Abstract
Background
Biofilm formation is an important virulence factor expressed by Acinetobacter baumannii. It shields and protects microbial cells from host immune responses, antibiotics, and other anti-infectives. Its effects on Acinetobacter baumannii infection treatments notwithstanding, important environmental factors that influence its formation have not been fully investigated.
Methods
Biofilm formation was assessed using the qualitative modified Congo red assay and quantitative microtiter plate methods. The combined effect of temperature, medium and shear force was determined by measuring adherence (OD570 nm) in microtiter plate after incubation at 26°C, 30°C, and 37°C when biofilm-grown cells were cultured in the presence of minimal nutrient medium (EAOB) and nutrient-rich medium (TSB) without or with agitation at 50 rpm. Antibiotics susceptibility of meropenem, imipenem, and ciprofloxacin were tested with Kirby-Bauer disc method. P<0.05 was considered statistically significant in all the tests.
Results
A noticeable variation in adherence was observed among the isolates cultured with both media. Biofilm forming capacity of the isolates range from 0.09–0.33. The majority of the isolates had their relative biofilm-forming capacity significantly (p<0.05) higher than the positive control, Acinetobacter baumannii ATCC 19606. The biofilm biomass during growth in nutrient-rich medium (TSB) without shaking was significantly different (p<0.05; Tukey’s test) among the three temperatures tested compared with when it was cultured in EAOB without shaking. A positive correlation was observed between biofilm formation and resistance to imipenem (r=0.2889; p=0.05). There was a statistically significant difference among the median of the three source groups (p<0.05) compared with the median between the source groups.
Conclusion
This observation extended further the view that A. baumannii biofilm formation is enhanced when nutrient-poor medium is used at room temperature (26°C) with or without agitation compared to growth at 37°C.
Keywords: Acinetobacter baumannii, biofilm, incubation temperature, growth medium, agitation, isolate source, antibiotic, resistance, hospital-acquired, infections
Introduction
Acinetobacter baumannii is considered a threat to microbial infection treatment in hospitals and other related settings because it is resistant to almost all known last resort antibiotics and other anti-infective agents.1 The survival in desiccated environments and tolerance to human immune response when aggregated into biofilm have, nonetheless, promoted scientific research interest in the last decade.2 Microbial biofilm life-form is different from their planktonic counterpart in that it increases the structural fitness advantage needed to resist the access of anti-infective to their target sites, creating physiochemical concentration gradients across the constituting microbes, thereby triggering differential gene expression and regulation among the microbes.3 Biofilm is an independent aggregate of bacteria and it appears to be the most common form of bacterial life.4 Biofilms are found growing everywhere across the surface and they constitute a huge problem when they are located inside the human body.5,6 Free floating bacteria can easily be killed by human white blood cells, antibiotics, and anti-infectives but, when in biofilm life-form, the bacteria are completely protected.7
Alterations in physicochemical factors within microbial environment enable changes in some phenotypic and genotypic features that promote microbial virulence and biofilm formation.8 Biofilm formation is triggered when bacterial cells produce quorum chemicals and conduct cell-to-cell communication in a cell density-dependent manner during change in environmental factors such as temperature, quality of growth medium, and the dynamic state of the environment which either encourages or discourages initial bacterial attachment.9–11 Moreover, change in environmental factors can modify the microbial physiological conditions within the constituting microbial cells in biofilm and influence microbial interaction and other cell properties.12
Microbial adhesion to substratum depends on the physicochemical properties of the substratum which in turn could be modified by temperature and nutrient composition of the growth medium matrix.13,14 For instance, microbial polysaccharides undergo transition from a disordered state at higher temperature to an ordered state at lower temperature, and this, therefore, can affect aggregation of bacterial cells into biofilm.15 The physical strength and density of microbial biofilm could equally be influenced by fluid shear: static or dynamic conditions.16 The hydrodynamics in biofilm formation influences not only the rate of attachments and detachments of microbial cells but also biofilm structure in most bacteria cells.17
A. baumannii is among the non-fermentative Gram-negative microbes and has its gene expression and regulation influenced by the existing ecological niche through selective adaptation.18 Some virulence-associated genes have been shown to vary in both the level and extent of expression and regulation at various microbial environments and life forms.19–21 For example, biofilm-associated protein (Bap) and pili assembly gene (Csu) are vital gene products required for biofilm production in most bacteria and have been reported to be lost or variant in some strains of A. baumannii due to the environmental adaptation.22,23 Other microbial species have been shown to form biofilm at specific environments such as aquaculture environment, human bronchial cavity and object surfaces.24–26
A. baumannii causes diverse infections at different anatomical sites such as hospital-acquired infections, skin and soft tissue infections, nosocomial meningitis, cystic fibrosis, periodontitis, septicemia, and urinary tracts infections.27–29 Some of these infections may be caused by a single bacterial species, but more often are caused by a complex and diverse community of microorganisms and result into chronic infection due to the establishment of biofilm life form thereby, leading to high levels of morbidity and mortality, gross economic loss, and a prolonged stay in hospital.30
Although A. baumannii is an important pathogen in the intensive care units of hospital, and have been isolated from wound and chronic infections, the processes leading to its biofilm formation have not been completely explained. The present study thus aimed to investigate the ability of A. baumannii isolated from hospital environments to form biofilm using a microtitre plate adherence and modified Congo red assays. In addition, the combined influence of growth medium, incubation temperature, and fluid shear in A. baumannii were also examined.
Methods
Ethics
Exemption (BREC reference number EXM063/19) of the the project protocol was granted by the Biomedical Research Ethics Committee (BREC) of the University of KwaZulu-Natal).
Isolation And Identification
Effluent water samples from three sampling points (final effluent collection point, main ward collection point, and pathology lab collection point) from Applebosh (Hospital A): a rural primary healthcare center and Greys (Hospital B): an urban primary healthcare center, respectively, were filtered through a 0.45µm membrane filter (Millipore, Billerica, MA, USA) and incubated at 37ºC for 24 hours on Acinetobacter selective agar: Leed Acinetobacter Medium (LAM) agar.31 Isolates were sub-cultured onto a general-purpose medium (Muller-Hinton agar), incubated at 42ºC for 24 hours and streaked back on LAM agar for confirmation. Preliminary colony selection from plates was carried out by Gram staining, oxidase, and catalase production tests.
Selection Criteria
A total of 118 mucoid pink colonies which appeared as short pink rods, catalase positive and oxidase negative, were further screened for the presence of plasmids using alkaline lysis method. The plasmid harboring strains (n=71) were stored in 15% glycerol-Luria-Bertani broth frozen stock and kept at −80ºC for further molecular analysis. All isolates were maintained on Tryptic Soy Agar (TSA) plates and stored at 4ºC for short-term use. The type strain A. baumannii ATCC 19606 (American Type Culture Collection, Manassas, VA, USA) was used as a reference strain in all assays.32
Qualitative Biofilm Examination: Modified Congo Red Assay (CRA)
The biofilm formation ability of the isolates (n=71) was first determined by the modified Congo red assay method as previously reported.33 The composition of the growth medium was as follows: brain heart infusion broth (37 g/L), sucrose (50 g/L), agar bacteriological (10 g/L) were autoclaved separately with Congo red dye (8 g/L). The solidified medium plate was punched with 48-punch pin dipped onto 200 µL of overnight broth cultures of different bacterial strains in a 96-well microtiter plate and incubated at 37°C. The presence of black, brown, and red colonies around each punch point indicated strong, moderate, and weak biofilm production of the individual isolates, respectively. Type strains Staphylococcus aureus ATCC 12600, which was previously reported as a strong biofilm producer, and Staphylococcus epidermidis ATCC 12228 were used as positive and negative reference strains, respectively.33
Determination Of Antimicrobial Susceptibilities
The antimicrobial susceptibility of imipenem, meropenem, and ciprofloxacin, which were previously associated with biofilm formation in Acinetobacter baumannii, were tested by Kirby-Bauer disc diffusion method (Oxoid, UK). The concentrations of the discs (expressed in µg) were: imipenem (10), meropenem (10), and ciprofloxacin (5). The petri dishes were incubated at 37°C for 24 hours. The diameters of the inhibition zone were measured and used to classify the bacteria as susceptible, intermediate, or resistant to each antibiotic. The results were interpreted according to the CLSI guidlines.34
Measurement Of Microbial Adherence
The isolates were cultured overnight in tryptic soy broth (TSB) at 37°C for 24 hours and the biofilm-forming ability, measured as the level of adherence to microtiter plate, was determined by modified microtiter plate assay.24 In brief, 10 µL of cell suspension containing 1.0 McFarland prepared in TSB/enriched Anacker and Ordal’s broth (EAOB) was inoculated into 96-well round-bottom polystyrene plates. The suspension was made up with 90 µL of growth medium (TSB and EAOB) and 100 µL of sterile distilled water.35 The plates were incubated for 24 hours at different temperatures (26°C, 30°C, 37°C) without or with agitation at 50 rpm. After incubation, the plates were washed thrice with 250 µL sterile distilled water and 200 µL of methanol for 5 minutes. The plates were allowed to dry in an inverted position and the attached biofilm was stained with 150 µL of 2% Hugo’s crystal violet solution for 10 minutes. The stain was washed out using gentle running tap water. The remaining stain was solubilized with 150 µL 33% (v/v) glacial acetic acid and the absorbance at 570 nm was considered as a measure of the biofilm biomass.36 The experiments were done in triplicate for each strain. Growth media (TSB and EAOB, respectively) without bacterial cells were used as negative control, while A. baumannii ATCC 19606 was used as a positive control.32 Biofilm production was interpreted as the level of absorbance and the values were averaged and interpreted.37 The optical density (OD) cut-off value was established as three standard deviations (SD) above the mean OD of the negative control: ODC=average OD of negative control (un-inoculated growth media). The isolates were classified as follows: OD≤ODC=non-adherent, ODc<OD≤(2×ODC)=weakly adherent; (2×ODC)<OD≤(4×ODC)=moderately adherent and (4×ODC)<OD=strongly adherent.24,37
A second method of expressing biofilm-forming ability for each Acinetobacter baumannii isolate was to calculate the relative biofilm capacity to the average value of all the isolates as follows:
Relative biofilm capacity,
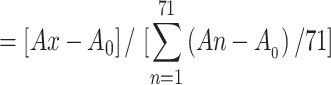 |
where Ax=absorbance at 570 nm for isolates x, and A0=absorbance for un-inoculated growth medium.38
Statistical Analysis
The statistical significant difference (p<0.05) as a result of the change in the physicochemical factors (temperature, medium, and agitation) in the microtiter adherence assay and the biofilm formation capacity of the isolates from different sources were determined by repeated measure one-way analysis of variance by ranks (ANOVA) and non-parametric Kruskal-Wallis test, respectively, using Sigmaplot 14.0 software and confirmed with GraphPad Prism 5.0 (GraphPad Software, San Diego, CA, USA). The means between the groups were separated by the Tukey’s pairwise multiple comparison test. Linear correlation and Fisher’s exact test were used to determine the relationship between the biofilm forming capacity of the isolates and their resistance to one or more of the three antibiotics tested; and this was confirmed by comparing the median of each group with Mann–Whitney U-test. Statistically significant difference is assumed when p<0.05 in all the tests.
Results
Qualitative Biofilm Assay (CRA)
On modified CRA, biofilm-forming strains formed black colonies, whereas non-biofilm forming strains developed strong red colonies (Figure 1). Moderate biofilm producing isolates showed light red to brown pigmentation (Figure 1). Both reference strains A. baumannii ATCC 19606 and S. aureus ATCC 12600 were observed to be biofilm formers, whereas S. epidermidis ATCC 12228 was observed to be a non-biofilm producing strain. Among the tested A. baumannii isolates (n=71), 16 (22.54%) were strong biofilm producers (complete black to slight black), and 32 (45.07%) were moderate biofilm formers (brown), while 23 (32.39%) were non-biofilm formers (strong red) (Table 1).
Figure 1.
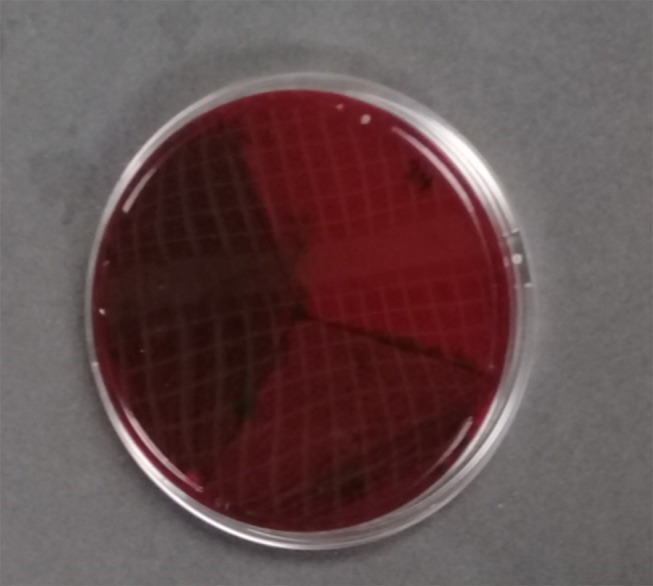
Description of the colony colours when cultured on CRA. Black colonies indicated strong biofilm producers; Brown/slight dark colonies indicated weak biofilm producers; and Dark red colonies indicated non-biofilm producers.
Table 1.
Biofilm Formation of Acinetobacter baumannii Isolates (n=71) Following Incubation at 26°C, 30°C and 31°C Under Dynamic or Static Conditions in Nutrient-Rich (TSB) or Nutrient-Poor (EAOB) Media, Respectively
| Physicochemical factor | Non-Adherent | Weak Adherent | Moderate Adherent | Strong Adherent | Total | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| N (%) | Average | N (%) | Average | N (%) | Average | N (%) | Average | N (%) | Average | |
| OD ± SD | OD ± SD | OD ± SD | OD ± SD | OD ± SD | ||||||
| 26°C EAOB dynamic | 0 | 0 | 8 (11.3) | 0.09 ± 0.01 | 38 (53.5) | 0.17 ± 0.02 | 25 (35.2) | 0.25 ± 0.02 | 71 (100) | 0.19±0.02 |
| 26°C EAOB static | 0 | 0 | 1 (1.4) | 0.09 ± 0.01 | 31 (43.7) | 0.18 ± 0.02 | 39 (54.92) | 0.25 ± 0.01 | 71 (100) | 0.22 ±0.01 |
| 26°C TSB dynamic | 0 | 0 | 0 | 0 | 65 (91.5) | 0.25 ± 0.01 | 6 (8.5) | 0.30 ± 0.01 | 71 (100) | 0.26 ±0.01 |
| 26°C TSB static | 0 | 0 | 0 | 0 | 48 (67.6) | 0.27 ± 0.01 | 23 (32.4) | 0.29 ± 0.01 | 71 (100) | 0.28 ±0.01 |
| 30°C EAOB dynamic | 0 | 0 | 10 (14.1) | 0.10 ± 0.01 | 53 (74.6) | 0.19 ± 0.02 | 8 (11.3) | 0.28 ± 0.02 | 71 (100) | 0.18±0.02 |
| 30°C EAOB static | 3 (4.23) | 0.11 ± 0.01 | 28 (39.4) | 0.18 ± 0.02 | 40 (56.3) | 0.25 ± 0.01 | 71 (100) | 0.22 ±0.02 | ||
| 30°C TSB dynamic | 0 | 0 | 2 (2.8) | 0.10 ± 0.02 | 69 (97.2) | 0.25 ± 0.02 | 0 | 0 | 71 (100) | 0.24 ±0.02 |
| 30°C TSB static | 0 | 0 | 26 (36.6) | 0.22 ± 0.02 | 45 (63.4) | 0.26 ± 0.01 | 0 | 0 | 71 (100) | 0.24 ±0.01 |
| 37°C EAOB static | 3 (4.2) | 0.10 ± 0.00 | 26 (36.6) | 0.16 ± 0.01 | 42 (59.27) | 0.25 ± 0.02 | 0 | 0 | 71 (100) | 0.21 ±0.01 |
| 37°C EAOB static | 0 | 0 | 11 (15.5) | 0.15 ± 0.01 | 54 (76.1) | 0.23 ± 0.01 | 4 (5.6) | 0.30 ± 0.01 | 71 (100) | 0.21 ±0.02 |
| 37°C TSB dynamic | 2 (2.8) | 0.10 ± 0.02 | 23 (32.4) | 0.20 ± 0.01 | 46 (64.8) | 0.26 ± 0.01 | 0 | 0 | 71 (100) | 0.23 ±0.02 |
| 37°C TSB static | 3 (4.2) | 0.10 ± 0.01 | 56 (78.9) | 0.21 ± 0.02 | 12 (16.9) | 0.28 ± 0.01 | 0 | 0 | 71 (100) | 0.22 ±0.02 |
Notes: Biofilm formation assay data is the mean of three independent experiments carried out in triplicate SD after growth in minimal (EAOB) and rich (TSB) media at 26°C, 30°C, and 37°C under dynamic and static conditions, respectively.
Microtiter Adherence Assay
The adherence of the tested isolates (n=71) obtained with polystyrene round-bottom microtiter plates following incubation at 26°C, 30°C, and 37°C for 24 hours under static or dynamic conditions in nutrient-rich (TSB) or nutrient-poor (EAOB) media was determined (Table 2; Figures 2A–D), respectively. A noticeable variation in adherence was observed among the isolates cultured with EAOB, with values ranging from 0.09±0.01 (isolate FG116) to 0.32±0.02 (isolate FG121), at 26°C, from 0.10±0.00 (isolate PL448) to 0.30±0.02 (isolate FA012) at 30°C, from 0.10±0.01 (isolate MW437) to 0.31±0.01 (isolates FA001, MW181 and FA012) and among the isolates cultured with TSB, with values ranging from 0.22±0.02 (isolate MW416) to 0.30±0.02 (isolate FG121, MW179, FA012, and FA034) at 26°C, from 0.13±0.06 (isolate FG148) to 0.31±0.02 (isolates MW179 and FA035) at 30°C, and from 0.10±0.02 (isolate FA088) to 0.33±0.01 (isolate FA35) at 37°C, respectively (Table 1). The isolates with their adherence and relative biofilm-forming capacity ≤0.1 (media OD570 nm), respectively, were considered non-biofilm formers (Table 3). Hence, 8/71 (11.27%) of the isolates were observed to be non-biofilm formers in the presence of either EAOB or TSB. All the isolates showed noticeable adherence only in nutrient-poor (EAOB) medium at 26°C without agitation, while 98.59% (70/71) were able to adhere with both nutrient-poor and nutrient-rich TSB. None of the isolates (0%) and 1/71 (1.41%) of the isolates adhered in EAOB or TSB alone, respectively (Table 2).
Table 2.
Biofilm Formation, Biofilm Forming Capacity At Different Temperatures, Growth Media, And Antibiotic Susceptibility Of Acinetobacter baumannii Isolates
| Isolate ID | Source | Biofilm-Forming Capacity OD570 nm | Congo Red | Antibiotic Sucsceptibility | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| EAOB | TSB | ||||||||||
| 26°C | 30°C | 37°C | 26°C | 30°C | 37°C | 37°C | IMP | MEM | CIP | ||
| Media | - | 0.05±0.00 | 0.06±0.01 | 0.08±0.01 | 0.07±0.01 | 0.12±0.02 | 0.13±0.01 | ||||
| ATCC 19606 | + | 0.21±0.03 | 0.22±0.02 | 0.22±0.03 | 0.25±0.01 | 0.21±0.02 | 0.16±0.04 | S | S | I | |
| FA008 | Final effluent | 0.27±0.00 | 0.26±0.00 | 0.28±0.01 | 0.30±0.00 | 0.26±0.02 | 0.25±0.02 | Black | R | R | R |
| PL482 | Pathology lab | 0.25±0.02 | 0.27±0.01 | 0.21±0.00 | 0.26±0.01 | 0.25±0.04 | 0.25±0.03 | Brown | R | R | R |
| MW422 | Mainward | 0.22±0.01 | 0.21±0.01 | 0.15±0.01 | 0.25±0.00 | 0.22±0.01 | 0.14±0.02 | Red | R | R | R |
| PL450 | Pathology lab | 0.21±0.01 | 0.23±0.01 | 0.21±0.02 | 0.28±0.00 | 0.27±0.01 | 0.19±0.04 | Brown | R | R | R |
| FA007 | Final effluent | 0.27±0.00 | 0.28±0.01 | 0.30±0.01 | 0.29±0.00 | 0.25±0.02 | 0.25±0.02 | Black | R | R | R |
| PL476 | Pathology lab | 0.20±0.02 | 0.21±0.02 | 0.20±0.00 | 0.28±0.00 | 0.27±0.00 | 0.18±0.01 | Brown | R | R | R |
| MW181 | Mainward | 0.25±0.01 | 0.25±0.00 | 0.31±0.00 | 0.30±0.00 | 0.27±0.00 | 0.21±0.04 | Brown | I | R | R |
| MW186 | Mainward | 0.29±0.04 | 0.28±0.00 | 0.30±0.01 | 0.27±0.01 | 0.29±0.01 | 0.20±0.03 | Brown | R | R | S |
| FA108 | Final effluent | 0.21±0.01 | 0.22±0.01 | 0.23±0.05 | 0.30±0.03 | 0.25±0.03 | 0.23±0.02 | Brown | R | R | R |
| FA113 | Final effluent | 0.29±0.01 | 0.28±0.00 | 0.21±0.05 | 0.28±0.00 | 0.23±0.04 | 0.25±0.01 | Brown | R | R | R |
| MW419 | Mainward | 0.23±0.02 | 0.23±0.00 | 0.22±0.01 | 0.27±0.01 | 0.22±0.01 | 0.18±0.02 | Red | R | R | R |
| FG148 | Final effluent | 0.24±0.01 | 0.20±0.11 | 0.22±0.01 | 0.25±0.02 | 0.13±0.06 | 0.16±0.02 | Red | R | S | S |
| FG359 | Final effluent | 0.22±0.01 | 0.25±0.04 | 0.26±0.02 | 0.28±0.00 | 0.27±0.01 | 0.18±0.03 | Brown | R | S | S |
| FA38 | Final effluent | 0.28±0.02 | 0.24±0.01 | 0.23±0.01 | 0.27±0.01 | 0.25±0.06 | 0.28±0.01 | Brown | R | S | S |
| PL470 | Pathology lab | 0.18±0.01 | 0.21±0.00 | 0.19±0.02 | 0.27±0.00 | 0.25±0.01 | 0.15±0.01 | Red | R | R | I |
| FA68 | Final effluent | 0.19±0.02 | 0.18±0.04 | 0.22v0.01 | 0.29±0.01 | 0.27±0.00 | 0.25±0.01 | Brown | R | R | R |
| FA001 | Final effluent | 0.24±0.01 | 0.22±0.01 | 0.31±0.00 | 0.28±0.01 | 0.26±0.01 | 0.25±0.01 | Black | R | I | S |
| FG377 | Final effluent | 0.29±0.00 | 0.29±0.00 | 0.25±0.01 | 0.29±0.00 | 0.27±0.01 | 0.25±0.03 | Red | R | R | S |
| FG127 | Final effluent | 0.11±0.02 | 0.14±0.03 | 0.14±0.02 | 0.29±0.01 | 0.27±0.00 | 0.21±0.00 | Red | R | R | R |
| PL520 | Final effluent | 0.12±0.00 | 0.12±0.02 | 0.13±0.01 | 0.28±0.01 | 0.27±0.01 | 0.19±0.01 | Red | R | R | R |
| FA56 | Final effluent | 0.25±0.01 | 0.27±0.01 | 0.21±0.02 | 0.26±0.01 | 0.22±0.00 | 0.27±0.01 | Black | R | R | R |
| PL517 | Pathology lab | 0.11±0.01 | 0.10±0.00 | 0.14±0.01 | 0.29±0.01 | 0.27±0.01 | 0.16±0.02 | Brown | R | R | S |
| MW440 | Mainward | 0.19±0.03 | 0.16±0.09 | 0.11±0.03 | 0.28±0.01 | 0.24±0.02 | 0.13±0.01 | Red | R | S | S |
| FA009 | Final effluent | 0.27±0.01 | 0.27±0.01 | 0.30±0.00 | 0.30±0.00 | 0.28±0.01 | 0.27±0.01 | Black | R | R | R |
| MW187 | Mainward | 0.24±0.00 | 0.25±0.02 | 0.25±0.01 | 0.29±0.00 | 0.27±0.00 | 0.22±0.02 | Brown | R | I | S |
| FG361 | Final effluent | 0.31±0.09 | 0.25±0.01 | 0.23±0.00 | 0.28±0.00 | 0.23±0.01 | 0.23±0.01 | Brown | R | R | I |
| PL480 | Final effluent | 0.19±0.01 | 0.18±0.03 | 0.24±0.02 | 0.27±0.00 | 0.24±0.02 | 0.23±0.00 | Red | R | R | I |
| PL467 | Pathology lab | 0.24±0.00 | 0.25±0.02 | 0.25±0.01 | 0.29±0.00 | 0.27±0.00 | 0.22±0.02 | Red | R | R | R |
| PL530 | Pathology lab | 0.22±0.01 | 0.24±0.01 | 0.22±0.01 | 0.27±0.01 | 0.25±0.00 | 0.21±0.02 | Red | R | S | S |
| FA42 | Final effluent | 0.21±0.00 | 0.24±0.01 | 0.28±0.01 | 0.29±0.00 | 0.25±0.03 | 0.22±0.01 | Brown | R | I | R |
| PL448 | Final effluent | 0.13±0.01 | 0.10±0.00 | 0.15±0.01 | 0.26±0.01 | 0.25±0.00 | 0.09±0.00 | Red | R | I | I |
| FA30 | Final effluent | 0.17±0.02 | 0.22±0.01 | 0.22±0.04 | 0.27±0.01 | 0.22±0.01 | 0.27±0.01 | Brown | R | R | R |
| FA41 | Final effluent | 0.22±0.04 | 0.17±0.02 | 0.22±0.02 | 0.28±0.01 | 0.25±0.01 | 0.28±0.01 | Brown | R | I | I |
| FA89 | Final effluent | 0.23±0.03 | 0.24±0.01 | 0.29±0.01 | 0.29±0.00 | 0.25±0.01 | 0.23±0.00 | Black | R | I | S |
| FA99 | Final effluent | 0.24±0.01 | 0.25±0.01 | 0.29±0.02 | 0.28±0.01 | 0.19±0.01 | 0.17±0.01 | Red | R | R | S |
| PL460 | Pathology lab | 0.19±0.01 | 0.17±0.01 | 0.15±0.01 | 0.24±0.01 | 0.22±0.00 | 0.21±0.02 | Red | R | I | S |
| MW420 | Mainward | 0.19±0.02 | 0.23±0.01 | 0.21±0.01 | 0.30±0.00 | 0.23±0.01 | 0.21±0.02 | Red | R | S | S |
| FA88 | Final effluent | 0.16±0.00 | 0.17±0.05 | 0.20±0.03 | 0.29±0.01 | 0.23±0.00 | 0.10±0.02 | Red | R | I | I |
| MW184 | Mainward | 0.14±0.01 | 0.13±0.04 | 0.16±0.01 | 0.30±0.00 | 0.27±0.01 | 0.19±0.01 | Brown | R | I | S |
| FA29 | Final effluent | 0.18±0.03 | 0.25±0.01 | 0.20±0.01 | 0.29±0.00 | 0.25±0.02 | 0.24±0.03 | Brown | R | R | I |
| PL494 | Pathology lab | 0.19±0.00 | 0.21±0.01 | 0.29±0.01 | 0.29±0.00 | 0.29±0.01 | 0.31±0.01 | Black | R | S | S |
| PL466 | Pathology lab | 0.16±0.01 | 0.18±0.01 | 0.16±0.03 | 0.27±0.01 | 0.24±0.02 | 0.22±0.01 | Red | R | R | I |
| FA34 | Final effluent | 0.22±0.01 | 0.25±0.02 | 0.24±0.01 | 0.30±0.01 | 0.26±0.01 | 0.26±0.10 | Black | R | R | R |
| MW416 | Mainward | 0.22±0.01 | 0.21±0.03 | 0.18±0.02 | 0.22±0.02 | 0.20±0.01 | 01.9±0.03 | Brown | R | I | I |
| MW447 | Mainward | 0.18±0.01 | 0.14±0.00 | 0.17±0.05 | 0.26±0.01 | 0.26±0.01 | 016±0.00 | Red | R | R | I |
| FA18 | Final effluent | 0.22±0.01 | 0.22±0.01 | 0.25±0.02 | 0.28±0.00 | 0.20±0.01 | 0.21±0.03 | Brown | R | R | R |
| PL508 | Pathology lab | 0.18±0.02 | 0.17±0.01 | 0.15±0.01 | 0.28±0.01 | 0.24±0.02 | 0.22±0.01 | Brown | R | R | R |
| FA14 | Final effluent | 0.26±0.01 | 0.25±0.02 | 0.22±0.01 | 0.27±0.01 | 0.25±0.03 | 0.28±0.00 | Brown | R | R | R |
| PL449 | Pathology lab | 0.17±0.02 | 0.17±0.01 | 0.14±0.01 | 0.26±0.00 | 0.24±0.00 | 0.14±0.01 | Red | R | R | R |
| FA002 | Final effluent | 0.19±0.02 | 0.23±0.01 | 0.21±0.01 | 0.30±0.00 | 0.23±0.01 | 0.21±0.02 | Black | R | S | S |
| FA006 | Final effluent | 0.24±0.00 | 0.26±0.00 | 0.30±0.02 | 0.29±0.00 | 0.26±0.01 | 0.25±0.02 | Black | R | S | I |
| PL446 | Pathology lab | 0.28±0.01 | 0.24±0.01 | 0.23±0.01 | 0.28±0.02 | 0.22±0.05 | 0.26±0.01 | Brown | R | R | S |
| PL487 | Pathology lab | 0.25±0.02 | 0.26±0.01 | 0.30±0.01 | 0.29±0.00 | 0.25±0.01 | 0.20±0.02 | Brown | R | S | S |
| FG121 | Final effluent | 0.32±0.02 | 0.28±0.00 | 0.20±0.03 | 0.29±0.02 | 0.30±0.01 | 0.21±0.02 | Black | R | R | I |
| FA12 | Final effluent | 0.30±0.00 | 0.30±0.02 | 0.31±0.01 | 0.30±0.01 | 0.29±0.01 | 0.28±0.01 | Black | I | I | I |
| MW192 | Mainward | 0.25±0.03 | 0.25±0.01 | 0.25±0.00 | 0.28±0.01 | 0.27±0.00 | 0.22±0.01 | Brown | R | R | I |
| FA73 | Final effluent | 0.20±0.03 | 0.21±0.09 | 0.18±0.03 | 0.28±0.00 | 0.22±0.02 | 0.24±0.05 | Black | R | I | I |
| FA21 | Final effluent | 0.18±0.00 | 0.19±0.01 | 0.15±0.01 | 0.25±0.02 | 0.22±0.01 | 0.17±0.01 | Brown | S | S | S |
| MW179 | Mainward | 0.31±0.01 | 0.29±0.01 | 0.12±0.02 | 0.30±0.03 | 0.31±0.02 | 0.21±0.01 | Brown | R | R | I |
| MW190 | Mainward | 0.16±0.03 | 0.13±0.01 | 0.19±0.00 | 0.29±0.01 | 0.26±0.01 | 0.25±0.01 | Brown | R | I | I |
| FA14 | Final effluent | 0.26±0.01 | 0.25±0.02 | 0.22±0.01 | 0.27±0.01 | 0.25±0.03 | 0.28±0.00 | Brown | I | I | I |
| PL479 | Pathology lab | 0.15±0.02 | 0.16±0.00 | 0.17±0.01 | 0.27±0.00 | 0.26±0.00 | 0.22±0.01 | Red | R | S | S |
| MW400 | Mainward | 0.25±0.03 | 0.26±0.02 | 0.26±0.01 | 0.28±0.00 | 0.22±0.01 | 0.21±0.05 | Black | R | R | R |
| FA35 | Final effluent | 0.28±0.01 | 0.29±0.01 | 0.27±0.01 | 0.28±0.03 | 0.31±0.02 | 0.33±0.01 | Black | R | R | S |
| FG116 | Final effluent | 0.09±0.01 | 012±0.01 | 0.11±0.02 | 0.28±0.01 | 0.25±0.01 | 0.25±0.09 | Brown | R | S | S |
| FG378 | Final effluent | 0.23±0.02 | 0.26±0.01 | 0.21±0.01 | 0.27±0.00 | 0.21±0.03 | 0.20±0.01 | Red | R | S | S |
| FA28 | Final effluent | 0.28±0.01 | 0.24±0.02 | 0.23±0.00 | 0.27±0.00 | 0.23±0.01 | 0.28±0.00 | Brown | R | R | S |
| MW437 | Mainward | 0.16±0.01 | 0.14±0.01 | 0.10±0.01 | 0.23±0.01 | 0.20±0.01 | 0.26±0.01 | Brown | R | S | S |
| MW397 | Mainward | 0.25±0.01 | 0.24±0.01 | 0.27±0.00 | 0.29±0.00 | 0.23±0.02 | 0.24±0.05 | Black | R | I | S |
| FA106 | Final effluent | 0.17±0.01 | 0.23±0.04 | 0.17±0.03 | 0.27±0.01 | 0.24±0.01 | 0.15±0.01 | Red | R | R | S |
| PL471 | Pathology lab | 0.22±0.01 | 0.23±0.01 | 0.22±0.01 | 0.27±0.01 | 0.25±0.01 | 0.27±0.01 | Red | R | R | R |
Notes: Biofilm formation assay is the mean of three independent experiments carried out in triplicate±SD following growth in minimal (EAOB) and rich (TSB) at 26°C, 30°C, and 37°C without shaking, respectively.
Abbreviations: IMP, Imipenem; MEM, Meropenem; CIP, Ciprofloxacin; R, Resistance; I, Intermediate; S, Susceptible; Black, Strong biofilm formation; Brown, moderate biofilm formation; Red, weak biofilm formation using congo red assay method; EAOB, Anacker and Ordal’s broth; TSB, Tryptic soy broth.
Figure 2.
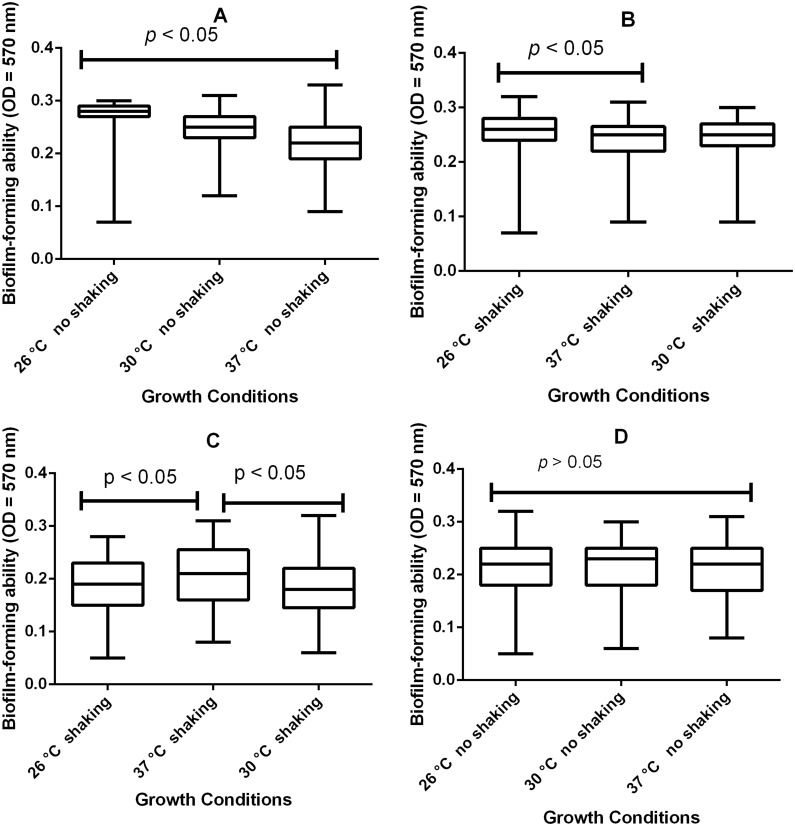
Biofilm formation of Acinetobacter baumannii at various growth conditions. (A) The measures of adherence to microtiter plates (OD570 nm) during growth at 26°C, 30°C, and 37°C in nutrient-rich medium (TSB) without shaking were significantly different (p<0.05; Tukey’s test) among the three temperatures tested, indicating that the variance in their means cannot be by chance but by the temperature. (B) Growth at the same nutrient-rich medium (TSB) with shaking only caused significant variation in the means (p=0.05) between groups 26°C and 37°C. (C) Biofilm-forming ability within the groups (26°C and 37°C) and (30°C and 37°C), respectively, were significantly different (p<0.05; Tukey’s comparison test) but not among the three groups when cultured in minimal medium (EAOB) with shaking. (D) There is no significant difference (p>0.05) among the three groups when cultured in EAOB without shaking, indicating that biofilm growth was optimized at this condition.
Table 3.
Relative Biofilm-Forming Capacity Of Individual Acinetobacter baumannii Isolates From Hospital Effluent Water
| Isolate ID | EAOB | TSB | ||||
|---|---|---|---|---|---|---|
| 26°C | 30°C | 37°C | 26°C | 30°C | 37°C | |
| ATCC 19606 | 0.97±0.002 | 0.98±0.002 | 1.00±0.002 | 0.83±0.003 | 0.91±0.001 | 1.00±0.002 |
| FA008 | 1.30±0.003 | 1.21±0.003 | 1.46±0.003 | 1.09±0.002 | 1.09±0.002 | 1.46±0.003 |
| PL482 | 1.17±0.003 | 1.27±0.003 | 0.96±0.002 | 0.92±0.002 | 1.02±0.002 | 0.96±0.002 |
| MW422 | 0.97±0.002 | 0.95±0.002 | 0.55±0.001 | 0.87±0.002 | 0.75±0.001 | 0.55±0.001 |
| PL450 | 0.96±0.002 | 1.03±0.002 | 0.93±0.002 | 0.97±0.003 | 1.21±0.002 | 0.93±0.002 |
| FA007 | 1.32±0.003 | 1.33±0.003 | 1.60±0.003 | 1.05±0.003 | 1.01±0.002 | 1.60±0.003 |
| PL476 | 0.86±0.002 | 0.93±0.002 | 0.89±0.002 | 1.02±0.003 | 1.17±0.002 | 0.89±0.002 |
| MW181 | 1.17±0.003 | 1.16±0.003 | 1.63±0.003 | 1.10±0.002 | 1.19±0.002 | 1.63±0.003 |
| MW186 | 1.45±0.003 | 1.33±0.003 | 1.56±0.003 | 0.97±0.003 | 1.32±0.002 | 1.56±0.003 |
| FA108 | 0.96±0.002 | 1.00±0.002 | 1.08±0.002 | 1.07±0.002 | 1.01±0.002 | 1.08±0.002 |
| FA113 | 1.39±0.003 | 1.36±0.003 | 0.93±0.002 | 0.98±0.002 | 0.85±0.002 | 0.93±0.002 |
| MW419 | 1.06±0.003 | 1.06±0.002 | 1.03±0.002 | 0.94±0.002 | 0.76±0.001 | 1.03±0.002 |
| FG148 | 1.13±0.003 | 0.89±0.002 | 1.02±0.002 | 0.87±0.003 | 0.06±0.000 | 1.02±0.002 |
| FG359 | 0.98±0.002 | 1.20±0.003 | 1.28±0.003 | 1.00±0.002 | 1.13±0.002 | 1.28±0.003 |
| FA038 | 1.38±0.003 | 1.11±0.003 | 1.10±0.002 | 0.93±0.003 | 0.99±0.002 | 1.10±0.002 |
| PL470 | 0.78±0.002 | 0.93±0.002 | 0.77±0.002 | 0.95±0.003 | 1.03±0.002 | 0.77±0.002 |
| FA068 | 0.81±0.002 | 0.73±0.002 | 0.98±0.002 | 1.03±0.003 | 1.16±0.002 | 0.98±0.002 |
| FA001 | 1.14±0.003 | 1.02±0.002 | 1.62±0.003 | 1.02±0.003 | 1.11±0.002 | 1.62±0.003 |
| FG377 | 1.43±0.003 | 1.43±0.003 | 1.22±0.002 | 1.03±0.003 | 1.15±0.002 | 1.22±0.002 |
| FG127 | 0.37±0.001 | 0.50±0.001 | 0.47±0.001 | 1.05±0.002 | 1.14±0.002 | 0.47±0.001 |
| PL520 | 0.39±0.001 | 0.41±0.001 | 0.41±0.001 | 1.00±0.003 | 1.16±0.002 | 0.41±0.001 |
| FA056 | 1.19±0.003 | 1.29±0.003 | 0.96±0.002 | 0.91±0.003 | 0.75±0.001 | 0.96±0.002 |
| PL517 | 0.33±0.001 | 0.28±0.001 | 0.42±0.001 | 1.03±0.003 | 1.15±0.002 | 0.42±0.001 |
| MW440 | 0.84±0.002 | 0.65±0.002 | 0.27±0.001 | 0.99±0.003 | 0.93±0.002 | 0.27±0.001 |
| FA009 | 1.29±0.003 | 1.28±0.003 | 1.54±0.003 | 1.09±0.003 | 1.24±0.002 | 1.54±0.003 |
| MW187 | 1.11±0.003 | 1.20±0.003 | 1.24±0.002 | 1.05±0.003 | 1.17±0.002 | 1.24±0.002 |
| FG361 | 1.55±0.004 | 1.14±0.003 | 1.07±0.002 | 0.97±0.003 | 0.88±0.002 | 1.07±0.002 |
| PL480 | 0.81±0.002 | 0.73±0.002 | 1.17±0.002 | 0.96±0.003 | 0.94±0.002 | 1.17±0.002 |
| PL467 | 1.11±0.003 | 1.20±0.003 | 1.24±0.002 | 1.05±0.003 | 1.17±0.002 | 1.24±0.002 |
| PL530 | 0.99±0.002 | 1.12±0.003 | 1.02±0.002 | 0.93±0.003 | 1.02±0.002 | 1.02±0.002 |
| FA042 | 0.95±0.002 | 1.10±0.003 | 1.42±0.003 | 1.05±0.003 | 1.04±0.002 | 1.42±0.003 |
| PL448 | 0.49±0.001 | 0.29±0.001 | 0.53±0.001 | 0.89±0.003 | 1.01±0.002 | 0.53±0.001 |
| FA030 | 0.72±0.002 | 1.00±0.002 | 1.00±0.002 | 0.97±0.003 | 0.76±0.001 | 1.00±0.002 |
| FA041 | 0.98±0.002 | 0.72±0.002 | 1.03±0.002 | 1.02±0.003 | 1.00±0.002 | 1.03±0.002 |
| FA089 | 1.08±0.003 | 1.10±0.003 | 1.50±0.003 | 1.03±0.002 | 1.05±0.002 | 1.50±0.003 |
| FA099 | 1.11±0.003 | 1.15±0.003 | 1.51±0.003 | 0.98±0.002 | 0.57±0.001 | 1.51±0.003 |
| PL460 | 0.82±0.002 | 0.68±0.002 | 0.54±0.001 | 0.82±0.003 | 0.78±0.001 | 0.54±0.001 |
| MW420 | 0.82±0.002 | 1.05±0.002 | 0.93±0.002 | 1.08±0.003 | 0.83±0.001 | 0.93±0.002 |
| FA088 | 0.62±0.001 | 0.69±0.002 | 0.86±0.002 | 1.05±0.003 | 0.87±0.002 | 0.86±0.002 |
| MW184 | 0.54±0.001 | 0.46±0.001 | 0.61±0.001 | 1.08±0.003 | 1.19±0.002 | 0.61±0.001 |
| FA029 | 0.76±0.002 | 1.16±0.003 | 0.86±0.002 | 1.03±0.003 | 1.01±0.002 | 0.86±0.002 |
| PL494 | 0.80±0.002 | 0.96±0.002 | 1.51±0.003 | 1.07±0.002 | 1.30±0.002 | 1.51±0.003 |
| PL466 | 0.67±0.002 | 0.73±0.002 | 0.57±0.001 | 0.93±0.003 | 0.93±0.002 | 0.57±0.001 |
| FA034 | 0.99±0.002 | 1.15±0.003 | 1.18±0.002 | 1.08±0.002 | 1.07±0.002 | 1.18±0.002 |
| MW416 | 1.01±0.002 | 0.92±0.002 | 0.74±0.001 | 0.69±0.002 | 0.64±0.001 | 0.74±0.001 |
| MW447 | 0.74±0.002 | 0.52±0.001 | 0.66±0.001 | 0.91±0.003 | 1.07±0.002 | 0.66±0.001 |
| FA018 | 1.01±0.002 | 1.00±0.002 | 1.21±0.002 | 1.01±0.00 | 0.59±0.001 | 1.21±0.002 |
| PL508 | 0.74±0.002 | 0.71±0.002 | 0.50±0.001 | 0.98±0.002 | 0.96±0.002 | 0.50±0.001 |
| FA014 | 1.22±0.003 | 1.18±0.003 | 0.98±0.002 | 0.93±0.002 | 0.99±0.002 | 0.98±0.002 |
| PL449 | 0.71±0.002 | 0.69±0.002 | 0.42±0.001 | 0.88±0.003 | 0.93±0.002 | 0.42±0.001 |
| FA002 | 0.82±0.002 | 1.05±0.002 | 0.93±0.002 | 1.08±0.003 | 0.83±0.001 | 0.93±0.002 |
| FA006 | 1.10±0.003 | 1.22±0.003 | 1.60±0.003 | 1.05±0.002 | 1.13±0.002 | 1.60±0.003 |
| PL446 | 1.34±0.003 | 1.09±0.003 | 1.05±0.002 | 0.99±0.003 | 0.81±0.001 | 1.05±0.002 |
| PL487 | 1.15±0.003 | 1.24±0.003 | 1.56±0.003 | 1.06±0.003 | 1.04±0.002 | 1.56±0.003 |
| FG121 | 1.57±0.004 | 1.35±0.003 | 0.89±0.002 | 1.02±0.003 | 1.40±0.003 | 0.89±0.002 |
| FA012 | 1.45±0.003 | 1.46±0.003 | 1.61±0.003 | 1.08±0.002 | 1.34±0.002 | 1.61±0.003 |
| MW192 | 1.16±0.003 | 1.17±0.003 | 1.20±0.002 | 0.98±0.003 | 1.15±0.002 | 1.20±0.002 |
| FA073 | 0.89±0.002 | 0.93±0.002 | 0.76±0.002 | 1.02±0.002 | 0.77±0.001 | 0.76±0.002 |
| FA021 | 0.77±0.002 | 0.82±0.002 | 0.53±0.001 | 0.84±0.003 | 0.77±0.001 | 0.53±0.001 |
| MW179 | 1.54±0.004 | 1.43±0.003 | 0.30±0.001 | 1.10±0.003 | 1.51±0.003 | 0.30±0.001 |
| MW190 | 0.62±0.001 | 0.44±0.001 | 0.82±0.002 | 1.05±0.002 | 1.09±0.002 | 0.82±0.002 |
| FA004 | 1.22±0.003 | 1.18±0.003 | 0.98±0.002 | 0.93±0.003 | 0.99±0.002 | 0.98±0.002 |
| PL479 | 0.57±0.001 | 0.61±0.001 | 0.65±0.001 | 0.95±0.003 | 1.11±0.002 | 0.65±0.001 |
| MW400 | 1.21±0.003 | 1.21±0.003 | 1.31±0.003 | 1.02±0.003 | 0.79±0.001 | 1.31±0.003 |
| FA035 | 1.39±0.003 | 1.43±0.003 | 1.38±0.003 | 1.00±0.003 | 1.51±0.003 | 1.38±0.003 |
| FG116 | 0.26±0.001 | 0.36±0.001 | 0.23±0.000 | 1.01±0.003 | 1.02±0.002 | 0.23±0.000 |
| FG378 | 1.06±0.003 | 1.23±0.003 | 0.91±0.002 | 0.97±0.003 | 0.70±0.001 | 0.91±0.002 |
| FA028 | 1.33±0.003 | 1.12±0.003 | 1.06±0.002 | 0.78±0.003 | 0.84±0.002 | 1.06±0.002 |
| MW437 | 0.65±0.002 | 0.50±0.001 | 0.19±0.000 | 1.04±0.003 | 0.64±0.001 | 0.19±0.000 |
| MW397 | 1.21±0.003 | 1.09±0.003 | 1.39±0.003 | 0.96±0.003 | 0.82±0.001 | 1.39±0.003 |
| FA106 | 0.70±0.002 | 1.04±0.002 | 0.67±0.01 | 9.54±0.003 | 0.96±0.002 | 0.67±0.001 |
| PL471 | 0.97±0.002 | 1.07±0.002 | 1.01±0.002 | 0.94±0.000 | 1.00±0.002 | 1.01±0.002 |
Note: Relative biofilm-forming capacity was determined using equation described by Van Houdt et al.38
Less than 25% of the isolates had their biofilm biomass below 0.27, 0.25, and 0.22 at 26°C, 30°C, and 37°C, respectively, and 0.22, 0.22,and 0.20 at 26°C, 30°C, and 37°C, respectively at static conditions when cultured in nutrient-poor and nutrient-rich media, respectively (Figures 2A–D). A statistically significant difference (p<0.05) in adherence was observed when the isolates were assayed under static/dynamic conditions in nutrient-poor and nutrient-rich media, indicating that the variations among the means of the group was as a result of variable incubation temperatures (Table 2). Agitation alone did not significantly affect adherence if the medium was unchanged (p>0.05). The majority of the isolates had their relative biofilm-forming capacity significantly (p<0.05) higher than the positive control, A.baumannii ATCC 19606 (Figures 3A–C). While A. baumannii ATCC 19606 preferred EAOB to TSB for adherence at 30°C, 48/71 (67.61%) of biofilm-forming isolates were strongly adherent in both TSB and/or EAOB, respectively. The relationship between adherence and/or biofilm-forming capacity and resistance to imipenem was only statistically significant (r=0.2889; alpha=0.05) at 30°C when cultured in EAOB without agitation (Table 3).
Figure 3.
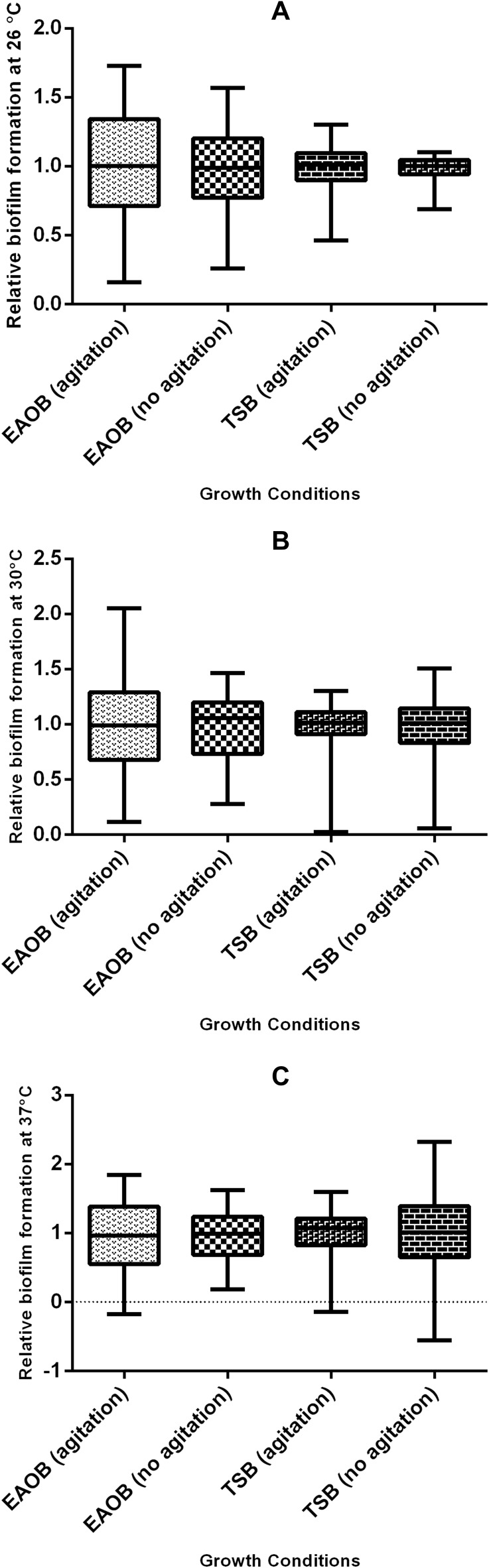
Description of the relative biofilm-forming capacity at different growth conditions (temperature, medium type, and hydrodynamics). There was wide variability in the relative biofilm-forming capacity of the isolates when cultured in nutrient-poor medium (EAOB), unlike in nutrient-rich medium (TSB). (A) More than 25% (above Q4) of the isolate of the isolates maintained their relative biofilm forming capacity above 1.23 (shaking) and 1.20 (no shaking) in EAOB, while it was 1.12 (with shaking) and 1.11 (no shaking) in TSB, respectively. (B) More than 50% (median value) of the isolates had their relative biofilm forming capacity above the positive reference strain (0.98; Table 2) when cultured in EAOB at 30°C without shaking. (C) More than 50% of the isolates had their capacity below 1.0 when cultured in EAOB at 37°C with or without shaking.
Distribution Of Biofilm-Formers In Various Hospital Sources And Wards
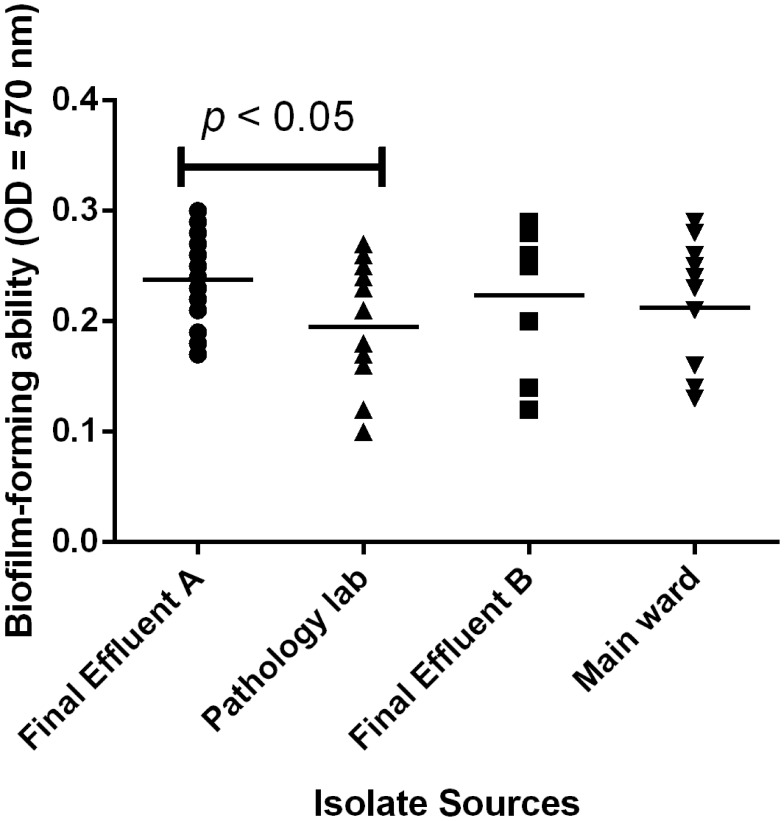
All the tested isolates were obtained from final effluent 36/71 (50.70%), the pathological lab 19/71 (26.76%), and the main hospital ward 16/71 (22.54%) from two tertiary care centers. The proportion of strong biofilm producers from the main ward was 2/16 (12.5%), pathological lab 1/19 (5.26%), and final effluent 13/36 (36.11%) using modified Congo red method (Figure 1). Biofilm biomass (OD570 nm) among the various sources were compared by non-parametric Kruskal-Wallis test. There was a statistically significant difference among the median of the groups (p-value=0.0492). However, comparing the median OD570 nm between the groups indicated that there was no significant difference between the final effluent and the main ward (p>0.05) (Dunn’s test). Analysis of the biofilm forming capacity of the isolates obtained from various sources revealed a statistically significant difference in OD570 nm between the pathology lab and final effluent (p<0.05) (Figure 4).
Figure 4.
The biofilm forming capacity of the isolates according to their sources. There was a statistically significant difference (p<0.05) in adherence among the groups. However, using Dunn’s multiple comparison test to separate the means, it was observed that only the sources from final effluent A and the pathology lab were significantly different (p=0.0492).
Discussion
A. baumannii is a continued threat to microbial infection treatment, especially in the intensive care units in hospitals.39 This is due to its resistance to almost all known antibiotics and survival in desiccated environments.40 Acinetobacter spp have also been isolated from diverse environments such as hospital environments, ventilators, and water systems.41–43 The presence of bacteria in hospital effluents have been identified as a reservoir of several resistance determinants and have been placed on a watch list as one of the major distributors of novel virulence, resistance, structural and regulatory genetic determinants which may impact on their ability to form biofilm.44 In the current study, we investigated for the first time the combined effect of physicochemical factors on the ability of Acinetobacter baumannii to produce biofilm.
The ability of A. baumannii to form biofilm appear to depend on several factors which can inter-affect each other. This was shown by a noticeable variation in adherence among the isolates when cultured in nutrient rich (TSB) and nutrient poor (EAOB) media. Although all the isolates were able to form biofilm in one or more cultural growth conditions, microbial biofilm formation appeared to be more favorable in the presence of nutrient-poor medium (EAOB) (Table 2). A previous study also reported that A. baumannii biofilm formation was strongly impaired in rich growth medium.44 In another study, minimal medium supplemented with glucose and amino acids was used to compare the early stage of biofilm to a mature biofilm and it was observed that a nutrient starved environment favors mature biofilm formation.46 This observation is extended further by showing that there seemed to be enhancement in biofilm formation when nutrient-rich medium was used at room temperature (26°C) with or without agitation compared to growth at 37°C. However, in the present study, isolates generally showed a preference for nutrient-poor medium when incubated at 30°C without agitations during biofilm cultivation (p<0.05) (Table 2). It is not surprising that agitation during microbial growth had a minimal influence on biofilm formation since it was previously suggested that it decreased molecular orderliness required for the formation of vital structural proteins required to form a mature biofilm,15 although it was previously reproted that a threshold value of shear stress enhanced biofilm formation.47
The maximum microbial adherence to 96-microtiter plates at 30°C was previously reported.48,49 Although another study reported an increase in the expression of virulence factors such as OmpA and PaaC which contribute to biofilm formation at 37°C, it further established that Acinetobacter biofilm formation is more favorable at 28°C due to upregulation of Csu operon.50 It is therefore interesting to observe in the current study that the combined effect of lower incubation temperature, minimal growth medium, and reduced shear force can optimize Acinetobacter biofilm formation. The findings of the present study gives credence to the opinion that biofilm formation is a physiological response of bacteria to external stress.51 The capacity for biofilm formation was shown to be higher in microtiter plates when compared with modified Congo red assay method. However, all the isolates (n=16) that showed strong biofilm formation in Congo red assay were also strong biofilm producers at 30°C with EAOB in microtiter plate assay. Previous studies have also demonstrated that the CRA method has low accuracy but is very cheap and easy to perform and visualize.33,52 The indication for black pigmentation in strong biofilm formers has been explained to be due to the production of exopolysaccharide (Figure 1A). This was in sharp contrast with moderate and weak biofilm formers which appeared brown and red, respectively (Figures 1B and C). It is also interesting to note from this study that the relative biofilm formation capacity of the weakest (0.06; isolate FG148) and the strongest (1.63; isolate MW181) biofilm formers were different by a factor of 27.17 indicating that the isolates have great in vitro biofilm-forming capacity (Table 3).37 This assumption was also explained further since the majority of the isolates have their relative biofilm forming capacity greater than that of the positive reference strain, A. baumannii ATCC 19606 at one or more growth conditions (Figures 3A–C).
Although previous studies have reported the association of biofilm formers with multidrug-resistance in A. baumannii, it appeared in the current study that there was similarity in the rate of resistance to meropenem and ciprofloxacin among biofilm-formers and non-biofilm (Table 4). However, a positive correlation exists between imipenem resistance and adherence to microtiter plates (p<0.05). This has also been reported by other researchers who have correlated resistance to imipenem to biofilm formation.53,54 In the present study, the association between biofilm formation in A. baumannii and resistance to ciprofloxacin and meropenem cannot be established, as was previously reported.55 However, it appeared that the rate of resistance in biofilm formers and non-biofilm formers were the same (Table 4). Intriguingly, in this study, a significant difference in adherence among the isolates from different sources was found, although the difference was not statistically significant between some groups (p<0.05). This finding is in contrast to a previous study which reported that the isolate source and the clonality contributed insignificantly to the capacity of bacterial cells to produce biofilm in vitro.56,57 Other studies have since reported variation in the biofilm forming capacity of bacterial cells isolated from different hospital wards and body location which, hence, paid credence to this finding.58,59 Hence, it is desired that extensive investigations be carried out in this regard.
Table 4.
Antimicrobial Profile Of Acinetobacter Baumannii Isolates (n=71) And Their Biofilm Forming Ability At 26°C, 30°C And 37°C, Respectively
| Antimicrobial agents/Culture Temp. | Strong biofilm Formers | Moderate biofilm Formers | Weak biofilm Formers | Non-biofilm Formers | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R | S | I | R | S | I | R | S | I | R | S | I | |
| 26°C | n % | n % | n % | n % | n % | n % | n % | n % | n % | n % | n % | n % |
| Imipenem | 21 (29.57) | 0 (0) | 2 (2.82) | 46 (64.79) | 1 (1.41) | 1 (1.14) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Meropenem | 11 (15.49) | 5 (7.04) | 7 (9.86) | 30 (42.25) | 10 (14.08) | 8 (11.27) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Ciprofloxacin | 9 (12.67) | 8 (11.27) | 6 (8.45) | 16 (22.54) | 20 (28.17) | 12(16.90) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| 30°C | ||||||||||||
| Imipenem | 0 (0) | 0 (0) | 0 (0) | 40 (56.34) | 0 (0) | 3 (4.23) | 27 (38.03) | 1 (1.41) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Meropenem | 0 (0) | 0 (0) | 0 (0) | 21 (29.58) | 8 (11.27) | 10 (14.08) | 18 (25.35) | 7 (9.86) | 5 (7.04) | 0 (0) | 0 (0) | 0 (0) |
| Ciprofloxacin | 0 (0) | 0 (0) | 0 (0) | 16 (22.54) | 15 (21.13) | 12 (16.90) | 9 (12.68) | 13 (18.31) | 6 (8.45) | 0 (0) | 0 (0) | 0 (0) |
| 37°C | ||||||||||||
| Imipenem | 0 (0) | 0 (0) | 0 (0) | 10 (14.08) | 0 (0) | 2 (2.82) | 54 (76.05) | 1 (1.41) | 1 (1.41) | 3 (4.23) | 0 (0) | 0 (0) |
| Meropenem | 0 (0) | 0 (0) | 0 (0) | 7 (9.85) | 2 (2.2.82) | 3 (4.23) | 34 (47.89) | 10 (14.08) | 10 (14.08) | 0 (0) | 1 (1.41) | 2 (2.82) |
| Ciprofloxacin | 0 (0) | 0 (0) | 0 (0) | 5 (7.04) | 4 (5.63) | 1 (1.41) | 20 (28.17) | 12 (16.90) | 12 (16.90) | 0 (0) | 1 (1.41) | 2 (2.82) |
Conclusions
We have shown the combined effect of incubation temperature, growth media, and flow state of microbial environment, as well as discussed the influence of carbapenems and isolation site on the ability of A. baumannii to form biofilm in vitro. Low incubation temperature significantly optimizes biofilm production in both nutrient-rich and nutrient-poor media without agitation. However, A. baumannii prefers minimal nutrient media to produce high biofilm biomass, although this property appears to be strain-dependent. An increase in adherence at limiting environmental conditions suggests that biofilm formation is an adaptation property in A. baumannii.
Acknowledgments
This study was supported in part by internal discretionary research funds awarded from the University of KwaZulu-Natal to Prof. Dr. Mohamed Ezzzat El Zowalaty. The authors would like to thank the University of KwaZulu-Natal for support through a scholarship awarded to Mr Emmanuel C. Eze. The authors would also like to thank Prof. Manormoney Pillay from School of Laboratory Medicine and Medical Sciences for her support and Dr H. Chenia from the School of Life Sciences for her in-part support to Mr Emmanuel C. Eze through the supply of consumables, reagents, laboratory equipment, and for her initial comments in the study.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Potron A, Poirel L, Nordmann P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents. 2015;45(6):568–585. doi: 10.1016/j.ijantimicag.2015.03.001 [DOI] [PubMed] [Google Scholar]
- 2.Gunn JS, Bakaletz LO, Wozniak DJ. What is on the outside matters: the role of the extracellular polymeric substance of Gram-negative biofilms in evading host immunity and as a target for therapeutic intervention. J Biol Chem. 2016;291(24):12538–12546. doi: 10.1074/jbc.R115.707547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciofu O, Rojo‐Molinero E, Macià MD, Oliver A. Antibiotic treatment of biofilm infections. APMIS. 2017;125(4):304–319. doi: 10.1111/apm.12673 [DOI] [PubMed] [Google Scholar]
- 4.Costerton JW, Cheng K, Geesey GG, et al. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41(1):435–464. doi: 10.1146/annurev.mi.41.100187.002251 [DOI] [PubMed] [Google Scholar]
- 5.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–1322. doi: 10.1126/science.284.5418.1318 [DOI] [PubMed] [Google Scholar]
- 6.Jamal M, Tasneem U, Hussain T, Andleeb S. Bacterial biofilm: its composition, formation and role in human infections. J Microbiol Biotechnol. 2015;4(3):1–15. [Google Scholar]
- 7.Davey ME, O’toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64(4):847–867. doi: 10.1128/mmbr.64.4.847-867.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eze EC, Chenia HY, El Zowalaty ME. Acinetobacter baumannii biofilms: effects of physicochemical factors, virulence, antibiotic resistance determinants, gene regulation, and future antimicrobial treatments. Infect Drug Resist. 2018;11:2277. doi: 10.2147/IDR.S169894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond GP. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol Immunol. 2001;25(4):365–404. [DOI] [PubMed] [Google Scholar]
- 10.Li YH, Tian XL. Quorum sensing and bacterial social interactions in biofilms: In: FJ de Bruijn, editor. Bacterial cooperation and competition. Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria. New Jersey: John Wiley & Sons; 2016:1195–1205. [Google Scholar]
- 11.Irie Y, Parsek MR. Quorum sensing and microbial biofilms In: Romeo T, editor. Bacterial Biofilms. Verlag Berlin: Springer; 2008:67–84. [DOI] [PubMed] [Google Scholar]
- 12.Horswill AR, Stoodley P, Stewart PS, Parsek MR. The effect of the chemical, biological, and physical environment on quorum sensing in structured microbial communities. Anal Bioanal Chem. 2007;387(2):371–380. doi: 10.1007/s00216-006-0720-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mediaswanti K. Influence of physicochemical aspects of substratum nanosurface on bacterial attachment for bone implant applications. J Nanotechnol. 2016;2016. doi: 10.1155/2016/5026184 [DOI] [Google Scholar]
- 14.Regina VR, Lokanathan AR, Modrzyński JJ, Sutherland DS, Meyer RL. Surface physicochemistry and ionic strength affects eDNA’s role in bacterial adhesion to abiotic surfaces. PLoS One. 2014;9(8):e105033. doi: 10.1371/journal.pone.0105033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrett TR, Bhakoo M, Zhang Z. Bacterial adhesion and biofilms on surfaces. Prog Nat Sci. 2008;18(9):1049–1056. doi: 10.1016/j.pnsc.2008.04.001 [DOI] [Google Scholar]
- 16.Rochex A, Godon -J-J, Bernet N, Escudié R. Role of shear stress on composition, diversity and dynamics of biofilm bacterial communities. Water Res. 2008;42(20):4915–4922. doi: 10.1016/j.watres.2008.09.015 [DOI] [PubMed] [Google Scholar]
- 17.Kim J, Kim H-S, Han S, et al. Hydrodynamic effects on bacterial biofilm development in a microfluidic environment. Lab Chip. 2013;13(10):1846–1849. doi: 10.1039/c3lc40802g [DOI] [PubMed] [Google Scholar]
- 18.Francino MP. The ecology of bacterial genes and the survival of the new. Int J Evol Biol. 2012;2012. doi: 10.1155/2012/394026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.López-Maury L, Marguerat S, Bähler J. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat Rev Genet. 2008;9(8):583. doi: 10.1038/nrg2398 [DOI] [PubMed] [Google Scholar]
- 20.Tung J, Gilad Y. Social environmental effects on gene regulation. Cell Mol Life Sci. 2013;70(22):4323–4339. doi: 10.1007/s00018-013-1357-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smoot LM, Smoot JC, Graham MR, et al. Global differential gene expression in response to growth temperature alteration in group A Streptococcus. Proc Natl Acad Sci. 2001;98(18):10416–10421. doi: 10.1073/pnas.191267598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright MS, Iovleva A, Jacobs MR, Bonomo RA, Adams MD. Genome dynamics of multidrug-resistant Acinetobacter baumannii during infection and treatment. Genome Med. 2016;8(1):26. doi: 10.1186/s13073-016-0279-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goh HS, Beatson SA, Totsika M, et al. Molecular analysis of the Acinetobacter baumannii biofilm-associated protein. Appl Environ Microbiol. 2013;79(21):6535–6543. doi: 10.1128/AEM.01402-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basson A, Flemming L, Chenia H. Evaluation of adherence, hydrophobicity, aggregation, and biofilm development of Flavobacterium johnsoniae-like isolates. Microb Ecol. 2008;55(1):1–14. doi: 10.1007/s00248-007-9245-y [DOI] [PubMed] [Google Scholar]
- 25.Moreau-Marquis S, Stanton BA, O’Toole GA. Pseudomonas aeruginosa biofilm formation in the cystic fibrosis airway. Pulm Pharmacol Ther. 2008;21(4):595–599. doi: 10.1016/j.pupt.2007.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu W, Forster T, Mayer O, Curtin JJ, Lehman SM, Donlan RM. Bacteriophage cocktail for the prevention of biofilm formation by Pseudomonas aeruginosa on catheters in an in vitro model system. Antimicrob Agents Chemother. 2010;54(1):397–404. doi: 10.1128/AAC.00669-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antunes L, Visca P, Towner KJ. Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis. 2014;71(3):292–301. doi: 10.1111/2049-632X.12125 [DOI] [PubMed] [Google Scholar]
- 28.Falagas M, Karveli E. The changing global epidemiology of Acinetobacter baumannii infections: a development with major public health implications. Clin Microbiol Infect. 2007;13(2):117–119. doi: 10.1111/j.1469-0691.2006.01596.x [DOI] [PubMed] [Google Scholar]
- 29.Camp C, Tatum OL. A review of Acinetobacter baumannii as a highly successful pathogen in times of war. Lab Med. 2010;41(11):649–657. doi: 10.1309/LM90IJNDDDWRI3RE [DOI] [Google Scholar]
- 30.Charnot-Katsikas A, Dorafshar AH, Aycock JK, David MZ, Weber SG, Frank KM. Two cases of necrotizing fasciitis due to Acinetobacter baumannii. J Clin Microbiol. 2009;47(1):258–263. doi: 10.1128/JCM.01250-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jawad A, Hawkey PM, Heritage J, Snelling AM. Description of Leeds Acinetobacter Medium, a new selective and differential medium for isolation of clinically important Acinetobacter spp., and comparison with Herellea agar and Holton's agar.. J Clin Microbiol. 1994. Oct;32(10):2353–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee HW, Koh Y, Kim J, et al. Capacity of multidrug‐resistant clinical isolates of Acinetobacter baumannii to form biofilm and adhere to epithelial cell surfaces. J Clin Microbiol Infect. 2008;14(1):49–54. doi: 10.1111/j.1469-0691.2007.01842.x [DOI] [PubMed] [Google Scholar]
- 33.Kaiser TDL, Pereira EM, Dos Santos KRN, Maciel ELN, Schuenck RP, Nunes APF. Modification of the Congo red agar method to detect biofilm production by Staphylococcus epidermidis. Diagn Microbiol Infect Dis. 2013;75(3):235–239. doi: 10.1016/j.diagmicrobio.2012.11.014 [DOI] [PubMed] [Google Scholar]
- 34.Patel J, Cockerill F, Bradford P, et al. Clinical and Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically Approved standard M07-A10, Wayne, PA; USA. 2016. [Google Scholar]
- 35.Stepanović S, Vuković D, Dakić I, Savić B, Švabić-Vlahović M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods. 2000;40(2):175–179. [DOI] [PubMed] [Google Scholar]
- 36.Qi L, Li H, Zhang C, et al. Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in Acinetobacter baumannii. Front Microbiol. 2016;7:483. doi: 10.3389/fmicb.2016.00483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stepanović S, Vuković D, Hola V, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by Staphylococci. APMIS. 2007;115(8):891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x [DOI] [PubMed] [Google Scholar]
- 38.Van Houdt R, Aertsen A, Jansen A, Quintana A, Michiels C. Biofilm formation and cell‐to‐cell signalling in Gram‐negative bacteria isolated from a food processing environment. J Appl Microbiol. 2004;96(1):177–184. doi: 10.1046/j.1365-2672.2003.02131.x [DOI] [PubMed] [Google Scholar]
- 39.Kollef MH, Niederman MS. Why is Acinetobacter baumannii a problem for critically ill patients? Intensive Care Med. 2015;41(12):2170–2172. doi: 10.1007/s00134-015-4096-3 [DOI] [PubMed] [Google Scholar]
- 40.Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51(10):3471–3484. doi: 10.1128/AAC.01464-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang C, Qiu S, Wang Y, et al. Higher isolation of NDM-1 producing Acinetobacter baumannii from the sewage of the hospitals in Beijing. PLoS One. 2013;8(6):e64857. doi: 10.1371/journal.pone.0064857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hrenovic J, Goic-Barisic I, Kazazic S, Kovacic A, Ganjto M, Tonkic M. Carbapenem-resistant isolates of Acinetobacter baumannii in a municipal wastewater treatment plant, Croatia, 2014. Euro Surveill. 2016;21(15):30195. doi: 10.2807/1560-7917.ES.2016.21.15.30195 [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, Marrs CF, Simon C, Xi C. Wastewater treatment contributes to selective increase of antibiotic resistance among Acinetobacter spp. Sci Total Environ. 2009;407(12):3702–3706. doi: 10.1016/j.scitotenv.2009.02.013 [DOI] [PubMed] [Google Scholar]
- 44.Kizny Gordon AE, Mathers AJ, Cheong EY, et al. The hospital water environment as a reservoir for carbapenem-resistant organisms causing hospital-acquired infections—a systematic review of the literature. Clin Infect Dis. 2017;64(10):1435–1444. doi: 10.1093/cid/cix132 [DOI] [PubMed] [Google Scholar]
- 45.Nucleo E, Steffanoni L, Fugazza G, et al. Growth in glucose-based medium and exposure to subinhibitory concentrations of imipenem induce biofilm formation in a multidrug-resistant clinical isolate of Acinetobacter baumannii. BMC Microbiol. 2009;9(1):270. doi: 10.1186/1471-2180-9-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kentache T, Abdelkrim AB, Jouenne T, De E, Hardouin J. Global dynamic proteome study of a pellicle-forming Acinetobacter baumannii strain. Mol Cell Proteomics. 2017;16(1):100–112. doi: 10.1074/mcp.M116.061044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomen P, Robert J, Monmeyran A, Bitbol A-F, Douarche C, Henry N. Bacterial biofilm under flow: first a physical struggle to stay, then a matter of breathing. PLoS One. 2017;12(4):e0175197. doi: 10.1371/journal.pone.0175197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pour NK, Dusane DH, Dhakephalkar PK, Zamin FR, Zinjarde SS, Chopade BA. Biofilm formation by Acinetobacter baumannii strains isolated from urinary tract infection and urinary catheters. FEMS Immunol Med Microbiol. 2011;62(3):328–338. doi: 10.1111/j.1574-695X.2011.00818.x [DOI] [PubMed] [Google Scholar]
- 49.Pruthi V, Al-Janabi A, Pereira BJ. Characterization of biofilm formed on intrauterine devices. Indian J Med Microbiol. 2003;21(3):161. [PubMed] [Google Scholar]
- 50.De Silva PM, Chong P, Fernando DM, Westmacott G, Kumar A. Effect of incubation temperature on antibiotic resistance and virulence factors of Acinetobacter baumannii ATCC 17978. Antimicrob Agents Chemother. 2018;62(1):e01514–e01517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lambert G, Bergman A, Zhang Q, Bortz D, Austin R. Physics of biofilms: the initial stages of biofilm formation and dynamics. New J Phys. 2014;16(4):045005. doi: 10.1088/1367-2630/16/4/045005 [DOI] [Google Scholar]
- 52.Hassan A, Usman J, Kaleem F, Omair M, Khalid A, Iqbal M. Evaluation of different detection methods of biofilm formation in the clinical isolates. Braz J Infect Dis. 2011;15(4):305–311. [PubMed] [Google Scholar]
- 53.Kumari AMS, Routray A, Yadav D, Madhavan R. Imipenem resistance and biofilm production in Acinetobacter. Drug Invent Today. 2013;5(3):256–258. doi: 10.1016/j.dit.2013.04.005 [DOI] [Google Scholar]
- 54.Bocanegra-Ibarias P, Pena-López C, Camacho-Ortiz A, et al. Genetic characterisation of drug resistance and clonal dynamics of Acinetobacter baumannii in a hospital setting in Mexico. Int J Antimicrob Agents. 2015;45(3):309–313. doi: 10.1016/j.ijantimicag.2014.10.022 [DOI] [PubMed] [Google Scholar]
- 55.Guo H, Xiang J. Influences of abaR gene on biofilm formation of Acinetobacter baumannii. Zhonghua Shao Shang Za Zhi. 2017;33(4):200–205. doi: 10.3760/cma.j.issn.1009-2587.2017.04.003 [DOI] [PubMed] [Google Scholar]
- 56.Wroblewska MM, Sawicka-Grzelak A, Marchel H, Luczak M, Sivan A. Biofilm production by clinical strains of Acinetobacter baumannii isolated frompatients hospitalized in two tertiary care hospitals. FEMS Immunol Med Microbiol. 2008;53(1):140–144. doi: 10.1111/j.1574-695X.2008.00403.x [DOI] [PubMed] [Google Scholar]
- 57.de Campos PA, Royer S, Da Fonseca Batistao DW, et al. Multidrug resistance related to biofilm formation in Acinetobacter baumannii and Klebsiella pneumoniae clinical strains from different pulsotypes. Curr Microbiol. 2016;72(5):617–627. doi: 10.1007/s00284-016-0996-x [DOI] [PubMed] [Google Scholar]
- 58.Thummeepak R, Kongthai P, Leungtongkam U, Sitthisak S. Distribution of virulence genes involved in biofilm formation in multi-drug resistant Acinetobacter baumannii clinical isolates. Int Microbiol. 2016;19(2):121–129. doi: 10.2436/20.1501.01.270 [DOI] [PubMed] [Google Scholar]
- 59.Abdi-Ali A, Hendiani S, Mohammadi P, Gharavi S. Assessment of biofilm formation and resistance to imipenem and ciprofloxacin among clinical isolates of Acinetobacter baumannii in Tehran. Jundishapur J Microbiol. 2014;7:1. doi: 10.5812/jjm [DOI] [PMC free article] [PubMed] [Google Scholar]