Abstract
Elucidation of the distinctive microbial taxonomic profiles of tropical fruit peels is the indispensable component of investigations aimed at the detection of microorganisms responsible for the post-harvest loss. The objective of the present work was to dissect the bacterial and fungal community of five tropical fruit peels (banana, guava, mango, papaya, and passion fruit) in wild (non-cultivated) and conventionally produced samples from Brazil. To that end, 16S rRNA–encoding gene and ITS rDNA amplicon analysis of the five tropical fruit peels were performed to discriminate the bacterial and fungal communities, respectively. The result showed that bacterial communities of the five types of fruit peels were by far more diversified than that of fungal communities, independent of the type of production system involved. Among the investigated fruits, non-cultivated papaya peels hosted the most diversified bacterial community while the least bacterial community diversity was found in the conventionally produced papaya fruit peels. The gene amplicon analysis clearly discriminated the bacterial community into their respective classes, while fungal communities were better classified in their phyla, yet with clearer component discrimination of fungal community based on the type of cultivation system practiced. Conventionally produced banana and non-cultivated passion fruit peels were characteristically dominated by fungal and bacterial groups, respectively. Overall, in conventionally produced fruit peels, bacterial community was mainly composed of Proteobacteria, Actinobacteria, and Bacilli. The result provided a broad microbial diversity profile that could be used as an important input for seeking alternative fruit spoilage control and post-harvest treatments.
Electronic supplementary material
The online version of this article (10.1007/s42770-019-00088-0) contains supplementary material, which is available to authorized users.
Keywords: Microbial diversity, Fruit peel, Cultivation system, Conventional, rDNA
Introduction
Tropical fruits are indisputably the most popular fruits consumed around the world. Banana (Musa acuminate), guava (Psidium guajava), mango (Mangifera indica), papaya (Carica papaya), and passion fruit (Passiflora edulis) are some of the tropical fruits making the top list destined for export and/or domestically consumed in countries in the tropics [1]. As fruit peels harbor diverse populations of microorganisms, tropical fruit post-harvest diseases such as anthracnosis pose serious problems to perishable fruits destined for exportation [2, 3]. Despite the increasingly striving modern post-harvest management technologies, post-harvest diseases are, however, still responsible for a reasonable yield loss of tropical fruits. Studies of disease prevalence and the extent of yield loss of conventional and non-cultivated fruits due to post-harvest diseases, on the other hand, have immense importance in deciphering economically important microbes. Secondary metabolites in the fruit peels contribute to fruit texture, quality, and storage [4], and in fact, fruit peels host different kinds of epiphytic and endophytic microorganisms [5]. Therefore, studies involving microbial diversity profiling and downstream identification of species of significant agricultural and medical importance (among the others) have enormous research interest. Furthermore, investigation of the dynamics of natural microbiota in fruit peels could be considered as one of the convenient strategies to unveil the fruit microbial community, as peel microbial colonization is easily detectable compared with systemic infections.
Unlike non-cultivated fruits (grown wild, with no human intervention), cultivation of conventionally produced fruits depends on the intensive application of chemical pesticides (such as fungicides) to prevent post-harvest loss mainly due to bacterial and fungal contaminations. Most of the fruit microbial diversity studies conducted so far are focused on the identification and screening of some specific microbial species colonizing conventionally produced fruits. On the other hand, gene amplicon analysis of microbial diversity colonizing fruit peels from conventionally and non-conventionally cultivated samples would provide a broad and rigorous understanding of fruit peel microbial communities for best post-harvest disease management practices. Furthermore, natural microbiota profiling of fruit peels provides detailed insights to establish the diversity and relative abundance of different microbial communities colonizing non-cultivated and conventionally produced fruit peels. Compared to non-cultivated fruits, the incidence of pathogen infestation and eventual loss of yield due to post-harvest disease is pervasively evident and demandingly appealing further studies in conventional fruit production. Equally important is the taxonomic identification of bacterial communities associated with fungal species strongly linked to fruit spoilage, which is poorly understood or scantily explored.
To unveil the molecular fingerprint of fruit peel microbial community, gene analysis is the most reliable and has been an increasingly applied tool in such endeavors of taxonomic profiling. As bacteria and fungi are the two major components of fruit peel microbiota, 16S rRNA encoding gene [6] and rDNA internal transcribed spacer (ITS) [7] DNA region are the most commonly used molecular fingerprints to differentiate bacterial and fungal taxa, respectively. Besides, the gene amplicon analysis of fruit peel microbiota in conventionally cultivated fruits as compared with those from non-conventionally (wild) produced fruit peel microbiota would generate valuable data of the microbial community at the post-harvest stage. Considering these aspects, the objective of the present study was to conduct a comparative analysis of fruit peel microbial community in five tropical fruits taken from conventional farms and non-cultivated areas.
Materials and methods
Plant material
All fruit peel samples of five tropical fruits (Banana, Musa cavendish var. Nanica; guava, Psidium guajava var. Pedro Sato; mango, Mangifera indica var. Tommy Atkins; papaya, Carica papaya var. Solo; and passion fruit, Passiflora edulis var. Gigante Amarelo) were taken once (without seasonal variation as a parameter) from areas in the Federal District Region, which surrounds the capital Brasilia, Brazil. All conventionally produced fruit samples were collected once from CEASA (Central Supply of Horticultural Products) in March 2014, while in the same way, non-cultivated samples were collected once from wild areas within five distinguished sites (Supplemental Table 1). Collection of non-cultivated samples was made in November and December of 2013. For all the five fruit peel samples, data were randomly collected in such a way that three replicates were considered for both samples from conventional farms and from non-cultivated collection areas. Ripened fruits just before harvest were carefully collected for all the five fruits for robust discrimination of bacterial and fungal diversities colonizing fruit peels that could ultimately result in post-harvest spoilage. Sample collection was as follows: fruit samples were collected, put in plastic bags, and brought to the laboratory where the peels were immediately removed for the DNA extraction. Similar procedures were carried out with the purchased fruits, where random sample collections were considered at the supermarket stage.
DNA extraction and gene amplicon analysis
DNA extraction and PCR
The fruit peel samples were collected in three replicates from conventional farms and non-cultivated areas. For all the five tropical fruits considered in this study, fruit peels were collected in such a way that 15 representative fruits were randomly taken from both conventional and non-cultivated samples, stored under freezing temperature (− 18 °C) until DNA extraction. Accordingly, 5-g peel samples were homogenized in liquid nitrogen followed by 7 mL CTAB DNA extraction buffer. Such protocol [8] had the chloroform/alcohol cleaning performed twice, and the final genomic DNA samples with a minimum concentration of ≥ 5 ng/μL were used for further downstream applications.
Bacterial 16S rRNA gene was amplified using the universal primer pair 534f-CS1 (5′-CCAGCAGCCGCGGTAAT-3′) and 783r-CS2 (5′-GGTCTACC MGGGTATCTAATCCKG-3′) [9]. Fungal rDNA ITS region was amplified with ITS1-CS1 (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and 5.8S-CS2 (5′-GGTCTAACTTTYRRCAAYGGATCWCT-3′) primer pair [10]. Both primers were linked to CS1 (ACACTGACGAC ATGGTTCTACA) and CS2 (TACGGTAGCAGAGACTT) adaptors (Fluidigm Co., USA). PCR reactions and library construction with Fluidigm were performed at Genome Quebec Co., Canada.
Amplicon sequencing and analysis
PCR products with their specific barcodes and adapters (Fluidigm Co., USA) were also sequenced by Illumina MiSeq platform at Genome Quebec Co., Canada, to generate the metagenome data. The library constructed from the index PCR data of the bacterial 16S rRNA gene was subjected to sequence assembly, followed by metagenome assay using Galaxy software (https://usegalaxy.org) and the remaining bioinformatics analysis by CloVR pipeline (http://clovr.org) [11]. For fungal ITS data, the QIIME pipeline (http://qiime.org) [12] was applied. Chimera was checked by USEARCH 6.1 [13]. QIIME software was also used to assess the relative abundance of OTUs, microbial diversity (Shannon–Wiener, Chao and Simpson diversity indexes), and principal coordinate analysis (PCoA) followed by the Bray–Curtis dissimilarity in the five different kinds of tropical fruit peels. OTU clusters were defined by a 97% identity threshold, and the data sets were submitted to rarefaction analysis before diversity analysis. Furthermore, the genetic features, represented by the abundance of OTU, were chosen by hierarchical feature engineering (HFE) [14] and then the feature selection was performed and their effects were verified by the random forest and the principal component analysis (PCA). The quality filtering was applied at a Phred score ≥ Q20. The taxonomy base analysis was accessed by Ribosomal Database Project (RDP) classifier supplemented with BLAST and Mothur v 1.32.1 [15]. Differences in the relative abundance of OTUs among samples and diversity indexes were estimated by analysis of variance (ANOVA) after checking the normality distribution with the Shapiro–Wilk test. Metagenome sequences were registered at DNA Data Bank of Japan (DDBJ).
Results
The total number of sequences obtained from all fruit samples were 2,131,220 (71,040 ANR (average number of reads)) for 16S rRNA encoding gene and 2,321,071 (77,369 ANR) for rDNA ITS. These reads were subjected to filtering based on a Phred quality score of ≥ 20. It resulted in the assignment of 2,028,998 16S rRNA (95%) sequences to bacteria and 2,042,542 rDNA ITS (88%) sequences to fungi and their discrimination at class and phylum taxonomic levels, respectively.
Fruit peel bacterial community
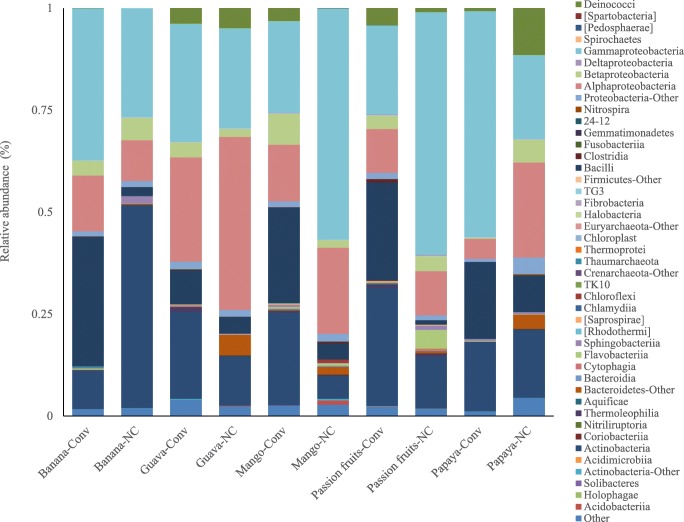
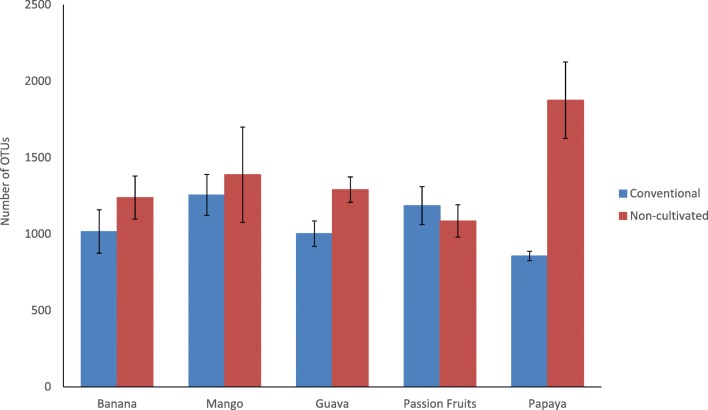
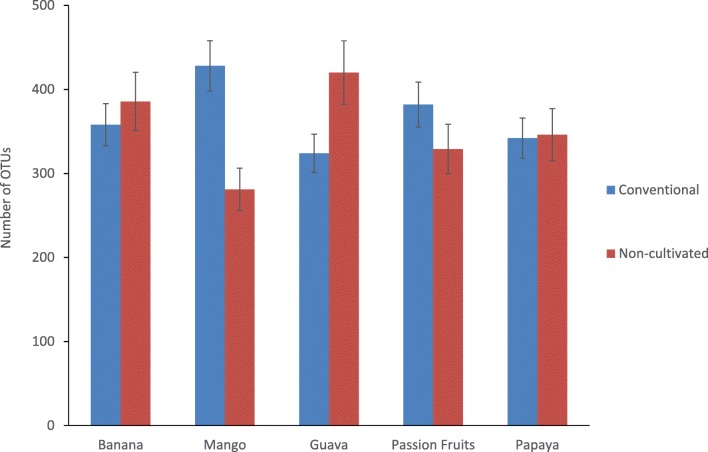
The bacterial community analysis indicated that fruit peel samples from both cultivated and non-cultivated areas harbored Gammaproteobacteria, Alphaproteobacteria/Actinobacteria, and Acidobacteria in order of their relative abundance (Fig. 1). Deinococci was detected in all but the non-cultivated banana in both conventional and non-cultivated samples in different abundances. Overall, most classes were detected in all the samples with different relative abundance. Unlike the other bacterial communities, the relative abundance of Bacilli was affected by production system and fruit type. A similar trend was detected in Agaricomycetes, Dothideomycetes, Sordaryomycetes, and Ustilaginomycetes relative abundance (Supplemental Table 2). The number of OTUs observed in non-cultivated papaya sample was by far greater than the one observed in the conventional sample and all the other samples as well (Fig. 2). Except in papaya, the number of OTUs in conventional and non-conventional fruit peel samples was less than 1500 units and was comparable as observed from the rarefaction curve.
Fig. 1.
Distribution of bacteria classes on five kinds of tropical fruit peels taken from conventional and non-cultivated areas
Fig. 2.
Number of OTUs within the 16S RNA gene sequences on five kinds of tropical fruit peels taken from conventional and non-cultivated areas
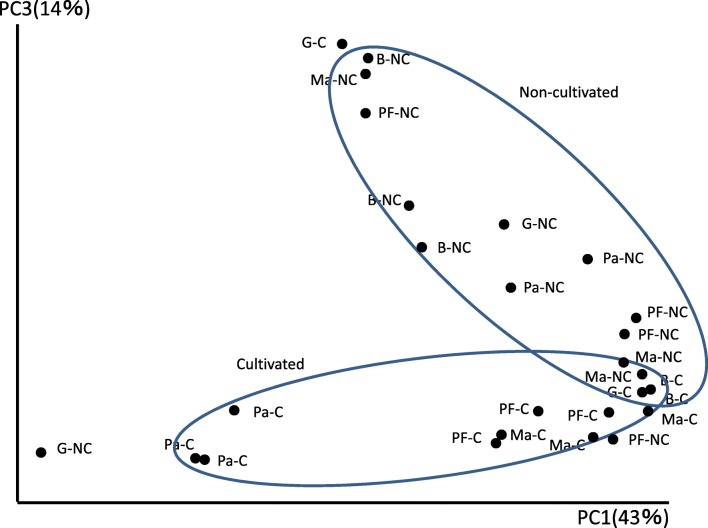
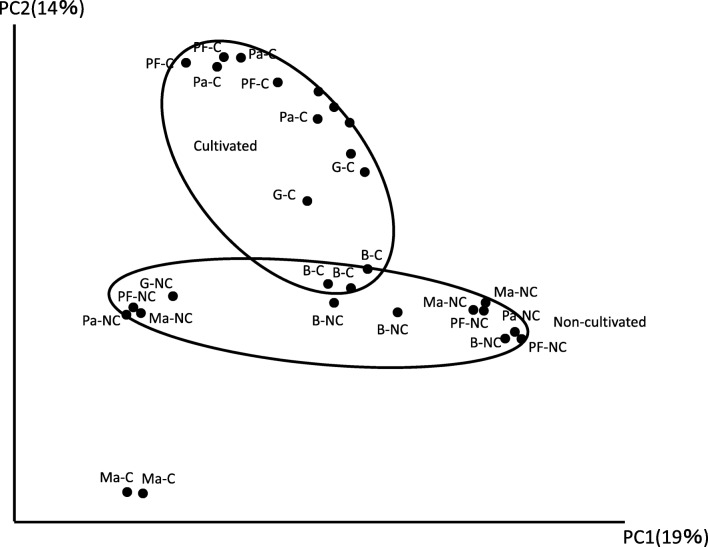
As revealed by the diversity indexes, bacterial community diversity for each fruit type in both conventional and non-cultivated areas had no significant difference, except in papaya in which the diversity was increased in non-cultivated fruit peel sample. The bacterial diversity detected by the three index analyses was, however, well differentiated in non-cultivated fruit peels of papaya, and in fact, this diversity was the highest compared with all samples (Table 1 and Supplemental Table 2). The diversity index analysis and the number of OTUs observed in non-cultivated papaya were complementary and worth further detailed discrimination of bacterial community at the lowest taxonomic level. Bacterial community PCoA had an overlapping distribution for all the samples (Fig. 3 and Supplemental Table 3). As the proportion of the bacterial components were dispersed along the component coordinate, the proportion of bacterial classes were evidently distributed indiscriminately in both conventional and non-cultivated fruit peels in all the fruits investigated. Bacterial community relative abundance in the non-cultivated fruit peels was distinguished as compared to samples from conventional counterparts (Supplemental Fig. S1). Accordingly, a diversity of bacterial species was found in most of the fruit peels in both conventionally produced and non-cultivated samples. One of the results worth mentioning here was the higher relative abundance of Streptomyces mashuensis in non-cultivated passion fruit peels (Supplemental Table 4).
Table 1.
Mean (standard error) of bacterial diversity according to Shannon–Wiener, Chao, and Simpson index on five kinds of tropical fruit peels taken from conventional and non-cultivated areas
| Chao | Shannon | Simpson | ||||
|---|---|---|---|---|---|---|
| Conventional | Non-cultivated | Conventional | Non-cultivated | Conventional | Non-cultivated | |
| Banana | 5479.7 (528.8) | 6771.3 (496.5) | 2.6 (0.3) | 3.7 (0.4) | 0.32 (0.01) | 0.08 (0.01) |
| Mango | 6296.1 (726.9) | 6757.9 (987.6) | 4.0 (0.4) | 3.0 (0.3) | 0.07 (0.01) | 0.32 (0.01) |
| Guava | 5001.3 (499.7) | 5684.7 (658.5) | 3.8 (0.3) | 3.6 (0.3) | 0.05 (0.01) | 0.08 (0.01) |
| Passion fruits | 6731.0 (235.9) | 6112.0 (350.9) | 4.1 (0.5) | 3.3 (0.4) | 0.05 (0.01) | 0.18 (0.01) |
| Papaya | 6284.5 (232.7) | 8300.0 (852.0) | 2.1 (0.3) | 4.2 (0.5) | 0.42 (0.01) | 0.08 (0.01) |
Fig. 3.
Principal coordinate analyses based on 16S rRNA gene sequences on five kinds of tropical fruit peels taken from conventional and non-cultivated areas. B—banana, G—guava, Ma—mango, Pa—papaya, PF—passion fruits, C—conventional, NC—non-cultivated
Fruit peel fungal community
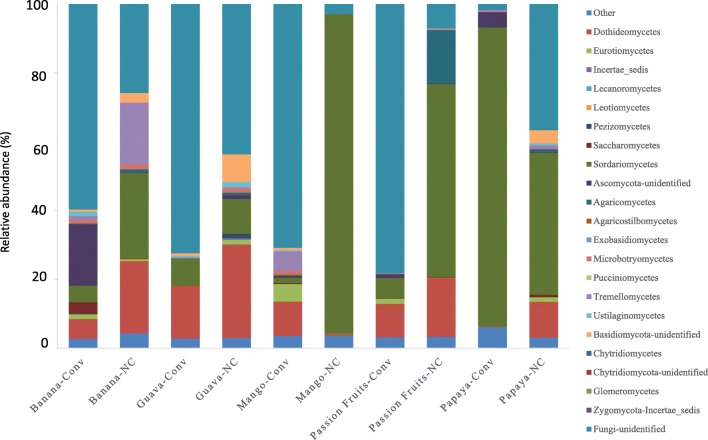
For all samples from both conventional and non-cultivated areas, the main fungal communities detected were Sordariomycetes and Dothideomycetes in their order of relative abundance (Fig. 4). The relative abundance of unidentified Basidiomycota was negligible in all conventionally produced fruit samples of all fruit types. The relative abundance of this phylum was higher in non-cultivated guava, papaya, and banana fruit peel samples. The effect of both system and fruit type was mostly observed in Agaricomycetes, Dothideomycetes, and Sordaryomycetes (Supplemental Table 2).
Fig. 4.
Distribution of fungal classes on five kinds of tropical fruit peels taken from conventional and non-cultivated areas
The number of fungal OTUs was the highest in conventionally produced mango when compared to all the other four fruit types, independent of the cultivation practice involved (Fig. 5). Despite the highly abundant fungal OTUs in both conventional and non-cultivated samples in all the fruit types, the abundance in conventionally produced samples was comparable to that of non-cultivated samples for all the five fruit types except in mango and guava, where a significant difference was observed for conventionally produced and non-cultivated fruit peel samples. Among the fungal diversity index analysis applied, Shannon diversity index detected a remarkable distinction in the diversity of the fungal community in conventional and non-cultivated peel samples of mango and papaya (Table 2 and Supplemental Table 5). While fungal community diversity was by far higher in conventionally produced mango peels, as compared with the diversity in non-cultivated samples, the diversity in papaya was otherwise. However, using the diversity index analyses, the diversity of the fungal community in guava peels was slightly greater than in papaya collected from non-cultivated samples. The results from the diversity index analysis for the rest of the fruit types vis-à-vis their respective sample source was generally comparable.
Fig. 5.
Number of OTUs within the ITS rDNA sequences on five kinds of tropical fruit peels taken from conventional and non-cultivated areas
Table 2.
Mean (standard error) of fungal diversity according to Shannon–Wiener, Chao, and Simpson index on five kinds of tropical fruit peels taken from conventional and non-cultivated areas
| Chao | Shannon | Simpson | ||||
|---|---|---|---|---|---|---|
| Conventional | Non-cultivated | Conventional | Non-cultivated | Conventional | Non-cultivated | |
| Banana | 726.8 (53.8) | 814.7 (79.5) | 3.8 (0.3) | 4.6 (0.4) | 0.77 (0.06) | 0.89 (0.09) |
| Mango | 776.1 (82.9) | 618.9 (82.6) | 5.0 (0.4) | 1.1 (0.1) | 0.92 (0.07) | 0.23 (0.02) |
| Guava | 741.1 (72.9) | 834.9 (87.6) | 3.1 (0.3) | 5.3 (0.4) | 0.76 (0.07) | 0.94 (0.08) |
| Passion fruits | 812.0 (73.9) | 718.0 (75.9) | 4.0 (0.5) | 2.6 (0.2) | 0.85 (0.06) | 0.69 (0.07) |
| Papaya | 657.5 (63.7) | 757.8 (81.0) | 1.2 (0.2) | 4.2 (0.4) | 0.24 (0.02) | 0.88 (0.08) |
The most distinguished fungal diversity output was detected by PCoA, by which a clear separation of a fungal constituent was established (Fig. 6 and Supplemental Table 5). Fungal components for all the five fruit types were well differentiated into conventional and non-cultivated clusters based on their constituent fungal phyla (Supplemental Fig. S2). Except in mango, all conventionally cultivated fruits showed similar composition in their fungal community. Detailed insight into fungal relative abundance fingerprint demonstrated the dominant genera characterizing each of the fruit types and their production systems involved. It followed that the fungal genera fingerprint indicated that most of the fungal communities belong to the pathogenic groups as was the case in the bacterial community (Supplemental Table 6). Another microbial genus of scientific interest is the characteristic abundance of Penicillium in conventionally produced banana fruit peel. Moreover, this same sample hosted the highest number of fungal genera fingerprints abundantly occupied by fungal pathogens.
Fig. 6.
Principal coordinate analyses based on ITS rDNA sequences on five kinds of tropical fruit peels taken from conventional and non-cultivated areas. B—banana, G—guava, Ma—mango, Pa—papaya, PF—passion fruits, C—conventional, NC—non-cultivated
Discussion
The microbiome of plants is a major factor in plant health and yield as the diversity and load of microorganisms are critical components of post-harvest disease management. Therefore, shifts in the plant microbiome diversity can alter the reaction to biotic and abiotic stresses and the quality of end product [16]. From the consumer end point of view, diagnosis of the presence of microbes, differentiation of their diversity, and quantification constitutes maintenance of fruit health to the desired levels [17]. In the present study, relative abundance, diversity, and composition of microbial communities were found to be affected by the type of fruit peel and the production system involved. Bacterial concentration of fruit peels was distinct in abundance and almost twice more abundant than that of fungal phyla, making the bacterial community crucially important for post-harvest management measures, including decisions to determine the right type of biological control to neutralize the most adversary microorganisms. Among the compared fruits, banana and guava contained a relatively higher number of OTUs as compared with the others. The estimated microbial diversity for both bacteria and fungi indicated that the diversity was generally higher in non-cultivated fruits than in the conventionally cultivated ones. The overall data suggest a strong presence of antagonistic microbes to pathogens in non-cultivated fruits, especially with higher relative abundance in passion fruit peel.
Bacterial community
Actinobacteria along with Alpha-, Beta-, and Gamma-proteobacteria were dominating all the fruit peels investigated. These taxa are known in colonizing flowers of vegetables and fruit peels of different tropical fruits including watermelon [16, 18, 19]. Proteobacteria and Actinobacteria were also reported to be among the main endophytic prokaryotes in rice root [20]. The relatively higher abundance of Bacillus species in conventionally cultivated fruits (percentage relative abundance as seen in the band size of the bar graph, Fig. 1) showed a similar trend for all five fruit types. This is an indication of the importance of Bacilli in conventional farms, where routine pesticide chemicals are intensively applied. Also, the presence of Bacillus, Cytophagia, and Alpha-, Beta-, and Gamma-proteobacteria across the investigated samples, independent of the cultivation and fruit types, was an indication that these bacterial phyla are retained as the major components for fruit spoilage as reported in other fruits [21, 22] or lead to the colonization by the secondary microbes. The diversity of the bacterial community is dependent on the type of fruit, growing location, fertilizer used, the pesticide used, and other farming inputs applied [23]. The relatively higher relative abundance of Bacillus species in conventionally produced fruit peels in all the fruit types could be attributed to the possible inclusion of Bacilli in the organic pesticides used during farming practice [19].
The application of agrochemicals in conventional farming (as could be the case in our conventionally produced samples) could have favored the elimination of natural enemies to Bacillus species that intrinsically exist in the non-conventionally produced fruit peels of tropical fruits. In addition, it is of worth noticing the higher relative abundance of Actinobacteria species in conventionally produced fruit samples (except in banana). This could be attributed to one of the important traits of Actinobacteria, their well-known source of bioactive compounds [24] that could have helped them degrade agrichemicals applied as pesticides, thereby providing them a competitive survival advantage over the others. Fruit peels from wild banana samples could be used as a source of different species of Actinobacteria (as the maximum abundance was harbored in this sample), some of which bioactive compounds could be targeted for industrial applications including saccharification. Banana fruit peels were reported as important sources of bioactive compounds of industrial importance [25, 26] as was saccharification using bioactive compounds of actinobacterial species too from a recent report [27]. Non-cultivated papaya fruit peels were dominated by the maximum diversity of the bacterial community, which could be considered as a bacterial reservoir for other studies aimed at the identification of bacterial species of agronomic and industrial importance. The in-depth taxonomic fingerprint identification of bacterial community revealed the importance of wild passion fruit peels as one of the main sources of S. mashuensis, for further search of bioactive compounds of antibacterial role. Further analysis of sequenced amplicons revealed bacterial species abundance fingerprints characterizing each of the five fruit types in their respective production systems (Supplemental Table 5).
Fungal community
The relative abundance of Ascomycota (Sordariomycetes and Dothideomycetes) was relatively low in conventionally produced fruits unlike their dominance in non-cultivated fruit peels of banana, guava, mango, and passion fruit. Ascomycetes are generally known to colonize fruit peels [22, 28]. These results were contrary to a study of fungal diversity from strawberry matured fruit, where Basidiomycota and Ascomycota were the dominant taxa, despite the application of routine chemical pesticides [5]. The reduced relative abundance of overall fungal diversity in the conventionally produced fruits could be attributed to the intensive use of broad-spectrum fungicidal chemicals. Similar reports were made from strawberry flower and leaves, in which the application of chemical fungicide was reported to lower fungal diversity [29, 30]. As Basidiomycetes were detected with more abundance in fruit peels with no history of agronomic management practice, this intact fungal community could be of great scientific interest in searching for unique species with characteristic products applicable to a broad range of industry.
Among the industrial applications, enzymes from Basidiomycetes are used for the degradation of various kinds of polysaccharides in the paper industry and bioremediation studies [31]. Besides, the relatively higher abundance of Basidiomycetes could also be further exploited for the identification of some species of medical and nutritional importance. Nutritive and medical importance of Basidiomycetes has already been reported from edible mushroom species [32]. In the present study, probably of great importance is the dominance of Penicillium genera in the conventionally produced banana peel samples. As the production and availability of this fruit is at the lowest economic cost, it could be of great research interest for isolation and commercialization of new variant compounds of medical importance. Of comparable importance is the discrimination of bacterial communities at species level although the discrimination of fungal communities was just possible at phylum and genus levels in the present study. The use of 16S rRNA gene for the discrimination of bacterial community at species level was demonstrated in another similar study [33], while the use of ITS rRNA gene for the discrimination of fungal communities at genera level is more reliable than at species level [34]. As the present study was based on the analysis of sequenced DNA amplicons, a culture-based microbiological investigation should be conducted to determine the load of fruit peel microorganisms, thereby supplement the present analysis. Such a comprehensive and thorough investigation of microbes colonizing fruit peels is also very important in improving post-harvest disease control.
In conclusion, we demonstrated the microbial diversity and the complexity in discriminating into their respective taxa in five common tropical fruit peel samples. Even though the same gene region sequence was analyzed across the investigated samples, bacterial communities colonizing fruit peels were better studied and documented than their counter fungal communities. The present study was able to differentiate the bacterial community at the class level and that of fungal diversity at the phylum level. In addition to the other importance, medical and industrial importance of the identified microbes indicates the need for further discrimination of the diversity beyond species level. The molecular data generated from sequenced amplicons were better in revealing clearer discrimination of microbial communities into their respective phyla in both conventional and non-cultivated samples. As the present work was entirely based on gene amplicon sequence analysis, the results obtained and the conclusions thereof need further study involving culture-based fruit peel microbial diversity profiling.
Electronic supplementary material
(PPTX 58 kb)
(PPTX 58 kb)
(DOCX 11 kb)
(DOCX 12 kb)
(DOCX 11 kb)
(DOCX 11 kb)
(DOCX 11 kb)
(DOCX 10 kb)
Acknowledgments
This project was supported by the Alexander von Humboldt-Foundation—AvH (Germany) and National Council for Scientific and Technological Development—CNPq (Brazil). Thanks to the Cell Biology Department, Universidade de Brasilia, for allowing us to use their facilities during the DNA extraction.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
11/30/2019
The original version of this article unfortunately contained two mistakes in the “Materials and methods” section, subsection “DNA extraction and PCR” of the article. The correct information is given below.
Change history
11/30/2019
The original version of this article unfortunately contained two mistakes in the ���Materials and methods��� section, subsection ���DNA extraction and PCR��� of the article. The correct information is given below.
References
- 1.Scherrer J (2018) The Most popular fruit in the world. Bandeiras News
- 2.Onyeani CA, Osunlaj SO, Owuru OO, Sosanya OS. Mango fruit anthracnose and the effects on mango yield and market values in southwestern Nigeria. Asian J Agric Res. 2012;6(4):171–179. doi: 10.3923/ajar.2012.171.179.. [DOI] [Google Scholar]
- 3.Udayanga D, Manamgoda DS, Liu X, Chukeatirote E, Hyde KD. What are the common anthracnose pathogens of tropical fruits? Fungal Divers. 2013;61(1):165–179. doi: 10.1007/s13225-013-0257-2. [DOI] [Google Scholar]
- 4.Wang Z, Cui Y, Vainstein A, Chen S, Ma H. Regulation of fig (Ficus carica L.) fruit color: metabolomic and transcriptomic analyses of the flavonoid biosynthetic pathway. Front Plant Sci. 2017;8:1990. doi: 10.3389/fpls.2017.01990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdelfattah A, Wisniewski M, Droby S, Schena L. Spatial and compositional variation in the fungal communities of organic and conventionally grown apple fruit at the consumer point-of-purchase. Hortic Res. 2016;3(1):16047. doi: 10.1038/hortres.2016.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mandal D, Sequenc NG, De Mandal S, Panda AK, Bisht SS, Kumar NS (2015) Microbial ecology in the era of next generation sequencing. Next Gener Seq Appl:1–6. 10.4172/jngsa.S1-001
- 7.Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson KH. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol. 2013;22(21):5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- 8.Diniz LEC, Sakiyama NS, Lashermes P, Caixeta ET, Oliveira ACB, Zambolim EM, Loureiro ME, Pereira AA, Zambolim L. Analysis of AFLP markers associated to the Mex-1 resistance locus in Icatu progenies. Crop Breed Appl Biotechnol. 2005;5(4):387–393. doi: 10.12702/1984-7033.v05n04a03. [DOI] [Google Scholar]
- 9.Rastogi G, Tech JJ, Coaker GL, Leveau JHJ. A PCR-based toolbox for the culture-independent quantification of total bacterial abundances in plant environments. J Microbiol Methods. 2010;83(2):127–132. doi: 10.1016/j.mimet.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Buée M, Reich M, Murat C, Morin E, Nilsson RH, Uroz S, Martin F. 454 pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytol. 2009;184(2):449–456. doi: 10.1111/j.1469-8137.2009.03003.x. [DOI] [PubMed] [Google Scholar]
- 11.Angiuoli SV, Matalka M, Gussman A, Galens K, Vangala M, Riley DR, Arze C, White JR, White O, Fricke WF. CloVR: a virtual machine for automated and portable sequence analysis from the desktop using cloud computing. BMC Bioinformatics. 2011;12(1):356. doi: 10.1186/1471-2105-12-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 14.Oudah M, Henschel A. Taxonomy-aware feature engineering for microbiome classification. BMC Bioinformatics. 2018;19(1):227. doi: 10.1186/s12859-018-2205-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, van Horn DJ, Weber CF. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saminathan T, García M, Ghimire B, Lopez C, Bodunrin A, Nimmakayala P, Abburi VL, Levi A, Balagurusamy N, Reddy UK. Metagenomic and metatranscriptomic analyses of diverse watermelon cultivars reveal the role of fruit associated microbiome in carbohydrate metabolism and ripening of mature fruits. Front Plant Sci. 2018;9:4. doi: 10.3389/fpls.2018.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narayanasamy P. Microbial Plant Pathogens-Detection and Disease Diagnosis. Dordrecht: Springer Netherlands; 2010. Detection of Fungal Pathogens in Plants; pp. 5–199. [Google Scholar]
- 18.Reiter B, Sessitsch A. Bacterial endophytes of the wildflower Crocus albiflorus analyzed by characterization of isolates and by a cultivation-independent approach. Can J Microbiol. 2006;52(2):140–149. doi: 10.1139/w05-109. [DOI] [PubMed] [Google Scholar]
- 19.Leff JW, Noah F. Bacterial communities associated with the surfaces of fresh fruits and vegetables. PLoS One. 2013;8(3):e59310. doi: 10.3389/fpls.2018.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sengupta S, Ganguli S, Singh PK. Metagenome analysis of the root endophytic microbial community of Indian rice ( O. sativa L.) Genomics Data. 2017;12:41–43. doi: 10.1016/j.gdata.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barth M, Hankinson TR, Zhuang H, Breidt F. Compendium of the microbiological spoilage of foods and beverages. New York, NY: Springer New York; 2009. Microbiological spoilage of fruits and vegetables; pp. 135–183. [Google Scholar]
- 22.Ezra D, Kirshner B, Hershcovich M, Shtienberg D, Kosto I. Heart rot of pomegranate: disease etiology and the events leading to development of symptoms. Plant Dis. 2015;99(4):496–501. doi: 10.1094/PDIS-07-14-0707-RE. [DOI] [PubMed] [Google Scholar]
- 23.Schmid F, Moser G, Müller H, Berg G. Functional and structural microbial diversity in organic and conventional viticulture: organic farming benefits natural biocontrol agents. Appl Environ Microbiol. 2011;77(6):2188–2191. doi: 10.1128/AEM.02187-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suriya J., Bharathiraja S., Manivasagan P., Kim S.-K. Marine Enzymes Biotechnology: Production and Industrial Applications, Part II - Marine Organisms Producing Enzymes. 2016. Enzymes From Rare Actinobacterial Strains; pp. 67–98. [DOI] [PubMed] [Google Scholar]
- 25.Joana Gil-Chávez G, Villa JA, Fernando Ayala-Zavala J, Basilio Heredia J, Sepulveda D, Yahia EM, González-Aguilar GA. Technologies for extraction and production of bioactive compounds to be used as nutraceuticals and food ingredients: an overview. Compr Rev Food Sci Food Saf. 2013;12(1):5–23. doi: 10.1111/1541-4337.12005. [DOI] [Google Scholar]
- 26.Sagar NA, Pareek S, Sharma S, Yahia EM, Lobo MG. Fruit and vegetable waste: bioactive compounds, their extraction, and possible utilization. Compr Rev Food Sci Food Saf. 2018;17:512–531. doi: 10.1111/1541-4337.12330. [DOI] [PubMed] [Google Scholar]
- 27.Muthusamy Sanjivkumar, Selvan Silambarasan Tamil, Arunachalam Palavesam, Grasian Immanuel. Bioconversion and bioethanol production from agro-residues through fermentation process using mangrove-associated actinobacterium Streptomyces olivaceus (MSU3) Biofuels. 2017;10(2):167–179. doi: 10.1080/17597269.2017.1309853. [DOI] [Google Scholar]
- 28.Shen Y, Nie J, Li Z, Li H, Wu Y, Dong Y, Zhang J. Differentiated surface fungal communities at point of harvest on apple fruits from rural and peri-urban orchards. Sci Rep. 2018;8(1):2165. doi: 10.1038/s41598-017-17436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinto Cátia, Pinho Diogo, Sousa Susana, Pinheiro Miguel, Egas Conceição, C. Gomes Ana. Unravelling the Diversity of Grapevine Microbiome. PLoS ONE. 2014;9(1):e85622. doi: 10.1371/journal.pone.0085622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh S, Gupta R, Kumari M, Sharma S. Nontarget effects of chemical pesticides and biological pesticide on rhizospheric microbial community structure and function in Vigna radiata. Environ Sci Pollut Res. 2015;22(15):11290–11300. doi: 10.1007/s11356-015-4341-x. [DOI] [PubMed] [Google Scholar]
- 31.Rytioja J, Hildén K, Yuzon J, Hatakka A, de Vries RP, Mäkelä MR. Plant-polysaccharide-degrading enzymes from Basidiomycetes. Microbiol Mol Biol Rev. 2014;78(4):614–649. doi: 10.1128/MMBR.00035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rathee S, Rathee D, Rathee D, Kumar V, Rathee P. Mushrooms as therapeutic agents. Brazilian J Pharmacogn. 2012;22:459–474. doi: 10.1590/S0102-695X2011005000195. [DOI] [Google Scholar]
- 33.Earl JP, Adappa ND, Krol J, Bhat AS, Balashov S, Ehrlich RL, Palmer JN, Workman AD, Blasetti M, Sen B, Hammond J, Cohen NA, Ehrlich GD, Mell JC. Species-level bacterial community profiling of the healthy sinonasal microbiome using Pacific Biosciences sequencing of full-length 16S rRNA genes. Microbiome. 2018;6(1):190. doi: 10.1186/s40168-018-0569-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blaalid R, Kumar S, Nilsson RH, Abarenkov K, Kirk PM, Kauserud H. ITS1 versus ITS2 as DNA metabarcodes for fungi. Mol Ecol Resour. 2013;13(2):218–224. doi: 10.1111/1755-0998.12065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PPTX 58 kb)
(PPTX 58 kb)
(DOCX 11 kb)
(DOCX 12 kb)
(DOCX 11 kb)
(DOCX 11 kb)
(DOCX 11 kb)
(DOCX 10 kb)