Abstract
Plexiform neurofibromas (PNs), which may be present at birth in up to half of children with type 1 neurofibromatosis (NF1), can cause serious loss of function, such as quadriparesis, and can undergo malignant transformation. Surgery is the first line treatment although the invasive nature of these tumors often prevents complete resection. Recent clinical trials have shown promising success for some drugs, notably selumetinib, an inhibitor of MAP kinase kinase (MEK). We have developed three-dimensional (3D) cell culture models of immortalized cells from NF1 PNs and of control Schwann cells (SCs) that we believe mimic more closely the in vivo condition than conventional two-dimensional (2D) cell culture. Our goal is to facilitate pre-clinical identification of potential targeted therapeutics for these tumors. Three drugs, selumetinib (a MEK inhibitor), picropodophyllin (an IGF-1R inhibitor) and LDN-193189 (a BMP2 inhibitor) were tested with dose-response design in both 2D and 3D cultures for their abilities to block net cell growth. Cell lines grown in 3D conditions showed varying degrees of resistance to the inhibitory actions of all three drugs. For example, control SCs became resistant to growth inhibition by selumetinib in 3D culture. LDN-193189 was the most effective drug in 3D cultures, with only slightly reduced potency compared to the 2D cultures. Characterization of these models also demonstrated increased proteolysis of collagen IV in the matrix by the PN driver cells as compared to wild-type SCs. The proteolytic capacity of the PN cells in the model may be a clinically significant property that can be used for testing the ability of drugs to inhibit their invasive phenotype.
Keywords: Plexiform neurofibroma, NF1, 3D culture model, Matrix proteolysis, Targeted therapeutics
1. Introduction
Plexiform neurofibromas (PNs) are a hallmark of neurofibromatosis type I (NF1), a genetic disease characterized by an autosomal dominant mutation in the NF1 gene. The encoded protein, neurofibromin, as well as other proteins in this class (Ras GTPase activating proteins, RasGAPs) function as negative regulators of Ras. The NF1 mutation results in a single functional NF1 allele in the afflicted individual. Mice, and presumably humans, nullizygous for NF1 do not survive gestation (Brannan et al., 1994). Neurofibromin expression is prominent in brain, spinal cord, peripheral nerve, and adrenal gland with highest abundance in neurons, Schwann cells (SCs) and oligodendrocytes. This expression pattern is consistent with the proliferation of SCs in neurofibromas associated with neurons in the peripheral nervous system (Daston et al., 1992).
PNs arise from large peripheral nerves. SCs or SC precursor cells are thought to be the tumor cells of origin (Zhu and Parada, 2002; Cichowski and Jacks, 2001; Muir et al., 2001). The initial event that predates and appears to be required for tumor growth is loss of SC heterozygosity for neurofibromin (NF1(−/−)) (Kluwe et al., 1999; Cichowski et al., 1999). As they grow these complex tumors become populated by other cell types typical of the environment immediately surrounding the nerve: fibroblasts, vascular endothelial cells, and an influx of mast cells (Kluwe et al., 1999; Le and Parada, 2007) that bear a single NF1 allele (NF1(+/−)). Under tumor conditions the interactions between these various cell types are heightened largely due to the loss of one or both NF1 alleles that confer multiple gain-in-function phenotypes including cytokine and growth factor production and an increased response to specific stimuli (Yang et al., 2012; Ingram et al., 2000).
PNs are present at birth in 25–50% of children with NF1 (Prada et al., 2012). Currently there is no standard drug therapy available although recent clinical trials have shown promising success with the MAP kinase kinase (MEK) inhibitor selumetinib (Dombi et al., 2016). Problematically, only 10% of the compounds that pass through the standard pre-clinical protocol for drug research, i.e., 2D cell culture and subsequent animal testing, are ultimately successful in human clinical trials (Edmondson et al., 2014). Our goal is to provide a bio-engineered 3D system that predicts translational effectiveness at a much higher frequency than is currently achieved through 2D testing. The 3D system consists of a matrix that provides physical support and mechanical cues that alter cell behavior (Mason et al., 2012). In addition cells in 3D are exposed and react to concentration gradients, imposed by the structure of the matrix, of endocrine signaling molecules, bioactive molecules stored and released by the matrix or, potentially, any therapeutics added to the system that will further affect cell behavior (Lee et al., 2008). Traditional 2D cultures, spread out and flat on plastic dishes, are likely to be missing the interactions stimulated by the supporting infrastructure. These differences in exposure to mechanical forces and matrix-mediated signaling molecules have led to the following observations over the last 10 years: 1) the layer of extracellular matrix (ECM) in 3D allows the cells to retain their morphology and spatial arrangement to other cell types mimicking the in vivo condition (Gurski et al., 2009; Feder-Mengus et al., 2008); 2) tumor cells grow more slowly in 3D reflecting tumor growth in vivo (Chitcholtan et al., 2013; Chignola et al., 2000); 3) tumor cells in 3D show increased energy production (Yamaguchi et al., 2013) and a difference in gene expression profiles as compared to 2D (Cheema et al., 2008; Kaur et al., 2012); and 4) tumor cells grown in 3D show different sensitivities to chemotherapeutic or targeted drug therapies (Li et al., 2010; Weaver et al., 2002; Imamura et al., 2015; Chambers KF et al., 2014).
We have developed 3D models that utilize a reconstituted basement membrane (rBM) based on ECM secreted from Engelbreth-Holm-Swarm mouse sarcoma cells: Matrigel with reduced growth factor content and free of phenol red dye. The major components of Matrigel are laminin (60%), collagen type IV (30%), entactin (8%) and heparan sulfate proteoglycan (Kleinman and Martin, 2005). These components are also found in the endoneurium surrounding the SC-axon unit of the peripheral nervous system. Collagen type IV, detected in abundance, is a major constituent of mammalian ECM (Platt et al., 2003). Laminin, a protein made up of 3 chains, is present at a high concentration at the inner surface of the endoneurium close to the SC (Suri and Schmidt, 2010). Proteoglycans commonly expressed in the nervous system are part of the ECM or are associated with cell membranes (Hartmann and Maurer, 2001). Under normal physiological conditions SCs bear integrins that bind to laminin allowing adhesion of the cell to the ECM, which is a necessary step in myelination (Berti et al., 2006). There is thus the potential for a working relationship between the PN driver cell and the rBM.
Culture in 2D has contributed much to our understanding of the molecular biology of the cell but fails to take into account the presence in vivo of multiple cell types in addition to the complexity of the tumor environment. There is thus an emerging paradigm that 3D models provide a superior pathomimetic recapitulation of tumor biology (Kimlin et al., 2013). The goal of this study was to develop robust 3D NF1 PN models utilizing NF1(−/−) cells originally derived from PNs and controls of SCs with wild-type neurofibromin expression, to provide new platforms for identification of targeted therapeutics that will be effective for translation into the clinic. Toward this goal we examined differences in morphology and matrix proteolysis between wild-type SCs and NF1 PN cells in 3D culture. We used our 3D model to test a MEK inhibitor, selumetinib, that has recently been successful in inhibiting PN growth in clinical trials in addition to testing two developmental drugs that block other pathways that have not yet been targeted clinically for NF1 patients.
2. Materials and methods
2.1. Materials
Addgene, Cambridge MA: pCMV-dR8.2 dvpr (Cat. #8455). Agilent Technologies, Santa Clara CA: pVPack-VSV-G (Cat. #217567); pVPack-GP (Cat. #217566). Axon Medchem LLC, Reston VA: LDN-193189 hydrochloride (Cat. #1509; Batch 7). Clontech, Mountain View CA: pLVX–IRES-tdTomato (Cat. #631238). Cell Biolabs, Inc., San Diego CA: ViraDuctin Lentivirus Transduction Kit (Cat. #LTV-200). Corning, Bedford MA: (reconstituted basement membrane preparation, rBM) Matrigel, growth factor-reduced and phenol red-free (Cat. #356231; lot #4097006, #4153012, #5005006); collagen I, high concentration, rat tail (Cat. #354249; lot #4191001); fetal bovine serum (FBS) (Cat. #35010CV); solution, 50 × (Cat. #30–001-Cl). Denville Scientific, Holliston MA: 30 mm cell culture plates (Cat. #T1103). Invitrogen, Thermo Fisher Scientific, Waltham MA: Hyclone DMEM/high glucose (Cat. #SH30243.01); Dye-quenched (DQ)-collagen IV, fluorescein conjugate (Cat. #D12052; lot #1586272); Hoechst dye 33,342 (Cat. #H3570, lot #1608494); lipofectamine 2000 (Cat. #11568019; lot #1401254). Promega, Madison WI: CellTiter-Glo Luminescent Cell Viability Assay (Cat. #G7573; lot #0000156347). Sigma-Aldrich, St. Louis MO: mycoplasma detection kit (Cat. #MP0025). Thermo Fisher Scientific, Rochester NY: Nunc 96-well optical bottom plate sterile white, for Glo-max assays (Cat. #165306; lot #1173754); Corning 96-well flat bottom plate for tissue culture (Cat. #07–200–20); Tocris Bioscience, Bristol UK: picropodophyllin (Cat. #2956; lot #2B/110105).
2.2. Cell lines and genetic analysis
The following immortalized cell lines were described in (Li et al., 2016): ipn02.3 2λ, ipn97.4 and ipn02.8 were originally derived from human normal (wild-type) non-tumor nerves; cell lines ipNF95.6, ipNF95.11b C, and ipNF95.11b C/T were derived from NF1 PNs. Lines ipNF95.11b C and ipNF95.11b C/T were derived from the same PN but immortalized independently. Mutation analysis was performed (Li et al., 2016) on ipNF95.11b C/T and ipNF95.11b C by PCR amplification of NF1 gene intron 41 followed by mutation-specific restriction digest/polyacrylamide electrophoresis. Results showed that both lines had lost the wild-type allele with no evidence of a normal allele. Sequencing was performed (Li et al., 2016) on ipNF95.6 that showed both the germline and somatic NF1 alleles have nonsense mutations although at different sites. These PN lines are thus regarded as two-hit tumor cell lines (not heterozygous) (Li et al., 2016). All cell lines were maintained as monolayers at 37° C, 5% CO2 by passing every 2 or 3 days in growth media (DMEM/high glucose + 10% FBS). Cell lines were routinely checked to ensure that they were free of mycoplasma contamination.
2.3. Red Fluorescent Protein (RFP) lentiviral transduction
The producer cell line HEK293T was transfected with the packaging plasmids pCMV-dR8.2 dvpr (Stewart et al., 2003), pVPack-VSV-G, and pVPack-GP along with pLVX-IRES-tdTomato for expression of RFP using lipofectamine 2000 transfection reagent. The producer cells were incubated for 24 h at 32 °C after which the viral broth was collected. Transduction of SCs, ipn02.3 2λ, ipn02.8, and ipn97.4 and NF1 PN cells, ipNF95.6, ipNF95.11b C, and ipNF95.11b C/T (at low cell confluence, ~20%) was performed using ViraDuctin Lentivirus Transduction Kit according to the manufacturer’s instructions. This protocol was repeated for a 2nd and 3rd day using additional viral broth collected from the producer cells. A 30 to 90% transduction was achieved depending on the cell line. Cells were incubated with the selection agent G418 for 6 days at concentrations that ranged from 600 to 2400 mg/ml depending on the cell line. This was followed by sorting for the population of cells (20% of the total number) emitting red fluorescence at the highest intensity on a Sony SH800 cell sorter (Microscopy, Imaging and Cytometry Resources Core at Karmanos Cancer Institute, Wayne State University). Samples from these mixed populations were appropriately diluted and clones were isolated and expanded from ipn02.8-RFP, ipn97.4-RFP and ipNF95.11b C-RFP.
2.4. 2D culture
The conditions under which testing of the effect of drugs on cell viability in 2D culture were as follows: cells were plated in 96-well plates with no additional coatings applied. The plating densities (in cells/well) of the cell lines used to test the MEK inhibitor varied to account for individual growth rate and plating efficiency and were as follows: wild-type cell lines, ipn02.8-RFP (1800) and ipn97.4-RFP (3000); PN cell lines, ipNF95.11b C-RFP (2500) and ipNF95.6-RFP (2000). For testing LDN-193189 and picropodophyllin, the plating density of the wild-type cell line, ipn02.3, and the PN cell lines, ipNF95.11b C and ipNF95.6 was 1500 cells/100 μl growth media/well. Assay media for drug testing was DMEM, high glucose, supplemented with 10% FBS and 1% penicillin streptomycin solution.
2.5. 3D culture
Cell viability assays for drug testing in 3D were performed in 96-well plates that we pre-coated with 30 μl rBM (Matrigel) in each well. This provides a layer of about 1 mm of thickness. If the layer is too thin the cells migrate to the plastic and assume the morphology of a 2D monolayer. Because of the additional space for growth provided by the 3rd dimension, the plating density for 3D was higher than 2D. Specifically plating numbers for MEK testing were varied according to growth rate as follows: wild-type cells lines, ipn02.8-RFP at 2500 cells/well and ipn97.4-RFP at 3300 cells/well; PN cell lines, ipNF95.11b C-RFP and ipNF95.6-RFP at 8000 cells/well. Plating numbers for testing the effects of LDN-193189 and picropodophyllin were 4000 cells/well for all cell lines. The higher 3D plating density likely increases the presence of cell-secreted factors that promote the formation of networks or aggregates. The volume provided by rBM also allows the cells to assume a morphology that more closely mimics in vivo conditions (Barcellos-Hoff et al., 1989); see Fig. 1. In the presence of rBM the 3D growth media supports viability at a reduced FBS concentration of 2% as compared to 2D growth media supplemented at 10%. The 3D assay media was further supplemented with 2% rBM and 1% penicillin streptomycin solution.
Fig. 1.
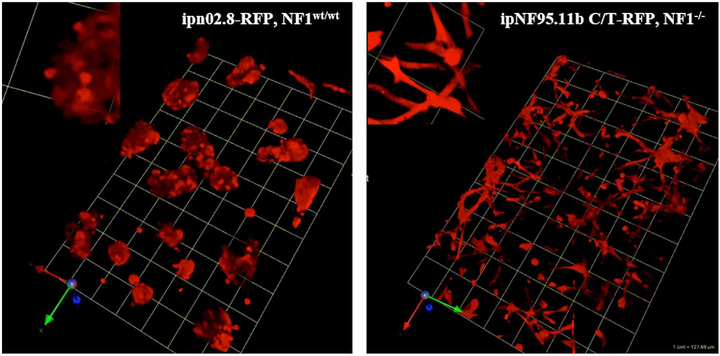
Comparison of cell morphology between structures formed in 3D by wild-type SCs and NF1 PN cells. RFP variants of wild-type SCs and NF1 PN cells were established in rBM + collagen I (60:40). After six days, the live 3D cultures were imaged on confocal microscope. The images shown are tilted views of the z-stacks that have been reconstructed in 3D to show the population of multicellular structures formed. The RFP marks the volume occupied by the cells. One grid unit = 128 μm. Left panel: wild-type SC line, ipn02.8-RFP, inset: magnification of selected area; right panel: NF1 PN cell line, ipNF95.11b C/T-RFP, inset: magnification of selected area.
For confocal imaging, cells were cultured in 3D using plastic coverslips as described (Sameni et al., 2012) with some modifications. A single 1 × 2 cm sterile coverslip was placed in a 35-mm culture dish, chilled on ice and layered with 60 μl of ice-cold matrix. The matrix consisted of either rBM (8 mg/ml) or rBM:collagen I. The rBM:collagen I mixture was prepared on ice as follows: the rBM was diluted to 8 mg/ml with unsupplemented media (DMEM/high glucose); the collagen I was diluted to 8 mg/ml with PBS and the pH was elevated to 7.4 with 1 N NaOH. The rBM and collagen I, respectively, were combined by volume according to the ratio, 60:40. For proteolysis experiments, the DQ-collagen IV substrate was added for a final concentration of 25 μg/ml to the matrix prior to layering. The matrix was then placed on the lowest shelf of the incubator (where humidity is the highest) at 37 °C, 5% CO2 for 20 min to insure gel formation. Cells were trypsinized and resuspended in assay media for a final concentration of 8000 cells/60 μl (unless otherwise noted). The cell suspension (60 μl) was gently spotted across the surface of the solidified matrix producing a sheet of overlaying fluid. Surface tension held the cell suspension onto the matrix as long as touching the plastic was avoided during application. The cells were incubated at 37° C, 5% CO2 for 45–60 min until they adhered to the gel, after which 2 ml of 3D growth media was added.
2.6. Confocal imaging
Images of transfected RFP or Hoechst stained cells as well as proteolytic release of DQ-collagen IV substrate were obtained using a Zeiss laser scanning confocal microscope, LSM 780 equipped with a Zeiss W Plan-Apochromat 20 ×/1.0 NA (numerical aperture) or a Zeiss Achroplan 10 ×/0.3W Ph1 objective. Prior to microscopy the assay media was replaced by imaging media (DMEM, phenol red free to minimize interference with the fluorescent signal, supplemented with 2% FBS). Depending on the experiment, either four randomly selected sections of sample or a single large section was scanned using a water-dipping objective (10 × or 20 ×). Fluorescence of RFP was collected at an excitation wavelength of 550 nm and an emission gating of 551–595 nm. Fluorescein signals (from degraded DQ-collagen IV) were collected at an excitation wavelength of 490 nm and an emission gating of 495–571 nm. Blue fluorescence from the Hoechst dye was collected at an excitation wavelength of 405 nm and an emission gating of 413–517 nm. Z-stacks of the images at 5.67 μm intervals (proteolysis assays), 9.67 μm intervals (volumetric assays), or 2.5–4 μm (morphology assays) were reconstructed with Volocity software (Perkin Elmer) that yielded a numerical evaluation of the intensity and total volume of the signal or calculated the number of nuclei. Intensity of the proteolytic signal (green channel) per cellular structure volume (red channel) or cell number (blue channel) was quantified.
2.7. Cell morphology
Fluorescent emission of cytoplasmic RFP captured by the confocal microscope to define the shape and volume of the cells. Cells were plated on a coverslip at 8000 cells/60 μl of assay media/slip (unless otherwise noted) and scanned after 6 days of growth. Assay media were changed on day 4. Optical sections were collected using a 20 ×/1.0 NA objective. To ensure representative data were collected, a single large section of the sample was scanned by setting the microscope to capture six adjacent fields for an area of approximately 1270 × 889 μm. The setting for the vertical axis was determined by the height of the cell population.
2.8. Proteolysis
For each proteolysis experiment, samples were plated on one of two matrices, rBM or rBM/collagen I, with incorporation of DQ-collagen IV. The protocol for confocal microscopy using cover slips described in the section 3D culture was followed. Each of the five cell lines were tested along with a negative control to define the background signal, i.e., 60 μl of rBM supplemented with DQ-collagen IV at 25 μg/ml without cells. Cells and control were incubated as described for 6 days with a media change on day 4. Four randomly selected fields on a single coverslip were imaged for each sample with parameters described under Confocal Imaging. The images were reconstructed with Volocity software. Proteolysis was determined using a volumetric RFP assay: with the software the fluorescent signals were converted to numerical values for the intensity of the signal from the fluorescein (green) channel and the volume from the RFP (red) channel for a selected group of cells. Proteolysis was expressed as intensity/volume. The results of the experiments for either rBM or rBM:collagen I were analyzed using oneway ANOVA using Kruskal-Wallis Test followed by Dunn’s Multiple Comparison Test in the software analysis program, GraphPad Prism.
2.9. Drug treatment
These assays were designed for a 96-well plate format in 2D and 3D culture using either tissue culture treated, polystyrene plates (MTT assay) or 96 well optical-bottom plates with white upper structure (CellTiter-Glo luminescent assay). Steps of the 3D culture protocol were performed on ice or at 4 °C. Plates and an 8-channel reservoir were chilled for a minimum of 10 min. Using an 8-channel pipette, 30 μl per well of ice cold rBM was dispensed into the plate. In order to reduce the meniscus and create an even spread of the rBM in the wells, plates were subsequently centrifuged at 2000 × g, 4 min, 4 °C in a microplate swinging bucket rotor. This permitted a uniform plating of cells across the surface. The plates were incubated at 37 °C until the rBM formed a gel. Cells were resuspended at the appropriate density (described in section: 3D culture) in 3D assay media (2% rBM, 2% FBS, 1% penicillin-streptomycin in DMEM/high glucose) and plated at 100 μl per well in duplicate plates. Cells were prepared in parallel for 2D culture by resuspension at densities described (2D culture) in DMEM/high glucose supplemented with 10% FBS and 1% penicillin-streptomycin. The cell suspension was dispensed at 100 μl per well on duplicate plates. Empty cells at the perimeters of the plates were filled with sterile water to minimize media evaporation.
Twenty-four hours after plating the drug was applied at the final concentrations indicated. A vehicle treated control (DMSO) was applied to each cell line at the same dilution used to deliver the drug, i.e., for selumetinib testing, 1:1000 dilution of DMSO; for LDN-193189 and PPP testing, 1:250 dilution of DMSO. For LDN-193189 and PPP testing, 50 μM doxorubicin was applied as a positive control for net growth inhibition and an ATP standard curve (1 μM, 100 nM and 10 nM) was prepared in culture media as a positive control for the assay reagent. After an additional 48 h of incubation net cell viability was assayed. The net viability of the cells per sample was determined using tetrazolium dye, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium, (MTT) for selumetinib and the CellTiter-Glo Luminescent Cell Viability Assay for LDN-193189 and PPP.
2.9.1. MTT Assay
After 48 h of drug incubation, 20 μl of 5 mg/ml MTT solution was added to the culture medium of all wells. In addition, 8 μl of FBS was added to each 3D well to achieve a final concentration of 10% FBS. Cells were incubated for 1 to 3 h until a purple colored formazan product was formed. The medium was removed by flipping the plates over and tapping gently. The formazan product was incubated overnight at 37 °C in a 10% SDS in 0.01% HCl solution. This solution solubilized the formazan and also the rBM. The optical density was measured at 570 nm with a background absorbance read at 690 nm (Epoch microplate reader, Biotek Instruments, Winooski, Vermont).
2.9.2. CellTiter-Glo Luminescent Cell Viability Assay
After 48 h of drug incubation the CellTiter-Glo Reagent was added according to the manufacturer’s instructions with the exception that for the 3D wells solubilization of the gel was ascertained by microscope before proceeding to measurement of the luminescent signal (Glomax™, 96 microtiter plate luminometer, Promega, Madison, Wisconsin).
2.9.3. Analysis
To facilitate comparisons of drug effects between cell lines the primary data were normalized to percent of control and then converted from the number of viable cells left after treatment to percent inhibition (100% – % of control). These values were used to construct dose-response curves. Using GraphPad Prism, the logarithm of the concentration of the drug was plotted against the % inhibition and was analyzed as a nonlinear regression according to the equation, log(inhibitor) vs. response–Variable slope (four parameters). Examination of the graphs dictated the use of the least squares (ordinary) method of fitting except in the case of an extreme outlier, where a robust fit was used. The IC50 values obtained are reported in Table S1 in the supplementary data.
2.10. Data archiving and availability
Original data from this study have been deposited at www.synapse.org and will be available on publication.
3. Results
3.1. Morphology of SCs and NF1 PN cells in 3D culture
To facilitate the visualization of the SC and NF1 PN cells in the optically transparent rBM matrix, we stably transduced all the cell lines for this study with RFP. Tumor cells in 3D culture often grow in aggregates that take on a rounded or spherical shape as opposed to the flat morphology induced by growth on plastic (Gurski et al., 2009). The normal microenvironment of SCs is dominated by collagen IV and laminin (Carey et al., 1983; Haraida et al., 1992). In vivo growth of PNs results in increased fibroblast infiltration/proliferation and production of collagen I (Yang et al., 2012; Yang et al., 2006). We therefore used either rBM (predominantly collagen IV and laminin) or a 60:40 (v:v) mix of rBM plus collagen I for this study. The SCs and NF1 PN cells cultured in 3D showed two distinct morphologies. To illustrate these phenotypes, Fig. 1 shows wild-type SCs, ipn02.8(clone8)-RFP and NF1 PN cells, ipNF95.11b C/T-RFP. The individual wild-type SC cells were spherical and typically formed hollow cup-shaped aggregates embedded in the matrix (a morphology representative of the wild-type SC lines cultured in this study). In contrast, the NF1 PN cells grew as layered sheets of cells within the matrix where the individual cells were highly elongated, having two or more tendril-like outgrowths (representative of all PN cell lines cultured). For cell morphology images of wild-type ipn97.4-RFP SCs and NF1 PN cells, ipNF95.11b C-RFP and ipNF95.6, see Supplementary Fig. 1.
3.2. NF1 PN cell lines have an increased capacity for matrix proteolysis
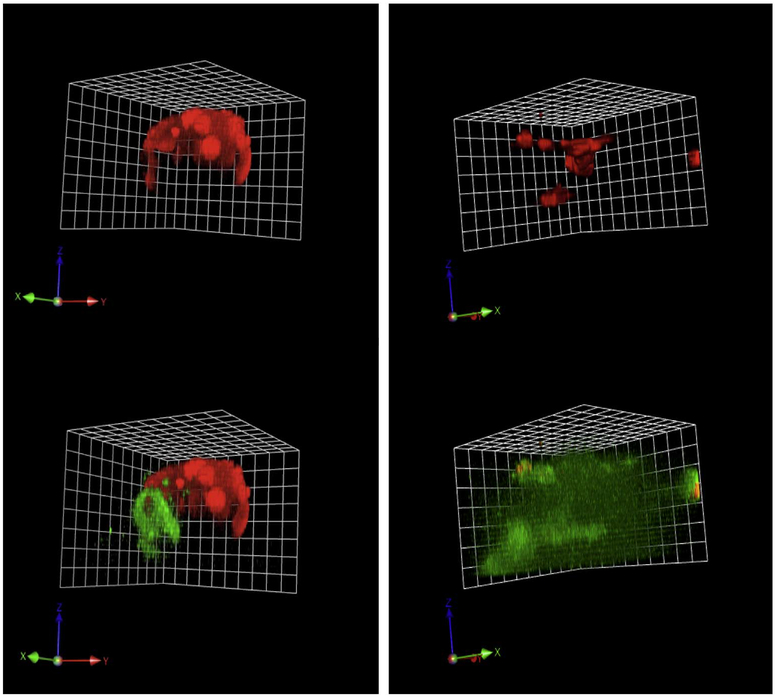
One of the clinical problems of NF1 PNs is that their invasive quality can prevent the possibility of a complete, curative resection. The ability to invade the matrix is promoted by tumor cell secretion of matrix metalloproteinases (MMPs). This class of proteases is located at the cell surface (Sevenich and Joyce, 2014) and at the leading edge of tumors (Cavallo-Medved et al., 2009) enabling the processing of substrates. Although it is the work of multiple proteinases interacting with companion proteins that create a favorable environment for invasion and metastasis, the degradation of collagen IV is part of the process. MMP2 (gelatinase A) enhances the cleavage of collagen IV by MMP9 (gelatinase B) (Kraniak et al., 2018). We therefore assayed the ability of NF1 PN cells to degrade their ECM in comparison to that of wild-type SCs to determine whether this invasive phenotype could be assayed in vitro. We used a live-cell proteolysis assay that reports collagen IV degradation (Sameni et al., 2003) and adapted it for use in 3D cultures of wild-type SCs and NF1 PN cells. There was notably higher capacity for collagen IV degradation (matrix proteolysis) in the NF1 PN cell lines tested compared to the minimal level of activity produced by non-NF1 SCs (Fig. 2).
Fig. 2.
Pre-malignant phenotype of NF1 PN cells: increased matrix proteolysis. RFP variants of wild-type SCs and NF1 PN cells, were established in rBM mixed with dye-quenched (DQ) collagen IV. After six days, the live 3D cultures were imaged on confocal microscope and z-stacks of images were collected and reconstructed with Volocity software to show the structures. The RFP (red) marks the volume occupied by the cells. The cleaved collagen IV (green) shows the matrix proteolysis. The reconstructed 3D images from the confocal microscopy are angled views and representative of four rBM experiments for each cell line. Example movies of the 3D reconstructions are in supplementary data. One grid unit = 21.3 μm. Upper left, wild type cells, ipn97.4-RFP embedded in rBM (red channel only); Lower left, same cell structure as upper left but showing matrix proteolysis (overlaid with green channel). Upper right, PN cells, ipNF95.6-RFP embedded in rBM (red channel only); Lower right, same cell structure as in upper right but showing matrix proteolysis (overlaid with green channel).
In addition to providing morphological analysis of cell structures in 3D cultures (Fig. 1 and Fig. S1), the stable transduction to express RFP was used to develop a novel cell volume assay for proliferation in 3D (Fig. S2) that was then used to quantify the proteolytic phenotype (Fig. 3). To confirm the validity of using RFP volume to report cell number, RFP volumetric data were compared to cell counts based on quantifying the number of stained nuclei. Cells were plated for 3D culture on plastic coverslips and live cell imaging was performed with the confocal microscope after one and six days of growth. Cellular DNA was stained with Hoechst dye 33,342 prior to imaging. RFP fluorescence localized in the cytoplasm and blue fluorescence emitted from the stained DNA in the nuclei were collected in z-stacked images. Reconstruction and quantification of data from both channels by Volocity software yielded structure volume and cell number (count of nuclei). Both structure volume and cell number were compared to determine fold change in the number of viable cells for each parameter over time (Fig. S2) and found to be comparable using either measuring system for all the cell lines tested. One consistent result from this analysis is that the wild-type SCs have a higher growth rate in 3D cultures than do the NF1 PN cells.
Fig. 3.
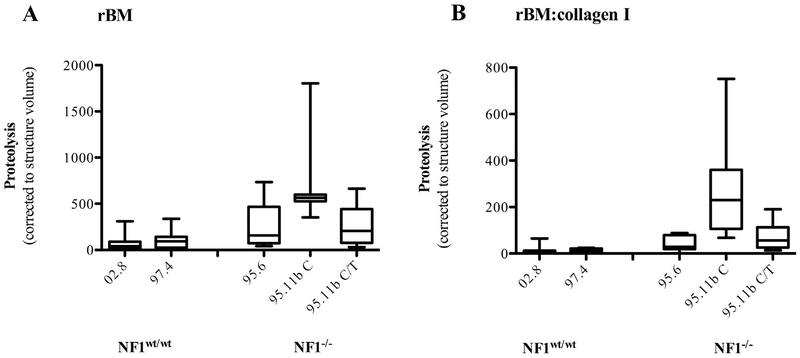
Quantification of collagen IV proteolysis by wild-type SCs and NF1 PN cell lines. A. Indicated cell populations were grown in rBM that incorporates DQ-collagen IV for six days. B. Indicated cell populations were grown in rBM:collagen I that incorporates DQ-collagen IV for six days. Data are box-and-whisker plots of either four (A) or two (B) experiments and show the degradation of the collagen IV in the matrix (green fluorescence) corrected to the cell number as assayed by structure volume (RFP: see Fig. S2). Statistical analysis of the results revealed that the proteolytic activities of the three PN cell lines grown in rBM were significantly higher (p < 0.05) when compared to both wild-type cells. When grown in rBM:collagen I, PN cell line ipNF95.11b C-RFP continues to show significantly (p < 0.05) higher proteolysis. The other two PN lines demonstrate the same trend for enhanced proteolysis although the increases compared to the control lines do not reach statistical significance. 02.8 = ipn02.8-RFP, 97.4 = ipn97.4-RFP, 95.6 = ipNF95.6-RFP, 95.11b C = ipNF95.11b C-RFP, 95.112b C/T = ipNF95.11b C/T-RFP.
Normalization of the proteolytic results to structure volume demonstrated that all of the NF1 PN cell lines tested had a significantly higher (p < 0.05) ability to degrade the collagen IV in the rBM matrix than the control SCs (Fig. 3). We also tested the effect of matrix composition on proteolysis by comparing results from 3D cultures growing in rBM with those growing in rBM mixed with collagen I (60:40, v:v). The data revealed a similar trend for the NF1 PN cell lines to again have higher proteolytic capacity, although the difference reached significance (p < 0.05) only in the ipNF95.11b C-RFP line (Fig. 3). Examples of 3D reconstructions are included as movie files in supplementary data.
3.3. Analysis of potential targeted agents for NF1 PN treatment in 2D and 3D cultures
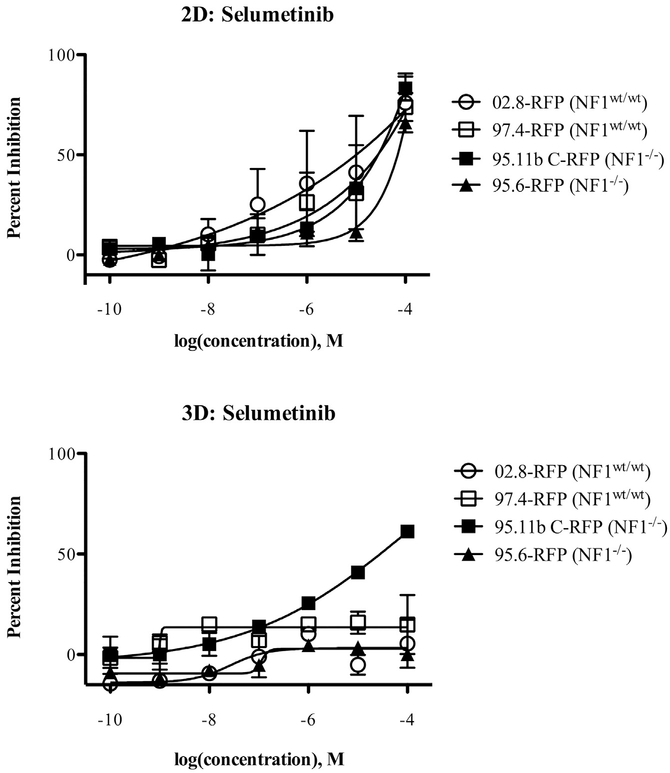
To begin use of the models for testing potential targeted therapeutics, we tested the effects of selumetinib (MEK inhibitor) against 2D and 3D cultures of the RFP variants of two wild-type SC lines (ipn97.4 and ipn02.8) and two NF1 PN cell lines (ipNF95.6 and ipNF95.11b C) using a standard MTT assay for net cell viability. The results show that selumetinib is able to inhibit the net cell growth of all the cell lines tested when they are grown in 2D cultures. The inhibitory effect on the wild-type cell lines has a trend toward being more potent, with ipn02.8 being the most sensitive and ipNF95.6 least sensitive in 2D culture (Fig. 4).
Fig. 4.
Selumetinib effects on net cell growth of wild-type SCs and NF1 PN cells in 2D and 3D cultures. The cell populations indicated were plated with or without rBM for 24 h and then treated for 48 h using a dose-response analysis (2.5 nM, 12.8 nM, 64 nM, 320 nM, 1.6 μM, 8 μM, 40 μM) in comparison to DMSO vehicle control. Net viability was assessed using an MTT assay. Results are expressed as the mean ± SE of two replicates. 02.8-RFP = ipn02.8-RFP, 97.4-RFP = ipn97.4-RFP, 95.11b C-RFP = ipNF95.11b C-RFP, 95.6-RFP = ipNF95.6-RFP.
Under 3D conditions, both of the wild-type lines became resistant to the selumetinib-induced growth inhibition. The NF1 PN line ipNF95.6 also was resistant in 3D culture, but the PN line ipNF95.11b C retained sensitivity to selumetinib (Fig. 4).
To begin testing novel targeted agents that have not previously been developed for NF1 treatment we changed our assay design to make it more amenable to higher throughput and to align it with a screening initiative for NF1 PN treatments that has been performed by the Neurofibromatosis Therapeutic Acceleration Program (NTAP). These changes were to assay net cell viability by ATP content (Cell-Titer Glo) rather than oxido-reductase enzymatic capacity (MTT) and to use two NF1 PN cell lines – ipNF95.11b C and ipNF95.6 – with comparison to the control, wild-type SC line ipn02.3.
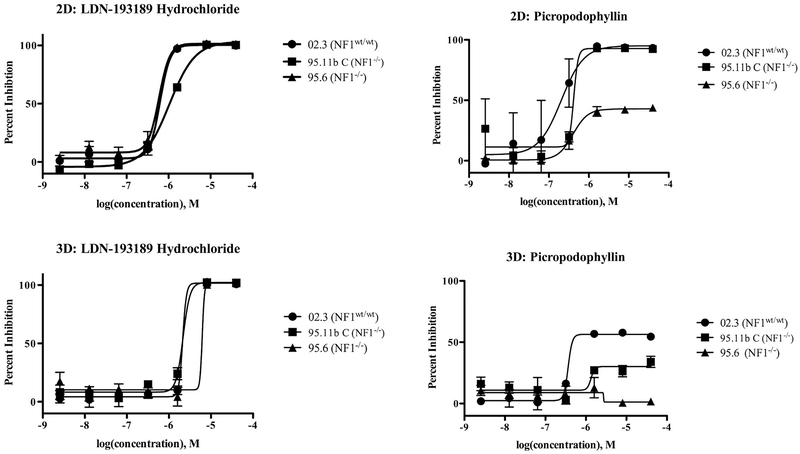
We assayed the effects of two drugs, LDN-193189 hydrochloride and picropodophyllin (PPP), for their effects on net cell viability of SCs and NF1 PN cells under both 2D and 3D growth conditions (Fig. 5). The bone morphogenetic protein 2 (BMP2) inhibitor, LDN-193189 hydrochloride, has been shown to decrease motility and invasion but not growth over a 24 h exposure in NF1-associated malignant peripheral nerve sheath tumor (MPNST) cells (Sun et al., 2013). Besides the association with NF1 pathology this drug was selected for its ability to promote or repress proliferation and migration in a number of cell types (Lehrberg and Gardiner, 2015; Yao et al., 2014). PPP inhibits insulin-like growth factor 1 receptor (IGF-1R) activity. We selected the IGF1-R as a novel and rational potential target for two reasons. First, because IGF-1R is required for Schwann cell development and response to injury through inhibition of Schwann cell apoptosis (D’Ercole et al., 1996; Ogata et al., 2006; Freude et al., 2008). Second, because IGF-1R is strongly expressed in patient samples of MPNST and PNs (Friedrich et al., 2007).
Fig. 5.
LDN-193189 hydrochloride and PPP differentially inhibit cell growth of wild-type SCs and NF1 PN cells under 2D and 3D conditions. The protocol is described in Fig. 4 except that data were collected by using CellTiter-Glo Luminescent Cell Viability Assay. Results are expressed as the mean ± SE of two replicates. 02.3 = ipn02.3 2λ-RFP, 95.11b C = ipNF95.11b C-RFP, 95.6 = ipNF95.6-RFP.
To determine effects on cells cultured in 2D and 3D, 48 h treatments of PPP and LDN-193189 were performed. The results for PPP show that both the control cell line (02.3) and the two PN cell lines were inhibited in their growth across an approximately similar range of concentrations when the cells were grown in 2D (Fig. 5). One notable distinction here is that while PPP was able to completely block growth of both wild-type ipn02.3 SCs and ipNF95.11b C PN cells, it was only able to partially inhibit (approximately 50%) growth of ipNF95.6 PN cells. The distinctions between the different cell lines’ sensitivities to PPP became much more evident when the cells were growing in 3D. All the cell lines showed very significant resistance. The ipn02.3 SCs retained some sensitivity to growth inhibition by PPP, but the maximal effect just cleared 50% inhibition of growth. The two PN cell lines were much more resistant, with only a marginal effect remaining in ipNF95.11b C, but the ipNF95.6 cells continued to grow even at the highest concentrations of PPP tested. IC50 values for both drugs are reported in supplementary material, Table S1.
Results for LDN-193189 hydrochloride showed that all three cell lines were inhibited in their growth in 2D over a similar concentration range, with complete inhibition of growth recorded at 1.6 μM for ipn02.3 SCs and ipNF95.6 PN cells, but that ipNF95.11b C PN cells were slightly less sensitive, requiring 8 μM for full inhibition (Fig. 5). There was a slight shift in the dose-response curve to the right when the cells were allowed to grow in 3D, but all the cell lines were fully inhibited at 8 μM of LDN-193189, which shows that there is little resistance induced to this agent by the presence of the richer microenvironment.
4. Discussion
Culture in 2D has contributed much to our understanding of the molecular biology of the cell but fails to take into account the presence in vivo of multiple cell types and the complexity of the tumor environment. These differences can result in data that is misleading and may falsely predict clinical outcome. For example, studies have shown differences in response to drugs between 2D and 3D culture. Although sufficient data is not yet available determining 3D to be more predictive of clinical outcome, there is an emerging paradigm that 3D models may provide a superior pathomimetic recapitulation of tumor biology (Kimlin et al., 2013; Santo et al., 2017). The goal of this study was to develop robust 3D models of NF1 PNs and SCs with wild-type neurofibromin expression to provide new platforms for identification of targeted therapeutics that will be effective for translation into the clinic for slow growing peripheral nerve sheath tumors.
We transduced five cell lines, two SC and three PN, with RFP and defined their morphology in 3D in rBM with confocal microscopy (Fig. 1). The SCs derived from normal nerves are rounded and formed structures that are cup-like with a hollow center. The individual PN cells exhibit an elongated cell body and form stellate structures that branch to neighboring cell aggregates. This is reminiscent of structural differences in breast cancer epithelial cells grown in 2D versus 3D with the morphological differences being associated with invasiveness or malignancy (Benton et al., 2009; Kenny et al., 2007). Similarly, the results of the present study suggest that SCs and PN cells have distinctly different morphologies that may be associated with a relatively more aggressive/invasive phenotype for the PN cells.
To further investigate the potentially more aggressive phenotype of PN cells we assayed the ability of 3D cultures of SCs and NF1 PN cells to degrade collagen IV in their matrix. The wild-type cell lines derived from normal nerves showed very low levels of proteolysis, a factor that supports a non-invasive phenotype. In contrast is the distinctive PN cell morphology of elongated cell body, multiple processes and overall network formation was associated with significantly higher matrix proteolysis. The ability to migrate and invade is closely allied to proteolysis of the ECM. MMPs (collagenases and gelatinases), located in the peri-cellular space of the tumor, drive cancer progression by degrading the structural components of the extracellular matrix and break down basement membranes, for example, of blood vessels to gain access to the circulatory system (Brinckerhoff et al., 2000). These MMPs can be secreted by invading inflammatory cells, endothelial cells, fibroblasts and tumor cells (Kessenbrock et al., 2010). However migration is a complex function. MT1-MMP and MT3-MMP regulate the attachment and detachment of tumor cells to extracellular matrix molecules like hyaluronic acid, collagen I, laminin and fibrin. This attach/stretch/detach activity increases cell motility (Senbanjo and Chellaiah, 2017; Kajita et al., 2001) and ultimately invasion. These complex interactions can be investigated in 3D culture models with the use of confocal microscopy, interactions not possible to observe with 2D models.
The increase in matrix proteolytic activity (capacity to degrade collagen IV) observed in the 3D cultures of NF1 PN cells as compared to control cultures of SCs may be highly significant and translatable to human tissues (Kraniak et al., 2018). First, these results confirm original observations that found that SCs derived from human neurofibromas were highly invasive and secreted much more matrix metalloprotease-2 (a gelatinase) than control human SCs (Muir, 1995). Second, the clinical experience is that these tumors often penetrate into adjacent tissues and thus it is very difficult to achieve a complete, curative resection (Friedrich et al., 2005). It is possible that this fluorescence-based assay developed here could be used to screen for drugs that may effectively block this phenotype of infiltrating nerve lesions in future studies.
The proteolytic assay used here is adapted from that developed by Bonnie Sloane and colleagues (Sameni et al., 2003; Moin et al., 2012). In order to normalize the proteolytic activity to cell number, we used a volumetric assay of the expression of RFP as a surrogate for cell counting. Previously, for example, this normalization has been done by counting the number of stained nuclei (Sameni et al., 2009). There are two notable limitations of using DNA dyes. The first is that they are toxic and therefore cannot be used to track changes in cell number in the same 3D culture over time. The second problem is that it takes significant image analysis time to accurately discern and quantify the nuclei from the reconstructed 3D images. The volumetric assay of RFP overcomes both of these obstacles because it is relatively non-invasive, and thus compatible with repeated application, and requires much less image analysis. We initially confirmed that the rate of cell proliferation reported for both slowly and rapidly growing NF1 PN cells and SCs was comparable by counting nuclei and measuring volume of RFP (Fig. S2). This approach is also supported by recent results showing that volumetric assay of fluorescence is the best way to assay 3D structures that are composed of labeled cells (Leary et al., 2016).
Notably, our results show that wild-type SC growth in 3D is faster than that of NF1 PN cell growth in 3D. In vivo SCs that have myelinated the axon do not proliferate. However, if the nerve is injured and the axon is lost, the SCs dedifferentiate into repair cells that are proliferative (Glenn and Talbot, 2013). When the SCs are isolated from the nerve tissue, they proliferate in a 2D culture on plastic. Our study shows that this proliferation is maintained in 3D culture even though the serum content of the media is greatly reduced. It is possible that the wild-type SCs are proliferative in 3D culture due to the absence of the nerve. We hypothesize that if the 3D model better represented the in vivo condition by including functional axonal mimics then the SCs would reduce their proliferation rate and potentially redifferentiate into myelinating cells. This highlights a limitation of our study, which is that our models did not include multiple cell types in a co-culture design. Although beyond the scope of this initial study, future work will focus on creating a multicellular 3D model that we expect to provide further distinction between wild-type SCs and NF1-mutant SCs.
Although it was unexpected that the growth rate of the NF1 PN cells in our 3D culture system is slower than that of the wild-type SCs, this may in fact be an accurate representation of PN growth in humans in vivo. Human tumor growth rate in situ, as measured in doubling time, depends on the histological type of the tumor and varies widely from less than one week to over a year (Fisher et al., 2008). Clinically, PN have the most rapid growth in young children (Dombi et al., 2007). In this study the net viable mass of PN cells in 3D cultures doubled in about a week. A rough estimate of doubling time on plastic is 10 to 48 h (depending on the cell line). Hence, the slower growth of NF1 PN in 3D is consistent with behavior of PN in vivo.
The underlying loss of neurofibromin in NF1 tumors has long suggested that targeting activated Ras and its downstream pathways might be an effective therapeutic approach (Dilworth et al., 2006). Oncogenic Ras has proved a particularly challenging drug target, and the non-mutated, wild-type Ras proteins that are activated in NF1 have been less studied in this context (Mattingly, 2013; Brock et al., 2016). The MAP kinase pathway downstream of activated Ras is a promising target for NF1 tumors (Mattingly et al., 2006). Intriguing pre-clinical data in animal models of NF1 neurofibromas (Jessen et al., 2013) led to the recently reported clinical trial in which the MEK inhibitor selumetinib produced partial responses in 17 of 24 children with PNs (Dombi et al., 2016). Despite these extremely encouraging results, it is important to realize that responses to single targeted therapeutics are often transient, including in the animal models of NF1 tumors, due to development of resistance (Watson et al., 2014). Given this, it may be that the 3D culture allows more predictive modeling of the human NF1 PN behavior. Specifically, the results from the 2D cultures of wild-type SCs and NF1 PN showed no discrimination in their sensitivity to the MEK inhibitor. The 3D cultures, however, revealed a particular sensitivity for one of the NF1 PN cell lines whereas both the wild-type SC lines tested were profoundly resistant to selumetinib. The fact that only one of the NF1 PN cell lines tested was sensitive to selumetinib in 3D could reflect patient variability in the origin of the tumors.
The goal of this project was to develop pathomimetic 3D models of NF1 PNs that would allow identification of additional targeted therapeutic approaches at a much higher throughput than is available in the animal models and with much greater likelihood of predicting clinical utility than is available from traditional 2D cell culture. We therefore developed a higher throughput assay of net cell viability based on ATP content that can be applied to 3D cultures using Cell-Titer Glo. PPP, which inhibits IGF-1R, blocks cell proliferation in 2D cultures but is less active in 3D notably for PN lines. The efficacy of PPP to block proliferation of ipn02.3-RFP cells may be attributed to a PPP-induced mitotic block (Waraky et al., 2014; Wu et al., 2013) (an IGF-1R independent mechanism). This mechanism of action would be consistent with the observation that ipn02.3-RFP demonstrated increased proliferation over PN cell lines in 3D culture.
There is a lack of differential activity between 2D and 3D for the BMP-2 inhibitor, LDN-193189, which may be explained by lack of pathway significance or off-target effects of the drug. The action of LDN-193189 against cultures of NF1 MPNST cells is, however, highly synergistic with the effects of selumetinib (Ahsan et al., 2016), which suggests that it is a promising agent to assay in combination studies in the PN 3D cultures with a view to further development of therapeutic approaches for these tumors. In conclusion, the methodology developed here represents a significant advance for a culture system that has the promise to improve the predictability of pre-clinical in vitro systems particularly for slow growing peripheral nerve tumors like PN.
Supplementary Material
Acknowledgements
Funding for this research was made possible by the Neurofibromatosis Therapeutic Acceleration Program (NTAP) (2001805609) at Johns Hopkins University. Confocal microscopy and cell sorting was performed at the Microscopy, Imaging and Cytometry Resources Core of Wayne State University that is supported, in part, by NIH Center grant P30CA22453. We thank the director and staff of the core for their assistance. We express our appreciation to Markiesha Baines for uploading raw files and preparing and uploading accompanying annotation files onto the Synapse.org website for public viewing.
Abbreviations:
- 2D
two dimensional
- 3D
three dimensional
- BMP2
bone morphogenetic protein 2
- DQ
dye-quenched
- ECM
extracellular matrix
- FBS
fetal bovine serum
- IGF-1R
insulin-like growth factor 1 receptor
- NF1
type 1 neurofibromatosis
- PN
plexiform neurofibroma
- PPP
picropodophyllin
- rBM
reconstituted basement membrane
- RFP
red fluorescent protein
- SC
Schwann cell
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.expneurol.2017.10.012.
References
- Ahsan S, Ge Y, Tainsky MA, 2016. Combinatorial therapeutic targeting of BMP2 and MEK-ERK pathways in NF1-associated malignant peripheral nerve sheath tumors. Oncotarget 7, 57171–57185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcellos-Hoff MH, Aggeler J, Ram TG, Bissell MJ, 1989. Functional differentiation and alveolar morphogenesis of primary mammary cultures on reconstituted basement membrane. Development (Cambridge, England) 105, 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton G, George J, Kleinman HK, Arnaoutova IP, 2009. Advancing science and technology via 3D culture on basement membrane matrix. J. Cell. Physiol 221, 18–25. [DOI] [PubMed] [Google Scholar]
- Berti C, Nodari A, Wrabetz L, Feltri ML, 2006. Role of integrins in peripheral nerves and hereditary neuropathies. NeuroMolecular Med. 8, 191–204. [DOI] [PubMed] [Google Scholar]
- Brannan CI, Perkins AS, Vogel KS, Ratner N, Nordlund ML, Reid SW, Buchberg AM, Jenkins NA, Parada LF, Copeland NG, 1994. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 8, 1019–1029. [DOI] [PubMed] [Google Scholar]
- Brinckerhoff CE, Rutter JL, Benbow U, 2000. Interstitial collagenases as markers of tumor progression. Clin. Cancer Res. 6, 4823–4830. [PubMed] [Google Scholar]
- Brock EJ, Ji K, Reiners JJ Jr., Mattingly RR, 2016. How to target activated Ras proteins: direct inhibition vs. induced mislocalization. Mini Rev. Med. Chem 16, 358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey DJ, Eldridge CF, Cornbrooks CJ, Timpl R, Bunge RP, 1983. Biosynthesis of type IV collagen by cultured rat Schwann cells. J. Cell Biol. 97, 473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallo-Medved D, Rudy D, Blum G, Bogyo M, Caglic D, Sloane BF, 2009. Livecell imaging demonstrates extracellular matrix degradation in association with active cathepsin B in caveolae of endothelial cells during tube formation. Exp. Cell Res. 315, 1234–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers KF, Mosaad EM, Russell PJ, Clements JA, Doran MR, 2014. 3D cultures of prostate cancer cells cultured in a novel high-throughput culture platform are more resistant to chemotherapeutics compared to cells cultured in monolayer. PLoS One 9, e111029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheema U, Brown RA, Alp B, MacRobert AJ, 2008. Spatially defined oxygen gradients and vascular endothelial growth factor expression in an engineered 3D cell model. Cell. Mol. Life Sci. 65, 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chignola R, Schenetti A, Andrighetto G, Chiesa E, Foroni R, Sartoris S, Tridente G, Liberati D, 2000. Forecasting the growth of multicell tumour spheroids: implications for the dynamic growth of solid tumours. Cell Prolif. 33, 219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitcholtan K, Asselin E, Parent S, Sykes PH, Evans JJ, 2013. Differences in growth properties of endometrial cancer in three dimensional (3D) culture and 2D cell monolayer. Exp. Cell Res. 319, 75–87. [DOI] [PubMed] [Google Scholar]
- Cichowski K, Jacks T, 2001. NF1 tumor suppressor gene function: narrowing the GAP. Cell 104, 593–604. [DOI] [PubMed] [Google Scholar]
- Cichowski K, Shih TS, Schmitt E, Santiago S, Reilly K, McLaughlin ME, Bronson RT, Jacks T, 1999. Mouse models of tumor development in neurofibromatosis type 1. Science (New York, N.Y.) 286, 2172–2176. [DOI] [PubMed] [Google Scholar]
- Daston MM, Scrable H, Nordlund M, Sturbaum AK, Nissen LM, Ratner N, 1992. The protein product of the neurofibromatosis type 1 gene is expressed at highest abundance in neurons, Schwann cells, and oligodendrocytes. Neuron 8, 415–428. [DOI] [PubMed] [Google Scholar]
- D’Ercole AJ, Ye P, Calikoglu AS, Gutierrez-Ospina G, 1996. The role of the insulinlike growth factors in the central nervous system. Mol. Neurobiol 13, 227–255. [DOI] [PubMed] [Google Scholar]
- Dilworth JT, Kraniak JM, Wojtkowiak JW, Gibbs RA, Borch RF, Tainsky MA, Reiners JJ Jr., Mattingly RR, 2006. Molecular targets for emerging anti-tumor therapies for neurofibromatosis type 1. Biochem. Pharmacol 72, 1485–1492. [DOI] [PubMed] [Google Scholar]
- Dombi E, Solomon J, Gillespie AJ, Fox E, Balis FM, Patronas N, Korf BR, Babovic-Vuksanovic D, Packer RJ, Belasco J, et al. , 2007. NF1 plexiform neurofibroma growth rate by volumetric MRI: relationship to age and body weight. Neurology 68, 643–647. [DOI] [PubMed] [Google Scholar]
- Dombi E, Baldwin A, Marcus LJ, Fisher MJ, Weiss B, Kim A, Whitcomb P, Martin S, Aschbacher-Smith LE, Rizvi TA, et al. , 2016. Activity of Selumetinib in Neurofibromatosis type 1-related Plexiform Neurofibromas. N. Engl. J. Med 375, 2550–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson R, Broglie JJ, Adcock AF, Yang L, 2014. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol 12, 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder-Mengus C, Ghosh S, Reschner A, Martin I, Spagnoli GC, 2008. New dimensions in tumor immunology: what does 3D culture reveal? Trends Mol. Med 14, 333–340. [DOI] [PubMed] [Google Scholar]
- Fisher MJ, Basu S, Dombi E, Yu JQ, Widemann BC, Pollock AN, Cnaan A, Zhuang H, Phillips PC, Alavi A, 2008. The role of [18F]-fluorodeoxyglucose positron emission tomography in predicting plexiform neurofibroma progression. J. Neuro-Oncol. 87, 165–171. [DOI] [PubMed] [Google Scholar]
- Freude S, Leeser U, Muller M, Hettich MM, Udelhoven M, Schilbach K, Tobe K, Kadowaki T, Kohler C, Schroder H, et al. , 2008. IRS-2 branch of IGF-1 receptor signaling is essential for appropriate timing of myelination. J. Neurochem 107, 907–917. [DOI] [PubMed] [Google Scholar]
- Friedrich RE, Schmelzle R, Hartmann M, Mautner VF, 2005. Subtotal and total resection of superficial plexiform neurofibromas of face and neck: four case reports. J. Craniomaxillofac. Surg 33, 55–60. [DOI] [PubMed] [Google Scholar]
- Friedrich RE, Keiner D, Hagel C, 2007. Expression of insulin-like growth-factor-1 receptor (IGF-1R) in peripheral nerve sheath tumors in neurofibromatosis type 1. Anticancer Res. 27, 2085–2090. [PubMed] [Google Scholar]
- Glenn TD, Talbot WS, 2013. Signals regulating myelination in peripheral nerves and the Schwann cell response to injury. Curr. Opin. Neurobiol 23, 1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurski LA, Jha AK, Zhang C, Jia X, Farach-Carson MC, 2009. Hyaluronic acid-based hydrogels as 3D matrices for in vitro evaluation of chemotherapeutic drugs using poorly adherent prostate cancer cells. Biomaterials 30, 6076–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraida S, Nerlich AG, Bise K, Wiest I, Schleicher E, 1992. Comparison of various basement membrane components in benign and malignant peripheral nerve tumours. Virchows Arch. A Pathol. Anat. Histopathol 421, 331–338. [DOI] [PubMed] [Google Scholar]
- Hartmann U, Maurer P, 2001. Proteoglycans in the nervous system—the quest for functional roles in vivo. Matrix Biol. 20, 23–35. [DOI] [PubMed] [Google Scholar]
- Imamura Y, Mukohara T, Shimono Y, Funakoshi Y, Chayahara N, Toyoda M, Kiyota N, Takao S, Kono S, Nakatsura T, et al. , 2015. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol. Rep 33, 1837–1843. [DOI] [PubMed] [Google Scholar]
- Ingram DA, Yang FC, Travers JB, Wenning MJ, Hiatt K, New S, Hood A, Shannon K, Williams DA, Clapp DW, 2000. Genetic and biochemical evidence that haploinsufficiency of the Nf1 tumor suppressor gene modulates melanocyte and mast cell fates in vivo. J. Exp. Med 191, 181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen WJ, Miller SJ, Jousma E, Wu J, Rizvi TA, Brundage ME, Eaves D, Widemann B, Kim MO, Dombi E, et al. , 2013. MEK inhibition exhibits efficacy in human and mouse neurofibromatosis tumors. J. Clin. Invest 123, 340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajita M, Itoh Y, Chiba T, Mori H, Okada A, Kinoh H, Seiki M, 2001. Membranetype 1 matrix metalloproteinase cleaves CD44 and promotes cell migration. J. Cell Biol. 153, 893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, Mao S, Li Q, Sameni M, Krawetz SA, Sloane BF, Mattingly RR, 2012. RNA-Seq of human breast ductal carcinoma in situ models reveals aldehyde dehydrogenase isoform 5A1 as a novel potential target. PLoS One 7, e50249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-Hoff MH, Petersen OW, et al. , 2007. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol. Oncol 1, 84–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessenbrock K, Plaks V, Werb Z, 2010. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell 141, 52–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimlin LC, Casagrande G, Virador VM, 2013. In vitro three-dimensional (3D) models in cancer research: an update. Mol. Carcinog 52, 167–182. [DOI] [PubMed] [Google Scholar]
- Kleinman HK, Martin GR, 2005. Matrigel: basement membrane matrix with biological activity. Semin. Cancer Biol. 15, 378–386. [DOI] [PubMed] [Google Scholar]
- Kluwe L, Friedrich R, Mautner VF, 1999. Loss of NF1 allele in Schwann cells but not in fibroblasts derived from an NF1-associated neurofibroma. Genes Chromosom. Cancer 24, 283–285. [DOI] [PubMed] [Google Scholar]
- Kraniak J, Mattingly RR, Sloane BF, 2018. In: Janetka J (Ed.), Roles of Pericellular Proteases in Tumor Angiogenesis: Therapeutic Implications, ch14. Wiley. [Google Scholar]
- Le LQ, Parada LF, 2007. Tumor microenvironment and neurofibromatosis type I: connecting the GAPs. Oncogene 26, 4609–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary E, Rhee C, Wilks B, Morgan JR, 2016. Accurate quantitative wide-field fluorescence microscopy of 3-D spheroids. BioTechniques 61, 237–247. [DOI] [PubMed] [Google Scholar]
- Lee J, Cuddihy MJ, Kotov NA, 2008. Three-dimensional cell culture matrices: state of the art. Tissue Eng. Part B Rev. 14, 61–86. [DOI] [PubMed] [Google Scholar]
- Lehrberg J, Gardiner DM, 2015. Regulation of axolotl (Ambystoma Mexicanum) limb Blastema cell proliferation by nerves and BMP2 in organotypic slice culture. PLoS One 10, e0123186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Chow AB, Mattingly RR, 2010. Three-dimensional overlay culture models of human breast cancer reveal a critical sensitivity to mitogen-activated protein kinase kinase inhibitors. J. Pharmacol. Exp. Ther 332, 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Chang LJ, Neubauer DR, Muir DF, Wallace MR, 2016. Immortalization of human normal and NF1 neurofibroma Schwann cells. Lab. Investig 96, 1105–1115. [DOI] [PubMed] [Google Scholar]
- Mason BN, Califano JP, Reinhart-King CA, 2012. Matrix stiffness: a regulator of cellular behavior and tissue formation In: Bhatia S (Ed.), Engineering Biomaterials for Regenerative Medicine. Springer, New York, NY. [Google Scholar]
- Mattingly RR, 2013. Activated Ras as a therapeutic target: constraints on directly targeting Ras isoforms and wild-type versus mutated proteins. ISRN Oncol. 2013, 536529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattingly RR, Kraniak JM, Dilworth JT, Mathieu P, Bealmear B, Nowak JE, Benjamins JA, Tainsky MA, Reiners JJ Jr., 2006. The mitogen-activated protein kinase/extracellular signal-regulated kinase kinase inhibitor PD184352 (CI-1040) selectively induces apoptosis in malignant schwannoma cell lines. J. Pharmacol. Exp. Ther 316, 456–465. [DOI] [PubMed] [Google Scholar]
- Moin K, Sameni M, Victor BC, Rothberg JM, Mattingly RR, Sloane BF, 2012. 3D/4D functional imaging of tumor-associated proteolysis: impact of microenvironment. Methods Enzymol. 506, 175–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir D, 1995. Differences in proliferation and invasion by normal, transformed and NF1 Schwann cell cultures are influenced by matrix metalloproteinase expression. Clin. Exp. Metastasis 13, 303–314. [DOI] [PubMed] [Google Scholar]
- Muir D, Neubauer D, Lim IT, Yachnis AT, Wallace MR, 2001. Tumorigenic properties of neurofibromin-deficient neurofibroma Schwann cells. Am. J. Pathol 158, 501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata T, Yamamoto S, Nakamura K, Tanaka S, 2006. Signaling axis in schwann cell proliferation and differentiation. Mol. Neurobiol 33, 51–62. [DOI] [PubMed] [Google Scholar]
- Platt CI, Krekoski CA, Ward RV, Edwards DR, Gavrilovic J, 2003. Extracellular matrix and matrix metalloproteinases in sciatic nerve. J. Neurosci. Res 74, 417–429. [DOI] [PubMed] [Google Scholar]
- Prada CE, Rangwala FA, Martin LJ, Lovell AM, Saal HM, Schorry EK, Hopkin RJ, 2012. Pediatric plexiform neurofibromas: impact on morbidity and mortality in neurofibromatosis type 1. J. Pediatr 160, 461–467. [DOI] [PubMed] [Google Scholar]
- Sameni M, Dosescu J, Moin K, Sloane BF, 2003. Functional imaging of proteolysis: stromal and inflammatory cells increase tumor proteolysis. Mol. Imaging 2, 159–175. [DOI] [PubMed] [Google Scholar]
- Sameni M, Cavallo-Medved D, Dosescu J, Jedeszko C, Moin K, Mullins SR, Olive MB, Rudy D, Sloane BF, 2009. Imaging and quantifying the dynamics of tumor-associated proteolysis. Clin. Exp. Metastasis 26, 299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sameni M, Anbalagan A, Olive MB, Moin K, Mattingly RR, Sloane BF, 2012. MAME models for 4D live-cell imaging of tumor: microenvironment interactions that impact malignant progression. J. Vis. Exp 60, 1–5 (e3661). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santo VE, Rebelo SP, Estrada MF, Alves PM, Boghaert E, Brito C, 2017. Drug screening in 3D in vitro tumor models: overcoming current pitfalls of efficacy readouts. Biotechnol. J 12. [DOI] [PubMed] [Google Scholar]
- Senbanjo LT, Chellaiah MA, 2017. CD44: a multifunctional cell surface adhesion receptor is a regulator of progression and metastasis of cancer cells. Front. Cell Dev. Biol 5, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevenich L, Joyce JA, 2014. Pericellular proteolysis in cancer. Genes Dev. 28, 2331–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, EY Yu, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, et al. , 2003. Lentivirus-delivered stable gene silencing by RNAi in primary cells RNA (New York, NY) 9, 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Haddad R, Kraniak JM, Horne SD, Tainsky MA, 2013. RAS/MEK-in-dependent gene expression reveals BMP2-related malignant phenotypes in the Nf1-deficient MPNST. Mol. Cancer Res. 11, 616–627. [DOI] [PubMed] [Google Scholar]
- Suri S, Schmidt CE, 2010. Cell-laden hydrogel constructs of hyaluronic acid, collagen, and laminin for neural tissue engineering. Tissue Eng. A 16, 1703–1716. [DOI] [PubMed] [Google Scholar]
- Waraky A, Akopyan K, Parrow V, Stromberg T, Axelson M, Abrahmsen L, Lindqvist A, Larsson O, Aleem E, 2014. Picropodophyllin causes mitotic arrest and catastrophe by depolymerizing microtubules via insulin-like growth factor-1 receptor-independent mechanism. Oncotarget 5, 8379–8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson AL, Anderson LK, Greeley AD, Keng VW, Rahrmann EP, Halfond AL, Powell NM, Collins MH, Rizvi T, Moertel CL, et al. , 2014. Co-targeting the MAPK and PI3K/AKT/mTOR pathways in two genetically engineered mouse models of Schwann cell tumors reduces tumor grade and multiplicity. Oncotarget 5, 1502–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver VM, Lelievre S, Lakins JN, Chrenek MA, Jones JC, Giancotti F, Werb Z, Bissell MJ, 2002. beta4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell 2, 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Sooman L, Wickstrom M, Fryknas M, Dyrager C, Lennartsson J, Gullbo J, 2013. Alternative cytotoxic effects of the postulated IGF-IR inhibitor picropodophyllin in vitro. Mol. Cancer Ther. 12, 1526–1536. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Deng D, Sato Y, Hou YT, Watanabe R, Sasaki K, Kawabe M, Hirano E, Morinaga T, 2013. Silicate fiber-based 3D cell culture system for anticancer drug screening. Anticancer Res. 33, 5301–5309. [PubMed] [Google Scholar]
- Yang FC, Chen S, Clegg T, Li X, Morgan T, Estwick SA, Yuan J, Khalaf W, Burgin S, Travers J, et al. , 2006. Nf1 +/− mast cells induce neurofibroma like phenotypes through secreted TGF-beta signaling. Hum. Mol. Genet 15, 2421–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang FC, Staser K, Clapp DW, 2012. The plexiform neurofibroma microenvironment. Cancer Microenviron. 5, 307–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, Wang Y, Zhang P, Chen H, Xu Z, Jiao J, Yuan Z, 2014. BMP2-SMAD signaling represses the proliferation of embryonic neural stem cells through YAP. J. Neurosci 34, 12039–12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Parada LF, 2002. The molecular and genetic basis of neurological tumours. Nat. Rev. Cancer 2, 616–626. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.