Abstract
Recalcitrant characteristics and insolubility in water make the disposal of synthetic polymers a great environmental problem to be faced by modern society. Strategies towards the recycling of post-consumer polymers, like poly (ethylene terephthalate, PET) degradation/depolymerization have been studied but still need improvement. To contribute with this purpose, 100 fungal strains from hydrocarbon-associated environments were screened for lipase and esterase activities by plate assays and high-throughput screening (HTS), using short- and long-chain fluorogenic probes. Nine isolates were selected for their outstanding hydrolytic activity, comprising the genera Microsphaeropsis, Mucor, Trichoderma, Westerdykella, and Pycnidiophora. Two strains of Microsphaeropsis arundinis were able to convert 2–3% of PET nanoparticle into terephthalic acid, and when cultured with two kinds of commercial PET bottle fragments, they also promoted weight loss, surface and chemical changes, increased lipase and esterase activities, and led to PET depolymerization with release of terephthalic acid at concentrations above 20.0 ppm and other oligomers over 0.6 ppm. The results corroborate that hydrocarbon-associated areas are important source of microorganisms for application in environmental technologies, and the sources investigated revealed important strains with potential for PET depolymerization.
Electronic supplementary material
The online version of this article (10.1007/s42770-019-00093-3) contains supplementary material, which is available to authorized users.
Keywords: Bioeconomy, Enzymes, Polymer biodegradation
Introduction
Among the vast list of ecological problems caused by industrial activities, pollution by synthetic polymers is one of great concern. Ranging from thermoplastics to synthetic fibers, human-made polymers are found in an extensive variety of highly consumable products. Despite the many advantages and comfort their use brings to human society, they may also lead to the death of sea animals, persistent organic pollutants (POPs) secretion, landfill accumulation, and aesthetical and economical losses. In its 1990 report, the Environmental Protection Agency (EPA-USA) recognized plastic recycling as one of the most promising methods for reducing waste [1].
Synthetic polymers such as poly (ethylene terephthalate), PET, are esterification products originated from petroleum and designed for environmental resistance, with very low rates of (bio) degradation. In 2014, plastic production reached 311 million tons, which is expected to double in 20 years and almost to quadruplicate by around 2050 [2]. Brazil has a high recycling rate of PET, being considered one of the most developed in the world in this sense. According to 2016 PET recycling census, 51% of the 537 thousand tons of polymer used in the country were recycled [3]. In the USA, from about 2.5 million tons consumed in 2016, only 28.4% were collected for recycling [4].
PET fragment hydrolysis is a recycling strategy, although microorganisms can achieve the same results under greener conditions. PET depolymerization is desired because it yields terephthalic or isophthalic acid (and small oligomers, bis(2-hydroxyethyl terephthalate (BHET), and 2-hydroxyethyl methyl terephthalate (MHET)) plus ethylene glycol [5, 6], with lower energy consumption and environmental damage [7, 8]. Searching microbial strains capable of producing active ester hydrolases against PET films is an important step in a worldwide recycling [9–12].
Lipases and esterases represent the most widely used variety of enzymes in biotechnological applications and organic chemistry. Their use as a model for plastic biodegradation and chemical analyses has been reported in many studies [13–16]. In nature, they are present in many catalytic processes and despite also produced by animals and plants, microorganisms are considered the most important source for industrial application [17]. Microbial enzymes are economically very attractive mainly due to their high cost-benefit ratio [18].
Bacteria and yeasts are reported as important lipase-producers, such as Bacillus [19, 20], Pseudomonas [21], Burkholderia [22], and Candida [23]. However, its outstanding ability of adaptation to adverse conditions makes the filamentous fungi ideal agents for biodegradation studies. Firstly, mycelial growth confers an advantage over unicellular cells, especially regarding the colonization of insoluble substrates. Fungal hyphae branch rapidly into the substrate, digesting it through the extracellular enzymes and allowing bacterial attack access. Secondly, they are able to grow under stressing conditions such as low pH values and scarce nutrient availability [24].
Up to date, some of the most recognized fungal genera reported as lipase and esterase producers are Trichoderma [25]; Aspergillus [26]; Penicillium [27]; Alternaria [28], and Fusarium [29]. They were reported in soil samples collected from different regions [25, 30, 31], plant species of Cerrado and Atlantic forest [32]; pig manure [33]; crude fragment of a chicken slaughterhouse [34], and oil-contaminated soil [35]. Investigations suggest that oil contaminated areas may be of scientific value in obtaining strains for application in environmental biotechnology, such as Aspergillus sp. [36]; Trichoderma and Penicillium [37]; Rhodococcus cercidiphylli [38], and Bacillus subtilis [39].
The activity of lipases and esterases can be evaluated by qualitative and/or quantitative methods. Qualitative assays use color-changing when the esterified substrate is transformed into a short-chain fatty acid. Phenol red is one of the most indicated dye and triglyceride tributyrin a widely used substrate [40, 41]. For esterase plate assay, α-naphthyl acetate is enzymatically hydrolyzed to α-naphthol, which couples to a diazonium salt (Fast Blue RR salt), forming a brown diazo dye complex [42, 43]. Conventional quantitative tests such as chromatography, spectrophotometry, and tensiometry are time-consuming. High-throughput screening (HTS) methods are fast and can be accomplished by fluorescence analysis using synthetic chromogenic substrates (e.g., p-nitrophenyl, resorufin, umbelliferyl esters) [44]. The measurement of released chromophore after enzymatic reaction is a tool that allows the selection of useful organisms, in a wide range of biotechnological applications.
This research is part of a PET recycling program aiming the production of terephthalic acid and ethylene glycol by fungal strains isolated from different substrates. The isolation sources were considered to identify which one was able to reveal the most promising strains. Plate assays and HTS technique were employed for lipase and esterase pre-screening. The production of terephthalic acid by selected strains was evaluated on PET nanoparticles; thereafter, microbial performance on real condition (PET fragments of commercial bottles) was measured through PET loss weight, scanning electron microscopy (SEM) and Fourier-transform infrared spectroscopy (FTIR) analysis, and lypolitic/esterolytic activities after the fermentation process.
Material and methods
Microorganisms
One hundred fungal strains from different sources (Table 1) were screened. They were preserved by ultra-freezing method (− 80 °C) at the Laboratory of Environmental Microbiology (LMA), located at the Center of Environmental Studies, CEA, São Paulo State University (Unesp), Rio Claro, SP, Brazil. The best performing strains were also deposited at the Brazilian Collection of Environmental and Industrial Microorganisms, CBMAI/CPQBA/UNICAMP, Paulínia, SP, Brazil.
Table 1.
Source, locality of isolation, and fungal taxa screened for lipase and esterase activities
| Source of isolation | Locality | Number of isolates/source | Taxa represented (number of isolates analyzed) |
|---|---|---|---|
| Fresh water | 22° 41′ 48″ S; 47° 08′ 59″ W | 41 | Paecilomyces (5), Cladosporium (1), Penicillium (6), Curvularia (2), Trichoderma (9), Cylindrocladium (1), Aspergillus (2), Pycnidiophora (2), Phoma (1), Mucor (2), Pleosporales (1), Microsphaeropsis (4), Cochliobolus (1), Westerdykella (3), non-sporulating fungus (1) |
| Treated waste water | 22° 44′ 26″ S; 47° 07′ 33″ W | 10 | Westerdykella (1), Penicillium (2), Pseudallescheria (1), Curvularia (1), Trichoderma (4), Pleosporales (1) |
| Ant cuticle | 22° 23′ 58″ S; 47° 32′ 38″ W | 8 | Penicillium (2), Mucor (2), Fusarium (4) |
| Fresh water | 22° 41′ 28″ S; 47° 07′ 22″ W | 7 | Penicillium (1), Paecilomyces (1), Aspergillus (2), Phoma (1), Trichoderma (2) |
| Fresh water | 22° 44′ 25″ S; 47° 07′ 33″ W | 7 | Penicillium (3), Trichoderma (4) |
| Soil (citrus plantation) | 22° 08′ 49.4″ S; 47° 10′ 47.6″ W | 8 | Aspergillus (2), Fusarium (5), Penicillium (1) |
| Sewage sludge | 22° 44′ 02″ S; 47° 07′ 03″ W | 6 | Penicillium (2), Trichoderma (1), Fusarium (1), Pycnidiophora (1), Phoma (1) |
| Treated effluent | 22° 41′ 28″ S; 47° 07′ 22″ W | 5 | Aspergillus (2), Trichoderma (1), Coletotrichum (1), Pleosporales (1) |
| Fresh water | 22° 44′ 23″ S; 47° 07′ 40″ W | 5 | Penicillium (1), Trichoderma (2), Pycnidiophora (1), Paraconiothyrium (1) |
| Clean PET Fragment | 22° 23′ 58″ S; 47° 32′ 38″ W | 2 | Cladosporium (1), Penicillium (1) |
| Air | 23° 51′ 62,4″ S; 46° 62′ 40,1″ W | 1 | Penicillium (1) |
Agar plate methods for enzymatic activities
Lipase
A 3 mm inoculum from a 7-day old colony was transferred to a selective medium with the following composition per liter: phenol red (0.01%) with 1% lipidic substrate tributyrin, 10 mmol.L−1 of CaCl2 and 2% agar; pH was adjusted to 7.3–7.4, using 0.1 N NaOH. Fungal colonies that produced yellow halo after 7 days at 28 °C were considered lipase positive [45].
Esterase
Fungal colonies maintained for 7 days at 28 °C in Potato Dextrose Agar (PDA, 20 g L−1, Sigma-Aldrich) were used to evaluate esterase activity. To reveal the esterase activity of isolates, 20 mL of 0.4% HEPES/NaOH buffered agarose at 50–60 °C with 320 μL of Fast Blue RR (80 mg.mL−1 in dimethysulfoxide) and 320 μL α-naphthyl acetate (20 mg.mL−1 in acetone) were added onto the plate. Colonies with positive results showed brown staining precipitates in a period of up to 15 min [42].
Once the enzymatic activity plate assays are qualitative, they were performed in duplicate. However, discrepant results were repeated to ensure the data reliability.
High-throughput screening fluorescence assay
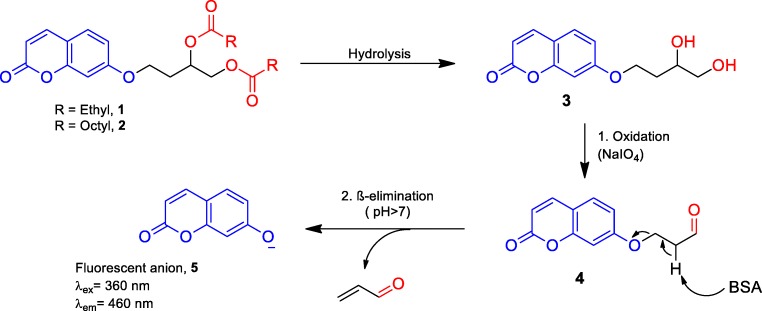
Fungal taxa evaluated by HTS were cultivated for 7 days at 28 °C in PDA. Fungal suspensions (1 mg.mL−1) were prepared in borate buffer (pH 7.4). The assays were performed on 96-well flat bottom microtiter plates, as proposed by Mantovani, Oliveira, and Marasaioli [46]; for esterase activity, Probe 1-short-chain ester, namely, 4-[(2-oxo-2H-1-benzopyran-7-yl)oxy]butane-1,2-diylacetate, was used, and probe 2-long-chain ester, 4-[(2-oxo-2H-1-benzopyran-7-yl)oxy]butane-1,2-diyldioctanoate, for lipase activity (Fig. 1).
Fig. 1.
Chain reaction for the fluorescence-based HTS assays with probes 1 and 2
The assays were composed of microbial analysis (to measure the hydrolytic activity), positive control (using the probe product after reaction 4-[(2-oxo-2H-1-benzopyran-7-yl)oxy]butane-1,2-diol), and negative control (containing only buffer and fluorogenic reagent).
Fluorescence analyses were performed at 0, 24, 48, and 96 h in a 2300 EnSpireTM Multimodal Reader (PerkinElmer), with excitation filter at λex = 360 nm and emission filter at λem = 460 nm at 25 °C. At the end of the reaction, the conversion was calculated according to Eq. 1.
The enzymatic hydrolysis process of fluorogenic probes 1 (short-chain ester) and 2 (long-chain ester) is shown in Fig. 1.
Fluorescence analysis for PET nanoparticle biodegradation
Fungi with promising conversion results obtained from HTS assays were evaluated regarding the potential of PET nanoparticles biodegradation based on a methodology developed by Chaves et al. [47]. Briefly, fungal cell suspensions (100 μL, 1.0 mg.mL−1 in borate buffer (pH 7.8), resulted from colonies with 7 day-growth were incubated with a solution of PET nanoparticles (30 μL, 0.11 mg.mL−1) for 15 days at 30 °C and at 200 rpm. The terephthalic acid formed after PET hydrolysis was converted to the fluorescent 2-hydroxyterephthalate (HOTP) by adding (in this sequence) H2O2 (30 μL, 2% v/v), EDTA (20 μL, 3 mmol.L−1), and FeSO4.7H2O (20 μL, 3 mmol.L−1) and incubating for 25 min at room temperature. The fluorescence data were analyzed in a 2300 EnSpire Multimodal Reader (Perkin Elmer), with excitation wavelength λex = 328 nm and emission wavelength λem = 421 nm.
The study of each isolate was delimited per line in the 96-well plate. In order to allow comparison between the types of assay, microbial assays were performed in triplicate, positive and negative controls; duplicate and microbial control were executed only once. Thus, the positive control determined the maximum fluorescence intensity for the assay, the negative control evaluated the spontaneous hydrolysis of the PET polymer, and the microbial control provided the microorganism background fluorescence. The conversions were obtained according to Eq. 2.
PET depolymerization test (adapted from Sharon, Sharon [10])
PET bottles of Crystal® mineral water (500 mL) and Sprite® soft drink diet (2000 mL) were denomited as PET 1 and PET 2, respectively. They were commercially obtained and pretreated, besides undergoing labels and caps removal. Fragments of approximately 4.5 × 2.5 cm (500 mg) were cut with scissors and washed with tap water and neutral detergent, and rinsed in sterilized distilled water. The pieces were immersed in 2% hypochlorite for 1 h, rinsed in sterilized distilled water, and then kept for 30 min under ultraviolet light in a laminar flow closet (lamp output of 492 W at a distance of 50 cm). The fragments were then oven dried at 50 °C until water evaporation.
For evaluation of PET depolymerization potential, flasks with 40 mL of potato dextrose broth (PDB) containing three 0.5 cm2 PDA fragments (of 7-day-old fungal colonies, incubated at 28 °C) and two PET fragments (previously weighed in analytical scale) were used. The flasks were incubated on a rotary bench shaker Tecnal® model TE-420 at 100 rpm for 14 days at 28 °C. Cultures were centrifuged at 4472g in sterilized 50 mL-Falcon tubes and filtered through a 0.45 μm membrane to separate mycelium and PET fragments from the fermented broth.
PET fragments were subjected to a new disinfection process as previously mentioned, until no mycelial structure was observed. Control reactions (without inoculum) were carried out based on the same conditions used in each experiment. The percentage of weight loss of the PET fragments and SEM images were evaluated, as well as the lipolytic activity of the fermented broth.
PET fragments Fourier-transformed infrared
The fragments spectra were read in a Fourier-transform infrared spectrophotometer, Shimadzu, model FTIR-8300 in the range of 4000–400 cm−1 using 32 accumulations and 2 cm−1 resolutions.
Scanning electron microscopy (adapted from Sepperumal, Markandan, Paljara [48])
Scanning electron microscopy analyses were performed using a scanning electron microscope Hitachi model TM 3000. The medium accelerating voltage was 15 Kv (analy mode 1-30 Kv). The surface of the PET fragment was covered with gold by using the Balzers SCD 050 apparatus at a 25 mA for 40 s under 80 Pa pressure.
Fermented broth analysis
Total protein assays
Protein concentrations were measured with Bradford method [49]. Bovine serum albumin (BSA), protein standard, was solubilized in distilled water to the concentration of 1 mg.mL−1 and, this solution was used for preparation of a calibration curve, which aims to 3.33–33.3 μg.mL−1.
Evaluation of lipase activity
The lipolytic activity was evaluated by titrimetry using standard NaOH (0.05 mol.L−1) according to Burkert, Maugeri, and Rodrigues [50]. These assays were performed in triplicate and the lipolytic activity expressed in unit of lipolytic activity (UA), corresponding to the amount of enzyme capable of releasing 1 μmol of fatty acids per minute of reaction.
Evaluation of esterase activity
The activity was quantified using the potential enzymatic hydrolysis of 1.5 mMol.L−1 α-naphthyl acetate (in phosphate buffer 300 mM containing isopropyl alcohol) to α-naphtol, coupling to diazonium salt Fast Blue RR (0.1 mg.mL−1 in dimetilsulfoxide and ethyl alcohol), forming a diazo complex. The quantification was performed against α-naphtol calibration curve in concentration between 10 to 250 μmol.L−1 [51].
Terephthalic acid and other oligomers quantification by high-performance liquid chromatography with UV detector (HPLC-UV) (adapted from De Castro et al. [7])
The terephthalic acid (PTA), bis-hydroxyethyl terephthalate (BHET), and methyl hydroxyethyl terephthalate (MHET) samples contents were partitioned liquid-liquid extraction using 2 mL of fermented broth with ammonium sulfate (0.5 g) and hexane; ethyl acetate and dichloromethane (5:4:1), 3 mL (two times) after agitation for 40 s. To the end, the organic phase was separated and evaporated in a nitrogen stream at 40 °C and the residues were resuspended in 100 μL of 0.5% formic acid by agitation in the vortex for 1 min. Three microliters were injected into the equipment.
HPLC separations were performed by a Shim-pack VP-ODS C18 column (4.6 mm, 250 mm, 5 μm) with C18 pre-column (Shimadzu, USA). The mobile phase consisted of solvent A (formic acid 0.5% (v/v), acetonitrile (90:10)) and solvent B (formic acid 0.5% (v/v), acetonitrile (60:40)) with a flow rate of 0.5 mL min−1. The separation was accomplished by gradient elution, as shown in Table S1 in supplementary material. Retention times were 10.450 (PTA); 12.505 (BHET); and 38.0 min (MHET). Benzoic acid was used as internal standard. The analytes were detected at 254 nm. The method range of linearity was 3 to 300 ppm for PTA and, 0.15 to 30 ppm for BHET and MHET. The correlation coefficient was higher than 0.99.
Results
Qualitative and quantitative assays for lipase and esterase activity
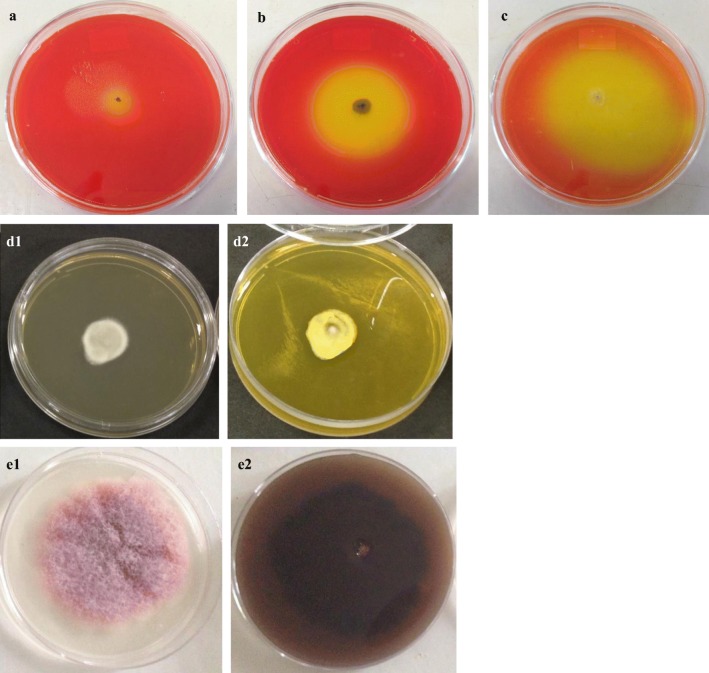
Plate assays revealed different responses concerning the ability to produce and secrete lipases: 19% of the strains showed low ability while, 35% were intermediate and, 39% high. Fifty-one strains were esterase positive based on the capacity to convert alpha-naphtyl acetate to alpha-naphtol. Combining both results, 21 isolates were considered the most promising ones to produce both enzymes. Figure 2 illustrates the evaluation criteria.
Fig. 2.
Plate assay for lipase and esterase activity. a, b, and c. strain with low (Aspergillus sp. LMA 904), intermediate (Curvularia sp. LMA 1206), and high (Penicillium sp. LMA 1285) production of lipase, respectively. Culture medium with Tributiryn as sole source of carbon. d and e. Plate reactions after revelation for esterase activity with α-naphthyl acetate and fast blue RR. d1–d2: negative result: 7-day-old Microascaceae representative, LMA 192; e1–e2: positive result: 7-day-old Fusarium sp. LMA 1144
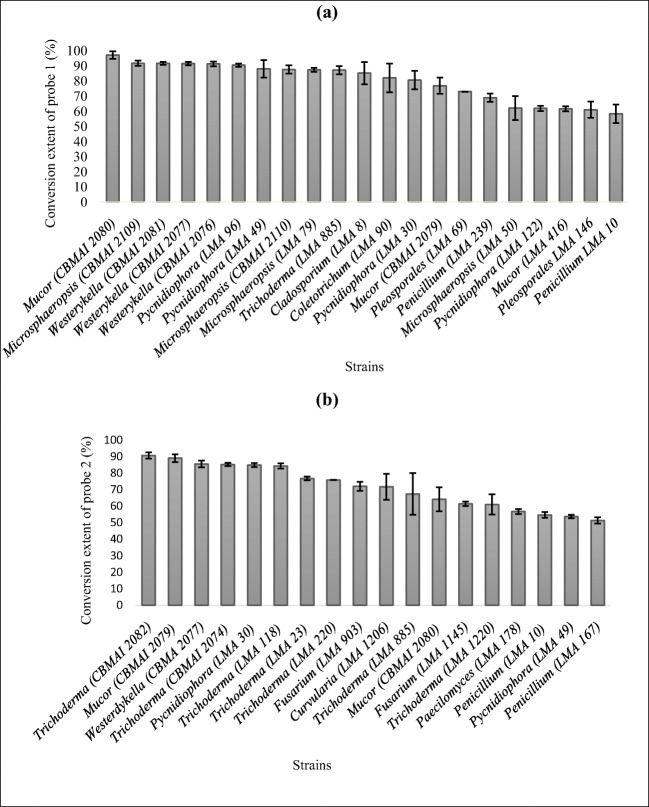
Twenty-one strains presented conversion extent of the short-chain probe [1] and 18 converted the long-chain probe [2] into umbelliferone, both at levels higher than 50% (Fig. 3a, b). The taxa belong to Mucor, Microsphaeropsis, Westerdykella, Pycnidiophora, Trichoderma, Cladosporium, Fusarium, Curvularia, and Paecilomyces. The results in Fig. 3 were obtained after 96 h. Tables S2 and S3 (supporting information) show data from other periods of analysis.
Fig. 3.
Conversion extent of probes by the most promising fungal strains: a. Probe 1 b. Probe 2. Data expressed as average ± standard deviation (n = 4). Results for conversions after 96 h
All promising strains recovered in this study came from hydrocarbon-associated substrates. M. arundinis CBMAI 2109 and W. dispersa (CBMAI 2076, CBMAI 2077, and CBMAI 2081) came from water samples under the influence of an oil refinery. Mucor fragilis CBMAI 2080 revealed higher activity against short-chain probes and converted 64.1% ± 7.28 of the long-chain probe. This strain was obtained from leafcutter ant body, also a well-known source of hydrocarbons (Table 2).
Table 2.
Conversion extent of short-chain ester probe of selected isolates
| Code | Identification/source | Plate assay* | Conversion of probe 1 (n = 4, % media ± standard deviation, SD) | |||
|---|---|---|---|---|---|---|
| 0 h | 24 h | 48 h | 96 h | |||
| CBMAI 2080 | Mucor fragilis/ant cuticle | + | 0.00 ± 0.00 | 54.09 ± 1.36 | 81.83 ± 1.07 | 97.00 ± 1.12 |
| CBMAI 2109 | Microsphaeropsis arundinis/fresh water | + | 92.64 ± 0.35 | 91.08 ± 1,42 | 86.38 ± 0.88 | 91.61 ± 0.92 |
| CBMAI 2081 | Westerdykella dispersa/fresh water | + | 93.78 ± 0.57 | 91.52 ± 1.49 | 84.59 ± 0.84 | 91.50 ± 1.46 |
| CBMAI 2077 | Westerdykella dispersa/fresh water | + | 81.52 ± 10.49 | 91.80 ± 1.62 | 84.05 ± 1.55 | 91.37 ± 2.06 |
| CBMAI 2076 | Westerdykella dispersa/fresh water | + | 67.87 ± 7.85 | 86.67 ± 2.73 | 84.45 ± 0.84 | 91.17 ± 0.89 |
*Esterase activity revealed by alpha naphthyl acetate (20 mg mL−1 in acetone) and fast blue RR (80 mg mL−1 in dimethysulfoxide). See in supplementary material the positive results of the strains
Similar results were observed in the octanoate ester probe (Table 3) by T. longibrachiatum CBMAI 2082, W. dispersa CBMAI 2077, P. dispersa LMA 30, and T. asperellum CBMAI 2074 an isolated from water samples collected next to an oil refinary. M. ramosissimus CBMAI 2079 was found on the cuticle of leafcutter ants.
Table 3.
Conversion extent of long-chain ester probe of selected isolates
| Code | Identification/source | Plate assay (tributiryn)* | Conversion of probe 2 (n = 4, % media ± standard deviation, SD) | |||
|---|---|---|---|---|---|---|
| 0 h | 24 h | 48 h | 96 h | |||
| CBMAI 2082 | Trichoderma longibrachiatum/fresh water | +++ | 1.63 ± 0.35 | 89.97 ± 0.50 | 89.94 ± 2.51 | 90.71 ± 1.87 |
| CBMAI 2079 | Mucor ramosissimus/ant cuticle | +++ | 0.00 ± 0.00 | 73.58 ± 5.09 | 86.09 ± 2.86 | 89.08 ± 2.43 |
| CBMAI 2077 | Westerdykella dispersa/fresh water | ++ | 83.58 ± 2.02 | 86.01 ± 1.80 | 85.56 ± 2.34 | 83.03 ± 1.63 |
| CBMAI 2074 | Trichoderma asperellum/fresh water | +++ | 0.88 ± 0.05 | 85.69 ± 1.26 | 85.67 ± 1.48 | 85.27 ± 1.03 |
| LMA 30 | Pycnidiophora dispersa/fresh water | +++ | 82.16 ± 1.16 | 86.07 ± 0.79 | 88.84 ± 1.19 | 82.59 ± 0.96 |
*(+++) fungi that changed completely the selective medium color (from pink to yellow), (++) fungi that partially altered color, classified as intermediate producers. Tributyrin as sole source of carbon. Fig. S2 containing the results of plate assays for the related strains in this table (see in supplementary material)
Analyzing the data of plate assays versus fluorescent probes, T. longibrachiatum CBMAI 2082 had positive results in all tests, with a higher activity for probe 2 than for probe 1 (90.71% ± 1.87 and 40.88% ± 0.58, respectively). Supporting material shows results for esterase and lipase plate assays of the above-cited strains, Figs. S1 and S2.
Fungal activity on PET nanoparticles and commercial bottle fragments
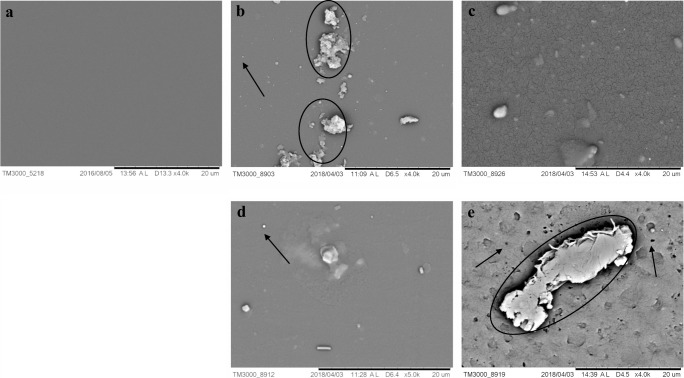
Our screening of filamentous fungi revealed nine strains with high hydrolytic activity, which were selected for subsequent tests. From these, only M. arundinis CBMAI 2109 was able to convert 2.0% ± 0.4 of the PET nanoparticle to the terephthalic acid (monomer), as confirmed by the calculations from Eq. 2, which compares the fluorescence of the positive control (total fluorescence of HOTP) with that of the assay [52]. This strain also converted 43.3% ± 2.7 of long-chain ester probe in umbelliferone. SEM analysis endorsed the action of this isolate on PET (Fig. 4b, c and Fig. 5b, c).
Fig. 4.
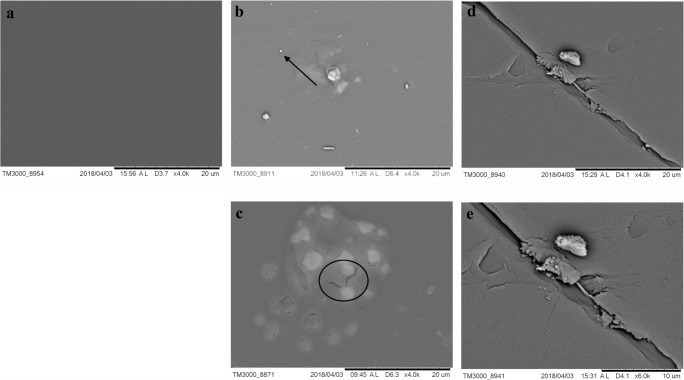
SEM images of PET fragments of commercial bottle PET 1. a. Intact PET fragment after 14 days in PDB (abiotic control). b and c. PET fragment after 14 days of fermentation with M. arundinis CBMAI 2109. In d, PET surface with cracking. d and e. PET fragment after 14 days of fermentation with M. arundinis CBMAI 2110 (PDB, 28 °C, 150 rpm). Scale bar 20 μm. Arrows indicate micropitting, Circles indicate mycelial structure adhered even after cleaning procedure
Fig. 5.
SEM images of PET fragments of commercial bottle PET 2 after fermentation. a. Intact PET fragment after 14 days in PDB (abiotic control). b and c. PET fragment after 14 days of fermentation with M. arundinis CBMAI 2109. d and e. PET fragment after 14 days of fermentation with M. arundinis CBMAI 2110. e. Shows a zoomed image indicating strong adherence of fungal mycelia after cleaning and PET surface with cracking (PDB, 28 °C, 150 rpm). Scale bar 20 μm. Arrows indicate pittings, circle highlights microcracking
Another isolate of M. arundinis (CBMAI 2110) obtained from the same region (fresh water, 22° 41′ 48″ S; 47° 08′ 59″ W) presented high conversion for probe 1 (84.0 ± 0.88%) and low conversion rate for probe 2 (19.3 ± 0.5%) after 96 h. The result of the PET nanoparticle assay showed 2.7 ± 0.9% of terephthalic acid produced from the polymer. SEM images from PET fragments after the fermentation test CBMAI 2110 showed pieces with evidence for significant results (Fig. 4d, e and Fig. 5d, e.
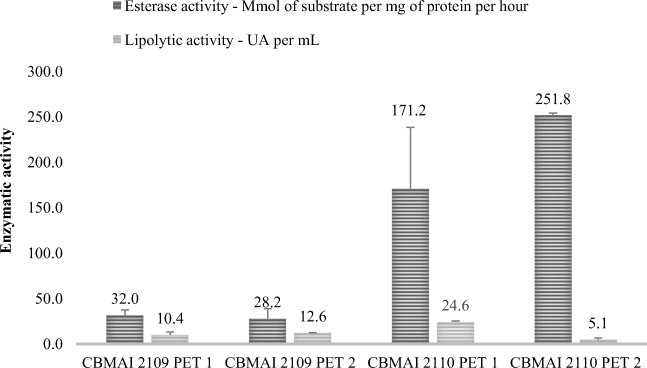
Both strains of M. arundinis, CBMAI 2109 and CBMAI 2110, showed promising results in the assays with fragments of different commercial PET bottles (Fig. 6). The lipase and esterase activities showed a moderated positive correlation; when there was an increase in lipolytic activity, the same was observed for esterase activity (r = + 0.66) in all analyzed samples (PET 1 and PET 2). In addition to the production capacity of hydrolytic enzymes, these isolates promoted the depolymerization of polyethylene terephthalate and the weight loss of commercial fragments of PET bottles after 14 days (Fig. 7).
Fig. 6.
Lipase and esterase activity after 14 days of fermentation in PDB with isolates M. arundinis (CBMAI 2109 and CBMAI 2110). Study on PET from different commercial bottles. Data for lipase activity were expressed in units of activity (UA) and for esterase are expressed in mmol of α-naphthol/mg of protein. h−1 (average ± standard deviation, n = 3)
Fig. 7.
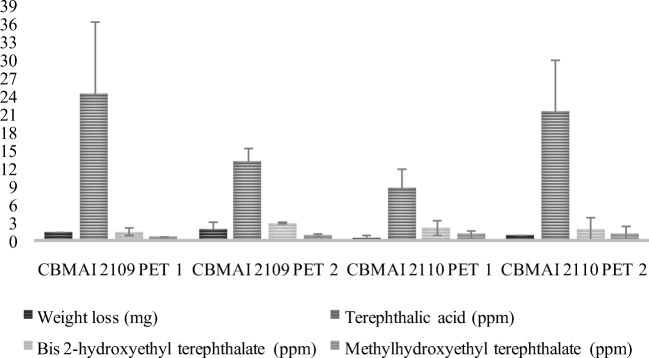
Weight loss of PET fragments and terephthalic acid, bis-2 hydroxyethyl terephthalate (BHET), and methylhydroxyethyl terephthalate (MHET) content in PDB after 14 days of fermentation with the isolates M. arundinis (CBMAI 2109 and CBMAI 2110). Study on PET from different bottles. Data were expressed mg.L−1 or ppm (media ± standard deviation, n = 3). For quantification was used relative area (analyte area/benzoic acid ratio—IS area)
The terephthalic acid monomer (PTA), as well as the smaller oligomers, BHET and MHET, was quantified in concentrations above 0.6 ppm for all PET by-products, in the fermented broth. M. arundinis CBMAI 2109 showed higher concentrations of terephthalic acid in PET 1 (24.4 ± 12.1 ppm) and CBMAI 2110 in PET 2 (21.4 ± 8.6 ppm) Fig. 7.
Fig. 8.
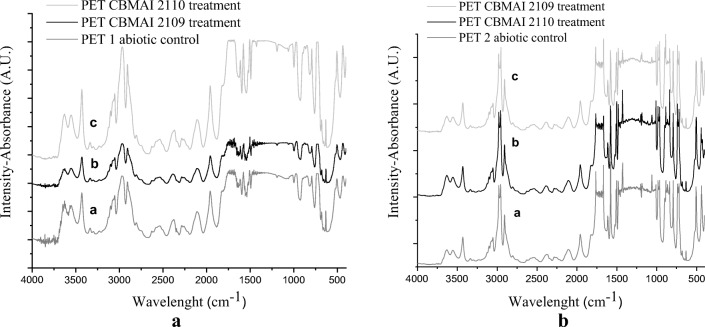
FTIR graphs of PET 1 (A) and PET 2 (B) after 14 days of fermentation with the isolates M. arundinis (CBMAI 2109 and CBMAI 2110)
Figure 8 A and B compare the infrared spectra of PET 1 and PET 2 samples, respectively, relating to the treated (CBMAI 2109 and CBMAI 2110) and untreated (abiotic control) fragments. The carbonyl indices showed no significant difference between the treatments; however, in the microbiologically treated materials, differences in absorption intensity were observed at specific points in the spectrum, as well as the appearance or disappearance of absorption bands, indicating possible molecular modification in the evaluated region
Infrared spectrum sites with significant changes were found in the regions of 3080 to 3050 cm−1, relative to the symmetrical stretching of −CH; from 2900 to 2800 cm−1, due to symmetrical C–H stretching; 1575 cm−1 for vibrations with draw C = C of aromatic ring. The different intensities of bands 1960 and 794 cm−1 indicate vibrations of two hydrogens on para-substituted aromatic rings.
Discussion
Substrates × fungal activity
Among the 100 isolates studied (Table 1), 34% belong to Eurotiales, mainly Penicillium and Aspergillus, 33% Hypocreales (Trichoderma and Fusarium), and 28% Pleosporales and Mucorales, (5%) minor group comprising Glomerellales, Microascales, Capnodiales, and non-esporulating fungus. The main groups are reported as stress-tolerants and able to grow under conditions of water and nutrient limitations, high temperature, and low oxygen availability [53]. Eurotiales and Hypocreales have been under extensive research in the evaluation of extracellular enzymatic profile, because they usually present expressive results of enzyme production as lipase and esterase, for example [54].
The literature reports that the substrate may induce microbial gene expression [52, 55]. Gargouri et al. [56] concluded that the presence of hydrocarbons stimulates microorganisms to increase cell surface hydrophobicity. Other investigations also observed high hydrolytic activities (lipase/esterase) in strains isolated from hydrocarbon-associated environments [57]. This hypothesis is based on the theory of directed evolution when the substrate of contact to which a strain or a wild enzyme is exposed, leading to genotypic alterations that allow the assimilation of previously unassimilated molecules, due to the increase in the enzymatic activities carried out by modifications in certain proteins [47]. Thereby, Crecy et al. [58] obtained strains of Metarhizium anisopliae with a recognized thermotolerance increment, desired for the application of the strain as a biocontrol agent.
Based on our results, it is suitable to say that substrates where hydrocarbons are present tend to be adequate for PET depolymeration researches. In this study, fungal strains of higher interest (Tables 2 and 3) were obtained from water samples under the influence of an oil refinery and leafcutter ant body. The presence of hydrocarbons on ant cuticles is well known. They provide an important ant-to-ant communication signal [59], besides working as a protective barrier against dehydration of the cuticle [60]. Cervantes-Gonzalez et al. [61] verified a positive influence of aromatic, heteroaromatic, and alkane hydrocarbons regarding the lipase activity of Pseudomonas sp. obtained from oil-polluted soil. Kanwar and Goswani [62] showed that lipases from P. pseudomallei were produced using pure n-hexadecane, as the sole source carbon.
Qualitative and quantitative assays for lipase and esterase activity
Several authors report the use of different substrates to distinguish lipases from esterases. Substrates commonly used for identification or isolation of lipase-producing microorganisms were tween 80 and phenol red [63], tributyrin [64, 65], olive oil/tributyrin/phenol red [66], and trypropionin, tributyrin vinyl laurate, tryoctanoin, and olive oil [67]. Studies to detect esterase usually recommend alpha naphthyl acetate and short-chain vinyl esters (acetate and propionate) [67].
Consistent with scientific information, this investigation employed tributyrin and phenol red for lipase production and alpha-naphthyl acetate for esterase. Also, a large number of publications recommend these methods because they allow a better visualization and interpretation of the results. The present study corroborates with this observation and states that the assays were very satisfactory enabling the identification of 18 “true lipases” which acted on the lipophilic substrate, while they were not active against alpha naphthyl acetate (more hydrophilic substrate).
The enzymatic activities of the selected strains were confirmed by HTS method, as a refinement of the results. Techniques able to detect fluorescent signals are more sensitive than the colorimetric ones to distinguish lipase-producing microorganisms from esterase ones [68]. In this method, enzymatic hydrolysis results in a potentially fluorescent compound which signal may be detected following oxidation and β-elimination chain reactions, producing umbelliferone. Long-chain ester (octanoate) fluorogenic probes indicate lipase production and the short-chain (ethanoate) ones signalize esterase activity [46]. Another study found esterase production using substrates with less than six carbons [69] and lipase employing 8-carbon substrates [70]. According to Lopes et al. [71], the reliability in the classification of lipolytic enzymes is only defined through the analysis of activity determination by multiple methods; what justifies why different methodologies were used here to differentiate lipases and esterases production in our strains.
Mucor fragilis
CBMAI 2080 showed the best results regarding the short-chain fluorescent probe activity (Table 2), highlighting this strain as an important esterase producer for further studies, and contradicting Singh [53] who reports Mucorales as an order of limited enzymatic activities. Fonzi and Sypherd [72] ascribed Mucor sp. as resistance to Trichodermin (antifungal) thanks to its effective production of esterases. Our strain showed positive results in plate assays and the potential for conversion of 8-carbon probe. Reports on esterases capable of converting long-chain substrates with low water solubility are already available [73, 74]. Papers on Mucorales representatives report their enzymes produced as esterase [74] and, sometimes as lipases [75]. Indeed, there are few studies using this group of fungi, which we assumed as good candidates for biotechnological applications, needing further studies. A search in an American Type Culture Collection, ATCC, recovered no Mucor strain with evidenced esterase activity. This is the novel information from the present research.
Trichoderma longibrachiatum
CBMAI 2082 was found as a promising extracellular lipase producer (Table 3) with the long-chain esters probe assay. Other species, such as T. viride [25] and T. reesei [76] are widely recognized by its lipase production. A strain of T. longibrachiatum with lipase activity was isolated from oil contaminated soil [37]. While CBMAI 2082 showed 89.97% of conversion of the lipophilic probes after 24 h of analysis (Table 3), Lima et al. [77] obtained conversion percentages of 0.4 and 4.3% in isolates of Hyphomycetes and Dematiaceous, respectively, working with fluorogenic probes to assess lipases activity at the same time of experiment. This demonstrates how promising our strain can be considered.
Fungal activity on nanoparticles and commercial bottle fragments
Two types of PET bottles were investigated: (1) from “Plantbottle” technology that uses 20% less PET and 30% PET made from sugarcane (ethyleneglycol portion), namely, PET 1 [78]; (2) made by conventional method (PET 2). The aim was to test the performance of our strains to degrade both materials.
In general, PET biodegradation studies firstly evaluate lipase, esterase, and cutinase activities of microbes considered potential degraders [10, 79]. So, the initial selection performed in this work highlighted the activity of nine strains, which belong to Mucorales and Hyphomycetes. The latter are already widely recognized as efficient enzyme producers, but the former were previously considered of limited activity by Singh [53]. This investigation, however, also showed strains of Mucor spp. of expressive potential for enzymatic screening.
M. Arundinis
CBMAI 2109 also presented an excellent result on PET, despite its intermediate enzymatic activity. It showed a relative action in long-chain ester probe (43.3% ± 2.7), even though Reddy and Reddy [80] reported expressive lipase production for other strains belonging to the same fungal group. Yoshida et al. [81] evaluated the enzymatic activity of PETase over different substituents for para-nitrophenyl (C2, C4, C6, and C8) and observed that this ester hydrolase showed optimal activity in PET and the lowest catalytic activity in pre-related substrates. Future studies will be carried out with this strain in an attempt to improve cultivation conditions and obtain greater enzymatic efficiency.
To establish the extent of microbial degradation, the monitoring of seven characteristics are usually recommended: surface functional group, hydrophobicity/hydrophilicity, crystallinity, surface topography, mechanical properties, and molecular weight distribution [11]. The degree of M. arundinis action on PET bottle fragments was evaluated by surface topography, weight loss, biodeterioration, and biofragmentation.
Figures 4 a and 5 a evidence the SEM micrography of a PET 1 and 2 fragments after cleaning (abiotic control), subjected to the established fermentation conditions and no fungal inoculum. Homogenous and unspotted surfaces can be noted, demonstrating that no alterations occurred. On the other hand, stains and small spots similar to micropittings and pores can be observed in Figs. 4 and 5 (b, c, d, and e). Comparable images were reported by Nowak et al. [82] and Watanabe et al. [83] in their studies with low density polyethylene (LDPE). In Fig. 4 (b and e) and Fig. 5 (d and e), the penetration of hyphae structures into the PET was verified even after the cleaning procedure. Similar result was also found by Volke-Seplveda [84] in LPDE and by Sharon and Sharon [10] and Yoshida et al. [81] in PET investigations.
Previous studies employed gravimetric analyses, such as weight loss, to evaluate PET consumption [81, 82]. M. arundinis CBMAI 2109 reduced PET mass by 0.5% and CBMAI 2110, 0.16%, after 14 days (Fig. 6). Nowak et al. [82] observed weight loss percentage of 0.08% in PET using Penicillium funiculosum, after 84 days, grown in minimal mineral medium (sucrose-free). This study, the medium employed potato dextrose broth (PDB) where a greater amount of carbon source is present, compared to other medium used for PET degradation studies, might have favored the mycelial growth of the fungus and the consequent release of hydrolytic enzymes (Fig. 7). In experiments with dye discoloration employing Phanerochaete chrysosporium, Sem, Pakshirajan, and Santra [85] reported that the maximum fungal growth increased with the raising of the initial glucose concentration as well as the lignin peroxidase activity.
Figure 7 also contains information about monomer (terephthalic acid) and smaller oligomers (BHET and MHET) comprising the potential of the produced enzymes by strains CBMAI 2109 and CBMAI 2110 to promote depolymerization. These results were confirmed when FTIR analysis was performed in PET 1 and 2, demonstrating modifications in important points of the spectra when biotic samples were compared to abiotic control. FTIR PET spectra obtained by Sepperumal et al. [86] in soil experiment, presented the following results: appearance of new absorption peaks at 2930 cm−1 and 2869 cm−1 reveals C–H bond stretching (CH 2), 2725 cm−1 and 2520 cm−1 has been assigned to C=O bond stretching (carbonyl group), and 2118 cm−1 and 877 cm−1 to C=C bond stretching (Benzene ring).
In our FTIR spectra, it can be observed in PET 1, for both treatment, the disappearance of the peak at 2353 cm−1 and reduction in intensity of the peaks 1635 cm−1 and 877 cm−1, related to H-bond of hydroxyl groups and aromatic CH out-of-plane bend, respectively [87]. For PET 2, CBMAI 2109 treatment promoted the appearance of new absorption peaks in 2956 cm−1 and 1186 cm−1, attributed to asymmetry stretching aliphatic vibration of C–H of the methylene (–CH 2– in ethylene glycol segment) and aromatic C–H in-plane bend [87, 88]. While, both strains led to disappearance of peak at 2330 cm−1(characteristic of methyl C–H asymmetric) and reduction in peak intensity at 1060 and 862 cm−1 (that to indicate 1,4-disubstitution in aromatic ring). The peak intensities were increased in 1480 cm−1 (methylene C–H bend); 984 cm−1 and 964 cm−1 (trans-unsaturation CH=CH); and 837 cm−1 (C–H out-of-plane bending of aromatic ring para-substituted). For Ioakeimidis et al. [89], the creation of new functional groups consisting only of carbon and hydrogen represents a totally new surface, of unknown degradation. The abovementioned data indicate that the biotreatments with the highlighted lineages for both types of PET were able to promote changes in the aromatic and aliphatic portions of the polymer.
Few studies report the use of whole-cell filamentous fungi (not only pure enzymes) for PET biodegradation. However, most papers evaluate the action of the isolated enzymes in the polymer. An enzyme S238F/W159H engineering developed by Austin et al. [90] promoted the liberation of proximately 190 ppm and 175 ppm of TPA and MHET, respectively. The two strains assayed in this study mobilized PET to PTA inquantities of approximately 20 ppm.
Considering the initial proposal of this study, nine filamentous fungi with high hydrolytic activity were obtained. In addition to another strain that did not fit the criteria, the isolate M. arundinis CBMAI 2110 also showed good results in nanoparticles and commercial fragment of PET bottles such as maintenance of lipase/esterase activities after fermentation process and demonstrating a good potential for PET depolymerization.
This is the first report of application of M. arundinis strains (CBMAI 2109 and CBMAI 2110) in studies of PET biodegradation/depolymerization from commercial bottles (real condition). Chaves et al. [47] obtained promising results with strains of these fungi on PET nanoparticles and reported the importance of subsequent studies with these strains. The analyses showed that the isolates can be considered sources of enzymes with potential application in PET degradation.
M. arundinis belongs to the order Pleosporales where other representatives, also involved in studies of polymer biodegradation, are reported with important results. The strain Ss1-3, Alternaria solani, isolated from soil in northern Jordan, for example, demonstrated the ability to use polyester polyurethane as the sole source of carbon and nitrogen. This potential was evidenced by the weight loss in polyurethane cubes, as well as the reduction of tension to the traction and the percentage of stretching to be broken. Cracks in the surface of the material were observed by scanning electron microscopy and changes in the chemical structure of the polymer were evidenced by FTIR [91].
Another Pleosporales, Phoma herbarium, was able to grow in culture media containing highly pure lignin extracted from Abies sp. (Pinaceae), besides using synthetic lignin as the only carbon source. It was also observed the presence of extracellular enzymes in the culture medium, possibly responsible for the loss of mass and changes in the chemical structure of the polymer. Such evidences pointed to a promising activity of the strain as a biodegrader [48], as it was observed in this study with M. arundinis, but on the substrate PET.
Kumari, Tiwari, and Yadav [92], working on LDPE biodegradation (low density polyethylene), emphasized the importance of using microbial consortia to improve the process. It is possible that a consortium of M. arundinis strains may help to achieve even better results for depolymerization of PET.
Conclusions
Our data showed that environments associated with hydrocarbons are indeed a promising and still few-prospected source for the search of strains with recognized biotechnological potential. In this study, nine strains showed high conversions for long- and short-chain fluorogenic probes esters, and other 21 isolates were selected for further investigations as potential esterase and lipase producers.
From the set of microorganisms here investigated, the taxon Microsphaeropsis arundinis was considered efficient to promote the depolymerization of PET. It is the first time this fungal species of Pleosporales is introduced for polymer biodegradation application. Strains CBMAI 2109 and CBMAI 2110 are the most promising ones. A deeper approach to qualify what type of enzyme (lipase, esterases, or cutinases) is produced by such strains is recommended as well as an attempt to use them in a consortium.
Few previous studies on PET depolymerization have undertaken broad approaches like in this investigation. Indeed, the combination of techniques like plate assays, high-throughput screening (HTS), fluorescence analysis, FTIR, SEM, enzyme activities, and high-performance liquid chromatography with UV detector (HPLC-UV) are highly desirable to obtain reliable conclusions.
Finally, it was concluded that unrevealed species of filamentous fungi isolated from substrates associated with hydrocarbons are capable of mobilizing terephthalic acid and other oligomers, and they can be used to recycle plastic bottles among other containers, decontaminating the environment and bringing better quality of life.
Electronic supplementary material
(DOCX 1898 kb)
Acknowledgments
The authors thank PETROBRAS for their authorization to publish this study, and to MSc. Tulio de Lucca Capelini for technical support. We thank the Center for Environmental Studies (CEA) for providing site and infrastructure to carry out part of this work.
Funding information
The authors received financial support from PETROBRAS (2012/00327-7). One of the authors (L.M.P.) received a support grant from the Coordination for the Improvement of Higher Education Personnel (CAPES).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.United States Environmental Protection Agency (USEPA). The facts on recycling plastics. 1990 (February) . https://nepis.epa.gov/Exe/ZyNET.exe/10001ABI. Accessed 10 January 2018
- 2.Ellen MacArthur Foundation. The New Plastics Economy: Rethinking the future of plastics. Ellen MacArthur Found. 2016;(January):120. 10.1103/Physrevb.74.035409
- 3.Associação Brasileira da Indústria do PET (ABIPET). 10° Censo da reciclagem do PET no Brasil. 2016 (November). http://www.abipet.org.br/index.html?method=mostrarDownloads&categoria.id=3. Accessed 10 January 2018
- 4.National Association for PET Container Resources (NAPCOR). Report on Postconsumer PET Container Recycling Activity in 2012. 2013:13. http://www.napcor.com/pdf/NAPCOR_2012RateReport.pdf. Accessed 10 January 2018
- 5.Di Souza L, Torres MCM, Ruvolo Filho AC. Despolimerização do poli (tereftalato de etileno) - PET: efeitos de tensoativos e excesso de solução alcalina. Polímeros. 2008;18(4):334–341. doi: 10.1590/S0104-14282008000400013. [DOI] [Google Scholar]
- 6.Pellis A, Gamerith C, Ghazaryan G, Ortner A, Herrero E, Guebitz GM. Bioresource technology ultrasound-enhanced enzymatic hydrolysis of poly (ethylene terephthalate ) Bioresour Technol. 2016;218:1–5. doi: 10.1016/j.biortech.2016.07.106. [DOI] [PubMed] [Google Scholar]
- 7.de Castro AM, Carniel A, Nicomedes Junior J, da Conceição GA, Valoni É. Screening of commercial enzymes for poly(ethylene terephthalate) (PET) hydrolysis and synergy studies on different substrate sources. J Ind Microbiol Biotechnol. 2017;44(6):835–844. doi: 10.1007/s10295-017-1942-z. [DOI] [PubMed] [Google Scholar]
- 8.Ronkvist ÅM, Xie W, Lu W, Gross RA. Cutinase-catalyzed hydrolysis of poly (ethylene terephthalate) Macromolecules. 2009;42(14):5128–5138. doi: 10.1021/ma9005318. [DOI] [Google Scholar]
- 9.Tokiwa Y, Calabia BP, Ugwu CU, Aiba S. Biodegradability of plastics. Int J Mol Sci. 2009;10(9):3722–3742. doi: 10.3390/ijms10093722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharon C, Sharon M. Studies on biodegradation of polyethylene terephthalate: a synthetic polymer. J Microbiol Biotechnol Res Sch Res Libr J Microbiol Biotech Res. 2012;2(2):248–257. [Google Scholar]
- 11.Restrepo-Flórez JM, Bassi A, Thompson MR. Microbial degradation and deterioration of polyethylene - a review. Int Biodeterior Biodegrad. 2014;88:83–90. doi: 10.1016/j.ibiod.2013.12.014. [DOI] [Google Scholar]
- 12.Wei R, Zimmermann W. Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: how far are we? Microb Biotechnol. 2017;10(6):1308–1322. doi: 10.1111/1751-7915.12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma R, Chisti Y, Chand U, Banerjee UC. Production, purification, characterization, and applications of lipases. Biotechnol Adv. 2001;19(8):627–662. doi: 10.1016/S0734-9750(01)00086-6. [DOI] [PubMed] [Google Scholar]
- 14.Hasan F, Shah AA, Hameed A. Industrial applications of microbial lipases. Enzym Microb Technol. 2006;39(2):235–251. doi: 10.1016/j.enzmictec.2005.10.016. [DOI] [Google Scholar]
- 15.Trincone A. Marine biocatalysts: enzymatic features and applications. Mar Drugs. 2011;9(4):478–499. doi: 10.3390/md9040478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zafar U, Houlden A, Robson GD. Fungal communities associated with the biodegradation of polyester polyurethane buried under compost at different temperatures. Appl Environ Microbiol. 2013;79(23):7313–7324. doi: 10.1128/AEM.02536-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adrio JL, Demain AL. Microbial enzymes: tools for biotechnological processes. Biomolecules. 2014;4(1):117–139. doi: 10.3390/biom4010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sayali K, Sadichha P, Surekha S. Microbial esterases : an overview. Int J Curr Microbiol App Sci. 2013;2(7):135–146. [Google Scholar]
- 19.Mazzucotelli CA, Ponce AG, Kotlar CE, Moreira M del R. Isolation and characterization of bacterial strains with a hydrolytic profile with potential use in bioconversion of agroindustial by-products and waste. Food Sci Technol 2013;33(2):295–303. 10.1590/S0101-20612013005000038
- 20.Lee LP, Karbul HM, Citartan M, Gopinath SCB, Lakshmipriya T, Tang TH. Lipase-secreting Bacillus species in an oil-contaminated habitat: promising strains to alleviate oil pollution. Biomed Res Int. 2015;2015:1–9. doi: 10.1155/2015/820575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khannous L, Jrad M, Dammak M, Miladi R, Chaaben N, Khemakhem B, Gharsallah N, Fendri I. Isolation of a novel amylase and lipase-producing Pseudomonas luteola strain: study of amylase production conditions. Lipids Health Dis. 2014;13(1):9. doi: 10.1186/1476-511X-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boekema BKHL, Beselin A, Breuer M, Hauer B, Koster M, Rosenau F, Jaeger KE, Tommassen J. Hexadecane and tween 80 stimulate lipase production in Burkholderia glumae by different mechanisms. Appl Environ Microbiol. 2007;73(12):3838–3844. doi: 10.1128/AEM.00097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vakhlu J, Kour A. Yeast lipases: enzyme purification, biochemical properties and gene cloning. Electron J Biotechnol. 2006;9(1):69–85. doi: 10.2225/vol9-issue1-fulltext-9. [DOI] [Google Scholar]
- 24.Wiseman, A. Introduction to principles. In: Wiseman, A (ed.,) Handbook of enzyme biotechnology 3rd edition. Padstow, Cornwall, U.K: Ellis Horwood Ltd., T.J Press; 1995: 3–8
- 25.Kashmiri MA, Adnan A, Butt BW. Production, purification and partial characterization of lipase from Trichoderma Viride. Biotechnology. 2006;5(10):878–882. doi: 10.1016/j.procbio.2005.10.017. [DOI] [Google Scholar]
- 26.Contesini FJ, Calzado F, Valdo J, et al. Fungal metabolites. Reference series in phytochemistry. Springer. 2017:1–28. 10.1007/978-3-319-25001-4
- 27.Schneider WDH, Gonçalves TA, Uchima CA, Couger MB, Prade R, Squina FM, Dillon AJP, Camassola M. Penicillium echinulatum secretome analysis reveals the fungi potential for degradation of lignocellulosic biomass. Biotechnol Biofuels. 2016;9(1):66. doi: 10.1186/s13068-016-0476-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iftikhar T, Abdullah R, Iqtedar M, Kaleem A, et al. Production of lipases by Alternaria sp .(MBL 2810) through optimization of environmental conditions using submerged fermentation technique. Int. J. Biosci. 2015;6655:178–186
- 29.Rodrigues C, Cassini STA, Antunes PWP, Pinotti LM, Keller RP, Gonçalves RF. Lipase-producing fungi for potential wastewater treatment and bioenergy production. Afr J Biotechnol. 2016;15(18):759–767. doi: 10.5897/AJB2015.14666. [DOI] [Google Scholar]
- 30.Nwuche CO, Ogbonna JC. Isolation of lipase producing fungi from palm oil mill effluent (POME) dump sites at Nsukka. Braz Arch Biol Technol. 2011;54(1):113–116. doi: 10.1590/S1516-89132011000100015. [DOI] [Google Scholar]
- 31.Cihangir N, Sarikaya E. Investigation of lipase production by a new isolate of Aspergillus sp. World J Microbiol Biotechnol. 2004;20(2):193–197. doi: 10.1023/B:WIBI.0000021781.61031.3a. [DOI] [Google Scholar]
- 32.Lisboa HCF, Biasetto CR, de Medeiros JB, et al. Endophytic fungi producing of esterases: evaluation in vitro of the enzymatic activity using pH indicator. Braz J Microbiol. 2013;44(3):923–926. doi: 10.1590/S1517-83822013005000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pereira MG, Vici AC, Facchini FDA, Tristão AP, Cursino-Santos JR, Sanches PR, Jorge JA, Polizeli MLTM. Screening of filamentous fungi for lipase production: Hypocrea pseudokoningii a new producer with a high biotechnological potential. Biocatal Biotransformation. 2014;32(1):74–83. doi: 10.3109/10242422.2013.873417. [DOI] [Google Scholar]
- 34.Moro S, Pacheco V, Júnior AC, Morgado AF. Isolation and screening of filamentous fungi producing extracellular lipase with potential in biodiesel production. Adv Enzym Res. 2015;(December):101–114
- 35.Hombalimath VS, Udapudi BB, Patil LR, Shet AN, Yaraguppi DA, Tennalli G. Isolation and characterization of lipolytic microorganisms from oil contaminated soil. Int J Adv Sci Eng Technol. 2012;2(3):293–297. [Google Scholar]
- 36.Colla LM, Ficanha AMM, Rizzardi J, Bertolin TE, Reinehr CO, Costa JAV. Production and characterization of lipases by two new isolates of Aspergillus through solid-state and submerged fermentation. Biomed Res Int. 2015;2015:1–9. doi: 10.1155/2015/725959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiama C. Isolation and characterization of hydrocarbon biodegrading fungi from oil contaminated soils in Thika, Kenya. 2015. [Google Scholar]
- 38.Yu D, Margesin R. Partial characterization of a crude cold-active lipase from Rhodococcus cercidiphylli BZ22. Folia Microbiol (Praha) 2014;59(5):439–445. doi: 10.1007/s12223-014-0318-2. [DOI] [PubMed] [Google Scholar]
- 39.Iqbal SA, Rehman A. Characterization of lipase from Bacillus subtilis I-4 and its potential use in oil contaminated wastewater. Braz Arch Biol Technol. 2015;58(5):789–797. doi: 10.1590/S1516-89132015050318. [DOI] [Google Scholar]
- 40.Mobarak-Qamsari E, Kasra-Kermanshahi R, Moosavi-Nejad Z. Isolation and identification of a novel, lipase-producing bacterium, Pseudomnas aeruginosa KM110. Iran J Microbiol. 2011;3(2):92–98. [PMC free article] [PubMed] [Google Scholar]
- 41.Veerapagu M, Sankara Narayanan A, Ponmurugan K, Jeya KR. Screening selection identification production and optimization of bacterial lipase from oil spilled soil. Asian J Pharm Clin Res. 2013;6(SUPPL.3):62–67. 10.20546/ijcmas.2016.503.087
- 42.Reyes-duarte D, Ferrer M, García-arellano H. Lipases and Phospholipases. 2012;861:101–113. doi: 10.1007/978-1-61779-600-5. [DOI] [PubMed] [Google Scholar]
- 43.Sánchez-Carbente M del R, Batista-García RA, Sánchez-Reyes A, et al. The first description of a hormone-sensitive lipase from a basidiomycete: structural insights and biochemical characterization revealed Bjerkandera adusta BaEstB as a novel esterase. Microbiology open 2017;6(4):1–15. 10.1002/mbo3.463 [DOI] [PMC free article] [PubMed]
- 44.Ramírez L, Arrizon J, Sandoval G, Cardador A, Bello-Mendoza R, Lappe P, Mateos-Díaz JC. A new microplate screening method for the simultaneous activity quantification of feruloyl esterases, tannases, and chlorogenate esterases. Appl Biochem Biotechnol. 2008;151(2–3):711–723. doi: 10.1007/s12010-008-8319-8. [DOI] [PubMed] [Google Scholar]
- 45.Singh R, Gupta N, Goswami VK, Gupta R. A simple activity staining protocol for lipases and esterases. Appl Microbiol Biotechnol. 2006;70(6):679–682. doi: 10.1007/s00253-005-0138-z. [DOI] [PubMed] [Google Scholar]
- 46.Mantovani SM, De Oliveira LG, Marsaioli AJ. Esterase screening using whole cells of Brazilian soil microorganisms. J Braz Chem Soc. 2010;21(8):1484–1489. doi: 10.1590/S0103-50532010000800011. [DOI] [Google Scholar]
- 47.Chaves MRB, Lima MLSO, Malafatti-Picca L, et al. A practical fluorescence-based screening protocol for polyethylene terephthalate degrading microorganisms. J Braz Chem Soc. 2018;29(6):1278–1285. 10.21577/0103-5053.20170224
- 48.Bi R, Lawoko M, Henriksson G. Phoma herbarum , a soil fungus able to grow on natural lignin and synthetic lignin ( DHP ) as sole carbon source and cause lignin degradation. J Ind Microbiol Biotechnol 2016;43(8):1175–1182. 10.1007/s10295-016-1783-1 [DOI] [PubMed]
- 49.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1–2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 50.Burkert JFM, Maugeri F, Rodrigues MI. Optimization of extracellular lipase production by Geotrichum sp. using factorial design. Bioresour Technol. 2004;91(1):77–84. doi: 10.1016/S0960-8524(03)00152-4. [DOI] [PubMed] [Google Scholar]
- 51.He X. A continuous spectrophotometric assay for the determination of diamondback moth esterase activity. Arch Insect Biochem Physiol. 2003;54(2):68–76. doi: 10.1002/arch.10103. [DOI] [PubMed] [Google Scholar]
- 52.Meier MJ, Paterson ES, Lambert IB. Use of substrate-induced gene expression in metagenomic analysis of an aromatic hydrocarbon-contaminated soil. Appl Environ Microbiol. 2015;82(3):897–909. doi: 10.1128/AEM.03306-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh H. Mycoremediation: fungal bioremediation. Hoboken, New Jersey: John Wiley & Sons, Inc.; 2006. [Google Scholar]
- 54.Mehta A, Bodh U, Gupta R. Fungal lipases: a review. J Biotech Res. 2017;8(1):58–77. [Google Scholar]
- 55.Uchiyama T, Abe T, Ikemura T, Watanabe K. Substrate-induced gene-expression screening of environmental metagenome libraries for isolation of catabolic genes. Nat Biotechnol. 2005;23(1):88–93. doi: 10.1038/nbt1048. [DOI] [PubMed] [Google Scholar]
- 56.Gargouri B, Mhiri N, Karray F, Aloui F, Sayadi S. Isolation and characterization of hydrocarbon-degrading yeast strains from petroleum contaminated industrial wastewater. Biomed Res Int. 2015;2015:1–11. doi: 10.1155/2015/929424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ugochukwu KC, Agha NC, Ogbulie JN. Lipase activities of microbial isolates from soil contaminated with crude oil after bioremediation. Afr J Biotechnol. 2008;7(16):2881–2884. doi: 10.5897/AJB08.093. [DOI] [Google Scholar]
- 58.de Crecy E, Jaronski S, Lyons B, Lyons T, Keyhani N. Directed evolution of a filamentous fungus for thermotolerance. BMC Biotechnol. 2009;9:74. doi: 10.1186/1472-6750-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vander Meer RK, Morel L. Nestmate recognition in ants. In: Vander Meer RK, Breed M, Winston M, Espelie L. Pheromone communication in social insects. Westview Press: Boulder: Colo; 1998: 70–103
- 60.Menzel F, Blaimer BB, Schmitt T. How do cuticular hydrocarbons evolve? Physiological constraints and climatic and biotic selection pressures act on a complex functional trait. Proc R Soc B Biol Sci. 2017;284(1850):20161727. doi: 10.1098/rspb.2016.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cervantes-gonzález E, Zambrano-monroy B, Ovando-medina VM, Briones-gallardo R, Ventura-suarez A. Influence of aromatic , heteroaromatic , and alkane hydrocarbons on the lipase activity of Pseudomonas sp . in Batch Culture. 2014;23(5):1507–1513
- 62.Kanwar L, Goswami P. Isolation of a Pseudomonas lipase produced in pure hydrocarbon substrate and its application in the synthesis of isoamyl acetate using membrane-immobilised lipase. Enzym Microb Technol. 2002;31(6):727–735. doi: 10.1016/S0141-0229(02)00191-6. [DOI] [Google Scholar]
- 63.Rai B, Shrestha A, Sharma S, Joshi J. Screening, optimization and process scale up for pilot scale production of lipase by Aspergillus niger. Biomed Biotechnol. 2014;2(3):54–59. 10.12691/bb-2-3-3
- 64.Gupta P. Studies on lipase isolated from suitable microbial sources. World Journal of Pharmacy and Pharmaceutical Sciences.2016; 6(1): 1555–1566. 10.20959/wjpps20171-8450
- 65.Kumar D, Kumar L, Nagar S, Raina C, Parshad R, Gupta VK. Screening, isolation and production of lipase/esterase producing Bacillus sp. strain DVL2 and its potential evaluation in esterification and resolution reactions. Arch Appl Sci Res. 2012;4(4):1763–1770. [Google Scholar]
- 66.Ramnath L, Sithole B, Govinden R. Classification of lipolytic enzymes and their biotechnological applications in the pulping industry. Can J Microbiol. 2017;63(October 2016):1–14. doi: 10.1139/cjm-2016-0447. [DOI] [PubMed] [Google Scholar]
- 67.Chahinian H, Nini L, Boitard E, Dubès J-P, Comeau L-C, Sarda L. Distinction between esterases and lipases: a kinetic study with vinyl esters and TAG. Lipids. 2002;37(7):653–662. doi: 10.1007/s11745-002-0946-7. [DOI] [PubMed] [Google Scholar]
- 68.Gonçalves CDCS, Marsaioli AJ. Monitorando atividades enzimáticas com sondas fluorogênicas. Quim Nova. 2014;37(6):1028–1036. doi: 10.5935/0100-4042.20140142. [DOI] [Google Scholar]
- 69.Kulkarni N, Gadre RV. Production and properties of an alkaline, thermophilic lipase from Pseudomonas fluorescens NS2W. J Ind Microbiol Biotechnol. 2002;28(6):344–348. doi: 10.1038/sj.jim.7000254. [DOI] [PubMed] [Google Scholar]
- 70.Sicart R, Collin MP, Reymond JL. Fluorogenic substrates for lipases, esterases, and acylases using a TIM-mechanism for signal release. Biotechnol J. 2007;2(2):221–231. doi: 10.1002/biot.200600181. [DOI] [PubMed] [Google Scholar]
- 71.Lopes DB, Fraga LP, Fleuri LF, Macedo GA. Lipase and esterase: to what extent can this classification be applied accurately? Ciência e Tecnol Aliment. 2011;31(3):603–613. doi: 10.1590/S0101-20612011000300009. [DOI] [Google Scholar]
- 72.Fonzi WA, Sypherd PS. Trichodermin esterase activity and trichodermin resistance in Mucor racemosus. Antimicrob Agents Chemother. 1986;29(4):570–575. doi: 10.1128/AAC.29.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moskowitz GJ, Shen T, West IR, Cassaigne R, Feldman LI. Properties of the esterase produced by Mucor miehei to develop flavor in dairy products. J Dairy Sci. 1977;60(8):1260–1265. doi: 10.3168/jds.S0022-0302(77)84020-4. [DOI] [Google Scholar]
- 74.Lopez-Lopez O, Cerdan M, Siso M. New extremophilic lipases and esterases from metagenomics. Curr Protein Pept Sci. 2014;15(5):445–455. doi: 10.2174/1389203715666140228153801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alves MH, Campos-Takaki GM, Figueiredo Porto AL, Milanez AI. Screening of Mucor spp. for the production of amylase, lipase, polygalacturonase and protease. Braz J Microbiol. 2002;33(4):325–330. doi: 10.1590/S1517-83822002000400009. [DOI] [Google Scholar]
- 76.Rajesh EM, Arthe R, Rajendran R, Balakumar C, Pradeepa N, Anitha S. Investigation of lipase production by Trichoderma Reesei and optimization of production parameters 2010; 9(7):1177–1189
- 77.Lima MLSO, Chaves MRB, do Nascimento RMC, Gonçalves CCS. Marsaioli AJ. Simultaneous multienzymatic screening with fluorogenic probes. J. Braz. Chem. Soc.2018; 29( 5), 1149–1156. 10.21577/0103-5053.20170188
- 78.Byun, Y, Kim, YT. Utilization of Bioplastics for Food Packaging Industry. [s.l.] Elsevier Ltd, 2013
- 79.Mueller RJ. Biological degradation of synthetic polyesters-enzymes as potential catalysts for polyester recycling. Process Biochem. 2006;41(10):2124–2128. doi: 10.1016/j.procbio.2006.05.018. [DOI] [Google Scholar]
- 80.Reddy AS, Reddy SM. Lipase activity of two seed-borne fungi of sesamum (Sesamum indicum Linn.) Folia Microbiol (Praha) 1983;28(6):463–466. doi: 10.1007/BF02879683. [DOI] [Google Scholar]
- 81.Yoshida S, Hiraga K, Takehana T, et al. Response to comment on “A bacterium that degrades and assimilates poly(ethylene terephthalate).” Science (80- ). 2016;353(6301):759–759. 10.1126/science.aaf8625 [DOI] [PubMed]
- 82.Nowak B, Pajak J, Drozd-Bratkowicz M, Rymarz G. Microorganisms participating in the biodegradation of modified polyethylene films in different soils under laboratory conditions. Int Biodeterior Biodegrad. 2011;65(6):757–767. doi: 10.1016/j.ibiod.2011.04.007. [DOI] [Google Scholar]
- 83.Watanabe T, Ohtake Y, Asabe H, Murakami N, Furukawa M. Biodegradability and degrading microbes of low-density polyethylene. J Appl Polym Sci. 2009;111:551–559. doi: 10.1002/app.29102. [DOI] [Google Scholar]
- 84.Volke-Seplveda T, Saucedo-Castaeda G, Gutirrez-Rojas M, Manzur A, Favela-Torres E. Thermally treated low density polyethylene biodegradation by Penicillium pinophilum and Aspergillus niger. J Appl Polym Sci. 2002;83(2):305–314. doi: 10.1002/app.2245. [DOI] [Google Scholar]
- 85.Sen K, Pakshirajan K, Santra SB. Modelling the biomass growth and enzyme secretion by the white rot fungus Phanerochaete chrysosporium in presence of a toxic pollutant. J Environ Prot (Irvine, Calif). 2012;3(January):114–119. 10.4236/jep.2012.31014 [DOI] [PubMed]
- 86.Sepperumal U, Markandan M, Palraja I. Micromorphological and chemical changes during biodegradation of polyethylene terephthalate ( PET ) by Penicillium sp. J Microbiol Biotechnol Res. 2013;3(4):47–53. [Google Scholar]
- 87.Coates J. Interpretation of infrared spectra, A practical approach Encycl Anal Chem 2006:1–23. 10.1002/9780470027318.a5606
- 88.Al-Azzawi F. Degradation studies on recycled Polyethylene terephthalate 2015. London Metropolitan University. http://ethos.bl.uk/OrderDetails.do?uin=uk.bl.ethos.681923. Accessed 15 may 2018
- 89.Ioakeimidis C, Fotopoulou KN, Karapanagioti HK, Geraga M, Zeri C, Papathanassiou E, Galgani F, Papatheodorou G. The degradation potential of PET bottles in the marine environment: an ATR-FTIR based approach. Sci Rep. 2016;6(October 2015):1–8. doi: 10.1038/srep23501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Austin HP, Allen MD, Donohoe BS, Rorrer NA, Kearns FL, Silveira RL, Pollard BC, Dominick G, Duman R, el Omari K, Mykhaylyk V, Wagner A, Michener WE, Amore A, Skaf MS, Crowley MF, Thorne AW, Johnson CW, Woodcock HL, McGeehan JE, Beckham GT. Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc Natl Acad Sci. 2018;201718804:E4350–E4357. doi: 10.1073/pnas.1718804115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ibrahim IN, Maraqa A, Hameed KM, Saadoun IM, Maswadeh HM, Nakajima-kambe T. Polyester-polyurethane Biodegradation by Alternaria solani , isolated from Northern Jordan.Adv Environ Biol 2009;3(2):162–170
- 92.Kumari G, Tiwari A, Yadav M. LDPE-biodegradation using microbial consortium by the incorporation of cobalt ferrite nanoparticle as the enhancer for biodegradation. Int J Adv Eng Res Dev Sci J Impact Factor (SJIF. 2017;4(6):4–72
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 1898 kb)