Abstract
The aim of the present study was to evaluate the genetic diversity of Ehrlichia canis in naturally infected dogs from six mesoregions of Rio de Janeiro state. E. canis was diagnosed with a real-time polymerase chain reaction (qPCR) targeting a 93 base pair (bp) fragment of the 16S rDNA gene. To evaluate the genetic diversity of the parasite, we amplified a positive sample from each mesoregion by distinct conventional PCR assays with targets in the gp19 (414 bp), gp36 (814 bp), and p28 (843 bp) genes. A total of 267 samples were collected from dogs in Rio de Janeiro state. Among the samples analyzed, 42.3% (n = 113/267) were 16S rDNA-qPCR positive. When performing PCR for the gp19 and gp36 genes, 100% (n = 113/113) and 5.3% (n = 6/113) of the samples amplified fragments of 414 bp and 814 bp, respectively. The six PCR-positive samples for the gp36 gene also amplified the p28 gene fragment. The characterization based on the gp19 gene demonstrated that it is highly conserved. In protein analysis (TRP36), all samples showed a tandem repeat protein (TRP) that comprised 11 replicates. Seven high-entropy amino acid sites were distributed throughout the gp36 gene. Eleven high-entropy amino acid sites were found throughout the p28 gene. There is a positive selection pressure in both genes (p ≤ 0.05). Comparing and characterizing an organism are useful for providing important information about the host’s immune response and identifying new antigenic targets, as well as essential characteristics for the development of vaccines and new diagnostic tools.
Keywords: Genetic diversity, Molecular markers, Epitopes, Canine monocytic ehrlichiosis
Introduction
Ehrlichia canis [1] is an obligate intracellular bacterium and the etiological agent of canine monocytic ehrlichiosis (CME). This organism is a parasite of cells in the mononuclear phagocytic system, which mainly includes monocytes, and it can cause high morbidity in susceptible dog populations [2]. Ehrlichia canis presents a ubiquitous distribution, and this distribution is associated with the availability of the vector tick Rhipicephalus sanguineus sensu lato (s.l). In the state of Rio de Janeiro, the prevalence ranges from 5 to 16% [3, 4]. In the state of São Paulo, the prevalence is 27.6% [5]; the frequency of dogs infected with E. canis can be as high as 65% [6]. In other countries, such as Argentina, India, Nigeria, and Iran, the percentage of dogs positive for E. canis ranges from 5.8 to 22.9% [7–10]. The most studied genetic sequence is the 16S subunit of ribosomal deoxyribonucleic acid (16S rDNA) [11–13]. However, this is a conserved gene with an identity that ranges from 99.4 to 100% in Southern Africa, North America, Asia, and Europe. Studies of immunoreactive proteins have demonstrated that E. canis has serial repeat regions (tandem repeat protein; TRP) in its genome [14, 15]. These glycoproteins are immunoreactive and have species-specific antigens [15] that are closely related to the parasite-host interaction. The 36-kDa membrane glycoprotein (TRP36) contains a major antibody epitope in the tandem repeat region. The protein TRP36 has been associated with functional host-pathogen interactions, such as adhesion and internalization, actin nucleation, and immune evasion [16, 17]. In some sequences, this variation includes 12 to 15 replicates [14, 18]. The gene encoding the 19-kDa membrane glycoprotein (gp19) is more conserved and contains epitope regions [19].
The 28-kDa outer surface protein (P28) is encoded by a multigenic locus, which has at least 22 alleles of the p28 gene (p28-1 to p28-22) on the chromosomes of E. canis and E. chaffeensis [18]. The recombinant p28 protein is effective in serological diagnosis of E. canis [19]. In addition, the p28 gene is conserved in all E. canis sequences from North America, and this conservation permits the development of vaccine antigens [20].
Brazilian sequences identified in this study were used to confirm genetic differences with E. canis sequences from other parts of the world [14]. Few sequences of the gp19, gpP36, and p28 genes of E. canis are available in the public database, limiting the performance of robust analysis of genetic diversity. Thus, the present study aims to contribute to identification of circulating genotypes of E. canis in the state of Rio de Janeiro and improve the understanding of the epidemiological chain in the state of Rio de Janeiro. The aim of this study was to perform molecular characterization of E. canis using the gp19, gp36, and p28 genes in whole-blood samples from naturally infected dogs living in six mesoregions from the state of Rio de Janeiro.
Material and methods
Samples and DNA extraction
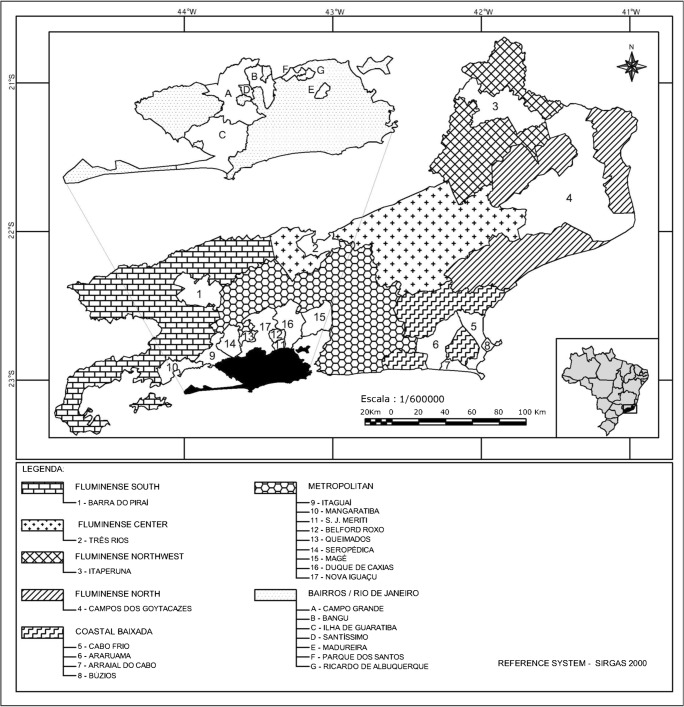
This study was performed using whole-blood samples obtained from naturally infected dogs from the six mesoregions of Rio de Janeiro state. According to IBGE [21], these mesoregions are classified as Metropolitan (Met), Fluminense North (FluNor), Fluminense Northwest (FluNW), Coastal Baixada (CoastBaix), Fluminense Center (FluCen), and Fluminense South (FluSouth). Samples from the first four regions were obtained through partnerships with Clinical Pathology laboratories, and samples from the Center and South Fluminense regions were obtained from Non-Governmental Organizations (NGOs) (Fig. 1). The 95% confidence level, an expected prevalence of 16%, and a 5% margin of error were established for the sample size determination of dogs. The minimum number of required animal samples was 206 according to the equation described by Sampaio [22]. Sampling was performed in a non-probabilistic manner. Three milliliters (mL) of blood were collected by cephalic venipuncture from each animal, then placed in tubes containing the anticoagulant ethylene diamine tetraacetic acid (EDTA), stored in a refrigerated container and transported to the Laboratory of Hemoparasites and Vectors of the Federal Rural University of Rio de Janeiro for further analysis. DNA extraction was conducted with a commercial kit (Wizard Genomic DNA Purification Kit—Promega, USA) from 200 μL of whole blood following the manufacturer’s recommendations. The samples were quantified in a spectrophotometer (NanoDrop 2000®, USA) and then aliquoted at a concentration of 100 ng/μL. The aliquots were frozen in a freezer at − 20 °C until molecular analysis.
Fig. 1.
Whole-blood samples obtained from naturally infected dogs from the six mesoregions of Rio de Janeiro state
Standard control
A whole-blood sample from a dog with E. canis inclusion in monocytes was used as a positive control. The sample was amplified using the gp19 gene with a 414-bp target [13]. Nuclease-free water (Ambion®, ThermoFisher Scientific, Inc., Waltham, MA, USA) was used as a negative control. The sample was sequenced by the Sanger method and deposited in the GenBank database (ID: MG584542). The DNA concentration was checked using a fluorometer (Qubit™, ThermoFisher Scientific, Inc., Waltham, MA, USA). The concentration served as the starting point for obtaining the amount of molecules per microliter. As a positive control in the polymerase chain reaction (PCR) reactions, the penultimate dilution (100 copies) of the detection limit was used. The controls were performed in duplicate in all PCR reactions.
Screening of positive samples for Ehrlichia canis by qPCR
Real-time polymerase chain reaction (qPCR) targeting a 93-bp fragment of the 16S rDNA gene was carried out with the purpose of screening positive E. canis samples according to the reaction described in Table 1. In the present study, these qPCR characteristics were evaluated according to the recommendations of Bustin [23]. The analytical sensitivity of qPCR was evaluated by using serial decimal dilutions of the amplicon cloned into a plasmid. Plasmid DNA concentrations were verified using fluorimetry (Qubit™, ThermoFisher Scientific, Inc., Waltham, MA, USA). The plasmid copy numbers versus Cq values were plotted to determine the analytical sensitivity of the qPCR. The number of copies ranged from 1 to 1 × 106 per μL, with seven separate dilution series performed for each point of the curve in triplicate. Linear regression, along with the coefficient R2 obtained after determination of each point of the curve, can be used to evaluate whether the qPCR assay has been optimized. Each reaction’s efficiency was determined using the following formula: [Efficiency = 10(− 1/slope) – 1] [24].
Table 1.
Primers used for amplification and sequencing of 16S rDNA-, g19-, gp36-, and p28-specific genes for detection of Ehrlichia canis in blood samples from dogs
| Target | Primers | Size (bp) | Reagents | Thermocycler | Reference |
|---|---|---|---|---|---|
| 16S rDNA |
F-5′TATAGCCTCTGGCTATAGGAAATTGTTA′3 R-5′ACCATTTCTAATGGCTATTCCGTACTA′3 5′6-FAMTGGCAGACGGGTGAGTAATGGTAGG-TAMRA-3′ (probe) |
93 bp |
TaqMan Universal Master Mix (ThermoFisher Scientific, Inc., Waltham, MA, USA), 1 × Primer, 0.3 μM (each) DNA, 3 μl Fv, 12 μl |
50 °C, 2 min 95 °C, 10 min 45 cycles, 95 °C, 15 s; 60 °C, 1 min; |
Baneth et al. 2008 |
| gp19 |
F 5′-ATTAGTGTTGTGGTTATGCAA-3′ R 5′-TACGCTTGCTGAATATCATGA-3′ |
414 bp |
Buffer (Tris-HCl 200 mM, pH 8.4, KCl 500 mM),1 × MgCl2, 2.5 mM dNTPs, 0.2 mM Primers, 0.6 μM (each) Taq, 0.125 U DNA, 5 μl Fv, 25 μl |
94 °C, 3 min 35 cycles, 94 °C, 1 min; 55 °C, 1 min; 72 °C, 1.5 min; 72 °C, 5 min |
Chen et al. 2010 |
| p28 |
F 5′-ATGAATTGCAAAAAAATTCTTATA-3′ R 5′-TTAGAAGTTAAATCTTCCTCC -3′ |
843 bp |
Buffer (Tris-HCl 200 mM, pH 8.4, KCl 500 mM), 1 × MgCl2, 2.5 mM dNTPs, 0.2 mM Primers, 0.6 μM (each) Taq, 1.2 U DNA, 5 μl Fv, 25 μl |
95 °C, 5 min 30 cycles, 95 °C, 30 s; 55 °C, 1 min; 72 °C, 2 min; 72 °C, 5 min. |
Nakaghi et al. 2010 |
| gp36 |
F 5′-GTATGTTTCTTTTATATCATGGC-3′ R 5′-GGTTATATTTCAGTTATCAGAAG-3′ |
840 bp |
Buffer (Tris-HCl 200 mM, pH 8.4, KCl 500 mM), 1 × MgCl2, 2.5 mM dNTPs, 0.2 mM Primers, 0.6 μM (each) Taq: 0.125 U DNA, 5 μl Fv, 25 μl |
94 °C, 3 min 35 cycles, 94 °C, 1 min; 55 °C, 1 min; 72 °C, 1.5 min; 72 °C, 5 min |
Chen et al. 2010 |
F, forward; R, reverse; bp, base pairs; min, minutes; sec, seconds; Fv, final volume; Taq polymerase and dNTPS-Invitrogen® (Thermo Fisher Scientific)
The specificity of the assay was evaluated using Anaplasma phagocytophilum DNA obtained from cellular cultures; Anaplasma platys, Babesia vogeli, and Hepatozoon canis were obtained from the blood samples of naturally infected dogs with high parasitemia in the acute phase, with infection diagnosed by microscopy and confirmed by a specific molecular assay.
Amplification of the gp19, gp36, and p28 genes of Ehrlichia canis
To perform the molecular characterization, we selected positive samples from each mesoregion (Met, FluSouth, FluNor, FluNW, CoastBaix, and FluCen) using specific primers for E. canis targeting 843 bp of the p28 gene, 414 bp of the gp19 gene, and 840 bp of the gp36 gene. The conditions of the PCR reactions and the sequences of the primers are listed in Table 2.
Table 2.
Ehrlichia canis sequences of the gp19, gp36, and p28 genes available from GenBank used in the present study
| Sequences | |||||
|---|---|---|---|---|---|
| gp36 | Access code | gp19 | Access code | p28 | Access code |
| Ehrlichia canis—Jake, USA | DQ085427 | Ehrlichia canis—Jake, USA | DQ858221 | Ehrlichia canis—Jaboticabal, Brazil | EF014897 |
| Ehrlichia canis—Duque de Caxias, Brazil | KF233413 | Ehrlichia canis—Louisiana | DQ858224 | Ehrlichia canis—North Carolina, USA | AF082749 |
| Ehrlichia canis—Taiwan | EF551366 | Ehrlichia canis—Florida | DQ858225 | Ehrlichia canis—North Carolina, USA | AF082748 |
| Ehrlichia canis—Nigeria | JN982341 | Ehrlichia canis—Mexico | DQ858226 | Ehrlichia canis—Demon, USA | AF082747 |
| Ehrlichia canis—Nigeria | JN982338 | Ehrlichia canis—São Paulo | DQ860145 | Ehrlichia canis—Oklahoma, USA | AF082746 |
| Ehrlichia canis—Duque de Caxias, Brazil | KF233414 | Ehrlichia canis—Taiwan | EF587270 | Ehrlichia canis—Louisiana, USA | AF082745 |
| Ehrlichia canis—Czech Republic | KC479021 | Ehrlichia canis—Taiwan | EU139492 | Ehrlichia canis—Florida, USA | AF082750 |
| Ehrlichia canis—Czech Republic | KC479020 | Ehrlichia canis—USA | HW577579 | Ehrlichia canis—Philippines | JQ663860 |
| Ehrlichia canis—Czech Republic | KC479019 | Ehrlichia canis—Nigeria | JN982337 | Ehrlichia ruminantium | CR925678 |
| Ehrlichia canis—USA | KT357370 | Ehrlichia ewingii | AF287966 | ||
| Ehrlichia canis—USA | KT357369 | Ehrlichia chaffeensis | CP000236 | ||
| Ehrlichia canis—Taiwan | EF651794 | Anaplasma platys | GU357493 | ||
| Ehrlichia canis—Taiwan | HQ009756 | Anaplasma phagocytophilum | EU008082 | ||
| Ehrlichia canis—Taiwan | EU13949 | Anaplasma marginale | AY786994 | ||
| Ehrlichia canis—Taiwan | EF560599 | Neorickettsia risticii | HQ906682 | ||
| Ehrlichia canis—Thailand | KT363877 | Neorickettsia risticii | HQ906682 | ||
| Ehrlichia canis—Thailand | KT363876 | ||||
| Ehrlichia canis—Thailand | KT363875 | ||||
| Ehrlichia canis—Nigeria | JN622143 | ||||
| Ehrlichia canis—USA | DQ146155 | ||||
| Ehrlichia canis—Taiwan | HM188566 | ||||
| Ehrlichia canis—Florida, USA | DQ146152 | ||||
| Ehrlichia canis—Louisiana, USA | DQ146151 | ||||
| Ehrlichia canis—São Paulo, Brazil | DQ146154 | ||||
| Ehrlichia canis—North Carolina, USA | DQ146153 | ||||
| Ehrlichia canis—Oklahoma, USA | DQ085428 | ||||
| Ehrlichia canis—Monte Negro, Brazil | JX312082 | ||||
| Ehrlichia canis—Cuiaba, Brazil | JX312079 | ||||
| Ehrlichia canis—Belem, Brazil | JX29924 | ||||
| Ehrlichia canis—Londrina, Brazil | JX312080 | ||||
| Ehrlichia canis—Demon, USA | DQ085429 | ||||
Sequencing, phylogenetic analysis, and analysis of sequence entropy
The produced amplicons were purified using a Clean Sweep PCR Purification kit (ThermoFisher Scientific) and sequenced with Sanger [25] methods on an ABI 3730 DNA analyzer (Applied Biosystems®). The purified PCR products were sequenced in both directions using the primers described in Table 1. The sequences were assembled and edited. The phylogeny and amino acid sequences derived from the DNA sequences were transcribed and analyzed using the CLC Genomics WorkBench v.7.9.1 [26] program and deposited on GenBank under accession number MG584539 to MG584544 (gp19 gene), MG584545 to MG584549 (gp36 gene), and MG584550 to MG584556 (p28 gene). Basic Local Alignment Search Tool (BLAST) was used to evaluate the identity of new fragments with E. canis sequences available in GenBank. The sequences obtained in this study for the gp19, gp36, and p28 genes were aligned with other sequences from different parts of the world to characterize the genetic diversity of E. canis samples from Brazil. All access codes and origin countries of the gp19, gp36, and p28 genes sequences used for the alignment are listed in Table 2. Alignments were performed using the Clustal W algorithm [27]. Phylogenetic groupings were performed using the Neighbor-Joining method. The Kimura 2-parameter model was used to calculate the evolutionary distance. The model was selected based on Akaike Information Criterion (AIC) using jModelTest2 software [28]. The combination of phylogenetic clusters was evaluated using a bootstrap test with 1000 pseudo replications to analyze different phylogenetic reconstructions.
To understand the degree of variation in TRP36 and P28, we calculated the entropy of amino acid sequence alignments using BioEdit software version 7.0.9.0 [29]. In addition, positive selection tests based on codons (Z test, MEGA 7) [30] were used to estimate the number of synonymous and non-synonymous substitutions per site (dN/dS ratio) of the amino acids for TRP36 and P28 proteins to determine if these proteins led to positive selection.
Procedures performed on animals in this study were approved by the Ethics Committee on Animal Use of the UFRRJ (CEUA/UFRRJ) under procedure number 072/2014. These procedures comply with the basic and ethical principles for research involving the use of animals. All procedures were performed by a team of trained veterinarians.
Results
Molecular detection of Ehrlichia canis DNA in dog blood samples
Before evaluating the genetic diversity of E. canis, samples were submitted to molecular detection by 16S rDNA-qPCR. A total of 267 blood samples were collected from dogs from the state of Rio de Janeiro. Among the samples analyzed, 42.3% (n = 113/267) were qPCR positive for the 16S rDNA gene of E. canis. The average value of Cq observed in positive samples was 34.1 ± 5.1, ranging between 18 and 40 cycles. The detection limit of the qPCR was ten copies of the plasmid per microliter containing a 16S rDNA gene from E. canis. The determination coefficient of the seven dilutions tested in the standard curve was 99.9%, and the efficiency was 95.7%. When performing gp19-PCR and gp36-PCR, 100% (n = 113/113) and 5.3% (n = 6/113) of the samples were positive, respectively. The six PCR-positive samples for the 16S rDNA gene also amplified the p28 gene. Only one PCR-positive sample for the three genes (gp19, gp36, and p28) in each of the six mesoregions was selected and subjected to amino acid and nucleotide sequence analysis.
The frequency in each mesoregion for the 113 animals positive for E. canis by the 16S rDNA gene was 59.29% (n = 67/113) in the Metropolitan mesoregion, 13.27% (n = 15/113) in South Fluminense, 15.04% (n = 17/113) in Northern Fluminense, 5.3% (n = 6/113) in Center Fluminense, 4.42% (n = 5/113) in Northwest Fluminense, and 2.65% (n = 3/113) in Coastal Baixada.
Alignment analysis of nucleotide and amino acid sequences
Samples amplified with the gp19 gene from the mesoregions of Rio de Janeiro state showed 100% identity with E. canis sequences from Brazil and other countries of the world. The genetic profile of the E. canis sequences was highly conserved according to the gp19 gene sequences. In the analysis of the partial sequence of the polypeptide, which was deduced from the nucleotide sequence of the gp19 gene, no mutation point was observed. When the six-mesoregion sequences were compared with the Jake reference sequence (DQ085427), the gp36 gene demonstrated 99.80% identity, except for the E. canis-FluNW and E. canis-FluSouth sequences, which showed 100% identity. The variation in the percentage of identity was 99.9% to 100% for the p28 gene compared with that for the Jaboticabal reference sequence (EF014897).
Among the six amplified samples for the gp36 gene, it was not possible to sequence samples from the central Fluminense mesoregion due to the difficulty in acquiring amplicons with adequate concentrations for sequencing using the primers described by Chen [13]. Sequences from the United States of America (USA; DQ146151-DQ14153, DQ085427-DQ085429) demonstrated that the gp36 gene comprises two regions. There was an initial region of 429 bp and a serial repeat region (TRP) of approximately 303 bp. Although there was variation in the final size of the sequenced samples, they were aligned and produced a final fragment of 732 bp. In the initial region up to position 92, the alignment of the gp36 gene sequences obtained from five sequences from different mesoregions had 100% identity, even though they originated in distinct areas of Rio de Janeiro state. However, from position 92, we can observe variations in nucleotides resulting in nine-point mutations (positions 92 to 331). The sequences of this study showed 98% identity when compared with sequences from the south (JX312079) and central-western (JX312080) regions and 97% with the sequence from the northeastern (JX312082) region of Brazil. The identity of the sequences found in the present study in relation to those of other countries, such as the USA, Taiwan, Czech Republic, Thailand, and Nigeria, ranged from 99.88 to 100%.
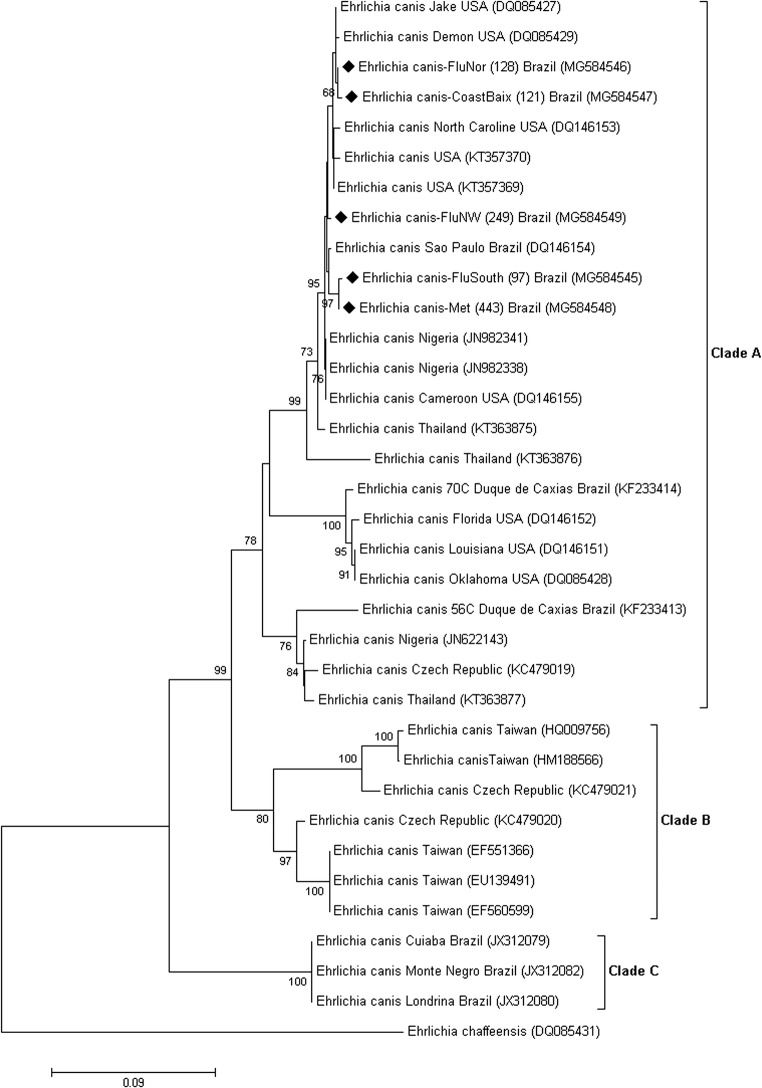
In cluster analysis, the formation of three clades (clades A, B, and C) supported by a high bootstrap value (≥ 98%) was observed. All sequences from the present study were grouped together with sequences from the USA in clade A, suggesting genetic similarity between Brazilian and American sequences. Clade B was represented by samples from the Czech Republic and Taiwan. Clade C comprised only Brazilian sequences from the states of Pará (JX429924), Paraná (JX312080), Mato Grosso (JX312079), and Rondônia (JX312082) (Fig. 2).
Fig. 2.
The formation of three clades (clades A, B, and C) supported by a high bootstrap value
The serial number of E. canis TRP36 from all sequences from the state of Rio de Janeiro was completely conserved and encoded nine amino acids (TEDSVSAPA). The number of variations was 11 copies (Table 3). Eight polymorphism points were observed in the amino acid chain, and five of them were between amino acids 31 and 111, a site with greater variability. Five polymorphic points were observed between amino acids 10 to 46 compared with other sequences (KF233413 and KF233414) of the same state. Compared with the Jake reference sequence (DQ085427), all sequences of the present study contained an aspartic acid (D) at position 31 (Table 3).
Table 3.
Amino acid differences in the variable region and number of repeats in the TRP region of the partial sequence of the polypeptide deduced from TRP36 among Ehrlichia canis sequences and obtained from dogs from the mesoregions of the state of Rio de Janeiro
| Sequences | Variable regiona | TRP* | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 19 | 23 | 31 | 46 | 70 | 72 | 78 | 79 | 86 | 111 | 145-240 | |
| Ehrlichia canis—Jake, USA (DQ085427) | M | N | H | G | E | A | A | D | M | V | V | TEDSVSAPA [12] |
| Ehrlichia canis—FluSouth (97) (MG584545) | . | . | . | D | R | V | . | N | V | . | M | TEDSVSAPA [11] |
| Ehrlichia canis—Met (443) (MG584548) | . | . | . | D | R | V | . | . | V | . | M | TEDSVSAPA [11] |
| Ehrlichia canis—FluNW (249) (MG584549) | . | . | . | D | . | . | . | N | . | A | . | TEDSVSAPA [11] |
| Ehrlichia canis—FluNor (128) (MG584546) | . | . | . | D | . | . | . | . | . | . | . | TEDSVSAPA [11] |
| Ehrlichia canis—CoastBaix (121) (MG584547) | . | . | . | D | . | . | G | . | . | . | . | TEDSVSAPA [11] |
| Ehrlichia canis—Caxias, Brazil (KF233413) | K | G | N | S | G | V | . | N | . | . | M | TEDSVSAPA [8] |
| Ehrlichia canis—Caxias, Brazil (KF233414) | K | . | . | . | . | V | . | N | . | . | M | TEDSVSAPA [5] |
| Ehrlichia canis—Cuiabá, Brazil (JX312079) | . | G | N | . | G | V | . | N | . | A | M | ASVVPEAE [13] |
| Ehrlichia canis—Montenegro, Brazil (JX312082) | . | G | N | . | G | V | . | N | . | A | M | ASVVPEAE [10] |
| Ehrlichia canis—Londrina, Brazil (JX312080) | . | G | N | . | G | V | . | N | . | A | M | ASVVPEAE [15] |
| Ehrlichia canis—São Paulo, Brazil (DQ146154) | . | . | . | . | A | V | . | N | . | . | M | TEDSVSAPA [10] |
| Ehrlichia canis—Oklahoma, USA (DQ085428) | . | . | . | . | . | . | . | . | . | . | . | TEDSVSAPA [6] |
| Ehrlichia canis—Demon, USA (DQ085429) | . | . | . | . | . | . | . | . | . | . | . | TEDSVSAPA [9] |
| Ehrlichia canis—Louisiana, USA (DQ146151) | . | . | . | . | . | . | . | N | . | . | . | TEDSVSAPA [9] |
| Ehrlichia canis—Florida, USA (DQ146152) | . | . | . | . | . | . | . | . | . | . | . | TEDSVSAPA [6] |
| Ehrlichia canis—North USA (DQ146153) | . | . | . | . | . | . | . | N | . | . | . | TEDSVSAPA [9] |
| Ehrlichia canis—Cameroon (DQ146155) | . | . | . | . | . | V | G | N | . | A | M | TEDSVSAPA [9] |
| Ehrlichia canis—USA (KT357369) | . | . | . | . | . | . | . | N | . | . | . | TEDSVSAPA [11] |
| Ehrlichia canis- USA (KT357370) | . | . | . | . | . | . | . | N | . | . | . | TEDSVSAPA [9] |
| Ehrlichia canis—Taiwan (EF551366) | . | G | S | D | . | V | G | . | . | . | M | TEDSVSAPA [11] |
| Ehrlichia canis—Taiwan (EF560599) | . | G | S | . | G | . | . | . | . | A | M | TEDSVSAPA [9] |
| Ehrlichia canis—Taiwan (EU139491) | . | . | S | . | G | V | . | . | . | A | M | TEDSVSAPA [9] |
| Ehrlichia canis—Taiwan (HQ009756) | . | G | S | . | . | . | . | . | . | A | M | TEDSVSAPA [10] |
| Ehrlichia canis—Taiwan (HM188566) | . | G | S | . | G | V | . | . | . | A | M | TEDSVSAPA [9] |
| Ehrlichia canis—Thailand (KT363877) | . | . | . | . | . | V | . | N | . | A | M | TEDSVSAPA [10] |
| Ehrlichia canis—Thailand (KT363876) | . | . | . | . | V | . | N | . | A | M | TEDSVSAPA [10] | |
| Ehrlichia canis—Thailand (KT363875) | . | . | . | . | . | V | . | N | . | A | M | TEDSVSAPA [11] |
| Ehrlichia canis—Nigeria (JN622143) | . | . | . | . | . | V | . | N | . | A | M | TEDSVSAPA [10] |
| Ehrlichia canis—Nigeria (JN982341) | . | . | . | . | . | V | . | N | . | A | M | TEDSVSAPA [10] |
| Ehrlichi acanis—Nigeria (JN982338) | . | . | . | . | . | V | . | N | . | A | M | TEDSVSAPA [10] |
| Ehrlichia canis—Czech Republic (KC479021) | . | G | . | D | G | V | . | N | . | A | M | TEDSVSAPA [9] |
| Ehrlichia canis—Czech Republic (KC479020) | . | G | . | . | G | V | . | N | . | A | K | TEDSVSAPA [9] |
| Ehrlichia canis—Czech Republic (KC479019) | . | G | . | . | G | V | . | N | . | A | K | TEDSVSAPA [9] |
*TRP tandem repeat protein. aPosition based on the sequence of E. canis (DQ085427). The dots “.” represent conserved regions
In cluster analysis of the p28 gene, the formation of two clades (clades A and B) supported by a high bootstrap value (≥ 93%) was observed. All sequences from the present study were grouped in clade A, along with other sequences from Brazil (EF014897) and the Philippines (JQ663860), suggesting genetic similarity between these sequences. Clade B was represented by US samples (Fig. 3). The identity of the E. canis p28 gene among the sequences obtained in the present study ranged from 99.98 to 100% with the Jaboticabal isolate (EF014897). Compared with the sequences in the present study, the P28 protein of the Jaboticabal sample was divergent for approximately ten amino acids. Among the analyzed samples, there were differences in some amino acids. Compared with the reference sequence Jaboticabal (EF014897), all sequences of the present study showed an Isoleucine (I) at position 206 of the polypeptide except for the E. canis-met (24C) sequence, which showed a threonine (T) substitution at this position. The sequences E. canis-FluNw and FluNor showed S156D substitution; the sequences E. canis-Met (24C), FluCen and FluSouth showed S156N substitution, and the sequence E. canis-Met (443) showed S156G (Table 4). Considering the similarity of the amplified sequences with the p28 gene, the sequences of the present study were aligned with a sample of E. chaffeensis, the similarity between the sequences was 79%. To establish position of E. canis within the Rickettsiales order using the p28 gene, we made an inference between the samples from the present study and the subjects of the order. The similarity between E. canis of the present study and other organisms of the order varied from 33 to 80%, which demonstrated the distance between these bacteria. In the phylogenetic reconstruction, all positive samples for E. canis were grouped in a clade together with E. canis Jaboticabal (EF014897) and the USA. The formation of other clades with other genera and species was also observed (Fig. 4).
Fig. 3.
The formation of two clades (clades A and B) supported by a high bootstrap value
Table 4.
Amino acid differences in the partial sequence of the polypeptide deduced from P28 among Ehrlichia canis sequences obtained from dogs from the mesoregions of the state of Rio de Janeiro
| Sequences | Amino acid positiona | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 58 | 60 | 72 | 80 | 84 | 96 | 156 | 206 | 252 | 259 | |
| Ehrlichia canis—SP, Brazil (EF014897) | S | P | Q | T | I | S | S | I | E | S |
| Ehrlichia canis—FluNW (294) MG584554 | R | S | N | S | T | N | D | . | G | N |
| Ehrlichia canis—FluNor (128) MG584556 | R | S | N | S | T | N | D | . | G | N |
| Ehrlichia canis—Met (24C) MG584550 | R | S | N | S | T | N | N | T | G | N |
| Ehrlichia canis—FluCen (254) MG584553 | R | S | N | S | . | N | N | . | . | . |
| Ehrlichia canis—Met (443) MG584555 | R | S | . | S | T | N | G | . | G | N |
| Ehrlichia canis—CoastBaix (121) MG584552 | . | . | . | . | . | . | . | . | G | . |
| Ehrlichia canis—FluSouth (86) MG584551 | . | . | . | . | . | . | N | . | G | N |
aPosition based on the sequence of E. canis (EF014897). The dots “.” represent conserved regions
Fig. 4.
The formation of other clades with other genera and species
Entropy analysis of amino acid sequences
The result of the entropy analysis of the deduced amino acid alignments of the TRP36 and P28 sequences demonstrated that seven high entropy regions were detected along the TRP36 sequence, and 11 high entropy amino acid sites were found along the P28 sequence. The entropy at the deduced amino acid sites of TRP36 was lower than that of P28 sequence. An entropy value greater than 0.4 indicated that a particular site was not conserved. In the present study, the entropy value ranged from 0.25 to 0.65 for TRP36 and from 0.15 to 1 for the P28 sequences. In the amino acids deduced from P28, there was a peak with an approximate value of 1.0 in the position between 150 and 160. This value indicated regions with high genetic variability in this gene. When we analyzed the deduced amino acids of TRP36, we observed the formation of seven peaks with values above 0.4 between positions 40 and 110, representing approximately 70 amino acids. The analyses of P28 showed greater variation than did those of TRP36.
Codon-based tests to check for positive selection in E. canis samples (Z test, MEGA5) were also performed. In the analysis of the gp36 gene, positive selection pressure was identified among the sequences from the Fluminense North region with sequences from the Fluminense South region, from Coastal Baixada, and those from Fluminense South, as well as sequences from the Metropolitan region with Fluminense North, sequences from the Northwest region with Fluminense South, and sequences from the Metropolitan region with Coastal Baixada and Fluminense Northwest (p ≤ 0.05). When the p28 gene was used, there was selection pressure among the sequences from the Fluminense Center region with sequences from the Fluminense Northwest region, sequences from the Metropolitan region and sequences from the Fluminense Northwest, as well as sequences from the Fluminense North region with those from the Metropolitan and Northwest regions with Fluminense North (p ≤ 0.05).
Discussion
The 100% identity between the Brazilian and North American sequences of E. canis demonstrates the high degree of conservation for the gp19 gene. Ferreira [4] found similar results when analyzing samples from the Metropolitan region of Rio de Janeiro. Chen [13] found that the E. canis gp19 gene is highly conserved by analyzing 153 samples from dogs with clinical signs at a Veterinary Hospital in Taiwan/China with PCR. Other studies have also mentioned conservation of the gp19 gene in sequences from the USA, Israel, Brazil, and Taiwan [14, 18, 19]. According to Brum [31], the similarity between the geographically distinct samples suggests that the TRP19 protein can be used for diagnostic immunoenzymatic assays, as well as in vaccine programs, since this protein is specific for E. canis and does not have cross-reactivity with other species in the genus Ehrlichia. Preparations based on Serine-Threonine-Glutamate and Cysteine (STE and Cy) have demonstrated high sensitivity and specificity in the detection of antibodies against E. canis in naturally infected dogs. McBride [32] demonstrated that amino-terminal STE-rich patch recombinant antigen was more reactive with serum from an E. canis-infected dog than those tested with synthetic non-glycosylated peptide as antigen.
Three genotypes of E. canis are circulating in the state of Rio de Janeiro. The amino acid sequence TEDSVSAPA present in E. canis sequences from the USA (clade A) was also present in all samples in the present study, supporting other research conducted in Rio de Janeiro state [4]. All sequences in this study presented 11 replicates, higher than the number of replicates found by Ferreira [4], with 5 to 8 replicates for an 840-bp fragment. The ASVVPEAE sequence of Clade C comprised the sequences of E. canis from the north, central-western (Mato Grosso) and southern regions of Brazil [4, 6, 33]. On the other hand, clade A was comprised only sequences from southeastern region of Brazil (from Sao Paulo and Rio de Janeiro).
The samples from the present study presented genetic diversity among the sequences of the Rio de Janeiro mesoregion and among the world sequences using the gp36 and p28 genes. The variability of sequences using the gp36 gene was also observed by Ferreira [4], who identified genetic differences among sequences from the metropolitan region of Rio de Janeiro. The data based on the phylogenetic reconstruction, using the gp36 gene, revealed that samples from closer geographical regions were more closely related; for example, the coastal, Fluminense North and Metropolitan regions presented 100% similarity. In the analysis of amino acids, mutations were observed in all sequences, similar to findings by other authors [4, 6, 14]. The differences in some amino acids may be sufficient to cause conformational differences in proteins, which have structural, functional, and antigenic implications [33].
The study regions contain neighboring municipalities that favor the movement of dogs between sites, which is how vector dispersion (e.g., ticks) occurs [3]. Aguiar [6] posited that the differences demonstrated in proteins involved in the immune response are due to greater selection pressure. Phenotypic alterations can influence the adaptation of the bacterium to vertebrate and invertebrate hosts, as well as their evasive mechanisms and this mechanism used by the bacterium can lead to mutations in its genetic code. The total blockade of CD4 T lymphocyte activities is considered the main form of E. canis escape of the host immune response. This blockade favors the continuous release of TNF-α by cytotoxic cells, and the host’s constant attempts to eliminate the etiological agent may result in pancytopenia [34].
Although the use of the p28 gene may represent a useful tool for the diagnosis of E. canis, few sequences are available in GenBank for this gene. This limitation means that a more robust analysis of the genetic variability of the studied sequences cannot be conducted. The sequences of the p28 gene from E. canis samples from the mesoregions of Rio de Janeiro state revealed a high degree of identity with the samples analyzed from Jaboticabal (99 to 100%) and samples from North Carolina, Oklahoma, Louisiana, and the Philippines (99%). These results show a significant degree of conservation among the Brazilian, American, and Asian samples. The p28 gene is useful for the production of vaccines and standardization of immunodiagnostic methods since there is high genetic conservation of this gene in E. canis from Brazil and the USA (EF014897; AF082750) [19, 35]. Despite the similarity with the Jaboticabal sequence, significant changes in amino acid sequences were observed with approximately 4% of amino acids showing divergences. In a previous study, Alves [36] described the occurrence of 8 polymorphism points using the deduced amino acid sequence of the p28 gene. These results agree with those observed by Nakaghi [35] and Aguiar [37] that found variable numbers of polymorphism-identified divergences in the number of amino acids of the analyzed sequences. For Nakaghi [35], these differences correspond to the differences between the amino acid sequences of the proteins and the factors associated with these divergences and may be associated with the selection pressure exerted on the microorganisms and their hosts.
In this study, when comparing the E. canis sequences from the six mesoregions of the Rio de Janeiro state with the E. chaffeensis sequence (DQ085431) of the USA, it was possible to observe a similarity of 79%. In this study, using the p28 gene, the identity of E. canis was 79% with E. chaffeensis from the state of Rio de Janeiro. In this study, we determined that the p28 gene is a good tool for the molecular characterization of E. canis; similar results have been observed in other studies [5]. Comparing and characterizing an organism are useful for providing important information about the host’s immune response and serve as a basis for the identification of new molecular markers for the specific diagnosis of CME. These factors depend on an understanding of the differences that can occur in geographically dispersed strains of E. canis. The genetic variability of this bacterium should be considered because it directly influences the development of new diagnostic techniques and vaccines.
Conclusions
The present study represents a robust genetic analysis of the gp19, gp36, and p28 genes of E. canis in naturally infected dogs. The genetic diversity found in the sequences of the gp36 and p28 genes in E. canis in the state of Rio de Janeiro are useful for a better understanding of the epidemiological chain of this agent. The mechanisms responsible for these genetic divergences in E. canis remain unknown. However, since these polymorphisms occur in genes encoding proteins, vertebrate host immune pressure and adaptation to the invertebrate host may play an important role in the survival of this bacterium.
Acknowledgments
This study was financially supported by the National Council for Scientific and Technological Development (CNPq) of Brazil, the “Carlos Chagas Filho” Foundation for Research Support of the state of Rio de Janeiro (FAPERJ) and Coordination of Improvement of Higher Education Personnel (CAPES) funds.
Compliance with ethical standards
Procedures performed on animals in this study were approved by the Ethics Committee on Animal Use of the UFRRJ (CEUA/UFRRJ) under procedure number 072/2014. These procedures comply with the basic and ethical principles for research involving the use of animals. All procedures were performed by a team of trained veterinarians.
Conflict of interest
The author(s) declared that there is no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Donatien A, Lestoquard F. Existence En Algerie D’une Rickettsia Du Chein. Bull Soc Pathol Exot. 1935;28:418–419. [Google Scholar]
- 2.Huxsoll DL, Hildebrandt PK, Nims RM, Walker JS. Tropical canine pancytopenia. Journal of American Veterinary Medical Association. 1970;157(11):1627–1632. [PubMed] [Google Scholar]
- 3.Macieira DB, Messick JB, Cerqueira AM, Linhares GFC, Freire IMA, Almeida NKO, Almosny NRP. Prevalence of Ehrlichia canis infection in thrombocytopenic dogs from Rio de Janeiro. Brazil Veterinary Clinical Pathology. 2005;34:44–48. doi: 10.1111/j.1939-165X.2005.tb00008.x. [DOI] [PubMed] [Google Scholar]
- 4.Ferreira RF, AMF C, Castro TX, Ferreira EO, FPG N, Barbosa AV, Macieira DB, NRP A. Genetic diversity of Ehrlichia canis strains from naturally infected dogs in Rio de Janeiro. Rev Bras Parasitol Vet. 2014;23:301–308. doi: 10.1590/S1984-29612014055. [DOI] [PubMed] [Google Scholar]
- 5.Nakaghi ACH, Machado RZ, Ferro J, Labruna MB, Chryssafidis AL, André MR, Baldani CD. Sensitivity evaluation of a single-step PCR assay using Ehrlichia canis p28 gene as a target and its application in diagnosis of canine ehrlichiosis. Rev Bras Parasitol Vet. 2010;19:75–79. doi: 10.1590/S1984-29612010000200001. [DOI] [PubMed] [Google Scholar]
- 6.Aguiar DM, Zhang X, Melo ALT, Pacheco TA, Meneses AMC, Zanutto MS, Horta MC, Santarém VA, Camargo LMA, Mcbride JW, Labruna MB. Genetic diversity of Ehrlichia canis in Brazil. Vet Microbiol. 2013;164:315–321. doi: 10.1016/j.vetmic.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 7.Eiras DF, Craviotto MB, Vezzani D, Eyal O, Baneth G. First description of natural Ehrlichia canis and Anaplasma platys infections in dogs from Argentina. Comp Immunol Microbiol Infect Dis. 2013;36(2):169–173. doi: 10.1016/j.cimid.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 8.Mittal M, Kundu K, Chakravarti S, Mohapatra JK, Nehra K, Sinha VK, Sanjeeth BS, Churamani CP, Kumar A. Canine Monocytic Ehrlichiosis among working dogs of organised kennels in India: a comprehensive analyses of clinico-pathology, serological and molecular epidemiological approach. Prev Vet Med. 2017;1(147):26–33. doi: 10.1016/j.prevetmed.2017.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamani J, Lee CC, Haruna AM, Chung PJ, Weka PR, Chung YT. First detection and molecular characterization of Ehrlichia canis from dogs in Nigeria. Res Vet Sci. 2013;94(1):27–32. doi: 10.1016/j.rvsc.2012.07.031. [DOI] [PubMed] [Google Scholar]
- 10.Maazi N, Malmasi A, Shayan P, Nassiri SM, Salehi TZ, Fard MS. Molecular and serological detection of Ehrlichia canis in naturally exposed dogs in Iran: an analysis on associated risk factors. Braz J Vet Parasitol. 2014;23(1):16–22. doi: 10.1590/S1984-29612014002. [DOI] [PubMed] [Google Scholar]
- 11.Murphy GE, Ewing SA, Whitworth LC, Fox JC, Kocan AAA. A molecular and serologic survey of Ehrlichia canis, E. chaffensis, and E. ewingii in dogs from Oklahoma. Vet Parasitol. 1998;79:325–339. doi: 10.1016/S0304-4017(98)00179-4. [DOI] [PubMed] [Google Scholar]
- 12.Baneth G, Harrus S, Ohnona FS, Schlesinger Y. Longitudinal quantification of Ehrlichia canis in experimentel infection with comparison to natural infection. Vet Microbiol. 2008;136:321–325. doi: 10.1016/j.vetmic.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 13.Chen YH, Lee CC, Tsang CL, Chung YT. Detection and characterization of four novel genotypes of Ehrlichia canis from dogs. Vet Microbiol. 2010;146:70–75. doi: 10.1016/j.vetmic.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Luo T, Keysary A. Genetic and antigenic diversities of major immunoreactive proteins in globally distributed Ehrlichia canis strains. Clin Vaccine Immunol. 2008;15:1080–1088. doi: 10.1128/CVI.00482-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cárdenas AM, Doyle CK, Zhang X, Nethery K, Corstvet RE, Walker DH, Mcbride JW. Enzyme-linked Immunosorbent assay with conserved immunoreactive glycoproteins TRP36 and gp19 has enhanced sensitivity and provides species-specific immunodiagnosis of Ehrlichia canis infection. Clin Vacc Immun. 2007;14:123–128. doi: 10.1128/CVI.00361-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doyle CK, Nethery KA, Popov VL, McBride JW. Differentially expressed and secreted major immunoreactive protein orthologs of Ehrlichia canis and E. chaffeensis elicit early antibody responses to epitopes on glycosylated tandem repeats. Infec Immun. 2006;74:711–720. doi: 10.1128/IAI.74.1.711-720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mcbride JW, Walker DH. Molecular and cellular pathobiology of Ehrlichia infection: targets for new therapeutics and immunomodulation strategies. Expert Rev Mol Med. 2011;31:1–3. doi: 10.1017/S1462399410001730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long W, Zhang XF, Qi H, Standaert S, Walker D, Yu XJ. Antigenic variation of Ehrlichia chaffeensis resulting from differential expression of the 28-Kilodalton protein gene family. Infect Immun. 2002;70:1824–1831. doi: 10.1128/IAI.70.4.1824-1831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mcbride JW, Yu XJ, Walker DH. Molecular cloning of the gene for a conserved major immunoreactive 28-Kilodalton protein of Ehrlichia canis: a potential serodiagnosis antigen. Clin Diagn Lab Immun. 1999;6:392–399. doi: 10.1128/cdli.6.3.392-399.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mcbride JW, YU X, Walker DH. A conserved, transcriptionally active p28 multigene lócus of Ehrlichia canis. Gene. 2000;254:245–252. doi: 10.1016/S0378-1119(00)00256-0. [DOI] [PubMed] [Google Scholar]
- 21.IBGE (2010) Municípios. Disponível em: https://ww2.ibge.gov.br. Último acesso em: 14 de dezembro de 2017
- 22.Sampaio IBM. Estatística aplicada à experimentação Animal. 2. Brazil: Belo Horizonte; 2002. [Google Scholar]
- 23.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 24.Svec AC, Tichopa DB, Novosadova V, Michael W, Pfaffl MW, Kubistaa M. How good is a PCR efficiency estimate: recommendations for precise and robust qPCR efficiency assessments. Biomolecular Detection and Quantification. 2015;3:9–16. doi: 10.1016/j.bdq.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 27.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;11(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;30:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 30.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA 6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brum FA (2010) Cloning and Expression of the gp19 protein of Ehrlichia canis. 2010. 10f. Dissertação (Mestrado: Biologia) Universidade Federal de Pelotas, Pelotas
- 32.Mcbride JW, Doyle CK, Zhang X, Cardenas AM, Popov VL, Nethery KA, Woods ME. Identification of a glycosylated Ehrlichia canis 19-Kilodalton major immunoreactive protein with a species-specific serine-rich glycopeptide epitope. Infect Immun. 2007;75:74–82. doi: 10.1128/IAI.01494-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zweygarth A, Cabezas-Cruz A, Marinda C, Oosthuizen P, Matjila KL, Marzena B, Schöl H, Ferrolho J, Grubhoffer L, Passos LMF (2014) In vitro culture and structural differences in the major immunoreactive protein TRP36 of geographically distant Ehrlichia canis sequences. Ticks and Tick-borne Diseases:1–9 [DOI] [PubMed]
- 34.Hasegawa MY (2005) Dinâmica da infecção experimental de cães por Ehrlichia canis: Aspectos clínicos, laboratoriais e resposta imune humoral e celular. 136f. Tese (Doutorado em Medicina Veterinária) Programa de Pós Graduação em Clínica Veterinária Faculdade de Medicina Veterinária e Zootecnia da Universidade de São Paulo-FMVZ, SP
- 35.Nakaghi ACH, Machado RZ, Costa MT, André MR, Baldani CD. Canine Ehrlichiosis: clinical, hematological, serological and molecular aspects. Ciência Rural. 2008;38:766–770. doi: 10.1590/S0103-84782008000300027. [DOI] [Google Scholar]
- 36.Alves RN, Rieck SE, Ueira-Vieira C, Labruna MB, Beletti ME. Isolation, in vitro propagation, genetic analysis, and immunogenic characterization of an Ehrlichia canis strain from southeastern Brazil. J Vet Sci. 2014;15(2):241–248. doi: 10.4142/jvs.2014.15.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aguiar DM, Hagiwara MK, Labruna MB. In vitro isolation and molecular characterization of an Ehrlichia canis strain from São Paulo, Brazil. Braz J Microbiol. 2008;39(3):489–493. doi: 10.1590/S1517-83822008000300014. [DOI] [PMC free article] [PubMed] [Google Scholar]