Abstract
Purpose
Ubiquitin-conjugating enzyme E2S (UBE2S) is important for the development and progression of several types of cancer. However, neither the role of UBE2S in pancreatic cancer nor its mechanism is clear.
Methods
We analyzed three GEO datasets to obtain 150 differentially expressed genes (DEGs) between pancreatic ductal adenocarcinoma (PDAC) and non-cancerous samples. Moreover, GSEA and mutation analysis were also done for UBE2S. The UBE2S expression in PDAC was measured by immunohistochemistry and qRT-PCR. Colony formation, scratch wound-healing and tumor growth assays were conducted to examine the effect of UBE2S on PDAC cells. Migration was detected using Transwell assay. UBE2S knockdown pancreatic cells were treated with proteasome inhibitor MG132. Immunofluorescence was undertaken for interaction between UBE2S and VHL. The expression of Snail and Twist1 and the changes of HIF-1α/STAT3 pathway were detected by Western blotting.
Results
The mRNA of UBE2S was significantly upregulated in human pancreatic cancer compared to normal tissues. Immunohistochemistry confirmed that the protein level of UBE2S increased in tissue microarrays (TMAs) and was associated with lymph nodes metastasis and distant metastasis.
Conclusion
UBE2S could enhance EMT by the VHL/HIF-1α/STAT3 pathway via the ubiquitin-proteasome system. Co-expression of CDC20 may represent a novel and promising therapeutic target for the patients with PDAC.
Keywords: pancreatic cancer, UBE2S, VHL/HIF-1α/STAT3 signaling, the ubiquitin-proteasome system, EMT
Introduction
Pancreatic cancer is one of the most frequent malignant tumors and ranks the top seven causes of cancer-induced death worldwide. Pancreatic ductal adenocarcinoma (PDAC) accounts for around 90% of pancreatic cancer. Besides, it has been reported that pancreatic cancer, surpassed only by lung cancer, will become the second leading cause of deaths in western countries by 2030. Previous studies show that pancreatic cancer is known for its high recurrence rate, extreme liability for metastasis1,2 and drug-resistance, with few sensitive markers predicting the occurrence of pancreatic cancer and patients’ prognosis. In the past decade, high-throughput sequencing techniques have been widely used to scan genetic alterations at the genomic level, bringing about better identification of differential expression of genes (DEGs) and metabolic pathways involved in the carcinogenesis and progression of cancer.
Ubiquitin Conjugating Enzyme E2S (UBE2S) is the member of ubiquitin-conjugating enzyme family, which adopts ubiquitin from the E1 complex and drives its covalent parts to other proteins.3 As an important component of anaphase in complex/cyclosome (APC/C) and a cell-cycle-regulated ubiquitin ligase controlling progression through mitosis, UBE2S can elongate K11-linked polyubiquitin chain on APC/C substrates. As a result, UBE2S regulates the 26 S proteasome-mediated degradation by the proteasome and promotes mitotic exit, playing pivotal roles in cell division.4,5 Moreover, UBE2S directly interacts with ANAPC2 and ANAPC4.6 There is evidence that UBE2S interacts with CDC20, FZR1/CDH1 and VHL.7 CDC20 is responsible for the development of ubiquitin ligase activity of APC/C and plays a pivotal role in substrate specificity on the complex. In addition, researches have indicated that high expression of UBE2S in various tumors, compared with normal tissues, is related to poor prognosis of esophageal cancer and glioma, suggesting that UBE2S may be an important factor in promoting tumor proliferation, invasion and metastasis.8–10 Nevertheless, the clinical significance and role of UBE2S in PDAC remain unknown, and its underlying mechanism has not been clarified as well.
It is known that epithelial-mesenchymal transition (EMT) involves with loss of cell–cell adhesion and apical-basal polarity and development of mesenchymal features such as migratory and invasive abilities. EMT is required in cancer progression.11,12 Along with the advancement of technology, mechanisms of the transformation and progression of pancreatic cancer have been frequently identified, facilitating potential therapeutic targets for personalizing treatment. What is more, tumor cells induced by certain stimulus lead to EMT process, which causes the generation of multiple, distinct cellular subpopulations including cells with stem-cell-like characteristics. EMT program is regulated by the convergence of various signals which induces EMT transcription factors (EMT-TFs) such as Twist1 and Slug that the process depends on.13 Also, epithelial cell adhesion molecule E‐cadherin plays a predominant role in EMT, and other signalling like WNT signalling is critical to induce the programme. More importantly, EMT can induce metastasis, the major cause of cancer death. Previous researches indicated that hypoxia-inducible factor 1α (HIF-1α) plays a significant role in cancer metastasis. In normoxic condition, knockout of HIF-1α was shown to deteriorate the growth of tumor in vitro.14
The activity of ubiquitination is thought to be regulated by the sequential actions of the three kinds of enzymes: ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2) and ubiquitin ligase (E3). Among these enzymes, there have been about 40 members in E2 family found, some of which have been also shown to have prognostic value in human. For instance, high UBE2C expression is thought to be associated with poor survivals in breast cancer15 and ovarian carcinoma.16
Overall, the aim of the present study was to discover the molecular mechanisms regulating the interaction between EMT and UBE2S in human pancreatic cancer. Here we provide the first evidence that the expression of UBE2S promoted pancreatic cancer cell EMT and the interaction between UBE2S and VHL via the ubiquitin-proteasome system, suggesting that UBE2S significantly enhanced the VHL/HIF-1α/STAT3-induced EMT and metastasis in vitro and in vivo by attenuating the activity of the promoter.
Materials And Methods
Microarray
Three gene expression datasets GSE15471, GSE16515 and GSE28735 were obtained from GEO Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo). The probes were annotated according to the annotation information in the GEO. The GSE15471 dataset contained 39 PDAC and matching normal pancreatic tissue samples. GSE33006 contained 16 PDAC samples and 16 non-cancerous samples. GSE28735 matched 36 pairs of PDAC and adjacent non-tumor tissues.
Identification Of DEGs
The differentially expressed genes (DEGs) between HCC and non-cancerous samples were identified with GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r). Genes with logFC (fold change) >1 and adj. P-value <0.01 were considered as significant.
Construction Of PPI Network
The PPI network was predicted by Retrieval of Interacting Genes (STRING; http://string-db.org) (version 10.0). Cytoscape (version 3.7.0) was used to visualize the PPI networks.
GEPIA Dataset
We used GEPIA, a standard processing pipeline, to investigate the mRNA expression levels and related patients’ clinical features of UBE2S in PDAC and normal tissues.
Gene Enrichment Analysis
To identify potential function of UBE2S, Gene Set Enrichment Analysis (GSEA) algorithm was conducted to detect a series of biological processes. RNA-sequencing dataset of PDAC cohort which was downloaded from TCGA (https://tcga-data.nci.nih.gov/tcga/) and GSE16515 were divided into two groups according to median UBE2S expression. In validation data set GSE15471, samples were also divided into two groups based on the expression of UBE2S.
Oncomine Database
Oncomine (https://www.oncomine.org/) was used to analyze the transcription levels of the UBE2S in pancreatic cancers, with a student’s t-test to generate a P-value. Briefly, the gene was searched in the Oncomine database and GEPIA using the threshold values as follows: fold of change, 2; P-value of 0.05; gene rank in the top 10% among all mentioned genes in our study.
Tissue Microarrays And Immunohistochemistry
Tissue microarrays (TMAs) were obtained from Zuocheng Biotech Company (China), which contained 87 pancreatic cancer tissues and matched non-tumor tissues. The TMAs were included with well-documented information of the resources, which included with patients’ age, gender, tumor size and position, the grade of pathology, metastasis, tumor stage and follow-up information (by December 2011) (Table 1). There were 87 patients in total, which included with 56 males and 31 females, with a median age of 62 years old (ranged from 38 to 85 years old). The overall survival (OS) time of patients ranged from 0.9 to 49.5 months, with a median time of 7.7 months.
Table 1.
Relationship Between UBE2S Expression Level And Clinicopathologic Variables In 87 PDAC Tissues
| Clinicopathologic Parameters | Number | UBE2S Immunostaining | P | |
|---|---|---|---|---|
| Weak Positive | Strong Positive | |||
| Gender | ||||
| Male | 56 | 30 | 26 | 0.184 |
| Female | 31 | 12 | 19 | |
| Age (years)# | ||||
| <62 | 40 | 21 | 19 | 0.74 |
| ≥62 | 47 | 23 | 24 | |
| Tumor size (cm) | ||||
| ≤4 | 49 | 26 | 23 | 0.598 |
| >4 | 38 | 18 | 20 | |
| Tumor location | ||||
| Head, neck | 51 | 24 | 27 | 0.599 |
| Body, tail | 36 | 19 | 17 | |
| Tumor differentiation | ||||
| Poor | 30 | 13 | 17 | 0.256 |
| Well | 57 | 32 | 25 | |
| Lymph nodes metastasis | ||||
| No auth sub rev ms | 55 | 40 | 15 | 0.000 * |
| N1 | 32 | 8 | 24 | |
| Distant metastasis | ||||
| M0 | 72 | 48 | 24 | 0.002 * |
| M1 | 15 | 3 | 12 | |
| Invasion depth | ||||
| T1+T2 | 67 | 40 | 27 | 0.052 |
| T3+T4 | 20 | 7 | 13 | |
| Clinical stage | ||||
| Early stages (≤IIa) | 59 | 30 | 29 | 0.700 |
| Advanced stages (>IIa) | 28 | 13 | 15 | |
| Nervous invasion | ||||
| Negative | 50 | 27 | 23 | 0.321 |
| Positive | 37 | 16 | 21 | |
Notes: #Median age. *P<0.05 indicates a significant association among the variables.
The immunohistochemistry was performed as previously documented. The TMAs were incubated with a rabbit polyclonal antibody against UBE2S (1:400 dilutions; AbcamInc., USA) at 4°C overnight and incubated with horseradish peroxidase (HRP) (Gene Tech GT Vision III Detection Kit, Shanghai, China) at room temperature for 4 hrs. Finally, we used PBST for washing about 30 mins.
Cell Lines And RNA Interference (RNAi)
Pancreatic cancer cell lines PANC-1 and CFPAC-1 were both obtained from ATCC. PANC-1 cells were cultured in DMEM (Gibco, USA) which was supplemented with 10% fetal bovine serum (FBS). CFPAC-1 cells were cultured in RPMI 1640 (Gibco, USA) which was supplemented with 10% FBS. The cells were grown in 5% CO2-saturated humidity at 37°C and sub-cultured by digesting with trypsin-EDTA.
Pancreatic cell lines PANC-1 and CFPAC-1 were plated in 6-well plates at an appropriate density (40–60% cell density at time of transfection) before transfection with 3 μL UBE2S-RNAi (40 pmol) in a certain amount of serum-free medium, along with the presence of 2 μL lipofectamine 2000, according to the manufacturers’ instructions. The cells were incubated with transfection mixture about 4–6 hrs and that was replaced with fresh medium after transfection. siRNAs for UBE2S and one negative control siRNA were synthesized by Genepharm Technologies (Shanghai, China). The effects of gene silencing were confirmed by Western blotting 48 hrs after transfection.
Western Blot Analysis
Cells were lysed in NP-40 lysis buffer and the concentration of the protein was determined by Bradford assay (Beyotime Biotechnology, Shanghai, China). To denature the samples, cell lysate in sample buffer was boiled to 100°C for 5 mins. Proteins (20 μg for each sample) from each cell line were subjected to Western blot. SDS-PAGE were used to fractionate the total proteins which were followed by transferring onto a polyvinylidene fluoride membrane. The transferred proteins were checked with Ponceau S staining before blocking. The membranes were blocked with 5% Skimmed milk for 1 hr in TBST buffer containing 0.1% Tween-20 and incubated with the indicated primary antibodies at 4°C overnight. Then the membrane was washed using TBST for 10 mins each, three times totally, incubated with the recommended secondary antibody in blocking buffer at room temperature for 1 hr, and detected by enhanced chemiluminescence detection system. The data were normalized with β-Actin. The primary antibodies used for Western blot analyses were anti-UBE2S (Cell Signaling Technology) and anti-β-Actin (Cell Signaling Technology).
Realtime RT-PCR (qRT-PCR) Analysis
Total RNAs from samples were extracted with Trizol (Invitrogen, USA). 500 ng of total RNA was used to synthesize cDNA with Takara RT-PCR Master Mix kit (Takara, Japan) according to the instructions. The mRNA levels were determined by qRT-PCR with SYBR® Premix Ex Taq™ (Takara, Japan) on a VIIA7 real-time PCR machine. Results were normalized to GAPDH. All the primers used in the study were purchased from GenePharma.
Immunofluorescence Analysis
The procedures were performed as described previously. Briefly, overexpressed UBE2S cells were cultured on cover glass. For endogenous UBE2S, after laser micro-irradiation, cells were placed in ice-cold pre-extraction buffer as described. Images of the immunostaining were obtained by Nikon confocal microscope.
Cell Proliferation Assay
Cells were seeded into the 96-well plates (5000 cells per well) with clear flat bottom and cultured for 24 hrs. PANC-1 and CFPAC-1 cells were transfected with plasmid or RNAi and cultured for 24, 48, 72 hrs, respectively. Stably overexpressed UBE2S cells were treated following the same procedure. At the end of the treatment, the cells were fixed and added with 10 µL of WST-8 solution to each well, then protected from the light, and finally incubated for 1–4 hrs at 37°C. Optical density (OD) values were measured at 450 nm using a microtiter plate reader (VERSMax).
Scratch Assay
The scratch assay was performed by seeding the cells onto 6-well plate per well. The distance of cell migration was measured under a light-microscope with at least three random views.
Cell Migration And Cell Invasion Assay
The potentials of cell migration were evaluated with a transwell assay as described previously. Briefly, the experiment was conducted with MilliCell chambers (Millipore, Bedford, MA, USA). PANC-1 and CFPAC-1 cells harvested from transfection were manifested as described before. The cells on the upper layer of membrane were scraped off, whose number was counted under a microscope in five random fields at a magnification of 200 folds.
Protein Degradation Assay
MG132 (carbobenzoxy-Leu-Leu-leucinal) was purchased from Selleck (Selleck Chemicals, TX, USA). To confirm whether UBE2S regulated related protein degradation through the ubiquitin-proteasome system, we measured the expression level of VHL/HIF-1α/STAT3 axis following the treatment of MG132 under either silencing or overexpressing UBE2S.
Xenograft Mouse Model
To determine the tumorigenesis in vivo, 5*106 PANC1 cells were transfected with siRNA. The control cells in 100 μL PBS were subcutaneously inoculated into the left shoulder of 4 to 5-week-old BALB/c nude mice. The sizes of the tumor were measured with calipers, and volumes of the tumors were calculated according to the equation: V (mm3) =½ [width2 (mm2) × length (mm)]. The growth curves of the tumors were plotted against mean tumor volume at the different time points. The mice were sacrificed 28 days after inoculation. There were five mice in each group. The size of the tumor was measured every 3 days.
Statistical Analysis
SPSS 16.0 software (Chicago, IL, USA) was used for all the statistical analyses. The relationship between UBE2S expression and clinicopathological parameters was statistically determined using the Pearson χ2 test. The patients were monitored if they were still alive or to prevent the patients’ lost to follow-up until the end of the research. OS was assessed using a Kaplan–Meier method. A P-value of <0.05 was used to consider as the significance.
Results
Higher UBE2S Expression Correlates With Unfavorable Prognosis In PDAC
To identify the candidate genes in the carcinogenesis and progression of PDAC, three microarray datasets GSE15471, GSE16515 and GSE28735 were downloaded from GEO database. 150 differentially expressed genes (DEGs) were identified; in addition, to detect the potential relationship among these DEGs and UBE2S, we used STRING app in Cytoscape and mapped the DEGs into STRING to construct protein–protein interaction network (PPI) (Figure 1A and B). Besides, the relative expression of UBE2S can be seen in the 3 databases (Figure 1C–G). In the Oncomine database, UBE2S was more highly expressed in pancreatic carcinoma tissues than in normal tissues from two groups based on tumor stage (99 cases) (Figure 1C). Furthermore, RNA-seq data from the TCGA PDAC group also indicated that UBE2S expression was higher in PDAC tissues than in normal tissues (Figure 1D–G).
Figure 1.
Higher UBE2S expression correlates with unfavorable prognosis in PADC. (A) Differentially expressed genes identified in the analysis of three datasets. (B) The protein–protein interaction network (PPI) of 150 DEGs was constructed and the module analysis was performed using STRING and Cytoscape. (C) The box plot of UBE2S mRNA level in PDAC patients’ tissues and normal tissues from GEPIA. **P<0.01. (D–E) UBE2S was upregulated in pancreatic cancer tissues compared with normal or adjacent pancreatic tissues in two datasets. ***P<0.001. (F–G) Box plots of the UBE2S mRNA expression based on different tissue types and pathological stages, from Oncomine. (H) Representative immunohistochemically stained sections of peritumor tissues and pancreatic cancer. (I) Significantly higher rates of high UBE2S expression in PDAC tissues compared with peritumor tissues (P<0.01). (J) Relative expression of UBE2S in PDAC was significantly higher than that in matched peritumor tissues. **P<0.01. (K) Verification of expression of UBE2S in each of the paired PDAC samples (1~4). (L) Higher expression of UBE2S possessed a worse prognosis. (P=0.046).
To examine UBE2S expression patterns in PDAC, UBE2S protein expressions were examined by immunohistochemical staining on tissue microarrays containing 87 cases of PDAC and matched adjacent-normal pancreatic tissues. As shown in Figure 1H and I, UBE2S protein expression was significantly up-regulated (P<0.01) in pancreatic cancer compared to adjacent-normal pancreatic tissues. The expression pattern of UBE2S in 4 paired fresh samples of PDAC was respectively detected by qPCR and Western blot. As shown in Figure 1J and K, relative expression of UBE2S in PDAC was significantly higher than that in matched peritumor tissues. As shown in Table 1, UBE2S expression was correlated with lymph nodes metastasis and distant metastasis, but not with gender, age, tumor size, tumor location, tumor differentiation and clinical stage. Furthermore, Kaplan–Meier survival analysis showed that patients with high UBE2S expression had a shorter overall survival (Figure 1L). To determine the biological effects of UBE2S on PDAC tumorigenesis, UBE2S expression in PDAC cell lines was measured. Furthermore, protein and mRNA were extracted from fresh samples which were 4 groups of pancreatic cancer patients, and the results manifested that PDAC patients with higher UBE2S expression had shorter overall survival.
UBE2S Promotes Pancreatic Cancer Proliferation In Vivo And Vitro
As demonstrated in Figure 2, compared with the other PDAC cell lines, PANC-1 and CFPAC1 cells showed a higher UBE2S expression, while BxPC-3 cells manifested a lower UBE2S expression (Figure 2A and B). To investigate the role of UBE2S in pancreatic cancer cell proliferation, UBE2S in PANC-1 and CFPAC-1 cell lines were expressed and knocked down using RNAi. In addition, UBE2S was expressed or knocked down in both cell lines; the protein expression was verified by Western blot (Figure 2C). Besides, SW1990 cells which expressed lower levels of UBE2S were infected with pLVX-Puro-UBE2S lentivirus or pretty blank pLVX-Puro lentivirus (vector), and the expression was significantly up-regulated in transfected SW1990 cells compared to the controls (Figure 2D). Then, this study performed the proliferation assays contained. Solid evidence showed a significant difference between growth curves of UBE2S knockdown cells and stably over-expressed UBE2S cells when compared with controls (Figure 2E and F). Moreover, over-expression of UBE2S promoted cell proliferation (Figure 2G), while silencing UBE2S expression inhibited cell proliferation (Figure 2H).
Figure 2.
UBE2S promoted pancreatic cancer proliferation in vivo and vitro. (A–B) UBE2S was up-regulated in cancer cell lines (BXPC3, PANC1, CAPAN2, SW1990, CFPAC1) compared with normal pancreatic ductal epithelial cell line (HPNE). (C) The two designed siRNA presented satisfied inhibition efficiency in both CFPAC1 and PANC1 cell lines. (D) UBE2S protein expression in SW1990 cells infected with pLVX-Puro-UBE2S lentivirus. (E) UBE2S knockdown inhibits proliferation. (F) UBE2S overexpression promotes proliferation. (G) UBE2S overexpression promotes the clone formation of SW1990 cells. (H) UBE2S knockdown inhibits the clone formation of PANC1 and CFPAC1 cells. (I) UBE2S promotes subcutaneous tumour growth in nude mice. (J) The tumor growth rate of the si-NC group was significantly faster than that of the si-UBE2S group. *P<0.05, **P<0.01.
To further investigate the contribution of UBE2S to the pancreatic cancer proliferation in vivo, BALB/c nude mice were subcutaneously injected in the right hind limb with PANC1-siControl cells or PANC1-siUBE2S cells, and then the mice were assigned into different groups. Figure 2I and J demonstrated that the volume and size of the tumors from PANC1-siUBE2S cells were smaller than those of the tumors originating from the control group, suggesting that the knockdown of UBE2S expression rendered a lower ability of proliferation in vivo.
UBE2S Regulates Pancreatic Cancer Cell Metastasis And Invasion By Epithelial-Mesenchymal Transition
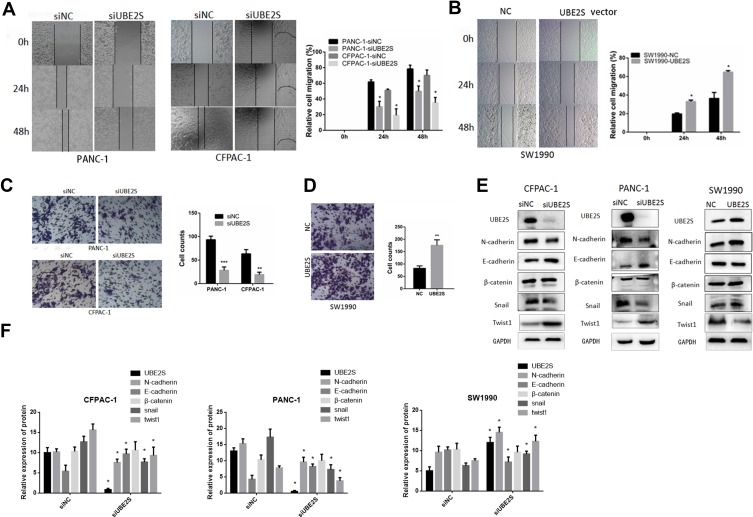
To further investigate the molecular mechanism by which UBE2S regulates PDAC cell metastasis and invasion, we performed migration and invasion experiment using PANC-1 and CFPAC1 cells transfected with siRNA (Figure 3A). Conversely, stably overexpressed UBE2S significantly attenuated metastasis and invasion (Figure 3B). Compared with the control group, cell proliferation analysis demonstrated that over-expression of UBE2S generally exerted a positive effect on the cell growth at 24 and 48 hrs, showing that UBE2S enhanced the oncogenic activities in vitro. Besides, a wound-healing assay and transwell metastasis assay were used to examine the effect of UBE2S on pancreatic cancer cell invasion and metastasis (Figure 3C and D). Silencing UBE2S reduced the number of migrating cells as compared to the control group (Figure 3C), indicating lower migratory abilities following its knockdown.
Figure 3.
UBE2S enhances the migratory and invasive ability of PDAC cells by EMT. (A) Effects of knockdown of UBE2S on PANC1 and CFPAC1 cells wound healing and representative images of distance (mm) of wound healing were shown. (B) Effects of overexpression of UBE2S on SW1990 cells wound healing and representative images of distance (mm) of wound healing were shown. (C) Effects of knockdown of UBE2S on PANC1 and CFPAC1 cell migration and invasion was measured and representative images of migrated and invaded cells were shown. (D) Effects of overexpression of UBE2S on SW1990 cell migration and invasion were measured and representative images of migrated and invaded cells were shown. (E) The effects of knockdown and overexpression of UBE2S on EMT markers were determined. (F) Densitometric analysis shows the effects of knockdown and overexpression of UBE2S on EMT markers of PDAC cells. *P<0.05, **P<0.01, ***P<0.001.
The GSEA data indicated that generally higher expressions of UBE2S for the most part were perhaps negatively correlated with epithelial-to-mesenchymal transition. Afterwards, this study evaluated the effects of UBE2S overexpression or silencing on the expression of related EMT factors in PANC-1, CFPAC1 and SW1990 cells. Additionally, it was demonstrated that the expression level and activity of Twist1 and Snail were inversely correlated in knockdown cells (Figure 3E and F) but had almost no significant correlation with other EMT inducers such as Slug and β-catenin (Figure 3E and F). Besides, over-expressing UBE2S reduced the expression level of the same EMT-inducing transcription factors, especially Twist1, as compared to the control group (Figure 3E and F), indicating that UBE2S enhanced EMT stimulated by those factors through a VHL-independent mechanism.
Over-Expression Of UBE2S Enhances The EMT In Pancreatic Cancer Cells Via The Vhl/Hif-1α/Stat3 Pathway
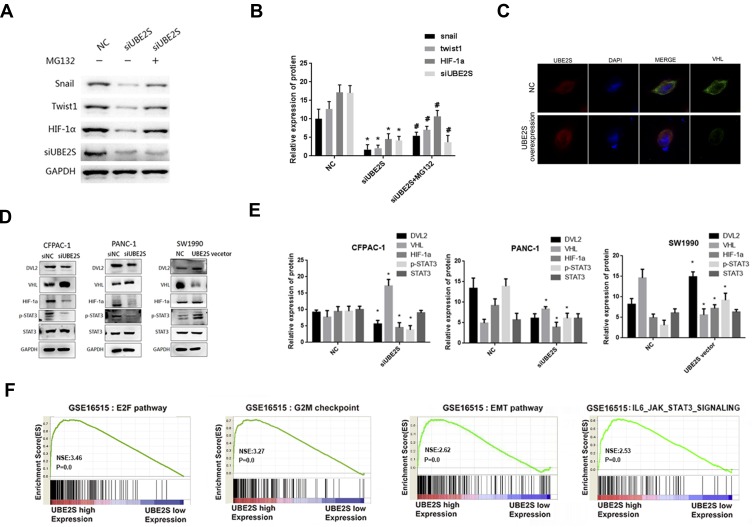
To investigate whether UBE2S negatively regulates EMT process through ubiquitin-proteasome degradation, PANC-1 and SW1990 cells which were transduced with the siRNA and pLVX-Puro-UBE2S lentivirus were treated with the proteasome inhibitor MG132, and related protein levels were analyzed. Furthermore, MG132 treatment largely abolished the inhibitory effect of UBE2S on snail, Twist1 and HIF-1α protein levels in cells (Figure 4A). The Western blotting results indicated that UBE2S inhibited the expression of related EMT factors such as Snail and Twist1 via the ubiquitin-proteasome system (Figure 4B). These data, therefore, suggested that UBE2S promoted PDAC cell metastasis and invasion by up-regulating EMT process via standard ubiquitination reactions.
Figure 4.
UBE2S enhances the EMT in pancreatic cancer cells via the VHL/HIF-1α/STAT3 pathway. (A) The expression levels of Snail, Twist1 and HIF-1α in the presence of MG132 were detected by Western blot analysis. (B) Densitometric analysis shows the expression levels of Snail, Twist1 and HIF-1α in the presence of MG132. *P<0.05, #P<0.05. (C) Reciprocal immunoprecipitation with VHL antibodies brought down UBE2S. (D) The expressions of DVL2, VHL, HIF-1α, p-STAT3 and STAT3 were tested by Western blotting. (E) Densitometric analysis shows the effects of knockdown and overexpression of UBE2S on DVL2, VHL, HIF-1α, p-STAT3 and STAT3. *P<0.05. (F) “E2F pathway”, “G2M checkpoint”, “EMT pathway” and 4 “IL6_JAK_STAT3 signing pathway” were enriched in patients with UBE2S-higher expression versus patients with UBE2S-lower expression.
As manifested in Figure 4C, this study performed an immunofluorescence with VHL and UBE2S, and reciprocal immunoprecipitation with VHL antibodies brought down UBE2S, suggesting a significant interaction between UBE2S and VHL in cells. The immunofluorescence revealed that UBE2S was co-located with VHL in pancreatic cancer cell. It also indicated that the knockdown and over-expression of UBE2S perhaps regulated the expression of related proteins by forming ubiquitin-related complexes. Moreover, UBE2S over-expression also prohibited the protein level and ubiquitination of VHL, causing the change of downstream proteins. Because of the role of UBE2S in regulating the VHL expression and the genes of Ubiquitin Mediated Proteolysis pathway, it was suggested that UBE2S perhaps regulated VHL expression via ubiquitination.
UBE2S silencing also significantly reduced the expression level and activity of p-STAT3, while the protein STAT3 did not display a difference (Figure 4D). Besides, the condition turned inversely in the over-expressed UBE2S cells. Furthermore, the HIF-1α expression demonstrated an opposite effect and revealed a correlation with VHL both in silencing and over-expressed cells in PDAC (Figure 4D and E). These results indicated that VHL/HIF-1α/STAT3 signaling was a possible downstream effector of UBE2S in PDAC. Moreover, the GSEA results also demonstrated that UBE2S could affect the downstream proteins through the IL6-JAK-STAT3 pathway. In summary, UBE2S perhaps controlled the JAK2-STAT3 pathway through VHL (Figure 4F), thereby promoting PDAC proliferation and EMT processes.
UBE2S Facilitates EMT Via The Ubiquitin-Proteasome System And Is Co-Expressed With CDC20
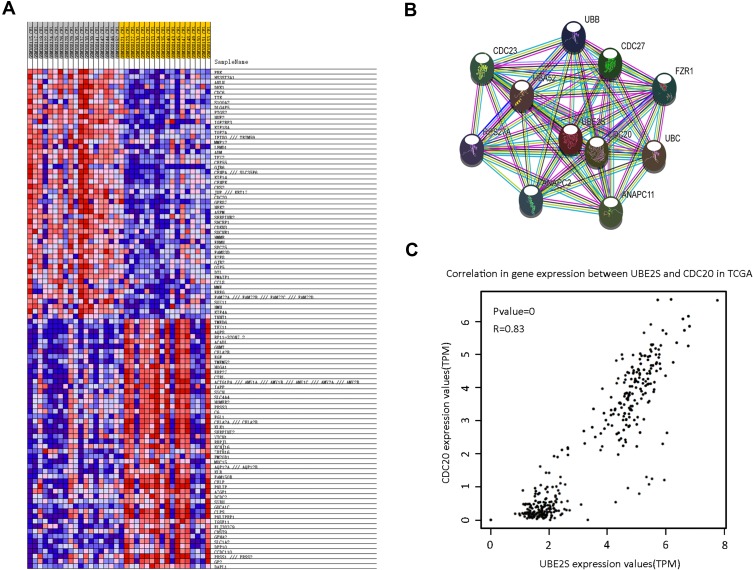
To evaluate the clinical importance of UBE2S in pancreatic cancer metastasis, we analyzed the co-expression between UBE2S expression levels and other related genes with GSE15471 dataset using GSEA. Furthermore, bioinformatics analysis and screening were used in the co-expression heat map of GSEA (Figure 5A). A series of proteins which had strong interaction with UBE2S protein were found through STRING (Figure 5B). Afterwards, highly expressed CDC20 in various types of malignant tumors was screened out by GEPIA and Oncomine database. Hence, solid evidence showed that there was a positive correlation between protein expression levels of UBE2S and CDC20 (Figure 5C). Besides, higher CDC20 expression was significantly correlated with poor disease-free survival in the group of human pancreatic cancer patients from the TCGA database. Moreover, similar results were obtained from four different groups of pancreatic cancer patients from the public Oncomine database.
Figure 5.
UBE2S co-expressed with CDC20. (A) The co-expression heat map from GSEA. The result indicated that there may be a positive correlation between UBE2S and CDC20. (B) Protein–protein interaction network of UBE2S based on STRING database with number of first shell interactors limited to 10 and using a medium confidence (0.400). (C) The correlation analysis of UBE2S and CDC20 in TCGA.
Estimated Mechanism
On one hand, the statistics indicated that UBE2S down-regulated the expression level of VHL trough Ubiquitin, forming Ubiquitin complexes with VHL and HIF-1α, and regulating transcription factor STAT3. Afterwards, it changed the downstream protein related with EMT. Another possibility was that UBE2S, as ubiquitin-modified enzyme, directly acted on STAT3. On the other hand, many studies demonstrated that ANAPC2 and ANAPC4 could form an important ubiquitin ligase APC/C with UBE2S, and APC/C played a vital role in cell cycle regulation. Moreover, CDC20 could regulate APC/C, activating other significant factor in the EMT progression such as E-cadherin by ubiquitin to affect the occurrence, development, invasion and metastasis of pancreatic cancer.
Discussion
Pancreatic cancer is a life-threatening disease worldwide, which could be the second leading cause of death by 2030.10 The main reason for high mortality is that the vast majority of patients present no obvious signs or symptoms, and another reason is the lack of sensitive biomarkers. Therefore, it is necessary and urgent to discover new therapeutic strategies and drugs.
It has been demonstrated that ubiquitination plays a crucial role in cancer development as previously mentioned. Here, we assumed that overexpression of UBE2S promotes pancreatic cancer cell EMT and enhances their tumorigenicity via the VHL-HIF-1α-STAT3 pathway. This study examined the gene expression profiles of GSE15471, GSE16515 and GSE28735 in several tumor and normal tissue samples to identify the molecular mechanism of PDAC and to seek some biomarkers beneficial to treatment. Moreover, the previous data mining identified 150 differentially expressed genes and the up-regulated gene UBE2S was selected. This study then analyzed the correlation between UBE2S expression and its clinical association with clinicopathological features and prognosis correlation TMAs of PDAC. Additionally, datasets from GEO and TCGA database also demonstrate that UBE2S highly expressed in pancreatic cancer patients’ tissues. Although UBE2S plays a role in tumor proliferation and metastasis, the functions of accumulated UBE2S in pancreatic cancer remain largely unknown. Chen et al9 found that UBE2S can promote the invasion and metastasis of esophageal squamous cell carcinoma. Besides, it was further reported by Kasamatsu et al17 that UBE2S binds to the anaphase-promoting complex/cyclosome (APC/C) to control the protein level of p21 through the ubiquitin proteasome system, thereby controlling the cell cycle of oral squamous cell carcinoma. It is generally established that ubiquitination is essential for plenty of biological processes, such as signal transduction, intracellular trafficking and protein degradation.18 Furthermore, when the ubiquitin proteasome system is overly activated, the ubiquitin degradation of p53 protein will be excessive, and thus its expression will be too low to act as tumor suppressor gene. Hence, it will exert influence on tumor occurrence and development, such as EMT.18
Then, to investigate the mechanism underlying the effect of UBE2S on the pancreatic cancer patients’ prognosis and therapeutic effect of medicine, functional experiments confirmed that over-expression of UBE2S induced pancreatic cancer cell migration, invasion and proliferation in vitro. The xenograft murine model experiments further evaluated the contribution of UBE2S to the pancreatic cancer proliferation in vivo. Besides, GSEA analysis indicated that EMT signaling and IL6-JAK-STAT3 pathway were facilitated in PDAC with high expression of UBE2S. Afterwards, new evidence provided has directly manifested that UBE2S promotes PDAC cells to accept EMT and metastasis/invasion by targeting the downstream epithelial markers such as Snail, Twist1 and other important proteins like STAT3, indicating that restoring the expression of UBE2S regulated the EMT. As a result, this study further confirmed the biological role of UBE2S in pancreatic cancer, especially in EMT progress. For instance, Snail1 functions as a consequence of its coordinated and continuous ubiquitination by several F-box-specific E3 ligases in normal epithelial cells, but its degradation is prevented in cancer cells and in activated fibroblasts.19
Emphasis was also put on the ubiquitin complex formed by UBE2S and VHL as well as its control over related downstream protein. It is generally accepted that the deletion and mutation of VHL lead to the development of many types of tumors.20 VHL, along with other members such as elongin B form E3 ubiquitin ligase complex, and the complex, binding to HIF-1α, are degraded by the ubiquitin-proteasome system. VHL deletion causes the activation of HIF-1α, thereby controlling biological behaviors such as cell proliferation and EMT progressions, which explains the reason why VHL promotes tumors’ development. Jung et al reported that UBE2S can control the stability of the tumor suppressor gene VHL.7 Solid evidence has indicated that under normoxic conditions, UBE2S can reduce the level of VHL protein and activate HIF-1α, hence promoting the proliferation and EMT progressions of renal cell carcinoma and melanoma cells.21 Literature retrieval manifested that UBE2S is involved in ubiquitination and subsequent degradation of VHL, resulting in an accumulation of HIF1-α.14 To be specific, high expression of UBE2S in tumor cells allows VHL to be recognized for degradation, leading to the increased release of HIF-1α and permits the entry of HIF-1α into nuclei and activation of downstreaming gene transcription. This mechanism is therefore contributing to increased proliferation, angiogenesis and metastasis of the tumor cells.22 Thus, it can be assumed that UBE2S may be identified as a regulatory gene of HIF-1α, and its protein directly targets VHL for degradation. Furthermore, in PDAC, there is a certain negative correlation between the expression of UBE2S and VHL at the cellular level.14 Though it is demonstrated that UBE2S regulates the VHL-HIF-1α pathway in tumors and participates in tumor development such as cervical cancer, the accurate significance and mechanism remain unknown.23 In this study, in PDAC UBE2S, our results of immunofluorescent fluorescence and other experiments fully explained control the VHL-HIF-1α pathway by forming a ubiquitination complex.
Hence, it was assumed that UBE2S perhaps controls the JAK2-STAT3 pathway through VHL, and thereby promotes PDAC proliferation and EMT progression. To investigate the molecular mechanism of UBE2S on EMT-related factors’ expression and HIF-1α, we treated PANC-1 and SW1990 cells, which had been transduced with the siRNA and pLVX-Puro-UBE2S lentivirus; MG132 treatment largely abolished the inhibitory effect of UBE2S on snail, Twist1 and HIF-1α in cells. What is more, this study performed an immunofluorescence with VHL and UBE2S, suggesting a significant interaction between UBE2S and VHL in cells and co-location of UBE2S with VHL in pancreatic cancer cell. Considering the direct interaction between them, it was tentatively proposed that UBE2S regulates Snail/Twist1 expression and functions indirectly or directly with HIF-1α via a complex molecular mechanism. Recent researches have indicated that VHL reduces downstream signal transduction by inhibiting the JAK-STAT pathway.24 For instance, Russell et al found that in Chuvash polycythemia, VHL binds to SOCS1 to form E3 ubiquitin ligase, and then it further binds to JAK2 and its phosphorylated protein, thereby reducing related protein levels and inhibiting STAT5 activation.24 Wu et al reported that in renal cell carcinoma, VHL can control JAK kinase activity through SOCS protein and then further control tumor metastasis, and the whole procedure is done without HIF-1α.21 The study by Kanno et al showed that VHL controls the oncogenic activities of glioma stem cells by inhibiting the JAK2-STAT3 signaling pathway.25 Together, previous studies indicate that JAK-STAT can control the transduction of downstream signals without HIF-1α in the VHL-controlled signal transduction pathway.24
Interestingly, the result of GSEA showed that CDC20 has a certain positive correlation with UBE2S. More specifically, CDC20 was considered to be involved in the mammalian cell cycle mechanism activating the anaphase-promoting complex.26 CDC20 is important for cell division, whose activity might be regulated by a balance between ubiquitination and de-ubiquitination. APC activation is required for initiation of anaphase and exit from mitosis, and CDC20 is highly expressed in many tumor cells where it leads to chromosomal instability. Kidokoro27 discovered that inactivation of p53 observed in various cancer tissues is perhaps the cause of the CDC20 up-regulation. Thus, high CDC20 expression has been reported in various malignancies, including hepatocellular carcinoma, breast cancer, gastric cancer, colorectal cancer and pancreatic cancer cells. At present, there are clinical trials on relevant targeted CDC20 drugs. Craney reported that the phosphorylation of a specific serine residue in the APC/C coactivator CDC20 prevents delivery of UBE2S to the APC/C, allowing UBE2S to bind to the APC/C and catalyze ubiquitin chain elongation.28 More importantly, previous results of bioinformatic analysis indicated that the co-expression of UBE2S and CDC20 perhaps offers new possibility of early diagnosis, dynamic monitoring of curative effect and new drug target of pancreatic cancer.
To our knowledge, our study is the first to manifest that UBE2S is associated with oncogenic activities in PDAC by VHL/HIF-1α/STAT3 signal via the ubiquitin-proteasome system, based on which the possible mechanism can be presented as follows (Figure 6): on one hand, the statistics of the experiment demonstrated that UBE2S down-regulated the expression level of VHL through ubiquitin, forming ubiquitin complexes with VHL and HIF-1α, thereby regulating transcription factor STAT3. Afterwards, it changed the downstream protein related to EMT. Another possibility is that UBE2S, as ubiquitin-modified enzyme, directly acted on STAT3. On the other hand, many researches suggested that ANAPC2 and ANAPC4 could form an important ubiquitin ligase APC/C with UBE2S, and APC/C played a vital role in cell cycle regulation. Moreover, CDC20 could regulate APC/C, activating other significant factors in the EMT progression, such as E-cadherin by ubiquitin to affect the occurrence, development, invasion and metastasis of pancreatic cancer. Our immunofluorescence analysis indicated that UBE2S perhaps modulates Snail and Twist1 without binding directly to their promoters. On the contrary, UBE2S perhaps forms part of a complex regulating VHL as well as HIF-1α. However, further studies are required to identify and characterize this regulatory mechanism.
Figure 6.
Schematic diagram of molecular mechanism of UBE2S regulating oncogenic activities by VHL/HIF-1α/STAT3 signal via the ubiquitin-proteasome system in PDAC.
In conclusions, we identified UBE2S as an important protein by data mining, and reported UBE2S expression in pancreatic cancer. We also analyzed its clinical association of such expression with clinicopathological features and prognostic correlation by TMAs. Furthermore, it was confirmed that UBE2S functions in pancreatic cancer cell lines. In vitro experiments, it was demonstrated that UBE2S overexpression promoted but UBE2S knockdown suppressed cell proliferation and migration via modulation of VHL/HIF-1α/STAT3 signaling pathway. Co-expression of UBE2S and VHL up-regulated the expression of EMT signal factors and its downstream effectors such as Snail and Twist1. UBE2S, together with CDC20, a popular drug target gene recently, could also form a novel biomarker of clinic dynamic monitoring of curative effect. Overall, this study provides compelling evidence that UBE2S is a potential prognostic factor and it functions via the ubiquitin-proteasome system in PDAC.
Funding Statement
This work was supported by National Natural Science Foundation of China grants 81472792.
Ethical Approval
All animal experiments were approved by the Animal Research Ethics Committee of Nanjing Medical University and performed in accordance with the guidelines for animal care of Lianyungang NO.1 people Hospital’s Institutional Animal Care and Use Committee. Nude mice (BALB/c nu/nu) (Shanghai SLAC Laboratory Animal Co. Ltd) were housed in the animal facility of the Experimental Animal Center of Nanjing Medical University.
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This study was approved by the Ethics Committee of Lianyungang NO.1 people Hospital.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet TJ, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 2.Whittle MC, Izeradjene K, Rani PG, et al. RUNX3 controls a metastatic switch in pancreatic ductal adenocarcinoma. Cell. 2015;161:1345–1360. doi: 10.1016/j.cell.2015.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheng Y, Hong JH, Doherty R, et al. A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen. Mol Cell Proteomics. 2012;11:329–341. doi: 10.1074/mcp.O111.013706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang LF, Zhang ZG, Yang J, McLaughlin SH, Barford D. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature. 2015;522:450–454. doi: 10.1038/nature14471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garnett MJ, Mansfeld J, Godwin C, et al. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat Cell Biol. 2009;11:1363–1369. doi: 10.1038/ncb1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown NG, Vanderlinden R, Watson ER, et al. Dual RING E3 architectures regulate multiubiquitination and ubiquitin chain elongation by APC/C. Cell. 2016;165:1440–1453. doi: 10.1016/j.cell.2016.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jung CR, Hwang KS, Yoo JS, et al. E2-EPF UCP targets pVHL for degradation and associates with tumor growth and metastasis. Nat Med. 2006;12:809–816. doi: 10.1038/nm1440 [DOI] [PubMed] [Google Scholar]
- 8.Tedesco D, Zhang JH, Trinh L, et al. The ubiquitin-conjugating enzyme E2-EPF is overexpressed in primary breast cancer and modulates sensitivity to topoisomerase II inhibition. Neoplasia. 2007;9:601–613. doi: 10.1593/neo.07385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen MF, Lee KD, Lu MS, et al. The predictive role of E2-EPF ubiquitin carrier protein in esophageal squamous cell carcinoma. J Mol Med (Berl). 2009;87:307–320. doi: 10.1007/s00109-008-0430-3 [DOI] [PubMed] [Google Scholar]
- 10.Smith BD, Smith GL, Hurria A, Hortobagyi GN, Buchholz TA. Future of cancer incidence in the United States: burdens upon an aging, changing nation. J Clin Oncol. 2009;27:2758–2765. doi: 10.1200/JCO.2008.20.8983 [DOI] [PubMed] [Google Scholar]
- 11.Marcucci F, Stassi G, De Maria R. Epithelial-mesenchymal transition: a new target in anticancer drug discovery. Nat Rev Drug Discov. 2016;15:311–325. doi: 10.1038/nrd.2015.13 [DOI] [PubMed] [Google Scholar]
- 12.Thiery JP, Acloque H, Huang RYJ, Nieto MA, EMT also contributes to tissue repair BICA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. [DOI] [PubMed] [Google Scholar]
- 13.Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells – what challenges do they pose? Nat Rev Drug Discovery. 2014;13:497–512. doi: 10.1038/nrd4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LaGory EL, Giaccia AJ. The ever-expanding role of HIF in tumour and stromal biology. Nat Cell Biol. 2016;18:356–365. doi: 10.1038/ncb3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620 [DOI] [PubMed] [Google Scholar]
- 16.Berlingieri MT, Pallante P, Guida M, et al. UbcH10 expression may be a useful tool in the prognosis of ovarian carcinomas. Oncogene. 2007;26:2136–2140. doi: 10.1038/sj.onc.1210010 [DOI] [PubMed] [Google Scholar]
- 17.Yoshimura S, Kasamatsu A, Nakashima D, et al. UBE2S associated with OSCC proliferation by promotion of P21 degradation via the ubiquitin-proteasome system. Biochem Biophys Res Commun. 2017;485:820–825. doi: 10.1016/j.bbrc.2017.02.138 [DOI] [PubMed] [Google Scholar]
- 18.Herman AG, Hayano M, Poyurovsky MV, et al. Discovery of Mdm2-MdmX E3 ligase inhibitors using a cell-based ubiquitination assay. Cancer Discov. 2011;1:312–325. doi: 10.1158/2159-8290.CD-11-0104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambies G, Miceli M, Martínez-Guillamon C, et al. TGF2-activated USP27X deubiquitinase regulates cell migration and chemoresistance via stabilization of snail1. Cancer Res. 2019;79:33–46. doi: 10.1158/0008-5472.CAN-18-0753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaelin WG; In TVH-LHCSWFDAYATUCFOTDPARFTVH-LGV. Molecular basis of the VHL hereditary cancer syndrome. Nat Rev Cancer. 2002;2:673–682. doi: 10.1038/nrc885 [DOI] [PubMed] [Google Scholar]
- 21.Wu KL, Miao H, Khan S. JAK kinases promote invasiveness in VHL-mediated renal cell carcinoma by a suppressor of cytokine signaling-regulated, HIF-independent mechanism. Am J Physiol Renal Physiol. 2007;293:F1836–F1846. doi: 10.1152/ajprenal.00096.2007 [DOI] [PubMed] [Google Scholar]
- 22.Kim WY, Kaelin WG. Role of VHL gene mutation in human cancer. J Clin Oncol. 2004;22:4991–5004. doi: 10.1200/JCO.2004.05.061 [DOI] [PubMed] [Google Scholar]
- 23.Park KS, Kim JH, Shin HW, et al. E2-EPF UCP regulates stability and functions of missense mutant pVHL via ubiquitin mediated proteolysis. BMC Cancer. 2015;15:800. doi: 10.1186/s12885-015-1786-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russell RC, Sufan RI, Zhou B, et al. Loss of JAK2 regulation via a heterodimeric VHL-SOCS1 E3 ubiquitin ligase underlies chuvash polycythemia. Nat Med. 2011;17:845. doi: 10.1038/nm.2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanno H, Sato H, Yokoyama TA, Yoshizumi T, Yamada S. The VHL tumor suppressor protein regulates tumorigenicity of U87-derived glioma stem-like cells by inhibiting the JAK/STAT signaling pathway. Int J Oncol. 2013;42:881–886. doi: 10.3892/ijo.2013.1773 [DOI] [PubMed] [Google Scholar]
- 26.Weinstein J, Jacobsen FW, Hsu-Chen J, Wu T, Baum LG. A novel mammalian protein, p55CDC, present in dividing cells is associated with protein kinase activity and has homology to the saccharomyces cerevisiae cell division cycle proteins Cdc20 and Cdc4. Mol Cell Biol. 1994;14:3350–3363. doi: 10.1128/MCB.14.5.3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kidokoro T, Tanikawa C, Furukawa Y, Katagiri T, Nakamura Y, Matsuda KC. CDC20, a potential cancer therapeutic target, is negatively regulated by p53. Oncogene. 2008;27(11):1562–1571. doi: 10.1038/sj.onc.1210799 [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi M, VanderLinden R, Weissmann F, et al. Cryo-EM of mitotic checkpoint complex-bound APC/C reveals reciprocal and conformational regulation of ubiquitin ligation. Mol Cell. 2016;63:593–607. doi: 10.1016/j.molcel.2016.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]