Abstract
Fecal samples from 27 pigs were longitudinally analyzed for Teschovirus A (TV-A), Sapelovirus A (SV-A), and Enterovirus G (EV-G) RNA presence. Suckling piglet fecal samples were negative for the three enteric picornaviruses. However, these picornaviruses were detected in 22/27 weaned pig fecal samples. This study provides new data on TV-A, SV-A, and EV-G infection dynamics.
Electronic supplementary material
The online version of this article (10.1007/s42770-018-0018-1) contains supplementary material, which is available to authorized users.
Keywords: Swine, Picornaviridae, Porcine enterovirus, Fecal shedding, Infection dynamics
Introduction
The Picornaviridae family is divided into 40 genera and includes viruses of importance in human and animal health. The viruses of this family are small, non-enveloped, and have a single-stranded positive-sense RNA (ssRNA) genome [1]. Porcine enteric picornaviruses were previously classified into the Enterovirus genus. Studies on the biological and molecular features of porcine enteroviruses led to their reclassification into Teschovirus A (TV-A), Sapelovirus A (SV-A), and Enterovirus G (EV-G), each belonging to three distinct genera [1, 2].
Porcine enteric picornaviruses have been reported in domestic pigs and wild boars in Asia [3–6], Europe [7–10], North America [11, 12], Central America [13, 14], and South America [15, 16]. Pigs of all ages are susceptible to infection with TV-A, SV-A, and EV-G, which are viruses that circulate worldwide in asymptomatic domestic pigs [10]. However, depending on the virus serotype and infection conditions, these picornaviruses may be considered important etiological agents of enteric, respiratory, reproductive, or neurological disorders in young and adult animals [17].
To date, little information on the epidemiology of TV-A, SV-A, and EV-G infections is available, and there are no longitudinal studies based on the enteric picornavirus infection dynamics and fecal excretion. Considering that suckling and weaned pigs are included in the producing categories with a higher susceptibility to enteric virus infections; pigs belonging to these age groups were the target of this investigation. The aim of this longitudinal study was to evaluate the dynamics of TV-A, SV-A, and EV-G natural infection by the analysis of fecal excretion of these three viruses during two life periods (suckling and weaning) of young pigs.
This study is in agreement with the ethical principles compiled by the Brazilian College of Animal Experimentation (COBEA) and was approved by the Animals Use Ethics Committee (CEUA/protocol number 22/2013) of the Universidade Federal do Paraná–Palotina sector.
Samples included in this study were derived from pig feces collected from a single multi-site pig farm located in Guarapuava City (25° 23′ 43″ S, 51° 27′ 29″ W), the central-southern region of Paraná state, Brazil, and stored at − 80 °C.
The farm consisted of three sites. Site 1 housed breeder, gestating, and farrowing sows, as well as suckling piglets up to 3 weeks of age. During the fourth week of age, piglets were weaned and moved to site 2, where they were housed until they were 8 weeks old. The distance between sites 1 and 2 was of approximately 80 m. Site 3 was a grower-to-finish unit where 9 to 24-week-old animals were raised. Animals were raised with good nutritional and sanitary practices, including “all-in, all-out” management. Reproductive management was done by artificial insemination.
Fecal samples were collected directly from the rectum according to the age of the animals. Solid, pasty, and liquid consistency pig fecal samples (n = 54) from 24 litters that were sampled during the suckling (2 to 3 weeks old, n = 27) and resampled later during the weaning (4 to 6 weeks old, n = 27) period were included in this survey. Piglets were born from 12 non-vaccinated sows and 12 sows vaccinated for porcine Rotavirus A (RVA). Additionally, normal consistency fecal samples from second (n = 1) and fourth (n = 2) parity sows that had farrowed four of the animals sampled in this study also were investigated for the viruses. Fecal samples of these sows were collected at the moment of the parturition.
The nucleic acids were extracted from fecal suspensions using a combination of phenol/chloroform/isoamyl alcohol (25:24:1) and silica/guanidinium isothiocyanate extraction methods [18, 19]. Fecal samples BRA/UEL1/11, BRA/UEL-WB20/13, and BRA/UEL4/13, previously known as positive for TV-A, SV-A, and EV-G [15, 16] and an aliquot of ultrapure autoclaved water were included as positive and negative controls, respectively, during the nucleic acid extraction and the following procedures. The presence of TV-A, SV-A, and EV-G ssRNA in fecal samples was determined using previously described RT-nested PCR assays [16, 17].
Considering the likely co-infection of porcine enteric picornaviruses and rotavirus, all the samples included in this study were screened by RT-PCR assays for porcine RVA VP4 and VP7 genes [20, 21], and the RVB NSP2 [22], RVC VP6 [23], and RVH VP6 genes [24].
Two TV-A, three SV-A, and three EV-G nested PCR amplicons of high quality on agarose gels were selected for sequencing analysis. For the RVA genotype determination, amplicons of G (VP7) and P (VP4) genes were also sequenced. Sequence quality analyses and consensus sequences were assembled using Phred and CAP3 software, respectively (http://asparagin.cenargen.embrapa.br/phph/). Only sequences with base quality ≥ 20 were used. Similarity searches were performed with sequences deposited in GenBank using the Basic Local Alignment Search Tool—BLAST software (http://blast.ncbi.nlm.nih.gov/Blast.cgi). A phylogenetic tree based on nucleotide (nt) was obtained using the maximum likelihood statistical method based on the kimura 2-parameter model (MEGA 7), which provided statistical support via bootstrapping with 1000 replicates. Sequence alignment and identity matrix were performed using BioEdit software version 7.1.11.
The 27 fecal samples from piglets during the suckling period were negative for the three porcine enteric picornaviruses evaluated in this study. Among the 27 feces that were collected from the same animals during the weaning age, five were negative for all of these viruses. Eighteen fecal samples from the weaned pigs were positive for TV-A, 15 for SV-A, and 22 for EV-G. Single EV-G excretion was detected in only three fecal samples. Double (TV-A and EV-G / SV-A and EV-G) and triple (TV-A, SV-A, and EV-G) virus excretions were detected in four, one, and 14 fecal samples, respectively. The results according to the age groups at which animals were sampled and the feces consistency at the moment of collection are presented in Table 1 and Table S1. Fecal samples from the three tested sows were negative for the porcine enteric picornaviruses.
Table 1.
Single and mixed Teschovirus A, Sapelovirus A, and Enterovirus G fecal excretion detected by nested-PCR assays in a longitudinal survey from pigs at suckling and weaned ages
| Porcine enteric picornaviruses | Age group / fecal consistency | TOTAL (n = 54) | |||||
|---|---|---|---|---|---|---|---|
| Suckling (2–3 weeks old) (n = 27) |
Weaned (4–6 weeks old) (n = 27) |
||||||
| Solid | Pasty | Liquid | Solid | Pasty | Liquid | ||
| (n = 15) | (n = 9) | (n = 3) | (n = 4) | (n = 19) | (n = 4) | ||
| TV-A | – | – | – | – | – | – | – |
| SV-A | – | – | – | – | – | – | – |
| EV-G | – | – | – | 1 | 2 | – | 3 |
| TV-A + SV-A | – | – | – | – | – | – | – |
| TV-A + EV-G | – | – | – | – | 3a | 1 | 4 |
| SV-A + EV-G | – | – | – | 1b | – | – | 1 |
| TV-A + SV-A + EV-G | – | – | – | 1 | 11c | 2 | 14 |
| TOTAL | 0 | 0 | 0 | 3 | 16 | 3 | 22 |
TV-A, Teschovirus A; SV-A, Sapelovirus A; EV-G, Enterovirus G; −, negative samples
Rotavirus A RNA genotypes aG9P[23], bG9P[X], cG5P[13] were also identified in one fecal sample each
Porcine RVA was the only RV species detected in the fecal samples evaluated herein. A total of eight fecal samples were positive for RVA, of which two were from suckling piglets (2 weeks old) and six were from weaned pigs. Six and two animals were born from non-vaccinated and vaccinated sows, respectively. RVA-positive fecal samples from suckling piglets were presenting with both solid and pasty consistency; feces of the weaned pigs were of a pasty consistency, except for a unique RVA-positive solid fecal sample from this age group. It was not possible to determine the RVA genotypes in the fecal samples of the suckling piglets and for two of the weaned pigs. The RVA genotypes identified in four of the weaned pig fecal samples were G5P[13], G9P[23], and G9P[X]. RVA G5P[13] was detected in two weaned pig fecal samples, of which one was positive for the three porcine enteric picornaviruses and the other was negative for all of them. RVA G9P[23] was detected in co-infection with TV-A and EV-G, and RVA G9P[X] was detected in co-infection with SV-A and EV-G.
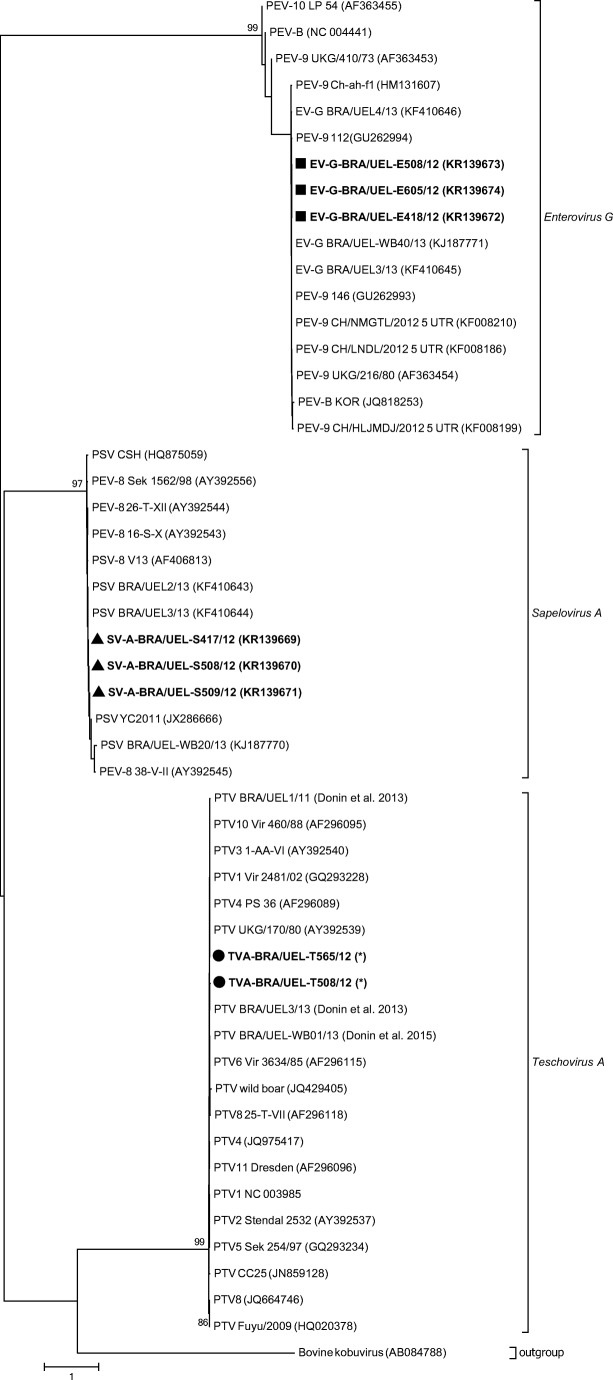
Sequencing analysis confirmed the specificity of TV-A, SV-A, and EV-G amplicons. The TV-A nt sequences described in this study (BRA/UEL-T508/12 and BRA/UEL-T565/12) are shorter than 200 bp and therefore were not submitted to the GenBank (supplementary material). The TV-A sequences herein were 97.4% similar to each other and presented nt similarities varying from 96.1 to 99.3% with TV-A strains of Brazil and other countries. The SV-A nt sequences in this study (BRA/UEL-S417/12, BRA/UEL-S508/12, and BRA/UEL-S509/12; GenBank accession numbers KR139669, KR139670, and KR139671, respectively) were 100% identical to each other and showed 96.1% to 99% similarity with SV-A nt sequences available in the GenBank, including Brazilian strains. Finally, the EV-G nt sequences herein (BRA/UEL-E418/12, BRA/UEL-E508/12, and BRA/UEL-E605/12; GenBank accession numbers KR139672, KR139673, and KR139674, respectively) were 99.6% to 100% identical to each other and had 88.4% to 99.6% nt similarity with other Brazilian and foreign EV-G strains. The phylogenetic tree shows the genetic relationship of the porcine enteric picornavirus strains in this study with the related viruses (Fig. 1).
Fig. 1.
Genetic relationship of Teschovirus A (TV-A), Sapelovirus A (SV-A), and Enterovirus G (EV-G) of Brazilian strains and other published sequences which represent the related viruses. Maximum likelihood phylogenetic tree construction using the Kimura 2-parameter model based on the partial 5′ non-translated region of the enteric picornavirus strains. GenBank accession numbers of representative sequences are TV-A (●BRA/UEL-T508/12 and ●BRA/UEL-T565/12), SV-A (▲BRA/UEL-S417/12, ▲BRA/UEL-S508/12, and ▲BRA/UEL-S509/12), and EV-G (∎BRA/UEL-E418/12, BRA/UEL-E508/12, and ∎BRA/UEL-E605/12) in this study, and other Brazilian strains presented herein are indicated between parentheses. Bootstrap values (1000 replicates) higher than 70% are shown. Asterisk refers to TV-A nt sequences < 200 bp; these TV-A sequences are available as supplementary material
This first longitudinal analysis for porcine enteric picornavirus fecal shedding showed that animals in the suckling age were not excreting TV-A, SV-A, and EV-G in feces, but there was virus shedding detected in the same sampled pigs at weaning age. These are novel data that contribute to the understanding of the infection dynamics of the porcine enteric picornaviruses.
A possible explanation for the negative results from suckling piglets evaluated in this study is that these animals might not be experiencing productive infection at the moment of the fecal collection. It is also likely that they have ingested good quality colostrum, with high titers of specific antibodies against TV-A, SV-A, and EV-G, providing lactogenic immunity and protection for litters against early infection with these viruses. The negative results from all the piglets at suckling age in this study also suggested that the investigated viruses might not be circulating in this site of the herd, indicating that the sanitary conditions of the farm unit might have influenced the porcine enteric picornavirus infection rates in the farrow-to-weaning pig herd site.
Regarding the weaned animals, post-weaning is a critical period in a pig’s life due to nutritional, environmental, and social changes. These facts and the decline in their passive antibodies may lead to poor animal health condition, making pigs more susceptible to infections. Additionally, the mixing of animals of different litters is a common management practice at nursery age [25]. These might be an explanation for the increasing rate of detection of porcine enteric picornavirus infections in animals at this age.
In this study, porcine enteric picornavirus infections were shown to increase with the age of the animals, suggesting that the pig-to-pig route is an important via of TV-A, SV-A, and EV-G transmission. The role of sows as sources of porcine enteric picornavirus infection transmission to piglets was not evaluated in this study. However, normal consistency fecal samples from sows (n = 3) that had farrowed four of the animals sampled in this study were investigated for the three viruses, which were not detected. The TV-A, SV-A, and EV-G infections and excretion from sows during the gestational, farrowing, and lactation periods and of their offspring during the suckling age should be continuously monitored to assess the potential sow-to-piglet transmission of the porcine enteric picornaviruses. Additionally, it is important to evaluate other sources of virus shedding by sows, such as milk and urine.
Studies conducted in Hungary [7], Czech Republic [10], China [26], and Vietnam [27] reported pigs at different ages were all susceptible to TV-A, SV-A, and EV-G infections; however, the prevalence of these viruses varied within the age groups. A previous Brazilian porcine enteric picornavirus-based study reported suckling piglets of distinct Brazilian geographical regions as positive for TV-A and EV-G infections, while SV-A was not detected in this age group [15]. Although in that study suckling piglets were positive for the viruses, porcine enteric picornavirus infections were less frequent in 1- to 3-week-old animals than in pigs of 4 to 8 weeks of age [15].
Studies have reported that TV-A is the most abundant porcine enteric picornavirus in pig feces, followed by SV-A, while EV-G has been reported as the least common enteric picornavirus [8]. The results in this study are in agreement with previous porcine enteric picornavirus-based studies conducted in Brazil that revealed that TV-A was detected most frequently (45%, 18/40), followed by EV-G (40%, 16/40); SV-A was the least (17.5%, 7/40) common virus present in the fecal samples evaluated and was not detected in the feces of suckling piglets [15]. In another Brazilian study in which fecal samples from wild boars were evaluated, the results revealed that EV-G was most frequently (11/22, 50%) detected, followed by TV-A (10/22, 45.5%) and SV-A (4/22, 18.2%) [16].
Studies have reported different clinical manifestations associated with TV-A, SV-A, and EV-G infections, including diarrhea [11, 28]. Although the overall detection rates for the investigated viruses were detected from pasty to liquid consistency fecal samples in this study (19/23, 82.6%), it was not possible to correlate the virus’s presence with diarrhea. Van Dung et al. [27] detected EV-G in fecal samples from animals with and without diarrhea; however, it was not possible to associate the presence of the virus with the occurrence of enteritis because there was no significant difference in the detection rates according to fecal consistency. Additionally, histopathological evaluation should be performed to confirm and classify the enteritis; however, intestinal samples of these animals were not available. In the Brazilian survey, the porcine enteric picornaviruses were more frequently found in diarrheic fecal samples, although it was not possible to determine an association between illness and diarrhea due to the small sample evaluated [15]. Nevertheless, these viruses cannot be excluded as a causative agent of diarrhea. On the other hand, TV-A, SV-A, and EV-G RNA detection in solid fecal samples could indicate that porcine enteric picornaviruses were not always associated with the occurrence of diarrhea in this study. To properly assess the association of the virus’s presence with the occurrence of diarrhea, it is necessary to design a specific investigation, including the evaluation of infections with other porcine enteric viruses, such as rotaviruses, caliciviruses, picobirnavirus, Aichivirus C, Senecavirus A, and Torque teno sus virus, which are endemic in Brazilian pig herds [24, 29–38].
Porcine rotaviruses were also investigated, and only RVA was detected in eight of the fecal samples, which were from suckling (n = 2) and weaned (n = 6) animals born from porcine RVA vaccinated and non-vaccinated sows. Porcine RVA genotype combinations G5P[13] and G9P[23] identified in the fecal samples analyzed in this study have already been reported in pigs from Brazil [34, 35, 39]. Porcine RVB, RVC, and RVH RNA were not detected in the fecal samples evaluated herein. Diarrhea is the result of the combination of several factors, including infectious agents, host immunity, and management procedures [40]. A porcine RVA infection is considered to be one of the most important causative agents of diarrhea in piglets [34]. The non-RVA vaccination history of most of the sows, which delivered these piglets, should predispose their piglets to virus infection, especially during the neonatal period. However, RVA was detected in 2-week-old and 5- to 6-week-old pigs; the normal and pasty fecal consistency in these animals suggests that this infection was not necessarily associated with the occurrence of diarrhea, likely due to the good management practices conducted on the farm, limiting the occurrence of the predisposing factors associated with diarrhea.
To the best of authors’ knowledge, this study represents the first longitudinal survey for porcine enteric picornavirus fecal excretion. The results indicated that TV-A, SV-A, and EV-G early infection was not a common event and that horizontal transmission, primarily from pig-to-pig contact, is important for the maintenance of virus circulation within a pig herd. The TV-A, SV-A, and EV-G infection dynamics require continuous attention in pig herds since intestinal health is directly related to productivity parameters in the pork industry.
Electronic supplementary material
Teschovirus A, Sapelovirus A, Enterovirus G and rotavirus detection according to the age of the animals and sow parturition order. (DOCX 15 kb)
Funding
This study was funded by CNPq (grant number 305062/2015-8). The authors thank the following Brazilian Institutes for financial support: the National Council of Technological and Scientific Development (CNPq), the Brazilian Federal Agency for Support and Evaluation of Graduate Education (CAPES), Financing of Studies and Projects (FINEP), and the Araucaria Foundation (FAP/PR). Alfieri, AA; Alfieri, AF; and Leme, RA are recipients of CNPq fellowships. Lorenzetti, E is recipient of FAP/PR.
Compliance with ethical standards
This study is in agreement with the ethical principles compiled by the Brazilian College of Animal Experimentation (COBEA) and was approved by the Animals Use Ethics Committee (CEUA/protocol number 22/2013) of the Universidade Federal do Paraná–Palotina sector.
Conflict of interest
The authors declared no potential conflicts of interest relative to the research, authorship, and/or publication of this article.
Footnotes
Raquel A. Leme and Danilo R. Silva contributed equally to this work.
References
- 1.ICTV-International Committee on Taxonomy of Viruses, 2017. Virus taxonomy: 2017 release. http://www.ictvonline.org/virusTaxonomy.asp. Accessed 1 June 2018
- 2.Kaku Y, Sarai A, Murakami Y. Genetic reclassification of porcine enteroviruses. J Gen Virol. 2001;82:417–424. doi: 10.1099/0022-1317-82-2-417. [DOI] [PubMed] [Google Scholar]
- 3.Lan D, Ji W, Yang S, Cui L, Yang Z, Yuan C, Hua X. Isolation and characterization of the first Chinese porcine sapelovirus strain. Arch Virol. 2011;156:1567–1574. doi: 10.1007/s00705-011-1035-7. [DOI] [PubMed] [Google Scholar]
- 4.Lin W, Cui S, Zell R. Phylogeny and evolution of porcine teschovirus 8 isolated from pigs in China with reproductive failure. Arch Virol. 2012;157:1387–1391. doi: 10.1007/s00705-012-1315-x. [DOI] [PubMed] [Google Scholar]
- 5.Moon HJ, Song D, Seon BH, Kim HK, Park SJ, An DJ, Kim JM, Kang BK, Park BK. Complete genome analysis of porcine enterovirus B isolated in Korea. J Virol. 2012;86:10250. doi: 10.1128/JVI.01548-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W, Yang S, Shen Q, Ren L, Shan T, Wei J, Cui L, Hua X. Complete genome sequence of a novel porcine enterovirus strain in China. J Virol. 2012;86:7008–7009. doi: 10.1128/JVI.00711-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boros A, Pankovics P, Reuter G. Characterization of a novel porcine enterovirus in domestic pig in Hungary. Infect Genet Evol. 2011;11:1096–1102. doi: 10.1016/j.meegid.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 8.Buitrago D, Cano-Gomez C, Aguero M, Fernandez-Pacheco P, Gomez-Tejedor C, Jimenez-Clavero MA. A survey of porcine picornaviruses and adenoviruses in fecal samples in Spain. J Vet Diagn Investig. 2010;22:763–766. doi: 10.1177/104063871002200519. [DOI] [PubMed] [Google Scholar]
- 9.La Rosa G, Muscillo M, Di Grazia A, Fontana S, Iaconelli M, Tollis M. Validation of rt-PCR assays for molecular characterization of porcine teschoviruses and enteroviruses. J Vet Med B Infect Dis Vet Public Health. 2006;53:257–265. doi: 10.1111/j.1439-0450.2006.00955.x. [DOI] [PubMed] [Google Scholar]
- 10.Prodělalová J. The survey of porcine teschoviruses, sapeloviruses and enteroviruses B infecting domestic pigs and wild boars in the Czech Republic between 2005 and 2011. Infect Genet Evol. 2012;12:1447–1451. doi: 10.1016/j.meegid.2012.04.025. [DOI] [PubMed] [Google Scholar]
- 11.Anbalagan S, Hesse RA, Hause BM. First identification and characterization of porcine enterovirus G in the United States. PLoS One. 2014;9:e97517. doi: 10.1371/journal.pone.0097517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bangari DS, Pogranichniy RM, Gillespie T, Stevenson GW. Genotyping of porcine teschovirus from nervous tissue of pigs with and without polioencephalomyelitis in Indiana. J Vet Diagn Investig. 2010;22:594–597. doi: 10.1177/104063871002200415. [DOI] [PubMed] [Google Scholar]
- 13.Deng MY, Millien M, Jacques-Simon R, Flanagan JK, Bracht AJ, Carrillo C, Barrette RW, Fabian A, Mohamed F, Moran K, Rowland J, Swenson SL, Jenkins-Moore M, Koster L, Thomsen BV, Mayr G, Pyburn D, Morales P, Shaw J, Burrage T, White W, McIntosh MT, Metwally S. Diagnosis of porcine teschovirus encephalomyelitis in the Republic of Haiti. J Vet Diagn Investig. 2012;24:671–678. doi: 10.1177/1040638712445769. [DOI] [PubMed] [Google Scholar]
- 14.Ventura A, Gonzalez W, Barrette R, Swenson S, Bracht A, Rowland J, Fabian A, Moran K, Mohamed F, O'Hearn E, Jenkins-Moore M, Toms D, Shaw J, Morales P, Pyburn D, Carrillo C, Mayr G, McIntosh M, Deng M. Virus and antibody diagnostics for swine samples of the Dominican Republic collected in regions near the border to Haiti. ISRN Virology. 2013;2013:1–7. doi: 10.5402/2013/425831. [DOI] [Google Scholar]
- 15.Donin DG, Leme RA, Alfieri AF, Alberton GC, Alfieri AA. First report of porcine teschovirus (PTV), porcine sapelovirus (PSV) and enterovirus G (EV-G) in pig herds of Brazil. Trop Anim Health Prod. 2014;46:523–528. doi: 10.1007/s11250-013-0523-z. [DOI] [PubMed] [Google Scholar]
- 16.Donin DG, Leme RA, Alfieri AF, Alberton GC, Alfieri AA. Molecular survey of porcine teschovirus, porcine sapelovirus, and enterovirus G in captive wild boars (Sus scrofa scrofa) of Paraná state, Brazil. Pesqui Vet Bras. 2015;35:403–408. doi: 10.1590/S0100-736X2015000500003. [DOI] [Google Scholar]
- 17.Krumbholz A, Wurm R, Scheck O, Birch-Hirschfeld E, Egerer R, Henke A, Wutzler P, Zell R. Detection of porcine teschoviruses and enteroviruses by LightCycler real-time PCR. J Virol Methods. 2003;113:51–63. doi: 10.1016/S0166-0934(03)00227-1. [DOI] [PubMed] [Google Scholar]
- 18.Alfieri AA, Parazzi ME, Takiuchi E, Medici KC, Alfieri AF. Frequency of group A rotavirus in diarrhoeic calves in Brazilian cattle herds, 1998–2002. Trop Anim Health Prod. 2006;38:521–526. doi: 10.1007/s11250-006-4349-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, Das BK, Bhan MK. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992;30:1365–1373. doi: 10.1128/jcm.30.6.1365-1373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B, Fang ZY. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990;28:276–282. doi: 10.1128/jcm.28.2.276-282.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gouvea V, Allen JR, Glass RI, Fang ZY, Bremont M, Cohen J, McCrae MA, Saif LJ, Sinarachatanant P, Caul EO. Detection of group B and C rotaviruses by polymerase chain reaction. J Clin Microbiol. 1991;29:519–523. doi: 10.1128/jcm.29.3.519-523.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alfieri AA, Alfieri AF, Beuttemmüller EA. Porcine rotaviruses: etiology, infection and control topics [in Portuguese] Semin Cienc Agrar. 1999;20:90–97. doi: 10.5433/1679-0359.1999v20n1p90. [DOI] [Google Scholar]
- 24.Molinari BL, Lorenzetti E, Otonel RA, Alfieri AF, Alfieri AA. Species H rotavirus detected in piglets with diarrhea, Brazil, 2012. Emerg Infect Dis. 2014;20:1019–1022. doi: 10.3201/eid2006.130776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kummer R, Gonçalves MAD, Lippke RT, Marques BMFPP, Mores TJ. Factors associated with nursery pig performance. Acta Sci Vet. 2009;37:195–209. [Google Scholar]
- 26.Yang S, Wang Y, Shen Q, Zhang W, Hua X. Prevalence of porcine enterovirus 9 in pigs in middle and eastern China. Virol J. 2013;10:99. doi: 10.1186/1743-422X-10-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Dung N, Anh PH, Van Cuong N, Hoa NT, Carrique-Mas J, Hien VB, Campbell J, Baker S, Farrar J, Woolhouse ME, Bryant JE, Simmonds P. Prevalence, genetic diversity and recombination of species G enteroviruses infecting pigs in Vietnam. J Gen Virol. 2014;95:549–556. doi: 10.1099/vir.0.061978-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, Chen F, Zhou Q, Li W, Song Y, Pan Y, Zhang X, Xue C, Bi Y, Cao Y. Complete genome sequence of a novel porcine Sapelovirus strain YC2011 isolated from piglets with diarrhea. J Virol. 2012;86:10898. doi: 10.1128/JVI.01799-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alfieri AA, Alfieri AF, Freitas JC, Silva CA, Freire RL, Barros AR, Barreiros MAB, Müller EE. Ocurrence of Escherichia coli, Rotavirus, Picobirnavirus, and Cryptosporidium parvum in a postweaning dirrhoea focus in swine. Semin Cienc Agrar. 1994;15:5–7. doi: 10.5433/1679-0359.1994v15n1p5. [DOI] [Google Scholar]
- 30.Barry AF, Alfieri AF, Alfieri AA. Detection and phylogenetic analysis of porcine enteric calicivirus, genetically related to the Cowden strain of sapovirus genogroup III, in Brazilian swine herds. Pesqui Vet Bras. 2008;28:82–86. doi: 10.1590/S0100-736X2008000100013. [DOI] [Google Scholar]
- 31.Leme RA, Alfieri AF, Alfieri AA. Torque teno sus virus (TTSuV) infection at different stages of pig production cycle. Pesqui Vet Bras. 2013;33:840–846. doi: 10.1590/S0100-736X2013000700002. [DOI] [Google Scholar]
- 32.Leme RA, Oliveira TE, Alfieri AF, Headley SA, Alfieri AA. Pathological, immunohistochemical and molecular findings associated with Senecavirus A-induced lesions in neonatal piglets. J Comp Pathol. 2016;155:145–155. doi: 10.1016/j.jcpa.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 33.Leme RA, Zotti E, Alcantara BK, Oliveira MV, Freitas LA, Alfieri AF, Alfieri AA, Senecavirus A. An emerging vesicular infection in Brazilian pig herds. Transbound Emerg Dis. 2015;62:603–611. doi: 10.1111/tbed.12430. [DOI] [PubMed] [Google Scholar]
- 34.Lorenzetti E, Stipp DT, Possatti F, Campanha JET, Alfieri AF, Alfieri AA. Diarrhea outbreaks in suckling piglets due to rotavirus group C single and mixed (rotavirus groups A and B) infections. Pesqui Vet Bras. 2014;34:391–397. doi: 10.1590/S0100-736X2014000500001. [DOI] [Google Scholar]
- 35.Molinari BL, Possatti F, Lorenzetti E, Alfieri AF, Alfieri AA. Unusual outbreak of post-weaning porcine diarrhea caused by single and mixed infections of rotavirus groups A, B, C, and H. Vet Microbiol. 2016;193:125–132. doi: 10.1016/j.vetmic.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 36.Possatti F, Lorenzetti E, Alfieri AF, Alfieri AA. Genetic heterogeneity of the VP6 gene and predominance of G6P[5] genotypes of Brazilian porcine rotavirus C field strains. Arch Virol. 2016;161:1061–1067. doi: 10.1007/s00705-016-2750-x. [DOI] [PubMed] [Google Scholar]
- 37.Ribeiro J, Leme RA, Alfieri AF, Alfieri AA. High frequency of Aichivirus C (porcine kobuvirus) infection in piglets from different geographic regions of Brazil. Trop Anim Health Prod. 2013;45:1757–1762. doi: 10.1007/s11250-013-0428-x. [DOI] [PubMed] [Google Scholar]
- 38.Silva PF, Alfieri AF, Barry AF, Leme RA, Gardinali NR, van der Poel WH, Alfieri AA. High frequency of porcine norovirus infection in finisher units of Brazilian pig-production systems. Trop Anim Health Prod. 2015;47:237–241. doi: 10.1007/s11250-014-0685-3. [DOI] [PubMed] [Google Scholar]
- 39.Silva FD, Espinoza LR, Tonietti PO, Barbosa BR, Gregori F. Whole-genomic analysis of 12 porcine group A rotaviruses isolated from symptomatic piglets in Brazil during the years of 2012–2013. Infect Genet Evol. 2015;32:239–254. doi: 10.1016/j.meegid.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 40.Wittum TE, Dewey CE, Hurd HS, Dargatz DA, Hill GW. Herd and litter-level factors associated with the incidence of diarrhea morbidity and mortality in piglets 1-3 days of age. J Swine Health Prod. 1995;3:99–104. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Teschovirus A, Sapelovirus A, Enterovirus G and rotavirus detection according to the age of the animals and sow parturition order. (DOCX 15 kb)