Abstract
Radiotherapy is the main treatment strategy for nasopharyngeal carcinoma. A major factor affecting radiotherapy outcome is the accuracy of target delineation. Target delineation is time-consuming, and the results can vary depending on the experience of the oncologist. Using deep learning methods to automate target delineation may increase its efficiency. We used a modified deep learning model called U-Net to automatically segment and delineate tumor targets in patients with nasopharyngeal carcinoma. Patients were randomly divided into a training set (302 patients), validation set (100 patients), and test set (100 patients). The U-Net model was trained using labeled computed tomography images from the training set. The U-Net was able to delineate nasopharyngeal carcinoma tumors with an overall dice similarity coefficient of 65.86% for lymph nodes and 74.00% for primary tumor, with respective Hausdorff distances of 32.10 and 12.85 mm. Delineation accuracy decreased with increasing cancer stage. Automatic delineation took approximately 2.6 hours, compared to 3 hours, using an entirely manual procedure. Deep learning models can therefore improve accuracy, consistency, and efficiency of target delineation in T stage, but additional physician input may be required for lymph nodes.
Keywords: nasopharyngeal cancer, deep learning, automatic delineation
Introduction
Nasopharyngeal carcinoma (NPC) is one of the most common cancers in the nasopharynx. In 2015, an estimated 833 019 new cases of NPC and 468 745 deaths due to NPC were reported in China alone.1 The main treatment strategy for NPC is radiotherapy, which has a 5-year survival rate of about 80%, with or without chemotherapy.2 The most important factor for precise and effective radiotherapy in patients with NPC is accurate target delineation. However, accurate target delineation is time-consuming: Manual target delineation of a single head and neck tumor typically requires 2.7 hours, while delineation of tumor volume and adjacent normal tissues in NPC requires more than 3 hours.3,4 In fact, delineation must be repeated many times when treating locally advanced NPC due to tumor volume shrinkage and anatomical changes during treatment. Target delineation accuracy is also strongly dependent on the training and experience of the radiation oncologist and can vary widely.5-7 Therefore, it would be useful to develop a fully automatic delineation method to improve the consistency and accuracy of delineation, as well as to relieve the workload for doctors.
Deep learning is a method of machine learning based on artificial neural networks. Deep learning methods have been shown to perform better than traditional machine learning algorithms in many computer vision tasks, especially object detection in images, regression prediction, and semantic segmentation.8-10 Convolutional neural network (CNN) is a deep learning model with the ability to learn from labeled data, and it has shown impressive accuracy in prediction and detection in medical applications.11-15 For example, multiple-instance learning using chest X-ray images can detect tuberculosis with an area under the curve of 0.86.13 An alternating decision tree model using data from structural imaging, age, and scores on the Mini-Mental State Examination predicted treatment response in patients with late-life depression with 89% accuracy.15 Convolutional neural networks have also been used to segment organs and substructures during targeted treatments. A deep learning model based on CNNs has been used to segment liver images and optimize surface evolution, showing a dice similarity coefficient (DSC) of nearly 97%.12 A modified U-shaped CNN (U-Net) was used to segment retina thickness and yielded a mean DSC of 95.4% ± 4.6%.16 Segmentation based on deep learning has also been used in treating pulmonary nodules, liver metastases, and pancreatic cancer.17-19
Studies show that CNNs can be useful for delineating tumor targets for radiotherapy in brain, rectal, and breast cancer.20-24 A deep learning model called DeepMedic was used to segment brain tumors with a DSC of 91.4%.20 Another study using U-Net to segment brain tumors achieved a DSC of 86%.21 Deep learning models have also been used in rectal cancer to accurately delineate the clinical target volume (CTV), organs at risk, and the target tumor with a DSC of 78% to 87%.22,24 A CNN model called DD-ResNet was developed to delineate CTVs for breast cancer radiotherapy using big data and was shown to perform better than other deep learning methods, with a DSC of 91%.23 Given the above studies, we reasoned that CNN may also be useful for delineating NPC targets for radiotherapy. However, segmentation of NPC is more complex than other tumor types because of ambiguous and blurred boundaries between the tumor and normal tissues.
Here, our study was novel with 4 main contributions. First, few literature reported on delineation of primary tumor and lymph nodes in planning computed tomography (CT) images for radiotherapy with CNNs, especially considering the large data set used. Thus, we used a modified version of U-Net to segment CT images from 502 patients with NPC and delineate radiotherapy targets.25 Second, the performance of deep learning model was observed from early stage to advanced stage. Both DSC and Hausdorff distance (HD) value were shown in different stage and demonstrated a downward trend from early stage to advanced stage for both primary tumor and lymph node. Third, normalization technique was used in preprocess the input data for CT images. It could improve the accuracy of target volume delineation on segmentation of NPC, using deep learning methods. Finally, deep learning model can be used to delineate the nasopharynx gross tumor volume (GTVnx) with high accuracy. But in the delineation of the lymph node gross tumor volume (GTVnd), it needs to be intervened by experts, especially in N3 patients.
Materials and Methods
Data Sets
All experimental procedures involving human CT images were approved by the West China Hospital Ethics Committee. CT images were obtained from 502 patients with NPC admitted to the hospital over a period of 5 years. The patients were randomly divided into 3 groups: a training set (302 patients), validation set (100 patients), and testing set (100 patients). Tumor clinical stage was determined according to the American Joint Committee on Cancer (AJCC) staging system (seventh edition). Demographic data are shown in Table 1. There was no difference in the relative proportions of primary tumors (T stage) or lymph node (N stage) among the training, validation, and testing sets.
Table 1.
Baseline Characteristics of the 502 NPC Patients.a
| Characteristics | Training Set (%) | Validation Set (%) | Testing Set (%) |
|---|---|---|---|
| n = 302 | n = 100 | n = 100 | |
| Median age (range) | 46.9 (18-73) | 52.3 (12-67) | 50.7 (25-72) |
| Sex | |||
| Male | 195 (64.6%) | 73 (73%) | 69 (69%) |
| Female | 107 (35.4%) | 27 (27%) | 31 (31%) |
| T classification | |||
| T1 | 73 (24.2%) | 23 (23%) | 18 (18%) |
| T2 | 76 (25.2%) | 25 (25%) | 32 (32%) |
| T3 | 100 (33.1%) | 33 (33%) | 20 (20%) |
| T4 | 53 (17.5%) | 19 (19%) | 40 (40%) |
| N classification | |||
| N0 | 39 (12.9%) | 13 (13%) | 15 (15%) |
| N1 | 98 (32.5%) | 33 (33%) | 29 (29%) |
| N2 | 155 (51.3%) | 52 (52%) | 43 (43%) |
| N3 | 10 (3.3%) | 2 (2%) | 13 (13%) |
| Overall stage | |||
| I | 20 (6.6%) | 6 (6%) | 4 (4%) |
| II | 36 (11.9%) | 12 (12%) | 15 (15%) |
| III | 97 (32.1%) | 34 (34%) | 34 (34%) |
| IV | 149 (49.3%) | 48 (48%) | 47 (47%) |
Abbreviation: NPC, nasopharyngeal carcinoma.
a Tumor and lymph node stage were judged by the seventh edition of the American Joint Committee on Cancer (AJCC) stage criteria.
In total, 20 676 CT slices were collected from 502 CT scans. The number of CT slices was 13 310 slices for training set, 3673 slices for validation set, and 3693 slices for testing set. Computed tomography slices were extracted from Digital Imaging and Communications in Medicine (DICOM) files, with the image resolution of 512 × 512 and slice thickness of 3 mm. The gray levels converted by HU value from the DICOM files ranged from 0 to 3071. The target regions on CT slices were independently determined by 2 senior radiation oncologists and labeled nasopharyngeal primary tumor target or metastatic lymph node target.
Preprocessing
To make the image more suitable for segmentation, it needs to be preprocessed with the following steps:
The determination of region of interest (ROI): An original DICOM image has the data size of 512 × 512. In the training phase, the original CT image can cause heavy computing workload, due to large useless regions. The subregion has included the target and main anatomical structure in CT image. Therefore, a 224 × 224 region was cropped from original CT image as the ROI. The cropping operation is shown in Figure 1.
Computed tomography image normalization: In deep learning experiment, different CT scans equipment may have different configuration. According to eliminate differences, normalizing operations were deployed in these CT images and the formula was as follows:
Figure 1.
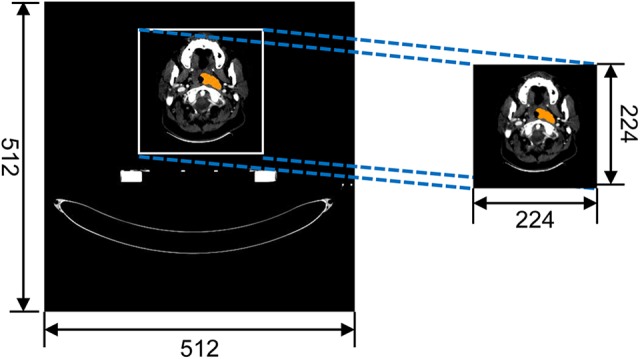
Cropping the region of interest.
| 1 |
where Pixel is the source pixel data in CT images; Pixelnorm is normalized CT images pixel data; and Pixelmin and Pixelmax are the minimum and maximum gray value of source CT images, respectively.
Deep Learning Model for Delineation
In the field of image segmentation, CNNs showed researchers excellent performance on image segmentation tasks. Fully convolutional neural network (FCN) is the first CNN algorithm model in image segmentation using deconvolution layers.8 The CT image was decoded to an input patch of 224 × 224 matrix. The FCN architecture can predict an output with an interesting region image. A more elegant FCN model was named U-Net, which extract a large number of feature channels in the upsampling part.
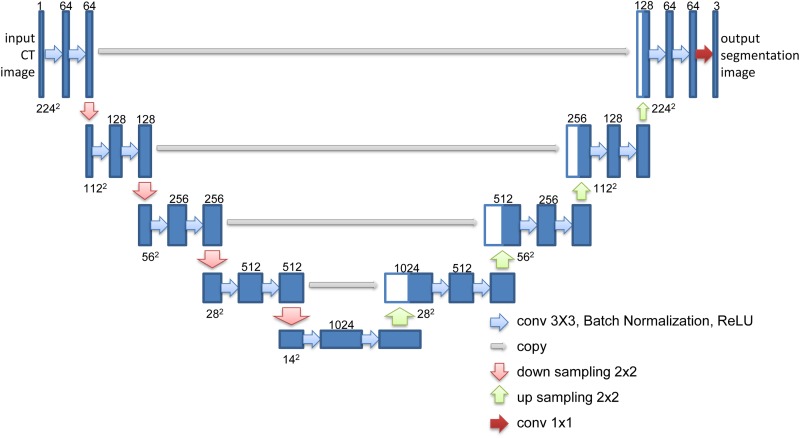
However, in the segmentation of nasopharyngeal CT image, FCN and U-Net only predict a lower spatial resolution than source CT image. The resolution of output images cannot be used here because of unsuitable output size. Therefore, a modified version of the U-Net model was proposed, in which downsampling layers and upsampling layers have similar learning ability. Each convolution layer is a convolution operation with padding followed by a batch normalization and Rectified Linear Unit activation function.26 The output feature maps of each convolution layer are the same input feature maps in the whole of this model. In downsampling path, the input CT image with the size of 224 × 224 was downsampled the spatial dimension to 14 × 14. Conversely, the upsampling path upsampled the feature maps from 14 × 14 to 224 × 224. The upsampling layers concatenated output feature maps with feature maps of downsampling layers. The network diagram is shown in Figure 2. The number of kernels about this model has shown in each output of convolutional layer.
Figure 2.
U-Net architecture.
U-Net model is implemented by Google TensorFlow framework, which is a famous machine learning library, and then accelerated by NVIDIA@ Compute Unified Device Architecture.27,28 The CT image data set was divided into training set, validation set, and testing set. The training set (302 patients) was used to optimize the parameters of the U-Net. The original 2-dimensional CT images were the inputs and the corresponding segmentation probability maps about the GTVnx and GTVnd were the outputs. The validation set was used to tune the deep learning model in training phase. The testing set was divided into T1, T2, T3, T4, N1, N2, and N3, according to seventh edition of AJCC for NPC.29
After data preprocessing, the U-Net architecture was defined in TensorFlow Machine Learning Library using Python Application Programmable Interface. Because of overfitting, the Dropout was set in every convolution layer.30 The initialization of parameters was configured by Xavier function and truncated normal distribution whose standard deviation is 0.1.31 In the phase of model training, the learning rate was set to 0.01 in Adam optimizer.32 The number of iterations was 40 with cross-entropy descent of the whole validation set.
Evaluation of Deep Learning Model
The CT images from the test set were used to evaluate the predictive performance of the U-Net model. The loss values were recorded for each patient in the validation set. The loss value illustrates how the model was trained in the training phase. U-Net performance was evaluated using the DSC value and HD, which quantify the results of GTVnx and GTVnd.
The values of DSC were defined in Equation 2 as follows:
| 2 |
where P denotes the segmented area for prediction, L denotes the segmented area for reference, and is the intersection of the 2 areas. DSC value is defined between 0 and 1, with 0 representing predicting miss and 1 shows that the predicting result is perfect.
Hausdorff distance (HD) was defined in Equation 3 as follows:
| 3 |
where and are 2 finite point sets, and
| 4 |
where is the normal form on the points of P and L (ie, the L 2 or Euclidean norm). describes the point that is farthest from any point of L and calculates the distance from p to its nearest neighbor in L. The HD is the maximum of and and expresses the largest degree of mismatch between P and L. The overlap between P and L increases with smaller .
Results
U-Net Model Training
The cross-entropy loss function is used to observe U-Net in the training situations. The loss value decreased with epoch number. After 20 epochs, the decrease in loss value slowed and the U-Net model stabilized. This was observed in both the training and validation set. Experiments were carried out on dual Intel Xeon E5-2643 v4 (3.4 GHz) and dual NVIDIA tesla K40m graphics card. The validation set, 100 of 502 patients, was deployed to evaluate the predicting performance of U-Net model. The reference segmentation maps labeled by the experienced radiation oncologists were used to calculate loss value of target function.
Delineation Results by U-Net Model in Testing Set
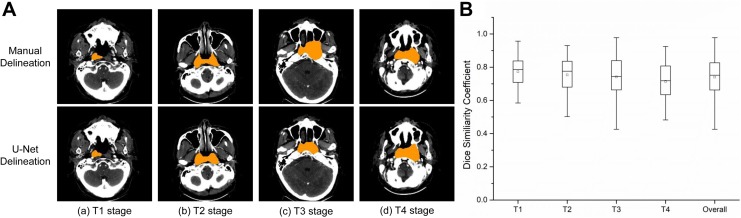
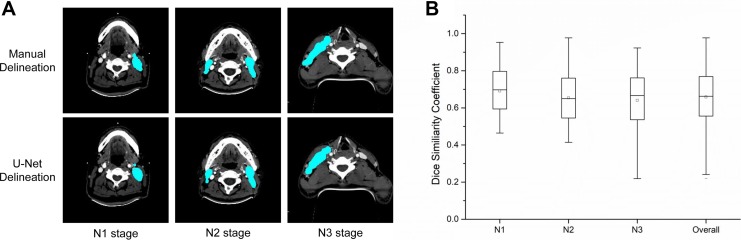
After training our model, we used CT images from the test set to perform target delineation. The DSC and HD results for GTVnx and GTVnd in the test set are summarized in Figure 3 and Table 2. The average normalized U-Net DSC values by T stage were 77.24% (T1), 75.38% (T2), 74.13% (T3), and 71.42% (T4), with an overall DSC of 74.00%. The average normalized HD values were 10.36 mm (T1), 11.37 mm (T2), 11.90 mm (T3), and 15.72 mm (T4), with an overall HD of 12.85 mm (Figure 3 and Table 2). The DSC and HD values were higher for T stage than N stage. The average normalized DSC values for N stage were 69.07% (N1), 65.32% (N2), and 64.03% (N3), with an overall DSC of 65.86% (Figure 4 and Table 2). The average normalized HD values for N stage were 31.08 mm (N1), 32.12 mm (N2), and 34.99 mm (N3), with an overall HD of 32.10 mm.
Figure 3.
Target delineation in T stage of NPC by U-Net model. A. Representative pictures from manual delineation and U-Net. The target region is shown in orange. B.DSC value in different T stage.
Table 2.
The DSC and HD Values for GTVnx and GTVnd Segmentation.
| Evaluation Metrics | Primary Tumor Stage | Lymph Nodes Stage | |||||||
|---|---|---|---|---|---|---|---|---|---|
| T1 | T2 | T3 | T4 | Overall | N1 | N2 | N3 | Overall | |
| DSC-norm (%) | 77.24 | 75.38 | 74.13 | 71.42 | 74.00 | 69.07 | 65.32 | 64.03 | 65.86 |
| DSC (%) | 76.58 | 73.18 | 71.49 | 68.80 | 71.78 | 65.64 | 59.87 | 59.42 | 61.05 |
| HD-norm (mm) | 10.36 | 11.37 | 11.90 | 15.72 | 12.85 | 31.08 | 32.12 | 34.99 | 32.10 |
| HD (mm) | 10.43 | 14.10 | 12.37 | 17.02 | 14.24 | 34.37 | 33.33 | 42.98 | 36.15 |
Abbreviations: DSC, dice similarity coefficient; DSC-norm, dice similarity coefficient with normalization; GTVnd, lymph node gross tumor volume; GTVnx, nasopharynx gross tumor volume; HD, Hausdorff distance; HD-norm, Hausdorff distance with normalization.
Figure 4.
Target delineation in N stage. A, Representative computed tomography scans showing the results of manual delineation and automated delineation using U-Net. The target region is shown in cyan. B, Normalized U-Net dice similarity coefficients by N stage.
There was good overlap in DSC and HD values for GTVnx between autosegmented contours and manual contours obtained by physicians. However, autosegmented contours did not show a good match in the lymph nodes, especially in patients with N3 lymph nodes. We also performed U-Net delineation without normalization to test the impact of normalization. The DSC and HD values without normalization were lower than those after normalization (Table 2).
Time Cost
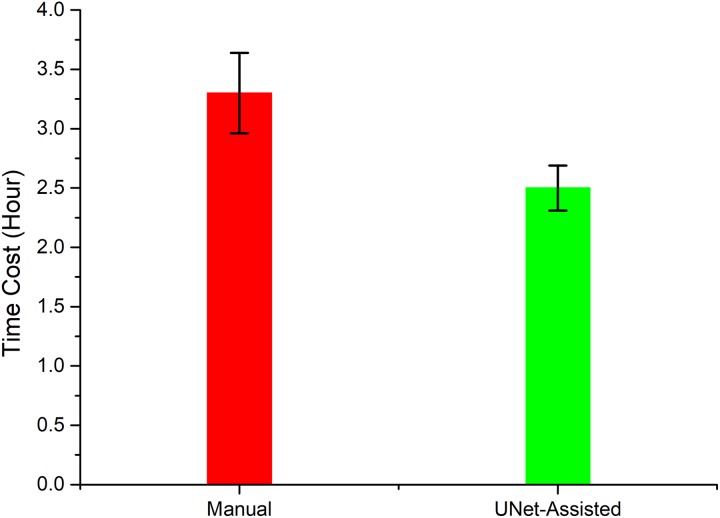
The time needed to train the U-Net model was about 18 hours using a DELL R730 server with dual Intel Xeon E5-2643 v4 (3.4 GHz) and dual NVIDIA tesla K40m graphics cards. The average time for automatic delineation of GTVnx and GTVnd with U-Net was about 40 seconds per patient. U-Net-assisted delineation required an average of 2.6 hours per patient, in contrast to manual delineation which required an average of 3 hours per patient. The comparison of 10 physicians was shown in Figure 5 between U-Net-assisted delineation and manual delineation.
Figure 5.
Comparison of total delineation time per patient for 10 physicians using manual delineation and U-Net-assisted delineation.
Discussion
Accurate target delineation is the most important step for precise and effective radiotherapy in patients with NPC, but is time-consuming and varies with the experience of the oncologist. In recent years, automatic target delineation using deep learning algorithms has been increasingly used by radiation oncologists. In this study, we used a modified version of a deep learning algorithm called U-net to automate segmentation and delineation of NPC tumors for radiotherapy. We show that U-Net is able to delineate NPC tumors with high accuracy and reduces the delineation time requirement for physicians.
Many studies reported that deep learning model can segment those obvious and clear tumors, such as lung cancer, hepatoma, and so on. But the contour and anatomical structure of NPC is more complex than other types of tumors. In this article, a modified deep learning model for automatic tumor target segment of nasopharyngeal cancer is carried out. On the one hand, our data showed that deep learning model had a better delineation accuracy in early stage, compared with advanced stage. Moreover, our deep learning model demonstrated lower DSC value and HD value in GTVnd, compared with GTVnx. Therefore, professional intervention is required because of unsatisfied delineation accuracy. On the other hand, considering impact of normalization technique, another U-Net without normalization was trained by same data set in order to compare with current results. The experimental results indicated that normalization technique could improve the delineation accuracy of deep learning model. The main reason is that original gray level values bring larger errors than normalized data after the model calculation in single precision floating point representation.
Consistency of target delineation is a key factor affecting clinical outcomes in patients with NPC. A study in which several oncologists manually delineated identical GTV contours of supraglottic carcinoma reported an interobserver overlap of only 53%.33 A comparison of CTV delineation among different radiation oncologists reported a DSC value of only 75%.34 Deep learning methods have been reported to perform better than other methods in many automatic delineation applications. Previous studies using nondeep learning methods reported mean DSC values of 60% to 80% for CTV delineation, whereas automatic delineation based on deep learning gave a mean DSC value of 82.6%.35-40 However, few studies have investigated their use in delineating complex targets such as NPC. Previous studies using nondeep learning methods reported DSC values of 69% and 75% for head and neck cancer, respectively.41,42 Delineation of lymph node is especially difficult: Studies show that the DSC value of lymph node delineation using atlas-based methods was only 46% in unilateral tonsil cancers.43 In comparison, we found that U-Net produced DSC values of 65.86% for overall N stage and 74.00% for overall T stage in patients with NPC. Previous studies did not differentiate delineation of primary tumor stage or lymph nodes stage. We found that U-Net produced higher DSC values in T stage than N stage and that the DSC value decreased in more advanced cancer. Moreover, we respectively analyzed the performance of U-Net model in different primary tumor stage (T1-T4) and lymph nodes stage (N1-N3), which helped us to find weakness of U-Net model in different stages.
Our study has several advantages compared to previous work. First, Sun et al investigated that the performance of deep learning models can be improved by increases of the data.44 The size of database used by most previous studies was smaller than ours. Not only that, but to ensure data set quality, manual target delineation was drawn by 2 radiation oncologists separately, who were trained according to the same professional guideline.45 Both of them have more than 15 years of experience in caring for patients with NPC. After that, 2 radiation radiologists were required to approve the contour drawn by each other. If inconsistent samples were identified, the third radiologist specializing in NPC imaging would consult in cases of disagreement. During the consultation, the third radiologist was required to discuss and reach an agreement with the 2 radiologists. To a limited extent, the inter-/intraoperator variability is addressed in our study. Second, we chose not to use data augmentation in our study, although this can enhance the performance of deep learning models, because we wished to investigate the performance of a deep learning model trained by a large, manually labeled data set curated by experienced radiation oncologists. As a result, we found clear evidence that deep learning methods show different accuracy of target delineation depending on cancer stage. Future work should examine the impact of including data augmentation on automatic delineation. Third, low-contrast visibility and high noise levels usually lead to ambiguous and blurred boundaries between GTVnx, GTVnd, and normal tissues in CT images. Variation in contrast among different slices may affect the robustness of the model. By comparing delineation with or without normalization, we clearly show that normalization improved U-Net performance.
Conclusion
We show that a modified U-Net model can delineate NPC tumor targets with higher consistency and efficiency than manual delineation, as well as reduce the amount of time required per patient. Delineation accuracy was better in early stage than advanced stage and better in primary tumor than in lymph nodes. The U-Net may be a useful tool for relieving physician workload and improving treatment outcomes in NPC.
Acknowledgments
The authors sincerely thank the radiation oncologists in Department of Biotherapy, Cancer Center, West China Hospital, Sichuan University, for the target delineation.
Abbreviations
- AJCC
American Joint Committee on Cancer
- CNNs
convolutional neural networks
- CT
computed tomography
- CTV
clinical target volume
- DSC
dice similarity coefficient
- FCN
fully convolutional neural network
- GTVnx
nasopharynx gross tumor volume
- GTVnd
lymph node gross tumor volume
- HD
Hausdorff distance
- NPC
nasopharyngeal carcinoma
- ROI
region of interest
Authors’ Note: Shihao Li and Jianghong Xiao contributed equally. Our study was approved by The West China Hospital Ethics Committee (approval no. WCC2018 YU-91). All patients provided written informed consent prior to enrollment in the study.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The work was supported by the National Natural Science Foundation of China (81672386) and Major Special Science and Technology Project of Sichuan Province (2018GZDZX0024).
ORCID iD: Shihao Li, PhD  https://orcid.org/0000-0002-6418-7818
https://orcid.org/0000-0002-6418-7818
Xuedong Yuan, PhD  https://orcid.org/0000-0003-0240-2831
https://orcid.org/0000-0003-0240-2831
References
- 1. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. [DOI] [PubMed] [Google Scholar]
- 2. Hua YJ, Han F, Lu LX, et al. Long-term treatment outcome of recurrent nasopharyngeal carcinoma treated with salvage intensity modulated radiotherapy. Eur J Cancer. 2012;48(18):3422–3428. [DOI] [PubMed] [Google Scholar]
- 3. Harari PM, Song S, Tomé WA. Emphasizing conformal avoidance versus target definition for IMRT planning in head-and-neck cancer. Int J Radiat Oncol Biol Phys. 2010;77(3):950–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Das IJ, Moskvin V, Johnstone PA. Analysis of treatment planning time among systems and planners for intensity-modulated radiation therapy. J Am Coll Radiol. 2009;6(7):514–517. [DOI] [PubMed] [Google Scholar]
- 5. Yamazaki H, Shiomi H, Tsubokura T, et al. Quantitative assessment of inter-observer variability in target volume delineation on stereotactic radiotherapy treatment for pituitary adenoma and meningioma near optic tract. Radiat Oncol. 2011;6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Feng M, Demiroz C, Vineberg K, Balter J, Eisbruch A. Intra-observer variability of organs at risk for head and neck cancer: geometric and dosimetric consequences. Int J Radiat Oncol Biol Phys. 2010;78(3):S444–S445. [Google Scholar]
- 7. Breen SL, Publicover J, De Silva S, et al. Intraobserver and interobserver variability in GTV delineation on FDG-PET-CT images of head and neck cancers. Int J Radiat Oncol Biol Phys. 2007;68(3):763–770. [DOI] [PubMed] [Google Scholar]
- 8. Shelhamer E, Long J, Darrell T. Fully convolutional networks for semantic segmentation. IEEE Transactions on Pattern Analysis & Machine Intelligence. 2017;4:640–651. [DOI] [PubMed] [Google Scholar]
- 9. Ren Y, Zhang L, Suganthan PN. Ensemble classification and regression-recent developments, applications and future directions. IEEE Comp Int Mag. 2016;11(1):41–53. [Google Scholar]
- 10. LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc IEEE. 1998;86(11):2278–2324. [Google Scholar]
- 11. Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu P, Wu F, Peng J, Liang P, Kong D. Automatic 3D liver segmentation based on deep learning and globally optimized surface evolution. Phys Med Biol. 2016;61(4):8676. [DOI] [PubMed] [Google Scholar]
- 13. Melendez J, van Ginneken B, Maduskar P, et al. A novel multiple-instance learning-based approach to computer-aided detection of tuberculosis on chest X-rays. IEEE Trans Med Imaging. 2015;34(1):179–192. [DOI] [PubMed] [Google Scholar]
- 14. Crown WH. Potential application of machine learning in health outcomes research and some statistical cautions. Value Health. 2015;18(2):137–140. [DOI] [PubMed] [Google Scholar]
- 15. Patel MJ, Andreescu C, Price JC, Edelman KL, Reynolds CF, Aizenstein HJ. Machine learning approaches for integrating clinical and imaging features in late-life depression classification and response prediction. Int J Geriatr Psychiatr. 2015;30(10):1056–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Venhuizen FG, van Ginneken B, Liefers B, et al. Robust total retina thickness segmentation in optical coherence tomography images using convolutional neural networks. Biomed Opt Express. 2017;8(7):3292–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fu M, Wu W, Hong X, et al. Hierarchical combinatorial deep learning architecture for pancreas segmentation of medical computed tomography cancer images. BMC Syst Biol. 2018;12(suppl 4):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Setio AAA, Ciompi F, Litjens G, et al. Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Transact Med Imaging. 2016;35(5):1160–1169. [DOI] [PubMed] [Google Scholar]
- 19. Ben-Cohen A, Diamant I, Klang E, Amitai M, Greenspan H. Fully convolutional network for liver segmentation and lesions detection. In: Carneiro G, et al. , eds. Deep Learning and Data Labeling for Medical Applications. DLMIA 2016, LABELS 2016. Lecture Notes in Computer Science, vol 10008 Cham: Springer; 2016:77–85. [Google Scholar]
- 20. Kamnitsas K, Ferrante E, Parisot S, et al. DeepMedic for brain tumor segmentation In Crimi A, Menze B, Maier O, Reyes M, Winzeck S, Handels H, eds. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2016. Lecture Notes in Computer Science, vol 10154 Cham: Springer; 2016:138–149. [Google Scholar]
- 21. Dong H, Yang G, Liu F, Mo Y, Guo Y. Automatic brain tumor detection and segmentation using U-net based fully convolutional networks In Valdés Hernández M, González-Castro V, eds. Medical Image Understanding and Analysis. MIUA 2017. Communications in Computer and Information Science, vol 723 Cham: Springer; 2017:506–517. [Google Scholar]
- 22. Men K, Dai J, Li Y. Automatic segmentation of the clinical target volume and organs at risk in the planning CT for rectal cancer using deep dilated convolutional neural networks. Med Phys. 2017;44(12):6377–6389. [DOI] [PubMed] [Google Scholar]
- 23. Men K, Zhang T, Chen X, et al. Fully automatic and robust segmentation of the clinical target volume for radiotherapy of breast cancer using big data and deep learning. Phys Med. 2018;50:13–19. [DOI] [PubMed] [Google Scholar]
- 24. Men K, Boimel P, Janopaul-Naylor J, et al. Cascaded atrous convolution and spatial pyramid pooling for more accurate tumor target segmentation for rectal cancer radiotherapy. Phys Med Biol. 2018;63(18):185016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation In Navab N, Hornegger J, Wells W, Frangi A, eds. Medical Image Computing and Computer-Assisted Intervention - MICCAI 2015. Lecture Notes in Computer Science, vol 9351 Cham: Springer; 2015:234–241. [Google Scholar]
- 26. Glorot X, Bordes A, Bengio Y. Deep sparse rectifier neural networks. In Proceedings of the Fourteenth International Conference on Artifical Intelligence and Statistics; April 11-13, 2011, Fort Lauderdale, Florida, USA; pp. 315–323. [Google Scholar]
- 27. Abadi M, Barham P, Chen J, et al. TensorFlow: A system for Large-Scale Machine Learning In Proceedings of the 12th USENIX conference on Operating Systems Design and Implementation (OSDI'16). USENIX Association, 2016, Berkeley, CA; pp. 265–283. [Google Scholar]
- 28. Nvidia C. Compute Unified Device Architecture Programming Guide. 2007. [Google Scholar]
- 29. Cuccurullo V, Mansi L. AJCC Cancer Staging Handbook: from the AJCC Cancer Staging Manual (7th edition). Eur J Nucl Med Mol Imaging. 2011;38:408–408. doi:10.1007/s00259-010-1693-9 [Google Scholar]
- 30. Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15(2):1929–1958. [Google Scholar]
- 31. Glorot X, Bengio Y. Understanding the difficulty of training deep feedforward neural networks. Journal of Machine Learning Research - Proceedings Track. 2010;9:249–256. [Google Scholar]
- 32. Kingma DP, Ba J. Adam: a method for stochastic optimization. arXiv Preprint arXiv. 2014: doi:10.1177/14126980. [Google Scholar]
- 33. Cooper JS, Mukherji SK, Toledano AY, et al. An evaluation of the variability of tumor-shape definition derived by experienced observers from CT images of supraglottic carcinomas (ACRIN protocol 6658). Int J Radiat Oncol Biol Phys. 2007;67(4):972–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Caravatta L, Macchia G, Mattiucci GC, et al. Inter-observer variability of clinical target volume delineation in radiotherapy treatment of pancreatic cancer: a multi-institutional contouring experience. Radiat Oncol. 2014;9:198 doi:10.1186/1748-717X-9-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Iglesias JE, Sabuncu MR. Multi-atlas segmentation of biomedical images: a survey. Med Imaging Anal. 2015;24(1):205–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Teguh DN, Levendag PC, Voet PW, et al. Clinical validation of atlas-based auto-segmentation of multiple target volumes and normal tissue (swallowing/mastication) structures in the head and neck. Int J Radiat Oncol Biol Phys. 2011;81:950–957. [DOI] [PubMed] [Google Scholar]
- 37. Qazi AA, Pekar V, Kim J, Xie J, Breen SL, Jaffray DA. Auto-segmentation of normal and target structures in head and neck CT images: a feature-driven model-based approach. Med Phys. 2011;38(11):6160–6170. [DOI] [PubMed] [Google Scholar]
- 38. Stapleford LJ, Lawson JD, Perkins C, et al. Evaluation of automatic atlas-based lymph node segmentation for head-and-neck cancer. Int J Radiat Oncol Biol Phys. 2010;77(3):959–966. [DOI] [PubMed] [Google Scholar]
- 39. Gorthi S, Duay V, Houhou N, et al. Segmentation of head and neck lymph node regions for radiotherapy planning using active contour-based atlas registration. IEEE J Selec Top Sig Pro. 2009;3(1):135–147. [Google Scholar]
- 40. Men K, Chen X, Zhang Y, et al. Deep deconvolutional neural network for target segmentation of nasopharyngeal cancer in planning computed tomography images. Front Oncol. 2017;7:315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tsuji SY, Hwang A, Weinberg V, Yom SS, Quivey JM, Xia P. Dosimetric evaluation of automatic segmentation for adaptive IMRT for head-and-neck cancer. Int J Radiat Oncol Biol Phys. 2010;77(3):707–714. [DOI] [PubMed] [Google Scholar]
- 42. Yang J, Beadle BM, Garden AS, Schwartz DL, Aristophanous M. A multimodality segmentation framework for automatic target delineation in head and neck radiotherapy. Med Phys. 2015;42(9):5310–5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang J, Beadle BM, Garden AS, et al. Auto-segmentation of low-risk clinical target volume for head and neck radiation therapy. Pract Radiat Oncol. 2014;4(6):e31–e37. [DOI] [PubMed] [Google Scholar]
- 44. Sun C, Shrivastava A, Singh S, Gupta A. Revisiting unreasonable effectiveness of data in deep learning era In 2017 IEEE International Conference on Computer Vision. New York, NY: IEEE; 2017:843–852. [Google Scholar]
- 45. Lee N, Harris J, Garden AS, et al. Intensity-modulated radiation therapy with or without chemotherapy for nasopharyngeal carcinoma: radiation therapy oncology group phase II trial 0225. J Clin Oncol. 2009;27(22):3684. [DOI] [PMC free article] [PubMed] [Google Scholar]