Abstract
Orf virus (ORFV) causes contagious ecthyma (CE), a highly transmissible, zoonotic disease of small ruminants. CE most commonly affects lambs and unvaccinated sheep. This work reports epidemiologic, clinicopathologic, and virologic findings in a CE outbreak in a vaccinated sheep flock in Uruguay and failure to detect ORFV in a commercial vaccine.
Keywords: Contagious pustular dermatitis, Ruminants, Phylogenetics, Live vaccine, Ovine
Orf virus (ORFV) is a Parapoxvirus (subfamily Chordopoxvirinae, family Poxviridae) with ovoid-shaped virions of ~ 260 by 160 nm and a linear double-stranded DNA genome of 134–139 kb. ORFV causes contagious ecthyma (CE), also known as contagious pustular dermatitis, a highly transmissible, zoonotic disease of worldwide distribution that predominantly affects small ruminants [1–3].
Pathologically, CE is characterized by pustular, proliferative, and necrotizing lesions in the mucous membranes and skin of the lips, mouth, nostrils, eyes, face, udder, and, less commonly, the limbs, esophagus, and forestomachs [4]. One of the main ORFV virulence factors is the vascular endothelial growth factor, which stimulates vascular and epidermal proliferation, favoring viral replication [1].
In sheep, CE most commonly affects lambs and unvaccinated adults. Morbidity is often high, resulting in significant economic losses, although mortality is rare, and is generally associated with secondary bacterial or parasitic infections or aspiration pneumonia [4].
Sheep production has economic and sociocultural importance in Uruguay. Two lyophilized live commercial vaccines to prevent CE are currently available in the country; however, there is limited research and genetic information on these vaccines [5] and wild ORFV strains [6] from Uruguay.
Although ORFV has a worldwide distribution, there are few studies on molecular characterization of this virus in South America [5, 6]. The objectives of this work are to describe the epidemiologic, clinical, pathologic, and molecular findings in an outbreak of CE by ORFV in a vaccinated sheep flock in Uruguay and to compare the viral strain involved in this outbreak with other wild and vaccine strains.
In January 2017, an outbreak of proliferative stomatitis, cheilitis, and nasal dermatitis was diagnosed in a commercial flock composed of 78 sheep in a farm in the department of Maldonado, Uruguay. All sheep had been vaccinated with a commercial CE vaccine, at least once in their life when they were 6 to 8 months of age, as indicated by the vaccine prospect. A total of seven sheep, four with labial and/or cutaneous lesions (sheep 1–4) ranging from 2 to 4 years in age and three adult sheep without integumentary lesions (sheep 5–7, unaffected controls), were selected for clinical and gross pathological examination. Duplicate skin or labial biopsies from the four affected sheep were either fixed for 48 h in 10% neutral buffered formalin for histology or preserved frozen at − 20 °C for molecular virology. For histologic examination, formalin-fixed biopsies were embedded in paraffin, sectioned at 4–5 μm, and stained with hematoxylin and eosin.
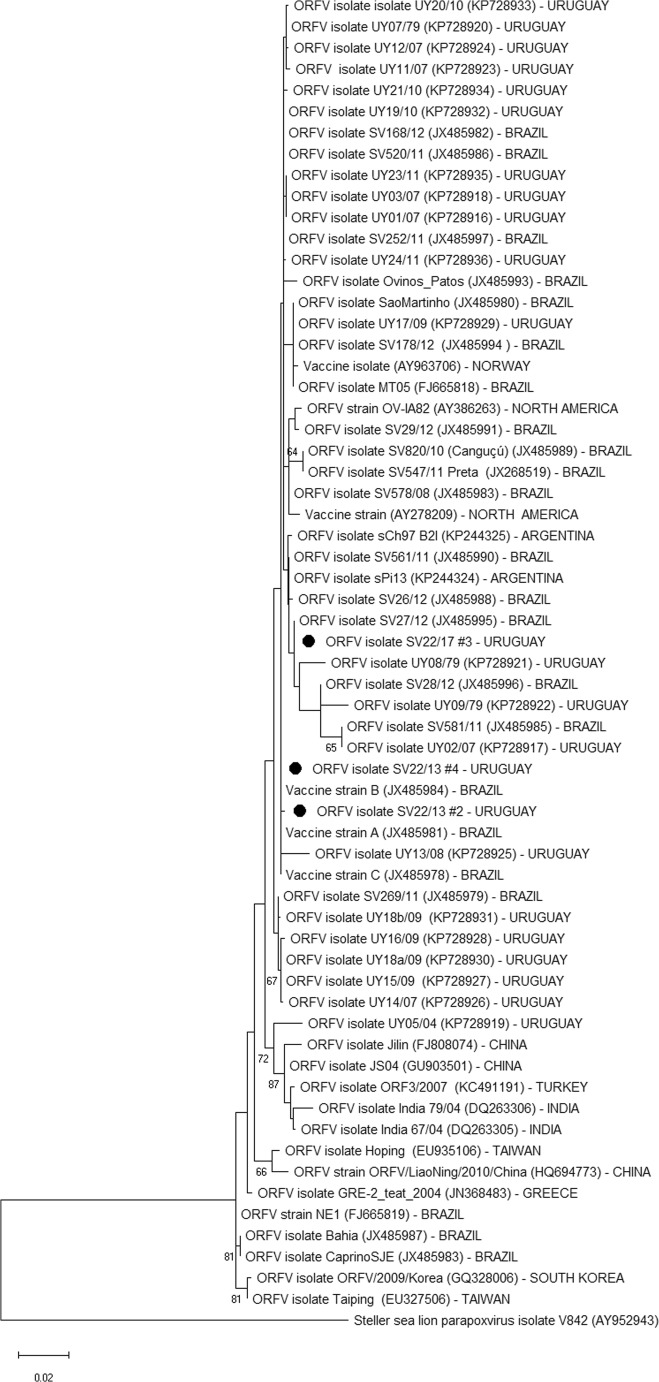
The frozen skin samples from three affected ewes (sheep 2–4) and the two commercially available vaccines purchased locally (vaccines A and B) were analyzed by PCR targeting the B2L gene of Parapoxvirus. Nucleotide sequencing and phylogenetic analysis of genomic amplicons were performed as described by Schmidt et al. [5]. A phylogenetic tree including the strains detected in this outbreak and other ORFV wild strains from Uruguay, Brazil, Argentina, North America, Greece, and Asia (China, India, Turkey, Taiwan, and South Korea) and vaccine strains from South America, North America, and Norway available in GenBank was constructed using MEGA5 software, using the maximum likelihood method based on the Kimura 2-parameter model, with 1000 bootstrap replicates [7]. The sequences obtained in this outbreak were also analyzed at the amino acid level and compared between them and with the sequences of the locally available vaccines.
Twenty of the 78 sheep developed CE (morbidity = 25.6%), only one of which died (mortality = 1.3%, lethality = 4.3%). The rectal temperature varied between 40.3 and 42 °C in four affected sheep (hyperthermia) and between 39.5 and 40.3 °C in three healthy controls (normothermia). All four clinically affected animals examined had moderate (sheep 1) to severe (sheep 2–4) nasal discharge, tachypnea, and weight loss.
Sheep 1 had moderate, multifocal, coalescing, proliferative, erosive, alopecic, and nodular lesions on the skin of the nasal plane. Sheep 2 had moderate, bilateral, coalescing, and extensive proliferative and necrotizing lesions, involving the lips, the mucosa of the nostrils, and the adjacent facial skin, and had extensive edema in the maxillary, mandibular, and nasal regions, resulting in facial swelling. Multiple, discrete, 2–5 mm irregular ulcers were observed on the hard palate and tongue. Sheep 3 presented with moderate, bilateral, proliferative, and crusty lesions on the lips, buccal commissure, and nasal plane. Sheep 4 had severe proliferative lesions with crusting, which extended caudally beyond the nasal plane. In addition, the skin surrounding the nostrils exhibited multiple epidermal nodules up to 5 mm in diameter (Fig. 1). The lesions partially obstructed the nostrils and anterior nasal cavity, causing dyspnea.
Fig. 1.
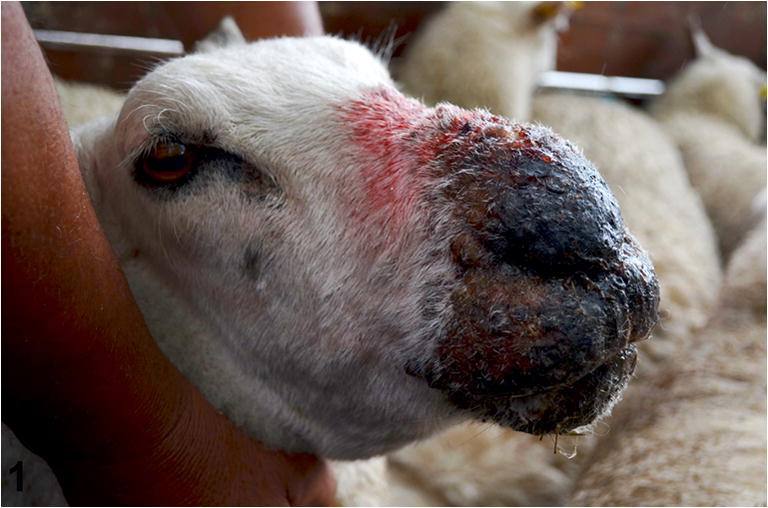
Ovine. Skin. Severe extensive bilateral proliferative and necrotizing cheilitis and dermatitis of the nasal plane and adjacent facial skin, with crusting, hyperpigmentation, edema, and swelling of the facial region resulting in partial obstruction of the nostrils. Note that the farmer handling the sheep is not wearing personal protective equipment (gloves)
Histologically, all affected sheep had moderate to severe proliferative, necrotizing, and pustular cheilitis and/or nasal dermatitis. The keratinocytes were swollen due to hydropic degeneration. Severe epidermal hyperplasia, moderate to severe neutrophilic infiltrate, and parakeratotic hyperkeratosis of the epidermis were also observed (Fig. 2). Single or multiple irregular intracytoplasmic eosinophilic inclusion bodies, morphologically resembling Parapoxvirus inclusions, were observed in the keratinocytes in three of the four sheep (Fig. 3).
Fig. 2.
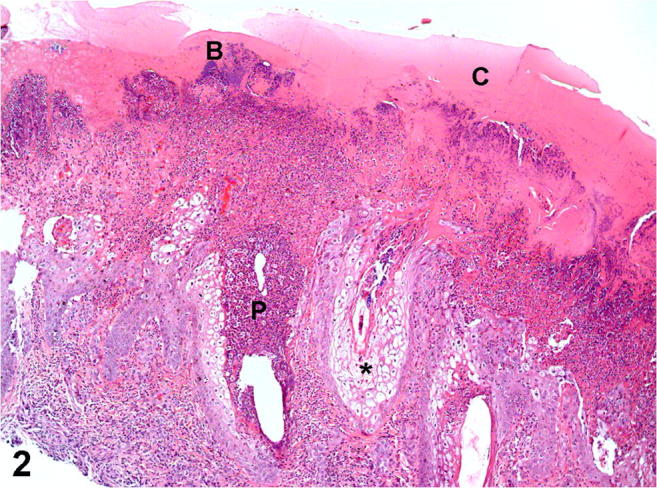
Ovine. Skin. Severe proliferative and necrotizing dermatitis with serocellular crusting (C), epidermal hyperplasia, severe intraepithelial neutrophilic infiltrate (pustules, P), ballooning degeneration of keratinocytes (asterisk), and secondary superficial bacterial colonization (B). H&E. Obj. × 10
Fig. 3.
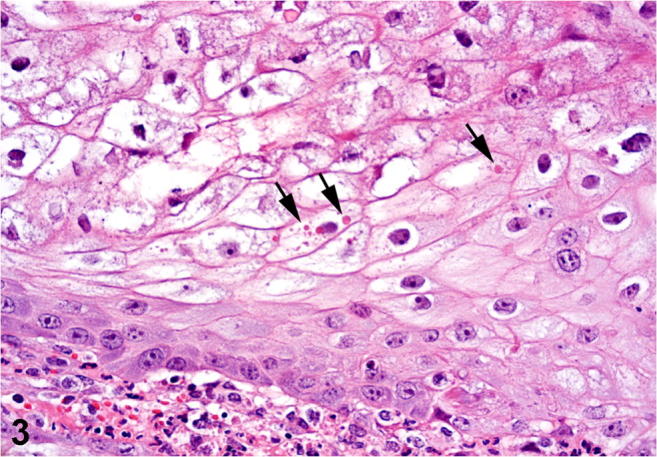
Ovine. Skin. Multiple eosinophilic inclusion bodies (arrows) compatible with Parapoxvirus within the cytoplasm of markedly swollen epidermal keratinocytes. H&E. Obj. × 40
Specific ORFV DNA was amplified by PCR from the three frozen skin biopsies. Amplicons of ~ 594 bp of the B2L major envelope protein obtained from the three samples were sequenced. The nucleotide identity was 99.7–100% among the samples and 97.6–100% when compared with sequences from other ORFV isolates/strains from sheep available in GenBank that were used to build the phylogenetic tree (Fig. 4). At the amino acid level, the identity between the strains obtained from the outbreak and the vaccine strains was 100%.
Fig. 4.
Phylogenetic relationships among ORFV strains based on the nucleotide sequence of the B2L gene. The phylogenetic tree was constructed using the maximum likelihood method, Kimura 2-parameter. Numbers on the tree branches represent the bootstrap calculated per 1000 replicates. Values > 60% are shown. The ORFV strains obtained from the sheep in this study are identified with black dots
Only one of the two commercially available vaccines analyzed was positive by PCR for the Parapoxvirus B2L gene. As shown in Fig. 4, two samples (SV22/17 #2 and #4) were grouped together and were close to the Uruguayan, Brazilian, and Argentine ORFV field strains. Furthermore, these strains were grouped in the same cluster as ORFV vaccine strains A (JX485981), B (JX485984), and C (JX485978) that are commercialized in Southern Brazil, Uruguay, and Argentina. This suggests that the strains found in this outbreak are very similar to the vaccine strains and that they have a common ancestor. The strain identified in the remaining skin biopsy (SV22/17 #3) was grouped in a different subcluster, which belongs to the same cluster as SV22/17 #2 and #4. Such subcluster segregation probably occurred due to a single nucleotide mutation and is likely of little epidemiologic importance. Additionally, the strains detected in the outbreak clustered separately of strains from Asia and Greece [8, 9], suggesting that South American strains have a common ancestor that differs with that of the Asian/Greek strains.
The diagnosis of CE was based on epidemiology, clinical signs, and macroscopic and histological lesions, which were typical of CE and similar to those described by other authors [4, 6, 10, 11], and was further supported with molecular identification of the virus in skin biopsies.
Uruguay has approximately 6.5 million sheep, which significantly contribute to the economy of the livestock sector [12]. The economic losses of this disease are not primarily related to deaths but to growth retardation and weight loss [11]. The proliferative, necrotizing, and/or ulcerative lesions are painful and they impair adequate food intake and, consequently, hamper weight gain [4, 11]. Secondary complications of severe host immunosuppression, including myiasis and bacterial infections, may result in death [1, 4]. In this outbreak, only one CE-affected sheep died. The carcass was not subjected to postmortem examination and the ultimate cause of death was thus undetermined.
In sheep, the immune response to ORFV is vigorous and involves neutrophils, dermal dendritic cells, CD4+ and CD8+ T cells, interferon gamma, B cells, and antibody production. CD4+ T cells, interferon gamma, and, to a lesser extent, CD8+ T cells are involved in partial protection against ORFV infection, while antibodies have little protective effect [13]. Several immunomodulatory viral virulence factors influence the host immune response. For instance, ORFV interferon resistance protein (OVIFNR) is produced in the early stages of infection and inhibits the interferon-induced shutdown of cellular protein translation. ORFV chemokine-binding protein (CBP) binds to chemokines that control monocyte/macrophage, lymphocyte, and neutrophil recruitment to the site of infection, such as monocyte chemotactic protein-1, macrophage inflammatory protein-1 alpha, RANTES (regulated upon activation, normal T cell expressed and secreted), and lymphotactin [1]. Additionally, ORFV interleukin-10 has an immunosuppressive function that aids viral replication. Thus, ORFV inhibits important cellular elements of the antiviral immune response [1].
The molecular characterization of ORFV strains from Uruguay was recently reported [6]. Phylogenetic analysis indicates that the national strains do not form a unique cluster and that most strains display similarities with the commercial vaccines available in the national market [6]. Although all sheep in the outbreak presented here had been vaccinated at least once in their life with a commercial vaccine, based on the history provided by the farmer and the veterinarian, this immunization did not appear to prevent the occurrence of the outbreak. Interestingly, there was no detectable ORFV DNA in one of the live commercial vaccines analyzed by PCR, suggesting inadequate vaccine production or preservation. In this sense, in November 2017, the Uruguayan Ministry of Livestock, Agriculture and Fisheries (MGAP) announced the recall of a specific batch/series of a commercial CE vaccine that had been released to the market after determining a drop in the viral titer in the potency test.
The ORFV strains detected in this outbreak were grouped separately from ORFV sequences obtained from goats (Bahia, Caprino SJE, and ORFV strain NE1), demonstrating greater relation with sheep strains. This is somewhat expected, since ORFV isolated from goats is molecularly divergent from sheep strains. In Uruguay, the goat population is scant and ORFV infection or CE has not been reported in this species. Other possible sources of infection are wild cevids, such as “guazubirá” (Mazama gouazoubira) or axis deer (Axis axis), although ORFV has yet to be found in these species in Uruguay.
CE has a worldwide distribution and severe disease as seen in this outbreak is uncommon in vaccinated adult sheep. Although ORFV can repeatedly infect sheep, reinfection is usually associated with reduced lesion size and time to resolution compared with primary infection [13]. The diagnostic investigation in this outbreak allowed for etiologic confirmation of severe CE caused by ORFV. Although ORFV vaccine strains have been implicated as sources of CE outbreaks [14], whether the outbreak described herein was caused by a vaccine or wild-type strain could not be determined based on partial B2L gene sequencing. CE appears to be a relatively common disease in Uruguay according to field observations by veterinary practitioners working with livestock; however, there is only one scientific report of this condition in the country [6]. It is worth mentioning that CE is a minor occupational zoonosis. In South America, human cases have been confirmed in Argentina [15], Brazil [5, 11], and Chile [15, 16], and cases have been linked to sheep and goats. To the best of our knowledge, no human cases have been recorded in Uruguay. Considering its potential public and animal health impact, further studies are needed to better characterize the epidemiology and economic impact of CE in Uruguay and to assess whether the commercially available vaccines provide protective immunity to local ORFV strains.
Acknowledgements
The authors thank Yissell Perdomo from the “Instituto Nacional de Investigación Agropecuaria (INIA)” for technical assistance with the histology techniques and Dr. Chrissy Eckstrand from the Washington Animal Disease Diagnostic Laboratory, Washington State University, for reviewing this manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Statement of animal rights
All cases described herein occurred spontaneously, with no experimentation, inoculation, or treatment of live animals. No animals were euthanized for this study.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Haig DM. Orf virus infection and host immunity. Curr Opin Infect Dis. 2006;19(2):127–131. doi: 10.1097/01.qco.0000216622.75326.ef. [DOI] [PubMed] [Google Scholar]
- 2.Khalafalla AI, El-Sabagh IM, Al-Busada KA, Al-Mubarak AI, Ali YH. Phylogenetic analysis of eight Sudanese camel contagious ecthyma viruses based on B2L gene sequence. Virol J. 2015;12:124. doi: 10.1186/s12985-015-0348-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nandi S, Ujjwal K, Chowdhury S. Current status of contagious ecthyma or orf disease in goat and sheep—a global perspective. Small Rumin Res. 2011;96(2–3):73–82. doi: 10.1016/j.smallrumres.2010.11.018. [DOI] [Google Scholar]
- 4.Mauldin EA, Kennedy JP. Integumentary System. In: Maxie MG, editor. Jubb, Kennedy and Palmer’s Pathology of Domestic Animals. 6. Amsterdam: Elsevier; 2016. pp. 617–618. [Google Scholar]
- 5.Schmidt C, Cargnelutti JF, Brum MC, Traesel CK, Weiblen R, Flores EF. Partial sequence analysis of B2L gene of Brazilian orf viruses from sheep and goats. Vet Microbiol. 2013;162(1):245–253. doi: 10.1016/j.vetmic.2012.10.031. [DOI] [PubMed] [Google Scholar]
- 6.Olivero N, Reolon E, Arbiza J, Berois M. Genetic diversity of Orf virus isolated from sheep in Uruguay. Arch Virol. 2018;163(5):1285–1291. doi: 10.1007/s00705-018-3717-x. [DOI] [PubMed] [Google Scholar]
- 7.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Billinis C, Mavrogianni VS, Spyrou V, Fthenakis GC. Phylogenetic analysis of strains of Orf virus isolated from two outbreaks of the disease in sheep in Greece. Virol J. 2012;9:24. doi: 10.1186/1743-422X-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karakas A, Oguzoglu TC, Coskun O, Artuk C, Mert G, Gul HC, Sener K, Özkul A. First molecular characterization of a Turkish orf virus strain from a human based on partial B2L sequence. Arch Virol. 2013;158(5):1105–1108. doi: 10.1007/s00705-012-1575-5. [DOI] [PubMed] [Google Scholar]
- 10.Maganga GD, Relmy A, Bakkali-Kassimi L, Ngoubangoye B, Tsoumbou T, Bouchier C, N’Dilimabaka N, Leroy EM, Zientara S, Berthetet N. Molecular characterization of Orf virus in goats in Gabon, Central Africa. Virol J. 2016;13:79. doi: 10.1186/s12985-016-0535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nobrega JE, Macêdo JT, Araújo JAS, Dantas AF, Soares MP, Riet-Correa F. Contagious ecthyma in sheep and goats in the semiarid of Paraiba, Brazil. Pesqui Vet Bras. 2014;28(3):135–139. doi: 10.1590/S0100-736X2008000300002. [DOI] [Google Scholar]
- 12.Ministerio de Ganadería, Agricultura y Pesca, República Oriental del Uruguay. Indicadores basados en la declaración jurada anual de existencias DICOSE-SNIG del año 2016. http://www.mgap.gub.uy/indicadores-basados-en-la-declaracion-jurada-anual-de-existencias-dicose-snig-2016. Accessed 21 Mar 2018
- 13.Haig DM, McInnes CJ. Immunity and counter-immunity during infection with the parapoxvirus orf virus. Virus Res. 2008;88(1–2):3–16. doi: 10.1016/S0168-1702(02)00117-X. [DOI] [PubMed] [Google Scholar]
- 14.Gilray JA, Nettleton PF, Pow I, Lewis CJ, Stephens SA, Mandeley JD, Reid HW. Restriction endonuclease profiles of orf virus isolates from the British Isles. Vet Rec. 1998;143(9):237–240. doi: 10.1136/vr.143.9.237. [DOI] [PubMed] [Google Scholar]
- 15.Peralta A, Flores Olivares CA, González Altamiranda E, Verna, A, Madariaga C, Odeón A, Cantón G. Ectima contagioso en humanos: confirmación y caracterización molecular de los primeros casos documentados en Argentina y Chile. XII Congreso Argentino de Virología, Sept 26–28, 2017. Buenos Aires, Argentina
- 16.Flores C, González E, Verna A, Peralta A, Madariaga C, Odeón A, Cantón G. Virus orf en humanos, confirmación molecular de un caso clínico en Chile. Rev Chil Infectol. 2017;34(6):607–609. doi: 10.4067/S0716-10182017000600607. [DOI] [PubMed] [Google Scholar]