Abstract
Burkholderia contaminans LTEB11 is a Gram-negative betaproteobacterium isolated as a contaminant of a culture in mineral medium supplemented with vegetable oil. Here, we report the genome sequence of B. contaminans LTEB11, identifying and analyzing the genes involved in its lipolytic machinery and in the production of other biotechnological products.
Electronic supplementary material
The online version of this article (10.1007/s42770-019-00076-4) contains supplementary material, which is available to authorized users.
Keywords: Lipases, Polyhydroxyalkanoates, Rhamnoplipids, Non-ribosomal peptide synthetases
The genus Burkholderia is widely distributed in the environment [1]. It is divided into two well-established clusters: the non-pathogenic cluster, which comprises beneficial plant symbionts, and the pathogenic cluster, which comprises opportunistic human, animal, and plant pathogens [1]. The species Burkholderia contaminans was described in 2009 and received this name because it was isolated as a contaminant of a Sargasso Sea DNA sample [2]. Currently, B. contaminans is classified in the Burkholderia cepacia complex (BCC), a group of at least 18 species that infect immunocompromised individuals, especially sufferers of cystic fibrosis [3, 4].
Despite their role as disease agents, B. contaminans and other BCC species have biotechnological applications. For example, they have been applied as biocontrol and bioremediation agents [5], for the production of biosurfactants [6] and for the production of extracellular lipases [7].
B. contaminans LTEB11 is a Gram-negative betaproteobacterium isolated in our laboratory as a contaminant of a fungal culture in mineral medium supplemented with vegetable oil. It was previously classified, erroneously, as both B. cepacia and B. lata. This strain produces an extracellular lipase (LipBC) that is active and highly stable in media containing organic solvents [8, 9]. Indeed, we have produced LipBC by submerged fermentation and by solid-state fermentation and applied it in esterification and transesterification reactions for biodiesel synthesis [10–12] and resolution of racemates [13]. Recently, genes encoding lipase LipBC (lipA) and foldase LifBC (lipB) were identified and co-expressed in Escherichia coli, with a recombinant Lip-LifBC complex being purified and characterized [14]. However, little is known about the genome of this bacterium and whether it might have other biotechnological applications. Here, we report the genome sequence of B. contaminans LTEB11, identifying and analyzing the genes involved in its lipolytic machinery.
Genomic DNA was isolated using phenol-chloroform extraction [15]. The whole-genome sequencing was performed on two different platforms: MiSeq Illumina (2,698,078 paired-end reads, 250 bp) and Ion Proton System (5,661,193 fragments, 125 bp long). The sequence data were de novo assembled using CLC Genomics Workbench 6.5.1 [16], Velvet 1.2.07 [17] and Masurca 2.3.2 [18] and the final assembly was optimized and finished using GFinisher [19]. The average nucleotide identity (ANI) [20] was calculated by a script developed by Kostas’s lab (http://enve-omics.gatech.edu/). BLASTn comparison of genomes was visualized by BRIG [21]. Coding sequences (CDS) and open reading frames (ORFs) were predicted using the RAST server [22]. Phylogenetic analysis was carried out with the neighbor-joining method and bootstrapping (1000 replicates) was used to estimate the confidence levels of phylogenetic reconstructions [23].
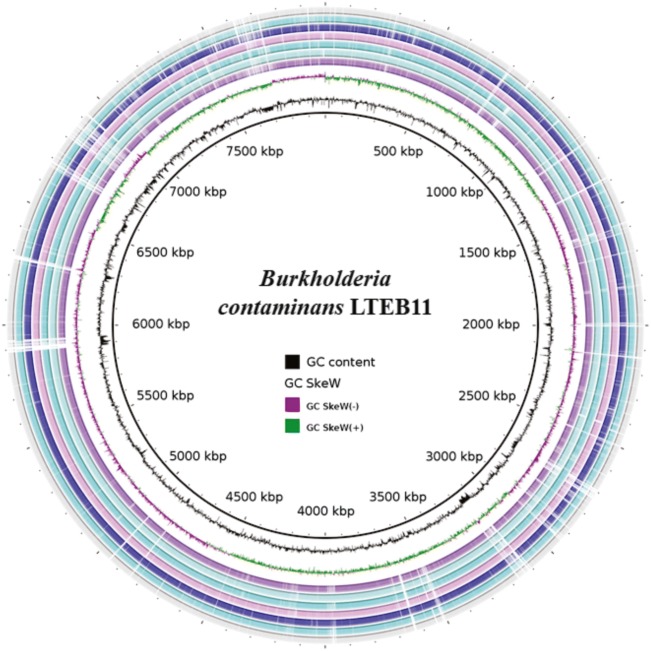
The B. contaminans LTEB11 genome was assembled in 7 contigs organized in three replicons of 3,548,326 bp (chromosome 1), 3,254,142 bp (chromosome 2), and 1,196,160 bp (chromosome 3) (Table 1). The estimated genome size is 7.9 Mb and the GC content is 66.5%. This genome size falls within the range of 7.4 to 9.7 Mb described for the genus Burkholderia [24]. Burkholderia contaminans LTEB11 showed ANI values greater than 97% when compared with B. contaminans strains LMG 23361, FFI-28, FFH2055, and MS14 (Table S1). Also, the highest nucleotide sequence identity of B. contaminans LTEB11 was for B. lata FL7530S1D0 (95%) and B. lata 383 (94%), values that indicate high genome relatedness (Fig.1).
Table 1.
Features of the genome of Burkholderia contaminans LTEB11
| Size (bp) | Contigs | Protein-coding genesa | tRNAs | G + C content (%) | |
|---|---|---|---|---|---|
| Chromosome 1 | 3,548,326 | 4 | 3366 | 56 | 66.7 |
| Chromosome 2 | 3,254,142 | 2 | 3074 | 6 | 66.6 |
| Chromosome 3 | 1,196,160 | 1 | 1113 | 3 | 65.8 |
| Total | 7,998,628 | 7 | 7553 | 65 | 66.3 |
aNote that the number of genes presented in Table 1 is different from the number of genes annotated in the genome in GenBank (accession number GCA_001865715.1). For the present paper, we annotated the genome using the RAST annotator, which considers pseudogenes in the final gene count. The PGAP annotator applied by GenBank disregards pseudogenes
Fig. 1.
Circular representation of the genome of B. contaminans LTEB11 and comparison with the whole-genome sequences of seven Burkholderia strains. Rings from the inside to outside: [1] GC content (black), [2] GC skew (purple and green), [3] BLASTn comparison with B. contaminans MS14, [4] BLASTn comparison with B. contaminans LGM23361, [5] BLASTn comparison with B. contaminans FFH2055, [6] BLASTn comparison with B. contaminans FFI-28, [7] BLASTn comparison with B. lata FL7530S1D0, [8] BLASTn comparison with B. lata 383, and [9] BLASTn comparison with B. cepacia ATCC 25416
In total, 7553 protein-coding genes were predicted (Table 1). Sixty genes were annotated as coding for α/β hydrolases, with 17 of these being classified as esterases or lipases, organized in different superfamilies (Table 2). Among these sequences, the lipAB operon, which codes for the lipase LipA (LipBC) and the lipase-specific foldase LipB (LifBC), was annotated in chromosome 2. In addition, we identified another operon, lipEF, in chromosome 1, with this operon coding for a lipase (LipE) and a foldase (LipF) that have 65% and 53% of identity, respectively, with LipA and LipB. Sequence analysis showed that LipA, LipE, and two more lipases (LipC and LipD) have the typical N-terminal signal sequence, suggesting that these lipases may be secreted by B. contaminans LTEB11. The genes encoding a Sec-translocase complex as well as a type II secretion system (T2SS) were annotated in chromosome 1. These systems are required for the secretion of lipases by Gram-negative bacteria [25].
Table 2.
Lipase and esterase genes identified in the genome of Burkholderia contaminans LTEB11 and compared with those of other strains of B. contaminans and other species of Burkholderia
%Represents identities of amino acid sequences. COG, clusters of orthologous group; Pfam, protein families database; Accession number, National Center for Biotechnology Information. NI, not identified
Comparative analysis showed that LipA, LipC, and LipD are also present in genomes of other isolates of B. contaminans (FFH2055, LMG 23361, MS14, FFI-28, and FFH 2055) and B. lata (383 and FL7530S1D0). However, in these genomes, the operon lipEF described here was only identified in B. lata FL (Table 2). Phylogenetic analysis classified LipA and LipE in the family I.2 of bacterial lipases, next to lipases of B. glumae, B. lata, and C. viscosum, whereas LipC and LipD were classified close to I.3 family members (Fig. S1).
In order to evaluate the lipolytic activity of B. contaminans LTEB11, a crude extract was obtained by submerged fermentation using olive oil 1% (v/v) as an inducer for lipase expression [9]. In addition, recombinant LipA (LipBC) was overexpressed in Escherichia coli and purified according to Alnoch et al. [14]. The activities of both crude extract and recombinant LipA were determined by the titrimetric method using a pHStat (as described in the Supplementary material). The crude extract of B. contaminans LTEB11 showed higher activity (90 U mg−1) against tributyrin than against olive oil (44 U mg−1) (Table 3). The same profile was observed for the recombinant LipA, with activities of 1330 U mg−1 against tributyrin and 790 U mg−1 against olive oil (Table 3). These results suggest that B. contaminans LTEB11 secretes LipA into the medium; however, other lipases or esterases might be produced and secreted during the cultivation. The high activity presented in the crude extract of B. contaminans LTEB11 and shown by recombinant LipA suggests that it would be interesting to characterize further the lipases and esterases produced by B. contaminans LTEB11.
Table 3.
Lipolytic activity of the crude extracts obtained from B. contaminans LTEB11 and recombinant LipA (LipBC) against triacylglycerols
| Lipolytic activity (U mg−1)a | ||
|---|---|---|
| Substrate | Crude extract | LipA |
| Tributyrin | 90 ± 14 | 1330 ± 86 |
| Olive oil | 44 ± 8 | 790 ± 29 |
aThe activity was determined by the titrimetric method using a pHStat, at pH 8.0 and 37 °C. Results are expressed as the average of triplicate assays ± the standard error of the mean
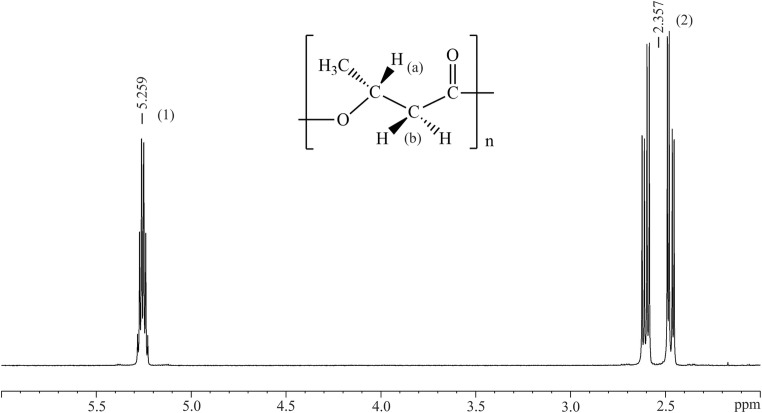
The genome of B. contaminans LTEB11 also contains the pha genes, phaC, phaA, phaB, coding for enzymes involved in the biosynthesis of polyhydroxyalkanoates (PHA); phaR, coding for the transcriptional regulator (PhaR) of the phasin gene phaP; and phaZ, the gene encoding the PHA depolymerase involved in PHA mobilization (Table S2) [26]. PHAs are classified, according to the carbon chain length of the monomers, as either short-chain or medium-chain, the best known PHAs being polyhydroxybutyrate (PHB) and the copolymer poly(3-hydroxybutyrate-co-3-hydroxyvalerate) [27]. PHB accumulation assays were performed according to Matias et al. [28], involving 72 h of cultivation in liquid ISP9 medium containing 2% (w/v) of glucose. After chloroform extraction, only poly-3-hydroxybutyrate (P3HB) was identified by [1]H-nuclear magnetic resonance (NMR) (Fig. 2).
Fig. 2.
[1]H-NMR spectrum of 3-hydroxybutyrate (P3HB) produced by Burkholderia contaminans LTEB11. (1) The multiplet at 5.25 ppm corresponds to 1H (a) in the asymmetric carbon; (2) The doublet of the quadruplet at 2.35 ppm corresponds to 2H (b) in the methylene group adjacent to an asymmetric carbon atom. PHB samples (10 mg) were dissolved in CDCl3 and subjected to analysis. 1H-NMR spectra were acquired for each sample at 600 MHz using an AscendTM 600 spectrometer (Bruker) equipped with a 5-mm QXI inverse probe and a sample case autosampler. PHB accumulation assays were performed according to Matias et al. [28]. Flask cultures containing 500 mL of liquid ISP9 medium (2% (w/v) of glucose) were incubated in a shaker at 30 °C for 72 h, 120 rpm. Culture samples were harvested by centrifugation, lyophilized, and pretreated with two acetone baths and treated with chloroform at 60 °C for 48 h under agitation. After the treatment, the contents of flasks were filtered through Whatman no. 1 filter paper and dried at room temperature until PHB film formation
The genome of B. contaminans LTEB11 also contains genes that code for enzymes required for the synthesis of rhamnolipids (rhlA, rhlB, and rhlC), biosurfactants of particular interest for cosmetic, pharmaceutical, and detergent manufacturers [29]. The genome also contains the ocf gene cluster that has been previously described in B. contaminans MS14, with an identity greater than 90% (Table 4). This cluster includes the ATP-binding cassette (ocfA) and the genes encoding non-ribosomal peptide synthetases (ocfD, ocfE, ocfF, ocfH, and ocfJ). These genes are required for the production of the antifungal compound occidiofungin, which is active against a broad range of plant and animal fungal pathogens [3].
Table 4.
Comparison of genes encoding for occidiofungin biosynthesis in Burkholderia contaminans LTEB11 and B. contaminans MS14
| Burkholderia contaminans LTEB11 | Burkholderia contaminans MS14 | ||
|---|---|---|---|
| Gene | Accession number | %a | Accession number |
| Orf1 - | WP_071335792.1 | 98 | ACN32485.1 |
| ambR1 | WP_071335793.1 | 98 | ACN32486.1 |
| ambR2 | WP_071335794.1 | 95 | ACI01437.2 |
| ocfA | WP_071335795.1 | 100 | ACJ24909.2 |
| ofcB | WP_071336295.1 | 97 | ACL81525.1 |
| ofcC | WP_039355063.1 | 99 | ACL81526.1 |
| ofcD | WP_071335796.1 | 98 | ACL81527.1 |
| ofcE | WP_083417853.1 | 98 | ACL81528.1 |
| ofcF | WP_071335798.1 | 99 | ACN32487.1 |
| ofcG | WP_071335799.1 | 95 | ACN32488.1 |
| ofcH | WP_071335800.1 | 99 | ACN32489.1 |
| ofcI | WP_071335801.1 | 98 | ADT64845.1 |
| ofcJ | WP_071335802.1 | 97 | ADT64846.1 |
| ofcK | WP_071335803.1 | 97 | ADT64847.1 |
| ofcL | WP_071335804.1 | 99 | ADT64848.1 |
| ofcM | WP_039362393.1 | 98 | ADT64849.1 |
| ocfN | WP_071335805.1 | 96 | ADT64850.1 |
aRepresents identities of amino acid sequences. Accession number, National Center for Biotechnology Information
The B. contaminans LTEB11 genome sequence reported here can underpin further studies into the production of new lipases and esterases and mechanisms involved in the regulation of lipase expression, as well as the potential of this bacterium to produce polyhydroxyalkanoates, rhamnolipids, and antifungal compounds with biotechnological relevance.
Nucleotide sequence accession numbers
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession MLFG00000000 and BioSample: SAMN04287748.
Electronic supplementary material
(DOCX 35 kb)
Acknowledgments
We thank Roseli Prado and Valter A. de Baura for technical support.
Funding information
This genome sequencing project was supported by the Brazilian Program of National Institutes of Science and Technology-INCT and the Brazilian Research Council-CNPq/MCT. Research scholarships were granted to Nadia Krieger, David Mitchell, Fábio Pedrosa, Guilherme Sassaki, and Emanuel Souza by CNPq. Robson Alnoch was granted a PhD scholarship by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Coenye T, Vandamme P. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ Microbiol. 2003;5(9):719–729. doi: 10.1046/j.1462-2920.2003.00471.x. [DOI] [PubMed] [Google Scholar]
- 2.Vanlaere E, Baldwin A, Gevers D, Henry D, de Brandt E, LiPuma JJ, Mahenthiralingam E, Speert DP, Dowson C, Vandamme P. Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int J Syst Evol Microbiol. 2009;59(1):102–111. doi: 10.1099/ijs.0.001123-0. [DOI] [PubMed] [Google Scholar]
- 3.Deng P, Wang X, Baird SM, Showmaker KC, Smith L, Peterson DG, Lu S. Comparative genome-wide analysis reveals that Burkholderia contaminans MS14 possesses multiple antimicrobial biosynthesis genes but not major genetic loci required for pathogenesis. Microbiologyopen. 2016;5(3):353–369. doi: 10.1002/mbo3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vandamme P, Dawyndt P. Classification and identification of the Burkholderia cepacia complex: past, present and future. Syst Appl Microbiol. 2011;34(2):87–95. doi: 10.1016/j.syapm.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Parke JL, Gurian-sherman D. Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Environ Prot. 2001;84(5):1229–1236. doi: 10.1111/j.1540-6296.2010.01192.x. [DOI] [PubMed] [Google Scholar]
- 6.Wattanaphon HT, Kerdsin A, Thammacharoen C, Sangvanich P, Vangnai AS. A biosurfactant from Burkholderia cenocepacia BSP3 and its enhancement of pesticide solubilization. J Appl Microbiol. 2008;105(2):416–423. doi: 10.1111/j.1365-2672.2008.03755.x. [DOI] [PubMed] [Google Scholar]
- 7.Villalobos MC, Goncalves AG, Noseda MN, Mitchell DA, Krieger N. A novel enzymatic method for the synthesis of methyl 6-O-acetyl-α-D-glucopyranoside using a fermented solid containing lipases produced by Burkholderia contaminans LTEB11. Process Biochem. 2018;73:86–93. doi: 10.1016/j.procbio.2018.07.023. [DOI] [Google Scholar]
- 8.Fernandes MLM, Saad EB, Meira JA, Ramos LP, Mitchell DA, Krieger N. Esterification and transesterification reactions catalysed by addition of fermented solids to organic reaction media. J Mol Catal B Enzym. 2007;44(1):8–13. doi: 10.1016/j.molcatb.2006.08.004. [DOI] [Google Scholar]
- 9.Salum TFC, Baron AM, Zago E, Turra V, Baratti J, Mitchell DA, Krieger N. An efficient system for catalyzing ester synthesis using a lipase from a newly isolated Burkholderia cepacia strain. Biocatal Biotransformation. 2008;26(3):197–203. doi: 10.1080/10242420701568674. [DOI] [Google Scholar]
- 10.Baron AM, Barouh N, Barea B, Villeneuve P, Mitchell DA, Krieger N. Transesterification of castor oil in a solvent-free medium using the lipase from Burkholderia cepacia LTEB11 immobilized on a hydrophobic support. Fuel. 2014;117:458–462. doi: 10.1016/j.fuel.2013.09.065. [DOI] [Google Scholar]
- 11.Salum TFC, Villeneuve P, Barea B, Yamamoto CI, Côcco LC, Mitchell DA, Krieger N. Synthesis of biodiesel in column fixed-bed bioreactor using the fermented solid produced by Burkholderia cepacia LTEB11. Process Biochem. 2010;45(8):1348–1354. doi: 10.1016/j.procbio.2010.05.004. [DOI] [Google Scholar]
- 12.Soares D, Pinto AF, Gonçalves AG, Mitchell DA, Krieger N. Biodiesel production from soybean soapstock acid oil by hydrolysis in subcritical water followed by lipase-catalyzed esterification using a fermented solid in a packed-bed reactor. Biochem Eng J. 2013;81:15–23. doi: 10.1016/j.bej.2013.09.017. [DOI] [Google Scholar]
- 13.Moure VR, Fabrício C, Frensch G, Marques FA, Mitchell DA, Krieger N. Enhancing the enantioselectivity of the lipase from Burkholderia cepacia LTEB11 towards the resolution of secondary allylic alcohols. Biocatal Agric Biotechnol. 2014;3(2):146–153. doi: 10.1016/j.bcab.2013.09.011. [DOI] [Google Scholar]
- 14.Alnoch RC, Stefanello AA, Paula Martini V, Richter JL, Mateo C, Souza EM, Mitchell DA, Muller-Santos M, Krieger N. Co-expression, purification and characterization of the lipase and foldase of Burkholderia contaminans LTEB11. Int J Biol Macromol. 2018;116:1222–1231. doi: 10.1016/j.ijbiomac.2018.05.086. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Maniatis T, Fritsch EF. 2ed. 1989. Cold Spring Harbor Cold Spring Harbor Laboratory Press NY. Molecular cloning: a laboratory manual. [Google Scholar]
- 16.Gnerre S, Maccallum I, Przybylski D, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 2011;108(4):1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimin AV, Marçais G, Puiu D, Roberts M, Salzberg SL, Yorke JA. The MaSuRCA genome assembler. Bioinformatics. 2013;29(21):2669–2677. doi: 10.1093/bioinformatics/btt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guizelini D, Raittz RT, Cruz LM, Souza EM, Steffens MBR, Pedrosa FO. GFinisher: a new strategy to refine and finish bacterial genome assemblies. Sci Rep. 2016;6(October):34963. doi: 10.1038/srep34963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci. 2009;106(45):19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA (2011) BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12. 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed]
- 22.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ussery DW, Kiil K, Lagesen K, Sicheritz-Ponten T, Bohlin J, Wassenaar TM. The genus Burkholderia: analysis of 56 genomic sequences. Genome Dyn. 2009;6:140–157. doi: 10.1159/000235768. [DOI] [PubMed] [Google Scholar]
- 25.Rosenau F, Tommassen J, Jaeger KE. Lipase-specific foldases. ChemBioChem. 2004;5(2):152–161. doi: 10.1002/cbic.200300761. [DOI] [PubMed] [Google Scholar]
- 26.Urtuvia V, Villegas P, González M, Seeger M. Bacterial production of the biodegradable plastics polyhydroxyalkanoates. Int J Biol Macromol. 2014;70:208–213. doi: 10.1016/j.ijbiomac.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Reddy CSK, Ghai R, Rashmi KVC. Polyhydroxyalkanoates: an overview. Bioresour Technol. 2003;87(2):137–146. doi: 10.1016/S0960-8524(02)00212-2. [DOI] [PubMed] [Google Scholar]
- 28.Matias F, Brandt CA, da Silva ES, de Andrade Rodrigues MF. Polyhydroxybutyrate and polyhydroxydodecanoate produced by Burkholderia contaminans IPT553. J Appl Microbiol. 2017;123(1):124–133. doi: 10.1111/jam.13469. [DOI] [PubMed] [Google Scholar]
- 29.Rahim R, Ochsner UA, Olvera C, Graninger M, Messner P, Lam JS, Soberón-Chávez G. Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol Microbiol. 2001;40(3):708–718. doi: 10.1046/j.1365-2958.2001.02420.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 35 kb)