Abstract
Microbes from hypersaline environments are useful in biotechnology as sources of novel enzymes and proteins. The current study aimed to characterize halophilic bacteria from the rhizosphere of halophytes (Salsola stocksii and Atriplex amnicola), non-rhizospheric, and brine lake-bank soils collected from Khewra Salt Mine and screening of these bacterial strains for industrially important enzymes. A total of 45 bacterial isolates from the rhizosphere of Salsola, 38 isolates from Atriplex, 24 isolates from non-rhizospheric, and 25 isolates from lake-bank soils were identified by using 16S rRNA gene analysis. Phylogenetic analysis showed that bacterial strains belonging to Bacillus, Halobacillus, and Kocuria were dominant in the rhizosphere of halophytes (Salsola and Atriplex), and Halobacillus and Halomonas were dominating genera from non-rhizospheric and lake-bank soils. Mostly identified strains were moderately halophilic bacteria with optimum growth at 1.5–3.0 M salt concentrations. Most of the bacterial exhibited lipase, protease, cellulase, amylase, gelatinase, and catalase activities. Halophilic and halotolerant Bacilli (AT2RP4, HL1RS13, NRS4HaP9, and LK3HaP7) identified in this study showed optimum lipase, protease, cellulase, and amylase activities at 1.0–1.5 M NaCl concentration, pH 7–8, and temperature 37 °C. These results indicated that halophilic and halotolerant bacteria can be used for bioconversion of organic compounds to useful products under extreme conditions.
Electronic supplementary material
The online version of this article (10.1007/s42770-019-00044-y) contains supplementary material, which is available to authorized users.
Keywords: Halophilic bacteria, 16S rRNA gene, Hydrolytic enzymes, Salsola stocksii, Atriplex amnicola
Introduction
The rhizosphere is considered one of the most diverse microbial habitats with respect to species richness and community size [1]. The plant root is divided into three compartments, the rhizosphere (soil close to the root surface), rhizoplane (root), and endosphere or histoplane (root interior), and each of these compartments was found to harbor a distinct microbiome. The rhizosphere of halophytes (Suaeda fruticosa, Kochia indica, Atriplex amnicola, and Salsola stocksii) harbors a variety of microorganisms (microbiome) that have the ability to promote plant growth by increasing the availability and uptake of carbon, nitrogen, and minerals from the soil [2, 3]. These microorganisms provide protection against plant pathogens and contribute significantly to the well-being and salinity tolerance of halophytes [4].
Halophiles provide a potential source of novel enzymes that function under salt stress conditions, such as lipases proteases, amylases, gelatinases, and xylanases with polyextremophilic properties [5]. Halophilic enzymes are capable of functioning under high salt concentrations, wide range of temperatures, and pH at which other proteins denature. Certain enzymes that halophiles synthesize are useful for bioremediation of pollutants in saline habitats [6] or are important biomolecules, for example exopolysaccharides and phytohormones [7]. The halophilic enzymes have also been used in many industries including pharmaceutical, textile, detergent, baking, paper, and pulp industries [8]. A number of halotolerant and halophilic bacteria and archaea such as Bacillus, Halobacillus, Halomonas, Salinibacter, Haloarcula, and Haloferax have been explored for their ability to hydrolyze enzymes like protease, amylase, and lipase [9]. Halophilic enzymes are also considered as important biocatalysts under low water conditions, such as hypersaline environments and non-aqueous media [10].
In Pakistan, Khewra Salt Mine provides a rich and extensive habitat for halophytes and halophilic microorganisms. The halophytes (S. fruticosa, K. indica, A. amnicola, and S. stocksii) growing here are not only producers of medicinal compounds that can be used to cure diseases such as cough, flu, and cold but also used as a food source [11]. Atriplex and Salsola are important biomass producers in barren lands of this area [12]. The present work endeavors to identify and characterize halophilic and halotolerant bacterial diversity isolated from the rhizosphere of halophytes (S. stocksii and A. amnicola), non-rhizosphere, and brine lake-bank soil samples collected from Khewra Salt Mines, Pakistan, and screening of industrially important enzymes (proteases, amylases, lipases, cellulases, gelatinase, and catalases) produced by these organisms. Here, we also determined the effect of salt concentration, pH, and temperature on lipase, protease, amylase, and cellulase activities of halophilic strains.
Material and methods
Soil and root samples collection
Khewra Salt Mine is the world’s second largest salt mine, located near Pind Dadan Khan Tehsil of Jhelum District, Punjab, Pakistan. Based on its origin, Khewra Salt Mine like other hypersaline bodies is classified as thalassic because it is derived from evaporation of sea water [13]. It has Na+ and Cl− dominating ions, and the pH is near neutral to slightly alkaline. Geographically, it is located about 32° 38′ North latitude, 73°10′ East longitude and an elevation of 313–360 above the sea level about 200 km from Islamabad (Fig. S1). We surveyed an area approximately 1.12 km from the Khewra Salt Mines. The sampling area was selected according to land use and vegetation cover. Vegetation of this area is classified as sub-tropical dry evergreen forest. Rhizospheric soil samples were collected by gently removing the plants and obtaining the soil attached to the roots. For non-rhizospheric saline soil samples, the upper 8–10 cm of mineral soil was collected. Brine lake-bank soil samples were collected from the bank of a salt lake. At each site, soil samples of approximately 500 g each from four different locations were collected in black sterile polythene bags. These samples were stored at 4 °C for further analysis.
Soil physicochemical parameters
Each soil sample (300 g) was thoroughly mixed and sieved through a pore size of 2 mm. Physical properties (pH, moisture content, salinity, and temperature) of the soil samples from different plants and non-rhizospheric soils samples were determined. Moisture (%), temperature, and texture class were measured by the Anderson method [14]; pH was measured by 1:2.5 (w/v) soil to water mixture; and electrical conductivity (dS/m) was measured by 1:1 (w/v) soil to water mixture at 25 °C [15]. Organic matter (Corg) was calculated by the Walkley-Black method [16]. Cation exchange capacity (CEC) is the capacity to retain and release cations (Ca2+, Mg2+, K+, and Na+) and sodium adsorption ratio (SAR) is the measure of the sodicity of the soil which is calculated as the ratio of the sodium to the magnesium and calcium.
Isolation of culturable halophilic bacteria
Halophilic medium (HaP) (tryptone 5 g/l, yeast extract 1 g/l, NaCl 88 g/l, 5 g/l KCl, 10 g/l MgSO4, 2 g/l K2HPO4, and pH 7.2) was used for the isolation and purification of bacteria present in saline environments [17]. Rhizosphere was fractionated into rhizosphere fraction (RS), rhizoplane fraction (RP), and root endosphere or histoplane bacterial fraction (HP) according to the method described by Malik et al. [18]. RS fraction indicates the soil adhering with the roots; RP fraction is the root surface; and HP is the interior of the roots. In the case of RS, the soil was mixed thoroughly, sieved, and then 1-g representative soil sample was taken. Bacterial fraction from RP was isolated by shifting 1 g of washed root to a falcon tube containing 9 ml saline along with some pebbles and incubated in a shaker for 30 min. For the isolation of HP, bacterium roots were sealed at both ends with wax after washing with water. Sealed roots were surface-sterilized by using 10% bleach for 10 min. After sterilization, waxed ends of the roots were removed and roots were macerated by using FastPrep® instrument (MP Biomedicals). The soil from each non-rhizospheric and lake-bank soils was mixed thoroughly, sieved, and then 1-g representative soil sample was taken. Serial dilutions (10–1–10–10) were made for all samples [19]. The dilutions from 10−3 to 10−6 were inoculated on HaP plates for counting colony-forming units (CFU) per gram of dry weight. Plates were incubated at 37 °C until the appearance of bacterial colonies. Bacterial colonies were counted and the number of bacteria per gram sample was calculated. The bacteria were purified by repeated sub-culturing of single colonies. Single colonies were selected, grown in HaP broth, and stored in 33% glycerol at − 80 °C for subsequent characterization.
Screening of the isolated bacterial strains with respect to their salt, pH, and temperature tolerance ability
Bacterial isolates were grown in the presence of different salt concentrations (1.5–4.5 M NaCl), pH ranges of 4–12, and temperature ranges of 4–42 °C by using HaP broth medium. Isolates were cultured in 250 ml flasks at 37 °C with continuous rotatory agitation at 150 rpm for 72 h. During incubation, bacterial growth in terms of optical density (OD 600) was measured after different time intervals (3, 6, 12, 24, 48, and 72 h).
Molecular characterization and phylogenetic analysis
Genomic DNA was isolated by CTAB method [20]. PCR amplifications of 16S rRNA were performed by using universal forward and reverse primers P1 (5′-GAGAGTTTGATCCTGGTCAGAACGAAC-3′) and P6 (5′-CGTACGGCTACCTTGTTACGACTTCACC-3′) for prokaryotes [21]. A PCR reaction of 50 μl was prepared by using Taq polymerase (5 U) 0.5 μl, Taq buffer (10×) 2 μl, MgCl2 (25 mM) 2.5 μl, dNTPS (2.5 mM) 2 μl, 2 μl each of forward and reverse primers (10 pmol), 36 μl of dd.H2O, and 3 μl of template DNA. First denaturation step was at 95 °C for 5 min followed by 35 cycles of 94 °C for 1 min, 55 °C for 1 min and 72 °C for 2 min, and a final extension step was at 72 °C for 10 min as described by Tan et al. [21]. PCR products were analyzed by using 1% agarose gel. PCR products were purified by using GeneJET PCR Purification Kit (K0702 - Thermo Fisher Scientific). Purified PCR products were sequenced by using forward and reverse primers (Eurofins, Germany).
Acquired sequences were assembled and analyzed with the help of Chromus Lite 2.01 sequence analysis software (Technelysium Pty Ltd., Australia). The gene sequences were compared to those deposited in the GenBank nucleotide database using the NCBI BLAST program. Sequences were aligned using Clustal X 2.1 program, and phylogenetic tree was constructed using neighbor-joining method [22]. Bootstrap confidence analysis was performed on 1000 replicates to determine the reliability of the distance tree topologies obtained [23]. The evolutionary distances were computed using the Maximum Composite Likelihood method [24] and are in the units of number of base substitutions per site. All positions containing gaps and missing data were eliminated from the dataset (complete deletion option). Phylogenetic analyses were conducted in MEGA7 [25]. There were a total of 1457 positions in the final dataset. Bacterial sequences were deposited in the GenBank database under the accession numbers of LT221118–LT221260.
Calculation of diversity indices
An operational taxonomic unit (OTU) was defined as a 16S ribosomal DNA (rDNA) sequence group in which sequences differed by less than 3%. Phylotype richness (S) was calculated as the total number of OTUs. Shannon and Simpson indices are diversity-measuring parameters which are commonly used to characterize species diversity in a community. Shannon index shows the uniformity of species and its abundance in OTUs while Simpson index is used to measure the number of species present in a community as well as the relative abundance of each species [26].
Enzyme assays for bacterial isolates
Protease activity was tested on the medium described by Kumar et al. [27]. Amylase and cellulose activities were identified by using 2% iodine solution and spotting single colony of the bacterial strains on CMC (carboxymethyl cellulose 1%) agar plates, respectively [28]. Catalase was identified by using H2O2 and pure culture colonies from agar plates [29]. Lipase activity was tested by using HaP medium with 1% butyrin and Tween 80 hydrolysis assay as described by Sierra [30]. Test for gelatin hydrolysis was performed by using the method described by Pitt and Dey [31]. Urea hydrolysis test was performed by using Christensen’s method [32]. The clear zones around the bacterial colonies after 4–12 days of incubation at 37 °C were considered as a positive result of protease, cellulase, and lipase activities.
Determination of optimal salt concentration, pH and temperature for enzymes activities
Four bacterial strains, two from rhizospheric soils (HL1RS13 and AT2RP4), one from non-rhizospheric soil of halophytes (NRS4HaP9), and one from lake-bank soils (LK3HaP7), were selected for determination of enzymes’ activity at salt concentrations 0–2.5 M NaCl, pH 6–11, and temperature 4–45 °C. The optimal NaCl concentrations, pH, and temperature for each of the enzyme’s activity were determined by using standard protocols as described above. The effect of pH and temperature on enzymes’ activity was determined by using optimal NaCl concentration. Buffer solutions including Tris-HCl buffer (pH 8), phosphate buffer (pH 7), and citrate-phosphate buffer (pH 5 and 6) were used in this study.
Statistical analysis
One-way ANOVA was applied to analyze microbial diversity differences among rhizospheric and non-rhizospheric soil samples, and significance at the 5% level was tested by the least significance difference test (LSDT) by using STATISTIX software (8.2 version).
Results
Physical and chemical properties of saline soil samples
Rhizospheric and non-rhizospheric soils were characterized by a great spatial variability and covered a significant variation in soil pH, salinity, organic matter, texture class, CEC, and SAR. Non-rhizospheric soils were more alkaline in nature as compared to rhizospheric soils with soil pH S. stocksii (8.21), A. amnicola (7.62), non-rhizospheric soils (8.25), and lake-bank soils (8.54). Electrical conductivity (dS/m) of the soil samples ranged from 5.67 to 6.55 dS/m, with the highest values in lake-bank soils and the lowest in rhizospheric soils of A. amnicola; soil temperature ranged from 21.02 to 25.23 °C and moisture contents from 24.11 to 36.12% (Table 1). The total organic matter content ranged from 29.23 to 36.27 g/Kg with the highest values in soil samples of S. stocksii and the lowest in brine lake-bank soils. CEC (cation exchange capacity) values ranged from 67.55 to 72.42 mg/dm3 and SAR (sodium adsorption ratio) values from 10.78 to 14.45 with the highest values in non-rhizospheric soils and the lowest values in brine lake-bank soils.
Table 1.
Physicochemical properties of rhizospheric and non-rhizospheric soil samples of halophytes (S. stocksii and A. amnicola) and brine lake-bank soil samples
| Parameters | S. stocksii | A. amnicola | Non-rhizospheric saline soil samples | Brine lake-bank soil samples |
|---|---|---|---|---|
| pH | 8.21ab | 7.62a | 8.25ab | 8.54b |
| EC1:1 (dS/m) | 6.14ab | 5.67a | 5.99ab | 6.55b |
| Moisture (%) | 27.31ab | 24.11a | 27.12b | 36.12ab |
| Temperature (°C) | 25.23a | 22.15b | 24.21ab | 21.02ab |
| Texture class | Silty loam | Sandy loam | Sandy loam | Sandy loam |
| OM (g Kg−1) | 36.27b | 31.95ab | 30.65a | 29.23a |
| P (mg kg−1) | 3.89ab | 3.21a | 3.85ab | 3.48ab |
| K (mg kg−1) | 0.76a | 0.58b | 0.65b | 0.49a |
| Ca (mg kg−1) | 1.72b | 1.68b | 1.41a | 1.38a |
| Mg (mg kg−1) | 1.48b | 1.05a | 1.56b | 1.19a |
| NO−3 (mg kg−1) | 12.95b | 13.54b | 10.21a | 10.67a |
| H + Al (mg kg−1) | 67.35b | 59.15a | 60.02a | 64.97b |
| V (mg kg−1) | 4.13b | 3.87a | 4.18b | 3.76a |
| CEC (mg dm−3) | 71.49b | 67.55a | 69.76ab | 72.42b |
| SAR | 13.14ab | 12.15a | 14.45b | 10.78a |
EC, electrical conductivity; OM, organic matter; P, phosphorous; K, potassium; Ca, calcium; Mg, magnesium; NO−3, nitrate ion; H + Al, potential acidity; V, base saturation index; CEC, cation exchange capacity; SAR, sodium adsorption ratio. Letters represent statistically significant values at 5% level
Quantification of bacterial populations
Halophilic bacteria were found to be abundant in the rhizosphere and roots of halophytes (S. stocksii and A. amnicola), non-rhizospheric, and brine lake-bank soils. The values of CFU (colony-forming unit) ranged from 32 × 106 to 66 × 106 with the highest number of bacteria isolated from non-rhizospheric soils and the lowest from the endosphere of S. stocksii (Fig. S2 and Table S1). The maximum number of bacterial isolates was obtained from the rhizosphere of A. amnicola (18 isolates) and minimum number of bacteria identified from brine lake-bank soils (13 isolates). Micrococcus, Enterobacter, Bacillus, Pseudomonas, and Aeromonas were common genera in all Salsola and Atriplex plants. Micrococcus and Bacillus were common genera in all non-rhizospheric and brine lake-bank soils (Fig. S2 and Table S1). At each site, certain bacterial species prevailed better than others.
Phenotypic characterization of bacterial isolates
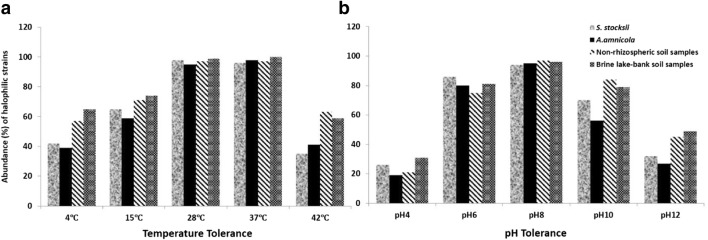
From the rhizosphere of Salsola and Atriplex plants, non-rhizospheric and lake soils, most of the strains could deter up to 3.0 M NaCl while some could grow up to the range of 4 M NaCl (Figs. 1 and 2). Growth of most of the strains was quite better at 1.5 M NaCl and generally growth decreased with the increase in salt concentration. Isolates from the rhizosphere of Atriplex plant samples and non-rhizospheric soil samples showed better growth at high salt concentrations (3.0–4 M NaCl) as compare to isolates obtained from the rhizosphere of Salsola plant samples and lake soil samples (Fig. 1a). On the basis of salt tolerance, 52% bacterial strains were halotolerant, 40% were moderately halophilic, and 8% were extremely halophilic from the rhizosphere of Salsola plants; 53% isolates were halotolerant, 34% were moderately halophilic, and 13% were extremely halophilic from the rhizosphere of Atriplex plants; 46% isolates were halotolerant, 37% were moderately halophilic, and 17% were extremely halophilic from non-rhizospheric soils; and 57% isolates were halotolerant, 34% were moderately halophilic, and 9% were extremely halophilic from lake-bank soils (Fig. 1b). Maximum bacterial growth was observed at pH 6, pH 8, and pH 10 as compared to pH 4 and pH 12 (Fig. 2b). More than 90% of the isolates were able to grow at 28 °C and 37 °C from all the rhizospheric and non-rhizospheric soils. Some bacterial strains could tolerate 4 °C and some could grow well even at 42 °C (Fig. 2a).
Fig. 1.
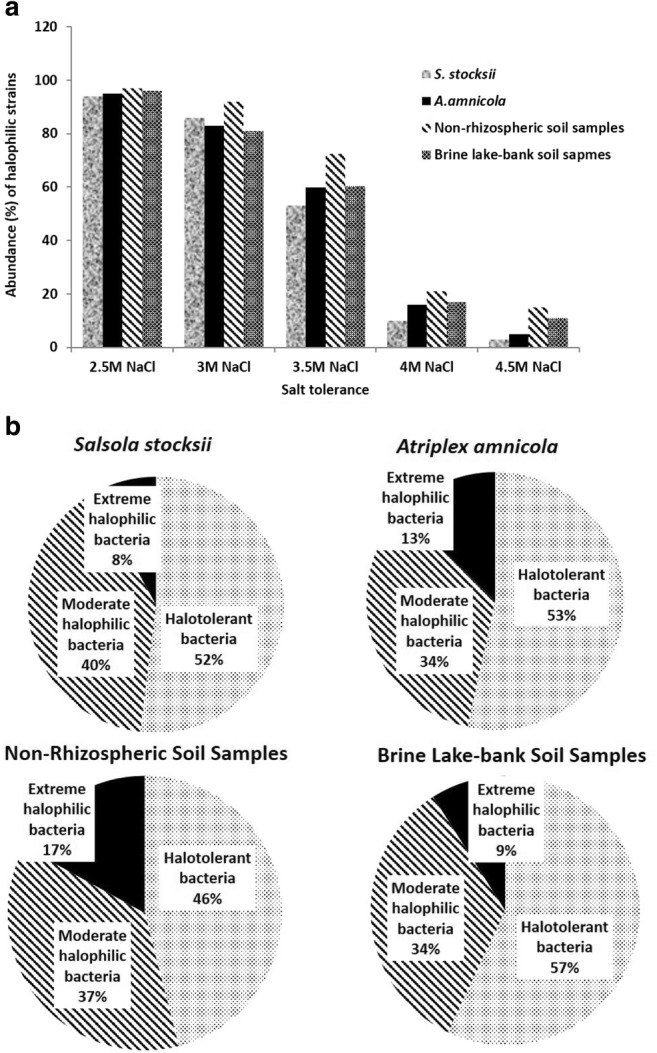
a Salt tolerance profile of halophilic bacterial isolates. b Comparison of halotolerant, moderately halophilic, and extremely halophilic bacteria from the rhizosphere of Salsola and Atriplex, non-rhizospheric, and lake soil samples
Fig. 2.
Phenotypic characterization of halophilic bacterial isolates from the rhizosphere of Salsola and Atriplex, non-rhizospheric, and lake soil samples. a Temperature tolerance profile. b pH tolerance profile
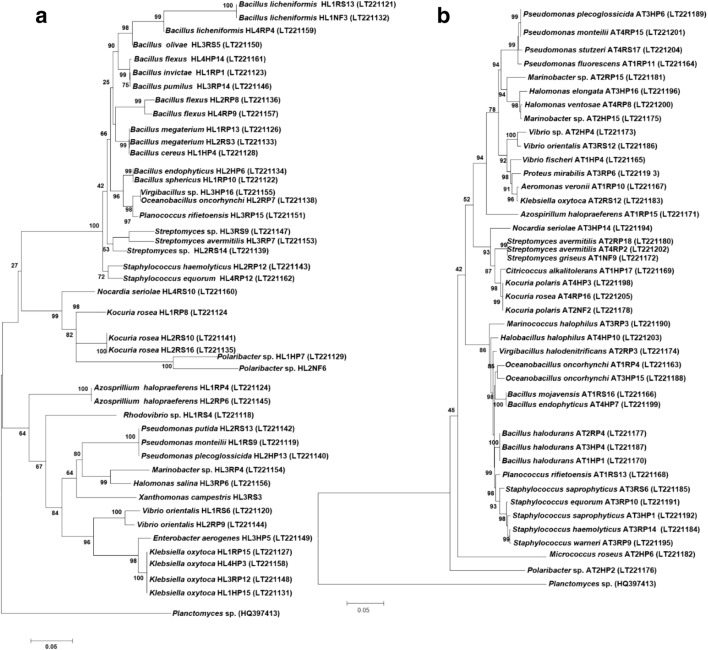
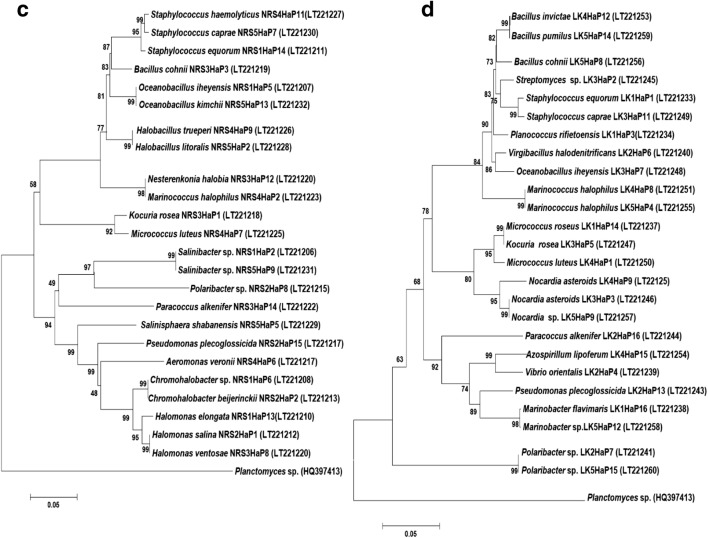
Phylogenetic analysis of halophilic bacteria based on 16S rRNA gene sequences
On the basis of phenotypic characterization, 45 strains were selected from the rhizosphere, rhizoplane, and endosphere of Salsola plant samples for 16S rRNA gene analysis. These isolates were represented by four phyla with the majority (48.89%) of the isolates belonging to the phylum Firmicutes. Bacterial isolates from Salsola plant samples were assigned to 19 genera. Members of the bacterial genera, Bacillus (28.89%), Pseudomonas (11.11%), Kocuria (6.67%), and Klebsiella (6.67%) represented 53.34% of all the isolates from Salsola plant samples (Table S2 and S3; Fig. 3a). From the rhizosphere, rhizoplane, and endosphere of Atriplex plant samples, 38 halophilic bacterial strains were represented by four bacterial phyla: Firmicutes (42.11%), Proteobacteria (42.11%), Actinobacteria (13.15%), and Bacteroidetes (2.63%). Bacterial isolates from Atriplex plant samples were assigned to 18 genera. About 18.42% bacterial isolates were identified as different species of the genus Bacillus, 13.16% isolates were related to Pseudomonas, and 7.89% were related to Kocuria (Table S2 and S4; Fig. 3b). The total 24 bacterial isolates from non-rhizospheric saline soil samples were grouped into four phyla: Firmicutes (44%), Proteobacteria (36%), Actinobacteria (16%), and Bacteroidetes (4%) and 17 bacterial genera. Members of the bacterial genera Staphylococcus (12%), Halomonas (12%), Bacillus (8%), Oceanobacillus (8%), Halobacillus (8%), and Streptomyces (8%) represented 56% of all the isolates from non-rhizospheric saline soil samples (Table S2 and S5; Fig. 3c). The phylogenetic analysis showed that isolates from lake-bank soils could be grouped into four phyla: Firmicutes (51.85%), Proteobacteria (37.04%), Actinobacteria (3.7%), and Bacteroidetes (7.41%). Bacillus and Staphylococcus were the major bacterial genera, comprising 14.82% and 11.11% of the total population, respectively (Table S2 and S6; Fig. 3d).
Fig. 3.
Phylogenetic tree based on partial 16S rRNA gene sequences of halophilic bacterial isolates from the rhizosphere of Salsola (a), Atriplex (b), non-rhizospheric soil samples (c), and brine lake-bank soil samples (d). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches
Calculation of diversity indices
Phylotype richness (S), Shannon diversity index (H), evenness (EH), and Simpson index (D) were calculated. Phylotype richness (S) of the bacterial communities from the rhizosphere of S. stocksii and A. amnicola was calculated as 32 and 30 and for non-rhizospheric and lake-bank soils calculated as 24 and 23, respectively (Table 2). Shannon diversity index (H) was 3.22 and 3.15 for bacterial communities from the rhizosphere of S. stocksii and A. amnicola and was 2.97 and 2.95 for non-rhizospheric and lake-bank soils, respectively. Evenness (EH) was maximum (0.91) in case of bacterial communities from non-rhizospheric soils. Simpson index (D) was 0.941 and 0.829 for bacterial communities from the rhizosphere of S. stocksii and A. amnicola, respectively. Simpson index (D) of the bacterial communities from non-rhizospheric and lake-bank soils was calculated as 0.912 and 0.821, respectively (Table 2). Shannon indices confirmed that microbial community from the rhizosphere of S. stocksii and A. amnicola had more diversity as compared to non-rhizospheric and lake-bank soils.
Table 2.
Phylotype richness, diversity indices, and evenness in microbial communities from the rhizosphere of S. stocksii and A. amnicola, non-rhizospheric soil samples, and brine lake-bank soil samples
| Soil sample | Total number of OTUs | Phylotype richness (S) | Shannon–Wiener indexa (H) | Evennessb (EH) | Simpson indexc (D) |
|---|---|---|---|---|---|
| S. stocksii rhizosphere | 47 | 32 | 3.22 | 0.86 | 0.941 |
| A. amnicola rhizosphere | 42 | 30 | 3.15 | 0.75 | 0.829 |
| Non-rhizospheric soil samples | 27 | 24 | 2.97 | 0.91 | 0.912 |
| Brine lake-bank soil samples | 27 | 23 | 2.95 | 0.85 | 0.821 |
aShannon–Wiener index was calculated as H = − SUM[(pi) × ln(pi)] where pi is the frequency of the species
bEvenness was calculated as Hmax = ln(S)
cSimpson index (D) was calculated as D = ∑(n / N)2 where n = the total number of organisms of a particular species and N = the total number of organisms of all species. The value of Simpson index ranges between 0 and 1
Screening of hydrolytic enzymes
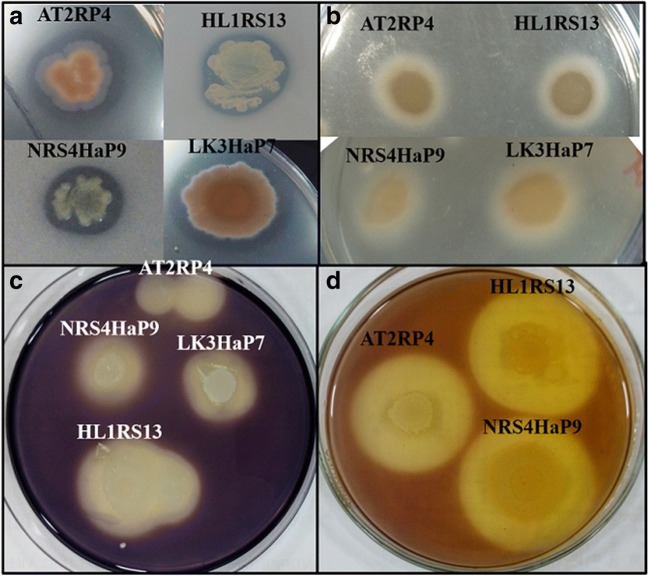
Halophilic bacterial strains with salt tolerance more than 2.5 M NaCl were selected for enzyme screening (protease, amylase, lipase, cellulase, urease, gelatinase, and catalases). In the case of enzyme profile, maximum strains showed catalase, lipase, and protease activities from all rhizospheric and non-rhizospheric soils. Out of the 45 strains identified from the rhizosphere of S. stocksii, more than 50% showed positive results for catalase, protease, gelatinase, and lipase activities (Table 3 and Fig. 4). About 84% strains showed gelatinase activity, 76% strains showed catalase activity, 74% strains showed lipase activity, and 47% strains showed positive results for proteolytic and amylase activities from the rhizosphere of A. amnicola (Table 3 and Fig. 4). More than 60% of bacterial strains from non-rhizospheric soils showed positive results for protease, lipase, gelatinase, and catalase. From brine lake-bank soils, most of the strains showed catalase, gelatinase, and lipase activities (Table 3 and Fig. 4). Mostly Bacillus, Halobacillus, Oceanobacillus, Virgibacillus, and Kocuria strains had the ability to degrade proteins, carbohydrates, and lipids.
Table 3.
Enzyme profile of halotolerant and halophilic bacterial isolates
| Soil samples | Abundance n (%) | Enzymatic activity n (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| Protease | Lipase | Cellulase | Amylase | Urease | Gelatinase | Catalase | ||
| S. stocksii | 45 (34) | 24 (53) | 31 (68) | 18 (40) | 15 (33) | 13 (29) | 26 (58) | 34 (75) |
| A. amnicola | 38 (29) | 18 (47) | 28 (74) | 16 (42) | 18 (47) | 9 (24) | 32 (84) | 29 (76) |
| Non-rhizospheric saline soils | 24 (18) | 18 (75) | 15 (62) | 14 (58) | 9 (37) | 5 (20) | 18 (75) | 20 (83) |
| Lake-bank soils | 25 (19) | 9 (36) | 15 (60) | 13 (52) | 7 (28) | 8 (32) | 21 (84) | 19 (76) |
Fig. 4.
Enzyme assays for halophilic bacterial isolates. Protease (a), lipase (b), amylase (c), and cellulase (d)
Effect of salt, pH, and temperature on lipase activity of bacterial strains
Four bacterial strains AT2RP4 (Bacillus halodurans), HL1RS13 (Bacillus licheniformis), NRS4HaP9 (Halobacillus trueperi), and LK3HaP7 (Oceanobacillus iheyensis) that showed better results as compared to others on plate assay were selected to study the effect of NaCl concentrations, pH, and temperature on lipase activity. The enzyme activity was quantified by using HaP medium with 0–2.5 M NaCl and 1% Tween 80. AT1RP7 and NRS4HaP9 showed halophilic lipase activity with maximum value of 2045 and 3456 U/mg at 1.5 M NaCl while HL1RS13 and LK3HaP7 showed halotolerant activity with maximum enzyme production (3512 and 3162 U/mg) at 1 M NaCl (Fig. 5a). All the strains showed maximum lipase activity at pH 8 and temperature 37 °C (Fig. 5b, c).
Fig. 5.
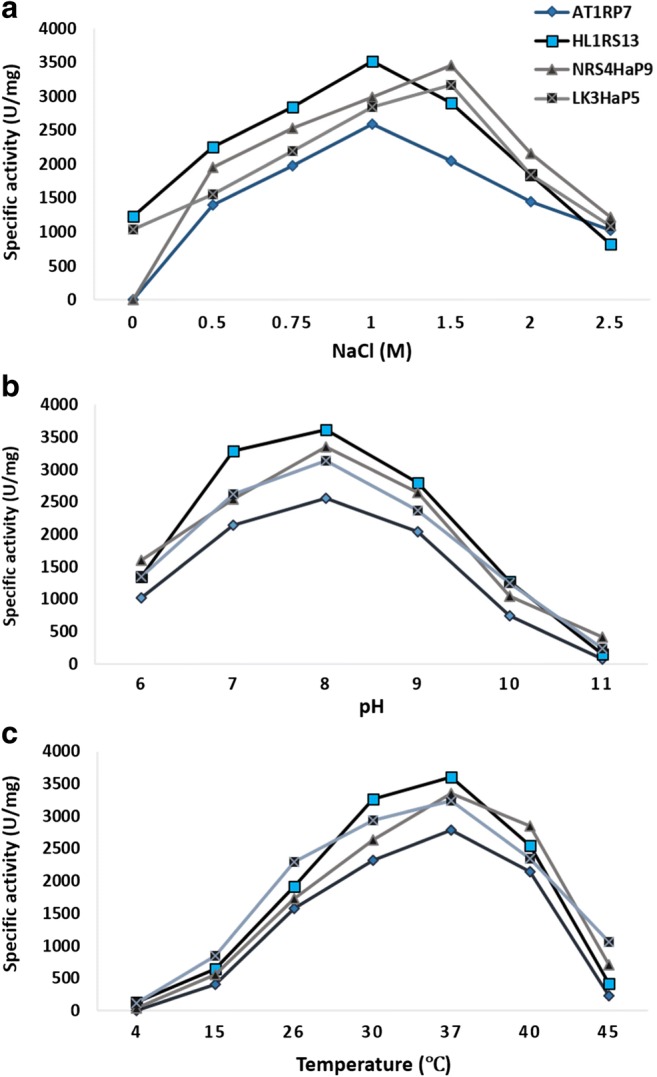
Effect of salt, pH, and temperature on lipase activity of halophilic strains AT2RP4, HL1RS13, NRS4HaP9, and LK3HaP7. Specific lipase activity (U/mg) was plotted against different concentrations of NaCl (0–2.5 M) (a), pH 6–11 (b), and temperature 4–45 °C (c). For each strains, the data represent an average of three independent experiments with standard error
Effect of salt, pH, and temperature on protease activity of bacterial strains
Bacterial strains AT2RP4, NRS4HaP9, and LK3HaP7 showed maximum protease activity 3045, 2956, and 2662 U/mg, respectively, at 1.5 M NaCl while HL1RS13 showed maximum enzyme activity 3023 U/mg at 1 M NaCl (Fig. 6a). Bacterial strains AT2RP4 and HL1RS13 isolated from rhizosphere of halophytes showed maximum protease activity at pH 7 while NRS4HaP9 and LK3HaP7 isolated from non-rhizospheric soils showed maximum activity at pH 8 (Fig. 6b). All the strains showed maximum protease activity at 37 °C (Fig. 6c).
Fig. 6.
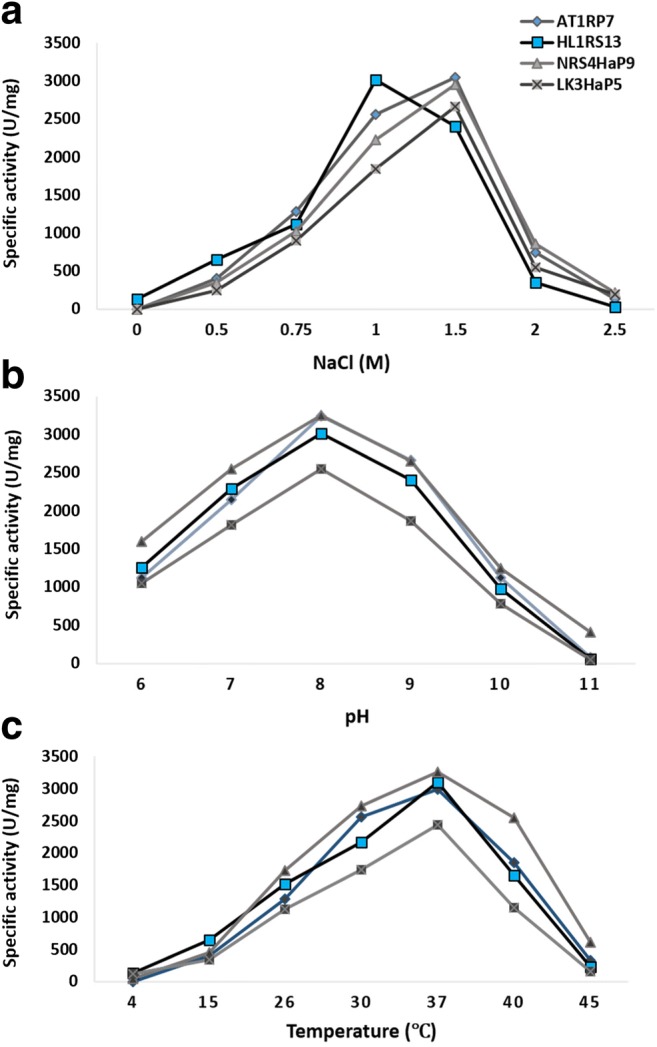
Effect of salt, pH, and temperature on protease activity of halophilic strains AT2RP4, HL1RS13, NRS4HaP9, and LK3HaP7. Specific protease activity (U/mg) was plotted against different concentrations of NaCl (0–2.5 M) (a), pH 6–11 (b), and temperature 4–45 °C (c). For each strains, the data represent an average of three independent experiments with standard error
Effect of salt, pH, and temperature on amylase activity of bacterial strains
To study the effect of NaCl, pH, and temperature on amylase activity of bacterial strains, quantitative amylase assay was carried by using starch as substrate and supernatant as enzyme source. Bacterial strains AT2RP4, NRS4HaP9, and LK3HaP7 showed maximum amylase activity 3104, 1976, and 2878 U/mg, respectively at 1.5 M NaCl while HL1RS13 showed maximum protease activity 3311 U/mg at 1 M NaCl (Fig. 7a). All the strains showed maximum lipase activity at pH 8 (Fig. 7b). Bacterial strains AT2RP4, NRS4HaP9, and LK3HaP7 showed maximum protease activity at 37 °C while HL1RS13 showed maximum protease activity at 30 °C (Fig. 7c).
Fig. 7.
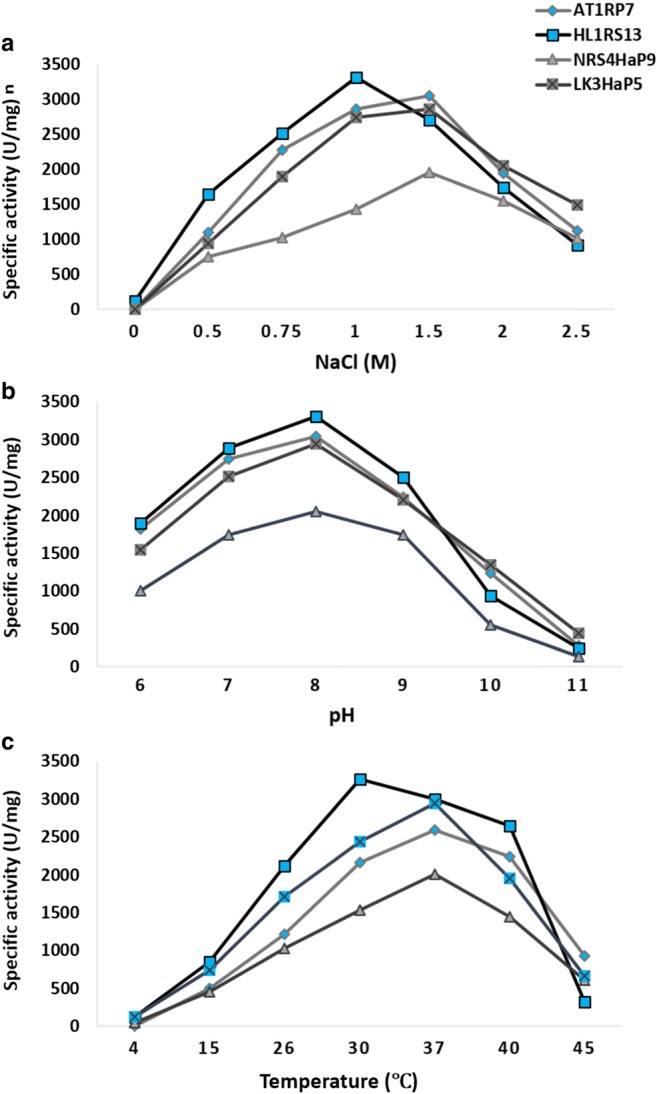
Effect of salt, pH, and temperature on amylase activity of halophilic strains AT2RP4, HL1RS13, NRS4HaP9 and LK3HaP7. Specific amylase activity (U/mg) was plotted against different concentrations of NaCl (0–2.5 M) (a), pH 6–11 (b), and temperature 4–45 °C (c). For each strains, the data represent an average of three independent experiments with standard error
Effect of salt, pH, and temperature on cellulase activity of bacterial strains
The maximum cellulase activity of halophilic bacterial strains HL1RS13, NRS4HaP9, and LK3HaP7 at 1.5 M NaCl was 2712, 3331, and 2202 U/mg, respectively. Bacterial strain AT2RP4 showed maximum cellulase activity 3251 U/mg at 1 M NaCl (Fig. 8a). All the strains showed maximum cellulase activity at pH 8 (Fig. 8b). Bacterial strains AT2RP4, NRS4HaP9, and LK3HaP7 showed maximum cellulase activity at 37 °C while HL1RS13 showed maximum cellulase activity at 30 °C (Fig. 8c).
Fig. 8.
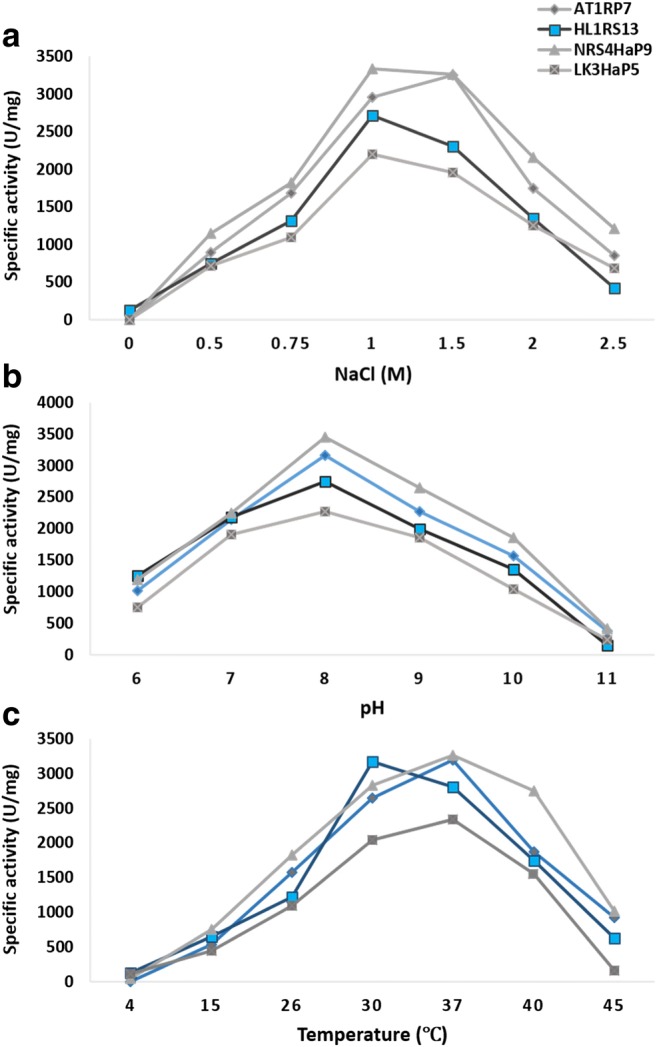
Effect of salt, pH, and temperature on cellulase activity of halophilic strains AT2RP4, HL1RS13, NRS4HaP9, and LK3HaP7. Specific cellulase activity (U/mg) was plotted against different concentrations of NaCl (0–2.5 M) (a), pH 6–11 (b), and temperature 4–45 °C (c). For each strains, the data represent an average of three independent experiments with standard error
Discussion
In the recent years, saline soils received a great attention due to the shortage of arable land and increasing demand of restorations of areas affected by secondary salinity [33]. Soil microbiome in salt-affected lands and halophyte microbiome have a great biotechnological potential that can be used to in some kind of restoration or conservation techniques of saline environments [34, 35]. In the present study, identification of halophilic and halotolerant bacterial diversity from the rhizosphere of halophytes (S. stocksii and A. amnicola), non-rhizospheric, and brine lake-bank soils was carried out. Here, we also described the screening and characterization of extracellular hydrolytic enzymes produced by these bacteria.
Various environmental factors such as soil salinity, pH, moisture, temperature, and stage of development of host plant influence the bacterial community of the plant [36]. It was observed that bacterial population density from different saline environments (rhizosphere of halophytes, non-rhizosphere, and lake-bank soil samples) showed similar pattern. Most of the strains were moderately halophiles but some were characterized as extremely halophilic bacteria. Moderately halophilic microorganisms form phylogenetically and ecologically very diverse group that includes a great variety of halotolerant and halophilic microorganisms.
Maximum bacterial strains were able to grow at pH range of 4–10 from all the soil samples. More than 90% of bacterial isolates grew well at 25 to 40 °C but some could also grow at 4 °C and 42 °C. Previous studies also reported that moderately halophiles and mesophiles are more abundant as compare to extremely halophilic and thermophilic bacteria in different soils [34, 37].
Bacterial strains belonging to the genus Bacillus were the most abundant in the rhizosphere of halophytes (Salsola and Atriplex) and non-rhizospheric soils. Bacillus-like halophilic bacteria have been previously isolated from various environments like deep-sea hypersaline sediments and saline soils [38, 39]. They have high agricultural, industrial, medical, and biotechnological importance [7, 40]. Halophilic Bacillus strains have a wide range of applications in bioenzyme production, biodefense, biofuel production, and bioremediation of organic toxic compounds [41, 42]. Members of the genus Staphylococcus (S. equorum, S. haemolyticus, and S. saprophyticus) were identified from the rhizosphere of Salsola and Atriplex and lake-bank soils. They are moderately halophilic strains and play an important role in plant growth and biodegradation of hemicelluloses and lipases [43, 44]. Bacterial isolates related to Kocuria and Streptomyces were identified from the rhizosphere of halophytes as well as from the non-rhizospheric soils. These bacteria have plant growth abilities like phosphate solubilization and can be used as biofertilizers [45, 46]. Polaribacter strains identified in this study are polar marine bacteria having gas vacuoles. It can grow from 5 to 25% NaCl concentrations. It was first isolated from sea ice and water from the Arctic regions [47].
Bacillus, Halobacillus, Virgibacillus, and Oceanobacillus are a good source of halophilic and thermophilic enzymes such as lipases, proteases, amylases, cellulases, gelatinases, and catalases [41, 48]. Halophilic and halotolerant Bacilli (AT2RP4, HL1RS13, NRS4HaP9, and LK3HaP7) identified in this study showed optimum lipase activity at 1 M NaCl concentration, pH 8, and 37 °C (Fig. 5). Previous studies also reported that moderate halophilic bacteria isolated from a various hypersaline environments showed lipase activity at a wide range of salt, pH, and temperature [49]. The optimum salt concentration, pH, and temperature for protease activity of halophilic Bacillus strains (AT2RP4, NRS4HaP9, and LK3HaP7) were 1.5 M NaCl, pH 7–8, and 37 °C (Fig. 6). Protease-producing alkaliphilic and halophilic bacteria such as Halobacillus karajensis and Halomonas meridiana have been previously isolated marine environments and food sources such as fish sauce [43, 50]. Like protease, maximum amylase activity of Bacillus strains (AT2RP4, NRS4HaP9, and LK3HaP7) was also observed in the presence of 1.5 M NaCl, pH 7–8, and 37 °C (Fig. 7). Halophilic Bacillus strains have ability to utilize starch [51]. Halophilic bacterial strains related to Halobacillus and Oceanobacillus are known to be a good source of α-amylases [52]. Cellulase activity of bacterial strains HL1RS13, AT2RP4, NRS4HaP9, and LK3HaP7 was found to be stable in the presence of 0.5–2.5 M NaCl at pH 8 and 37 °C (Fig. 8). Halophilic cellulases have been produced by different lignocellulose-hydrolyzing bacteria such as Bacillus, Halobacillus, Salibacillus, and Halomonas [53].
Conclusion
To best of our knowledge, the present study is the first report about screening and characterization of hydrolytic enzymes from halophilic and halotolerant bacteria isolated the rhizosphere and non-rhizospheric soils of halophytes (S. stocksii and A. amnicola). Bacillus, Halobacillus, and Kocuria were the dominating genera from the rhizosphere of Salsola and Atriplex, and Staphylococcus, Halobacillus, and Halomonas were the dominating genera from the non-rhizospheric soils. Mostly, bacterial strains showed positive activity for hydrolytic enzymes like lipase, protease, amylase, catalase, gelatinase, and cellulase. Halophilic and halotolerant bacteria identified in this study have the ability to produce a number of hydrolytic enzymes under extreme conditions of salinity, pH, and temperature. The cost-effective production of extracellular enzymes by halophilic and halotolerant bacterial strains identified from rhizospheric and non-rhizospheric soils of halophytes from Khewra Salt Mine have great biotechnological potential and can be used in various industries such as pharmaceutical, agriculture, detergent, paper, and pulp industries.
Electronic supplementary material
(DOCX 1238 kb)
Acknowledgments
We are highly thankful to the Higher Education Commission for research grant of a project entitled “Microbial diversity and metagenomic analysis of rhizosphere of plants growing in extremely halophytic and xerophytic environments” and the Pakistan Academy of Sciences for a project entitled “Identification and characterization of different proteins involved in osmoregulation of halophilic bacteria through culturable and metaproteomics approaches.”
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mendes R, Garbeva P, Raaijmakers JM. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev. 2013;37:634–663. doi: 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- 2.Ruppel S, Franken P, Witzel K. Properties of the halophyte microbiome and their implications for plant salt tolerance. Func Plan Biol. 2013;40:940–951. doi: 10.1071/FP12355. [DOI] [PubMed] [Google Scholar]
- 3.Hussain A, Arshad M, Zahir ZA, Asghar M. Prospects of zinc solubilizing bacteria for enhancing growth of maize. Pak J Agri Sci. 2015;52:915–922. [Google Scholar]
- 4.Bulgarelli D, Rott M, Schlaeppi K, ver Loren van Themaat E, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J, Gloeckner FO, Amann R, Eickhorst T, Schulze-Lefert P. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488:91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- 5.DasSarma S, DasSarma P. Halophiles and their enzymes: negativity put to good use. Curr Opin Microbiol. 2015;25:120–126. doi: 10.1016/j.mib.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dastgheib SM, Amoozegar MA, Khajeh K, Ventosa A. A halotolerant Alcanivorax sp. strain with potential application in saline soil remediation. Appl Microbiol Biotech. 2011;90:305–312. doi: 10.1007/s00253-010-3049-6. [DOI] [PubMed] [Google Scholar]
- 7.Liszka M, Clark M, Schneider E, Clark DS. Nature versus nurture: developing enzymes that function under extreme conditions. Ann Rev Chem Biomol Eng. 2012;3:77–102. doi: 10.1146/annurev-chembioeng-061010-114239. [DOI] [PubMed] [Google Scholar]
- 8.Sarwar MK, Azam I, Iqbal T. Biology and applications of halophilic bacteria and archaea: a review. E J Bio. 2015;11:98–103. [Google Scholar]
- 9.Bozic N, Ruiz J, Lopez-Santin J, et al. Production and properties of the highly efficient raw starch digesting α-amylase from a Bacillus licheniformis ATCC 9945a. Biochem Eng J. 2011;53:203–209. doi: 10.1016/j.bej.2010.10.014. [DOI] [Google Scholar]
- 10.Marhuenda-Ege FC, Bonete MJ. Extreme halophilic enzymes in organic solvents. Curr Opin Biotechnol. 2002;13:385–389. doi: 10.1016/S0958-1669(02)00338-5. [DOI] [PubMed] [Google Scholar]
- 11.Ajmal M, Qaiser M. Halophytes of Pakistan: characteristics, distribution and potential economic usages. Saline Ecosys. 2006;2:129–153. [Google Scholar]
- 12.Dagla HR, Shekhawat NS. In vitro multiplication of Haloxylon recurvum (Moq.)- a plant for saline soil reclamation. J Plant Biol. 2005;7:155–160. [Google Scholar]
- 13.Ahmad K, Hussain M, Ashraf M, Luqman M, Ashraf MY, Khan ZI. Indigenous vegetation of Soon valley at the risk of extinction. Pak J Bot. 2007;39:679–690. [Google Scholar]
- 14.Anderson JM, Ingram JS, editors. Tropical soil biology and fertility: a handbook of methods. 2. Wallingford: CAB International; 1993. pp. 93–94. [Google Scholar]
- 15.Adviento-Borbe MA, Doran JW, Drijber RA, Dobermann A. Soil electrical conductivity and water content affect nitrous oxide and carbon dioxide emissions in intensively managed soils. J Environ Qual. 2006;35:1999–2010. doi: 10.2134/jeq2006.0109. [DOI] [PubMed] [Google Scholar]
- 16.Walkley A, Black IA. An examination of Degtjareff method for determining soil organic matter and a proposed modification of the chromic acid titration method. Soil Sci. 1934;37:29–37. doi: 10.1097/00010694-193401000-00003. [DOI] [Google Scholar]
- 17.Schneegurt MA. Media and conditions for the growth of halophilic and halotolerant bacteria and archaea. In: Vreeland RH, editor. Advances in understanding the biology of halophilic microorganisms. Dordrecht: Springer; 2012. pp. 35–58. [Google Scholar]
- 18.Malik KA, Bilal R, Mehnaz S, Rasool G, Mirza MS, Ali S. Association of nitrogen-fixing, plant growth promoting rhizobacteria (PGPR) with kallar grass and rice. Plant Soil. 1997;194:37–44. doi: 10.1023/A:1004295714181. [DOI] [Google Scholar]
- 19.Somasegaran P. Handbook for Rhizobia: methods in legume- rhizobium technology. New York: Springer-Verlag, cop; 1994. [Google Scholar]
- 20.Winnepenninckx B, Backeljau T, de Wachter R. Extraction of high molecular weight DNA from molluscs. Trends Genet. 1993;9:407–412. doi: 10.1016/0168-9525(93)90102-N. [DOI] [PubMed] [Google Scholar]
- 21.Tan ZY, Xu XD, Wan ET, Gao JL, Romer EM, Chen WX. Phylogenetic and genetic relationships of Mesorhizobium tianshanense and related rhizobia. Int J Sys Bacteriol. 1997;47:874–879. doi: 10.1099/00207713-47-3-874. [DOI] [PubMed] [Google Scholar]
- 22.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 23.Varian H. Bootstrap tutorial. Math J. 2005;9:768–775. [Google Scholar]
- 24.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin AP. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl Environ Microbiol. 2002;68:3673–3682. doi: 10.1128/AEM.68.8.3673-3682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar KV, Srivastava S, Singh N, Behl HM. Role of metal resistant plant growth promoting bacteria in ameliorating fly ash to the growth of Brassica juncea. J Haz Mat. 2009;170:51–57. doi: 10.1016/j.jhazmat.2009.04.132. [DOI] [PubMed] [Google Scholar]
- 28.Gupta P, Samant K, Sahu A. Isolation of cellulose-degrading bacteria and determination of their cellulolytic potential. Inter J Microbiol. 2012;20:1):28–1):35. doi: 10.1155/2012/578925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacFadden JF. Biochemical tests for identification of medical bacteria. Baltimore: Williams and Wilkins; 1980. [Google Scholar]
- 30.Sierra G. A simple method for the detection of lipolytic activity of micro-organisms and some observations on the influence of the contact between cells and fatty acid substrates. A Van Leeuw J Microbiol. 1957;23:15–22. doi: 10.1007/BF02545855. [DOI] [PubMed] [Google Scholar]
- 31.Pitt TL, Dey D. A method for the detection of gelatinase production by bacteria. J Appl Microbiol. 1970;33:687–691. doi: 10.1111/j.1365-2672.1970.tb02251.x. [DOI] [PubMed] [Google Scholar]
- 32.Christensen WB. Urea decomposition as a means of differentiating Proteus and paracolon cultures from each other and from Salmonella and Shigella types. J Bacteriol. 1946;52:461–466. doi: 10.1128/jb.52.4.461-466.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Canfora L, Bacci G, Pinzari F, Lo Papa G, Dazzi C, Benedetti A. Salinity and bacterial diversity: to what extent does the concentration of salt affect the bacterial community in a saline soil? PLoS One. 2014;9:e106662. doi: 10.1371/journal.pone.0106662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mwirichia R, Cousin S, Muigai AW, Boga HI, Stackebrandt E. Bacterial diversity in the haloalkaline lake Elmenteita, Kenya. Curr Microbiol. 2011;62:209–221. doi: 10.1007/s00284-010-9692-4. [DOI] [PubMed] [Google Scholar]
- 35.Mukhtar S, Mirza MS, Awan HA, Maqbool A, Mehnaz S, Malik KA. Microbial diversity and metagenomic analysis of the rhizosphere of para grass (Urochloa mutica) growing under saline conditions. Pak J Bot. 2016;48(2):779–791. [Google Scholar]
- 36.Berg G, Smalla K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol. 2009;68:1–13. doi: 10.1111/j.1574-6941.2009.00654.x. [DOI] [PubMed] [Google Scholar]
- 37.Irshad A, Ahmad I, Kim SB. Culturable diversity of halophilic bacteria in foreshore soils. Braz J Microbiol. 2014;45:563–571. doi: 10.1590/S1517-83822014005000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sass AM, McKew BA, Sass H, Fichtel J, Timmis KN, McGenity TJ. Diversity of Bacillus-like organisms isolated from deep-sea hypersaline anoxic sediments. Saline Sys. 2008;4:1–11. doi: 10.1186/1746-1448-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larose C, Prestat E, Cecillon S, Berger S, Malandain C, Lyon D, Ferrari C, Schneider D, Dommergue A, Vogel TM. Interactions between snow chemistry, mercury inputs and microbial population dynamics in an arctic snowpack. PLoS One. 2013;8:e79972. doi: 10.1371/journal.pone.0079972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma A, Singh P, Kumar S, Kashyap PL, Srivastava AK, Chakdar H, Singh RN, Kaushik R, Saxena AK, Sharma AK. Deciphering diversity of salt-tolerant Bacilli from saline soils of eastern Indo-Gangetic Plains of India. Geomicrobiol J. 2015;32:170–180. doi: 10.1080/01490451.2014.938205. [DOI] [Google Scholar]
- 41.Porwal S, Kumar T, Lal S, Rani A, Kumar S, Cheema S, Purohit HJ, Sharma R, Singh Patel SK, Kalia VC. Hydrogen and polyhydroxybutyrate producing abilities of microbes from diverse habitats by dark fermentative process. Bioresour Technol. 2008;99:5444–5451. doi: 10.1016/j.biortech.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 42.Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukhtar S, Mirza MS, Mehnaz S, Mirza BS, Malik KA. Diversity of Bacillus-like bacterial community in the rhizospheric and non-rhizospheric soil of halophytes (Salsola stocksii and Atriplex amnicola) and characterization of osmoregulatory genes in halophilic Bacilli. Can J Microbiol. 2018;64:567–579. doi: 10.1139/cjm-2017-0544. [DOI] [PubMed] [Google Scholar]
- 44.Mukhtar S, Shahid I, Mehnaz S, Malik KA. Assessment of two carrier materials for phosphate solubilizing biofertilizers and their effect on growth of wheat (Triticum aestivum) Microbiol Res. 2017;205:107–117. doi: 10.1016/j.micres.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 45.Pontes AP, de Souza R, Granada CE, Passaglia LMP. Screening of plant growth promoting bacteria associated with barley plants (Hordeum vulgare L.) cultivated in South Brazil. Biota Neotrop. 2015;15:1–6. doi: 10.1590/1676-06032015010514. [DOI] [Google Scholar]
- 46.Wang K, Zhang L, Liu Y, Pan Y, et al. Kocuria dechangensis sp. nov., an actinobacterium isolated from saline and alkaline soils. Int J Syst Evol Microbiol. 2015;65:3024–3030. doi: 10.1099/ijs.0.000372. [DOI] [PubMed] [Google Scholar]
- 47.Vanderpas J, Bontems P, Miendje VY, Cadranel S. Follow-up of Helicobacter pylori infection in children over two decades (1988–2007): persistence, relapse and acquisition rates. Epidemiol Infect. 2013;32:1–9. doi: 10.1017/S0950268813001428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taprig T, Akaracharanya A, Sitdhipol J, Visessanguan W, Tanasupawat S. Screening and characterization of protease-producing Virgibacillus, Halobacillus and Oceanobacillus strains from Thai fermented fish. J appl pharm sci. 2013;3:025–030. [Google Scholar]
- 49.Gupta S, Sharma P, Dev K, Sourirajan A. Halophilic bacteria of Lunsu produce an array of industrially important enzymes with salt tolerant activity. Biochem Res Int. 2016;3:923–928. doi: 10.1155/2016/9237418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phrommao E, Rodtong S, Yongsawatdigul J. Identification of novel halotolerant bacillopeptidase F-like proteinases from a moderately halophilic bacterium, Virgibacillus sp. SK37. J Appl Microbiol. 2010;110:191–201. doi: 10.1111/j.1365-2672.2010.04871.x. [DOI] [PubMed] [Google Scholar]
- 51.Sumit K, Grewal J, Sadaf A, Hemamalini R, Sunil KK. Halophiles as a source of polyextremophilic α-amylase for industrial applications. AIMS Microbiol. 2016;2:1–26. doi: 10.3934/microbiol.2016.1.1. [DOI] [Google Scholar]
- 52.Kumar S, Karan R, Kapoor S, Singh SP, Khare SK. Screening and isolation of halophilic bacteria producing industrially important enzymes. Braz J Microbiol. 2012;43:1595–1603. doi: 10.1590/S1517-83822012000400044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Lourdes MM, Pérez D, García MT, Mellado E. Halophilic bacteria as a source of novel hydrolytic enzymes. Life (Basel, Switzerland) 2013;3:38–51. doi: 10.3390/life3010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 1238 kb)