CRISPR activation or inhibition screens (CRISPRa/CRISPRi), which are performed by fusing a catalytically inactive Cas9 (dCas9) to transcriptional activation or repression domains, respectively. Data analysis for each type of CRISPR screen is similar in principle. For simplicity, within this protocol we will refer to CRISPR knockout and CRISPR activation/inhibition screens as “CRISPR screens”, and use CRISPR knockout screens as an example to demonstrate data analysis. CRISPR screens have been highly effective at identifying genes that function in tumorigenesis8,9, metastasis10, response to immunotherapy11,12, and genes associated with drug response13,14,15.
To identify essential genes in a cell population, cells with CRISPR perturbation can be harvested in two conditions, one representing the initial sgRNA status (Day 0), and the other allowed to proliferate under certain experimental conditions for a set amount of time. To study gene-drug interactions, CRISPR screens can be conducted using three different cell populations: the day 0 population, a drug-treated population (treatment) and a control population (mock-drug control, typically treated with vehicle such as DMSO). At the end of the screen, genomic DNA from the transduced cells is extracted and the sgRNA-encoded regions where the virus had integrated into the host genome are sequenced using high-throughput sequencing. The read count of each sgRNA is a proxy for the proliferation characteristics of the cell with that specific knockout.
For many research groups, data analysis is the most challenging aspect of CRISPR screens. The primary goal of data analysis is to identify genes whose disruption leads to phenotype change (e.g., cell growth) under certain screening conditions, relative to a predefined control condition (e.g., before screening starts or cells without drug treatment). A secondary goal is to infer biological insights from those hits using functional analysis approaches, including Gene Ontology (GO), pathway enrichment analysis or Gene Set Enrichment Analysis (GSEA)16,17. We have previously developed two algorithms18,19 to analyse CRISPR screen data: MAGeCK (Model-based Analysis of Genome-wide CRISPR-Cas9 Knockout)18 and MAGeCK-VISPR (Visualization for CRISPR)19. Both algorithms use a negative binomial distribution to model variances of sgRNA read counts. MAGeCK RRA and MAGeCK MLE are the two main functions of MAGeCK which can be used for identifying CRISPR screen hits. MAGeCK RRA uses Robust Rank Aggregation (RRA) and MAGeCK MLE utilizes a maximum-likelihood estimation (MLE) for robust identification of CRISPR-screen hits (see further discussion in Experimental Design). MAGeCK-VISPR is a comprehensive quality control, analysis and visualization workflow for CRISPR/Cas9 screens. It incorporates MAGeCK and VISPR which together interactively explore results and quality control in a web-based frontend. In combination, MAGeCK and MAGeCK-VISPR allow users to perform read count mapping, normalization, quality control, and to identify positively and negatively selected genes in the screens.
Overview of the Protocol
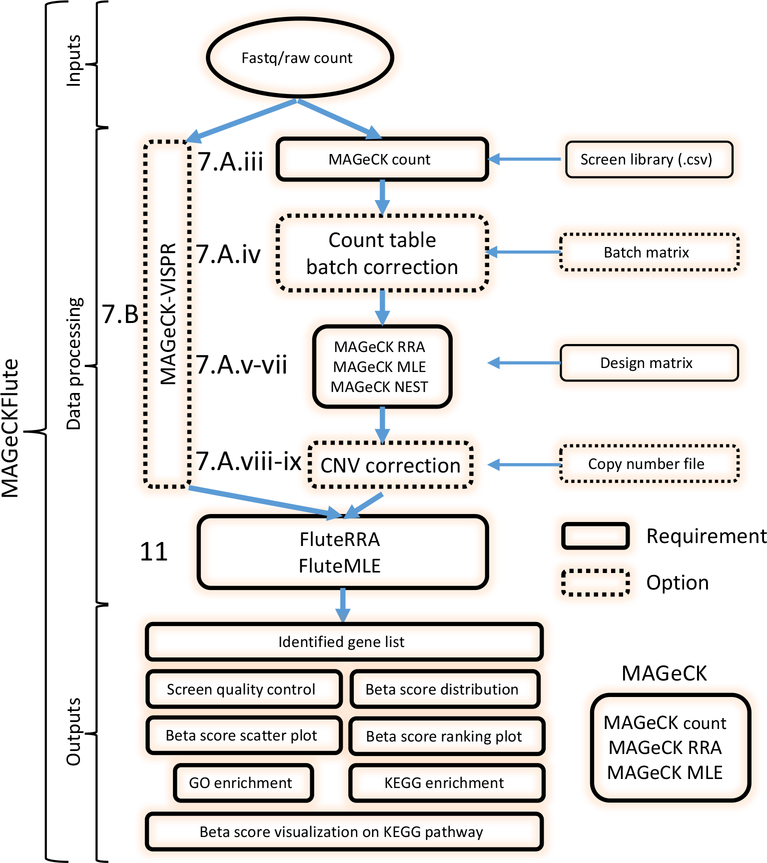
Here, we describe how to use MAGeCKFlute (Figure 1), a comprehensive CRISPR screen analysis pipeline that applies either MAGeCK or MAGeCK-VISPR to identify gene hits and then performs downstream functional analyses using FluteRRA or FluteMLE. MAGeCKFlute has functions to perform batch-effect removal, normalization, and copy-number correction. We chose the name MAGeCKFlute to invoke a pipeline, and as a metaphorical reference to the successful completion of a series of tests in Mozart’s popular opera of the same name. We give users the option of performing a step-by-step analysis of screen data with MAGeCK (Steps 7Ai-ix), or a comprehensive workflow with MAGeCK-VISPR (Steps 7Bi-vii) which also includes visualisation of the results. Within MAGeCK, we demonstrate how to identify gene hits with MAGeCK RRA (Steps 7Av) using publicly available data from a CRISPR screen performed in Glioblastoma stem-like (GBM) cells20. We also show how to use MAGeCK MLE (Steps 7Avi-vii) to identify gene hits from a screen with multiple conditions using an A375 dataset which is a breast melanoma cancer cell line, A375, treated with the BRAF protein kinase inhibitor vemurafenib (PLX)7. We also demonstrate how to remove batch effects using a CRISPR screen dataset which was generated in different batches20 (Step 7Aiv). In the final steps of the analysis, we show how to use FluteMLE to perform quality control at the beta score level for MAGeCK MLE results and how to use FluteRRA or FluteMLE to perform downstream GO term and Kyoto Encyclopedia of Genes and Genomes (KEGG)21 pathway enrichment analyses for MAGeCK RRA and MLE results (Step 11). With our example, we describe how to identify genes involved in drug pathways by comparing CRISPR screen results from different drug treatments.
Figure 1. A schematic representation of CRISPR/Cas9 screen analysis using MAGeCKFlute.
Procedure step numbers described in the main text are shown to the left of the corresponding box. The FASTQ files or raw read count files (Table 4), a screen library file (Table 2), and a Design matrix (Table 5) are required as input for initial analysis through both MAGeCK and MAGeCK-VISPR. The following input components are optional: count table batch correction (which requires an otherwise optional batch matrix file) and CNV analysis and correction. Users have the option of analysing CRISPR screen data step-by-step with the individual MAGeCK modules (Option A, right branch) or with MAGeCK-VISPR, which combines all MAGeCK modules and additional quality control and visualization functions in a single script (Option B, left branch). FluteRRA and FluteMLE use the results generated with MAGeCK or MAGeCK-VISPR for downstream analyses, including pathway enrichment using GO and KEGG. Outputs of FluteRRA or FluteMLE include the beta score distribution and beta-score scatter plots.
Comparison with other methods
Other algorithms such as RIGER22, RSA23, BAGEL24, ScreenBEAM25, and casTLE26 also perform parts of the CRISPR screen analysis pipeline. RIGER22 and RSA23 examine the rank distribution of all sgRNAs targeting a gene with regard to selection during the screen and calculate the statistical significance of all sgRNAs targeting a gene BAGEL is based on a Bayesian supervised learning method, ScreenBEAM uses Bayesian hierarchical model to assess gene-level activity from all relevant measurements, and casTLE combines measurements from multiple targeting reagents to estimate a maximum effect size. MAGeCKFlute differs from these algorithms because it uses a negative binomial model to address off-target sgRNAs and provides a comprehensive CRISPR screen workflow. The workflow is based on MAGeCK and MAGeCK-VISPR and includes read mapping, normalization, quality control, hit identification, and functional analysis. This protocol aims to enable wet-lab and computational investigators to analyse their CRISPR screen data, and to aid in understanding the biology behind the screen results.
Applications
MAGeCKFlute can be applied to remove batch effect, correct copy number bias and identify screening hits and perform downstream functional analysis for various CRISPR screens, such as CRISPR knockout, CRISPR activation, and CRISPR inhibition screens. MAGeCKFlute can map raw reads onto a CRISPR library, normalize read counts to allow comparison between different samples, identify genes that are positively or negatively selected under the screening conditions, and explore enriched GO terms and KEGG pathways for those selected genes. For CRISPR screening samples that have been treated with a drug, MAGeCKFlute can also be used to identify drug-associated genes. MAGeCKFlute generates figures and tables representing the results of CRISPR screen data analysis and can be paused at any step.
Limitations
Although the current protocol provides an integrated solution for analyzing pooled CRISPR screens, there are still limitations. For example, there are many different approaches, such as GOstats27, clusterProfiler28 and GESA17, to perform functional enrichment analysis, and currently it is unclear which model is most appropriate for analyzing screening results. MAGeCKFlute provides all three options mentioned above for enrichment analysis, and users are encouraged to test different models. Another limitation involves the quality of the screens. Certain samples may lose more than 50% of the cell population perhaps due to a long culture time or high-dose drug treatment that leads to strong selection. Such samples should be avoided, as they will influence the accurate identification of negatively selected hits. Alternatively, users may reduce the screening period to get higher quality screening data. Despite these limitations, our protocol provides a convenient approach to perform a comprehensive computational analysis for CRISPR screens.
Experimental Design
The basics of CRISPR screen data analysis with MAGeCK and MAGeCK-VISPR
MAGeCKFlute performs reads mapping and hit identification using mageck count and mageck test/mle, respectively, which are the main functions of MAGeCK and MAGeCK-VISPR (Table 1). The typical input of MAGeCKFlute is a FASTQ file or a raw-read-count table where columns are samples and rows are sgRNAs. CRISPR screen analysis usually contains two parts: sgRNA-level and gene-level analysis. The sgRNA-level analysis models read counts of individual sgRNAs independently. The fold change and p-value are calculated for each sgRNA, which is similar to RNA-Seq analysis. The gene-level analysis integrates the sgRNA-level fold change and p-values to identify interesting gene hits. MAGeCK first maps sequencing reads to the sgRNA design library29 and normalizes sgRNA read counts to adjust for sequencing depth.
Table 1 |.
Main functions of MAGeCKFlute
| Functions | Description of functions |
|---|---|
| mageck count | Map the raw FASTQ data to reference library file and count the reads for each sgRNA |
| mageck test | MAGeCK RRA (identifying CRISPR screen hits by calculating the RRA enrichment score to indicate the essentiality of a gene) |
| mageck mle | MAGeCK MLE (identifying CRISPR screen hits by calculating a ‘beta score’ for each targeted gene to measure the degree of selection after the target is perturbed) |
| VISPR | Visualisation of the result of MAGeCK. |
| mageck-vispr | Quality control at the FASTQ and raw count level. Includes all the functions of MAGeCK and VISPR |
| BatchRemove | Remove batch effect of CRISPR screen data at the raw read count level |
| mageck test/mle --cnv-norm parameter | Correct the bias caused by copy number when identifying hits with MAGeCK RRA and MAGeCK MLE. |
| mageck_nest.py | Improve hit identification and remove outlier sgRNAs |
| FluteRRA | Downstream analysis of the MAGeCK RRA results |
| FluteMLE | Quality control at the beta score level; normalization with essential genes; identify drug treatment related hits; functional analysis |
Quality control and read count table generation
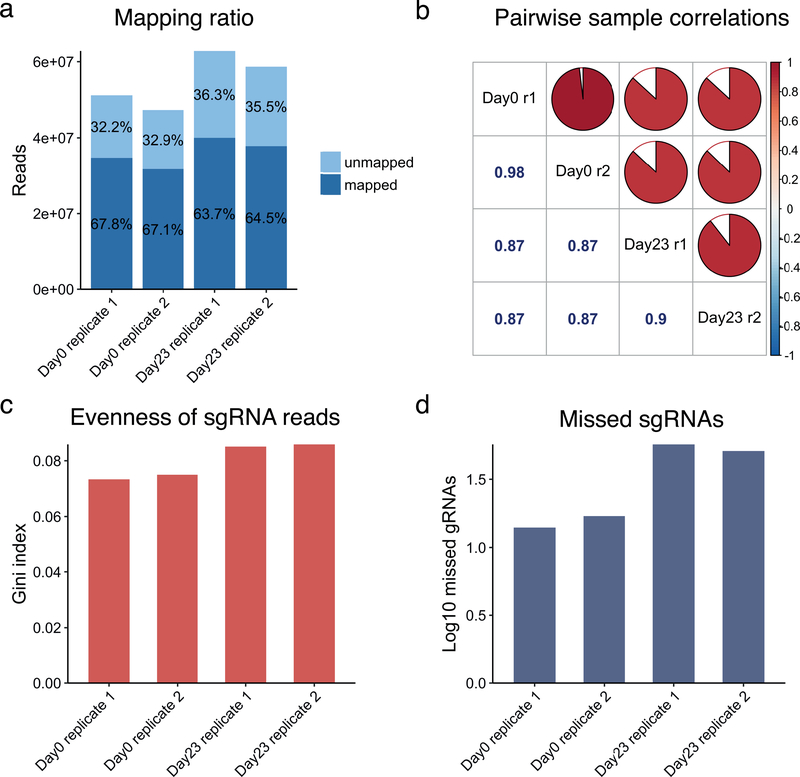
Aligning reads to a known sgRNA library and evaluating screen quality (Figure 2) are required before the identification of hits. MAGeCK and MAGeCK-VISPR align reads to a sgRNA library file, count the read number for each sgRNA, and output a set of quality control statistics, including:
The numbers of mapped reads (Figure 2a)
The percentage of reads mapped (Figure 2a)
The beta score correlation among samples (Figure 2b)
Gini index30 (measures the evenness of sgRNA read counts) (Figure 2c)
The number of sgRNAs to which zero reads mapped (Figure 2d)
Figure 2. Example Quality control assessment of CRISPR/Cas9 screen data.
All four samples analysed here are from a genome-wide CRISPR screen dataset generated from patient-derived Glioblastoma GBM stem-like cells (GSCs)20. These samples represent two conditions: Day0, initial time point of screen and Day23, after 23 days of culture. Replicate 1 and Replicate 2 are biological replicates. These results are generated by performing MAGeCK count. (a) Read counts and mapping percentages. The mapped read percentage should be greater than 65%. (b) pairwise sample correlations of read count, (c) Gini index, which measures read depth evenness within samples. (d) Number of missed sgRNAs.
A low percentage of mapped reads may indicate errors from oligonucleotide synthesis, sequencing errors, or contaminated samples. A high mapping rate suggests success in sample preparation and sequencing. A low number of missing sgRNAs is also a good indicator of high-quality samples. MAGeCK and MAGeCK-VISPR use the Gini index, a common measure of income inequality in economics30 to measure the evenness of sgRNA read counts. A high Gini index suggests that the sgRNA read count is distributed very heterogeneously across the target genes. This is potentially caused by unevenness in CRISPR oligonucleotide synthesis, low quality viral library packaging, poor efficiency in viral transfection, or over-selection during the screens.
Batch effect removal
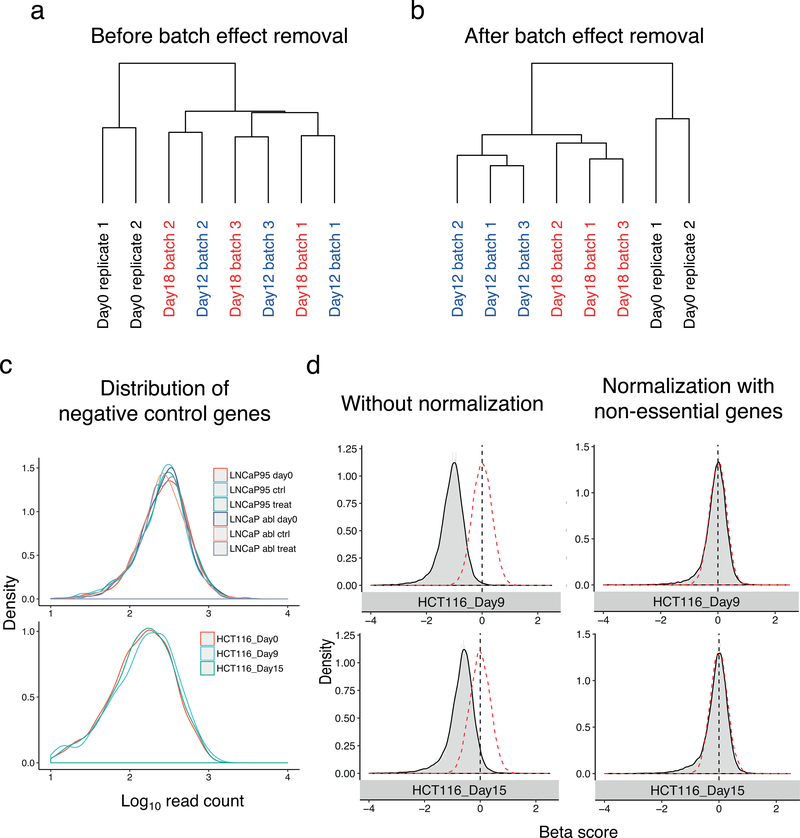
CRISPR screens that are performed or sequenced in multiple batches may harbor batch effects. If CRISPR screen data was generated with different reagents, sequencing platforms, at different times, or any other unintended variations in experimental conditions, batch effects could be observed in this data. In such cases batch effect removal becomes a necessary step for data analysis. One example is a public CRISPR screen in colon cancer which has strong batch effects20, where samples are clustered by batches instead of by conditions (Figure 3a). After correcting for batch effect in the dataset at the sgRNA count level using the ComBat31 function (incorporated into MAGeCKFlute), the biological replicates are properly clustered together (Figure 3b). This indicates that the batch effect has been removed.
Figure 3. Batch effect correction and normalization of read counts and beta scores from CRISPR screen data.
The data analysed here is a genome-wide CRISPR screen using HCT116 colorectal carcinoma cells8 harvested at several time points. Day 0, Day 12 and Day 18 were selected to demonstrate batch effect. Another two time points, Day 9 and Day 15, were selected to demonstrate negative normalization with non-essential genes. (a) Before and (b) after batch-effect correction of HCT116 CRIRSPR screen data using ComBat44. The sgRNAs from several time points (Day 0, Day 12, and Day 18) were harvested and sequenced. Each time point contains more than 1 replicate, but the replicates were generated independently. Different batches are shown in different colours, and replicates are marked by number 1, 2, 3. (c) Density plot of read counts from gRNAs corresponding to negative control genes AAVS1, CCR5, and ROSA26 (top) and to non–essential genes (bottom). Data shown here is from the HCT116 genome-wide CRISPR screen and a LNCap dataset (Supplementary Data 4) which is a genome-wide CRISPR screen data and includes 2 cell lines, LNCap95 and LNCap abl. (d) Beta score distribution of HCT116 CRISPR screen samples before (left) and after (right) normalization using non-essential genes. Samples were harvested at two data points (Day 9 and Day 15). The red dashed line represents a normal distribution with a mean of zero and the same standard deviation as the original beta score. Black dashed line indicates the mean of the simulated normal distribution. After a correct normalization, the mean of beta score should be close to zero.
Screen Hit Identification
The first method used to identify gene hits is MAGeCK RRA. MAGeCK RRA allows to compare two experimental conditions. It can identify sgRNAs and corresponding genes that are significantly selected between the two conditions. MAGeCK RRA ranks sgRNAs based on their p-values calculated from the negative-binomial model and uses a modified RRA algorithm named α-RRA to identify positively or negatively selected genes. MAGeCK RRA uses the RRA enrichment score to indicate the essentiality of a gene.
An alternative method that can model complex experimental designs is MAGeCK MLE, which can be used to analyse data from screens with multiple conditions, such as a typical drug screen which includes at least 3 conditions: a day 0 condition, a control condition (treated with vehicle such as DMSO), and a drug-treated condition. MAGeCK MLE also models the sgRNA knockout efficiency, which may vary depending on different sequence contents and chromatin structures. MAGeCK MLE calculates a ‘beta score’ for each targeted gene to measure the degree of selection upon genes perturbation similar to the “log fold-change” measurement in differential expression analysis.
MAGeCK-VISPR further incorporates all functions of MAGeCK and performs quality control and visualization of all results using VISPR, a web-based interactive framework. MAGeCK NEST32 adds additional features to MAGeCK-VISPR to improve hit calling. First, MAGeCK NEST can improve results using the Network Essentiality Scoring Tool33 (NEST) to integrate information from protein-protein interaction networks. Second, MAGeCK NEST adopts a maximum-likelihood approach to remove sgRNA outliers, which often have higher G-nucleotide counts. Users may consider using MAGeCK NEST to improve hit calling if there are many sgRNA outliers or if a high Gini30 index is observed in the screen data.
Read count normalization with negative control sequences or non-essential genes
It is often desirable to compare read counts between different conditions in a single experiment. For the purpose of normalization between different growth conditions, an ideal standard would be sgRNAs which target a completely inert genomic location in all of the starting cell populations, such that cell proliferation was not differentially affected under any of the experimental conditions. AAVS1 is a well-validated locus to host an exogenous gene sequence. It has an open chromatin structure and is transcription-competent. Most importantly, there are no known adverse effects on the cell resulting from the insertion or deletion of the AAVS1 locus34,35. sgRNAs targeting AAVS1 have similar behavior across samples (Figure 3c), suggesting AAVS1-targeting sgRNAs may be appropriate controls for read-count normalization. Using AAVS1-targeting sgRNAs as controls also mitigates the nuclease-induced toxicity of Cas9 and reduces the overall false positive rate.
Similar to sgRNAs targeting the AAVS1 locus, sgRNAs targeting non-essential genes can also be used to normalize read counts. We compiled a list of non-essential genes (Supplementary Data 1) for normalization of CRISPR screens, if AAVS1-targeting sgRNAs are not available. Starting from 927 non-essential genes whose knock-down has no significant effect in multiple CRISPR screens8, we removed genes that are expressed at low levels in multiple cell lines. We selected genes whose expression ranked in the 5th to 100th percentile (Supplementary Figure 1a) in 98.3% (1019 out of 1036) Cancer Cell Line Encyclopedia (CCLE)36 cell lines (Supplementary Figure 1b). 350 out of 937 non-essential genes passed these criteria (Supplementary Figure 1). The expression distributions of these 350 genes are consistent across hundreds of cancer cell lines (Supplementary Data 1). This suggests that sgRNAs targeting these genes are appropriate controls for normalization of CRISPR screens, if AAVS1-targeting sgRNAs are not available (Figure 3d). MAGeCKFlute also supports read normalization using sgRNAs from a list of non-essential genes, and we suggest including at least 200 non-essential genes in the library to ensure efficient normalization.
Copy number bias correction
The process of inducing double-strand breaks at targeted genomic sites in CRISPR screens triggers DNA damage response mechanisms and may cause cell-cycle arrest, especially for cells with high copy number regions targeted37. When an amplified region contains a targeted non-essential gene, the observed beta scores often appear more negative than expected (Supplementary Figure 3a). A negative beta score indicates that knock out of this gene may inhibit cell proliferation or cause cell death. Therefore, false positives are introduced in essential gene identification. There is an optional method (Step 7Aviii) presented in this protocol for correcting this copy-number related bias if the corresponding copy number file is provided by users (The corresponding copy number file for the example data is provided as Supplementary Data 3). In this method, the relationship between genomic copy number and observed essentiality is quantitatively modelled for each gene in each experiment. The copy number bias is then adjusted from the observed outcomes, generating corrected beta scores for all affected genes (Supplementary Figure 3b). This function had been incorporated to MAGeCKFlute pipeline and can be applied when performing MAGeCK RRA or MLE.
Beta score normalization with essential genes
Cells exposed to different conditions (with or without drug treatment) may have different proliferation rates. For example, CDK4/6 inhibitors affect the cell cycle and generally reduce cell proliferation38. Therefore, comparing cells that have a faster doubling time to more slowly proliferating cells may lead to biases in hit identification, since genes will appear to have a stronger selection in cell populations that are proliferating more rapidly. This is often the case when comparing samples with and without drug treatment, since many drugs affect cell proliferation. The ‘beta score’ for each gene indicates the type of selection a gene is undergoing: a positive beta score indicates positive selection and a negative beta score indicates negative selection. When different samples are cultured for the same time in CRISPR screens, those with shorter doubling time will have more cell cycles of selection, thus genes in faster-growing cells tend to give higher absolute beta scores (Supplementary Figure 2a). To overcome this bias, we generated a list of 625 refined, high-confidence core essential genes (Supplementary Data 2) to normalize the beta score (for details see Supplementary Method). MAGeCKFlute performs normalization of gene beta scores using the list of core essential genes (Step 11B), assuming they are equally negatively selected between two samples, even if the two samples have a different baseline proliferation rate. The beta scores of all genes are normalized based on the median beta score of this refined set of 625 essential genes. After normalization, the slope and X-intercept of the regression line of two samples are close to 1 and 0 respectively, which indicates that the normalization with essential genes in genome-wide screens makes the beta scores comparable across samples (Supplementary Figure 2b). For CRISPR screens with a certain treatment, we recommend users to conduct normalization with essential genes to make the beta score comparable between treatment and control samples.
Differential hit identification upon cell treatment
After beta score normalization with essential genes, the next step is to identify differential hits between treatment and control conditions, by subtracting their beta scores. This differential beta score is used to identify treatment-related screen hits. The cutoff can be specified in the FluteMLE function, with the default at 1 standard deviation from the differential beta score mean. We adopted a “quantile matching” approach to robustly estimate σ which is the standard deviation of the beta score β σ is chosen such that the (1-p) empirical quantile of the absolute values of β matches the (1-p/2) theoretical quantile of the prior normal distribution N (0, σ2. p stands for the quantile of the beta score β. p is set as 0.32 for 1 standard deviation and 0.05 for 2 standard deviations, which corresponds to 68% and 95% of the beta score falling within 1 and 2 standard deviations away of the mean, respectively. If we write the theoretical upper quantile of a normal distribution as QN(1 – p) and the empirical upper quantile of β as Q|β|(1 – p), then σ is calculated as:
Functional analysis of screen hits
The functional analysis of screen hits provides information about the biology of the cell system that was queried in the design of the screen. Several types of functional analyses have been widely used, including GO enrichment analysis27,39 and GSEA analysis17. As expected, in simple proliferation screens core components of housekeeping pathways (e.g., ribosome and spliceosome) are typically negatively selected5–7, and components of pathways predicted to be cell-type specific have been found to be essential in the predicted cell types40,41.
MAGeCKFlute incorporates several functional modules to explore the biological functions of screen hits. We included published enrichment functions derived from the clusterProfiler28, GOstats27 and GSEA packages, and added enrich.HGT to test the enrichment of molecular signatures based on the hypergeometric distribution. These functions allow users to specify the size of genes annotated by GO terms, KEGG pathways, MSigDB gene set collections, or user-defined gene sets, and test for their statistical overrepresentation in the screen hits. Sometimes users might be interested in the strong selection of protein complexes or pathways with small number of genes, so limiting the gene set size would allow such enrichment to be detected rather than being overwhelmed by weak selection of big pathways.
Materials
EQUIPMENT
Software
MAGeCK v0.5.6 or newer (https://mageck.sourceforge.net/)
MAGeCK-VISPR v0.5.3 or newer (https://bitbucket.org/liulab/mageck-vispr)
Conda 4.5.4 or newer (https://conda.io/docs/)
Python v3.4.3 or newer (https://www.python.org)
R v3.5.0 or newer (https://www.r-project.org) or RStudio (https://www.rstudio.com/)
sva package v3.7 or newer (http://bioconductor.org/packages/release/bioc/html/sva.html)
Hardware
A 64-bit computer running either Linux or Mac OS X; 4 GB of RAM (16 GB preferred)
Data
We selected five CRISPR screen datasets as example data sets, which are accessible at http://cistrome.org/MAGeCKFlute/.
A CRISPR screen dataset generated from patient-derived Glioblastoma (GBM) stem-like cells (GSCs; GEO accession number: GSE70038)20 (Dataset 1) In this screen, cells were harvested at two time points: Day 0 (initial time point of the screen) and Day23 (after 23 days of culture) Replicate 1 and Replicate 2 are biological replicates. This dataset is in FASTQ format. This data is used to demonstrate how to analyse screen data using MAGeCK RRA.
The second dataset is a CRISPR screen in a breast melanoma cancer cell line, A375, treated with the BRAF protein kinase inhibitor vemurafenib (PLX)7(Dataset 2). In this case, the cells were harvested at two time-points, 7 days and 14 days after treatment and were compared to a control (DMSO-treated) condition. The A375 dataset provides a raw read count table for each sgRNA. This data contains three conditions (Day0, DMSO, drug treatment) and is used to demonstrate how to analyse screen with more than two conditions using MAGeCK MLE.
HCT116 dataset – a genome-wide CRISPR screen using HCT116 colorectal carcinoma cells8. The sgRNAs from several time points were harvested and sequenced. This dataset was generated in different batches, and read count table is included in the demo data. We use this to demonstrate how to perform batch effect removal.
HL60 dataset – a genome-wide CRISPR screen using the acute myelocytic leukemia HL60 cell line 42 with copy number variation information. Cells were harvested at two time points: HL60_initial (initial time point of the screen) and HL60_final (after 12 doubling time). We use this screen to demonstrate copy number bias correction.
LNCap dataset (Supplementary Data 4) – a genome-wide CRISPR screen data with 2 cell lines, LNCap95 and LNCap abl43. Both of them contain 3 conditions (day0, DMSO, drug treatment) and include AAVS1-targeting sgRNAs as negative controls, so are used to demonstrate the normalization with AAVS1-targeting sgRNAs.
EQUIPMENT SETUP
Software setup
Most of the commands given in the protocol run in a typical Linux or Mac shell prompt, and all commands should be run in the same directory as the data files. The protocol also includes R scripts. Commands meant to be executed from the Linux or Mac shell (for example, bash or csh) are prefixed with a ‘$’ character. Commands meant to be run from either an R script or at the R interactive shell are prefixed with a ‘>‘ character.
Procedure
Install MAGeCKFlute Timing ~30 min
-
1.
Start an R session with a terminal or an integrated development environment, such as Rstudio:
$R
Install MAGeCKFlute, from either Liu lab using option A, or Bioconductor using Option B:
-
A.
Install MAGeCKFlute from Liu lab
-
(i)
Install MAGeCKFlute using the following commands:
>install.packages(“devtools”) >library(‘devtools’) >install_bitbucket(“liulab/MAGeCKFlute”)
-
B.
Install MAGeCKFlute from Bioconductor
-
(i)
Install MAGeCKFlute using the following commands:
>source(‘http://www.bioconductor.org/biocLite.R’) >biocLite(‘MAGeCKFlute’)
-
2.
Test whether the MAGeCKFlute package was installed successfully, using the following command:
>library(‘MAGeCKFlute’)
If no error occurs in the loading of the package, it means MAGeCKFlute was installed successfully.
Download and install MAGeCK and MAGeCK-VISPR Timing 20 min
-
3.MAGeCK and MAGeCK-VISPR can be installed with either conda (option A) or source code (option B).
-
A.Install MAGeCK and MAGeCK-VISPR with conda
-
(i)To install MAGeCK and MAGeCK-VISPR, installation of the Python variant included in the Miniconda Python distribution is required. Download Miniconda (http://conda.pydata.org/miniconda.html), and locate the download directory, then install Miniconda by executing the following command in a terminal:
-
A.
$bash path/to/file/Miniconda3-latest-Linux-x86_64.sh
Where ‘path/to/file’ is the directory containing the Miniconda installation file.
When the question below appears, answer “yes”:
Do you wish the installer to prepend the Miniconda3 install location to PATH …? [yes|no]
CAUTION: Python 2 is incompatible with MAGeCK and MAGeCK-VISPR.
-
(ii)
Afterwards, add a bioconda channel using the following command:
$conda config --add channels conda-forge
$conda config --add channels bioconda
CRITICAL STEP: Addition of this channel is essential as MAGeCK and MAGeCK-VISPR depend on it. It is important to add conda-forge and bioconda in the order specified above to ensure that bioconda is the highest priority. This allows setup to be run properly.
-
(iii)
Then, create an isolated software environment for MAGeCK-VISPR by executing the following in a terminal:
$conda create -n mageck-vispr mageck mageck-vispr python=3
-
(iv)
Activate the environment via:
$source activate mageck-vispr
-
B.
Install MAGeCK and MAGeCK-VISPR with source code
-
(i)
MAGeCK (version 0.3 and later) supports a standard Python installation procedure, with compiling and installation of the software. First, download the source code from the website (https://sourceforge.net/p/mageck/wiki/Home/) and locate the download location. Then unzip the files and go into the unzipped directory with the following commands:
$tar xvzf mageck-0.5.6.tar.gz $cd mageck-0.5.6
-
(ii)
Invoke python setup.py using the following command:
$python3 setup.py install
-
(iii)
MAGeCK-VISPR can be installed with source code. First, download the source code and go into the source code directory using the fowling command:
$git clone git@bitbucket.org:liulab/mageck-vispr.git $cd mageck-vispr
-
(iv)
Invoke python setup.py using the following command:
$python3 setup.py install
Install MAGeCK NEST (Optional) Timing 5 mins
-
4.
Download the source code from https://bitbucket.org/liulab/mageck_nest. After the “mageck_nest-3.0.tar.gz” file is downloaded, unzip the source code file by typing:
$cd path/to/file/ $tar -zxvf mageck_nest-3.0.tar.gz
The path/to/file/ is the path for the “mageck_nest-3.0.tar.gz” file.
-
5.
Then change the work directory to ‘mageck_nest-3.0’ as follows:
$cd mageck_nest-3.0
-
6.
Finally, install MAGeCK NEST using the following command:
$python3 setup.py install
Process CRISPR screen data with MAGeCK or MAGeCK-VISPR
-
7.CRISPR screen data can be processed with MAGeCK (option A) or using MAGeCK-VISPR (option B). In a typical use case, CRISPR screen data is processed with MAGeCK (option A) step-by-step. If users want to perform quality control and visualize the results, we recommend MAGeCK-VISPR (option B) instead. To illustrate the procedure, we use the two test CRISPR screen datasets described in the EQUIPMENT section.
-
A.Process CRISPR screen data step-by-step with MAGeCK. TIMING: 1.5 h
-
(i)Download and unzip the test data for both datasets using the following commands:
-
A.
$ wget http://cistrome.org/MAGeCKFlute/demo.tar.gz $ tar zxvf demo.tar.gz $ cd demo_data
-
(ii)
Generate a count table for Dataset 1 with the mageck count function, by firstly changing the working directory to a directory that contains raw fastq data and is able to store the output of mageck count as follows:
$cd path/to/demo_data/mageck_count
CRITICAL STEP The command mageck count aligns reads onto a sgRNA library and generates a read count table. The count table can be used directly in the downstream analysis. The command requires a known sgRNA library file (library.csv; included in the Dataset 1), in which the columns 1–3 are sgRNA names, sequences, and target genes respectively. The library file is either in .txt or in .csv format. For an example sgRNA library file, see Table 2.
Table 2 |.
Example of an sgRNA library file
| ID | sgRNA | Gene_symbol |
|---|---|---|
| s_1 | ACCTGTAGTTGCCGGCGTGC | A1BG |
| s_10 | CCCACAGACGCCTCAGTCTC | A2M |
| s_100 | CCGTGAGCAGGCAGTTCCGC | AATK |
-
(iii)
Run the mageck count on Dataset 1, type the following command with:
$mageck count -l library.csv -n GSC_0131 --sample-label day0_r1,day0_r2,day23_r1,day23_r2 --fastq GSC_0131_Day0_Rep1.fastq.gz GSC_0131_Day0_Rep2.fastq.gz GSC_0131_Day23_Rep1.fastq.gz GSC_0131_Day23_Rep2.fastq.gz
The meanings of the parameters in this command are as follows (see Box 1 for further parameters that could be used or see all the parameters by typing the command mageck count -h):
| -l | The provided sgRNA library file, including the sgRNA id, the sequence, and the gene it is targeting (Table 2). |
| -n | The prefix of the output files. |
| --sample-label | The sample labels, separated by a comma (,). Must be equal to the number of samples provided (in --fastq option). Default “sample1, sample2, …”. |
| --fastq | The sample fastq files (or fastq.gz files), separated by a space; use a comma (,) to indicate technical replicates of the same sample. |
Box 1. Additional mageck count parameters.
Several additional key parameters that may be used to run MAGeCK in step 7A(iii) are as follows:
| --trim-5 | Length of trimming the 5’ of the reads, default AUTO (MAGeCK will automatically determine the trimming length). |
| --sgrna-len | Length of the sgRNA. Default AUTO (MAGeCK will automatically determine the sgRNA length. Only use this if users turn on the “--unmapped-to-file” option. |
| --count-n | Count sgRNAs with Ns. By default, sgRNAs containing N will be discarded. |
| --unmapped-to-file | Save unmapped reads to file, with sgRNA lengths specified by --sgrna-len option. |
-
(iv)
(Optional) Batch effect removal. If any portion of the screen was performed in batches (at separate times or with different reagents), we recommend running Combat in the sva package44 to remove possible batch effects, as follows. To run the package, a BatchMatrix file (see Table 3 for format) is required. This file can be generated using a text editor and saved as .txt file, and each item in this file should be separated by a tab. In this file, columns 1–3 are sample names that correspond to a raw count table, batch covariate which could be numbers that represent batches, and another covariate besides batch (optional), respectively. Most commonly, the additional covariate in column 3 would be a number representing an experimental condition. The easiest approach is to put all needed files in the same folder. Alternatively, provide full paths for the files and scripts in the command. Here, using the HCT116 dataset as an example we demonstrate the general steps to remove batch effects from any screens which have been performed in batches. The batch-effect removed count table can be used as the input for MAGeCK RRA or MLE. To run the batch effect removal package, initiate R and type the following commands:
Table 3 |.
Example of a batch matrix file
| Sample name | Batch | Conditions |
|---|---|---|
| HCT116_2_T18A | 1 | 1 |
| HCT116_2_T18B | 2 | 1 |
| HCT116_2_T18C | 3 | 1 |
| HCT116_2_T12A | 1 | 2 |
| HCT116_2_T12B | 2 | 2 |
| HCT116_2_T12C | 3 | 2 |
| HCT116_1_T0 | 1 | 3 |
| HCT116_2_T0 | 2 | 3 |
$R > library(MAGeCKFlute) > BatchRemove(mat = “rawcount.txt”, batchMat = “BatchMatrix.txt”, prefix = “BatchCorrect”, -pca = T, -cluster = T, -outdir = “.”)
In this command,
| -mat | Matrix, or file path of data. |
| -batchMat | Matrix or file path of batch table, which has at least three columns, including Samples matched colname of mat, Batch, and Covariates. |
| -cov | Specify the covariates besides batch, such as treatment condition, which can be used to model the outcome. |
| -log2trans | Boolean, specifying whether to do log2 transition before batch removal. |
| -pca | Boolean, specifying whether to do principle component analysis before and after batch removal. |
| -cluster | Boolean, specifing whether to do cluster analysis before and after batch removal. |
| -prefix | Character, specifying prefix of output figures, only needed if cluster/pca is TRUE. |
| -outdir | Output directory on disk. |
-
(v)
Identify screen hits using MAGeCK RRA Use the mageck test subcommand to perform MAGeCK RRA for comparison between two conditions, such as an initial condition versus cells cultured for a period of time. The input of mageck test is a count table which can be generated by the mageck count command (Step 7Aiii) or other alignment tools such as bowtie45 or bwa46. To identify screen hits from Dataset 1, type the following:
$mageck test -k GSC_0131.count.txt -t day23_r1,day23_r2 -c day0_r1,day0_r2 -n GSC_0131_rra --remove-zero both --remove-zero-threshold 0
In this command,
| -k | Count table (Table 4, Step 7Aiii), provide a tab-separated count table. Each line in the table should include sgRNA name (1st column), target gene (2nd column) and read counts (3rd column) in each sample. |
| -t | Sample label or sample index (0 as the first sample according to python standard) in the count table that are to be treated as treatment experiments, separated by comma (,). If sample labels are provided (rather than a sample index), the labels must match the labels in the first line of the count table. This parameter is required, which means at least one sample should be assigned to this parameter. |
| -c | Sample label or sample index in the count table that are to be treated as control experiments, separated by comma (,). If no samples are specified by this parameter, controls will be defined as all the samples not specified by the -t parameter. This parameter is required, which means at least one sample should be assigned to this parameter.” |
| -n | The prefix of the output files. |
| --remove-zero | Remove sgRNAs whose mean value is zero in control, treatment, both control/treatment, or any control/treatment sample. Default: both (remove those sgRNAs that are zero in both control and treatment samples). |
| --remove-zero-threshold sgRNA normalized count threshold to be considered removed in the --remove-zero option. Default 0. | |
Table 4 |.
Example of count table
| sgRNA | Gene | day0_r1 | day0_r2 | day23_r1 | day23_r2 |
|---|---|---|---|---|---|
| s_48202 | RPL | 44 | 45 | 44 | 29 |
| s_47147 | RHBDD | 477 | 472 | 445 | 560 |
| s_48746 | RWDD2B | 487 | 405 | 644 | 587 |
Please use command mageck test –h to see additional parameters.
CRITICAL STEP: Note that rather than employing MAGeCK RRA in this step, it is possible instead to use MAGeCK MLE, as described in the next steps, Steps 7Avi-vii.
-
(vi)
(Optional) Identify screen hits using MAGeCK MLE if an experiment contains more than two conditions, for example a 3-condition design: day0, drug treatment, DMSO treatment, we recommend using MAGeCK MLE or MAGeCK-NEST (Box 2) instead of MAGeCK RRA to employ the previous step. To do this, access the directory of raw counts (generated with MAGeCK count in Step 7Aii) with the following command:
Box 2. Use MAGeCK NEST to improve the accuracy of hit calling.
MAGeCK NEST adds additional features to MAGeCK MLE to improve hit calling. First, MAGeCK NEST can improve results using Network Essentiality Scoring Tool (NEST) to integrate information from protein-protein interaction networks. Second, MAGeCK NEST adopts a maximum-likelihood approach to remove sgRNA outliers, which often have higher G-nucleotide counts. If the Gini index of the read count is high (such as greater than 0.2) or there are a lot of sgRNA outliners in the screen data, users can consider using MAGeCK NEST to improve hit calling. The input and output files of MAGeCK NEST are as same as for uses the parameter -e to specify negative control genes. To identify screen hits with MAGeCK NEST, use the following command:
$mageck_nest.py nest -k rawcount.txt -d designmatrix.txt -n nest_res --norm-method control -e negative_control_genes.txt
$ cd path/to/demo_data/mageck_mle
path/to/demo_data/ should point to either the demo data (Step 7Ai) or to the user’s own raw count data. In the following Step 7Avii, we use MAGeCK MLE to obtain gene hits from the screen in Dataset 2 (EQUIPMENT).
CRITICAL STEP The input of MAGeCK MLE function should be a raw count table in which columns are samples and rows are sgRNAs. Generally, this raw count table is generated from a FASTQ file using the MAGeCK count function in Step 7Aiii. Since Dataset 2 provides the raw count table instead of the FASTQ file, we used it directly as the input of MAGeCK MLE.
-
(vii)
(Optional) Run MAGeCK MLE with the following command:
$mageck mle --count-table rawcount.txt --design-matrix designmatrix.txt --norm-method control --control-sgrna nonessential_ctrl_sgrna_list.txt --output-prefix braf.mle
Here is a description of the key parameters of mageck mle (to see all the parameters, use the command mageck mle -h):
| --count-table | Provide a tab-separated count table. Each line in the table should include sgRNA name (1st column), target gene (2nd column) and the read count in each sample (3rd and subsequent columns). |
| --design-matrix | Provide a design matrix (For instructions to generate the design matrix, see Box 3), either as a file name or a quoted string of the design matrix. An example of design matrix is shown in Table 5. The row of the design matrix must match the order of the samples in the count table (if --include-samples is not specified), or the order of the samples is specified by the --include-samples option. |
| --norm-method | {none, median, total, control} Method for normalization, including “none” (no normalization), “median” (median normalization, default), “total” (normalization by total read counts), “control” (normalization by control sgRNAs specified by the --control-sgrna option). |
| --control-sgrna | A list of control sgRNAs. |
| --output-prefix | The prefix of the output file(s). Default is “sample1”. |
Box 3. Format of design matrix file.
The design matrix file (Table 5) is a binary matrix indicating which sample (contained in the first column) is affected by which condition (contained in the second and subsequent columns). Values under the headers are binary. The element in the design matrix, dij, equals to “1” if sample i is affected by condition j, and 0 if it is not. Each column of the design matrix file should be separated by a tab character. This file can be created with a text editing software and saved as a plain text file.
The following rules apply to the design matrix file:
The design matrix file must include a header line of condition labels.
The first column consists of the sample labels, which must match the sample labels in the read count file.
The non-header values in columns 2 and beyond must be either “0” or “1”.
The second column defines an initial condition that affects all samples and must be “1” for all rows (except the header row)
The design matrix file must contain at least one sample representing the ‘initial state’ (for example, day 0) that has only a single “1” in the corresponding row. That single “1” must be in the “initial condition” column (the second column). MAGeCK MLE will calculate the beta score by comparing the other conditions to the initial condition.
Table 5 |.
An example of a design matrix file
| samples | Initial condition | Mock treatment | Drug treatment |
|---|---|---|---|
| Day 0 | 1 | 0 | 0 |
| D7_R1 | 1 | 1 | 0 |
| D7_R2 | 1 | 1 | 0 |
| PLX7_R1 | 1 | 0 | 1 |
| PLX7_R2 | 1 | 0 | 1 |
CRITICAL STEP: We recommend that sgRNAs targeting negative control loci, such as AAVs1, CCR5, and ROSA26, be included in a custom sgRNA library. These sgRNAs can be used as negative controls to normalize screen data. If the library includes such sgRNAs, they can be specified in the command line with parameters, --norm-method control and --control-sgrna as shown above. If these negative control sgRNAs were not included in the library, sgRNAs targeting non-essential genes can be used as negative controls for normalization, as described in the parameter specifications above. We generated a non-essential gene list which includes 350 genes as described in “Experimental Design” section. Users can use the --control-sgrna parameter to provide sgRNAs corresponding to these genes to perform the normalization with these non-essential genes. The control sgRNA file should be a tab-separated table, with only a single column that includes the IDs of sgRNAs. It needs to prepared by the user and saved as a .txt file. The nonessential_ctrl_sgrna_list.txt file contains sgRNAs targeting the 350 non-essential genes specified in the ‘Experimental design’, which we use to perform negative control normalization.
CRITICAL STEP
-
(viii)
(Optional) Correct copy number bias MAGeCK RRA and MAGeCK MLE contain an optional method to correct copy number biases in the calculated RRA scores and beta scores, respectively. We recommend users to perform copy number bias correction if the copy number variation information is available for the cell line. Both MAGeCK RRA and MLE require a tab-delimited file containing copy number values for each gene across the cell line(s) associated with the experiment (provided as Supplementary Data 3). The copy number file contains 2 columns: gene name and copy number. The name of this file is incorporated into the analysis with the parameter --cnv-norm. For MAGeCK RRA, an additional parameter cell-line is required to specify the name of the cell line (from the copy number data file) to be used in the bias correction method. In order to perform copy number correction in MAGeCK MLE, the name of the cell line (from the copy number data file) must match sample labels in the design matrix file. The data analysed here is a HL60 dataset (see EQUIPMENT). To perform MAGeCK RRA with copy number bias correction, type the following:
$mageck test -k rawcount.txt -t HL60.final -c HL60.initial -n rra_cnv --cnv-norm cnv_data.txt –cell-line HL60_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE
-
(ix)
(Optional) We use the HL60 dataset (see EQUIPMENT) to demonstrate how to perform copy number correction with MAGeCK MLE. To perform MAGeCK MLE with copy number bias correction, type the following:
$mageck mle --count-table rawcount.txt --design-matrix designmatrix.txt --cnv-norm cnv_data.txt
-
B.
Process CRISPR screen data with MAGeCK-VISPR. TIMING: 1.5 h
-
(i)
Activate the MAGeCK-VISPR environment as follows. If MAGeCK-VISPR was installed with conda (Step 3Ai-iii), make sure to activate the corresponding conda environment (Step 3Aiv) before conducting any MAGeCK-VISPR related command:
$source activate mageck-vispr
-
(ii)
Chose a workflow directory and initialize the workflow with the fastq or fastq.gz files that contain the raw reads (downloaded in Step 7Ai). If the raw reads files are not in the working directory, remember to include a path when specifying the raw read files. MAGeCK will install a “README” file, a config file “config.yaml” and a Snakemake workflow definition (a “Snakefile”) to the given directory. Initialize the workflow as follows:
$mageck-vispr init workflow --reads path/to/file/*.fastq*
Here the term ‘workflow’ can be changed to any name. The parameter --reads is used to specify the fastq or fastq.gz files to be analysed. The path/to/file is the directory containing the fastq or fastq.gz files.
-
(iii)
(Optional) Alternatively, MAGeCK-VISPR supports analysis with raw counts table (e.g. rawcount.txt file in Dataset 2). To run MAGeCK-VISPR with raw counts, specify the raw count file in the “config.yaml” file instead of using the parameter –reads. A short video (Supplementary Video) describing how to edit the “config.yaml” file can be found at: https://www.youtube.com/watch?v=3maSxhy1JL0.
$mageck-vispr init workflow
-
(iv)
Configure the workflow using the following command.
$cd workflow
Next, specify the path to the library file (Table 2) and the normalization method by changing the “config.yaml” file. The sgRNAs used for normalization and batch information must be provided as well, if data were generated from different batches. Copy Number Variation (CNV) correction is recommended if CNV data is available.
-
(v)
To check whether the “config.yaml” files has been configured correctly, enter the filling command line in the terminal:
$snakemake –n
-
(vi)
Execute the workflow.
The execution of the workflow must specify how many CPU cores should be assigned to this process. In general, 3–4 cores should be sufficient, but an increase in cores will reduce the running time. The workflow can be performed, in this case with 8 cores, using the following command:
$snakemake --cores 8
-
(vii)
(Optional) Visualize results with VISPR
MAGeCK-VISPR also provides a web-based visualization framework (VISPR) for an interactive exploration of CRISPR screen quality control and analysis results. Once the workflow execution has finished, visualize the generated results and quality controls by issuing the following command into the terminal:
$vispr server results/*.vispr.yaml
This will generate the following output:
Loading data. Starting server. Open: go to http://127.0.0.1:5000 in your browser. Note: Safari and Internet Explorer are currently unsupported. Close: hit Ctrl-C in this terminal.
You can copy and paste the link (http://127.0.0.1:5000) into a browser (Chrome or Firefox recommended) to visualize the results.
Downstream analysis pipeline
-
8.
Open a new plotting script in the terminal or use the R interactive shell as follows:
$R
-
9.
Load the MAGeCKFlute package into the R environment:
>library(MAGeCKFlute)
-
10.
Set the working directory to the directory of the output files for MAGeCK, for example:
>setwd(‘path/to/file/’)
Where ‘path/to/file/’ is the location of the output files for MAGeCK.
-
11.Functional analysis can be carried out using results obtained from MAGeCK RRA (option A) or from MAGeCK MLE (option B).
-
A.Functional analysis for MAGeCK RRA results. TIMING: 1h
-
(i)To perform functional analysis for MAGeCK RRA results (from Dataset 1, generated in Step 7Av), enter the following command in the R environment:
-
A.
>FluteRRA(gene_summary = “path/to/file/rra.gene_summary.txt”, prefix=“FluteRRA”, organism=“hsa”)
The file “rra.gene_summary.txt” is included in the output files of MAGeCK RRA, which has been performed at step 7Av or 7B.
-
B.
Functional analysis of MAGeCK MLE results. TIMING: 1h
-
(i)
To perform essential gene normalization and functional analysis of MAGeCK MLE results (from Dataset 2, generated in Step 7Avii), enter the following command:
>FluteMLE(gene_summary=“path/to/file/mle.gene_summary.txt”, ctrlname=“dmso”, treatname=“plx”, organism=“hsa”, prefix=“FluteMLE”,-pathway_limit = c(3,50))
The file “mle.gene_summary.txt” is included in the output files of MAGeCK MLE which has been performed at step 7Avii or 7B.vi. FluteMLE performs essential gene normalization automatically, and custom essential gene list can be specified by the parameter – posControl.
The meanings of the parameters in this command are as follows (see Box 4 for further parameters that could be used or see all the parameters with the command help(“FluteMLE”) in R):
| -gene_summary | Either a file or a data frame whose column name contains ‘Gene’, ‘dmso.beta’ and ‘plx.beta’ which corresponding to the parameters -ctrlname and -treatname. In this specific MAGeCK MLE result (from Dataset 2, generated in Step 7Avii), ‘dmso.beta’ and ‘plx.beta’ correspond to the control (DMSO) and drug treatment conditions (PLX), respectively. |
| -ctrlname | A character vector, specifying the name of control samples. |
| -treatname | A character vector, specifying the name of treatment samples. |
| -organism | A character, specifying an organism, such as “hsa” or “Human”(default), and “mmu” or “Mouse”. |
| -prefix | A character, indicating the prefix of output file name. |
| -pathway_limit | A two-length vector (default: c(3, 50)), specifying the minimal and maximal size of gene sets for enrichent analysis. |
Box 4. Additional FluteMLE parameters.
Additional key parameters that may be used to run FluteMLE in step 11.B.ii:
| -top | An integer, specifying the number of top selected genes labeled in rank figure. |
| -bottom | An integer, specifying the number of bottom selected genes labeled in rank figure. |
| -interstGenes | A character vector, specifying the genes of interest labeled in rank figure |
| -pvalueCutoff | A numeric, specifying the FDR cutoff of enrichment analysis. |
| -adjust | One of “holm”, “hochberg”, “hommel”, “bonferroni”, “BH”, “BY”, “fdr”, “none”. |
| -enrich_kegg | One of “ORT”(Over-Representing Test), “GSEA”(Gene Set Enrichment Analysis), “DAVID”, “GOstats”, and “HGT”(HyperGemetric test), or index from 1 to 5, specifying the enrichment method used for kegg enrichment analysis. |
| -gsea | A boolean value that indicates whether GSEA analysis is performed. |
| -posControl | A file path or a character vector, specifying the positive control genes used for cell cycle normalization, if NULL, use build-in essential gene list. |
| -loess | Boolean, specify whether to include loess normalization in the pipeline. |
| -view_allpath | Boolean, specify if all pathway view figures are output. |
| -outdir | Output directory on disk. |
CRITICAL STEP: After the command line finishes processing, and if there is no error occurrence, users can check the current directory to verify the existence of the files listed in Table 9. A detailed description of the output files can be found in the Anticipated Results section. Before running FluteMLE, ensure that the “gene_summary.txt” file is in the current working directory, or that the file path name is inserted into the command.
Table 9 |.
Results of FluteMLE
| Files | Description |
|---|---|
| Prefix_Flute.mle_summary.pdf | The integration of the results |
| Distribution_of_BetaScores | The distribution of the beta score: all genes and essential genes under different normalization methods |
| Linear_Fitting_of_BetaScores | The linear fitting of the beta score with different normalization methods |
| MAplot | MA plot of the beta score |
| Scatter_Treat_Ctrl | Scatter plot and rank plot of the treatment and control samples |
| Enrichment_Treat-Ctrl | Functional enrichment analysis of the genes, of which the beta scores are significantly different between treatment and control samples |
| Pathview_Treat_Ctrl | The KEGG map of the enriched terms |
| Scatter_9Square | A 9 square scatter plot of the treatment and control beta scores |
| Enrichment_9Square | Functional enrichment analysis of the four separate groups of genes from the 9 square scatter plot, which relate to the experimental conditions |
| Pathview_9Square | The KEGG map of the enriched terms |
Troubleshooting
Troubleshooting advice can be found in Table 6.
Table 6 |.
Troubleshooting table
| Step | Problem | Probable reason | Solution |
|---|---|---|---|
| 2 | Package installation failed | R version is old or dependent package cannot be installed | Update R to 3.5.0 or newer. Try to start the R session from the root folder. The PATH variable can be set by typing into a terminal: For Linux $export PATH="/usr/bin:$PATH" For MacOS $export PATH="/usr/local/bin:$PATH" |
| 7Avii | MAGeCK crashed with an error related to the design matrix. Such as: “Error parsing the design matrix: 0 row sums for some samples.” |
Design matrix was not in a correct format | Follow the tutorial to generate a correct design matrix. https://bitbucket.org/liulab/mageck-vispr |
| 7Bv | MAGeCK-VISPR crashed with an error due to cannot find input files: “Error in configuration file (key=library, entry=xxx): File does not exist.” |
File names or paths of the input files is incorrect. | Double check the names and paths of the input files and make sure these files can be accessed. |
| 11B | FluteMLE crashed with an error due to cannot find samples: “Error in FluteMLE(gene_summary = ‘mle.gene_summary.txt’, treatname = ‘treatment.beta’, : No sample found!” |
The index specified with the parameters “treatname” or “ctrlname” does not fit the column names of “gene_summary.txt” file. | Ignore the suffix “.beta” when specifying the control names and treatment name with the parameter “treatname” and “ctrlname” |
Timing
Running this protocol on the example data provided will take ~3 h on a machine with eight processing cores and at least 8 GB of RAM. However, larger data sets with more samples or deeper sequencing runs may take longer, and timing will vary across different computers.
Steps 1–6, install MAGeCK, MAGeCK-VISPR and MAGeCKFlute: 20 min
Step 7Aiii, Generate a count table from a FASTQ file: 15 mins. ~6 million reads per min.
Step 7Aiv, Batch effect removal: 5 mins.
Step 7Av, Identify screen hits using MAGeCK RRA: 8 mins.
Step 7Avii, Identify screen hits using MAGeCK MLE: 1h
Step 7Bvi, Process CRISPR screen data with MAGeCK-VISPR:1.h.
Step 11Ai, Functional analysis of MAGeCK RRA results:30 mins
Step 11Bi, Functional analysis of MAGeCK MLE results:1.h.
Anticipated results
Results of MAGeCK count (Step 7Aiii)
The main output of mageck count includes a raw count table “count.txt”, a normalized count table “count_normalized.txt”, and a summary table of the mapping results “countsummary.txt”. The raw count table contains the raw sequencing read counts of each sgRNA for each sample, and the normalized count table records the normalized count, using the default median normalization method or an alternative method specified by the user. The “countsummary.txt” file includes the total reads, mapped reads, mapped percentage, zero count number and Gini index30 of each sample. The “zero count” number indicates the number of sgRNAs that have a read count of 0 in that sample. Screens with a large number of zero-count sgRNAs in the initial condition, after transfection, or after selection might indicate insufficient cell representation of the library complexity. MAGeCK count analysis on Dataset 1 yields approximately 65% mapped reads for both replicates collected at Day 0 and Day 23 (Figure 2a). The correlations between replicates are greater than 0.9 (Figure 2b). The Gini indices are less than 0.1 (Figure 2c), and the missing sgRNAs are less than 1% (Figure 2d).
Results of MAGeCK RRA (Step 7Av)
The main output of the MAGeCK RRA consists of the following files (Table 7):
Table 7 |.
results of MAGeCK RRA
| File name | Description |
|---|---|
| sgrna_summary.txt | The sgRNA rank results |
| gene_summary.txt | The gene rank results |
| log | The log information during the run |
| R | The R code that can be used to plot summary figures of the results |
The most important output of MAGeCK RRA is the file “gene_summary.txt”. For each gene, MAGeCK RRA outputs a score for both negative selection and positive selection. In either case, lower scores indicate a higher level of selection. MAGeCK RRA also outputs a p-value or FDR for the scores of each gene.
MAGeCK RRA directly performs some basic analysis at the sgRNA level and outputs the result to “sgrna_summary.txt”. This file contains normalized read counts for each sample, the mean counts in control and treatment sample(s), and the log fold-change, p-value and FDR of each sgRNA in the comparison. The details of each file can be found in the MAGeCK documentation using the following link: https://sourceforge.net/p/mageck/wiki/output/
Results of MAGeCK MLE and MAGeCK NEST (Step 7Avi and Box 2)
MAGeCK MLE and MAGeCK NEST generate files that are similar to MAGeCK RRA, such as a “log” file, a “gene_summary” file (including gene beta scores), and a “sgrna_summary” file (including sgRNA efficiency probability predictions) (Table 7). The “gene_summary” file includes the beta scores of the conditions specified in the design matrix except for the initial condition, and the associated statistics. The ‘p-value’ is calculated by randomly permuting sgRNA labels. The ‘fdr’ is the false discovery rate calculated by the Benjamini-Hochberg Procedure. Similarly, the ‘wald-p-value’ (and ‘wald-fdr’) is the p-value (and false discovery rate) calculated by the Wald test to determine whether the corresponding ‘beta score’ significantly differs from zero in the MAGeCK MLE model, respectively. A detailed description of the output files from MAGeCK MLE can also be found in MAGeCK documentation using the following link: https://sourceforge.net/p/mageck/wiki/output/
Results of MAGeCK-VISPR (Step 7B)
All the results from MAGeCK-VISPR will be written into the “result” folder. If there are no errors running MAGeCK-VISPR, users will see three subfolders in the “result” folder. The “count” subfolder includes all of the outputs from mageck count: raw count, normalized count and the summary of count files. The “QC” subfolder includes the quality control of the reads at the sequence level for each sample, which is generated by FASTQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). MAGeCK-VISPR also performs quality control at read count level, including mapping ratio (Figure 2a), correlation between samples (Figure 2b), evenness of sgRNA reads (figure 2c), and number of missing sgRNAs (Figure 2d). The quality control result can be visualized by VISRP (Step 7Bvii). The “test” subfolder contains the main results of MAGeCK RRA or MLE, including “gene_summary.txt” file, which can be used in the functional analysis in step 11.
Results of MAGeCKFlute (Step 11)
All pipeline results are written into a local directory “Prefix_Flute_Results/”, and all figures are integrated into a single report file “Prefix_Flute.mle_summary.pdf”.
FluteRRA (Step 11Ai)
MAGeCKFlute processes MAGeCK RRA results (“gene_summary” file) with the function FluteRRA, which compares between two conditions, such as Day23 vs day Day0 in Dataset 1. FluteRRA will perform functional analysis with both positively and negatively selected genes. GO enrichment analysis uses the clusterProfiler28 package as the default enrichment method. The Hypergeometric test is used as the default method for KEGG enrichment analysis. FluteRRA will output both figures similar to Figure 4c, d, e and tables of the significantly enriched terms. All the result files are listed in Table 8.
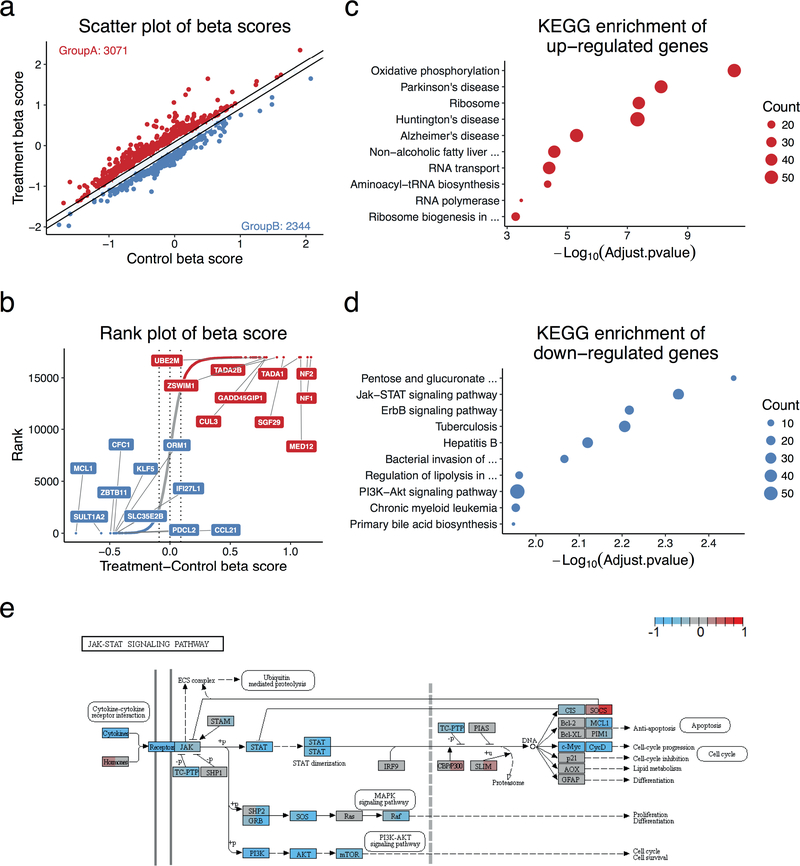
Figure 4. CRISPR/Cas9 screen analysis by MAGeCKFlute.
The data analysed here is a CRISPR screen in a breast melanoma cancer cell line, A375, treated with the BRAF protein kinase inhibitor vemurafenib (PLX)7. Data was processed with FluteMLE. (a) Scatterplot of treatment and control beta scores. The beta scores were normalized using the median of the beta scores of the core essential genes we compiled (Supplementary Data 2, Supplementary Method). The two diagonal lines indicate +/−1 standard deviation of the difference between treatment and control beta scores. Red dots (Group A) are genes whose beta score increased after treatment. Blue dots (Group B) are genes whose beta score decreased after treatment. (b) The genes are sorted based on the differential beta score, which is calculated by subtracting the control beta score from the treatment beta score. The colour scheme is the same as in panel a and dots between two diagonal lines are genes for which the beta score did not change significantly between different conditions. The top 10 enriched KEGG pathways with (c) positively (Red, Group A) and (d) negatively (Blue, Group B) selected genes. The p-value was calculated with the clusterProfile package that is based on the hypergeometric distribution. The size of each circle indicates the number of genes which are enriched in the corresponding function. (e) A visualization of treatment and control beta scores over the JAK-STAT signaling pathway generated by the Pathview package47. The left and right portion of a gene-box represent control and treatment beta scores, respectively. Red indicates a positive beta score, blue indicates a negative beta score, and grey marks genes are neither positively nor negatively selected. The dashed vertical line in this specific pathway indicates the nuclear membrane.
Table 8 |.
Results of FluteRRA
| Files | Description |
|---|---|
| Prefix_Flute.rra_summary.pdf | The integration of the results |
| bp.neg.png | The enrichment analysis of biology process using negatively selected genes. |
| bp.pos.png | The enrichment analysis of biology process using positively selected genes. |
| kegg.neg.png | The enrichment analysis of KEGG using negatively selected genes. |
| kegg.pos.png | The enrichment analysis of KEGG using positively selected genes. |
FluteMLE (Step 11Bi)
FluteMLE uses the “gene_summary” file as its input, which is the output of MAGeCK MLE. The results of FluteMLE will be written into a directory with the name “prefix_Flute_results” where ‘prefix’ is the prefix that users specify in the FluteMLE function. All the result files are listed in Table 9.
To illustrate the utility of MAGeCKFlute in the downstream analysis of MAGeCK MLE results, we used example data from a public CRISPR screen melanoma cancer cell line, A375 (see EQUIPMENT) We analysed the raw count using the mageck mle function, and performed normalization with non-essential genes with the parameter --norm-method and specified the non-essential gene list (Supplementary Data 1) with the parameter --control-sgrna. Normalization with the core essential genes (Supplementary Data 2) was performed using the FluteMLE function by default.
FluteMLE performs quality controls (QC) based on the beta score to ensure that the two conditions, control and treatment, are comparable. The QC results consist of 3 levels: distribution of the beta score for different conditions (Supplementary Figure 4a), linear fitting of the beta scores of essential genes (Supplementary Figure 4b), and MA plot (Supplementary Figure 4c). After the normalization, the beta score of most genes should be close to zero. Therefore, the mean beta score of all the genes should also be close to zero. We observed that the distribution of beta score in both treatment and control conditions were similar, making beta scores comparable between different conditions. (Supplementary Figure 4a, b) The MA plot can be used to visualize the differences between the beta scores generated in two samples, by transforming the data onto M (βT-βC) and A (βT+βC) scales; βT and βC are the beta score of the treatment and control samples, respectively. Since the beta scores for most genes will not change drastically in treatment condition compared to control condition, the M value for the majority of the genes in the MA plot should be close to zero (Supplementary Figure 4c). The distribution of the beta scores is in the folder “Distribution_of_BetaScores”. The folder “Linear_Fitting_of_BetaScores” contains figures showing the linear regression results of the beta scores for treatment and control samples. A summary table of the beta scores for each normalization is also generated in the “Scatter_Treat_Ctrl” folder.
After quality control, MAGeCKFlute will identify the differences between two treatment conditions (not including Day 0) such as samples treated with or without a particular drug. MAGeCKFlute will generate scatterplots (Figure 4a) and ranking plots (Figure 4b). After performing FluteMLE on Dataset 2, we observed 3,071 genes with decreased essentiality (GroupA, red dots) and 2,344 genes with increased essentiality (GroupB, blue dots) after drug treatment compared with control. These groups were used to generate as two independent gene lists for further downstream analysis. Ranking plot shows the changes of beta score between treatment and control conditions anduses the same criteria as scatterplots to identify beta score differences (Fig. 4b). Scatterplots and ranking plots can be found in the “Scatter_Treat_Ctrl” folder.
The functional annotation will be performed based on the two groups of genes selected in the scatterplot and ranking plot. In the FluteMLE function, the enrichment method and cutoff can be specified with the parameter -enrich_kegg and –pvalueCutoff, respectively. In this protocol, we performed all the enrichment analysis using the default enrichment method “HGT” (HyperGemetric test) and used the default cutoff 0.1. Enrichment results (Figure 4 c, d) will be generated in the folder “Enrichment_Treat-Ctrl”.
MAGeCKFlute utilizes the pathview package to perform data integration and visualization. The figures generated with pathview are included in the files “Pathview_9Square” and “Pathview_Treat_Ctrl”. MAGeCKFlute maps and renders data onto relevant pathway graphs, where each gene is coloured with two colours in a single pathway graph, based on the beta score under different conditions (Figure 4e). For each selected gene, the left half is coloured based on beta score of treatment samples and the right half is coloured according to the beta score of the control samples. This approach allows users to explore the variations of the beta scores within one pathway graph. For example, STAM is strongly negatively selected in the DMSO control condition and weakly positively selected in PLX drug treatment condition (Figure 4e). Therefore, this gene lost its essentiality upon PLX treatment, suggesting that it acts in a pathway targeted by PLX.
To identify treatment-related hits accurately, FluteMLE classifies genes into 4 groups (Supplementary Figure 4d), determined by differences in the beta scores between the treatment and control samples (MAGeCK MLE results of Dataset 2). Gene groups that exhibit different beta scores between treatment and control samples are coloured to represent these differences. Genes in the green group are strongly negatively selected (that is, cells whose the gene is disrupted are under-represented) in the control samples and are weakly selected (either positively or negatively) in the treatment samples. These genes lost their essentiality after treatment, and are potentially located in the pathways targeted by the treatment. The orange group contains genes that are weakly selected in the control and strongly positively selected in treatment (that is, cells whose gene is disrupted are over-represented). These are genes whose loss confers treatment resistance. Genes in the blue group are strongly positively selected in the control and weakly selected in the treatment. These genes may be either potential regulators of cell proliferation in general, or regulators of the treatment target (if the treatment target is an essential gene). Genes in the purple group are weakly selected in the control and strongly negatively selected in the treatment. These genes are potentially synthetically lethal in combination with the drug treatment. Figures and tables are located in the folder “Scatter_9Square”. The functional analysis and pathway visualization of the four groups are also performed by MAGeCKFlute, and the results are located in the folders named “Enrichment_9Square” and “Pathview_9Square”, respectively.
Supplementary Material
Acknowledgments
This project was supported by the Breast Cancer Research Foundation, the National Natural Science Foundation of China (81872290) (to X.S.L), the Startup fund from the Center for Genetic Medicine Research and the Gilbert Family Neurofibromatosis Institute (to W.L.).
Footnotes
Competing Financial Interests
T.X. and X.S.L are co-founders and M.B. and X.S.L. are on the Scientific Advisory Board of GV20 Oncotherapy. The authors declare no competing interests.
Availability
The source code of MAGeCKFlute (version 0.99.18) is freely available at https://bitbucket.org/liulab/mageckflute/ under the 3-clause Berkeley Software Distribution (BSD) open-source license. Questions or comments can be submitted through the MAGeCK Google group: https://groups.google.com/d/forum/mageck. The datasets used in this paper are presented in http://cistrome.org/MAGeCKFlute/.
Related links
Key references using this protocol
Li, W., et al. Genome Biology 15 (554) (2014). https://doi.org/10.1186/s13059–014-0554–4.
Li, W., et al. Genome Biology 16 (1) 281 (2015). https://doi.org/10.1186/s13059–015-0843–6.
Jeselsohn, R. et al. Cancer Cell 33 (2) 173–186 (2018). https://doi.org/10.1016/j.ccell.2018.01.004.
Xiao., et al. Proc Natl Acad Sci U S A. 115(31):7869–7878 (2018). https://doi.org/10.1073/pnas.1722617115.
Key data used in this protocol
Toledo, C.M., et al. Cell Rep 13, 2425–2439 (2015). https://doi.org/10.1016/j.celrep.2015.11.021.
Hart, T., et al. Cell 163, 1515–1526 (2015) https://doi.org/10.1016/j.cell.2015.11.015.
Shalem, O., et al. Science 343, 84–87 (2014) https://doi.org/10.1126/science.1247005
Wang, T., et al. Science 343, 80–84 (2014). https://doi.org/10.1126/science.1246981.
Chen, H., et al. Bioinformatics, bty450, (2018). https://doi.org/10.1093/bioinformatics/bty450
References
- 1.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159, 647–661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Konermann S, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mali P, et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang T, Wei Jj Fau - Sabatini DM, Sabatini Dm Fau - Lander ES & Lander ES Genetic screens in human cells using the CRISPR-Cas9 system. 343, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C & Yusa K Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol 32, 267–273 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hart T, et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 163, 1515–1526 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Wang T, et al. Identification and characterization of essential genes in the human genome. Science 350, 1096–1101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen S, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160, 1246–1260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manguso RT, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 547, 413–418 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burr ML, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 549, 101–105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurata M, et al. Using genome-wide CRISPR library screening with library resistant DCK to find new sources of Ara-C drug resistance in AML. Sci Rep 6, 36199 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han K, et al. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat Biotechnol 35, 463–474 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi J, et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol 33, 661–667 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li W, et al. Quality control, modeling, and visualization of CRISPR screens with MAGeCK-VISPR. Genome Biol 16, 281 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toledo CM, et al. Genome-wide CRISPR-Cas9 Screens Reveal Loss of Redundancy between PKMYT1 and WEE1 in Glioblastoma Stem-like Cells. Cell Rep 13, 2425–2439 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M & Goto S KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A 105(51), 20380–20385 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konig R, et al. A probability-based approach for the analysis of large-scale RNAi screens. Nat Methods 4, 847–849 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Hart T & Moffat J BAGEL: a computational framework for identifying essential genes from pooled library screens. Bioinformatics 17, 164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu J, Silva J & Califano A ScreenBEAM: a novel meta-analysis algorithm for functional genomics screens via Bayesian hierarchical modeling. Bioinformatics 32, 260–267 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgens Dw Fau - Morgens DW, Deans Rm Fau - Deans RM, Li A Fau - Li A & Bassik Mc Fau - Bassik MC Systematic comparison of CRISPR-Cas9 and RNAi screens for essential genes. Nat Biotechnol 34, 634–636 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falcon S & Gentleman R Using GOstats to test gene lists for GO term association. Bioinformatics 23, 257–258 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Yu G, Lg W, Y. H & Qy. H clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shalem O Fau - Shalem O, Sanjana Ne Fau - Sanjana NE & Zhang F Fau - Zhang F High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet 16, 299–311 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gini. “Concentration and dependency ratios” (in Italian). English translation in Rivista di Politica Economica. 87, 769–789 (1997). [Google Scholar]
- 31.Johnson WE, Li C Fau - Rabinovic A & Rabinovic A Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Chen CH, et al. Improved design and analysis of CRISPR knockout screens. Bioinformatics bty, 450 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang P, et al. Network analysis of gene essentiality in functional genomics experiments. Genome Biol 16(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeKelver RC, et al. Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res 20, 1133–1142 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hockemeyer D, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol 27, 851–857 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aguirre AJ, et al. Genomic Copy Number Dictates a Gene-Independent Cell Response to CRISPR/Cas9 Targeting. Cancer Discov 6, 914–929 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherr CJ & Roberts JM CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 13, 1501–1512 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Huang da W, Sherman Bt Fau - Lempicki RA & Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Wang T, et al. Gene Essentiality Profiling Reveals Gene Networks and Synthetic Lethal Interactions with Oncogenic Ras. Cell 168, 890–903 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tzelepis K, et al. A CRISPR Dropout Screen Identifies Genetic Vulnerabilities and Therapeutic Targets in Acute Myeloid Leukemia. Cell Rep 17, 1192–1205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang T, Wei Jj Fau - Sabatini DM, Sabatini Dm Fau - Lander ES & Lander ES Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen CH, et al. Improved design and analysis of CRISPR knockout screens. Bioinformatics bty 450(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leek JT, Johnson WE, Parker HS, Jaffe AE & Storey JD The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B, Trapnell C Fau - Pop M, Pop M Fau - Salzberg SL & Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H & Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo W & Brouwer C Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29, 1830–1831 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.