Abstract
Objective
Renal cancer is one of the most deadly urological malignancies. Currently, there is still a lack of effective treatment. Our purpose was to explore the mechanisms of miR-122-5p in renal cancer.
Methods
The expression levels of miR-122-5p and pyruvate kinase M2 (PKM2) in renal cancer cells were detected by RT-qPCR and Western blot analyses, respectively. Then, we measured the cell viability after knockdown of miR-122-5p and PKM2 using CCK-8 assay. Moreover, flow cytometry was used to investigate cell cycle and apoptosis of renal cancer cells. The cell migration of renal cancer cells transfected by miR-122-5p inhibitor and siPKM2 was then detected by wound healing assay. Furthermore, glucose consumption and lactate production were measured. Autophagy-related protein LCII/I was detected by Western blot.
Results
MiR-122-5p was upregulated in renal cancer cells compared to HK2 cells, especially in 786-O cells. We found that silencing miR-122-5p promoted PKM2 expression in 786-O cells. After transfection of siPKM2 or miR-122-5p inhibitor, the cell viability of 786-O cells was significantly reduced. Furthermore, the G1 phase of 786-O cells was significantly blocked, and the S phase was significantly increased. In addition, knockdown of miR-122-5p or PKM2 promoted renal cancer cell apoptosis and inhibited cell migration. Glucose consumption of 786-O cells was significantly increased after transfection by siPKM2. Silencing miR-122-5p significantly promoted the expression levels of LCII/I.
Conclusion
Our findings revealed that overexpressed miR-122-5p promotes renal cancer cell viability, proliferation, migration, glycolysis and autophagy by negatively regulating PKM2, which provide a new insight for the development of renal cancer therapy.
Keywords: PKM2, miR-122-5p, cell viability, glycolysis, renal cancer
Introduction
Despite much progress in the diagnosis and treatment, renal cancer remains one of the most deadly urological malignancies. Among the risk factors, smoking, obesity and hypertension are closely related to renal cancer.1 Early treatment of advanced and metastatic renal cancer is disappointing, such as chemotherapy, hormone therapy and radiation therapy.2 Lack of effective clinical diagnosis and treatment planning is one of the main causes of renal cancer mortality.3
An abundant and conserved microRNA (miRNA), miR-122-5p plays an important role in maintaining liver function, and its abnormal expression may contribute to the occurrence and development of various liver diseases by affecting hepatitis C virus RNA, liver metabolism and drug resistance and so on.4–8 Moreover, miR-122-5p is involved in several cancers such as colorectal cancer, melanoma, gastric cancer and lung cancer.9–12 Growing evidence has confirmed that miR-122-5p is upregulated in the tissue and serum of clear cell renal cell carcinoma (ccRCC). Previous research found that upregulated miR-122-5p induces epithelial–mesenchymal transition (EMT) by downregulating Dicer, which contributes to metastatic ccRCC.13 Furthermore, overexpressed miR-122-5p is correlated with poor prognosis of ccRCC patients. It has been found that miR-122-5p directly targets occludin in ccRCC cells, which affects malignant phenotypes in ccRCC.14 Another study demonstrated that miR-122-5p is highly expressed in ccRCC patients’ serum, furthermore, its high expression has correlation with metastasis and grade.15
Programming energy metabolism is major hallmark of cancers.16 Glycolysis is a metabolic pathway that converts glucose to pyruvate, ultimately leading to lactic acid production. Glycolysis is the main way of supplying energy to tumor cells.17 Glucose uptake and glycolysis are increased in cancer cells, which is also known as the Warburg effect.18 Metabolic reprogramming has a strong influence on tumor proliferation, apoptosis, metastasis and angiogenesis.19 A variety of oncogenes and tumor suppressor genes are involved in the regulation of metabolic pathways. Although this phenomenon was described by Otto Warburg more than 50 years ago, the molecular mechanism remains elusive.20 It has been confirmed that PKM2 plays a critical role in metabolic reprogramming.21 PKM2, one of the four isozymes of pyruvate kinase (PK), is mainly expressed in rapidly proliferating cell such as embryonic cells and cancer cells.22 Increasing research suggested that PKM2 plays a key role in cancer progression via metabolic pathways.23 Therefore, PKM2 may become a potential diagnostic or therapeutic target for cancer.
Further research on the molecular mechanisms of renal cancer could provide novel diagnostic or therapeutic targets for renal cancer. Thus, in our study, we further explored the role of miR-122-5p in renal cancer metabolism. Our findings provide a novel insight into the regulation of anaerobic glycolysis and the development of renal cancer.
Materials And Methods
Cell Culture
Human ccRCC cell lines (786-O and Caki-1), human renal adenocarcinoma cell line (Achn) and normal proximal tubular epithelial cell line (HK2) were obtained from Shanghai Cell Bank (China). All cells were cultured in RPMI 1640 medium and Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS), 100 kU/L, penicillin and 0.1 g/L streptomycin at 37°C in a humidified 5% CO2 incubator.
Quantitative Real-Time PCR (RT-qPCR)
Total RNA was extracted from cells using TRIol reagent (EZBioscience, USA) according to the manufacturer’s instructions. For detection of miRNA expression, cDNA was synthesized using the qScript miRNA cDNA Synthesis Kit (Quantabio, Beverly, MA). The RT-qPCR was performed with the Advance Universal SYBR Green Supermix reagents (BIO-RAD, USA) under the following thermocycling conditions: 95°C for 2 mins, and 40 cycles of 95°C for 5 s and 60°C for 30 s. For detection of mRNA expression, the cDNAs were synthesized using RevertAid First Strand cDNA synthesis Kit (Thermo, USA) under the following thermocycling conditions: 95°C for 5 mins, and 40 cycles of 95°C for 20 s and 62°C for 30 s, followed by 72°C for 3 mins. The primers of miR-122-5p, AKT3, mTOR, PKM2, PI3K, U6 and GADPH are shown in Table 1. The miRNA and mRNA expression levels were calculated using 2−ΔΔCT methods. U6 was used as an internal control of miRNA-122-5p, and GAPDH served as an internal control of mRNAs, respectively.
Table 1.
The Primers Of Genes For RT-qPCR
| Target Genes | Primer Sequence (5ʹ–3ʹ) |
|---|---|
| miR-122-5p | 5ʹ-ACACTCCAGCTGGGAACGCCATTATCACAC-3ʹ(forward) 5ʹ-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTATTTAGT-3ʹ(reverse) |
| AKT3 | 5ʹ-TGTGGATTTACCTTATCCCCTCA-3ʹ(forward) 5ʹ-GTTTGGCTTTGGTCGTTCTGT-3ʹ(reverse) |
| mTOR | 5ʹ-ATGCTTGGAACCGGACCTG-3ʹ(forward) 5ʹ-TCTTGACTCATCTCTCGGAGTT-3ʹ(reverse) |
| PKM2 | 5ʹ-CTCGCTTCGGCAGCACA-3ʹ(forward) 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ(reverse) |
| PI3K | 5′-ATACCGCGGACTGTGTTTCCAGTACACCT-3′(forward) 5′-GCCCTGCAGACTGACCGTGACATCCTC-3′(reverse) |
| U6 | 5ʹ-GGAGCGAGATCCCTCCAAAAT-3ʹ(forward) 5ʹ-GGCTGTTGTCATACTTCTCATGG-3ʹ(reverse) |
| GAPDH | 5ʹ-GGAGCGAGATCCCTCCAAAAT-3ʹ(forward) 5ʹ-GGCTGTTGTCATACTTCTCATGG-3ʹ(reverse) |
Detection Of Glucose Consumption And Lactate Production
For the glucose consumption assay, 1 nmol/μL glucose standard stock solution was plated into 96-well plates (Thermo, USA) as 0, 2, 4, 6, 8 and 10 nmol/well at 37°C for 30 mins in the dark. The supernatant of cells was incubated onto 96-well plates at 37°C for 30 mins in the dark. The glucose consumption was then measured using glucose colorimetric/fluorometric assay kit. For the lactate production assay, lactate standard was plated into 96-well plates as 0, 0.031, 0.063, 0.125, 0.25, 0.5, 1 and 2 μmol/mL. The cells were incubated with 50 μL Biotin-detection antibody solution at 37°C for 30 mins, followed by incubation with 100 μL HRP-streptavidin Conjugate (1:100) at 37°C for 30 mins in the dark. After washing, the cells were incubated with 90 μL TMB substrate at 37°C for 15–30 mins in the dark. After adding 50 μL stop buffer, the lactate production was measured using lactate (human) ELISA kit within 20 mins. The absorbance was measured at 450 nm using a microplate reader (Bio-Rad).
Western Blot
786-O cells were lysed using RIPA lysis (Beyotime, Shanghai, China) containing protease inhibitors (Bimake, USA). A BCA protein concentration assay kit (Beyotime, Shanghai, China) was used to measure protein concentrations of cell lysates. The cell lysates were separated by SDS-PAGE (Beyotime, Shanghai, China) and transferred onto PVDF membranes (Millipore, USA). Then, the membranes were blocked by blocking solution containing 5% skim milk powder or 5% BSA at room temperature for 2 hrs. The PVDF membranes were incubated with primary antibodies overnight, which were then incubated with secondary antibodies at room temperature for 2 hrs. The primary antibodies included PKM2 (1:1000, CST, USA), LC3 (1:1000, CST, USA), pyruvate dehydrogenase (PDH; 1:1000, CST, USA) and GAPDH (1:1000, CST, USA). The proteins were visualized using a ClarityTM Western ECL Substrate Kit (Bio-Rad, USA) and ChemiDocTM Touch Imaging System (Bio-Rad, USA).
Cell Transfection
MiR-122-5p inhibitor, siPKM2 or the corresponding negative controls was transfected into 786-O cells using lipofectamine reagent (Invitrogen, USA). The miR-122-5p inhibitor and siPKM2 were synthesized by Sai Lan Biological Technology Co., Ltd. (Zhejiang, China). The sequences of primers were as follows: negative control sense, 5ʹ-UUCUCCGAACGUGUC,ACGUTT-3ʹ; negative control antisense, 5ʹ-ACG UGA CAC GUU CGG AGA ATT-3ʹ; siPKM2-1, 5ʹ-CCAUAAUCGUC,CUCACCAATT-3ʹ; siPKM2-2, 5ʹ-UUGGUGAGGACGAU,UAUGGTT-3ʹ; human miR-122-5p inhibitor, 5ʹ-CAAACAC,CAUUGUCACACUCCA-3ʹ.
Cell Counting Kit-8 (CCK-8) Assay
The cell viability was measured using CCK-8 assay. 786-O cells were plated into 96-well plates and incubated overnight. A total of 10 μL CCK-8 reagent was added 96-well plates and incubated at 37°C for 1 hr in the dark. The OD value was read at 450 nm, and the cell viability was calculated.
Flow Cytometry
The cultured 786-O cells were washed with cold PBS twice and then were fixed with ice-cold methanol for 15 mins. Apoptotic cells were first marked with Annexin V-FITC/Propidium Iodide (PI) (KeyGene Biotech.), and then analyzed on Flow Cytometer (ACEA, Zhejiang, China) according to the manufacturer’s protocols. For cell cycle, cells were only stained with PI and analyzed on the Flow Cytometer.
Wound Healing Assay
786-O cells were incubated in 6-cm dishes overnight. After that, miR-122-5p inhibitor or siPKM2 was transfected into 786-O cells overnight. When the cells were grown to about 95%, serum-free medium was added. A total of 10 μL tip was used for cell scratch treatment (two parallel lines). Each group was photographed (100×) at 0 hr, and the distance between the scratches of each group was recorded. After 24 hrs, the cells were observed under an inverted microscope (Nikon, Japan) and photographed by Leica Application Suite software (100×). Furthermore, the scratch distance was used to calculate the cell migration viability of each group.
Cell Fraction Assay
786-O cells were lysed using membrane lysis buffer (10 mM HEPES, pH 8.0, 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT and 1% Igepal CA-630), then vortexed and centrifuged at 13,200 rpm for 5 mins. The supernatant was harvested as the cytoplasmic fraction. The pellets were re-suspended with RIPA buffer (Millipore), and sonicated twice on ice and centrifuged at 13,200 rpm for 15 mins. The supernatant was the nuclear fraction. The cytoplasmic fraction and the nuclear fraction were analyzed by Western blot.
Statistical Analysis
All data are presented as mean ± standard deviation (SD) at least three independent experiments. Statistical analyses were performed using GraphPad Prism 7.0 software (GraphPad Software, San Diego, CA, USA). The difference between two groups was analyzed using Student’s t-test. For pairwise multiple comparisons, while one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test was performed. P-value < 0.05 was considered to be statistically significant.
Results
miR-122-5p Is UpRegulated In Renal Cancer Cells
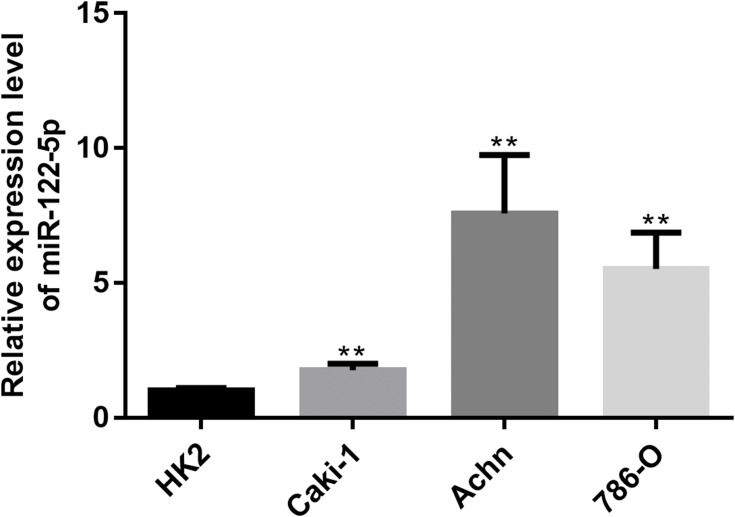
To explore the function of miR-122-5p in renal cancer, the expression of miR-122-5p in renal cancer cells was detected. The RT-qPCR results revealed that the expression of miR-122-5p in renal cancer cells was higher than in HK2 cells. Furthermore, we found that the expression levels of miR-122-5p in 786-O and Achn cells were higher than in Caki-1 cells (Figure 1).
Figure 1.
RT-qPCR results showed that miR-122-5p is upregulated in renal cancer cells compared to HK2 cells (**p<0.01).
Altered Energy Metabolism In Renal Cancer Cells
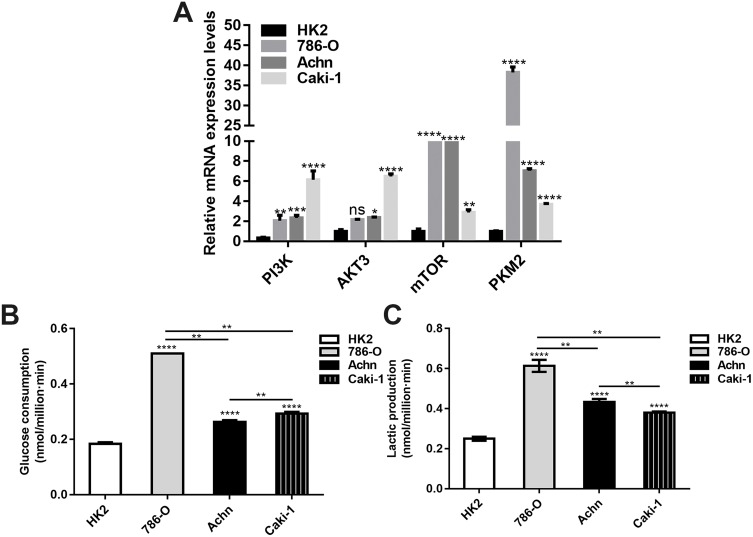
We investigated the mRNA expression of PI3K, AKT3, mTOR and PKM2 in renal cancer cells. The results showed that the expression levels of PI3K, AKT3, mTOR and PKM2 in all renal cancer cells were higher than in HK2 cells. There were significant differences in the expression levels of PI3K, AKT3, mTOR and PKM2 among different renal cancer cells (Figure 2A). In addition, we detected the energy metabolism in different renal cancer cells. As shown in Figure 2B and C, the glucose consumption and lactate production in all renal cancer cells were significantly higher than in HK2 cells. Moreover, 786-O cells had the highest glucose consumption and lactate production. Therefore, 786-O cell lines were chosen for subsequent analysis.
Figure 2.
Altered energy metabolism in renal cancer cells. (A) RT-qPCR results showed the relative mRNA expression levels of PI3K, AKT3, mTOR and PKM2 in renal cancer cells. (B, C) The glucose consumption and lactate production in renal cancer cells (*p<0.05; **p<0.01; ***p<0.001; ****p<0.0001).
miR-122-5p Could Inhibit PKM2 Expression In 786-O Cells
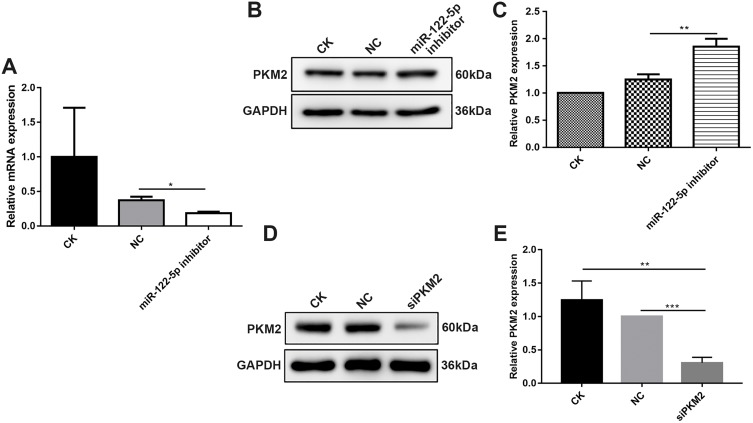
After 786-O cells transfected by miR-122-5p inhibitor, RT-qPCR results demonstrated that the mRNA expression of miR-122-5p after transfection was obviously lower than control group (Figure 3A), suggesting that 786-O cells were successfully transfected by miR-122-5p inhibitor. Then, we investigated the protein expression of PKM2 in 786-O cells transfected by miR-122-5p inhibitor. Western blotting analysis results showed that the protein expression of PKM2 in 786-O cells transfected by miR-122-5p inhibitor was significantly higher than in the control group (Figure 3B and C). Therefore, miR-122-5p could suppress PKM2 expression in 786-O cells.
Figure 3.
MiR-122-5p could inhibit PKM2 expression in 786-O cells. (A) RT-qPCR results showed that the mRNA expression of miR-122-5p in 786-O cells transfected by miR-122-5p inhibitor was obviously lower than of control group. (B, C) Western blot analysis results showed that the protein expression of PKM2 in 786-O cells transfected by miR-122-5p inhibitor was significantly higher than of control group. (D, E) The protein expression of PKM2 in 786-O cells transfected by siPKM2 was significantly lower than control group using Western blot analysis (*p<0.05; **p<0.01; ***p<0.001).
In addition, we tested the expression of PKM2 in 786-O cells transfected by siPKM2. Western blotting analysis results showed that the expression of PKM2 in 786-O cells transfected by si-PKM2 was significantly lower than control groups (Figure 3D and E). Therefore, 786-O cells were successfully transfected by siPKM2.
Knockdown Of miR-122-5p Or PKM2 Promotes Renal Cancer Cell Viability
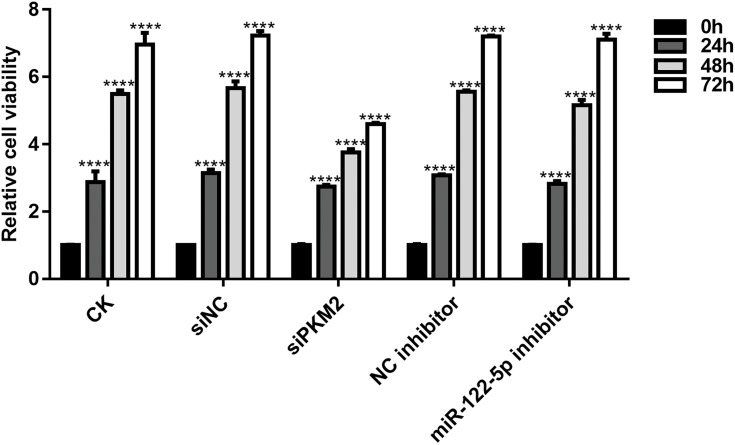
We further investigated whether miR-122-5p could affect renal cancer cell viability. As shown in Figure 4, CCK-8 assay results showed that the cell viability of 786-O cells was significantly reduced after transfection by siPKM2 or miR-122-5p inhibitor compared with control groups. Moreover, we found that the cell viability of 786-O cells transfected by miR-122-5p inhibitor was obviously higher than that of 786-O cells transfected by siPKM2. We have found that the expression of PKM2 was elevated after silencing miR-122-5p. Therefore, miR-122-5p could promote renal cancer cell viability by inhibiting PKM2.
Figure 4.
Knockdown of miR-122-5p or PKM2 suppresses renal cancer cell viability using CCK-8 assay (****p<0.0001).
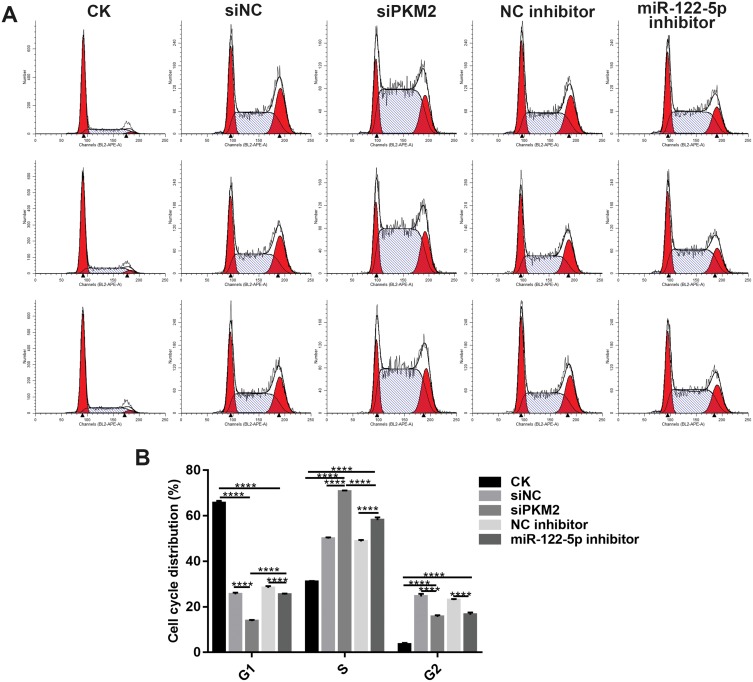
Knockdown Of miR-122-5p Or PKM2 Affects Cell Cycle Progression
We investigated whether miR-122-5p or PKM2 could affect cell cycle of renal cancer cells. After transfection by siPKM2, the G1 phase of 786-O cells was significantly blocked, and the S phase was significantly increased. After transfection of miR-122-5p inhibitor, the S phase of 786-O cells was significantly increased, and the G1 and G2 phases were significantly shortened. In addition, after 786-O cells transfected by miR-122-5p, PKM2 expression was up-egulated and the G1 phase was longer than 786-O cells transfected by siPKM2 (Figure 5A and B). Therefore, knockdown of miR-122-5p or PKM2 could affect cell cycle progression in renal cancer cells.
Figure 5.
Knockdown of miR-122-5p or PKM2 affects cell cycle progression of renal cancer cells. (A, B) The cell cycle of 786-O cells was detected with flow cytometry assay (****p<0.0001).
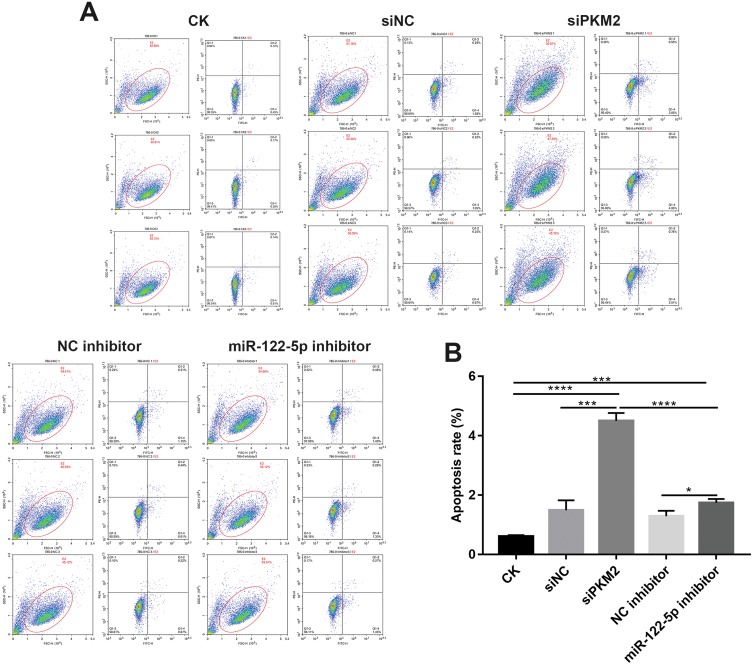
Knockdown Of miR-122-5p Or PKM2 Promotes Renal Cancer Cell Apoptosis
We found that the apoptosis of 786-O cells transfected by siPKM2 or miR-122-5p inhibitor was significantly increased compared to control groups (Figure 6A and B). Moreover, the apoptosis rate of 786-O cells transfected by siPKM2 was significantly higher than 786-O cells transfected by miR-122-5p inhibitor. Thus, knockdown of miR-122-5p or PKM2 could promote renal cancer cell apoptosis.
Figure 6.
Knockdown of miR-122-5p or PKM2 promotes renal cancer cell apoptosis. (A and B): The apoptosis of 786-O cells was detected with flow cytometry assay (*p<0.05; ***p<0.001; ****p<0.0001).
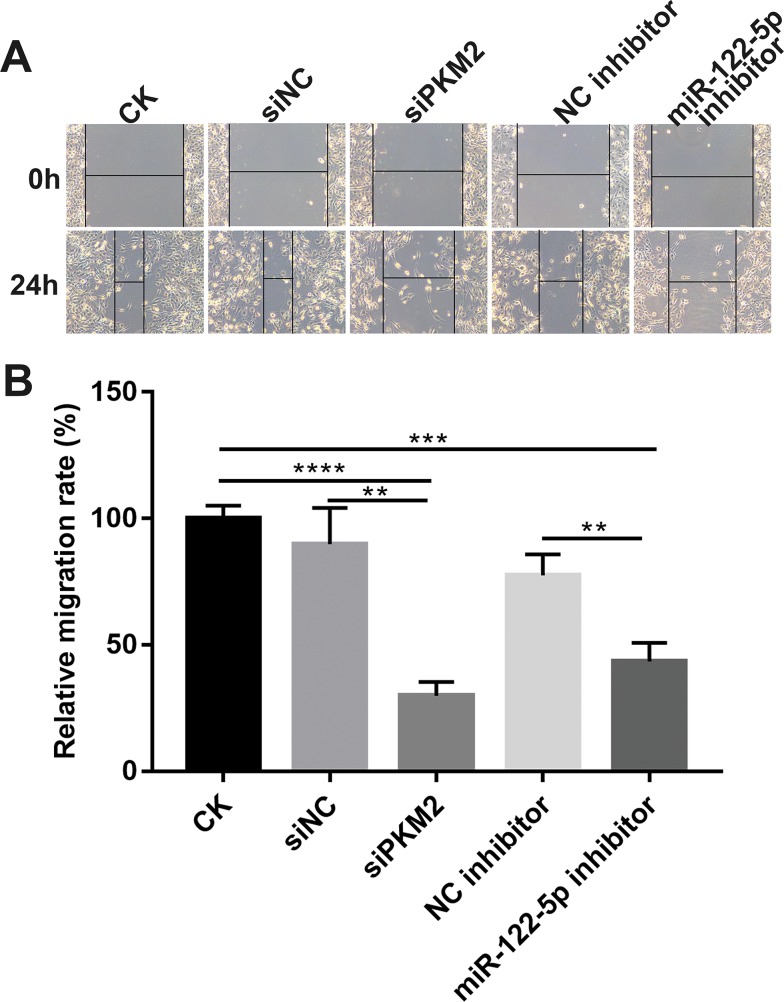
Knockdown Of miR-122-5p Or PKM2 Inhibits Renal Cancer Cell Migration
Wound healing assay results revealed that migration ability of 786-O cells was significantly suppressed after transfection by miR-122-5p inhibitor or siPKM2. The above results suggest that knockdown of miR-122-5p or PKM2 inhibits renal cancer cell migration (Figure 7A and B).
Figure 7.
Knockdown of miR-122-5p or PKM2 inhibits renal cancer cell migration. (A and B) The migration ability was detected in 786-O cells transfected miR-122-5p inhibitor or siPKM2 for 24 hrs (**p<0.01; ***p<0.001; ****p<0.0001).
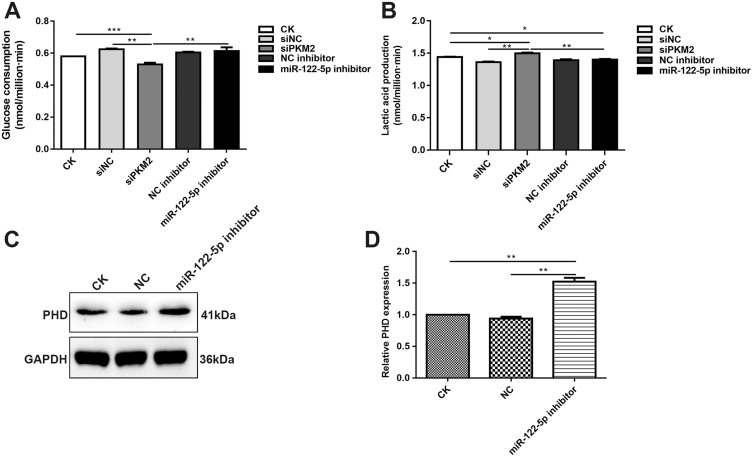
Inhibiting miR-122-5p Promotes Glycolysis Of Renal Cancer Cells By PKM2
We also detected whether miR-122-5p could affect the glycolysis of renal cancer cells. Our results showed that the glucose consumption was significantly decreased in 786-O cells transfected by siPKM2 compared with control group (Figure 8A). However, after inhibiting miR-122-5p, the glucose consumption of 786-O cells was elevated. Furthermore, we found that the glucose consumption of 786-O cells transfected by siPKM2 was obviously lower than 786-O cells transfected by miR-122-5p inhibitor. In addition, the lactate production was significantly increased in 786-O cells transfected by siPKM2 (Figure 8B). However, after transfection with miR-122-5p inhibitor, the lactate production of 786-O cells was inhibited compared with siPKM2 group. We also examined the expression of protein involved in glucose metabolism of renal cancer cells. The results showed that the expression of PDH was increased in 786-O cells transfected by miR-122-5p inhibitor (Figure 8C and D). These results indicated that inhibiting miR-122-5p might promote glycolysis of renal cancer cells by targeting PKM2.
Figure 8.
Inhibiting miR-122-5p promotes glycolysis of renal cancer cells by PKM2. (A) The glucose consumption of 786-O cells transfected by siPKM2 or miR-122-5p inhibitor. (B) The lactate production of 786-O cells transfected by siPKM2 or miR-122-5p inhibitor. (C, D) Western blot analysis results showed that the expression of PDH was significantly increased in 786-O cells transfected by miR-122-5p inhibitor (*p<0.05; **p<0.01; ***p<0.001).
miR-122-5p Could Be Involved In Autophagy Of Renal Cancer Cells
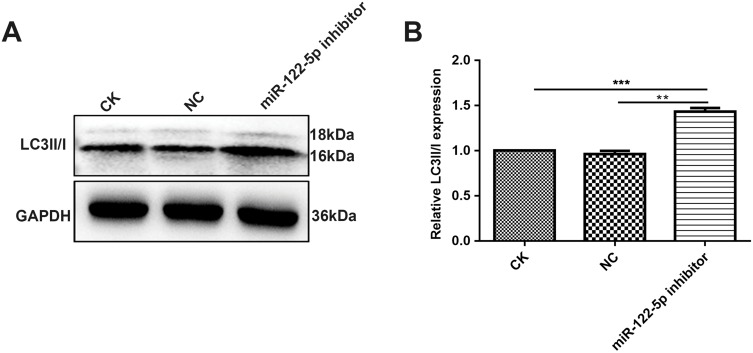
It has been well demonstrated that autophagy may have association with renal cancer.24 In this study, Western blot analysis results showed that LC3II/I was elevated in 786-O cells treated with miR-122-5p inhibitor (Figure 9A and B), suggesting that miR-122-5p could be involved in autophagy of renal cancer cells.
Figure 9.
MiR-122-5p could be involved in autophagy of renal cancer cells. (A, B) Western blot analysis results showed the expression levels of LC3II/I in 786-O cells treated with miR-122-5p inhibitor (**p<0.01; ***p<0.01).
Inhibiting miR-122-5p Promotes PKM2 Nuclear Translocation
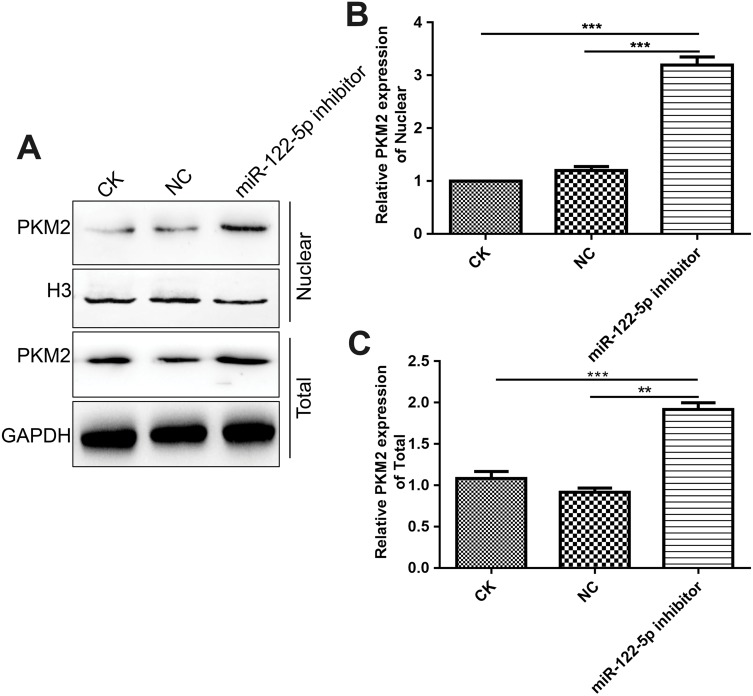
We further observed whether miR-122-5p could affect PKM2 nuclear translocation. In this study, we found that miR-122-5p inhibitor treatment increased the nuclear translocation of PKM2 in 786-O cells (Figure 10A–C), indicating that the activity of PKM2 might be affected by miR-122-5p in renal cancer cells.
Figure 10.
Inhibiting miR-122-5p promotes PKM2 nuclear translocation in 786-O cells. (A–C): The nuclear location of PKM2 in 786-O cells under miR-122-5p inhibitor treatment (**p<0.01; ***p<0.01).
Discussion
In the present study, we found that overexpressed miR-122-5p promotes cell viability, proliferation, migration, glycolysis and autophagy of renal cancer cells by negatively regulating PKM2.
Altered cell metabolism is critical for tumorigenesis, which is regulated by complex networks. In a fully oxidized environment, cancer cells increase glucose uptake along with increased lactic acid production.21 In our study, we found that glucose consumption and lactate production were increased in renal cancer cells. Metabolic reprogramming is a hallmark of cancer that is caused by changes in transcription, translation and post-translational events that collectively coordinate increased activity within cancer cells.25 Many genes involved in oxygen sensing, chromatin stabilization and cell metabolism are abnormally expressed in renal cancer cells.26 Despite the development of therapeutic methods, frequent recurrences and treatments have many side effects.27 Therefore, there is an urgent need to explore novel specific therapeutic targets.
MiRNAs have been found to be potential therapeutic targets for the treatment of renal cancer.28–32 MiRNAs have been found to play important roles in a variety of cellular processes. Among them, miR-122-5p has been shown to be involved in the progression of various cancers, including renal cancer.33,34 Our study found that miR-122-5p is highly expressed in renal cancer cells. After knockdown of miR-122-5p, the cell viability of 786-O cells was significantly reduced after transfection by si-miR-122-5p at 48 hrs. After knockdown of miR-122-5p, the S phase of 786-O cells was significantly increased, and the G1 and G2 phases were significantly shortened. Therefore, miR-122-5p could affect cell cycle of renal cancer cells. At the same time, the low expression of miR-122-5p promoted cell apoptosis and inhibited cell migration for renal cancer cells. Therefore, knockdown of miR-122-5p inhibited cell viability.
PI3K/AKT/mTOR signaling pathway in renal cell carcinoma has a critical role in oncogenesis. It has been confirmed that its dysregulation is related to poor prognosis in renal cell carcinoma.35 In the present study, we found that PI3K, AKT3 and mTOR were upregulated in renal cancer cells.
PKM2 is a crucial regulator in glycolysis, and its high expression contributes to the rapid proliferation of cancer cells.36–38 In our study, PKM2 was highly expressed in ccRCC cells. After knockdown of PKM2, the proliferation and migration abilities of renal cancer cells were significantly decreased. Furthermore, we found that apoptosis and autophagy were both significantly increased after inhibiting PKM2. Our results indicated that PKM2 affects the cell viability of renal cancer cells and participates in metabolic reprogramming in ccRCC cells, which was consistent with previous research. Previous study has confirmed that upregulated PKM2 promotes ccRCC cell proliferation, high glucose consumption and high lactate production.39–41 Furthermore, in this study, we found that PKM2 may become a new target for miR-122-5p. Because, after miR-122-5p knockdown, the expression level of PKM2 was significantly increased in 786-O cells. Previous studies have found that PKM2 may be a target of miR-122-5p through the public miRNA database (TargetScan), and the 3ʹ-untranslated region (3ʹ-UTR) of PKM2 contains a highly conserved binding site for miR-122-5p.42
Therefore, we found that overexpressed miR-122-5p promotes cell viability, proliferation, migration, glycolysis and autophagy of renal cancer cells by negatively regulating PKM2. Our findings shed novel light on molecular research in renal cancer, which provides a new solid foundation for the development of renal cancer therapy.
Conclusion
In our study, we found that miR-122-5p could inhibit the expression of PKM2 in renal cancer. Knockdown of PKM2 or miR-122-5p significantly inhibited renal cancer cell proliferation, migration and promoted apoptosis and autophagy. Furthermore, knockdown of PKM2 contributed to metabolic reprogramming. Therefore, upregulated miR-122-5p facilitates cell viability, proliferation, migration, glycolysis and autophagy of renal cancer cells by negatively regulating PKM2.
Funding Statement
This work was funded by Zhejiang Traditional Chinese Medicine Science and Technology Plan Project (2017ZA009) and Zhejiang Medical and Health Science and Technology Project (2018KY263).
Abbreviations
PKM2, pyruvate kinase M2; miRNA, microRNA; ccRCC, clear cell renal cell carcinoma; PK, pyruvate kinase; siRNA, small interfering RNA; RT-qPCR, quantitative real-time PCR.
Availability Of Data And Materials
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Author Contributions
Mi Zhou conceived and designed the study. Shuai Wang, Wei Zheng conducted most of the experiments and data analysis, and wrote the manuscript. Alin Ji, Dahong Zhang participated in collecting data and helped to draft the manuscript. All authors reviewed and approved the manuscript. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest in this work.
References
- 1.Capitanio U, Bensalah K, Bex A, et al. Epidemiology of renal cell carcinoma. Eur Urol. 2019;75:74–84. doi: 10.1016/j.eururo.2018.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lenis AT, Donin NM, Johnson DC, et al. Adjuvant therapy for high risk localized kidney cancer: emerging evidence and future clinical trials. J Urol. 2018;199:43–52. doi: 10.1016/j.juro.2017.04.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan HJ, Shirk JD, Chamie K, Litwin MS, Hu JC. Patient function and the value of surgical care for kidney cancer. J Urol. 2017;197:1200–1207. doi: 10.1016/j.juro.2016.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turato C, Fornari F, Pollutri D, et al. MiR-122 targets SerpinB3 and is involved in sorafenib resistance in hepatocellular carcinoma. J Clin Med. 2019;8:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Church RJ, Kullak-Ublick GA, Aubrecht J, et al. Candidate biomarkers for the diagnosis and prognosis of drug-induced liver injury: an international collaborative effort. Hepatology. 2019;69:760–773. doi: 10.1002/hep.29802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luna JM, Barajas JM, Teng KY, et al. Argonaute CLIP defines a deregulated miR-122-bound transcriptome that correlates with patient survival in human liver cancer. Mol Cell. 2017;67:400–10.e7. doi: 10.1016/j.molcel.2017.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamane D, Selitsky SR, Shimakami T, et al. Differential hepatitis C virus RNA target site selection and host factor activities of naturally occurring miR-122 3 variants. Nucleic Acids Res. 2017;45:4743–4755. doi: 10.1093/nar/gkw1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masaki T, Arend KC, Li Y, et al. miR-122 stimulates hepatitis C virus RNA synthesis by altering the balance of viral RNAs engaged in replication versus translation. Cell Host Microbe. 2015;17:217–228. doi: 10.1016/j.chom.2014.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandimali N, Huynh DL, Zhang JJ, et al. MicroRNA-122 negatively associates with peroxiredoxin-II expression in human gefitinib-resistant lung cancer stem cells. Cancer Gene Ther. 2019;26:292–304. doi: 10.1038/s41417-018-0050-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu X, Gao F, Wang J, et al. MiR-122-5p inhibits cell migration and invasion in gastric cancer by down-regulating DUSP4. Cancer Biol Ther. 2018;19:427–435. doi: 10.1080/15384047.2018.1423925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byrnes CC, Jia W, Alshamrani AA, Kuppa SS, Murph MM. miR-122-5p expression and secretion in melanoma cells is amplified by the LPAR3 SH3-binding domain to regulate Wnt1. Mol Cancer Res. 2019;17:299–309. doi: 10.1158/1541-7786.MCR-18-0460 [DOI] [PubMed] [Google Scholar]
- 12.Maierthaler M, Benner A, Hoffmeister M, et al. Plasma miR-122 and miR-200 family are prognostic markers in colorectal cancer. Int J Cancer. 2017;140:176–187. doi: 10.1002/ijc.30433 [DOI] [PubMed] [Google Scholar]
- 13.Fan Y, Ma X, Li H, et al. miR-122 promotes metastasis of clear-cell renal cell carcinoma by downregulating dicer. Int J Cancer. 2018;142:547–560. doi: 10.1002/ijc.v142.3 [DOI] [PubMed] [Google Scholar]
- 14.Jingushi K, Kashiwagi Y, Ueda Y, et al. High miR-122 expression promotes malignant phenotypes in ccRCC by targeting occludin. Int J Oncol. 2017;51:289–297. doi: 10.3892/ijo.2017.4016 [DOI] [PubMed] [Google Scholar]
- 15.Heinemann FG, Tolkach Y, Deng M, et al. Serum miR-122-5p and miR-206 expression: non-invasive prognostic biomarkers for renal cell carcinoma. Clin Epigenetics. 2018;10:11. doi: 10.1186/s13148-018-0444-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 17.Jia L, Huang S, Yin X, Zan Y, Guo Y, Han L. Quercetin suppresses the mobility of breast cancer by suppressing glycolysis through Akt-mTOR pathway mediated autophagy induction. Life Sci. 2018;208:123–130. doi: 10.1016/j.lfs.2018.07.027 [DOI] [PubMed] [Google Scholar]
- 18.Zhang K, Zhang M, Jiang H, Liu F, Liu H, Li Y. Down-regulation of miR-214 inhibits proliferation and glycolysis in non-small-cell lung cancer cells via down-regulating the expression of hexokinase 2 and pyruvate kinase isozyme M2. Biomed Pharmacother. 2018;105:545–552. doi: 10.1016/j.biopha.2018.06.009 [DOI] [PubMed] [Google Scholar]
- 19.Zhou Y, Niu W, Luo Y, et al. p53/Lactate dehydrogenase A axis negatively regulates aerobic glycolysis and tumor progression in breast cancer expressing wild-type p53. Cancer Sci. 2019;110:939–949. doi: 10.1111/cas.13928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nemazanyy I, Espeillac C, Pende M, Panasyuk G. Role of PI3K, mTOR and Akt2 signalling in hepatic tumorigenesis via the control of PKM2 expression. Biochem Soc Trans. 2013;41:917–922. doi: 10.1042/BST20130034 [DOI] [PubMed] [Google Scholar]
- 21.Wong N, Ojo D, Yan J, Tang D. PKM2 contributes to cancer metabolism. Cancer Lett. 2015;356:184–191. doi: 10.1016/j.canlet.2014.01.031 [DOI] [PubMed] [Google Scholar]
- 22.Dayton TL, Jacks T, Vander Heiden MG. PKM2, cancer metabolism, and the road ahead. EMBO Rep. 2016;17:1721–1730. doi: 10.15252/embr.201643300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu F, Ma F, Wang Y, et al. PKM2 methylation by CARM1 activates aerobic glycolysis to promote tumorigenesis. Nat Cell Biol. 2017;19:1358–1370. doi: 10.1038/ncb3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao Q, Bai P. Role of autophagy in renal cancer. J Cancer. 2019;10:2501–2509. doi: 10.7150/jca.29285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shanmugasundaram K, Nayak BK, Friedrichs WE, Kaushik D, Rodriguez R, Block K. NOX4 functions as a mitochondrial energetic sensor coupling cancer metabolic reprogramming to drug resistance. Nat Commun. 2017;8:997. doi: 10.1038/s41467-017-01106-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng W, Ji S, Zhang H, et al. mTOR activation is critical for betulin treatment in renal cell carcinoma cells. Biochem Biophys Res Commun. 2017;482:1030–1036. doi: 10.1016/j.bbrc.2016.11.153 [DOI] [PubMed] [Google Scholar]
- 27.Wang J, Sun X. MicroRNA-375 inhibits the proliferation, migration and invasion of kidney cancer cells by triggering apoptosis and modulation of PDK1 expression. Environ Toxicol Pharmacol. 2018;62:227–233. doi: 10.1016/j.etap.2018.08.002 [DOI] [PubMed] [Google Scholar]
- 28.Szabo Z, Szegedi K, Gombos K, et al. Expression of miRNA-21 and miRNA-221 in clear cell renal cell carcinoma (ccRCC) and their possible role in the development of ccRCC. Urol Oncol. 2016;34:533.e21-.e27. doi: 10.1016/j.urolonc.2016.06.011 [DOI] [PubMed] [Google Scholar]
- 29.Strauss P, Marti HP, Beisland C, et al. Expanding the utilization of formalin-fixed, paraffin-embedded archives: feasibility of miR-seq for disease exploration and biomarker development from biopsies with clear cell renal cell carcinoma. Int J Mol Sci. 2018;19. doi: 10.3390/ijms19030803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin C, Li Z, Chen P, et al. Oncogene miR-154-5p regulates cellular function and acts as a molecular marker with poor prognosis in renal cell carcinoma. Life Sci. 2018;209:481–489. doi: 10.1016/j.lfs.2018.08.044 [DOI] [PubMed] [Google Scholar]
- 31.Lei Z, Ma X, Li H, et al. Up-regulation of miR-181a in clear cell renal cell carcinoma is associated with lower KLF6 expression, enhanced cell proliferation, accelerated cell cycle transition, and diminished apoptosis. Urol Oncol. 2018;36:93.e23-93.e37. doi: 10.1016/j.urolonc.2017.09.019 [DOI] [PubMed] [Google Scholar]
- 32.Mu W, Hu C, Zhang H, et al. miR-27b synergizes with anticancer drugs via p53 activation and CYP1B1 suppression. Cell Res. 2015;25:477–495. doi: 10.1038/cr.2015.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Q, Liu HO, Liu YD, et al. Decreased expression of hepatocyte nuclear factor 4alpha (Hnf4alpha)/microRNA-122 (miR-122) axis in hepatitis B virus-associated hepatocellular carcinoma enhances potential oncogenic GALNT10 protein activity. J Biol Chem. 2015;290:1170–1185. doi: 10.1074/jbc.M114.601203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin Y, Wang J, Han J, Luo D, Sun Z. MiR-122 inhibits epithelial-mesenchymal transition in hepatocellular carcinoma by targeting Snail1 and Snail2 and suppressing WNT/beta-cadherin signaling pathway. Exp Cell Res. 2017;360:210–217. doi: 10.1016/j.yexcr.2017.09.010 [DOI] [PubMed] [Google Scholar]
- 35.Bodnar L, Stec R, Cierniak S, et al. Clinical usefulness of PI3K/Akt/mTOR genotyping in companion with other clinical variables in metastatic renal cell carcinoma patients treated with everolimus in the second and subsequent lines. Ann Oncol. 2015;26:1385–1389. doi: 10.1093/annonc/mdv166 [DOI] [PubMed] [Google Scholar]
- 36.Zhou Z, Li M, Zhang L, et al. Oncogenic kinase-induced PKM2 tyrosine 105 phosphorylation converts nononcogenic PKM2 to a tumor promoter and induces cancer stem-like cells. Cancer Res. 2018;78:2248–2261. doi: 10.1158/0008-5472.CAN-17-2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shiroki T, Yokoyama M, Tanuma N, et al. Enhanced expression of the M2 isoform of pyruvate kinase is involved in gastric cancer development by regulating cancer-specific metabolism. Cancer Sci. 2017;108:931–940. doi: 10.1111/cas.13211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Q, Tu J, Dou C, et al. HSP90 promotes cell glycolysis, proliferation and inhibits apoptosis by regulating PKM2 abundance via Thr-328 phosphorylation in hepatocellular carcinoma. Mol Cancer. 2017;16:178. doi: 10.1186/s12943-017-0748-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mendez-Lucas A, Li X, Hu J, et al. Glucose catabolism in liver tumors induced by c-MYC can be sustained by various PKM1/PKM2 ratios and pyruvate kinase activities. Cancer Res. 2017;77:4355–4364. doi: 10.1158/0008-5472.CAN-17-0498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi W, Keenan HA, Li Q, et al. Pyruvate kinase M2 activation may protect against the progression of diabetic glomerular pathology and mitochondrial dysfunction. Nat Med. 2017;23:753–762. doi: 10.1038/nm.4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang J, Kong W, Zhang J, et al. c-Myc modulates glucose metabolism via regulation of miR-184/PKM2 pathway in clear-cell renal cell carcinoma. Int J Oncol. 2016;49:1569–1575. doi: 10.3892/ijo.2016.3622 [DOI] [PubMed] [Google Scholar]
- 42.Xu Q, Zhang M, Tu J, Pang L, Cai W, Liu X. MicroRNA-122 affects cell aggressiveness and apoptosis by targeting PKM2 in human hepatocellular carcinoma. Oncol Rep. 2015;34:2054–2064. doi: 10.3892/or.2015.4175 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.