Abstract
BACKGROUND & AIMS:
Early-onset gastric cancer, which develops in patients younger than most gastric cancers, is usually detected at advanced stages, has diffuse histologic features, and occurs more frequently in women. We investigated somatic genomic alterations associated with the unique characteristics of sporadic diffuse gastric cancers (DGCs) from younger patients.
METHODS:
We conducted whole exome and RNA sequence analyses of 80 resected DGC samples from patients 45 years old or younger in Korea. Patients with pathogenic germline mutations in CDH1, TP53, and ATM were excluded from the onset of this analysis, given our focus on somatic alterations. We used MutSig2CV to evaluate the significance of mutated genes. We recruited 29 additional early-onset Korean DGC samples and performed SNP6.0 array and targeted sequencing analyses of these 109 early-onset DGC samples (54.1% female, median age, 38 years). We compared the SNP6.0 array and targeted sequencing data of the 109 early-onset DGC samples with those from diffuse-type stomach tumor samples collected from 115 patients in Korea who were 46 years or older (late onset) at the time of diagnosis (controls; 29.6% female, median age, 67 years). We compared patient survival times among tumors from different subgroups and with different somatic mutations. We performed gene silencing of RHOA or CDH1 in DGC cells with small interfering RNAs for cell-based assays.
RESULTS:
We identified somatic mutations in the following genes in a significant number of early-onset DGCs: the cadherin 1 gene (CDH1), TP53, ARID1A, KRAS, PIK3CA, ERBB3, TGFBR1, FBXW7, RHOA, and MAP2K1. None of 109 early-onset DGC cases had pathogenic germline CDH1 mutations. A higher proportion of early-onset DGCs had mutations in CDH1 (42.2%) or TGFBR1 (7.3%) compared with control DGCs (17.4% and 0.9%, respectively) (P < .001 and P = .014 for CDH1 and TGFBR1, respectively). In contrast, a smaller proportion of early-onset DGCs contained mutations in RHOA (9.2%) than control DGCs (19.1%) (P = .033). Late-onset DGCs in The Cancer Genome Atlas also contained less frequent mutations in CDH1 and TGFBR1 and more frequent RHOA mutations, compared with early-onset DGCs. Early-onset DGCs from women contained significantly more mutations in CDH1 or TGFBR1 than early-onset DGCs from men. CDH1 alterations, but not RHOA mutations, were associated with shorter survival times in patients with early-onset DGCs (hazard ratio, 3.4; 95% confidence interval, 1.5–7.7). RHOA activity was reduced by an R5W substitution—the RHOA mutation most frequently detected in early-onset DGCs. Silencing of CDH1, but not RHOA, increased migratory activity of DGC cells.
CONCLUSIONS:
In an integrative genomic analysis, we found higher proportions of early-onset DGCs to contain somatic mutations in CDH1 or TGFBR1 compared with late-onset DGCs. However, a smaller proportion of early-onset DGCs contained somatic mutations in RHOA than late-onset DGCs. CDH1 alterations, but not RHOA mutations, were associated with shorter survival times of patients, which might account for the aggressive clinical course of early-onset gastric cancer. Female predominance in early-onset gastric cancer may be related to relatively high rates of somatic CDH1 and TGFBR1 mutations in this population.
Keywords: Young, Stomach, Age, Genomic
Gastric cancer (GC), the third leading cause of cancer death worldwide, is a disease of older individuals, predominantly males.1 Among 2 major histologic subtypes of GC, diffuse-type GC (DGC) increased in incidence in the United States until 2000.2 Although the incidence of DGC remains consistently high, relatively small numbers of DGCs (n = 29–69) have been represented in whole exome sequencing (WES) studies performed thus far,3–7 and pathogenic mutations for DGC have not yet been fully discovered. Notably, the clinicopathologic features of DGCs differ according to age at diagnosis.8 Sporadic GC that is diagnosed in younger patients, termed early-onset (EO) GC, is notable for its strong enrichment of diffuse histology and female predominance, in addition to a high propensity for metastasis.8 The molecular basis for these unique phenotypes of EOGC has not been elucidated. In Korea, which has the highest rate of GC worldwide,1 15% of GCs develop before the age of 45 years.9 In the United States, increasing rates of GCs are noted in individuals younger than 40 years old.10 However, no studies have systematically evaluated somatic genomic alterations associated with the unique characteristics of EOGC due to its comparative rarity. Therefore, we undertook the largest WES study of DGCs to date in a population enriched with EODGC.
Materials and Methods
We conducted WES and RNA sequencing analyses with 80 resected tumors and matched germline samples that were collected from EO (≤45 years) patients with DGC in Korea (EODGC-WES), using HiSeq platforms (Illumina, San Diego, CA). The WES and RNAseq data of EODGC-WES were analyzed by the same centers at which The Cancer Genome Atlas (TCGA) stomach cancer samples were analyzed.6 To achieve a statistical power adequate for comparison analyses, we expanded the EODGC-WES cohort by recruiting 29 additional EO Korean DGC samples (EODGC, n = 109; Supplementary Table 1). EODGC tumors were compared with a control set of diffuse-type stomach cancers that were collected from 115 Korean patients who were ≥46 years at the time of diagnosis (late-onset [LO] DGC). We compared the SNP6.0 array (Affymetrix, Santa Clara, CA) and targeted DNA sequencing (Life Technologies, Carlsbad, CA) data between EODGCs and LODGCs (Figure 1A). The EODGCs and LODGCs were similar with regard to the proportion of metastatic disease at diagnosis and tumor purity as estimated by the histopathologic evaluation of top slides. EODGC-WES tumors were also compared for WES data with 61 LO (LO,≥46 years) diffuse tumors that originated in TCGA study (LODGC-TCGA). In LODGC-TCGA, 70% of the patients were Caucasians, including 26% females (median age, 63 years; range, 46–82 years). The CDH1 alteration was defined by one of the following somatic alterations in the CDH1 gene encoding E-cadherin: RNA splicing aberrations, homozygous deletions, promoter hypermethylation, single nucleotide variations (SNVs), and indels. All authors had access to the study data, and reviewed and approved the final manuscript. Methods are available in the Supplementary Material.
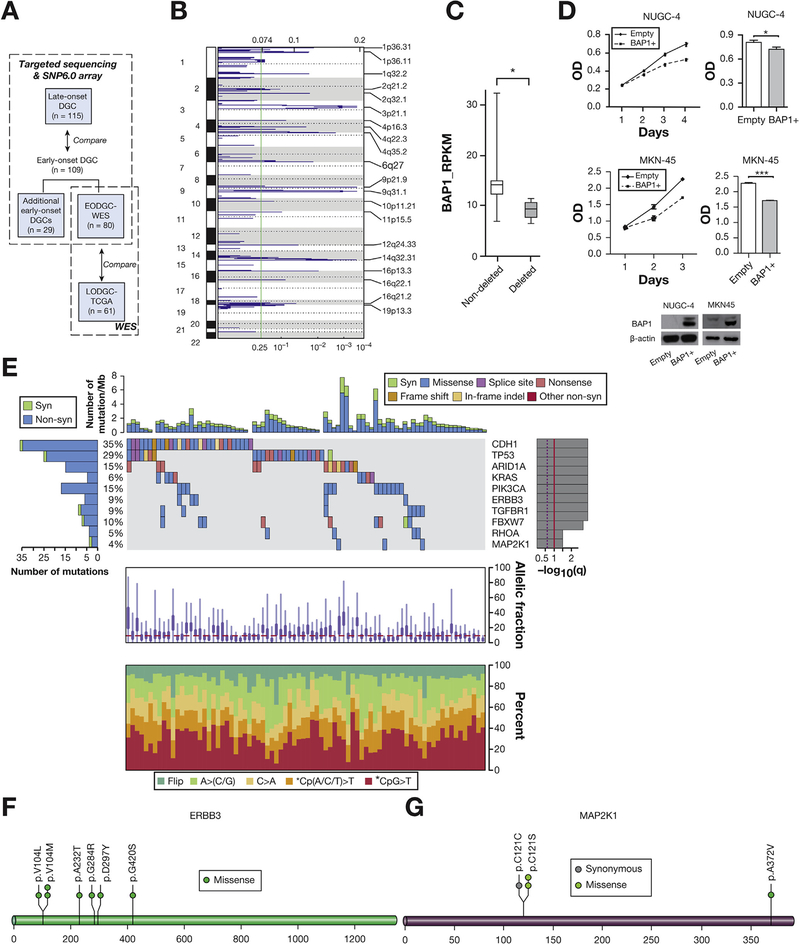
Figure 1.
Significant deletions and somatic mutations as identified by the WES data of EODGC-WES (A) Study scheme. (B) Significant deletions in EODGC-WES according to ReCapSeg and ISAR-GISITC analysis. (C) Underexpression of BAP1 messenger RNA in 6 3p21.1-deleted EODGC-WES tumors relative to 74 EODGC-WES tumors without 3p21.1 deletion. *q = .001 (D) The effect of BAP1 overexpression on the proliferation of NUGC-4 (top) and MKN-45 (middle) cells. Right histograms, final optical density (OD) on the last day of growth; mean ± SEM of at least 3 independent experiments; *P < .05; ***P < .001, t test. (E) MutSig2CV analysis. Significantly mutated genes in EODGC-WES are listed vertically by q value. Top histogram, mutation events per sample; left histogram, samples affected per mutation. The percentage of EODGC-WES samples with a non-silent mutation is displayed on the right. Heat map, mutation events; right histogram, significance level of mutations as determined by log10 transformation of MutSig2CV q value. A continuous red line indicates a cutoff for significance (q = .1). Lower charts, the mean fraction of tumor reads vs total number of reads per sample (top) and the spectrum of somatic mutations of samples in each column (bottom). (F) ERBB3 mutation sites in EODGC-WES (G) MAP2K1 mutation sites in EODGC-WES.
Results
Whole Exome Sequencing Analysis of Early-Onset Diffuse Gastric Cancers
We performed WES and RNA sequencing analyses of a total of 80 DGCs resected from Korean patients ≤45 years of age (EODGC-WES) (Figure 1A). The EODGC-WES cohort was 58.7% female in composition, with a median age of 38 years. We sequenced the whole exome of EODGC-WES at a mean coverage of 93.6-fold (range, 90.6–95.2).
According to ReCapSeg and ISAR-GISTIC analyses of the WES data, recurrent focal amplification of 10q26.13 at the locus for FGFR2 was the clearly dominant peak within EODGC-WES (q = 1 × 10−9, Supplementary Figure 1A). Chromosomal locus 3p21.1 (g.chr3:48,369,660–55,002,466) was the most significant deletion in EODGC-WES (q = .0003, Figure 1B). Among the 161 genes in this locus, we suggest that the deubiquitinase BAP1 may be a novel gastric tumor suppressor gene candidate, because BAP1 was 1 of 6 genes at this locus that were most significantly underexpressed in 3p21.1-deleted EODGC-WES tumors (Figure 1C and Supplementary Table 8) and BAP1 mutation was the 16th most recurrent mutation in EODGC-WES (Supplementary Table 9). All 4 of the BAP1 somatic mutations found in EODGC-WES were associated with loss of heterozygosity, and BAP1 over-expression suppressed the growth of DGC cells (Figure 1D).
MuTect and Indelocator analyses of the WES data of EODGC-WES identified 6196 nonsynonymous somatic mutations (including 5766 point mutations and 430 indels). Mean and median somatic mutation rates were 1.4 and 1.2 mutations/Mb, respectively (range, 0–7.7: Figure 1E). According to MutSig2CV analysis, somatic mutations in CDH1, TP53, ARID1A, KRAS, PIK3CA, ERBB3, TGFBR1, FBXW7, RHOA, and MAP2K1 were significantly recurrent in EODGC-WES (Figure 1E). Importantly, TGFBR1 mutations were female-specific in EODGC-WES (P = .014, χ2; Supplementary Figure 3). Consistent with their tumor suppressor roles, both TGFBR1 and FBXW7 possessed nonsense mutations (Supplementary Figure 4A). Five of the 7 ERBB3 mutations detected have been reported in other cancers (Figure 1F). While MAP2K1 (encoding MEK1) was mutated in 3 EODGC-WES cases, 2 mutations were the oncogenic C121S mutation (Figure 1G).11
Somatic CDH1 Mutations Were More Frequent in Early-Onset Diffuse Gastric Cancers Than in Late-Onset Diffuse Gastric Cancers
To compare EODGC and LODGC genomes with a higher statistical power, we expanded our EODGC-WES cohort by recruiting 29 additional EODGC samples from Korean patients (EODGC; 54.1% female, median age, 38 years; Figure 1A). We then compared SNP6.0 array–based copy number and targeted sequencing data between EODGC and a control set of DGCs that were obtained from 115 Korean patients ≥46 years of age (LODGC; 29.6% female, median age, 67 years). In contrast to previous studies suggesting unique copy number profiles of GCs in young patients,12 our study revealed that copy number profiles were similar between EODGCs and LODGCs (Supplementary Figure 5C and 5D). Focal amplification of FGFR2 was the most significant in both EODGCs and LODGCs, and focal amplifications in ERBB2, MYC, CCNE1, CCND1, and MET were recurrent in both cohorts (Figure 2A). Consistent with these results, when we classified EODGC tumors according to a molecular classification system proposed by TCGA marker paper (TCGA subgroup),6 the proportion of the chromosomal instability (CIN) subgroup was not significantly different between EODGCs and LODGCs (Figure 2B and Supplementary Figure 5D).
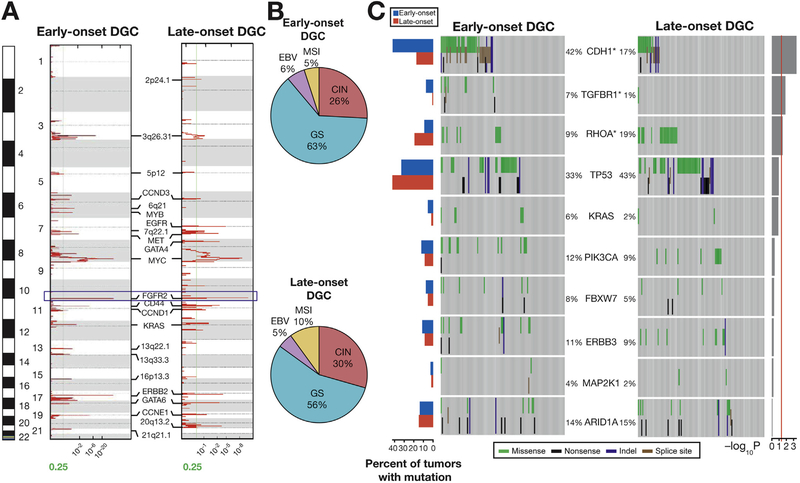
Figure 2.
SNP6.0 array-based copy number and targeted sequencing data in EODGCs and LODGCs (A) Significant focal amplifications in EODGCs (left; n = 109) and LODGCs (right; n = 115), according to the SNP6.0 array-based copy number analysis. Annotations include significant focal amplifications. Green lines indicate cutoffs for significance (q = .25). FGFR2 amplification (blue box) was the most significant focal amplification in both EODGCs and LODGCs. (B) CIN=was similar in proportion between EODGCs and LODGCs, suggesting an equivalent degree of aneuploidy between the 2 cohorts. (C) Targeted sequencing comparison analysis of 10 SNVs that were significant in the WES data of EODGC-WES. Left histogram, frequency of 10 SNVs in EODGCs (blue bar) and LODGCs (red bar); heatmaps, mutation events. The percentages of EODGCs and LODGC samples with non-silent somatic mutation are displayed at left and right sides of each gene name, respectively. *P < .05. Right histogram, log10 transformation of P value by χ2 test (EODGCs vs LODGCs). A continuous red line indicates a cutoff for significance (P = .05).
Using targeted sequencing analysis, we then compared somatic mutation rates of 10 SNVs, which were significant mutations according to the WES data of EODGC-WES (Figure 1E), between EOGDCs and LODGCs. According to targeted sequencing analysis, EODGC presented significantly higher somatic mutation rates of CDH1 (42.2% vs 17.4% in LODGC) and TGFBR1 (7.3% vs 0.9% in LODGC) than did LODGC (P < .001 and P = .014 for CDH1 and TGFBR1, respectively, χ2; Figures 2C and 3A–C). In contrast, somatic RHOA mutation was significantly less frequent in EODGC than in LODGC (9.2% vs 19.1%; P = .033, χ2). Thus, a pattern of frequent CDH1 and TGFBR1 mutations and less common RHOA mutation was identified as the characteristic somatic alteration for EODGC. Somatic TP53 mutations tended to be less frequent in EODGC than in LODGC (33.0% vs 43.5%; P = .11, χ2).
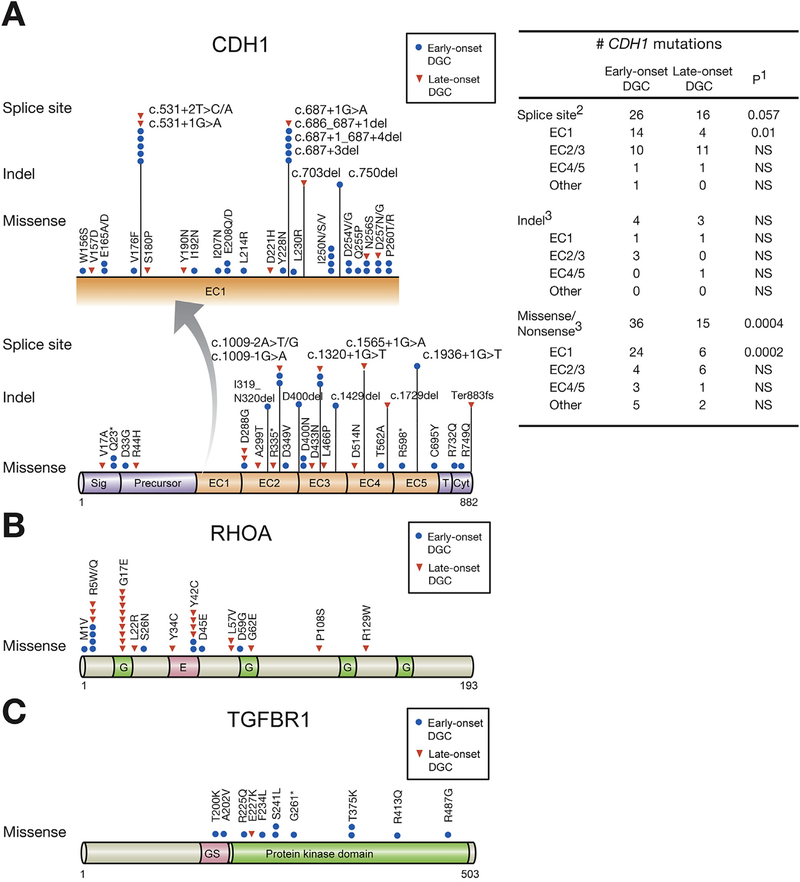
Figure 3.
Somatic mutations in CDH1, RHOA, and TGFBR1 of early- and LODGCs as identified by targeted sequencing. (A) CDH1 mutation sites. Left, CDH1 mutations in the E-cadherin EC1 domain, which are displayed in expanded view (arrow), were more frequent in EODGC (blue circle) than in LODGC (red triangle). “Hot spot” splice site mutations occurred. c.531+1G>A, and c.531+2T>C/A mutations led to exon 4 truncations, and c.687+1G>A, c.686_687+1del, c.687+1_687+4del, and c.687+3del mutations resulted in truncation of exon 5 in EC1 domain. Right, the number of somatic CDH1 mutation sites in EODGCs (n = 109) and LODGCs (n = 115) according to the targeted sequencing; 1P value by χ2 test. 2The number of CDH1 splicing aberrations that were identified either by targeted DNA sequencing or by reverse transcription polymerase chain reaction. 3The number of CDH1 mutations that were identified by targeted DNA sequencing (B) RHOA mutation sites (C) TGFBR1 mutation sites.
This study evaluates somatic CDH1 mutation rates in relation to patient age in a large-scale setting, and resulted in the identification of a striking difference in CDH1 mutation rates between EODGCs and LODGCs. We also observed that the somatic CDH1 mutation rate was higher in females than in males within EODGC (52.5% [31 of 59] vs 30.0% [15 of 50]; P = .018, χ2). Some CDH1 splicing aberrations were evidenced at the RNA level, but not at the DNA level (Supplementary Table 4). When splicing aberrations at the RNA level, homozygous deletions, promoter hypermethylation, as well as SNVs and indels, were also considered in determining the overall CDH1 alteration rate, EODGC demonstrated a higher CDH1 alteration rate than did LODGC (53.2% [58 of 109] vs 32.2% [37 of 115]; P = .0009, χ2). In addition, EODGC females had higher CDH1 alteration rate than did EODGC males (67.8% [40 of 59] vs 36.0% [18 of 50]; P = .0009, χ2; Supplementary Table 4). Thus, we propose that high prevalence of CDH1 alterations may be related to the predominance of females in this population. Notably, patients with pathogenic germline mutations in CDH1, TP53, and ATM were excluded from the onset of this analysis (Supplementary Table 2), given our focus on somatic alterations in sporadic EODGC.
We further investigated CDH1 alterations using transcriptome analyses, which revealed prominent CDH1 splicing aberrations impacting the expression of exons 4, 5, 8, and 9 (Supplementary Figure 6A and 6B). We identified unexpectedly frequent novel exon truncations and intron retentions in EODGC resulting from CDH1 splicing aberrations impacting the EC1 domain (exons 4 and 5), and this phenomenon was significantly more prominent in EODGC than in LODGC (12.8% [14 of 109] vs 3.5% [4 of 115]; P = .01, Figure 3A). In 5.5% of EODGC tumors (n = 6), c.chr16:531+2T>C/A and c.chr16: 531+1G>A mutations in the exon 4-intron 4 junction resulted in intron retentions (with premature termination codons) and 63-bp in-frame deletion transcripts due to the creation of a cryptic splice donor sites (Supplementary Table 4 and Figure 4B). We also identified a 42-bp (exon 5) in-frame deletion transcript in 6.4% (n = 7) of EODGCs using reverse transcription polymerase chain reaction (Figure 4B), and this fragment was also predicted to occur due to the creation of cryptic splice donor sites.
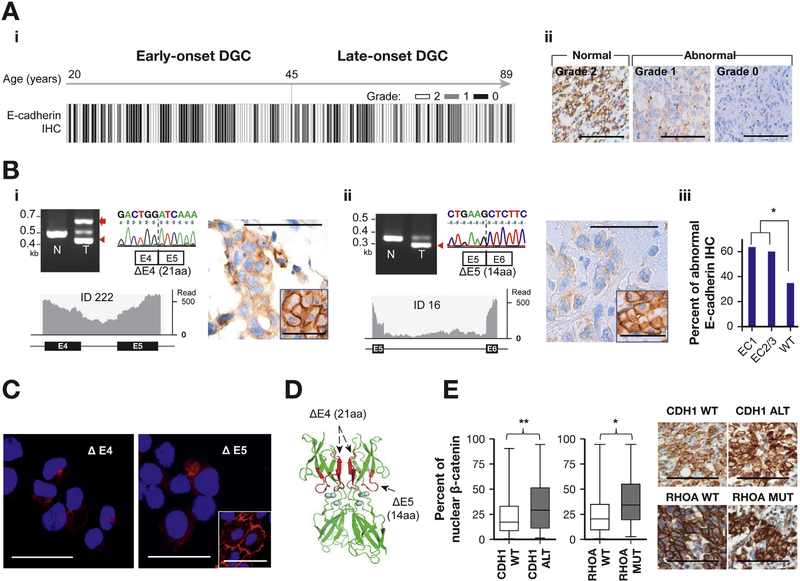
Figure 4.
Immunohistochemistry (IHC) and immunofluorescence (IF). (A) E-cadherin IHC (A-i) A total of 197 EODGC and LODGC tumors that were graded for E-cadherin IHC are ordered according to increasing age from left to right. Specimens with <6 high-power fields or 2000 cancer cells were not considered eligible for grading, and not all tumors were graded. IHC grade was based on the percentage of tumor cells with aberrant staining (<20%, grade 2 (normal, white bar); 20%-80%, grade 1 (gray bar); >80%, grade 0 (black bar). E-cadherin IHC was more frequently abnormal (grade 0 or 1) in EODGC than in LODGC (P = .001, χ2). (A-ii) Representative microscopic photographs for IHC grades. Bar = 100 μm. (B) Reverse transcription polymerase chain reaction capillary sequencing (upper left), RNA sequencing (lower left), and immunohistochemistry (right) results of representative splicing aberrations in CDH1. Red arrow, intron retention; red arrowheads, exon truncations; inset, adjacent normal tissue. Bar = 100 μm. (B-i) A 63-bp exon 4 truncation with an intron retention causing premature termination of translation. (B-ii) A 42-bp exon 5 truncation. (B-iii) E-cadherin immunostaining was more frequently abnormal in EODGCs with any CDH1 splicing aberrations (n = 21) than in those wild-type (WT) for CDH1 that were eligible for IHC grading (n = 46). (C) Representative E-cadherin IF images of NCC-S3 (Pdx-1-cre;Cdh1F/F ;Trp53F/F) DGC cells16,17 stably expressing CDH1 mutants with 63-bp exon 4 truncation (ΔE4) and 42-bp exon 5 truncation (ΔE5). Cells expressing CDH1 truncation mutants exhibited punctate (dot-like) or weak E-cadherin IF (red), whereas cells expressing WT CDH1 vector showed strong circumferential membranous E-cadherin IF (inset). The nucleus was visualized by 4′,6-diamidino-2-phenylindole stain (blue). Bar = 50 μm. (D) Three-dimensional computational modeling predicted the EC1 domain as an interface for a homodimerization intermediate, the “X-dimer.” Mutations (red) are near the calcium-binding domains (blue circles) that are required for E-cadherin’s functions. (E) Nuclear β-catenin positivity was higher in RHOA-mutated (MUT) tumors that were eligible for IHC grading (n = 30) than those WT for RHOA (Supplementary Tables 5 and 11). The ends of the whiskers represent minimum and maximum values. Right, representative photographs. *P < .05; **P < .01.
We then performed E-cadherin immunohistochemistry to evaluate the protein expression pattern of CDH1 splicing aberrations (Figure 4A). A majority (63.6%) of EODGC tumors with EC1 splicing aberrations (n = 11) demonstrated abnormal E-cadherin immunostaining= (grade 0 or 1: Figure 4B). We also evaluated nuclear β-catenin immunostaining, which represents the activation of WNT/β-catenin signaling. Among 10 genes that were significantly mutated in EODGC-WES, CDH1 alterations most significantly correlated with nuclear β-catenin immunostaining in a combined dataset of EODGCs and LODGCs (P = .005, Wilcoxon; Figure 4E).
We further investigated the functional effects of these “hot-spot” CDH1 splicing aberrations impacting the EC1 domain, which have not been characterized previously. According to the 3-dimensional computational modeling, exon truncations in the EC1 domain were predicted to hinder the homodimerization of E-cadherin (Figure 4D).13 The introduction of CDH1 mutants with truncated exons led to aberrant E-cadherin immunofluorescence (Figure 4C), which is consistent with the immunohistochemistry data of clinical samples (Figure 4B). These results may suggest the instability of mutant E-cadherin proteins, as described previously for CDH1 missense mutations.14 The introduction of CDH1 mutants with truncated exons was unable to induce the aggregation phenotype that was observed in wild-type CDH1-transduced GC cells (Figure 5B). Somatic missense mutations in the EC1 domain were also more prevalent in EODGC, suggesting that functional loss of the EC1 domain of E-cadherin shortens the latency of DGC (Figure 3A).
Figure 5.
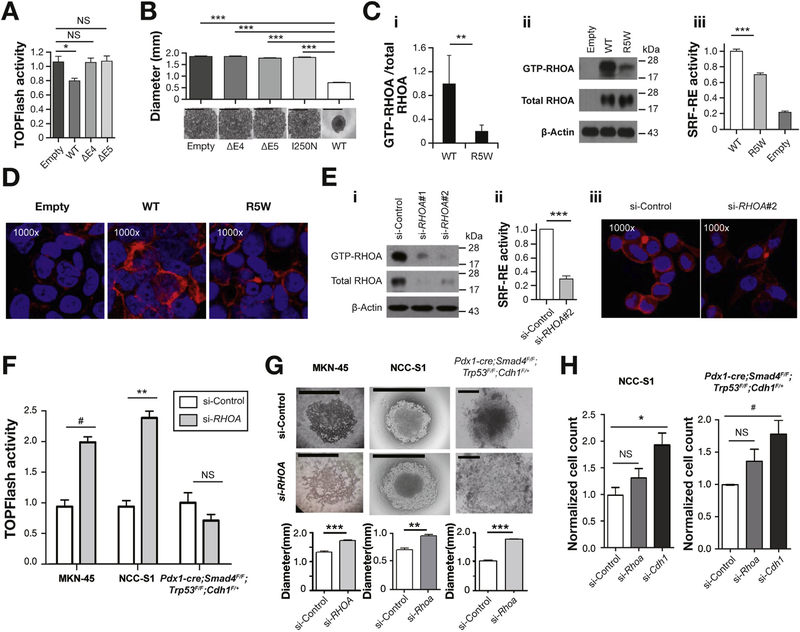
Functional assays. (A) TOPFlash β-catenin reporter activity remained unchanged after the ectopic expression of CDH1 mutants (ΔE4 and ΔE5) in NCC-S3 DGC cells (Pdx1-cre;Trp53F/F;Cdh1F/+),16,17 whereas it decreased after the ectopic expression of wild-type (WT) CDH1. ΔE4, 63-bp exon 4 truncation; ΔE5, 42-bp exon 5 truncation. (B) Slow aggregation assay of CHO-K1 cells. Aggregation was impaired in CHO-K1 cells overexpressing CDH1 splicing aberrations and a missense (I250N) mutation impacting the EC1 domain. Scale bar = 1 mm. (C) The ectopic expression of an R5W RHOA mutant vector blunted the increase in the GTP-bound RHOA fraction and SRF-RE reporter activity in 293FT cells, compared with the ectopic expression of the WT RHOA vector. (C-i) The mean ratio of GTP-RHOA to total RHOA as measured by densitometry in 3 independent experiments. (C-ii) A representative RhoTekin assay result. (C-iii) SRF-RE activity. (D) Blunted increase in rhodamine phalloidin-stained actin stress fiber immunofluorescence (IF) staining (shown in red) after R5W expression, compared with WT. (E) Suppression of RHOA activity in MKN-45 cells (a stimulus equivalent to an R5W mutation) using small interfering RNA–mediated RHOA knockdown. RHOA activity suppression was validated by (E-i) RhoTekin, (E-ii) SRF-RE, and (E-iii) actin stress fiber IF assays. (F) RHOA knockdown significantly increased TOPFlash β-catenin activity DGC cell lines (P = .005, paired t test), and this effect was prominent in MKN-45 and NCC-S116 cells. (G) Slow aggregation assays of DGC cells after RHOA knockdown. Top, representative photographs (scale bar = 0.5 mm); bottom, vertical axis denotes the mean diameter of cell aggregates. RHOA knockdown significantly impaired the aggregation of MKN-45, NCC-S1, and Pdx1-cre;Smad4F/F;Trp53F/F;Cdh1F/+ primary cultured DGC cells.16,17 (H) Effects of the knockdown of Rhoa and Cdh1 on migration activities of DGC cells that harbor no CDH1 mutations. Vertical axis denotes the normalized number of migrated cells per field. Mean values of at least 3 independent experiments. Cdh1 knockdown, but not Rhoa knockdown, significantly enhanced migration activities (P = .001 and P = .01, t test). #P < .05; *P < .01; **P < .001; ***P < .0001, t tests; Error bar, mean ± SEM.
The Effect of RHOA Inactivation on the Metastatic Phenotype of Diffuse Gastric Cancer Cells Was Not as Prominent as That of E-Cadherin Inactivation
As described, somatic mutations in RHOA and TP53 were less frequent in EODGC than in LODGC (Figure 2C). We were not surprised by the relative paucity of TP53 mutations in EODGC, given the association of TP53 mutations with aging,15 but we were surprised by the relative paucity of RHOA mutations in EODGC (9.2% [10 of 109] vs 19.1% [22 of 115] in LODGC; Figure 3B). In EODGC, R5W/Q substitutions were the most frequent RHOA mutation (Figure 3B).
The role of RHOA mutations in diffuse-type gastric carcinogenesis has not been fully elucidated.6,8 We discovered a clue to the functional effects of the RHOA mutations on DGC from our analysis of nuclear β-catenin immunostaining (Figure 4E and Supplementary Figure 6E). RHOA-mutated tumors (n = 30) exhibited more prominent nuclear β-catenin immunostaining than did wild-type tumors for RHOA (n = 164) (P = .011, Wilcoxon; Figure 4E and Supplementary Table 5). Independent of the CDH1 alteration status, the RHOA mutation status was significantly associated with nuclear β-catenin immunostaining (adjusted P = .01, linear regression; Supplementary Table 11). This immunohistochemistry data raised the possibility that RHOA mutations might partially overlap with CDH1 mutations in functional effect on diffuse-type gastric carcinogenesis, given that CDH1 mutations could not suppress the β-catenin reporter activity in DGC cells (Figure 5A). We first determined the RHOA activity of an R5W mutation, the most frequent RHOA mutation in EODGC, to evaluate whether it represents a gain- or loss-of-function mutation. Ectopic expression of the RHOA R5W mutation, compared with the ectopic expression of a wild-type RHOA construct in 293FT cells, led to blunted RHOA activation (Figure 5C and 5D). Thus, biochemical and transcriptional activities of RHOA were compromised by the R5W mutation. Based on this finding, we suppressed the RHOA activity of DGC cells using small interfering RNAs and an exoenzyme C3 transferase, to produce stimuli that are equivalent to the R5W mutation (Figure 5E and Supplementary Figure 7C). This suppression significantly increased β-catenin reporter activity in DGC cells (P = .005, paired t test), especially in 2 of the 3 cell lines tested (MKN-45 and NCC-S116,17; Figure 5F and Supplementary Figure 7F). RHOA knockdown significantly impaired the aggregation of all DGC cell lines evaluated (Figure 5G), which suggests a partial overlap in functional effect between RHOA and CDH1 inactivating mutations (Figure 5A and Figure 5B). In contrast, whereas Cdh1 knockdown enhanced the migration activity of wild-type DGC cells for Cdh1, RHOA inactivation did not significantly affect the migration activity of these cells (Figure 5H and Supplementary Figure 7H). These results may suggest that E-cadherin plays a more central role in suppressing DGC metastasis than does RHOA.
The Aggressive Clinical Course of Early-Onset Gastric Cancer Can Be Attributed to the High Prevalence of CDH1 Mutations and Relative Paucity of RHOA Mutations
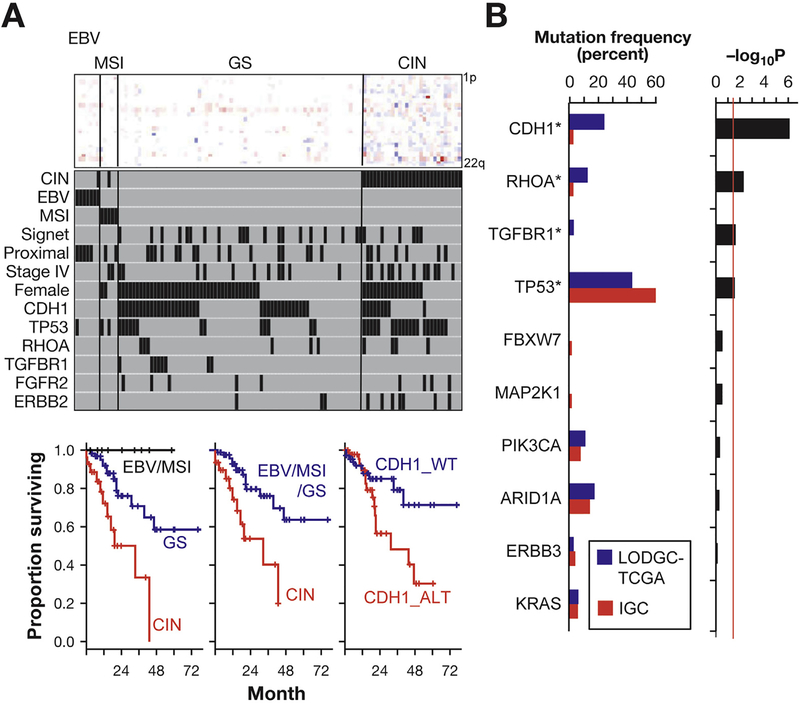
To evaluate the clinical relevance of the somatic alterations we identified, we classified EODGCs according to the TCGA subgroup (Figure 6A and Supplementary Figure 5B).6 Seven Epstein-Barr virus (EBV)-positive tumors, 5 microsatellite instability (MSI)-high tumors, 69 genomically stable (GS) tumors, and 28 CIN tumors were present in EODGC. The distribution of TCGA subgroup of EODGC did not differ significantly from that of LODGC (P = .34, χ2; Figure 2B). None of the 26 signet ring cell carcinomas in EODGC belonged to the EBV/MSI subgroup (P = .04, χ2; Figure 6A). The EBV subgroup in EODGC was enriched with the male sex (100% vs 42.2% in non-EBV; P = .003) and the proximal tumor location (71.4% vs 24.5% in non-EBV; P = .007). None of the 58 EODGC tumors with CDH1 alterations belonged to the EBV/MSI subgroup (P < .001, χ2). All 8 TGFBR1 mutations were found in the GS subgroup (P = .025, χ2).
Figure 6.
Somatic genomic alterations associated with clinicopathological features. (A) Top panel, heatmap for arm-level, copy number gains (red), and losses (blue) of 22 autosomes. Each column represents an EODGC tumor, grouped by TCGA subgroup, and each row represents a chromosome arm. Middle panel, black bars representing the presence of corresponding clinicopathologic feature or genomic alteration. Case ordering the same for top and middle panels. Bottom panels, Kaplan-Meier plots for the overall survival of 109 patients with EODGC according to the TCGA subgroup. (B) Mutation frequency in diffuse and intestinal tumors. TCGA diffuse tumors (LODGC-TCGA; n = 61, blue bar) and TCGA intestinal tumors (IGC; n = 157, red bar) were compared for somatic mutation rates of 10 SNVs that were recurrent in EODGC-WES. Left histogram, somatic mutation frequencies in LODGC-TCGA (blue) and IGC (red). *P < .05; right histogram, log10 transformation of P value by χ2 test (LODGC-TCGA vs IGC). A continuous red line indicates a cutoff for significance (P = .05). Diffuse tumors (LODGC-TCGA) exhibited higher frequencies of CDH1 mutations (24.6% vs 3.2%; P = 8 × 10−7) and TGFBR1 mutations (3.3% vs 0%; P = .022) than did intestinal tumors (IGC).
Then we compared patient survival according to the TCGA subgroup and somatic mutation status. CIN was associated with the highest hazard ratio (HR) and the most adverse prognostic factor in EODGC (HR, 3.7; 95% confidence interval [CI], 1.8–7.7) vs non-CIN; Figure 6A). At a median follow-up of 33.7 months, median overall survival times were not reached in EBV/MSI and GS, while the CIN subgroup had the median survival of 28.2 months (95% CI, 14.6–not reached; P = .0002, log-rank). The CDH1 alteration (SNVs, indels, splicing aberrations, homozygous deletions, and promoter hypermethylation) was the second most adverse prognostic factor (HR, 3.4; 95% CI, 1.5–7.7; P = .002, log-rank), and tended to be enriched in patients with distant metastasis (66.7% [16 of 24] vs 49.4% [42 of 85] in those without metastasis; P = .13, χ2; Figure 6A). In contrast, RHOA and TGFBR1 mutations were not associated with prognosis (HR, 0.7; 95% CI, 0.2–3.0 and HR, 1.7; 95% CI, 0.5–5.7, respectively; Supplementary Figure 8C and 8D). Mutation frequencies of RHOA and TGFBR1 did not differ between patients with distant metastasis and those without metastasis (8.3% vs 9.4% and 4.1% vs 8.2%, respectively, for RHOA and TGFBR1). FGFR2 and ERBB2 amplifications and TP53 mutation were also associated with poor prognosis (HR, 3.8; 95% CI, 1.5–9.4; HR, 2.5; 95% CI, 1.0–6.6 and HR, 2.6; 95% CI, 1.2–5.4, respectively; P = .0018, P = .05, and P = .008, respectively, log-rank). These results indicate that EODGC can be classified into 3 prognostic categories. EBV/MSI was the best prognosis group without CDH1 alterations. The GS subgroup, which was enriched with TGFBR1 mutations (P = .025, χ2), yielded intermediate prognosis. The CIN subgroup, which was enriched with TP53 mutations (P = 3.5 × 10−5, χ2), was associated with the worst prognosis. Prognostic implications of TCGA subgroups were not documented previously by the TCGA marker paper due to its short-term follow-up duration.6
EOGC is notable for its advanced stage presentation and for its strong enrichment of diffuse histology and female predominance. Given the equivalent proportion of CIN between EODGCs and LODGCs (Figure 2B), we suggest that EOGC’s aggressive clinical course is primarily due to its high frequency of the CDH1 alteration, which represents the second most significant adverse prognostic factor in EODGC. We again noted trends for the higher prevalence of CDH1 mutations and the lower rate of RHOA mutations in younger patients, when we compared the WES data between EODGC-WES and a TCGA dataset of non-hypermutated, diffuse GCs obtained from patients ≥46 years of age (LODGC-TCGA), 70% of whom were Caucasians.6 When compared with LODGC-TCGA, EODGC-WES showed trends for relatively high somatic mutation rates of CDH1 (35.0% vs 24.6% in LODGC-TCGA; P = .18) and TGFBR1 (8.8% vs 3.3% in LODGC-TCGA; P = .19) and a relatively low somatic mutation rate of RHOA (5.0% vs 13.1% in LODGC-TCGA; P = .09; Supplementary Figure 4C), whereas these 2 cohorts did not differ in tumor purity and ploidy as estimated by ABSOLUTE (Supplementary Figure 4B). Thus, sporadic EOGC was characterized by high prevalence of somatic CDH1 mutations and relative paucity of somatic RHOA mutations not only when compared to an older Korean population, but also when compared to LO populations of other ethnicities.
BRCA and Young Female Signatures Were Identified by Mutation Signature Analyses
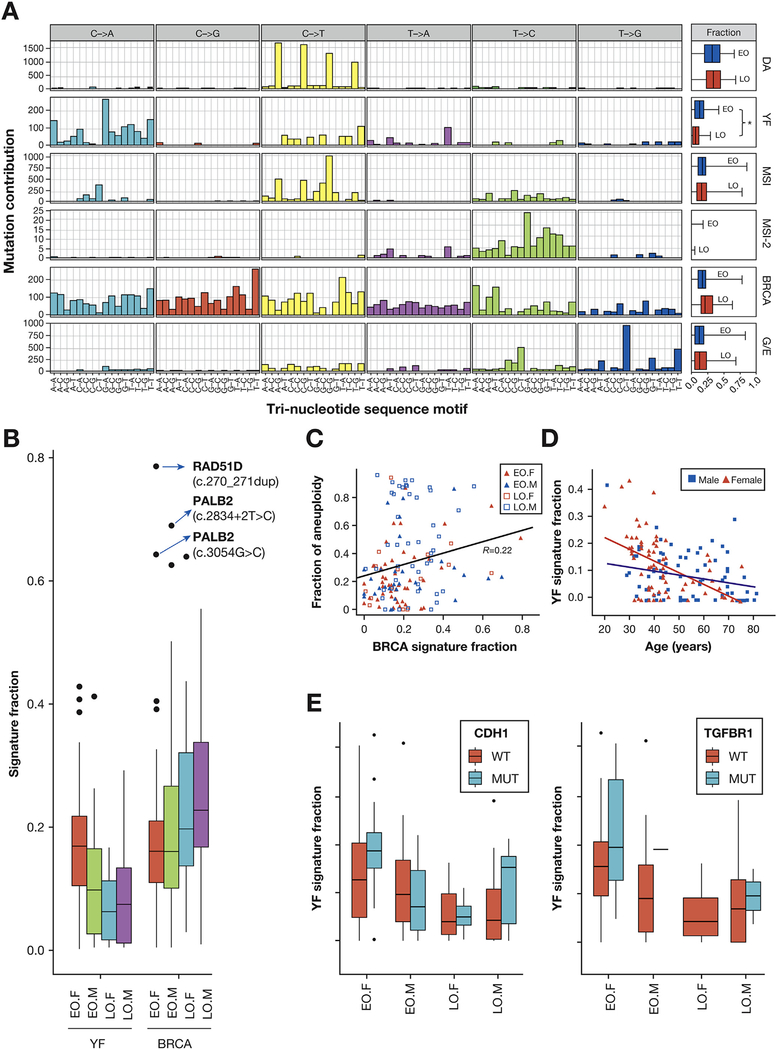
Aneuploidy, which was previously represented by the CIN subgroup (Figure 6A), was the most adverse prognostic factor in EODGC. As shown in Figure 2B, the CIN proportion of EODGC was comparable to that of LODGC, suggesting an equivalent degree of aneuploidy between the 2 cohorts. Mutation signature analysis revealed that germline mutations in homologous recombination pathway were associated with aneuploidy in EODGC. The homologous recombination defect-related BRCA mutation signature18 was 1 of 6 mutation signatures that were identified in the WES data of EODGC-WES and LODGC-TCGA (DGC, Figure 7A), and its signature fraction positively correlated with the degree of aneuploidy (R = 0.22; P = .01, Pearson, Figure 7C). The 3 tumors with the largest BRCA signature fraction, all of which were from aneuploidy EODGC-WES, presented germline mutations in either PALB2 or RAD51D (Figure 7B and Supplementary Tables 2 and 6). These data suggest that a small group of EODGC may be attributable to inherited DNA repair defects and these tumors manifest a distinct pattern of somatic alterations from other EODGC patients.
Figure 7.
Mutation signature analysis. (A) The spectrum of base substitutions for each mutation signature is displayed for mutated pyrimidines and adjacent 3′ and 5′ bases (motif; horizontal axis). Box plots in the far-right panels display median values of signature fractions in the WES data of EODGC-WES (EO, blue) and LODGC-TCGA (LO, red), with the ends of the whiskers representing minimum and maximum values. *P = .0002. (B) The mutation signature fraction according to sex and age group. Box plots in (B) and (E) display 5%, 25%, median, 75%, and 95%, with outliers shown as dots. The top 3 BRCA signature outliers (blue arrows), all of which were from EODGC-WES cases, harbored germline mutations in either RAD51D (n = 1, with loss of heterozygosity in the tumor) or PALB2 (n = 2), and tended to have a higher fraction of aneuploid genome than the other EODGC-WES tumors (P = .061, Wilcoxon). (C) Correlation between the BRCA mutation signature and aneuploidy (D) The YF signature according to sex and age (E) The YF signature according to mutational status. Within female EODGC-WES (EO.F), trends for the higher YF signature fraction were observed with CDH1- and TGFBR1-mutated (MUT) tumors than with wild-type (WT) tumors (P = .08 and P = .19, Wilcoxon).
We also asked whether other mutation signatures may be preferentially present in EODGC, most especially in female patients, as such a signature may provide insight into the etiology of more frequent DGC diagnoses in this population. We noted that the signature resembling neuroblastoma-associated COSMIC signature 18 with prevalent C>A/T was significantly enriched in EODGC-WES (P = .0002, vs LODGC-TCGA, Wilcoxon), especially in EODGC-WES females (P = .007, vs EODGC-WES males, Wilcoxon; Figure 7D). This mutation signature was therefore referred to as the young female (YF) signature. Further inquiry into YF signature demonstrated a significant strand bias of C>A/T mutations toward the transcribed strand (P = .001; Supplementary Figure 9C). The fraction of the YF signature was higher in CDH1- and TGFBR1-mutated DGCs than in wild-type DGCs (P = .016 and P = .018 Wilcoxon; Figure 7E). Therefore, the unidentified mutagenic process underlying this signature appears to be enriched in the YFs who develop mutations that are common in EODGC.
The Predominance of Diffuse Histology and Female Sex in Early-Onset Gastric Cancer May Be Related to Frequent Somatic Mutations in CDH1 and TGFBR1
In addition to advanced-stage presentation, EOGC is also notable for its strong enrichment of diffuse histology and female predominance. As described, somatic CDH1 and TGFBR1 mutation rates were higher in EODGCs from women than in EODGC from men (P = .031 and P = .007 χ2; Figure 6A). We also observed that CDH1 and TGFBR1 mutation rates were higher in diffuse tumors than in intestinal tumors within TCGA dataset. When the WES data were compared between LODGC-TCGA (n = 61) and a set of non-hypermutated intestinal tumors from TCGA (IGC: n = 157), somatic CDH1 and TGFBR1 mutations were more frequent in LODGC-TCGA than in IGC, a finding that had not been recognized previously (P = 8 × 10−7 and P = .022, respectively; Figure 6B and Supplementary Figure 11A; Supplementary Table 7).
Discussion
This is a comprehensive study of somatic genomic alterations associated with the unique clinicopathologic features of sporadic EODGC.8 This study is also the largest WES study of DGC to date, and resulted in the identification of previously unrecognized TGFBR1 and BAP1 alterations in DGC (Supplementary Figure 11A and 11C) and the characterization of hot-spot splice site mutations of CDH1 involving exon truncations and intron retentions. We documented the prominent difference in somatic CDH1 mutation rates between EODGCs and LODGCs in a large sample set and the clustering of CDH1 mutations at the E-cadherin EC1 domain in EODGC. Somatic CDH1 mutations documented in our sporadic EODGC cases provide a higher level of multi-omic information than did previous CDH1 mutation studies on sporadic DGC cases.19 Most of the earlier works on EOGC had been on familial cases and focused on the germline CDH1 mutation, which is a known risk factor for hereditary DGC.20–22 In contrast, our analysis from the onset excluded EODGC-WES cases with pathogenic germline mutations in CDH1, TP53, and ATM1 (Supplementary Table 2). As a result, there were no pathogenic germline CDH1 mutations among the entire 109 EODGC cases of this study, according to our genomic and functional analyses (Supplementary Tables 2 and 3; Supplementary Figure 6C). This study provides the largest somatic alteration data of sporadic EOGC.
Beyond identifying distinct somatic alterations in EODGC, we also observed distinct mutagenic processes. These include the findings of 3 tumors with a marked BRCA signature in patients without family history of GC, yet harboring pathogenic germline mutations in the homologous recombination repair pathway. Among our EODGC-WES cases, the BRCA mutation signature fraction was higher in the CIN subgroup (vs non-CIN, P = .008, Wilcoxon; Supplementary Figure 10A). But it was not associated with CDH1 alterations or platinum responsiveness after relapse according to our analysis of a limited sample set (Supplementary Figure 10C and Supplementary Table 10).
Our study found that the BAP1 locus, 3p21.1, was the most frequent deletion in EODGC-WES. Chromosomal locus 3p21.1 is deleted in mesothelioma, malignant melanoma, and cancers of the lung and breast.23 We also observed that the BAP1 mutation was the 16th most recurrent somatic mutation in EODGC-WES, and all 4 BAP1 somatic mutations were associated with loss of heterozygosity. Germline mutations in BAP1 predispose to melanocytic tumors and mesothelioma, and somatic mutations in BAP1 have been reported in intrahepatic cholangiocarcinoma,24 malignant melanoma, mesothelioma, and renal cell carcinoma. However, tumor-suppressive role of BAP1 has not been reported in GC. As in NCI-H226 cells,25 BAP1 expression suppressed the growth of our DGC cells, suggesting that BAP1 is a novel tumor suppressor candidate in DGC.
While neither the tumor content nor the proportion of metastatic disease differed between the compared age groups, our study identified a consistent pattern of frequent somatic CDH1 mutations and less common somatic RHOA mutations in sporadic EODGC, within the Korean population and within populations of other ethnicities. The unique mutation frequency pattern could not be explained by the rarity of MSI in young patients, given the lack of association between RHOA mutations and MSI (13.6% and 15.0% for MSI-low/microsatellite stable and MSI-high, respectively). We then investigated whether this mutation frequency pattern could be explained by a functional overlap between CDH1 and RHOA mutations, both of which are relatively uncommon in solid tumor types other than diffuse gastric and lobular breast cancers. Wang et al7 have shown that Y42C and L57V RHOA hotspot mutations cause defective RHOA signaling, promoting resistance to anoikis. Consistent with the Wang et al study, our study demonstrated that an R5W mutation reduced RHOA activity, and that the RHOA inactivation led to β-catenin activation in gastric epithelial cells. RHOA inactivation might possibly activate β-catenin signaling partially through destabilizing the E-cadherin/catenin complex, as was observed in colon cancer cells.26 Although the exact molecular mechanisms in DGC are not clear, functional consequences of RHOA inactivating mutations in DGC may overlap with those of CDH1 alterations regarding β-catenin activation as well as cell–cell adhesion loss,7,27 which might, to a degree, explain the relative paucity of RHOA mutations in EODGC harboring highly prevalent CDH1 alterations that might offset the necessity of RHOA mutations for diffuse-type gastric carcinogenesis. More importantly, the migration activity of our DGC cells became more prominent after the knockdown of CDH1, a central player in epithelial–mesenchymal transition, than after the knockdown of RHOA. Consistent with our genomics data that the CDH1 alteration, but not the RHOA mutation, was associated with poor prognosis of EODGC, these results indicate that a pattern of frequent CDH1 alterations and rare RHOA mutations may contribute to EOGC’s high propensity for metastasis.8
Intriguingly, Kakiuchi et al5 reported that G17E and Y42C RHOA mutations were biological gain-of-function mutations conferring a growth advantage on OE19 cells. Whereas these G17E and Y42C RHOA mutations were similar in frequency in our LODGC cohort (10.4%) and Kakiuchi et al’s (mostly LO) cohort (10.3%), they were rare in our EODGC (1.8%). A majority of RHOA mutations found in our EODGC cohort were inactivating28 R5W/Q mutations. Our experimental data of an R5W mutation, which cannot be directly compared with Kakiuchi et al’s data of G17E and Y42C mutations,5 demonstrated that biochemical and transcriptional activities of RHOA were significantly reduced by an R5W substitution, and that the impaired cell–cell aggregation was the most prominent phenotype of DGC cells after RHOA inactivation. Continued research may be necessary to evaluate whether R5W/Q substitutions might manifest oncogenic functions beyond the loss-of-function effects we described in this report. As of yet, functional and genomic data do not seem to support the feasibility of a therapeutic approach targeting RHOA mutations, which was proposed by Kakiuchi et al,5 in cases of EODGC.
The strong enrichment of diffuse histology (94% in our cases) and female predominance in EOGC can be attributed, at least partially, to high rates of somatic CDH1 and TGFBR1 mutations, which were shown to be significantly associated with the characteristics mentioned (Figure 6A and B), and might possibly involve common mutagens (Figure 7E). Our study found that somatic TGFBR1 mutations were enriched in EODGC females (Figure 6A) and diffuse histology-specific (Figure 6B). Previous studies in colon cancer cells have shown that transforming growth factor–β promotes cellular adhesions by extracellular matrix remodeling,29 but the TGFBR1 mutation itself has not yet been linked to diffuse gastric tumors. Further studies could explore whether and how TGFBR1 mutations lead to the development of DGC. Taken together, our study further elucidates the functional implications of somatic mutations that are recurrent in DGC, and the genomic basis of clinicopathologic characteristics of DGC that differ according to age.
Supplementary Material
EDITOR’S NOTES.
BACKGROUND AND CONTEXT
Genomic alterations associated with unique clinicopathological characteristics of early-onset gastric cancers are mostly unknown. Previous studies on this population were focused on familial cases and the germline CDH1 mutation.
NEW FINDINGS
Sporadic early-onset diffuse gastric cancers harbored less frequent RHOA mutations and more frequent CDH1 and TGFBR1 mutations than late-onset cancers.
LIMITATIONS
This study did not identify the specific mutagens that cause CDH1 and TGFRB1 mutations in young women.
IMPACT
This study elucidates the unique mutation profiles of early-onset gastric cancer and their clinical implications.
Acknowledgments
The biospecimens for this study were provided by members of National Biobank of Korea (Asan Bio-Resource Center, Keimyung Human Bio-Resource Bank, Biobank of Pusan National University, Biobank of Chonnam National University Hwasun Hospital, Biobank of Chungnam National University Hospital, and Ajou Human Bio-Resource Bank), which is supported by the Ministry of Health and Welfare, by Resource Banks at Dong-A University Medical Center and Kosin University Gospel Hospital and by the National Cancer Center of Korea. Adam J. Bass was supported by the Schottenstein Fund for Gastric Cancer Research and the DeGregorio Family Foundation.
The authors thank all the patients and their families who contributed to this study. The results published here are in part based on data generated by The Cancer Genome Atlas managed by the National Cancer Institute and National Human Genome Research Institute. Information about TCGA can be found at http://cancergenome.nih.gov.
The raw sequencing and SNP6.0 array data associated with this work were deposited to the European Genome-phenome Archive database under accession numbers EGAS00001001711 and EGAS00001001859.
Funding
This work was supported by the Multi-omic Research Program funded by the Ministry of Science, ICT, and Future Planning in Korea, and by National Cancer Center grants 1510111/1710809.
Abbreviations used in this paper:
- CIN
chromosomal instability
- DGC
diffuse gastric cancer
- EBV
Epstein-Barr virus
- EO
early onset
- GC
gastric cancer
- GS
genomically stable
- HR
hazard ratio
- LO
late onset
- MSI
microsatellite instability
- SNV
single nucleotide variation
- TCGA
The Cancer Genome Atlas
- WES
whole exome sequencing
- YF
young female
Footnotes
Supplementary Material
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at http://dx.doi.org/10.1053/j.gastro.2017.05.012.
Conflicts of interest
The authors disclose no conflicts.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359–E386. [DOI] [PubMed] [Google Scholar]
- 2.Wu H, Rusiecki JA, Zhu K, et al. Stomach carcinoma incidence patterns in the United States by histologic type and anatomic site. Cancer Epidemiol Biomarkers Prev 2009;18:1945–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen K, Yang D, Li X, et al. Mutational landscape of gastric adenocarcinoma in Chinese: implications for prognosis and therapy. Proc Natl Acad Sci U S A 2015;112:1107–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cristescu R, Lee J, Nebozhyn M, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 2015;21:449–456. [DOI] [PubMed] [Google Scholar]
- 5.Kakiuchi M, Nishizawa T, Ueda H, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet 2014;46:583–587. [DOI] [PubMed] [Google Scholar]
- 6.The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang K, Yuen ST, Xu J, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet 2014;46:573–582. [DOI] [PubMed] [Google Scholar]
- 8.Bautista MC, Jiang SF, Armstrong MA, et al. Impact of age on clinicopathological features and survival of patients with noncardia gastric adenocarcinoma. J Gastric Cancer 2014;14:238–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung HW, Noh SH, Lim JB. Analysis of demographic characteristics in 3242 young age gastric cancer patients in Korea. World J Gastroenterol 2010;16:256–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson WF, Camargo MC, Fraumeni JF, et al. Age-specific trends in incidence of noncardia gastric cancer in US adults. JAMA 2010;303:1723–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagle N, Emery C, Berger MF, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol 2011;29:3085–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buffart TE, Carvalho B, Hopmans E, et al. Gastric cancers in young and elderly patients show different genomic profiles. J Pathol 2007;211:45–51. [DOI] [PubMed] [Google Scholar]
- 13.Brasch J, Harrison OJ, Honig B, et al. Thinking outside the cell: How cadherins drive adhesion. Trends Cell Biol 2012;22:299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simões-Correia J, Figueiredo J, Lopes R, et al. E-Cadherin destabilization accounts for the pathogenicity of missense mutations in hereditary diffuse gastric cancer. PLoS One 2012;7:e33783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richardson RB. P53 mutations associated with aging-related rise in cancer incidence rates. Cell Cycle 2013; 12:2468–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park JW, Park DM, Choi BK, et al. Establishment and characterization of metastatic gastric cancer cell lines from murine gastric adenocarcinoma lacking Smad4, p53, and E-cadherin. Mol Carcinog 2015;54:1521–1527. [DOI] [PubMed] [Google Scholar]
- 17.Park JW, Jang SH, Park DM, et al. Cooperativity of E-cadherin and Smad4 loss to promote diffuse-type gastric adenocarcinoma and metastasis. Mol Cancer Res 2014;12:1088–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nik-Zainal S, Alexandrov LB, Wedge DC, et al. Mutational processes molding the genomes of 21 breast cancers. Cell 2012;149:979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corso G, Carvalho J, Marrelli D, et al. Somatic mutations and deletions of the E-cadherin gene predict poor survival of patients with gastric cancer. J Clin Oncol 2013;31:868–875. [DOI] [PubMed] [Google Scholar]
- 20.Yanjun X, Wenming C, Lisha Y, et al. Detection of CDH1 gene variants in early-onset diffuse gastric cancer in Chinese patients. Clin Lab 2014;60:1823–1830. [DOI] [PubMed] [Google Scholar]
- 21.Kim S, Chung JW, Jeong TD, et al. Searching for E-cadherin gene mutations in early onset diffuse gastric cancer and hereditary diffuse gastric cancer in Korean patients. Fam Cancer 2013;12:503–507. [DOI] [PubMed] [Google Scholar]
- 22.Bacani JT, Soares M, Zwingerman R, et al. CDH1/E-cadherin germline mutations in early-onset gastric cancer. J Med Genet 2006;43:867–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiesner T, Fried I, Ulz P, et al. Toward an improved definition of the tumor spectrum associated with BAP1 germline mutations. J Clin Oncol 2012;30:e337–e340. [DOI] [PubMed] [Google Scholar]
- 24.Moeini A, Sia D, Bardeesy N, et al. Molecular pathogenesis and targeted therapies for intrahepatic cholangiocarcinoma. Clin Cancer Res 2016;22:291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ventii KH, Devi NS, Friedrich KL, et al. BRCA1- associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res 2008;68:6953–6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodrigues P, Macaya I, Bazzocco S, et al. RHOA inactivation enhances Wnt signalling and promotes colorectal cancer. Nat Commun 2014;5:5458–5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sahai E, Marshall CJ. ROCK and Dia have opposing effects on adherens junctions downstream of Rho. Nat Cell Biol 2002;4:408–415. [DOI] [PubMed] [Google Scholar]
- 28.Rohde M, Richter J, Schlesner M, et al. Recurrent RHOA mutations in pediatric Burkitt lymphoma treated according to the NHL-BFM protocols. Genes Chromosomes Cancer 2014;53:911–916. [DOI] [PubMed] [Google Scholar]
- 29.Wang H, Radjendirane V, Wary KK, et al. Transforming growth factor beta regulates cell-cell adhesion through extracellular matrix remodeling and activation of focal adhesion kinase in human colon carcinoma Moser cells. Oncogene 2004;23:5558–5561. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.