Abstract
Coronaviruses are single-stranded positive-sense RNA viruses associated with important avian diseases. Their relatively high rates of mutation and recombination frequencies allow them to adapt to new hosts and ecological niches. Although Brazil has 18% of global avian species diversity, studies regarding the presence of avian viral diseases in wild birds in South America are scarce. In this study, we performed a retrospective analysis of the presence of CoVs in 746 wild birds. Oropharyngeal and cloacal swabs were obtained and placed together in vials containing VTM transport medium collected in different regions of Brazil between 2006 and 2013. Screening for viral nucleic acid was performed using conventional RT-PCR and pancoronavirus nested PCR. Positive samples were characterized by partial sequencing of the RNA-dependent RNA polymerase (RdRp) gene, and ensuing phylogenetic analysis was performed to investigate the association between virus epidemiology and bird migration routes. Coronavirus RNA were detected and sequenced from six samples, in which three were related to gammacoronaviruses group and the other three to deltacoronavirus group. Our study documents the presence of CoVs related to avian gamma- and deltacoronaviruses circulating in both urban- and poultry-farm regions of Brazil, implicating wild birds as potential carriers of CoVs which may represent a risk to poultry farms and public health in Brazil.
Electronic supplementary material
The online version of this article (10.1007/s42770-019-00065-7) contains supplementary material, which is available to authorized users.
Keywords: Gammacoronavirus, Deltacoronavirus, Wild birds, Brazil, South America
Introduction
Coronaviruses are RNA single-stranded positive polarity virus associated with important disease in avian poultry [1]. Infectious bronchitis virus (IBV), known as coronavirus of chickens, causes important economic losses to the poultry industry. Studies about the presence of CoVs in wild birds, especially IBV, in South America are scarce [2]. Coronaviruses (CoVs) possess the largest genomes among RNA viruses and have relatively high mutation rates and recombination frequencies when compared with other single-strand RNA viruses, which may allow them to adapt to new hosts and ecological niches [3–5]. These characteristics are exemplified by the two emerged highly pathogenic betacoronaviruses, CoV-SARS (severe acute respiratory syndrome) and the CoV-MERS (Middle East respiratory syndrome) [6].
Even though avian coronaviruses with zoonotic potential have not been reported yet, many species of wild and domestic birds can carry CoVs responsible for substantial economic loss within the poultry industry such as the gammacoronavirus infectious bronchitis virus (IBV). This virus affects the gastrointestinal, respiratory, urinary, and reproductive systems and may lead to mortality rates as high as 30% and sudden drops in egg production [7]. Moreover, many novel CoVs genetically distinct from IBV have been recently identified in different families of wild and domestic birds and mammals but their potential economic losses remain unknown. Recently, Durães-Carvalho et al. (2015), described the detection of delta- and betacoronaviruses in captive wild birds, from a rehabilitation center, originally captured in São Paulo and Mato Grosso do Sul state with close evolutionary relationships with CoVs isolated from mammals [8].
Wild migratory birds may facilitate the spread of viruses, as demonstrated by the surveillance of avian influenza virus [9, 10] and West Nile virus [11–13], but the circulation of CoV in avian migratory routes in South America, more specifically in Brazil, still need to be elucidated. The potential diversity of CoV in Brazilian bird fauna as consequence of the bird species diversity (18%), together with the large number of endangered species and the high intensity of poultry farms in Brazil, indicates that birds from Brazilian biomes are important subjects for CoV surveillance [2, 14].
In this study, we performed a retrospective analysis, molecular detection, and phylogenetic analysis of avian coronaviruses using oropharyngeal and cloacal swab samples from wild and domestic birds collected between 2006 and 2013 in different regions of Brazil to investigate the association between virus epidemiology, bird migration routes, and the proximity of urban areas and poultry farms.
Materials and methods
Sample collection
In total, 746 samples (Table 1) of birds captured between 2006 and 2013 in five expeditions to north, northeast, midwest, south, and southeast Brazil (Fig. 1) were analyzed. For selection, the migratory routes were investigated through a map analysis and experience of ornithologists [15]. The samples were collected by field team of the Laboratory Clinical and Molecular Virology, University of São Paulo (USP), São Paulo, Brazil, with support of the Wildlife Department of São Paulo (DEPAVE); University of Vale do Rio dos Sinos (UNISINOS); University of Pernambuco (UFRPE); National Center for Research and Conservation of Wild Birds (CEMAVE); Laboratory of Bird Ecology, Institute of Biosciences (UFMT); and Laboratory of Virology and Rickettsioses, Veterinary Hospital (UFMT). Samples were collected from wild birds after capture with mist nets, strategically placed between 30 and 250 cm of the ground, and by hand nets on an open field in public parks. The field works were made according to the permission of the Brazilian Institute of Environment and Renewable Natural Resources (IBAMA) and National Center for Research and Conservation of Wild Birds (CEMAVE) developed and followed protocols approved by the Ethics Committee on Animal Experimentation of the Institute of Biomedical Sciences, University of São Paulo. Sampling from oropharyngeal and cloacal swabs was obtained and placed together in vials containing VTM transport medium [16]. All birds were then released after sampling.
Table 1.
Surveillance of Coronavirus in resident and wild birds in Brazil by molecular tests. Bold represents positive samples
| Site | Date | Scientific name | Sample* | No. pos./No. birds |
|---|---|---|---|---|
| Ilha de Marajó, PA | Jul. 2006 | Cairina mochata | C and O | 0/42 |
| Gallus gallus | C and O | 0/36 | ||
| Meliagres gallopavo | C and O | 0/15 | ||
| Ajuruteua, PA | Nov.2006 | Actitis macularius | C and O | 0/5 |
| Calidris pusilla | C and O | 0/2 | ||
| Charadrius semipalmatus | C and O | 0/1 | ||
| Dendrocygna autumnalis | C and O | 0/18 | ||
| Arenaria interpres | C and O | 0/9 | ||
| Dendrocygna viduata | C and O | 0/56 | ||
| Ilha de Canelas, PA | Nov. 2006 | Actitis macularius | C and O | 0/10 |
| Calidris canutus | C and O | 0/1 | ||
| Calidris pusilla | C and O | 0/3 | ||
| Calidris minutilla | C and O | 0/1 | ||
| Sterna hirundo | C and O | 0/4 | ||
| Larus sp. | C and O | 0/1 | ||
| Nov. 2008 | Actitis macularius | C and O | 0/31 | |
| Arenaria interpres | C and O | 0/22 | ||
| Calidris alba | C and O | 0/2 | ||
| Limnodromus griseus | C and O | 0/9 | ||
| Tringa melanoleuca | C and O | 0/1 | ||
| Charadrius semipalmatus | C and O | 0/1 | ||
| Gelochelidon nilotica | C and O | 0/1 | ||
| Bahia de São Marco, MA | Apr. 2007 | Dendrocygna viduata | C and O | 0/27 |
| Netta erythrophthalma | C and O | 0/2 | ||
| Dendrocygna autumnalis | C and O | 0/48 | ||
| Gallinula chloropus | C and O | 0/2 | ||
| Amazonetta brasiliensis | C and O | 0/14 | ||
| Anas bahamensis | C and O | 0/13 | ||
| Meliagres gallopavo | C and O | 0/1 | ||
| Lagoa do Peixe National Park, RS | Nov. 2009 | Rynchops niger | C and O | 1/63 |
| Calidris fuscicolis | C and O | 1/63 | ||
| Haematopus palliatus | C and O | 0/6 | ||
| Himantopus melanurus | C and O | 0/2 | ||
| Pluvialis dominica | C and O | 0/7 | ||
| Arenaria interpres | C and O | 0/4 | ||
| Calidris canutus | C and O | 0/3 | ||
| Calidris alba | C and O | 1/6 | ||
| Calidris pusilla | C and O | 0/1 | ||
| Nycticriphes semicollaris | C and O | 0/1 | ||
| Fulica leucoptera | C and O | 0/1 | ||
| Puffinus gravis | C and O | 0/1 | ||
| Puffinus puffinus | C and O | 0/3 | ||
| Tringa flavipes | C and O | 0/2 | ||
| Sterna hirundo | C and O | 0/15 | ||
| Phalacrocorax brasilianus | C and O | 0/2 | ||
| Vanellus chilensis | C and O | 0/1 | ||
| Elaenia mesoleuca | C and O | 0/1 | ||
| Pluvialis squatarola | C and O | 0/1 | ||
| Tryngites subruficollis | C and O | 0/7 | ||
| Calidris melanotus | C and O | 0/1 | ||
| Sterna vitatta | C and O | 0/1 | ||
| São José do Egito Caatinga, PE | Oct. 2010 | Thaminophilus capstratus | C and O | 0/8 |
| Hylophilus amaurocephalus | C and O | 0/10 | ||
| Sarkesphorus cristapus | C and O | 0/8 | ||
| Hemitricus margaritaseiventer | C and O | 0/4 | ||
| Coryphospingus pileatus | C and O | 0/10 | ||
| Anopetia gounellei | C and O | 0/1 | ||
| Casiornis fuscus | C and O | 0/1 | ||
| Gyolophylox hellmayri | C and O | 0/3 | ||
| Tolmyias flaviventris | C and O | 0/2 | ||
| Heliomaster squamosus | C and O | 0/2 | ||
| Cychlars gujanensis | C and O | 0/1 | ||
| Cyanoloxia brissoni | C and O | 0/1 | ||
| Formicivora melanogastrer | C and O | 0/8 | ||
| Cantorchilus longirostris | C and O | 0/1 | ||
| Chlorostilbon lucidus | C and O | 0/1 | ||
| Coereba flaveola | C and O | 0/1 | ||
| Polioptila plumbea | C and O | 0/1 | ||
| Synalax frontalis | C and O | 0/1 | ||
| Pantanal Poconé, MT | Nov. 2012 | Ramphocelus carbo | C and O | 0/10 |
| Hemitriccus margaritaceiventer | C and O | 0/6 | ||
| Cnemotriccus fuscatus | C and O | 0/11 | ||
| Paroaria capitata | C and O | 0/2 | ||
| Picumnus albosquamatus | C and O | 0/3 | ||
| Gallus gallus | C and O | 0/9 | ||
| Furnarius rufus | C and O | 0/8 | ||
| Myiarchus swainsonii | C and O | 0/2 | ||
| Ibirapuera Park, SP | Jun. 2013 | Anser cygnoides | C and O | 3/81 |
| Total | 63 species | 6/746 |
*Sample type: C = cloacal swabs and O = oral swabs
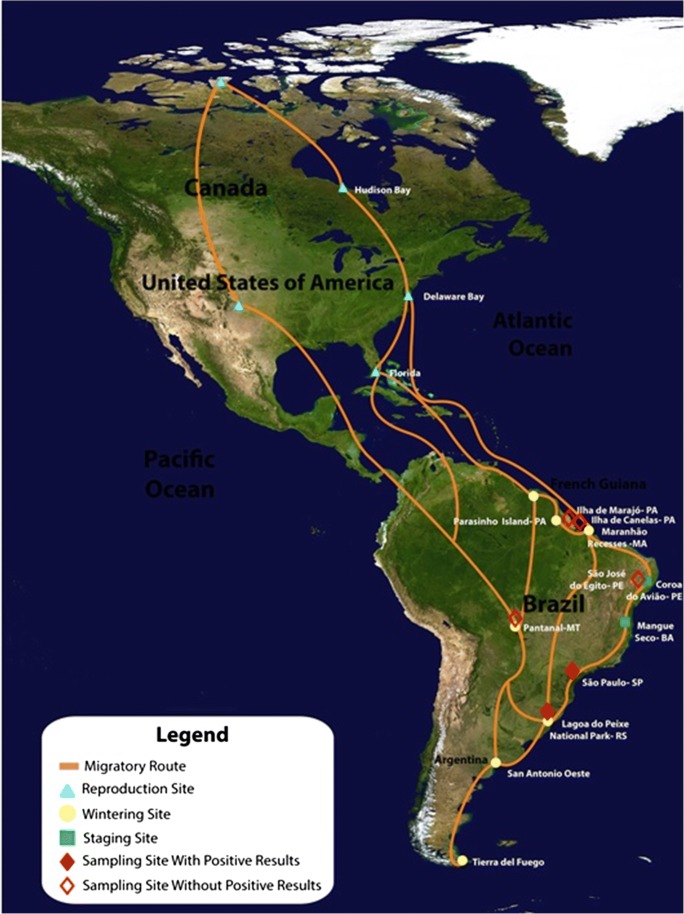
Fig. 1.
Location of sampling sites with positive results for avian CoVs and their relationship with the main bird migratory routes and congregation sites in North and South America. Congregation sites include reproduction, wintering, and tagging areas of boreal migrant birds (modified from http://www.icmbio.gov.br/cemave/)
RNA extraction and reverse transcription
Viral RNA was extracted using the MagMax TM-96 RNA Isolation Kit (Ambion, Austin, TX, USA) according to the manufacturer’s instructions. The extracted RNA was eluted in 80 μL of RNase-free water and stored at − 80 °C until processed. The cDNA was transcribed using the High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA, USA) following the manufacturer’s instructions. The RT reaction was performed by adding, to each reaction, 10 μL of total RNA, 2 μL of RT Buffer 10X, 2 μL of Random Primers (50 ng/μL) (Thermo Fisher Scientific, Waltham, MA, USA), 4.2 μL of DEPC water, and 50 U of MultiScribe™ Reverse Transcriptase (Thermo Fisher Scientific). The RT step conditions were 25 °C for 10 min, 37 °C for 120 min, and 85 °C for 5 min, and cooling to 4 °C.
Coronavirus detection
Polymerase chain reaction (PCR) followed by nested PCR were performed according to the protocol for the detection of avian CoVs developed by Chu et al. (2011) [17], which targets the RNA-dependent RNA polymerase (RdRp) gene in the ORF1 area of the viral genome. The resulting approximately 600-bp PCR fragment and 440-bp nested PCR fragment were separated by agarose gel [1.5%] electrophoresis and viewed under UV light after staining with ethidium bromide (25 μg/100 mL).
Purification and sequencing
Samples were purified using the ExoSap-IT enzyme (GE Healthcare, Chalfont St. Giles, UK) and GeneJET™ Gel Extraction Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The Sanger sequencing reaction was prepared using Big Dye Terminator v3.1 Cycle Sequencing Kit (Life Technologies, Foster City, CA, USA), followed by purification using BigDye X Terminator® Purification Kit (Applied Biosystems), and analysis in an automated ABI PRISM 3130XL DNA Sequencer (Applied Biosystems).
Genetic characterization and phylogenetic tree of the RdRp gene
Generated chromatograms containing nucleotide sequences were visualized using Bioedit Sequence Alignment Editor (v.7.1.5.0). Nucleotide sequences were aligned using CLC sequencer WorkBench version 7.6 (QIAGEN Aarhus, Denmark), and the nucleotide percentage similarities were calculated using the MegAlign program of the DNASTAR package (DNASTAR, Inc., Madison, WI, USA). The similarities with existing sequences were determined using the Basic Local Alignment Search Tool (BLAST). Phylogenetic analysis was performed using the neighbor-joining method (NJ) implemented in the MEGA 7.0.26 [18]. The nucleotide substitution model used for phylogenetic reconstruction was the p-distance model. The bootstrap values > 55% were obtained in the analysis of 10,000 replicates and are presented at the branching points.
Results
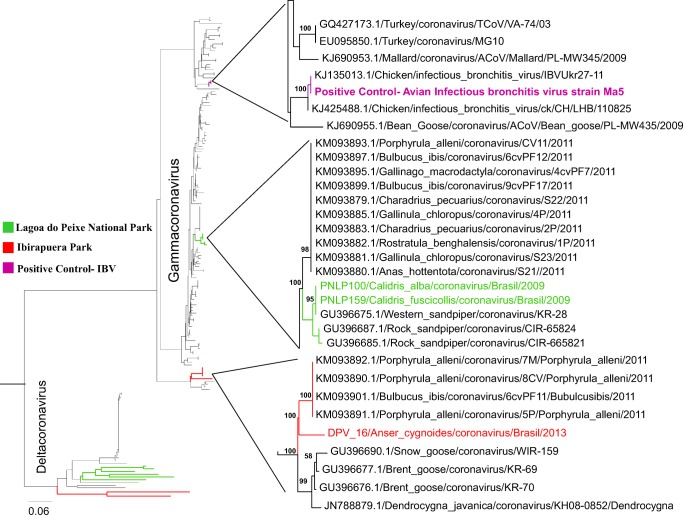
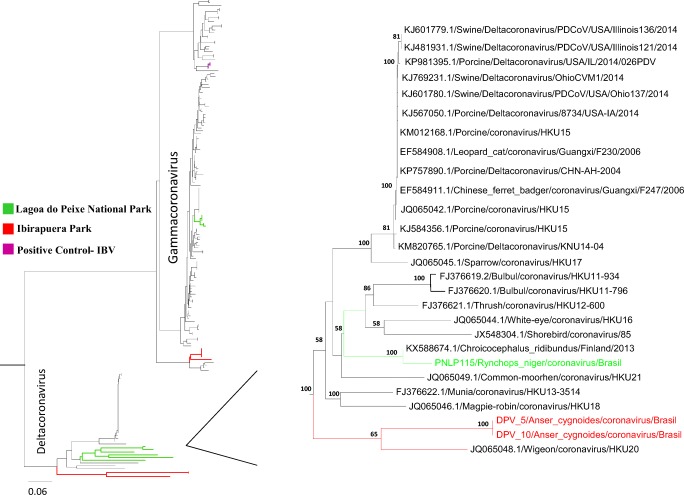
The CoV screening protocol, based in a nested PCR assay with degenerate consensus primers targeting the conserved region of RNA-dependent RNA polymerase (RdRp) gene, was selected for its greater sensitivity and CoV genera amplitude detection. Of the 746 bird samples collected from 2006 to 2013 in different regions of Brazil and analyzed by conventional RT-PCR, six samples (0.8% of total) from two different regions were confirmed by DNA sequencing (Table 2). Our study describes the detection of samples related to avian gamma- and deltacoronaviruses in birds collected in southeast and southern region of Brazil, more specifically in São Paulo and Rio Grande do Sul state (Table 3). The presence of CoV RNA was detected in three Chinese geese (Anser cygnoides) sampled in Ibirapuera Park, downtown São Paulo, southeast Brazil. One sample has shown similarities (93%) with sequences from viruses in the gammacoronavirus group, which are related to migratory birds only, whereas the other two samples (84%) belong to the deltacoronavirus group. Even though these sequences are closely related to those of wild bird CoVs, this group also includes ferret, pig, and leopard CoVs, demonstrating the potential for spillover events between coronaviruses from bird and mammal hosts. In the same way, in southern Brazil, three samples positive for CoVs were obtained from migratory birds captured in wintering areas at Lagoa do Peixe National Park, state of Rio Grande do Sul. Phylogenetic analysis showed that these samples (PNLP 100, PNLP 115, and PNLP 159) belonged to two CoV genera. Samples related to gammacoronaviruses were obtained from one Calidris alba and one Calidris fuscicollis individual, respectively, related a restricted to sandpiper species (Fig. 2). Samples related to deltacoronavirus were obtained from a black skimmer (Rynchops niger) carrying a strain that clustered, in phylogenetic analysis, closer to the clade of viruses found in Ibirapuera Park (Fig. 3).
Table 2.
Sampling location, total samples, number of positive samples obtained from birds collected across Brazil from 2006 to 2013
| Location | No. of samples | Positive samples | Year |
|---|---|---|---|
| Ilha de Marajó/PA | 93 | – | 2006 |
| Ajuruteua/PA | 91 | – | 2006 |
| Ilha de Canelas/PA | 67 | – | 2006–2008 |
| Bahia de São Marco/MA | 107 | – | 2007 |
| Lagoa do Peixe National Park/RS | 192 | 3 | 2009 |
| São José do Egito/PE | 64 | – | 2010 |
| Pantanal Poconé/MT | 51 | – | 2012 |
| Ibirapuera Park/SP | 81 | 3 | 2013 |
| Total | 746 | 6 | 2006–2013 |
PA Pará, SP São Paulo, MT Mato Grosso, BA Bahia, RS Rio Grande do Sul
Table 3.
Sample identification, bird species, place of capture, and coronavirus group of avian coronavirus sequences obtained from bird samples collected across Brazil from 2006 to 2013
| Sample | Species | Location | Group | Year | Accession number | (GenBank reference) | Similarity (%) |
|---|---|---|---|---|---|---|---|
| DPV 5 | Anser cygnoides | Ibirapuera Park/SP | Deltacoronavirus | 2013 | KU321643 | Wigeon coronavirus HKU20 strain HKU20-9243 (2012) | 84 |
| DPV 10 | Anser cygnoides | Ibirapuera Park/SP | Deltacoronavirus | 2013 | KU321644 | Wigeon coronavirus HKU20 strain HKU20-9243 (2012) | 84 |
| DPV 16 | Anser cygnoides | Ibirapuera Park/SP | Gammacoronavirus | 2013 | KU321645 | Avian coronavirus isolate 7 M/Porphyrula alleni (2011) | 93 |
| PNLP100 | Calidris alba | Lagoa do Peixe National Park/RS | Gammacoronavirus | 2009 | KU321640 | Rock Sandpiper Coronavirus (2010) | 99 |
| PNLP115 | Rynchops niger | Lagoa do Peixe National Park/RS | Deltacoronavirus | 2009 | KU321641 | Chroicocephalus ridibundus/Finland (2013) | 81 |
| PNLP159 | Calidris fuscicollis | Lagoa do Peixe National Park/RS | Gammacoronavirus | 2009 | KU321642 | Western Sandpiper Coronavirus/KR-28 (2010) | 99 |
SP São Paulo, RS Rio Grande do Sul
Fig. 2.
Phylogenetic analysis of partial RdRp gene sequence of avian gammacoronaviruses. Phylogenetic tree of partial sequences of the RdRp-encoding region constructed using the neighbor-joining method. In green, CoVs related to gammacoronaviruses sequences detected in samples from Lagoa do Peixe National Park, state of Rio Grande do Sul, southern Brazil. In red, CoVs related to gammacoronaviruses sequences detected in samples from Ibirapuera Park, São Paulo, southeast Brazil. Bootstrap values > 55% were obtained in the analysis of 10,000 replicates and are presented at the branching points
Fig. 3.
Phylogenetic analysis of partial RdRp gene sequence of avian deltacoronaviruses. Phylogenetic tree of partial sequences of the RdRp-encoding region constructed using the neighbor-joining method. In green, CoVs related to deltacoronavirus sequences detected in samples from Lagoa do Peixe National Park, state of Rio Grande do Sul, southern Brazil. In red, CoVs related to deltacoronavirus sequences detected in samples from Ibirapuera Park, São Paulo, southeast Brazil. Bootstrap values > 55% were obtained in the analysis of 10,000 replicates and are presented at the branching points
Discussion
The nested PCR assay with degenerate consensus primers was used to many studies. This assay has been used for the identification and differentiation of CoV in the four genera described to date (alpha-, beta-, gamma-, and deltacoronavirus) [17, 19, 20]. Similarly to our report, most of avian CoV diversity studies detected the same coronavirus genera [2, 17, 21, 22], with the exception of a report of betacoronavirus detection in distinct birds species from a rehabilitation center in Brazil [8]. Further studies are needed to understand the circulation of betacoronavirus in birds.
Ibirapuera is the main park in São Paulo, receiving on average 150,000 visitors in a single weekend. Wild birds make up a large portion of the species of animals that inhabit the park (ca. 142 species). The birds are kept free and cohabiting with the human population and other wild species (resident or visitors). There are another 21 species of wild animals, including bats and skunks, and the park also allows pets [23]. Such close contact between so many different species may cause interspecific transmission and recombination events, leading to the appearance of emerging viruses [24]. The three goose samples were collected from geese that live in this same environment in contact with wild birds that use the park as a stopping point. On the other hand, three migratory birds captured in wintering areas at Lagoa do Peixe National Park-RS have shown positive results of CoVs too. Sandpipers are boreal migrants that migrate south from North America in the winter [25]. A CoV related to deltacoronavirus group was obtained from a black skimmer (Rynchops niger), thus highlighting the possibility that migratory birds may be carrying these viruses across migratory routes in Brazil [9]. The Lagoa do Peixe region is an important migratory route [25], and it is located close (400 miles) from one of the most important region of poultry farm production of Brazil, more specifically region of Porto Alegre in Rio Grande do Sul State [26]. The proximity between the migratory routes of wild birds infected by gammacoronavirus shows the importance of a continuous surveillance and potential contact control between poultry and migratory birds as measures to avoid any chance of viral transmission between these animals.
The CoVs related to deltacoronaviruses group (DPV 5 and DPV 10) and gammacoronavirus group (DPV 16) from Ibirapuera Park showed the highest similarity to a CoV found by Lau et al. (2012) [27] in Guangdong province (84%), China, and by Lima et al. (2015) [28] (93% nt identity) in a migratory bird (Porphyrula alleni) from Madagascar in 2011, respectively (Table 2). The gammacoronaviruses found at Lagoa do Peixe National Park were more similar to those found by Muradrasoli et al. (2010) [29] in sandpipers (Calidris spp.) from the Bering Strait (99% nt identity). However, the deltacoronavirus was more similar to the strain isolated from a black-headed gull (Chroicocephalus ridibundus) by Hepojoki et al. (2017) in Finland (95% nt identity) (Table 3). Interestingly, this data were reported about sampling collected during the same years (2010–2013) by the same way, using RT-PCR targeting the RdRp gene to detect the presence of CoV in wild birds from Finland. The prevalence was found around 5.4% (51/939), which corroborates with our findings. Hepojoki et al. (2017) have shown positive samples to diverse gamma- and deltacoronavirus too [22].
In South America, there is a lack of information about coronavirus in wild birds. These data are interesting, once that, present birds infected in central park in Sao Paulo city and wild birds through migratory routes. In Hong Kong, Chu et al. (2011) detected a high prevalence (12.5%) in aquatic wild birds, reporting gamma and deltacoronavirus, being the first one more prevalent [17]. Similar studies screened before in Sweden (2016), with the number of sampling similar to our study, and reported a high prevalence (18.7%) in wild birds, with emphasis for Anseriformes that had the highest prevalence (51%), all belonging to the gammacoronavirus group [30]. On the other hand, Jordan et al. (2015) analyzed 303 samples from cloacal swabs, using universal CoV primers, and found only one duck coronavirus, belonging to the gammacoronavirus group too, which suggested a low prevalence in wild aquatic birds in USA, corroborating with our data [21]. As we know, migratory routes in Brazil are linked with flyways coming from North America and can confirm our findings [16]. Although our data have shown low prevalence when compared with studies before in Scandinavian Waterfowl, our data is clear when compared with North American data. In addition, it is the first time that samples related to Gamma and deltacoronavirus group in free wild birds were reported in Brazil.
The overall prevalence of CoV found in this study was 0.8% (6/746 birds), with 3.7% (3/81) from Ibirapuera Park and 1.6% (3/192) from migratory birds captured in Lagoa do Peixe National Park. Other studies report an overall prevalence of CoV in aquatic and migratory birds between 0.33 and 18.5% [8, 17, 21, 30, 31]. This data reinforces that CoVs are circulating across major migratory routes worldwide.
Our report points to the circulation of coronaviruses among migratory and resident birds sampled from distinct regions of Brazil. Coronavirus RNA were detected in birds species of Anatidae family (Chinese goose) in Ibirapuera Park, São Paulo, southeast Brazil. Anatidae birds were already described as hosts of coronavirus all over the globe. However, interestingly, we report for the first time the detection in two sandpiper species (Calidris alba and Calidris fuscicollis) from Lagoa do Peixe National Park, a wintering area in southern Brazil. Although our data indicate a low prevalence of coronavirus in birds sampled, the genetic similarity between coronavirus detected at birds from distinct continents may indicate that wild birds are possibly carrying coronavirus throughout migratory routes as observed for avian influenza virus. We report the detection of samples related to delta- and gammacoronaviruses in different species of birds, some sampled in a park located inside the largest city in South America, indicating the circulation of coronavirus in urban areas (Table 3). In addition, all birds tested in this study were in apparently healthy populations. Further studies are required to determine the species and full genome similarity with other avian CoVs. However, our findings indicate that CoVs may be circulating among one of the major avian migratory routes in the world.
Electronic supplementary material
(PDF 54.7 kb)
Acknowledgments
We thank the field logistic support of field team of Microbiology Department, University of São Paulo, Wildlife Department of São Paulo (DEPAVE), and Laboratory of Ornithology and Marine Animals (LOAM), Universidade do Vale do Rio dos Sinos. We thank Diogo Fiori Ribas, Edna Maria Gomes Cavalcante, and Hiroe. Positive control was kindly provided by Professor Paulo Brandão at the Department of Preventive Veterinary Medicine and Animal Health, School of Veterinary Medicine, University of São Paulo.
Funding
This work was supported by the Fundacão de Amparo a Pesquisa do Estado de São Paulo (FAPESP) (grant numbers 2009/05994-9, 2011/13821-7, 2011/11006-0, 2013/05485-2, 2014/03172-0); the Conselho Nacional de Desenvolvimento a Pesquisa (CNPq) (grant number 477912/2007-9]; CAPES/Newton Foundation (grant number 99999.005126/2015-00); Fundação de Amparo a Pesquisa do Estado do Rio Grande do Sul (FAPERGS No. 09/0574-7); and Wildlife Conservation Society (CWS No. 2008-05 and 2009-05).
Compliance with ethical standards
Conflict of interest
The authors declared that they have no conflict of interest.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. The procedures involving wild birds were approved by the Brazilian Society of Laboratory Animal Science (SBCAL) of the University of São Paulo, São Paulo, Brazil (protocol number 105/pg.74/book2) and licensed by the Chico Mendes Institute for Biodiversity Conservation (ICMBio/SISBIO), Ministry of the Environment (MMA), under protocol numbers 201/2006 CGFAU, 25895-1, 33602-1, 17565-1, and 14966-11.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bande F, Arshad SS, Omar AR, et al. Global distributions and strain diversity of avian infectious bronchitis virus: a review. Anim Health Res Rev. 2017;18:70–83. doi: 10.1017/S1466252317000044. [DOI] [PubMed] [Google Scholar]
- 2.Marandino A, Pereda A, Tomás G, et al. Phylodynamic analysis of avian infectious bronchitis virus in South America. J Gen Virol. 2015;96:1340–1346. doi: 10.1099/vir.0.000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michael MC, Lai DC. The molecular biology of coronaviruses. Adv Virus Res. 1997;65:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woo PCY, Lau SKP, Huang Y, Yuen K-Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med. 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- 5.Su S, Wong G, Shi W, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan JFW, To KKW, Tse H, et al. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21:544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavanagh D, Gelb J., Jr . Infectious Bronchitis. In: Saif YM, editor. Dis. Poult. 12. Oxford: Blackwell; 2008. pp. 117–135. [Google Scholar]
- 8.Durães-Carvalho R, Caserta LC, Barnabé ACS, et al. Coronaviruses detected in Brazilian wild birds reveal close evolutionary relationships with Beta- and Deltacoronaviruses isolated from mammals. J Mol Evol. 2015;81:21–23. doi: 10.1007/s00239-015-9693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Araujo J, de Azevedo SM, Gaidet N, et al. Avian influenza virus (H11N9) in migratory shorebirds wintering in the Amazon region, Brazil. PLoS One. 2014;9:e110141. doi: 10.1371/journal.pone.0110141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dusek RJ, Hallgrimsson GT, Ip HS et al (2014) North Atlantic migratory bird flyways provide routes for intercontinental movement of avian influenza viruses. PLoS One. 10.1371/journal.pone.0092075 [DOI] [PMC free article] [PubMed]
- 11.Ometto T, Durigon EL, de Araujo J, et al. West Nile virus surveillance, Brazil, 2008-2010. Trans R Soc Trop Med Hyg. 2013;107:723–730. doi: 10.1093/trstmh/trt081. [DOI] [PubMed] [Google Scholar]
- 12.Dusek RJ, McLean RG, Kramer LD, et al. Prevalence of West Nile virus in migratory birds during spring and fall migration. Am J Trop Med Hyg. 2009;81:1151–1158. doi: 10.4269/ajtmh.2009.09-0106. [DOI] [PubMed] [Google Scholar]
- 13.Reed KD, Meece JK, Henkel JS, Shukla SK. Birds, migration and emerging zoonoses: West Nile virus, Lyme disease, influenza A and enteropathogens. Clin Med Res. 2003;1:5–12. doi: 10.3121/cmr.1.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomazelli L, de Araujo J, de Ferreira CS, et al. Molecular surveillance of the Newcastle disease virus in domestic and wild birds on the north eastern coast and Amazon biome of Brazil. Rev Bras Ciência Avícola. 2012;14:01–07. doi: 10.1590/S1516-635X2012000100001. [DOI] [Google Scholar]
- 15.CEMAVE-ICMBio . Relatório Anual de rotas e áreas de concentraçào de aves migratórias no Brasil. Brasília: ICMBio; 2014. [Google Scholar]
- 16.Araujo J, Petry MV, Fabrizio T, et al. Migratory birds in southern Brazil are a source of multiple avian influenza virus subtypes. Influenza Other Respir Viruses. 2018;12:220–231. doi: 10.1111/irv.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu DKW, Leung CYH, Gilbert M, et al. Avian coronavirus in wild aquatic birds. J Virol. 2011;85:12815–12820. doi: 10.1128/JVI.05838-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagemeijer M, Rottier P, Haan C. Biogenesis and dynamics of the coronavirus replicative structures. Viruses. 2012;4:3245–3269. doi: 10.3390/v4113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Góes LGB, ACA C, de Carvalho C, et al. Genetic diversity of bats coronaviruses in the Atlantic Forest hotspot biome, Brazil. Infect Genet Evol. 2016;44:510–513. doi: 10.1016/j.meegid.2016.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jordan BJ, Hilt DA, Poulson R, et al. Identification of avian coronavirus in wild aquatic birds of the central and eastern USA. J Wildl Dis. 2015;51:218–221. doi: 10.7589/2014-03-070. [DOI] [PubMed] [Google Scholar]
- 22.Hepojoki S, Lindh E, Vapalahti O, Huovilainen A. Prevalence and genetic diversity of coronaviruses in wild birds, Finland. Infect Ecol Epidemiol. 2017;7:1408360. doi: 10.1080/20008686.2017.1408360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prefeitura (2008) Guia dos parques municipais de São Paulo. acessed January, 2018
- 24.Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scherer AL, Petry MV. Seasonal variation in shorebird abundance in the state of Rio Grande Do Sul, southern Brazil. Wilson J Ornithol. 2012;124:40–50. doi: 10.1676/11-034.1. [DOI] [Google Scholar]
- 26.Embrapa (2016) Embrapa Suinos e Aves. https://www.embrapa.br/suinos-e-aves/cias/mapas. Accessed 20 May 2016
- 27.Lau JHN, WPLSLCLCT a. K et al. Discovery of seven novel mammalian and avian coronaviruses in the genus Deltacoronavirus supports bat coronaviruses as the gene source of Alphacoronavirus and Betacoronavirus and avian coronaviruses as the gene source of Gammacoronavirus and Deltacoronavi. J Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Lima FES, Gil P, Pedrono M et al (2015) Diverse gammacoronaviruses detected in wild birds from Madagascar. Eur J Wildl Res:635–639 [DOI] [PMC free article] [PubMed]
- 29.Muradrasoli S, Bálint Á, Wahlgren J et al (2010) Prevalence and phylogeny of coronaviruses in wild birds from the bering strait area (Beringia). PLoS One. 10.1371/journal.pone.0013640 [DOI] [PMC free article] [PubMed]
- 30.Wille M, Muradrasoli S, Nilsson A, Järhult JD (2016) High prevalence and putative lineage maintenance of avian coronaviruses in Scandinavian waterfowl. PLoS One. 10.1371/journal.pone.0150198 [DOI] [PMC free article] [PubMed]
- 31.Lee Y-J, Kang H-M, Lee E-K, et al. Novel reassortant influenza A(H5N8) viruses, South Korea, 2014. Emerg Infect Dis. 2014;20:1087–1089. doi: 10.3201/eid2006.140233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 54.7 kb)