Abstract
Listeria monocytogenes is one of the most important foodborne pathogens and is a causal agent of listeriosis in humans and animals. The aim of this study was to determine the prevalence, serogroups, antibiotic susceptibility, virulence factor genes, and genetic relatedness of L. monocytogenes strains isolated from 500 poultry samples in Turkey. The isolation sources of 103 L. monocytogenes strains were retail markets (n = 100) and slaughterhouses (n = 3). L. monocytogenes strains were identified as serogroups 1/2a-3a (75.7%, lineage I), 1/2c-3c (14.56%, lineage I), 1/2b-3b-7 (5.82%, lineage II), 4a-4c (2.91%, lineage III), and 4b-4d-4e (0.97%, lineage III). Most of the L. monocytogenes strains (93.2%) were susceptible to the antibiotics tested. PCR analysis indicated that the majority of the strains (95% to 100%) contained most of the virulence genes (hylA, plcA, plcB, prfA, mpl, actA, dltA, fri, flaA inlA, inlC, and inlJ). Pulsed-field gel electrophoresis (PFGE) demonstrated that there were 18 pulsotypes grouped at a similarity of > 90% among the strains. These results indicate that it is necessary to prevent the presence of L. monocytogenes in the poultry-processing environments to help prevent outbreaks of listeriosis and protect public health.
Keywords: Listeria monocytogenes, Chicken meat, Virulence characterization, Antibiotic susceptibility, PFGE
Introduction
Listeria monocytogenes is a major foodborne pathogen that causes listeriosis in humans and animals. The clinical symptoms of this disease are serious and include meningitis, meningoencephalitis, septicaemia, abortion, and death [1]. It mainly affects pregnant women, infants, older adults, and immunocompromised individuals [2]. Interestingly, there has been a statistically significant increase in prevalence of confirmed listeriosis in the EU/EEA during last 5 years (periods 2013–2017) [3]. However, there is limited research on cases of listeriosis in humans in Turkey. Listeriosis is diagnosed in 32 cases (11 healthy children and childhood ages 3 patients) which 66% have reported meningitis between 1987 and 2001 [4]. Since this bacterium may survive under different environmental conditions, foods are highly susceptible to L. monocytogenes contamination. In the USA, it is reported that there are more than 2500 listeriosis cases per year, 99% of them are from food source, and 500 people have died [5]. Listeriosis is linked to the consumption of contaminated food such as unpasteurized milk, ready-to-eat meat products, ice cream, and seafood [6, 7]. Contamination may occur during different food-processing stages such as packing, transportation, and preparation. In fact, multiple factors are related with survival, proliferation, and widespread contamination of food [8].
It is very important to monitor antibiotic resistance especially Multi Drug Resistance (MDR) of pathogenic organisms for potential hazard assessment [9]. Although L. monocytogenes is susceptible to many antibiotics, sometimes, multi-drug resistance can be observed among the isolates [10]. Some factors may affect its antibiotic resistance mechanisms. These are gene transfer resulting from stress in the food chain, biofilm formation, or antimicrobial use [11]. Resistance to antibiotics in L. monocytogenes strains from different sources has also been previously documented [12].
Although approximately 5% prevalence of L. monocytogenes in food is observed, the overall rate of listeriosis is low in the European Union [13]. This may be due to variability in the virulence properties found among various strains. Further typing and characterization steps are a requirement to precisely determine the virulence factors caused by foodborne L. monocytogenes strains [9]. Serotyping is the primary method used for typing of L. monocytogenes [14]. There are 15 recognized serotypes of L. monocytogenes. These are mainly separated to four lineages such as division I (1/2b, 3b, 4b, 4d, and 4e), division II (1/2a, 1/2c, 3a, and 3c), division III (4a and 4c), and division IV serotypes (4a, 4c, and atypical 4b). Among these serotypes, serotypes 1/2a, 1/2b, and 4b are responsible for most human listeriosis and isolated from food and environmental samples. Serotypes 4a and 4c are rarely related with epidemics [15–17]. Because of the limitations of serotyping, modern molecular typing techniques such as randomly amplified polymorphic DNA (RAPD), pulsed-field gel electrophoresis (PFGE), and amplified fragment length polymorphism (AFLP) have been developed for characterization [18–20]. The use of whole genome sequencing is becoming more common as a sub-typing tool, although other methods are still important.
Pathogenicity of L. monocytogenes depends on various virulence factors [21]. There are many virulence genes identified in L. monocytogenes. Among these genes, hlyA is one of the essential pathogenic factors and is responsible for the escape from the phagosomes and invasion of host cells [22, 23]. The actA gene encodes the surface protein ActA and is associated with cell-to-cell spread [24]. The internalin genes are involved in internalization and adhesion of L. monocytogenes [25]. The prfA gene regulates and controls the expression of a number of virulence genes [22, 26]. The pathogenicity of L. monocytogenes depends mainly on the prfA virulence gene cluster [27].
The objectives of the present study were to characterize L. monocytogenes isolates from different poultry samples originated from retail points and slaughterhouses in Turkey in terms of antibiotic susceptibility, serogroups, virulence genes, and genetic relatedness.
Materials and methods
Sampling and isolation
A total of 500 poultry meat samples were collected; 443 fresh poultry meat samples (wing, drumstick, and breast parts) were collected between April of 2014 and June of 2015 from local markets and butchers located in Istanbul, Turkey, and 57 poultry samples including whole chicken, wing, drumstick, and breast were obtained from slaughterhouses located in two cities (Balıkesir and Bolu, Turkey). The collected samples (n = 500) were analyzed for the presence of L. monocytogenes. Isolation and identification of L. monocytogenes was conducted as described by ISO 11290-1 [28]. First, 25 g of samples was diluted in half-fraser broth (Oxoid, UK) and incubated at 30 °C for 24 h for initial enrichment. After that, 0.1 ml of this suspension was transferred into 10 ml fraser broth (Oxoid) and further incubated at 37 °C for 24–48 h. The suspensions were plated onto Chromogenic Listeria Agar (Oxoid) after the second enrichment and the plates were incubated at 37 °C for 48 h. Typical colonies (blue-green with an opaque halo) were purified on selective agar plates (Chromogenic Listeria Agar, Oxoid) incubated at 37 °C for 48 h. Next, a single colony was streaked onto Tryptone Soya Agar (TSA, Oxoid) and incubated at 37 °C for 24 h. Isolated strains were frozen in 20% (v/v) glycerol and then stored at − 80 °C for further analysis.
Confirmation of isolates as L. monocytogenes
The isolates were grown on TSA at 37 °C for 24 h. This culture was used as inoculum for biochemical analysis (catalase, Gram staining, oxidase production, and the CAMP test on 5% (v/v) sheep blood agar). Following this, bacterial DNA extraction was performed [29]. PCR targeting the monoA-B gene was performed to confirm the presumptive L. monocytogenes [30, 31].
Characterization of L. monocytogenes strains
Serogrouping
All L. monocytogenes strains were serogrouped by multiplex PCR (m-PCR) as described by Doumith et al. [32].
Antibiotic susceptibility and MIC profiles
L. monocytogenes strains were tested for antibiotic susceptibility by the agar disk diffusion method on Mueller-Hinton agar (Oxoid). The plates were incubated at 37 °C for 24 h using the EUCAST Disk Diffusion Method (EUCAST, 2016). Five different antibiotics were used: ampicillin (AMP, Oxoid-CT0002B, 2 μg), penicillin G (P, Oxoid-CT0152B, 1 U), erythromycin (E, Oxoid-CT0020B, 15 μg), meropenem (M, Oxoid-CT0774B, 10 μg), and trimethoprim-sulfamethoxazole (co-trimoxazole) (STX, Oxoid-CT0052B, 25 μg). Streptococcus pneumoniae ATCC 46919 was used as a quality control strain. Minimum inhibitory concentrations (MIC) of the antibiotics were tested using E-Test (Abbiodisk, Sweden) and the results were evaluated according to EUCAST breakpoint tables [33].
Detection of virulence genes
Virulence genes such as dltA, plcA, plcB, mpl, and actA were detected using the same PCR conditions described in Table 1 [36, 39–43]. The internalin genes (inlA, inlC, and inlJ) and other virulence factor genes (hylA, prfA, fri, and flaA) were determined using the modified PCR conditions shown in Table 1. The PCR products were resolved on 1–1.5% (w/v) agarose gels in 1× TAE buffer and then visualized using SafeView™ Classic stain (Applied Biological Materials, Canada) in Infinity Gel Imaging System (Vilber Lourmat, France). All experiments were repeated at least three times.
Table 1.
Modified PCR conditions used for the detection of virulence genes
| Gene | Amplicon (bp) | PCR conditions | Reference |
|---|---|---|---|
| hylA | 234 | Initial denaturation at 94 °C for 5 min, 40 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s. | Furrer et al. [34] and Mengaud et al. [35] |
| prfA | 571 | Initial denaturation at 94 °C for 5 min, 35 cycles of 94 °C for 1 min, 55 °C for 1 min, and 72 °C for 1 min. | Nishibori et al. [36] |
|
inlA inlC inlJ |
800 517 238 |
Initial denaturation at 94 °C for 5 min, 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min. | Liu et al. [37] |
| fri | 471 | Initial denaturation at 94 °C for 5 min, 35 cycles of 94 °C for 45 s, 47 °C for 45 s, and 72 °C for 1 min and the final cycle at 72 °C for 7 min. | Slama et al. [38] |
| flaA | 864 | Initial denaturation at 94 °C for 5 min, 35 cycles of 94 °C for 45 s, 47 °C for 45 s, and 72 °C for 1 min and the final cycle at 72 °C for 7 min. | Slama et al. [38] |
Pulsed-field gel electrophoresis
PFGE was performed using the standardized PulseNet PFGE protocol (http://www.cdc.gov/pulsenet/PDF/listeria-pfge-protocol-508c.pdf) for L. monocytogenes digested with AscI and ApaI. The PFGE patterns were analyzed using the BioNumerics software package (Ver.7.5; Applied Maths, Belgium). The TIFF images were normalized by aligning the peaks with the size of a standard Salmonella enterica serovar braenderup H9812 strain, which was loaded on three lanes in each gel. Dendrogram and matching unweighted pair group method with averages (UPGMA) analysis of the patterns were conducted using the Pearson or Dice coefficient with a 1.0% tolerance.
Results and discussion
In the present study, a total of 500 poultry samples were collected and analyzed for the presence of L. monocytogenes. All of the presumptive L. monocytogenes isolates (n = 103) obtained by the Chromogenic Listeria Agar were Gram (+), catalase (+), oxidase (−), and CAMP (+) motile rods. The results of monoA-B PCR were also used in parallel with the biochemical profiles to confirm the isolates as L. monocytogenes. One hundred (22.6%) and 3 (5.3%) samples from retail markets and slaughterhouses were positive for L. monocytogenes, respectively (Table 2). The overall frequency of L. monocytogenes strains in poultry samples was 21%. A higher occurrence of L. monocytogenes was obtained from retail markets (100/103). Based on the sample type, the majority were recovered from drumstick samples (71/103). Chicken meat is known to be one of the mainly important food for the L. monocytogenes contamination [44]. In the related literature, the reported prevalence rates of this bacterium in poultry meat products were 3.7% in Morocco [45], 8.9% in Algeria [46], 15.3% in Romania [47], 17.9% in Brazil [44], 38% in Greece [12], 2.1% [48], 45% in Turkey [49], and 58% in Brazil [50]. The differences in the prevalence may be due to the differences in geographical characteristics, sampling strategies, hygienic conditions, and microbiological analysis methods [51]. L. monocytogenes contamination of poultry meat may occur during production, processing, and storage [44]. Therefore, good thermal treatment, personnel hygiene, cleaning applications, processing, and storage conditions may prevent L. monocytogenes contamination during food processing [9].
Table 2.
Distribution of L. monocytogenes based on the sample type, sampling place, and date
| Sampling place | Sampling date | Type and number of samples | ||||
|---|---|---|---|---|---|---|
| Whole chicken | Breast parts | Wing | Drumstick | % (No.) of positive samples for L. monocytogenes | ||
| Slaughterhouse A | April 2014 | 8 | 1 | 1 | 1 | 9% (1/11) |
| May 2014 | 8 | 1 | 1 | 1 | 0% (0/11) | |
| October 2014 | 8 | 1 | 1 | 1 | 0% (0/11) | |
| Slaughterhouse B | July 2014 | 9 | 1 | 1 | 1 | 16.7% (2/12) |
| August 2014 | 9 | 1 | 1 | 1 | 0% (0/12) | |
| Retail markets | May 2014 | – | 6 | 20 | 39 | 1.5% (1/65) |
| August 2014 | – | 13 | 7 | 59 | 7.6% (6/79) | |
| March 2015 | – | 15 | 8 | 56 | 49% (39/79) | |
| April 2015 | – | 8 | 29 | 48 | 40% (34/85) | |
| June 2015 | – | 25 | 42 | 68 | 14.8% (20/135) | |
| Total | 42 | 72 | 111 | 275 | 21% (103/500) | |
Serogrouping of L. monocytogenes is useful for the initial identification of potential health risks that may be linked to food [52]. Therefore, the strains were serogrouped into five groups using serogroup-specific primers by m-PCR. The majority of the strains (75.7%) were characterized as serogroup IIa (1/2a or 3a). Most of them were originated from drumstick collected from retail markets (55%). Also, 15 (14.56%) and 6 strains (5.82%) were serogrouped as IIc (1/2c or 3c) and IIb (1/2b, 3b or 7), respectively. Serogroup IVa (4a or 4c) was observed in three strains (2.91%). In addition, the serogroup IV (4b, 4d, or 4e) was observed at a low prevalence (0.97%). Leong et al. [53] and Murugesan et al. [54] have shown that 100% of serogroups 1/2a-3a and 1/2c-3c are actually serotypes 1/2a and 1/2c, respectively. According to this data, it is assumed that many of the strains obtained from the present study are serotypes 1/2a and 1/2c. In fact, serotypes 1/2a, 1/2b, 1/2c, and 4b strains are known to cause most human listeriosis [10]; moreover, these serotypes can be recovered from food samples and patients [15]. Especially, serotype 1/2a strains account above 50% of the food and environment isolates [16]. Based on the serogrouping, all of the isolates obtained from poultry samples were in the serogroups that are associated with human listeriosis which may suggest a potential public health risk. Major epidemics of listeriosis occur because of serotype 4b strains [21]; however, there was only one strain within this serogroup in this study. In another study [51], 16 L. monocytogenes strains from food samples were found to belong to serogroup IVb (87.5%) within phylogenetic lineage I and serogroup IIa belonging to phylogenetic lineage II (12.5%). Similar to the findings of this study, predominant serogroup in their study was serogroup IIa. Cao et al. [21] also reported that the isolates from black-headed gull feces belonged to serotype 4a (44%), 1/2c (33%), and 1/2b (22%). Maury et al. [55] reported that isolates belonging to lineage I and lineage II are overrepresented among food and human isolates, respectively.
The formation of biofilm in food-processing environments constitutes serious safety problems in processed food and is difficult to remove [56]. Huang et al. [16] demonstrated that serotype 1/2a isolates formed biofilm with a higher intensity than serotype 4b isolates in laboratory medium at 37 °C most probably due to the production of higher amounts of extracellular polymeric substances. This information suggests that most of the strains (76%) in this study may have potential to form higher amounts of biofilms in food-processing environments.
Since the virulence properties of L. monocytogenes influence the development of listeriosis [23], the occurrence of 14 virulence genes was determined by monoplex or m-PCR. Table 3 presents the distribution of the virulence genes in 103 L. monocytogenes strains divided into different serogroups. All of the strains were found to contain hylA, plcA, plcB, and actA virulence genes. Among them, hlyA and actA genes are known to cause invasion of host cells and stimulation of cell-to-cell spread, respectively [23]. Internalin genes (inlA, inlC, and inlJ) were detected in 95.1% of the strains. Strains that produce a truncated internalinA are compromised in their virulence [57], implying that the five strains mentioned above would be compromised in virulence if they do not have the gene(s).
Table 3.
Virulence gene contents of isolated L. monocytogenes strains
| Virulence gene | No of positives (%) | Serogroup-positive sample no. (%) | ||||
|---|---|---|---|---|---|---|
| 1/2a-3a | 1/2c-3c | 4b-4d-4e | 1/2b-3b-7 | 4a-4c | ||
| hylA | 103 (100) | 78 (100) | 15 (100) | 1 (100) | 6 (100) | 3 (100) |
| inlA | 98(95.1) | 76 (97.4) | 12 (80) | 1 (100) | 6 (100) | 3 (100) |
| inlC | 98(95.1) | 76 (97.4) | 12 (80) | 1 (100) | 6 (100) | 3 (100) |
| inlJ | 98(95.1) | 76 (97.4) | 12 (80) | 1 (100) | 6 (100) | 3 (100) |
| plcA | 103(100) | 78 (100) | 15 (100) | 1 (100) | 6 (100) | 3 (100) |
| plcB | 103(100) | 78 (100) | 15 (100) | 1 (100) | 6 (100) | 3 (100) |
| prfA | 101 (98.0) | 76 (97.4) | 15 (100) | 1 (100) | 6 (100) | 3 (100) |
| mpl | 99 (96.1) | 74 (94.8) | 15 (100) | 1 (100) | 6 (100) | 3 (100) |
| actA | 103 (100) | 78 (100) | 15 (100) | 1 (100) | 6 (100) | 3 (100) |
| dltA | 102 (99.0) | 77 (98.7) | 15 (100) | 1 (100) | 6 (100) | 3 (100) |
| fri | 100 (97.0) | 76 (97.4) | 15 (100) | 1 (100) | 5 (83.3) | 3 (100) |
| flaA | 98 (95.1) | 73 (93.5) | 15 (100) | 1 (100) | 6 (100) | 3 (100) |
The remaining virulence genes were found in the range of 95 to 99%. The prfA gene which controls several virulence genes was detected in 101 strains (98%). Also, prfA and flaA are known as flagella-associated genes that are involved in biofilm formation [56]. The flaA gene was found in the 95% of the poultry strains. Most of the strains (97%) contained the oxidative resistance gene, fri. The stress regulator and oxidation resistance genes are needed for persistence and growth under oxidative stress conditions [16]. High incidence of virulence factor genes has also been detected by other researchers. Du et al. [9] demonstrated that food isolates contained internalin genes (inlA, inlC, and inlJ, 100%), hylA (100%), plcB (100%), actA (90.5%), and plcA genes (76.2%). In another study, all isolates from bovine carcasses were found to carry hylA, plcA, plcB, actA, inlA, inlC, and inlJ genes [58]. The actA gene is not a ubiquitous virulence factor of L. monocytogenes and the prevalence of this gene is variable [9]. It is important to highlight that the strains lacking inlC and inlJ genes are non-pathogenic [37]. It is noteworthy that the variations between the prevalence of virulence genes in different studies can be related to the use of several PCR target fragments within the genes [9].
Since antibiotic resistance is a worldwide public health concern [59], it is crucial to monitor the effect that antibiotic usage has on foodborne pathogens in order to understand resistance patterns. There is restricted information about the antibiotic resistance of L. monocytogenes strains isolated from chicken samples [44]. Ampicillin is generally combined with other antibiotics for the treatment of listeriosis. Sulfamethoxazole/trimethoprim has also been used as an alternative to ampicillin in the case of allergy [60]. Therefore, ampicillin (2 μg), erythromycin (15 μg), meropenem (10 μg), sulfamethoxazole/trimethoprim (25 μg), and penicillin G (1 U) were tested to detect antibiotic susceptibility of the isolates. Five tested antibiotics of L. monocytogenes isolates were susceptible (93.2%). These results were in agreement with Amajoud et al. [51] results. The presence of antibiotic resistance in L. monocytogenes isolates originated from food samples is low [58]. In fact, all samples from slaughterhouses (n = 57) were susceptible to the five antibiotics tested. Resistance was observed among strains obtained only from wing and drumstick samples from retail markets. Resistance to sulfamethoxazole/trimethoprim, penicillin G, and erythromycin was found among 5%, 3%, and 2% of these strains, respectively. Similarly, 2% of L. monocytogenes strains were found resistant to trimethoprim or tetracycline/trimethoprim, tetracycline, and erythromycin [61]. Sugiri et al. [62] also indicated that 17.2%, 6.9%, 6.9%, and 3.4% of the strains isolated from chicken carcasses were resistant to penicillin, ampicillin, erythromycin, and ampicillin/penicillin combination, respectively.
Antimicrobial resistance is thought to be related to inappropriate and overdose use of antibiotics in animals and humans [61, 63]. The widespread use of antibiotics has resulted in the improvement of antibiotic-resistant bacteria. In most countries, antibiotic resistance of L. monocytogenes is not prevalent, although in some countries such as Iran and India, it is seen relatively high [64–66]. Therefore, monitoring antibiotic resistance in L. monocytogenes is important. The use of antimicrobials as growth promoters in poultry farming is a reason for the presence of resistant bacterial strains in chicken meat [44]. According to the Organization for Economic Co-operation and Development (OECD), antibiotic-resistant bacteria problem may also occur in Turkey, Korea, and Greece due to the high antibiotic usage [67]. Also, it was concluded that there was an increase in the prevalence of antibiotic-resistant bacteria isolated between 2000 and 2017. However, it is difficult to compare the results obtained in previous studies with the current study because of the use of different methods, antibiotic types, and concentrations tested. In a recent study, 21 food isolates were found resistant to cefoxitin (100%), cefuroxime (85.7%), ampicillin (81%), clindamycin (52.4%), and cefotaxime (47.7%) by agar disk diffusion [9]. They also observed MDR to 3–8 of the 11 analyzed antibiotics. Amajoud et al. [51] demonstrated that food strains were resistant to sulfonamide, cefotaxime, fosfomycine, lincosamide, and nalidixic acid but they were susceptible to other tested antibiotics such as streptomycin, trimethoprim, penicillin G, chloramphenicol, vancomycin, rifampicin, kanamycin, fusidic acid, levofloxacin, ciprofloxacin, ampicillin, erythromycin, moxifloxacin, amikacin, tobramycin amoxicillin, gentamicin, and imipenem. In another study, only 0.6% of 351 food originated strains showed antibiotic resistance to eight tested antibiotics (erythromycin, ampicillin, chloramphenicol, tetracycline, vancomycin, streptomycin, penicillin G, and gentamicin) [68]. MDR in L. monocytogenes isolates was not observed. Threlfall et al. [69] indicated that 0.02% of L. monocytogenes strains originated from humans and food were resistant to erythromycin, trimethoprim, ciprofloxacin, and tetracycline. Gómez et al. [70] found that 34.5% of the isolates from meat-processing area and meat products showed resistance to one or two antibiotics. On the other hand, 2.9% of their isolates were classified as MDR. Ayaz and Erol [71] isolated L. monocytogenes from ground turkey samples in Turkey, 63 (80.8%) strains were resistant to penicillin G, and 53 (67.9%) strains were resistant to ampicillin. Similar to the results of this study, they also found that all the isolates tested were sensitive to the antimicrobials used for the treatment of listeriosis.
Similar to another report [72], antibiotic resistance in L. monocytogenes strains was found at low levels in this study. MDR isolates were not found among L. monocytogenes strains. The resistant strains (6.8%) were subjected to additional tests to determine MIC values for these antibiotics. These strains belonged to serogroup IIa (n = 2), IIb (n = 1), and IIc (n = 4). Five strains belonging to serogroups of IIb, IIa, and IIc had a higher MIC value for sulfamethoxazole/trimethoprim than the critical value (MICs, 0.16 to 0.32 μg/ml). Three strains were below the breakpoint level and classified as susceptible against penicillin G. On the other hand, only two strains within serogroups of IIa and IIb had a MIC of 0.12 μg/ml for erythromycin.
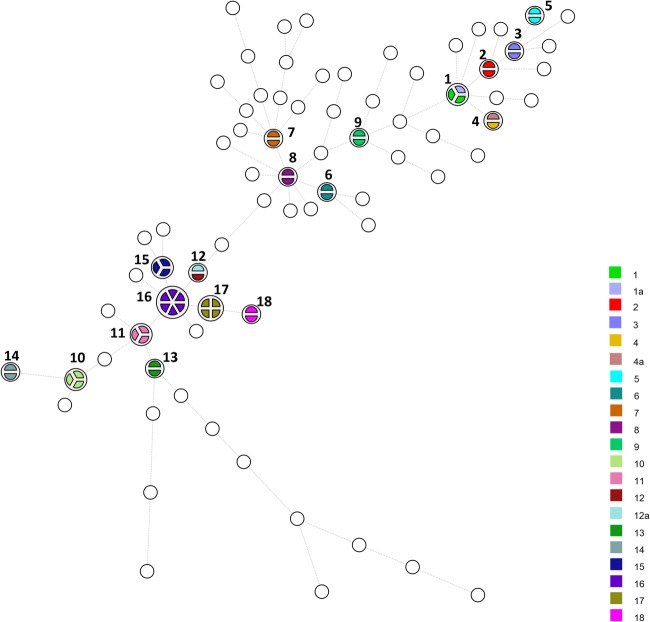
PFGE, which has a high discriminatory power [2], has been considered as a “gold standard” method, especially for tracking the source of contamination, although in some jurisdictions, whole genome sequencing is replacing it as the gold standard, particularly for disease epidemic investigation [73]. The strains were divided into 18 pulsotypes using AscI and ApaI when grouped at a similarity of > 90% (Figs. 1 and 2). The PFGE profiles showed a great variety among the isolates. Based on the PFGE patterns, 57 pulsotypes were composed of only one strain, while there was more than one strain in the remaining 18 pulsotypes. Pulsotypes 1&1a, 4&4a, and 12&12a were found to contain mixed PCR serogroups (IIa/ IIc or IIa/IVa). The strains within pulsotypes 2, 3, and 5 were in serogroup IIc (Fig. 1). The strains of the remaining pulsotypes belonged to serogroup IIa. The presence of high PFGE diversity among L. monocytogenes strains from chicken slaughterhouses has been investigated in a recent study [44]. After restriction by ApaI, those authors obtained 12 pulsotypes from 38 L. monocytogenes strains isolated from 195 samples of chicken cuts and carcasses sampled from a slaughterhouse in Bahia, Brazil. Based on their results, 11, 2, and 3 pulsotypes were in the serotype 1/2a, serotype 1/2b, and serotype 1/2c, respectively. Amajoud et al. [51] also found 8 different PFGE profiles among 16 L. monocytogenes strains obtained from 1096 food in Tetouan, Morocco, after restriction by AscI and ApaI and then grouped them into three clusters. The strains in cluster I belonged to PCR serogroup IVb and were isolated from traditional whey, pastries, mortadella, and mayonnaise. In cluster II, there were strains in the PCR serogroup IIa isolated from raw milk. In the third cluster, the strains isolated from pastries belonged to PCR serogroup IVb.
Fig. 1.
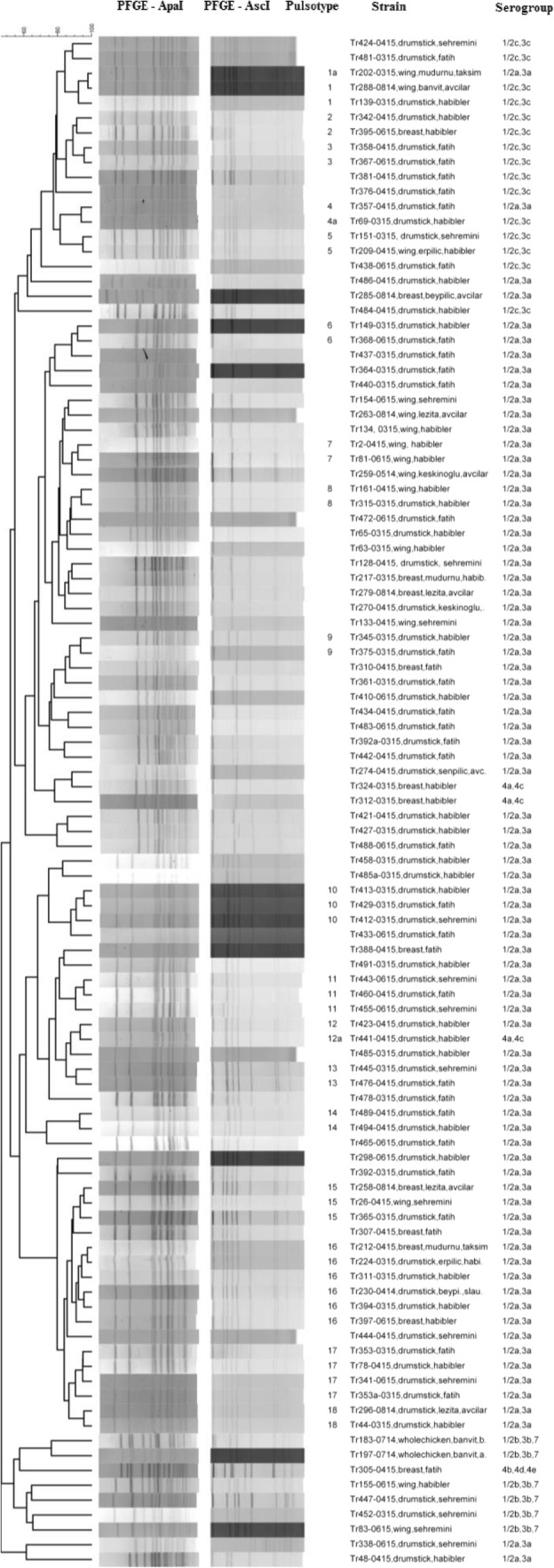
Dendrogram showing PFGE band patterns of 103 L. monocytogenes strains divided into 18 pulsotypes after AscI and ApaI digestion
Fig. 2.
Minimum spanning tree obtained from dendrogram analysis of all the isolates obtained from the survey. The white circles represent pulsotypes with no similarity at < 90%, while the colors indicate pulsotypes with > 90% similarity. Pulsotypes 1, 4, and 12 contain isolates with mixed serogroups
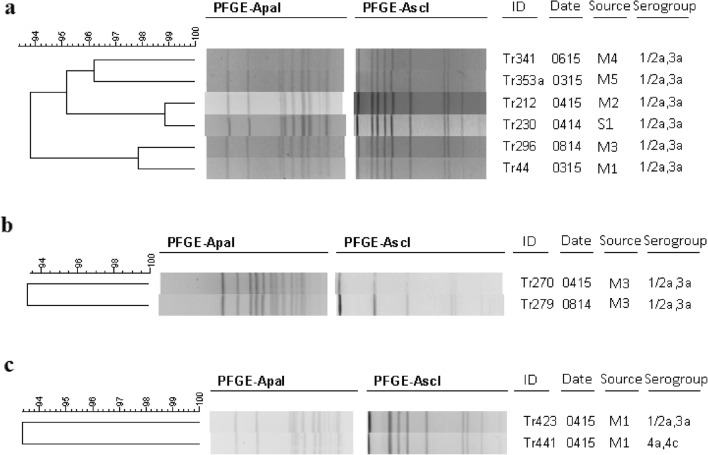
L. monocytogenes strains were considered as widespread when repeated isolation of strains of the same pulsotype at different geographic locations at the same time occurred and persistent when repeated isolation of strains of the same pulsotype at different sampling times occurs (Fig. 3). Some of the pulsotypes contained strains that were obtained from 2 to 4 markets, while one of the pulsotypes contained strains obtained at a slaughterhouse and 2 markets, representing widespread isolates. The presence of the same pulsotype among the strains collected from markets may be an indication of cross-contamination. On the other hand, some of the profiles obtained at the markets were not observed among the strains from the slaughterhouses suggesting that cross-contamination could also occur due to the customers or owners of market stalls.
Fig. 3.
Comparison of some PFGE profiles of different isolates showing the widespread nature of the isolates (a), the persistent nature of isolates (b), and isolates of different serogroups in the same pulsotype (c). Isolation sources: M1, Habibler (Istanbul); M2, Taksim (Istanbul); M3, Avcilar (Istanbul); M4, Sehremini (Istanbul); M5, Fatih (Istanbul); S1, Slaughterhouse A (Bolu)
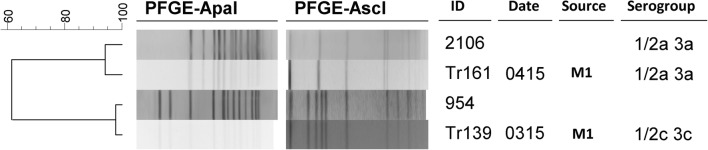
Contamination of the end product during processing may be related to the presence of persistent strains in the processing area [74]. Regarding the persistence, fifteen of the pulsotypes contained strains that were obtained at different sampling times. Their sampling time ranged from 1 to 7 months. After comparison of pulsotypes with the database, all strains from this study were distinguishable from the previous studies conducted in Ireland (n = 553), Romania (n = 55), Greece (n = 20), the Czech Republic (n = 8), the United Kingdom (n = 100), Austria (n = 32), and Australia (n = 200). At > 90% similarity, the isolates were similar to two strains characterized in Ireland (Fig. 4). Madden et al. [75] also showed the presence of 27 pulsotypes among isolates from food-processing facilities in Ireland. Among them, 9 pulsotypes were shared between different processing plants and 9 of them were persistent.
Fig. 4.
Comparison PFGE profiles of L. monocytogenes strains isolated from Turkey (Tr-coded strains) with isolates from Ireland
Conclusions
L. monocytogenes was found in poultry samples collected from three cities in Turkey. Most of the isolated strains belonged to serogroup IIa (lineage II) that are associated with major sporadic infection in humans. Also, the presence of virulence factor genes that are the determinants for the pathogenicity highlights the importance of this pathogen in food-processing environments. According to the results of the PFGE, poultry is a common source for widespread and persistent L. monocytogenes strains. Therefore, it is necessary to apply good hygiene conditions and food-processing technologies to prevent contamination of this foodborne pathogen for public health.
Funding sources
This study was funded by Istanbul University with the project number 56821. Additional fund was obtained from the Scientific and Technological Research Council of Turkey (TUBITAK 2214/A) to support Aysen COBAN on an International Doctoral Research Scholarship Program.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wehner S, Mannala GK, Qing X, Madhugiri R, Chakraborty T, Mraheil MA, Hain T, Marz M. Detection of very long antisense transcripts by whole transcriptome RNA-Seq analysis of Listeria monocytogenes by semiconductor sequencing technology. PLoS One. 2014;6:e108639. doi: 10.1371/journal.pone.0108639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oxaran V, In Lee SH, Chaul LT, Corassin CH, Barancelli GV, Alves VF, de Oliveira CAF, Gram L, De Martinis ECP. Listeria monocytogenes incidence changes and diversity in some Brazilian dairy industries and retail products. Food Microbiol. 2017;68:16–23. doi: 10.1016/j.fm.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 3.EFSA and ECDC Scientific report. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018;16:5500. doi: 10.2903/j.efsa.2018.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doganay M. Listeriosis: clinical presentation. FEMS Immunol Med Microbiol. 2003;35(3):173–175. doi: 10.1016/S0928-8244(02)00467-4. [DOI] [PubMed] [Google Scholar]
- 5.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferreira V, Wiedmann M, Teixeira P, Stasiewicz MJ. Listeria monocytogenes persistence in food-associated environments: epidemiology, strain characteristics, and implications for public health. J Food Prot. 2014;77:150–170. doi: 10.4315/0362-028X.JFP-13-150. [DOI] [PubMed] [Google Scholar]
- 7.Todd ECD, Notermans S. Surveillance of listeriosis and its causative pathogen, Listeria monocytogenes. Food Control. 2011;22:1484–1490. doi: 10.1016/j.foodcont.2010.07.021. [DOI] [Google Scholar]
- 8.Garner D, Kathariou S. Fresh produce-associated listeriosis outbreaks, sources of concern, teachable moments, and insights. J Food Prot. 2016;79:337–344. doi: 10.4315/0362-028X.JFP-15-387. [DOI] [PubMed] [Google Scholar]
- 9.Du X-J, Zhang X, Wang X-Y, Su Y-I, Li P, Wang S. Isolation and characterization of Listeria monocytogenes in Chinese food obtained from the central area of China. Food Control. 2017;74:9–16. doi: 10.1016/j.foodcont.2016.11.024. [DOI] [Google Scholar]
- 10.Soni DK, Singh M, Singh DV, Dubey SK. Virulence and genotypic characterization of Listeria monocytogenes isolated from vegetable and soil samples. BMC Microbiol. 2014;14:241. doi: 10.1186/s12866-014-0241-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen KJ, Walecka-Zacharska E, Chen JC, Katarzyna KP, Devlieghere F, Van Meervenne E, OSek J, Wieczorek K, Bania J. Listeria monocytogenes: an examination of food chain factors potentially contributing to antimicrobial resistance. Food Microbiol. 2016;54:178–189. doi: 10.1016/j.fm.2014.08.006. [DOI] [Google Scholar]
- 12.Sakaridis I, Soultos N, Iossifidou E, Papa A, Ambrosiadis I, Koidis P. Prevalence and antimicrobial resistance of Listeria monocytogenes isolated in chicken slaughterhouses in northern Greece. J Food Prot. 2011;74:1017–1021. doi: 10.4315/0362-028X.JFP-10-545. [DOI] [PubMed] [Google Scholar]
- 13.EFSA and ECDC The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2012. EFSA J. 2014;12:3547. doi: 10.2903/j.efsa.2014.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burall LS, Simpson AC, Datta AR. Evaluation of a serotyping scheme using a combination of an antibody-based serogrouping method and a multiplex PCR assay for identifying the major serotypes of Listeria monocytogenes. J Food Prot. 2011;74:403–409. doi: 10.4315/0362-028X. [DOI] [PubMed] [Google Scholar]
- 15.Chen JQ, Regan P, Laksanalamai P, Healey S, Hu Z. Prevalence and methodologies for detection, characterization and subtyping of Listeria monocytogenes and L. ivanovii in foods and environmental sources. Food Sci Human Wellness. 2017;6:97–120. doi: 10.1016/j.fshw.2017.06.002. [DOI] [Google Scholar]
- 16.Huang Y, Morvay AA, Shi X, Suo Y, Shi C, Knøchel S. Comparison of oxidative stress response and biofilm formation of Listeria monocytogenes serotypes 4b and 1/2a. Food Control. 2018;85:416–422. doi: 10.1016/j.foodcont.2017.10.007. [DOI] [Google Scholar]
- 17.Orsi RH, den Bakker HC, Wiedmann M. Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int J Med Microbiol. 2011;301:79–96. doi: 10.1016/j.ijmm.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Cocolin L, Stella S, Nappi R, Bozzetta E, Cantoni C, Comi G. Analysis of PCR-based methods for characterization of Listeria monocytogenes strains isolated from different sources. Int J Food Microbiol. 2005;103:167–178. doi: 10.1016/j.ijfoodmicro.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 19.Graves LM, Swaminathan B. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int J Food Microbiol. 2001;65:55–62. doi: 10.1016/S0168-1605(00)00501-8. [DOI] [PubMed] [Google Scholar]
- 20.Parisi A, Latorre L, Normanno G, Miccolupo A, Fraccalvieri R, Lorusso V, Santagada G. Amplified fragment length polymorphism and multi-locus sequence typing for high-resolution genotyping of Listeria monocytogenes from foods and the environment. Food Microbiol. 2010;27:101–108. doi: 10.1016/j.fm.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Cao X, Wang Y, Wang Y, Ye C. Isolation and characterization of Listeria monocytogenes from the black-headed gull feces in Kunming, China. J Infect Public Health. 2018;11:59–63. doi: 10.1016/j.jiph.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Dussurget O, Pizzaro-Cerda J, Cossart P. Molecular determinants of Listeria monocytogenes virulence. Annu Rev Microbiol. 2004;58:587–610. doi: 10.1146/annurev.micro.57.030502.090934. [DOI] [PubMed] [Google Scholar]
- 23.Ye K, Zhang X, Huang Y, Liu J, Liu M, Zhou G. Bacteriocinogenic Enterococcus faecium inhibits the virulence property of Listeria monocytogenes. LWT Food Sci Technol. 2018;89:87–92. doi: 10.1016/j.lwt.2017.10.028. [DOI] [Google Scholar]
- 24.Dussurget O, Cabanes D, Dehoux P, Lecuit M, Buchrieser C, Glaser P, Cossart P, European Listeria Genome Consortium Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol Microbiol. 2002;45:1095–1106. doi: 10.1046/j.1365-2958.2002.03080.x. [DOI] [PubMed] [Google Scholar]
- 25.Bonazzi M, Lecuit M, Cossart P. Listeria monocytogenes internalin and E-cadherin: from bench to bedside. Cold Spring Harb Perspect Biol. 2009;1:a003087. doi: 10.1101/cshperspect.a003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milohanic E, Glaser P, Coppée JY, Frangeul L, Vega Y, Vázquez-Boland JA, Kunst F, Cossart P, Buchrieser C. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol Microbiol. 2003;47:1613–1625. doi: 10.1046/j.1365-2958.2003.03413.x. [DOI] [PubMed] [Google Scholar]
- 27.Soni DK, Ghosh A, Chikara SK, Singh KM, Joshic C, Dubey SK. Comparative whole genome analysis of Listeria monocytogenes 4b strains reveals least genome diversification irrespective of their niche specificity. Gene Reports. 2017;8:61–68. doi: 10.1016/j.genrep.2017.05.007. [DOI] [Google Scholar]
- 28.ISO (2004) Microbiology of food animal feeding stuffs: Horizontal method for the detection and enumeration of Listeria monocytogenes. Part 1: detection method Amendment 1: modification of isolation media and haemolysis test, and inclusion of precision data. ISO 11290-1/A1, Geneva, Switzerland
- 29.Liu D, Ainsworth AJ, Austin FW, Lawrence ML. Use of PCR primers derived from a putative transcriptional regulator gene for species-specific determination of Listeria monocytogenes. Int J Food Microbiol. 2004;91:297–304. doi: 10.1016/j.ijfoodmicro.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 30.Bubert A, Köhler S, Goebel W. The homologous and heterologous regions within the iap gene allow genus- and species-specific identification of Listeria spp. by polymerase chain reaction. Appl Environ Microbiol. 1992;58:2625–2632. doi: 10.1128/AEM.58.8.2625-2632.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bubert A, Sokolovic Z, Chun SK, Papatheodorou L, Simm A, Goebel W. Differential expression of Listeria monocytogenes virulence genes in mammalian host cells. Mol Gen Genet. 1999;261:323–336. doi: 10.1007/pl00008633. [DOI] [PubMed] [Google Scholar]
- 32.Doumith M, Buchrieser C, Glaser P, Jacquet C, Martin P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J Clin Microbiol. 2004;42:3819–3822. doi: 10.1128/JCM.42.8.3819-3822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.EUCAST (2016) European committe on antimicrobial susceptibility testing. Breakpoints tables for interpretation of MICs and zone diameters, Version 6.0. Retrieved from http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_6.0_Breakpoint_table.pdf. Accessed 26 January 2019
- 34.Furrer B, Candrian U, Hoefelein C, Luethy J. Detection and identification of Listeria monocytogenes in cooked sausage products and in milk by in vitro amplification of haemolysin gene fragments. J Appl Microbiol. 1991;70:372–379. doi: 10.1111/j.1365-2672.1991.tb02951.x. [DOI] [PubMed] [Google Scholar]
- 35.Mengaud J, Vicente MF, Cossart P. Transcriptional mapping and nucleotide sequence of the Listeria monocytogenes hlyA region reveal structural features that may be involved in regulation. Infect Immun. 1989;57:3695–3701. doi: 10.1128/IAI.57.12.3695-3701.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishibori T, Cooray K, Xiong H, Kawamuro I, Fujita M, Mitsuyama M. Correlation between the presence of virulence-associated genes as determined by PCR and actual virulence to mice in various strains of Listeria spp. Microbiol Immunol. 1995;39:343–349. doi: 10.1111/j.1348-0421.1995.tb02211.x. [DOI] [PubMed] [Google Scholar]
- 37.Liu D, Lawrence ML, Austin FW, Ainsworth AJ. A multiplex PCR for species- and virulence-specific determination of Listeria monocytogenes. J Microbiol Methods. 2007;71:133–140. doi: 10.1016/j.mimet.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 38.Slama RB, Miladi H, Chaieb K, Bakhrouf A. Survival of Listeria monocytogenes cells and the effect of extended frozen storage (−20 °C) on the expression of its virulence gene. Appl Biochem Biotechnol. 2013;170:1174–1183. doi: 10.1007/s12010-013-0253-8. [DOI] [PubMed] [Google Scholar]
- 39.Jaradat ZW, Schutze GE, Bhunia AK. Genetic homogeneity among Listeria monocytogenes strains from infected patients and meat products from two geographic locations determined by phenotyping, ribotyping and PCR analysis of virulence genes. Int J Food Microbiol. 2002;76:1–10. doi: 10.1016/S0168-1605(02)00050-8. [DOI] [PubMed] [Google Scholar]
- 40.Leimeister-Wächter M, Domann E, Chakraborty T. Detection of a gene encoding a phosphatidylinositol-specific phospholipase C that is coordinately expressed with listeriolysin in Listeria monocytogenes. Mol Microbiol. 1991;5:361–366. doi: 10.1111/j.1365-2958.1991.tb02117.x. [DOI] [PubMed] [Google Scholar]
- 41.Kyoui D, Takahashi H, Miya S, Kuda T, Kimura B. Comparison of the major virulence-related genes of Listeria monocytogenes in Internalin A truncated strain 36-25-1 and a clinical wild-type strain. BMC Microbiol. 2014;14:15. doi: 10.1186/1471-2180-14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Promadej N, Fiedler F, Cossart P, Dramsi S, Kathariou S. Cell wall teichoic acid glycosylation in Listeria monocytogenes serotype 4b requires gtcA, a novel, serogroup-specific gene. J Bacteriol. 1999;181:418–425. doi: 10.1128/JB.181.2.418-425.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vazquez-Boland JA, Kochs C, Dramsi S, Ohayaon H, Geoffroy C, Mengaud J, Cossart P. Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect Immun. 1992;60:219–230. doi: 10.1128/IAI.60.1.219-230.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliveira TS, Varjão LM, Silva LNN, Pereira RCL, Hofer E, Vallim DC, Almeida RCC. Listeria monocytogenes at chicken slaughterhouse: occurrence, genetic relationship among isolates and evaluation of antimicrobial susceptibility. Food Control. 2018;88:131–138. doi: 10.1016/j.foodcont.2018.01.015. [DOI] [Google Scholar]
- 45.Khallaf M, Ameur N, Boraam F, Senouci S, Ennaji MM. Prevalence of Listeria monocytogenes isolated from chicken meat marketed in Rabat, Morocco. Int J Innov Sci Res. 2016;22:8–13. [Google Scholar]
- 46.Bouayad L, Hamdi TM, Naim M, Leclercq A, Lecuit M. Prevalence of Listeria spp. and molecular characterization of Listeria monocytogenes isolates from broilers at the abattoir. Foodborne Pathog Dis. 2015;12:606–611. doi: 10.1089/fpd.2014.1904. [DOI] [PubMed] [Google Scholar]
- 47.Dan SD, Tabaran A, Mihaiu L, Mihaiu M. Antibiotic susceptibility and prevalence of foodborne pathogens in poultry meat in Romania. J Infect Dev Ctries. 2015;9:35–41. doi: 10.3855/jidc.4958. [DOI] [PubMed] [Google Scholar]
- 48.Kahraman T, Aydin A. Prevalence of Salmonella spp., Escherichia coli O157:H7 and Listeria monocytogenes in meat and meat products in Turkey. Arch Leb. 2009;60:6–11. doi: 10.2376/0003-925X-60-6. [DOI] [Google Scholar]
- 49.Elmali M, Can HY, Yaman H. Prevalence of Listeria monocytogenes in poultry meat. Food Sci Technol. 2015;35:672–675. doi: 10.1590/1678-457X.6808. [DOI] [Google Scholar]
- 50.Ristori CA, Rowlands RE, Martins CG, Barbosa ML, Yoshida JT, Franco BD. Prevalence and populations of Listeria monocytogenes in meat products retailed in Sao Paulo, Brazil. Foodborne Pathog Dis. 2014;11:969–973. doi: 10.1089/fpd.2014.1809. [DOI] [PubMed] [Google Scholar]
- 51.Amajoud N, Lecrerq A, Soriano JM, Bracq-Dieve H, El Maadoudi M, Senhaji NS, Kounnoun A, Moura A, Lecuit M, Abrini J. Prevalence of Listeria spp. and characterization of Listeria monocytogenes isolated from food products in Tetouan, Morocco. Food Control. 2018;84:436–441. doi: 10.1016/j.foodcont.2017.08.023. [DOI] [Google Scholar]
- 52.Vongkamjan K, Benjakul S, Vu HTK, Vuddhakul V. Longitudinal monitoring of Listeria monocytogenes and Listeria phages in seafood processing environments in Thailand. Food Microbiol. 2017;66:11–19. doi: 10.1016/j.fm.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 53.Leong D, Alvarez-Ordóňez A, Jordon K. Monitoring occurrence and persistence of Listeria monocytogenes in foods and food processing environments in the Republic of Ireland. Front Microbiol. 2014;5:436. doi: 10.3389/fmicb.2014.00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murugesan L, Kucerova Z, Knabel SJ, LaBorde LF. Predominance and distribution of a persistent Listeria monocytogenes clone in a commercial fresh mushroom processing environment. J Food Prot. 2015;78:1988–1998. doi: 10.4315/0362-028X. [DOI] [PubMed] [Google Scholar]
- 55.Maury MM, Tsai YH, Charlier C, Touchon M, Chenal-Francisque V, Leclercq A, Criscuolo A, Gaultier C, Roussel S, Brisabois A, Disson O, Rocha EPC, Brisse S, Lecuit M. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat Genet. 2016;48:308–313. doi: 10.1038/ng.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li R, Du W, Yang J, Liu Z, Yousef AE. Control of Listeria monocytogenes biofilm by paenibacterin, a natural antimicrobial lipopeptide. Food Control. 2018;84:529–535. doi: 10.1016/j.foodcont.2017.08.031. [DOI] [Google Scholar]
- 57.Hilliard A, Leong D, O’Callaghan A, Culligan EP, Morgan CA, De Lappe N, Hill C, Jordan K, Cormican M, Gahan CGM. Genomic characterization of Listeria monocytogenes isolates associated with clinical listeriosis and the food production environment in Ireland. Genes. 2018;9:171. doi: 10.3390/genes9030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iglesias MA, Kroning IS, Decol LT, de Melo Franco BDG, Silva WP. Occurrence and phenotypic and molecular characterization of Listeria monocytogenes and Salmonella spp. in slaughterhouses in southern Brazil. Food Res Int. 2017;100:96–101. doi: 10.1016/j.foodres.2017.06.023. [DOI] [PubMed] [Google Scholar]
- 59.Doménech E, Jimenez-Belenguer A, Amoros JA, Ferrus MA, Escriche I. Prevalence and antimicrobial resistance of Listeria monocytogenes and Salmonella strains isolated in ready-to-eat foods in eastern Spain. Food Control. 2015;47:120–125. doi: 10.1016/j.foodcont.2014.06.043. [DOI] [Google Scholar]
- 60.Hof H. An update on the medical management of listeriosis. Expert Opin Pharmacother. 2004;5:1727–1735. doi: 10.1517/14656566.5.8.1727. [DOI] [PubMed] [Google Scholar]
- 61.Granier SA, Moubareck C, Colaneri C, Lemire A, Roussel S, Dao TT, Courvalin P, Brisabois A. Antimicrobial resistance of Listeria monocytogenes isolates from food and the environment in France over a 10-year period. Appl Environ Microbiol. 2011;77:2788–2790. doi: 10.1128/AEM.01381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sugiri YD, Gölz G, Meeyam T, Baumann MP, Kleer J, Chaisowwong W, Alter T. Prevalence and antimicrobial susceptibility of Listeria monocytogenes on chicken carcasses in Bandung, Indonesia. J Food Prot. 2014;77:1407–1410. doi: 10.4315/0362-028X.JFP-13-453. [DOI] [PubMed] [Google Scholar]
- 63.European Commission (2011) Commission recommendation on the research joint programming initiative ‘The microbial challenge-an emerging threat to human health.’ Brussels, Retrieved from https://era.gv.at/object/document/677/attach/CommRec_DOCUMENTDETRAVAIL_f.pdf. Accessed 27 January 2019
- 64.Akrami-Mohajeri F, Derakhshan Z, Ferrante M, Hamidiyan N, Soleymani M, Conti GO, Tafti RD. The prevalence and antimicrobial resistance of Listeria spp. in raw milk and traditional dairy products delivered in Yazd, Central Iran (2016) Food Chem Toxicol. 2018;114:141–144. doi: 10.1016/j.fct.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 65.Rezai R, Ahmadi E, Salimi B. Prevalence and antimicrobial resistance profile of Listeria species isolated from farmed and on-sale rainbow trout (Oncorhynchus mykiss) in Western Iran. J Food Prot. 2018;81:886–891. doi: 10.4315/0362-028X.JFP-17-428. [DOI] [PubMed] [Google Scholar]
- 66.Sharma S, Sharma V, Dahiya DK, Khan A, Mathur M, Sharma A. Prevalence, virulence potential, and antibiotic susceptibility profile of Listeria monocytogenes isolated from bovine raw milk samples obtained from Rajasthan, India. Foodborne Pathog Dis. 2017;14:132–140. doi: 10.1089/fpd.2016.2118. [DOI] [PubMed] [Google Scholar]
- 67.OECD (2018) Stopping antimicrobial resistance would cost just USD 2 per person a year. https://www.oecd.org/newsroom/stopping-antimicrobial-resistance-would-cost-just-usd-2-per-person-a-year.htm. Accessed 13 February 2019
- 68.Walsh D, Duffy G, Sheridan JJ, Blair IS, McDowell DA. Antibiotic resistance among Listeria, including Listeria monocytogenes, in retail foods. J Appl Microbiol. 2001;90:517–522. doi: 10.1046/j.1365-2672.2001.01273.x. [DOI] [PubMed] [Google Scholar]
- 69.Threlfall EJ, Skinner JA, McLauchlin J. Antimicrobial resistance in Listeria monocytogenes from humans and food in the UK, 1967-96. Clin Microbiol Infect. 1998;4:410–412. doi: 10.1111/j.1469-0691.1998.tb00087.x. [DOI] [PubMed] [Google Scholar]
- 70.Gómez D, Azon E, Marco N, Carraminana JJ, Rota C, Ariňo A, Yangüela J. Antimicrobial resistance of Listeria monocytogenes and Listeria innocua from meat products and meat-processing environment. Food Microbiol. 2014;42:61–65. doi: 10.1016/j.fm.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 71.Ayaz ND, Erol I. Relation between serotype distribution and antibiotic resistance profiles of Listeria monocytogenes isolated from ground Turkey. J Food Prot. 2010;73:967–972. doi: 10.4315/0362-028X-73.5.967. [DOI] [PubMed] [Google Scholar]
- 72.Byrne VV, Hofer E, Vallim DC, de Castro Almeida RC. Occurrence and antimicrobial resistance patterns of Listeria monocytogenes isolated from vegetables. Braz J Microbiol. 2016;47:438–443. doi: 10.1016/j.bjm.2015.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Popovich KJ, Snitkin ES. Whole genome sequencing-implications for infection prevention and outbreak investigations. Curr Infect Dis Rep. 2017;19:15. doi: 10.1007/s11908-017-0570-0. [DOI] [PubMed] [Google Scholar]
- 74.Mendonça KS, Michael GB, von Laer AE, Menezes DB, Cardoso MRI, Silva WP. Genetic relatedness among Listeria monocytogenes isolated in foods and food production chain in southern Rio Grande do Sul, Brazil. Food Control. 2012;28:171–177. doi: 10.1016/j.foodcont.2012.04.014. [DOI] [Google Scholar]
- 75.Madden RH, Hutchison M, Jordan K, Pennone V, Gundogdu O, Corcionivoschi N. Prevalence and persistence of Listeria monocytogenes in premises and products of small food business operators in Northern Ireland. Food Control. 2018;87:70–78. doi: 10.1016/j.foodcont.2017.12.020. [DOI] [Google Scholar]