Abstract
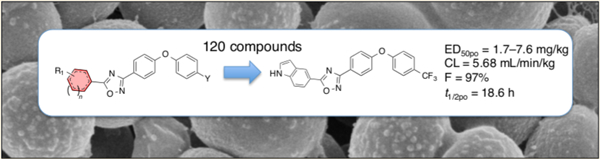
The structure–activity relationship (SAR) for the newly discovered oxadiazole class of antibiotics is described with evaluation of 120 derivatives of the lead structure. This class of antibiotics was discovered by in silico docking and scoring against the crystal structure of a penicillin-binding protein. They impair cell-wall biosynthesis and exhibit activities against the Gram-positive bacterium Staphylococcus aureus, including methicillin-resistant S. aureus (MRSA) and vancomycin-resistant and linezolid-resistant S. aureus. 5-(1H-Indol-5-yl)-3-(4-(4-(trifluoromethyl)phenoxy)phenyl)-1,2,4-oxadiazole (antibiotic 75b) was efficacious in a mouse model of MRSA infection, exhibiting a long half-life, a high volume of distribution, and low clearance. This antibiotic is bactericidal and is orally bioavailable in mice. This class of antibiotics holds great promise in recourse against infections by MRSA.
Graphical Abstract
INTRODUCTION
Staphylococcus aureus is a leading human bacterial pathogen that is a common source of infections in healthcare and community environments. The 2013 Centers for Disease Control and Prevention (CDC) report on antibiotic resistance prioritized methicillin-resistant Staphylococcus aureus (MRSA) as an ongoing serious threat, with 2011 records indicating that 11 285 of the 23 000 deaths caused by antibiotic-resistant bacteria and fungi in the United States were directly attributed to MRSA infections.1 Antibiotics that are approved for treatment of MRSA infections are vancomycin (a glycopeptide), linezolid (an oxazolidinone), daptomycin (a lipopeptide), and, more recently, ceftaroline (a β-lactam) and tedizolid (an oxazolidinone). Only linezolid and tedizolid are orally bioavailable among these agents.2 Furthermore, resistance to each of these antibiotics is known.3–8
We described recently the discovery of the oxadiazole class of antibiotics.9 The lead in this class came out of an in silico search for potential inhibitors for penicillin-binding protein 2a (PBP2a) of MRSA. PBPs are targets of β-lactam antibiotics. Inhibition of PBPs by β-lactams is bactericidal, as it interferes with biosynthesis of cell wall.10 Resistance to β-lactam antibiotics is widespread,11–14 but the importance of PBPs as targets for antibiotics has not diminished. We reasoned that PBPs remain worthy targets for antibiotics, and we sought to discover a new class of non-β-lactam inhibitors for these enzymes in this effort.
The in silico search and scoring of 1.2 million compounds from the ZINC library led to selection and purchase of the top-ranked compounds for screening with living bacteria.9 We set the bar high from the outset by screening compounds first against Escherichia coli and the ESKAPE panel of antibiotics, instead of against the recombinant protein. The ESKAPE panel is composed of Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species, a collection of bacteria that cause the majority of nosocomial infections.15 This strategy for screening easily eliminates any compound that would not have activity against bacteria, so the search was streamlined. The discovery produced the lead oxadiazole 1 (Scheme 1).9 We have explored in the present report the structural space for oxadiazole antibiotics by syntheses of derivatives. These compounds were in turn screened against the bacterial panel, from which a number exhibited good anti-MRSA activity. In another effort in streamlining the discovery process, the promising compounds went directly into the mouse MRSA peritonitis model for infection. This model has shown excellent correlation between the minimal-inhibitory concentration (MIC) and ED50 (the effective dose that rescues 50% of the animals from the infection) for 14 β-lactam antibiotics16 and for linezolid.17 This is a rapid animal model of infection that results in 100% fatality within 48 h. The compounds that would show efficacy would by necessity exhibit reasonable pharmacokinetic (PK) properties. This approach sped up lead optimization by identifying compounds with in vivo activity early. The compounds that resulted in survival of the animals were then further scrutinized for optimization by additional syntheses around the structural space and for attributes such as improved PK, decreased metabolism, and lack of toxicity to mammalian cells. The ring A of structure 1 (Scheme 1) produced excellent opportunities for these additional explorations. These efforts led to the SAR for the oxadiazoles as studied by 120 synthetic derivatives in ring A.
Scheme 1.
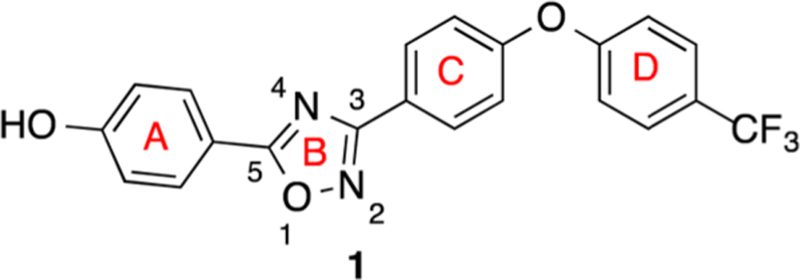
Structure of the Lead Oxadiazole 1
RESULTS AND DISCUSSION
Synthesis.
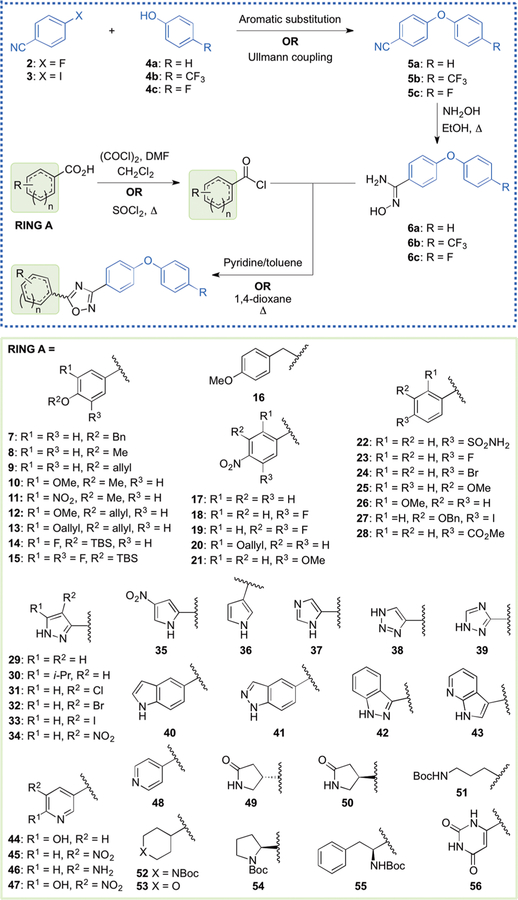
The focus of this SAR study is the variation of structure within ring A of the oxadiazole lead. This ring, attached to position 5 of the 1,2,4-oxadiazole moiety (1), proved versatile in generating many active antibiotics of this class. The diphenyl ether portion (rings C and D) was obtained by reaction of either 4-fluorobenzonitrile (2) or 4-iodobenzoni-trile (3) with the appropriate counterpart phenol (4a, 4b, or 4c, Scheme 2). Typically, coupling with 2 was done via nucleophilic aromatic substitution, and the reaction with 3 was achieved with an Ullmann coupling. Our variations to the aromatic rings of the diphenyl ether were minimal and involved only substitution at the 4-position of ring D with a fluoro or trifluoromethyl group. The para substitution of ring D, specifically with those two variations, proved beneficial for improved metabolic stability and lowered clearance. Nitriles 5a, 5b, and 5c were converted into their corresponding N′-hydroxybenzimidamides 6a, 6b, and 6c, respectively, using hydroxylamine in refluxing ethanol.
Scheme 2.
General Synthetic Route To Access the 1,2,4-Oxadiazoles, and the Starting Materials Used for Variations within Ring Aa
aCompounds 14, 15, and 28 were prepared from acyl chlorides, compound 40 from a methyl ester, and all others from the corresponding carboxylic acids. Aromatic substitution: K2CO3, DMF, 60–100 °C. Ullmann coupling: CuI, Cs2CO3, N,N-dimethylglycine·HCl, 1,4-dioxane 90 °C.
The left-hand portions of the oxadiazoles were accessed by starting (with a few exceptions) with the corresponding carboxylic acids, which would ultimately become ring A and the C5 of the oxadiazole ring (Scheme 2). These were converted to the corresponding acyl chlorides by reaction with either oxalyl chloride or thionyl chloride. The starting materials included several benzoic acid derivatives (7–28) and a variety of heteroatom-containing carboxylic acids (29–56). Biological analysis of the lead compound 1 and a few close analogues established that a hydrogen bond donor at the 4-position of ring A is generally beneficial for activity against S. aureus.9 Thus, the phenolic hydroxyl was retained using several different protected derivatives (7–16), while the 4-amino group (for anilines) was accessed by starting with the corresponding 4-nitrobenzoic acid derivatives (17–21), wherein the nitro functionality was later reduced. We also explored the effect of several other substituents at the 4-position of the phenyl ring, as exemplified by precursors 22–28. The heteroatom-containing starting materials included several pyrazoles (29–34), pyrroles (35, 36), imidazole 37, triazoles (38, 39), indole 40, indazoles (41, 42), pyrrolopyridine 43, pyridines (44–48), several aliphatic derivatives (49–53), protected amino acids (54, 55), and pyrimidine 56. While most of these precursors are commercially available, carboxylic acids 9, 12, 13, 20, 27, and 52, and acyl chlorides 14 and 15, had to be synthesized (procedures given in the Supporting Information). Once in hand, the acyl chlorides were allowed to react with 6a, 6b, or 6c in refluxing pyridine/toluene or 1,4-dioxane to produce the 1,2,4-oxadiazoles.
Many of these immediate oxadiazole products were subjected to further synthetic manipulation to broaden the structural diversity of the derivatives (protective group removal, nitro reduction, metal-catalyzed coupling, substitution on amine, etc.). These reactions are described in the Experimental Methods and Supporting Information.
Structure–Activity Relationship (SAR).
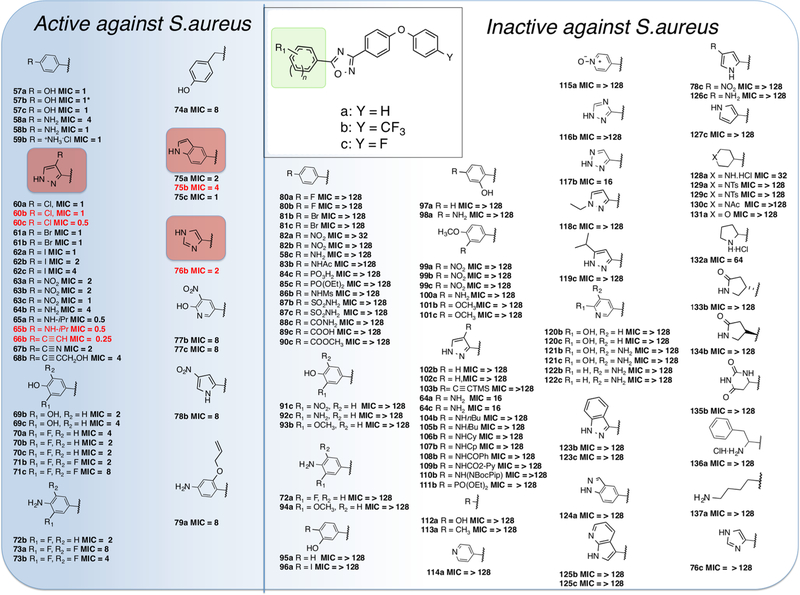
The SAR for the synthetic oxadiazole compounds was investigated using antibacterial screening against the aforementioned ESKAPE panel of bacteria plus E. coli. The 120 synthetic samples encompassed modifications in the lead at the 5-position of the 1,2,4-oxadiazole (ring A in Scheme 1), while keeping the 3-position constant as a 4-substituted diphenyl ether moiety (Figure 1). The oxadiazoles exhibit activity against Gram-positive bacteria. We expressly explored the activity against S. aureus for this study, for reasons that we outlined earlier. The SAR was evaluated by minimal-inhibitory concentration (MIC) measurements against S. aureus ATCC 29213, a standard methicillin-sensitive S. aureus (MSSA) strain for the purpose of screening. Active compounds (MIC ≤ 8 μg/mL) are enclosed within the blue shading in Figure 1. The substituents at the 4-position of ring D were hydrogen, trifluoromethyl, or fluorine (Figure 1), as indicated earlier. These modifications had little effect on the in vitro activity of compounds, except for some specific cases that are discussed below. However, the substitution with trifluoromethyl or fluorine at this position resulted in lower clearance and better metabolic stability, thus improving the PK properties.
Figure 1.
Antibacterial activities of the synthetic 1,2,4-oxadiazole derivatives. The functionality within the green box was altered to generate all the synthetic compounds in this series, whereas Y was limited to the three entities that are indicated. The MIC values (in μg/mL) measured for S. aureus ATCC 29213, with the active compounds placed within the shaded blue area (MIC ≤ 8 μg/mL). Compounds in red font underwent in vivo evaluation. Properties of the lead compounds (57a, 57b, and 58b) have been described previously.9 Compound 57b is identical to compound 1 and will be referred to as such in the remainder of the text.
Replacement of the phenol or aniline moieties in ring A with certain heterocyclic rings improved antibacterial activity. Introduction of 4-halogen-substituted pyrazoles (60a–c, 61a,b, 62a–c) maintained MIC values of ≤1 μg/mL. The pyrazolyl compounds also tolerated NO2 (63a–c) and NH2 (64b) substitution in this position; however, introduction of an isopropyl group on the amine (65a,b) caused the MIC to drop further to 0.5 μg/mL. The lowest observed MIC value of 0.25 μg/mL came from the ethynyl substituted derivative 66b. Other sp-hybridized functional group substitutions (67b and 68b) also maintained good activity.
Addition of a 3-hydroxyl group (69b,c) retained activity, and the addition of fluorine atoms in the 3- and 5-positions on the phenol (70a–c and 71b,c) and the aniline (72b, 73a,b) was possible without significant loss of activity, but an additional methylene spacer between the 1,2,4-oxadiazole and the phenol ring (74a) increased the MIC to 8 μg/mL. The other heterocyclic substitutions that retained good activity were the indolyl compounds (75a–c), the imidazolyl compound (76b), the substituted pyridinyl compounds (77b,c), and the nitro substituted pyrrolyl compound (78b).
Replacement of the hydrogen-bond donating phenol and aniline groups with aryl halogens (80a,b, 81b,c) resulted in loss of activity, as did replacement with other hydrogen-bond accepting aryl moieties (82a,b, 84c). Interestingly, the aniline derivative with a 4-F substitution on the biphenyl ether (58c) had no antibiotic activity despite the low MIC values for the 4-H and 4-CF3 substituted compounds (58a,b). All other substituted aryl systems failed to show activity (87b,c, 88c, 89c, 90c, 91c, 92c, 93b), including changing the hydroxyl group to the 2- or 3-positions (95a, 96a, 97a). No activity was seen for unsubstituted pyrazoles (102b,c), and the activity seen in the secondary amine series did not extend to larger straight chain alkyl or cyclic alkyl substitution (104b, 105b, 106b, 107b) or acylation (108b, 109b). Complete replacement of an aryl moiety with a simple hydroxyl (112a) or methyl (113a) abolishes activity and underlines the significance for activity of a spacing group between the 1,2,4-oxadiazole ring and the hydrogen-bond donating group. No activity was observed for any of the other heteroaromatic substituents that were introduced (114a, 115a, 116b, 117b, 118c, 119c, 120b,c, 121b,c, 122b,c, 123b,c, 124a, 125b,c, 78c, 126c, 127c). In a similar trend observed with the inactive 4-F diphenyl ether substituted aniline (58c), the 4-F diphenyl ether substituted imidazole derivative (76c) showed no activity compared to the active 4-CF3 derivative (76b). The effect of the 4-position diphenyl ether substitutions on activity in these cases is yet to be resolved. Several derivatives with saturated cyclic substitutions were also synthesized; however, activities were poor (≥32 μg/mL) (128a, 132a, 133b, 134b, 135b, 136a, 137a).
Activity Against Gram-Positive Organisms.
Compounds 60b, 60c, 65b, 66b, 75b, and 76b were evaluated against a panel of Gram-positive organisms. The pyrazoles showed activity against S. aureus MSSA (ATCC 29213) and MRSA (ATCC 27660, NRS119, VRS1, and VRS2) strains, including vancomycin-resistant strains (Table 1). The pyrazoles were not active against S. aureus NRS120 and other Gram-positive organisms. Replacement of the pyrazole with an indole (75b) broadened the spectrum of activity against Gram-positive organisms. The activity of the indole 75b was similar to that of the phenol derivative 1 (Table 1).
Table 1.
Minimal-Inhibitory Concentrations (MICs) of Oxadiazolesa
| MIC (μg/mL) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 60b | 60c | 65b | 66b | 75b | 76b | 1i | vancomycini | linezolidi | |
| S. aureus ATCC 29213b | 1 | 1 | 0.5 | 0.25 | 2 | 4 | 2 | 1 | 4 |
| S. aureus ATCC 27660c | 2 | 0.5 | 0.5 | 0.5 | 4 | 8 | 2 | 1 | 2 |
| S. aureus NRS100 (COL)c | >128 | >128 | >128 | >128 | 2 | 32 | 2 | 2 | 2 |
| S. aureus NRS119d | 1 | 2 | 2 | 32 | 2 | 4 | 2 | 2 | 32 |
| S. aureus NRS120d | 64 | 64 | >128 | >128 | 2 | 16 | 2 | 2 | 32 |
| S. aureus VRS1e | 1–2 | 1 | 1 | 1 | 1 | 1 | 2 | 1–2 | 2 |
| S. aureus VRS2f | 0.5–1 | 0.5 | 4 | 0.5 | 4 | 16 | 2 | 64 | 2 |
| S. epidermis ATCC 35547 | >128 | >128 | >128 | >128 | 4 | 32 | 2 | 16 | 1 |
| S. hemolyticus ATCC 29970 | >128 | >128 | >128 | >128 | 8 | 16 | 2 | 2 | 2 |
| S. oralis ATCC 9811 | >128 | >128 | >128 | >128 | 128 | >128 | 32 | 0.5 | 1 |
| S. pyogenes ATCC 49399 | >128 | >128 | >128 | >128 | 64 | 128 | 32 | 0.6 | 1 |
| B. cereus ATCC 13061 | >128 | >128 | >128 | >128 | 16 | 16 | 2 | 1 | 1 |
| B. licheniformis ATCC 12759 | >128 | >128 | >128 | >128 | 8 | 16 | 2 | 0.5 | 1 |
| E. faecalis ATCC 29212b | >128 | >128 | >128 | >128 | 4 | 16 | 2 | 2 | 2 |
| E. faecalis 201 (Van S)g | >128 | >128 | >128 | >128 | 8 | 16 | 2 | 1 | 2 |
| E. faecalis 99 (Van R)h | >128 | >128 | >128 | >128 | 16 | 16 | 2 | 128 | 1 |
| E. faecium 119–39A (Van S)g | >128 | >128 | >128 | >128 | 8 | 16 | 1 | 0.5 | 2 |
| E. faecium 106 (Van R)h | >128 | >128 | >128 | >128 | 8 | 16 | 2 | 256 | 1 |
| E. faecium NCTC 7171 | 16 | 32 | 32 | >128 | 2–4 | 8–16 | 2 | 0.5 | 2 |
The compounds were screened against E. coli and the ESKAPE panel of bacteria; they exhibited antibacterial activity against Gram-positive bacteria.
A quality-control strain to monitor accuracy of MIC testing.
mecA positive, resistant to methicillin, oxacillin, and tetracycline; susceptible to vancomycin and linezolid.
mecA positive, resistant to ciprofloxacin, gentamicin, oxacillin, penicillin, and linezolid.
Vancomycin-resistant MRSA (vanA) clinical isolate from Michigan.
Vancomycin-resistant MRSA (vanA) clinical isolate from Pennsylvania.
Vancomycin-susceptible clinical isolate.
Vancomycin-resistant clinical isolate.
Data from O’Daniel et al.,9 reproduced for the sake of comparison.
Plasma-Protein Binding.
Protein binding for compounds 60b, 60c, 65b, 66b, 75b, and 76b was determined in human plasma using equilibrium dialysis. Results are shown in Table 2. Protein binding of the pyrazoles 60b, 60c, 65b, 66b, and the imidazole 76b was lower than that of the indole 75b (98.2 ± 3.2%). Although plasma protein binding was high, 43% of the 1500 most frequently prescribed drugs have protein binding >90%,18 and 12 of the 100 most prescribed drugs have >98% plasma protein binding.19 Plasma protein binding of many antibiotics on the market, including daptomycin, oxacillin, teicoplanin, rifampicin, and clindamycin, is >91%.20–23
Table 2.
In Vitro and in Vivo Evaluation of Selected Oxadiazole Analogs
| PK Parametersa |
|||||
|---|---|---|---|---|---|
| antibiotic | human plasma protein binding (%) | AUC0–8h (μg min/mL) | CL (mL/min/kg) | XTT HepG2 IC50 (μg/mL) | mouse peritonitis (survived/total)b |
| 60b | 97.8 ± 0.3 | 910 | 22.0 | 24.1 ± 1.6 | 4/6 |
| 60c | 94.5 ± 2.1 | 446 | 44.9 | 18.2 ± 2.9 | 2/6 |
| 65b | 91.6 ± 0.3 | 1313 | 15.2 | 3.9 ± 0.8 | 1/6 |
| 66b | 96.4 ± 2.7 | 8261 | 2.4 | 9.8 ± 4.0 | 2/6 |
| 75b | 98.2 ± 3.2 | 1283 | 15.2 | 75.7 ± 7.3 | 5/6 |
| 76b | 93.5 ± 2.4 | 2054 | 9.7 | 31.5 ± 0.5 | 3/6 |
| 1 | 99.9 ± 0.1 | 2650c | 18.9c | 25.8 | ED50 = 40 mg/kgc |
PK parameters after a single iv dose at 20 mg/kg (n = 2 mice per 5 time points).
Mouse peritonitis, S. aureus ATCC27660 given ip at 5 e7 cfu/mL with 5% mucin. Compounds were given iv at 20 mg/kg at 30 min and 7.5 h after infection.
Data from O’Daniel et al.,9 reproduced for the sake of comparison. PK parameters for compound 1 at 50 mg/kg iv.
Fast Pharmacokinetic (PK) Studies.
To rapidly ascertain the PK properties of the compounds, fast PK studies were conducted. These studies involve administration of the compounds using a limited number of animals (n = 2 mice per time point) for a few time points. This allows us to rapidly compare the preliminary PK properties of the compounds, so that full PK studies are conducted only with the most promising compound(s). All compounds were administered intravenously (iv) with a single dose at 20 mg/kg. The alkyne substituted pyrazole 66b had the lowest clearance and the highest systemic exposure, as measured by area under the curve (AUC, Table 2). The highest clearance was observed for 60c, and as a result it had the lowest systemic exposure.
In Vitro Cytotoxicity.
We used the XTT assay with HepG2 cells to evaluate the in vitro toxicity of compounds 60b, 60c, 65b, 66b, 75b, and 76b (Table 2). The highest toxicity was observed for 65b and the lowest for the indole 75b. Compared to the lead 1, indole 75b was 5-fold less toxic.
In Vivo Efficacy.
Compounds 60b, 60c, 65b, 66b, 75b, and 76b were evaluated in the mouse peritonitis model of infection (Table 2). We used the ICR out-bred strain of mice that provides a heterogeneous population, similar to the human situation, thus ensuring the relevance of the antibacterial effect. This animal model of infection is widely used, it is easy to carry out, and the end points (death or survival) are rapidly assessed, making it less resource-intensive compared to other infection models. In addition, excellent correlation between MIC and ED50 has been shown for 14 β-lactam antibiotics using this model.16 The mouse peritonitis infection model continues to be an important model for evaluation of the efficacy of antibiotics against human pathogens.17 We use the iv route of administration in initial efficacy studies, as this allows us to test the efficacy without knowledge of the oral bioavailability of the lead. Evaluation was done at 20 mg/kg. The highest efficacy was observed for indole 75b (Table 2).
Minimal-Bactericidal Concentration (MBC).
The MBC of compound 75b was determined using S. aureus ATCC 29213, S. aureus ATCC 277660, and E. faecium NCTC 7171. For S. aureus ATCC 29213 (an MSSA strain), the MBC was the same as the MIC value, while for the two other strains, the MBC was 2-fold above the MIC values. These data indicated that compound 75b is bactericidal at concentrations that inhibit bacterial growth (Table 4).
Table 4.
Pharmacokinetic Parameters of 75b
| dose (mg/kg) | AUC0–24h (μg min/mL) | AUC0‑∞ (μg min/mL) | Cmax (μg/mL) | Tmax (h) | CL (mL/min/kg) | Vd (L/kg) | t1/2 | F (%) |
|---|---|---|---|---|---|---|---|---|
| 20 iv | 3140 | 3520 | 5.68 | 4.73 | t1/2α = 0.54 min | 97 | ||
| t1/2β = 9.6 h | ||||||||
| 20 po | 3060 | 4060 | 2.34 | 6 | t1/2abs = 0.83 h | |||
| t1/2dist = 15.9 h | ||||||||
| t1/2elim = 18.6 h |
Full PK Study.
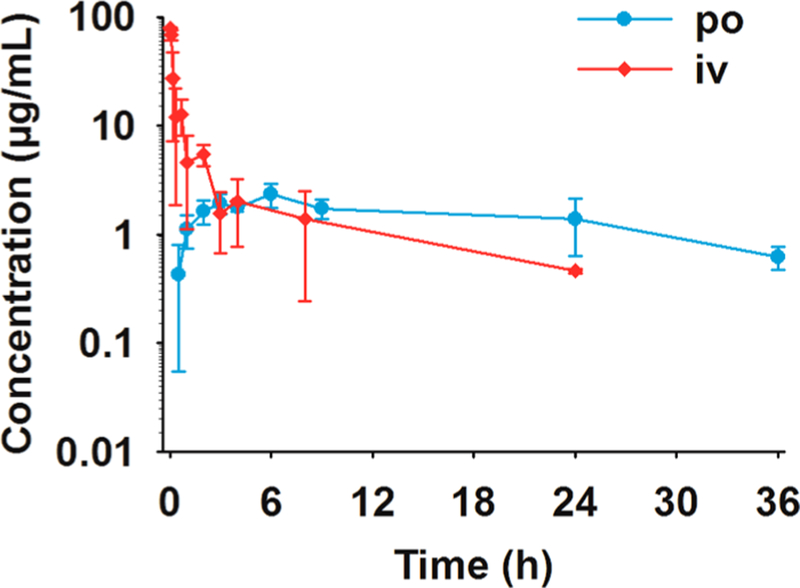
A full PK study was conducted with indole 75b after iv and oral (po) administration. This compound had the lowest in vitro toxicity and the highest efficacy in the mouse peritonitis infection model. Results are summarized in Figure 2 and Table 3. Antibiotic 75b was characterized by low clearance of 5.68 mL/min/kg (less than 10% of hepatic blood flow), a high volume of distribution of 4.73 L/kg, and a terminal half-life after iv administration of 9.6 h. After oral administration, maximum concentrations were observed at 6 h, after which time relatively high concentrations were sustained. The terminal half-life after oral administration was long (18.6 h). The oral bioavailability of 75b at 97% was high, and was similar to that of compound 1.9 Antibiotic 75b had 13-fold higher volume of distribution and 3-fold lower clearance than 1 and was more rapidly absorbed than 1 (t1/2abs of 0.8 h vs 3.3 h9). Thus, antibiotic 75b has superior PK properties compared to 1.
Figure 2.
Pharmacokinetics of 75b after single iv and po administration at 20 mg/kg to mice (n = 3 per time point).
Table 3.
In Vivo Efficacy of Compound 75b in the Mouse Peritonitis Model
| compd | route of administration | dose frequency | ED50 (mg/kg) |
|---|---|---|---|
| 75b | iv | 2 doses given at 30 min and 7.5 h after infection | 7.6 |
| 75b | po | 1.7 | |
| 1 | iv | 40a | |
| 75b | po | single dose given at 1 h after infection | 3.1 |
| 1 | po | 44a | |
| linezolid | po | 2.8 |
Data from O’Daniel et al.,9 reproduced for the sake of comparison.
In Vivo Efficacy of 75b.
Antibiotic 75b was evaluated in the mouse peritonitis infection model using S. aureus ATCC 27660 (MRSA) after iv and po administration (Table 3). The mean effective dose (ED50) values were 7.6 mg/kg and 1.7 mg/ kg after iv and po doses given at 30 min and 7.5 h after infection, respectively. The excellent oral efficacy was attributed to the sustained plasma concentrations of 75b following po administration. The ED50 value after iv administration of 75b is 6-fold better than that of 1. The ED50 of compound 75b was also evaluated after a single po dose given at 1 h after infection. Compound 75b has an excellent ED50 of 3.1 mg/kg, comparable to that of linezolid of 2.8 mg/kg (Table 3) and 14-fold better than that of 1. These data indicated that antibiotic 75b is superior than 1 and comparable in efficacy to linezolid.
CONCLUSION
The recent discovery of the oxadiazole class of anti-MRSA antibiotics provided the opportunity to explore the structural space for these cell-wall-active antibiotics. We have disclosed in the present work the SAR for this class by synthesis and evaluation of 120 structural variants, of which a few dozen exhibit antibacterial activity against S. aureus and MRSA strains. Certain heterocycles with the ability to donate hydrogen bonds are well-tolerated at ring A. Thus, the 4-phenol (57a–59b, 69b–71c) and 4-aniline (72b–73b) analogs are active against S. aureus, but substituents such as phosphates (84c), sulfonamides (87b, 87c), amides (88c), and carboxylic acids (89c) attenuate or abrogate activity. Hydrogen-bond-accepting substituents on ring A abolish activity (e.g., 80a–82b, 91c, 93b, 94a, 99a–101c). Replacing the phenyl moiety of ring A for an aromatic heterocyclic ring retains activity in some cases. Pyrazoles substituted with halogens (60a–62c), a nitro group (63a–63c), an isopropylamino group (65a, 65b), or sp-hybridized groups (66b–68b) are all active, as are indoles (75a–75c) and an imidazole (76b). Pyrazoles containing amino groups with larger substituents (104b–110b) lead to abolishment of antibacterial activity. Heteroaromatic systems such as pyridines (114a, 115a, 120b–122c), triazoles (116b, 117b), and pyroles (78c, 126c, 127c) generally abolish antibacterial activity, as do aliphatic heterocycles (128a–135b).
Although introducing a pyrazole at ring A generally results in compounds that are potently active in vitro with living bacteria, they are also generally cytotoxic. Thus, while pyrazole derivatives 65b and 66b are among the most active compounds reported here, they also exhibit the highest toxicity toward mammalian cells. Replacing the pyrazole with an indole circumvents these toxicity problems, while retaining antibacterial activity. One compound, antibiotic 75b, shows excellent efficacy in vivo with a long half-life, a high volume of distribution, and low clearance. Antibiotic 75b is bactericidal and is 97% orally bioavailable. This class of antibiotics holds great promise in treatment of infections by these difficult human pathogens.
EXPERIMENTAL METHODS
In Silico Screening.
A library of 1.2 million drug-like compounds from the ChemDiv subset of the ZINC database24 was prepared for high-throughput virtual screening against the X-ray structure of PBP2a (PDB ID: 1VQQ).25 The protein was prepared using the Schrödinger Preparation Wizard (Schrödinger, LLC, 2009). The top scoring 10% of the compounds by Schrödinger Glide were cross-docked with Glide-SP,26–28 Autodock,29 Gold-chemscore, Gold-goldscore, and Gold-PLP.30 The top-scoring 2000 poses from each were extracted and refined using Glide-XP mode.26–28 Finally, the best 2500 compounds were clustered according to structural similarity using hierarchical clustering. Binding poses of these clusters were inspected visually. From these, 29 compounds were selected and purchased from ChemDiv for in vitro activity experiments.
Syntheses.
The synthetic procedures for the six compounds chosen for in vivo evaluation are detailed below. These are representative of the methods that were used for the preparation of other derivatives. Purity of the final products was generally >95%, as confirmed by HPLC. Detailed conditions are provided in the HPLC section.
Methyl 1H-indole-5-carboxylate (40).
1H-Indole-5-carboxylic acid (0.843 g, 5.23 mmol), methyl iodide (3.21 g, 22.7 mmol), and NaHCO3 (1.76 g, 20.92 mmol) were stirred in DMF (24 mL) at room temperature for 3 days at which point water (50 mL) was added to the mixture forming a milky precipitate that was extracted with ethyl acetate (3 × 30 mL). The combined organic layer was washed with 5% LiCl (2 × 50 mL) and dried over anhydrous Na2SO4, and the suspension was filtered. The filtrate was concentrated to dryness in vacuo to produce an off white solid, which was purified by silica-gel chromatography (ethyl acetate/hexanes, 1:10) to give the desired product as a white solid (0.820 g, 90%). 1H NMR (400 MHz, CDCl3) δ 3.93 (s, 3H), 6.64 (m, 1H), 7.26 (m, 1H), 7.39 (dt, J = 8.6 Hz, 0.8 Hz, 1H), 7.91 (dd J = 8.6 Hz, 1.6 Hz, 1H), 8.42 (m, 1H), 8.48 (s, 1H). 13C NMR (100 MHz, CDCl3) δ 52.1, 104.2, 111.0, 122.2, 123.6, 124.0, 125.7, 127.7, 138.6, 168.5. HRMS (ESI): calcd for C10H10NO2 [M + H]+ 176.0706, found 176.0710.
5-(4-Chloro-1H-pyrazol-3-yl)-3-(4-(4-(trifluoromethyl)phenoxy)-phenyl)-1,2,4-oxadiazole (60b).
This compound was synthesized using the same procedure as for 63b and purified by silica-gel chromatography (EtOAc/hexanes, 1:4) to yield the product as an off-white powder (58%). Mp 193–195 °C. 1H NMR (400 MHz, CDCl3) δ 7.16–7.19 (m, 4H), 7.66 (d, J = 8.8 Hz, 2H), 8.02 (s, 1H), 8.17–8.20 (m, 2H), 13.58 (br, 1H). 13C NMR (100 MHz, DMSO-d6) δ 110.8, 119.8, 120.6, 122.6, 124.8, 125.1, 128.3 (q, J = 3.6 Hz), 130.1, 130.9, 134.1, 158.8, 159.9, 168.0, 170.2. HRMS (ESI): calcd for C18H11ClF3N4O2 [M + H]+ 407.0517, found 407.0540.
5-(4-Chloro-1H-pyrazol-3-yl)-3-(4-(4-fluorophenoxy)phenyl)-1,2,4-oxadiazole (60c).
This compound was synthesized using the same procedure as for 63b and was purified by silica-gel chromatography (EtOAc/hexanes, 1:6) to yield the product as an off-white powder (61%). Mp 212–214 °C. 1H NMR (400 MHz, CDCl3) δ 7.07–7.13 (m, 6H), 7.91 (s, 1H), 8.16 (d, J = 9.2 Hz, 2H). 13C NMR (100 MHz, DMSO-d6) δ 110.1, 116.8, 117.0, 117.9, 120.6, 121.8, 121.9, 129.3, 130.3, 133.5, 151.2, 151.3, 157.6, 160.0, 160.2, 167.4, 169.4. HRMS (ESI): calcd for C17H11ClFN4O2 [M + H]+ 357.0549, found 357.0544.
5-(4-Iodo-1H-pyrazole-3-yl)-3-(4-(4-trifluoromethyl)phenoxy)-phenyl)-1,2,4-oxadiazole (62b).
This compound was synthesized according to the procedure for 63b and was purified by silica-gel chromatography (EtOAc/hexanes, 1:3.5) to afford compound 62b as an off-white powder (64.0%). Mp 222–225 °C. 1H NMR (400 MHz, DMSO-d6) δ 7.29–7.36 (m, 4H), 7.80 (d, J = 8.8 Hz, 2H), 8.16 (d, J = 8.4 Hz, 2H), 8.31 (s, 1H). 13C NMR (100 MHz, DMSO-d6) δ 60.4, 119.1, 120.0, 122.1, 122.8, 124.1, 124.4, 125.5, 127.6 (q, J = 3.5 Hz), 129.4, 137.4, 138.3, 158.1, 159.2, 167.3, 170.4. HRMS (ESI): calcd for C18H11F3IN4O2 [M + H]+ 498.9873, found 498.9879.
5-(4-Nitro-1H-pyrazole-3-yl)-3-(4-(4-(trifluoromethyl)phenoxy)-phenyl)-1,2,4-oxadiazole (63b).
4-Nitropyrazole-3-carboxylic acid (34, 0.24 g, 1.50 mmol) was dissolved in SOCl2 (2.2 mL, 30.548 mmol), and the solution was stirred at reflux for 2 h. The excess SOCl2 was evaporated to dryness in vacuo, and the residue was taken up in toluene (15 mL) and pyridine (0.61 mL, 7.0 mmol), followed by the addition of (Z)-N′-hydroxy-4-(4-(trifluoromethyl)phenoxy)-benzimidamide (6b, 0.30 g, 1.0 mmol). The resultant mixture was stirred at reflux overnight. The solvent was evaporated to dryness in vacuo, and the residue was purified by silica-gel chromatography (CH2Cl2/MeOH, 100:1) to afford the title compound as a yellow powder (0.20 g, 48.0%). Mp 204–206 °C. 1H NMR (400 MHz, CDCl3) δ 7.15–7.18 (m, 4H), 7.65 (d, J = 8.4 Hz, 2H), 8.18–8.20 (m, 2H), 8.55 (s, 1H). 13C NMR (100 MHz, CDCl3) δ 120.0, 120.5, 122.2, 123.5, 125.0, 125.3, 126.2, 128.3 (q, J = 3.5 Hz), 130.2, 133.1, 134.8, 159.2, 159.8, 168.4, 169.3. HRMS (ESI): calcd for C18H10F3N5 NaO4 [M + Na]+ 440.0577, found 440.0579.
3-(3-(4-(4-(Trifluoromethyl)phenoxy)phenyl)-1,2,4-oxadiazol-5-yl)-1H-pyrazol-4-amine (64b).
Anhydrous THF (5 mL) was slowly added to a mixture of sulfur (0.22 g, 6.86 mmol) and sodium borohydride (74.4 mg, 1.96 mmol) in a round-bottom flask at room temperature. After stirring for 10 min, compound 63b (0.10 g, 0.24 mmol) in THF (2.0 mL) was added dropwise to the above mixture before heating it to 65 °C for 2.5 h. Upon cooling to room temperature, water (6 mL) and diethyl ether (6 mL) were added, and the mixture was stirred for 5 min. The layers were separated, and the aqueous portion was extracted with diethyl ether (3 × 12 mL). The combined organic layer was washed with brine and dried (Na2SO4) and was concentrated to dryness in vacuo. The residue was purified by silica-gel chromatography (EtOAc/hexanes, 1:2 to 2:1) to yield the title compound as a yellow foam (75.4 mg, 80.0%). Mp 184–187 °C. 1H NMR (400 MHz, CDCl3) δ 7.07–7.13 (m, 4H), 7.48 (s, 1H), 7.63 (d, J = 8.4 Hz, 2H), 8.08 (d, J = 8.4 Hz, 2H). 13C NMR (100 MHz, DMSO-d6) δ 118.3, 119.0, 119.2, 122.3, 124.1, 126.0, 126.3, 127.5 (q, J = 4.0 Hz), 129.5, 132.4, 158.6, 159.2, 167.3, 170.0. HRMS (ESI): calcd for C18H13F3N5O2 [M + H]+ 388. 1016, found 388.1010.
N-Isopropyl-3-(3-(4-(4-(trifluoromethyl)phenoxy)phenyl)-1,2,4-oxadiazol-5-yl)-1H-pyrazol-4-amine (65b).
To a solution of compound 64b (75.4 mg, 0.19 mmol) and acetone (17 μL, 0.23 mmol) in 5 mL of CH2Cl2 were added activated 3 Å molecular sieves and sodium triacetoxyborohydride (62.0 mg, 0.29 mmol). The mixture was stirred at room temperature for 7 days. The mixture was filtered through Celite, which was washed with EtOAc. The filtrate was concentrated to dryness, and the residue was purified by silica-gel chromatography (EtOAc/hexanes, 1:3) to afford the desired product as a light green powder (44 mg, 53%). Mp 158–161 °C. 1H NMR (400 MHz, CDCl3) δ 1.33 (s, 3H), 1.35 (s, 3H), 3.50–3.56 (m, 1H), 7.15–7.20 (m, 4H), 7.47 (s, 1H), 7.65 (d, J = 8.8 Hz, 2H), 8.18–8.21 (m, 2H). 13C NMR (100 MHz, CDCl3) δ 23.2, 47.8, 116.0, 119.1, 119.6, 122.8, 122.9, 125.9, 126.2, 127.6 (q, J = 3.7 Hz), 129.8, 135.7, 158.9, 159.6, 167.6, 170.6. HRMS (ESI): calcd for C21H19F3N5O2 [M + H]+ 430. 1485, found 430.1489.
5-(4-Ethynyl-1H-pyrazol-3-yl)-3-(4-(4-(trifluoromethyl)phenoxy)-phenyl)-1,2,4-oxadiazole (66b).
Compound 103b (0.12 g, 0.26 mmol), KF (31 mg, 0.53 mmol), and 10 mL of MeOH were placed in a round-bottom flask. The mixture was stirred at room temperature for 17 h. After the completion of the reaction, the solvent was removed in vacuo, and the residue was purified by silica-gel chromatography (EtOAc/hexanes, 1:4) to give the compound as an off-white powder (84.7 mg, 81%). Mp 206–209 °C. 1H NMR (500 MHz, CDCl3) δ 3.39 (s, 1H), 7.14–7.19 (m, 4H), 7.64 (d, J = 8.5 Hz, 2H), 8.06 (s, 1H), 8.21–8.23 (m, 2H). 13C NMR (100 MHz, CDCl3) δ 72.8, 83.1, 119.2, 119.8, 122.5, 122.9, 125.6, 126.0, 126.4, 127.6 (q, J = 3.6 Hz), 129.9, 159.1, 159.5, 168.5, 169.6. HRMS (ESI): calcd for C20H12F3N4O2 [M + H]+ 397.0907, found 397.0914.
5-(1H-Indol-5-yl)-3-(4-(4-(trifluoromethyl)phenoxy)phenyl)-1,2,4-oxadiazole (75b).
A solution of N′-hydroxy-4-(4-(trifluoromethyl)-phenoxy)benzimidamide (6b, 1.02 g, 3.44 mmol) in anhydrous THF (15 mL) was stirred under an argon atmosphere, and sodium hydride (60% in mineral oil, 0.172 g, 4.30 mmol) was added to the flask. The mixture was left to stir for 1 h at room temperature, and then a solution of methyl 1H-indole-5-carboxylate (40, 0.302 g, 1.72 mmol) in anhydrous THF (15 mL) was added and the mixture heated at reflux for 3.5 h. Once the solution had cooled to room temperature, water (50 mL) was added, and the resulting mixture was extracted with ethyl acetate (3 × 50 mL). The combined organic layer was dried over anhydrous Na2SO4 and then filtered, and the filtrate was evaporated to leave an orange residue. This was purified using column chromatography on silica gel (dichloromethane/hexanes, 9:1) to give the desired product as a white solid (0.190 g, 26%). Mp 138–141 °C. 1H NMR (400 MHz, CDCl3) δ 6.69 (m, 1H), 7.11 (d, J = 9.0 Hz, 2H), 7.14 (d, J = 8.9 Hz, 2H), 7.29 (m, 1H), 7.48 (dt J = 8.5 Hz, 1.6 Hz 1H), 7.61 (d, J = 9.0 Hz 2H), 8.03 (dd, J = 8.6 Hz, 1 Hz, 1.6 Hz 1H), 8.20 (d, J = 8.9 Hz, 2H), 8.6 (m, 1H), 8.6 (s, 1H). 13C NMR (100 MHz, CDCl3) δ 104.2, 111.9, 116.2, 118.9, 119.8, 122.1, 122.3, 123.5, 124.3 (q, J = 272.2 Hz), 125.9 (q, J = 32.9 Hz), 126.2, 127.5 (q, J = 3.6 Hz), 128.2, 129.7, 138.4, 158.6, 159.7, 168.3, 177.4. 19F NMR (376 MHz, CDCl3) δ 99.9 (s, 3F). HRMS (ESI): calcd for C23H14F3N3O2 [M + H]+ 422.1111, found 422.1078.
5-(1H-Imidazol-4-yl)-3-(4-(4-(trifluoromethyl)phenoxy)phenyl)-1,2,4-oxadiazole (76b).
The compound was synthesized according to the procedure used for 63b and purified by silica-gel chromatography (EtOAc/hexanes, 1:4 to 1:1.5) to afford the product as an off-white powder (72.0%). Mp 243–245 °C. 1H NMR (400 MHz, DMSO-d6) δ 7.29–7.31 (m, 4H), 7.80 (d, J = 8.8 Hz, 2H), 7.99 (s, 1H), 8.12 (d, J = 8.4 Hz, 2H), 8.24 (s, 1H), 13.00 (br, 1H). 13C NMR (100 MHz, DMSO-d6) δ 119.2, 119.8, 122.4, 122.5, 124.1, 124.4, 126.3, 127.7 (d, J = 3.0 Hz), 129.4, 138.3, 158.0, 159.2, 167.2, 172.2. HRMS (ESI): calcd for C18H12 F3N4O2 [M + H]+ 373.0907, found 373.0911.
3-(4-(4-Trifluoromethyl)phenoxy)phenyl)-5-(4-((trimethylsilyl)-ethynyl-1H-pyrazol-3-yl)-1,2,4-oxadiazole (103b).
Compound 62b (0.19 g, 0.38 mmol) was placed in a 10 mL round-bottom flask, and 5 mL of anhydrous THF was added to the flask. Ethynyltrimethylsilane (0.12 mL, 0.84 mmol), Pd(Ph3P)2Cl2 (21.4 mg, 0.03 mmol), CuI (9.00 mg, 0.05 mmol), and Et3N (0.14 mL, 0.99 mmol) were added to the above mixture. The resultant solution was heated in reflux for 5 h. After completion of the reaction, the solvent was evaporated to dryness in vacuo, and the residue was purified by silica-gel chromatography (EtOAc/hexanes, 1:6) to afford the product as a white foam (0.14 g, 79.5%). 1H NMR (400 MHz, CDCl3) δ 0.32 (s, 9H), 7.15–7.19 (m, 4H), 7.65 (d, J = 8.8 Hz, 2H), 8.18 (s, 1H), 8.22 (d, J = 8.8 Hz, 2H). 13C NMR (100 MHz, CDCl3) δ 0.1, 93.7, 100.8, 105.5, 119.2, 119.7, 122.6, 122.9, 125.6, 126.0, 126.4, 127.6 (q, J = 3.7 Hz), 129.9, 135.6, 138.5, 159.1, 159.4, 159.5, 168.5, 169.8. HRMS (ESI): calcd for C23H20F3N4O2Si [M + H]+ 469.1303, found 469.1336.
High Performance Liquid Chromatography (HPLC).
The system used was a PerkinElmer Series 200 Chromatography System (PerkinElmer, Waltham, MA) equipped with an autosampler, UV–vis detector, LC pump, NCI 900 Network Chromatography Interface, and 600 Series Link Chromatography Interface. The samples were analyzed on a Zorbax RX-C8 analytical column (5.0 μm, 4.6 mm id × 250 mm, Agilent Technologies, Santa Clara, CA). The mobile phase consisted of isocratic elution for 10 min with a 1:1 mixture of water/ 0.1% trifluoroacetic acid (TFA) and acetonitrile/0.1% TFA at a flow rate of 1.0 mL/min, with the effluent monitored by UV detection (detection window set to 250–255 nm).
Microbial Strains.
The ESKAPE organisms (E. faecium NCTC (ATCC 19734), S. aureus ATCC 29213, K. pneumonia ATCC 700603, A. baumannii ATCC 17961, P. aeruginosa ATCC 17853, E. aerogenes ATCC 35029) and E. coli ATCC 25922) in the initial screen, S. aureus ATCC 27660, S. epidermis ATCC 35547, S. hemolyticus ATCC 29970, S. oralis ATCC 9811, S. pyogenes ATCC 49399, B. cereus ATCC 13061, B. licheniformis ATCC 12759, and E. faecalis ATCC 29212 were purchased from the American Type Culture Collection (Manassas, VA). S. aureus strains NRS100, NRS119, NRS120, VRS1, and VRS2 were obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus (Chantilly, VA). E. faecalis strains 201 and 99, and E. faecium strains 119–39A and 106 were collected from Wayne State University School of Medicine.
Minimal-Inhibitory Concentration (MIC) Determination.
The procedure for MIC determination was the same as that previously reported.9
Minimal-Bactericidal Concentration (MBC) Determination.
The MBC of antibiotic 75b was determined by incubation of 1.5 × 105 cells of S. aureus ATCC29213, S. aureus ATCC 27660, and E. faecium NCTC7171 at MIC, 2× MIC, and 4× MIC. Aliquots of 10 μL (corresponding to 1.5 × 104 cells) were plated on agar plates and incubated for 48 h, and colonies were counted in the presence and absence of antibiotic 75c.9 The MBC was the concentration of antibiotic 75b that resulted in >1000-fold reduction in colonies.
Plasma Protein Binding.
Plasma protein binding was determined using human plasma and a rapid equilibrium dialysis device (Pierce Biotechnology, Thermo Scientific, Waltham, MA). Human plasma was thawed and centrifuged at 1200 g for 10 min to remove particulates. A 200 μL aliquot of human plasma was added to the sample chamber and 350 μL of 0.1 M phosphate buffered saline (pH 7.4) containing 0.15 mM sodium chloride was added to the adjacent chamber. A 2 μL aliquot of a stock solution of the compounds at a concentration of 1 mM in DMSO was diluted with human plasma to a final drug concentration of 10 μM and added to the sample chamber. The compounds were dialyzed at 37 °C in an orbital shaker for 6 h. Aliquots (50 μL) were taken from the buffer chamber (representing the free concentration) and from the plasma chamber (representing the total concentration) and mixed with 100 μL of internal standard in acetonitrile to a final concentration of 5 μM. Samples were analyzed by UPLC with UV detection at 285 nm. The plasma protein binding ratio (B%) was calculated according to the following equation
where Cp and Cf are the total plasma concentration and the free concentration of compound, respectively.
XTT Cytotoxicity Assay.
The XTT cytotoxicity assay was performed in triplicated using HepG2 cells (ATCC HB-8065), as previously described.9 The IC50 values were calculated with GraphPad Prism 5 (GraphPad Software, Inc., San Diego, CA).
Animals.
Female ICR mice (6–8 weeks old, ∼20-g body weight) were used for the PK and peritonitis studies. Animals were purchased from Harlan Laboratories, Inc. (Indianapolis, IN) and given Teklad 2019 Extruded Rodent Diet and water ad libitum. Mice were maintained in polycarbonate shoebox cages with 1/4 in. corncob (The Andersons Inc., Maumee, OH) and Alpha-dri (Sheperd Specialty Papers, Inc., Richland, MI) bedding under 12-h light/12-h dark cycle at 72 ± 2 °F. All procedures involving animals were approved by the University of Notre Dame Institutional Animal Care and Use Committee.
Fast Pharmacokinetic (PK) Studies.
For fast PK studies, the compounds were dissolved in 10% DMSO/25% Tween-80/65% water at a concentration of 5 mg/mL. The dosing formulations were sterilized by filtration through a 0.2 μm, 13 mm diameter PTFE membrane attached to an Acrodisc syringe filter (Pall Life Sciences, Ann Arbor, MI). Mice (n = 2 per time point) were given 100 μL of the test compound(s) intravenously (iv), equivalent to 20 mg/kg. Terminal blood was collected at 5 min, 40 min, 2 h, 4 h, and 8 h; blood was centrifuged at 1200 g for 10 min to harvest plasma.
Full PK Studies.
Antibiotic 75b was dissolved in 10% DMSO/25% Tween-80/65% water at a concentration of 5 mg/mL. Mice (n = 3 per time point per route of administration) were administered 100 μL of 75b (equivalent to 20 mg/kg) iv. A separate group of mice was given 100 μL of 75b (equivalent to 20 mg/kg) orally (po). Terminal blood was collected in heparin by cardiac puncture at 2, 5, 10, 20, and 40 min, and at 1, 2, 3, 4, 8, and 24 h after iv administration and at 0.5, 1, 2, 3, 4, 6, 9, 24, and 36 h after po administration. Blood was centrifuged at 1200 g for 10 min to obtain plasma. Plasma samples were stored at −80 °C until analysis.
Bioanalytical Method.
Plasma (50 μL aliquot) was mixed with 100 μL of acetonitrile containing internal standard (final concentration 8 μg/mL). After centrifugation at 10 000 g for 10 min, the supernatant was analyzed by ultraperformance liquid chromatography (UPLC) with UV detection at 285 nm. A Waters Acquity UPLC System (Waters Corporation, Milford, MA), consisting of a binary pump, an autosampler, a column heater, and a photodiode array detector, was used. An Acquity UPLC C18 1.7 μm, 2.1 mm id × 50 mm column was used. Elution was at 0.5 mL/min with 70% A/30% B for 2 min, followed by a 10 min linear gradient to 10% A/90% B, and then 70% A/30% B for 2 min, where A = 0.1% formic acid/water and B = 0.1% formic acid/acetonitrile. Monitoring was by UV detection at 285 nm. Calibration curves for each compound were prepared in control plasma containing internal standard. The concentrations in the PK samples were obtained using peak area ratio to the internal standard and the calibration curve regression analysis parameters. The methods were linear from 0.01 to 100 μg/mL; coefficients of determination R2 range from 0.98 to 0.99.
Pharmacokinetic Parameters.
The area under the curve (AUC), clearance (CL), volume of distribution (Vd), and terminal half-life were calculated using Phoenix WinNonlin 6.3 (Certara LP, St Louis, MO) noncompartmental analysis using uniform weighing. Half-lives were estimated from the linear portion of the initial or terminal phase of the concentration–time data by linear regression, where the slope of the line was the rate constant k and t1/2 = ln 2/k.
Mouse Peritonitis Studies.
The mouse peritonitis model was used with S. aureus ATCC 27660, as described previously.9 The final bacterial inocula contained 5 × 107 cfu/mL and 5% mucin (Sigma-Aldrich Chemical Co., St Louis, MO). Just prior to inoculation, bacteria at 108 cfu/mL were mixed 1:1 with 10% mucin. Mice (n = 6 per group) were given 0.5 mL of the bacterial inocula intraperitoneally. Mice were given two iv doses of the compounds at 30 min and 7.5 h after infection by tail vein injection. Vehicle and positive control (vancomycin at 5 mg/kg) groups were included. Mice were monitored for 48 h, at which time the number of surviving mice were counted.
ED50 Determination.
The effective dose that results in survival of 50% of the mice was determined using Probit analysis (XLSTAT, New York, NY). Groups of six mice per dose level were evaluated in the mouse peritonitis infection model at iv doses of 2.5, 5, 7.5, 10, 15, and 20 mg/kg and after po doses at 2.5, 5, 10, 20, and 40 mg/kg. The doses were given at 30 min and 7.5 h after infection. In addition, ED50 values were determined for compound 75b and linezolid after a single po dose given at 1 h after infection.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Grant AI090818 from the National Institutes of Health (to M.C. and S.M.).
ABBREVIATIONS USED
- AUC
area under the curve
- DMAP
dimethyl amino pyridine
- DMF
N,N-dimethylformamide
- DMSO
dimethyl sulfoxide
- Et3N
triethylamine
- MIC
minimum-inhibitory concentration
- MRSA
methicillin-resistant Staphylococcus aureus
- PK
pharmacokinetics
- TBS
tert-butyldimethylsilyl
- THF
tetrahydrofuran
- TLC
thin layer chromatography
- UPLC
ultraperformance liquid chromatography
- UV
ultraviolet
Footnotes
ASSOCIATED CONTENT
Supporting Information
General experimental procedures for MIC determination and further information on synthetic procedures. Spectral data for reported compounds and their MIC values against Gram-positive organisms of the ESKAPE panel. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- (1).Antibiotic Resistance Threats in the United States, 2013; Center for Disease Control, U.S. Government Printing Office: Washington, DC, 2013. [Google Scholar]
- (2).Kaka AS; Rueda AM; Shelburne SA; Hulten K; Hamill RJ; Musher DM. Bactericidal activity of orally available agents against methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother 2006, 58, 680–683. [DOI] [PubMed] [Google Scholar]
- (3).Appelbaum PC. The emergence of vancomycin-intermediate and vancomycin-resistant Staphylococcus aureus. Clin. Microbiol. Infect 2006, 12, 16–23. [DOI] [PubMed] [Google Scholar]
- (4).Tsiodras S; Gold HS; Sakoulas G; Eliopoulos GM; Wennersten C; Venkataraman L; M, R. C., Jr.; Ferraro MJ. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet 2001, 358, 207–208. [DOI] [PubMed] [Google Scholar]
- (5).Prystowsky J; Siddiqui F; Chosay J; Shinabarger DL; Millichap J; Peterson LR; Noskin GA. Resistance to linezolid: Characterization of mutations in rRNA and comparison of their occurrences in vancomycin-resistant enterococci. Antimicrob. Agents Chemother 2001, 45, 2154–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Silverman JA; Oliver N; Andrew T; Li T. Resistance studies with daptomycin. Antimicrob. Agents Chemother 2001, 45, 1799–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Sabol K; Patterson JE; Lewis JS; Owens A; Cadena J; Jorgensen JH. Emergence of daptomycin resistance in Enterococcus faecium during daptomycin therapy. Antimicrob. Agents Chemother 2005, 49, 1664–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Mendes RE; Tsakris A; Sader HS; Jones RN; Biek D; McGhee P; Appelbaum PC; Kosowska-Shick K. Characterization of methicillin-resistant Staphylococcus aureus displaying increased MICs of ceftaroline. J. Antimicrob. Chemother 2012, 67, 1321–1324. [DOI] [PubMed] [Google Scholar]
- (9).O’Daniel PI; Peng Z; Pi H; Testero SA; Ding D; Spink E; Leemans E; Boudreau MA; Yamaguchi T; Schroeder VA; Wolter WR; Llarrull LI; Song W; Lastochkin E; Kumarasiri M; Antunes NT; Espahbodi M; Lichtenwalter K; Suckow MA; Vakulenko S; Mobashery S; Chang M. Discovery of a new class of non-β-lactam inhibitors of penicillin-binding proteins with Gram-positive antibacterial activity. J. Am. Chem. Soc 2014, 136, 3664–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Testero SA; Fisher JF; Mobashery S. β-Lactam antibiotics In Burger’s Medicinal Chemistry, Drug Discovery and Development; Abraham DJ, Rotella DP, Eds.; Wiley and Sons: Hoboken, NJ, 2010; Vol. 7, pp 259–404. [Google Scholar]
- (11).Fisher JF; Meroueh SO; Mobashery S. Bacterial resistance to β-lactam antibiotics: compelling opportunism, compelling opportunity. Chem. Rev 2005, 105, 395–424. [DOI] [PubMed] [Google Scholar]
- (12).Fisher JF; Mobashery S. Enzymology of antibacterial resistance In Comprehensive Natural Products Chemistry II, Chemistry and Biology; Mander L, Liu HW, Eds.; Elsevier: Oxford, 2010; pp 443–487. [Google Scholar]
- (13).Llarrull LI; Testero SA; Fisher JF; Mobashery S. The future of the β-lactams. Curr. Opin. Microbiol 2010, 13, 551–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Leemans E; Fisher JF; Mobashery S. The β-lactam antibiotics: their future in the face of resistance In Antimicrobials: Novel and Old Molecules in the Fight Against Multi-resistant Bacteria; Marinelli F, Genilloud O, Eds.; Springer Verlag: Berlin, 2014; pp 59–85. [Google Scholar]
- (15).Boucher HW; Talbot GH; Bradley JS; Edwards JE; Gilbert D; Rice LB; Scheld M; Spellberg B; Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis 2009, 48, 1–12. [DOI] [PubMed] [Google Scholar]
- (16).Frimodt-Meller N; Bentzen MW; Thomsen VF. Experimental infection with Streptococcus pneumoniae in mice: correlation of in vitro activity and pharmacokinetic parameters with in vivo effect for 14 cephalosporins. J. Infect. Dis 1986, 154, 511–517. [DOI] [PubMed] [Google Scholar]
- (17).Ford CW; Hamel JC; Wilson DM; Moerman JK; Stapert D; Yancey RJ; Hutchinson DK; Barbachyn MR; Brickner SJ. In vivo activities of U-100592 and U-100766, novel oxazolidinone antimicrobial agents, against experimental bacterial infections. Antimicrob. Agents Chemother 1996, 40, 1508–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Kratochwil NA; Huber W; Müller F; Kansy M; Gerber PR. Predicting plasma protein binding of drugs: A new approach. Biochem. Pharmacol 2002, 64, 1355–1374. [DOI] [PubMed] [Google Scholar]
- (19).Smith DA; Di L; Kerns EH. The effect of plasma protein binding on in vivo efficacy: misconceptions in drug discovery. Nat. Rev. Drug Discovery 2010, 9, 929–939. [DOI] [PubMed] [Google Scholar]
- (20).Dykhuizen RS; Harvey G; Stephenson N; Nathwani D; Gould IM. Protein binding and serum bactericidal activities of vancomycin and teicoplanin. Antimicrob. Agents Chemother 1995, 39, 1842–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Kochansky CJ; McMasters DR; Lu P; Koeplinger KA; Kerr HH; Shou M; Korzekwa KR. Impact of pH on plasma protein binding in equilibrium dialysis. Mol. Pharmaceutics 2008, 5, 438–448. [DOI] [PubMed] [Google Scholar]
- (22).Lee BL; Sachdeva M; Chambers HF. Effect of protein binding of daptomycin on MIC and antibacterial activity. Antimicrob. Agents Chemother 1991, 35, 2505–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Sargent P. Oxacillin for injection USP. Oxacillin package insert, 2013.
- (24).Irwin JJ; Shoichet BK. ZINC A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model 2005, 45, 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Lim D; Strynadka NCJ. Structural basis for the [beta] lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat. Struct. Mol. Biol 2002, 9, 870–876. [DOI] [PubMed] [Google Scholar]
- (26).Friesner RA; Banks JL; Murphy RB; Halgren TA; Klicic JJ; Mainz DT; Repasky MP; Knoll EH; Shelley M; Perry JK; Shaw DE; Francis P; Shenkin PS. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem 2004, 47, 1739–1749. [DOI] [PubMed] [Google Scholar]
- (27).Halgren TA; Murphy RB; Friesner RA; Beard HS; Frye LL; Pollard WT; Banks JL. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem 2004, 47, 1750–1759. [DOI] [PubMed] [Google Scholar]
- (28).Friesner RA; Murphy RB; Repasky MP; Frye LL; Greenwood JR; Halgren TA; Sanschagrin PC; Mainz DT. Extra precision Glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein–ligand complexes. J. Med. Chem 2006, 49, 6177–6196. [DOI] [PubMed] [Google Scholar]
- (29).Morris GM; Huey R; Lindstrom W; Sanner MF; Belew RK; Goodsell DS; Olson AJ. Autodock4 and Autodocktools4: automated docking with selective receptor flexibility. J. Comput. Chem 2009, 30, 2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Verdonk ML; Cole JC; Hartshorn MJ; Murray CW; Taylor RD. Improved protein–ligand docking using GOLD. Proteins: Struct., Funct., Bioinf 2003, 52, 609–623. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.