Abstract
The aim of this paper is to conduct a systematic and theoretical analysis of estimation and inference for a class of functional mixed effects models (FMEM). Such FMEMs consist of fixed effects that characterize the association between longitudinal functional responses and covariates of interest and random effects that capture the spatial-temporal correlations of longitudinal functional responses. We propose local linear estimates of refined fixed effect functions and establish their weak convergence along with a simultaneous confidence band for each fixed-effect function. We propose a global test for the linear hypotheses of varying coefficient functions and derive the associated asymptotic distribution under the null hypothesis and the asymptotic power under the alternative hypothesis are derived. We also establish the convergence rates of the estimated spatial-temporal covariance operators and their associated eigenvalues and eigenfunctions. We conduct extensive simulations and apply our method to a white-matter fiber data set from a national database for autism research to examine the finite-sample performance of the proposed estimation and inference procedures.
Key words and phrases: Functional response, global test statistic, mixed effects, spatial-temporal correlation, weak convergence
1. Introduction
There has been an increasing interest in the analysis of massive functional data sets, many of which originate from brain imaging in large-scale longitudinal biomedical studies such as the Alzeimer’s Disease Neuroimaging Initiative (ADNI) (Evans and Group, 2006; Mueller et al., 2005; Greven et al., 2010; Yuan et al., 2014; Zipunnikov et al., 2014). In such studies, longitudinal functional data from n different subjects are usually observed at or are registered to a large number of locations in a common space, denoted by 𝒮, across multiple time points {tij : j = 1, …, Ti; i = 1, …, n}, where Ti is the total number of time points for the i–th subject. Here we use the term “functional data” for data that are measured densely in 𝒮, “spatial correlation” for correlations within the functional data, and “longitudinal data” and “temporal correlation” for data that are measured sparingly in {tij : j = 1, …, Ti, i = 1, …, n} to distinguish them.
The sheer size and complexity of longitudinal functional data poses substantial challenges to most existing statistical methods for analyzing univariate or multivariate longitudinal data (Diggle et al., 2002; Fitzmaurice et al., 2004). The major challenges include: (i) complexity of the temporal-spatial covariance structure, (ii) determining how to take advantage of the temporal-spatial smoothness, and (iii) theoretical justification of inference procedures. The first challenge is how to introduce random effects to characterize the spatial-temporal covariance structure of longitudinal functional responses. The second one is how to incorporate temporal-spatial smoothness into both estimation and inference procedures to improve statistical efficiency (Ramsay and Silverman, 2005). The third one is to systematically investigate the theoretical properties (e.g., consistency) of estimation and inference procedures for statistical models developed for longitudinal functional data.
Models for longitudinal functional data fall into a general functional mixed effects modeling framework, which serves to characterize functional data with various levels of hierarchical structures (Guo, 2002; Wu and Zhang, 2002, 2006; Morris and Carroll, 2006; Di et al., 2009; Greven et al., 2010; Zhou et al., 2010; Zhu et al., 2011; Shi and Choi, 2011; Cao et al., 2012; Chen and Müller, 2012; Horvath and Kokoszka, 2012; Meyer et al., 2015; Reiss et al., 2014; Scheipl et al., 2015; Zipunnikov et al., 2014; Staicu et al., 2015; Cederbaum et al., 2016). The term functional mixed effects models (FMEMs) for correlated functional data was introduced in Guo (2002), while Morris and Carroll (2006) and subsequent work by this group developed general functional mixed effects models with multiple levels of random effect functions as well as curve-to-curve deviations. Recently, a general framework of functional additive mixed models was introduced by (Scheipl et al., 2015). Moreover, several FMEMs have been developed for longitudinal functional data (Greven et al., 2010; Yuan et al., 2014; Zipunnikov et al., 2014; Di et al., 2014). To the best of our knowledge, most papers on functional mixed effects models focus on challenges (i) and (ii) above, while our focus in this paper is challenge (iii), the theoretical challenges.
To address challenge (iii), we provide a comprehensive theoretical analysis for a class of FMEMs. Our FMEM consists of a measurement model at each grid point s ∈ 𝒮 and a hierarchical factor model. The measurement model primarily includes fixed effects to characterize the varying association between longitudinal functional responses and the covariates of interest. The hierarchical factor model primarily uses random effects to capture the medium-to-long-range spatial covariance and the local covariance structure. Formally, we establish the weak convergence of the estimated varying association function, the uniform convergence rate of the spatial-temporal covariance estimator, the asymptotic distribution of a global test statistic for linear hypotheses of the regression coefficient functions, and an asymptotic simultaneous confidence band for each varying fixed effect function. The code and documentation for FMEM written in Matlab along with its documentation are freely accessible from the website http://www.nitrc.org/projects/fadtts.
2. FMEM: Functional Mixed Effects Model
2.1 Model Setup
Suppose that we observe longitudinal functional data and clinical variables from n independent subjects. Let Ti be the total number of longitudinal measurements for the i-th subject, i = 1, …, n, and tij be the j-th measurement time point for the i-th subject, so j = 1, …, Ti. Throughout this paper, we focus on a fixed number of time points and sparse longitudinal data, that is, maxi≤n Ti < T0 < ∞. Let sm represent a specific grid point of the functional template space 𝒮 for m = 1, …, M. Specifically, for the i-th subject at time tij, we observe functional data, denoted by yij(sm) = yi(tij, sm) for 1 ≤ m ≤ M, and a px dimensional covariate vector xi of interest, denoted by xij = xi(tij), at time tij. The xi may include time-independent as well as time-dependent covariates, such as age, gender, and genetic markers. For ease of notation, it is assumed throughout this paper that 𝒮 = [0, 1] and 0 = s1 ≤ ⋯ ≤ sM = 1, but our results can be easily extended to higher dimensions, when 𝒮 is a compact subset of a Euclidean space.
We consider a FMEM consisting of a measurement model and a hierarchical factor model. This model aims to extend conventional linear mixed-effects model to accommodate the additional spatial component. The measurement model associated with the FMEM characterizes the varying association between functional responses and their covariates at any s ∈ 𝒮 as
| (2.1) |
where μ(·, ·) is a known function, β(s) = (β1(s), …, βpβ (s))T is a pβ × 1 vector of the fixed-effect functions of s, and zij = zi(tij) = (zij1, …, zijpz)T is a pz × 1 vector of the random-effect covariates associated with the random effects bi(s). Here bi(s) = (bi1(s), …, bipz (s))T is a vector of the random effects that characterize the spatial temporal correlation structures across the functional domain space; whereas eij(s) is a spatial random process delineated from bi(s), i.e., after filtering out . Moreover, eij(s) and bi(s) are independent. In many applications, is a linear function of xij, similar to the setting of traditional linear mixed-effects model, so we focus on this special linear case in the paper. Extensions to nonlinear cases is discussed in Remark 1. Since marginally, for a fixed s, model (2.1) with is a standard linear mixed effects model, this motivates us to adopt standard notation for linear mixed effects models. Moreover, since zij may include time-independent, as well as time-dependent, covariates, the inclusion of allows us to capture a large portion of the variation in the spatial and temporal correlation structures.
The spatial random process eij in (2.1) is further decomposed into two parts,
| (2.2) |
where eij,G(s) is a smooth stochastic process representing the global dependency that depicts the medium-to-long-range spatial dependence, eij,L(s) is a measurement error representing local variability, and eij1,G(·) and eij2,L(·) are independent for any j1 and j2. Since eij,L(s) are measurement errors, we assume that eij1,L(s) and eij2,L(s′) are mutually independent whenever either j1 ≠ j2 or s ≠ s′. We also assume that, for any j1 ≠ j2, eij1,G(·) and eij2,G(·) are mutually independent. This assumption is equivalent to assume that the random effects bi(·) = (bi1(·), …, bipz (·))T explains all the within-subject correlation along the longitudinal direction, which is a common assumption in linear mixed-effects model. However, it does not exclude correlations along the functional direction as as eij,G(s) and eij,G(s′) are not required to be independent for s ≠ s′.
Moreover, bi(s), eij,L(s), and eij,G(s) are mutually independent and are independent and identical copies of SP(0, Σe,L), SP(0, Σb), and SP(0, Σe,G), respectively, where SP(μ, Σ) denotes a stochastic process vector with mean function (or function vector) μ(s) and covariance function (or function matrix) Σ(s, s′). Moreover, Σb(s, s′) is a pz × pz matrix with Σbkk′(s, s′) as the (k, k′)-th element, and the covariance structure of yi(s) = (yi1(s), …, yiTi(s))T, denoted by Σy,i(s, s′), is , where 1(·) is an indicator function.
2.2 Estimation Procedure
Our primary goal is to find efficient procedures for estimation and inference for β(·). Inspired by novel ideas from the literature (Yao et al., 2005; Greven et al., 2010; Zipunnikov et al., 2014), we develop a procedure to estimate β(·), Σbkk′(·, ·), Σe,G(·, ·), Σe,L(·, ·), and the eigenvalue-eigenvector pairs of Σbkk′(·, ·), and Σe,G(·, ·). Compared with the estimation methods of Greven et al. (2010) and Zipunnikov et al. (2014), our method is an improvement over the ordinary least square methods to estimate β(·) by incorporating spatial and/or temporal smoothness in longitudinal functional data. Explicitly, we incorporate the within-subject correlation among Ti longitudinal observations to gain statistical efficiency as stated in Theorem 1.
From hereafter, we focus on , but the proposed estimation procedure can be extended to a nonlinear mean function μ(xij, β(s)), which is discussed at the end of Section 2.2. There are four key steps in the estimation procedure as described below.
Step (I): Calculate an initial estimator β̂(s) of β(s) for each s ∈ 𝒮.
Step (II): Calculate estimates of the covariance operators Σbkk′(·, ·) and Σe,G(·, ·) and their spectral decompositions, and obtain the estimate of Σe,L(·, ·).
Step (III): Use the estimated covariance operators obtained from Step (II) to improve the estimate in step (I) with a refined estimator of β(s), denoted by β̃(s).
Step (IV): Obtain individual random effect functions .
Step (I): We employ a local linear smoother (Fan and Gijbels, 1996) to obtain an initial estimator of β(·) without incorporating spatial-temporal correlation. Specifically, we apply a Taylor expansion for β at s,
| (2.3) |
where sh1(sm − s) = (1, (sm − s)/h1)T and A(s) = [β(s) h1β̇(s)] is a px × 2 matrix. Here β̇(s) = (β̇1(s), …, β̇px(s))T is a px × 1 vector and β̇l(s) = dβl(s)/ds for l = 1, …, px. Let K(s) be a kernel function and Kh(s) = h−1K(s/h) be the rescaled kernel function with bandwidth h. We estimate A(s) by minimizing the following weighted least squares function:
| (2.4) |
Let a⊗2 = aaT for any vector a and C ⊗ D be the Kronecker product of two matrices C and D. For an M1 × M2 matrix C = (cjl), denote vec(C) = (c11, …, cM11, …, c1M2, …, cM1M2)T. Let Â(s) be the minimizer of (2.4). Then
| (2.5) |
where . Thus, we have
| (2.6) |
where Ipx is a px × px identity matrix. In practice, we may select the bandwidth h1 by using leave-one-curve-out cross-validation. Specifically, we pool the data from all n subjects and select a bandwidth h1 by minimizing the cross-validation score given by
| (2.7) |
where β̂(s, h1)(−i) is the local linear estimator of β(s) with the bandwidth h1 based on data excluding all the observations from the i-th subject.
Step (II): We use a two-step procedure to estimate Σb(s, s′) and Σe,G(s, s′). Let Σe(s, s′) be the covariance function of eij(s).
- (S1) First, we use a least squares method to estimate Σb(sm, sm′) and Σe(sm, sm′) for m, m′ = 1, …, M. Let . We estimate Σb(sm, sm′) and Σe(sm, sm′) by minimizing the following least squares function:
where Σj1≠j2 denotes the sum over all j1, j2 = 1, …, Ti such that j1 ≠ j2. The least squares method in (2.8) has been considered in the literature (Di et al., 2009; Greven et al., 2010; Cederbaum et al., 2016), where previous authors used penalized splines smoothing instead of local linear regression. Let and be the minimizers of (2.8). Then we have(2.8)
where ,(2.9) (S2) Next, for each (k, k′), with 1 ≤ k, k′ ≤ pz, we apply a local constant smoother to for sm, sm′ ∈ 𝒮 × 𝒮 and m, m′ = 1, …, M. This provides the final estimate for Σb(s, s′). Likewise, we can obtain an estimate of Σe,G(s, s′) through a local constant smoother, where the diagonal elements of , i.e. , are excluded from the estimation of Σe,G(s, s′).
Specifically, we estimate Σbkk′(s, s′) and Σe,G(s, s′) by minimizing the following weighted least squares functions:
| (2.10) |
| (2.11) |
The bandwidths h2 and h3 are selected through the leave-one-curve-out cross-validation method.
Finally, we perform the spectral decomposition of Σ̂bkk′(s, s′) and Σ̂e,G(s, s′) and then calculate Σ̂e,L(sm, sm) by using
Step (III): We incorporate the estimated covariance function to improve the local linear regression estimate of β(·). Similar but different ideas have been used to iteratively improve the mean estimation (Cederbaum et al. (2016); Di et al. (2014)). Letting Σyi,G(s, s′) be the covariance function of ui,G(s) = (ui1,G(s), …, uiTi,G(s))T, we obtain its estimator Σ̂yi,G(s, s′) based on Σ̂b(s, s′) and Σ̂e,G(s, s′) from step (II). Let Xi = (xi1 ⋯ xiTi) be a px × Ti matrix. We estimate A(s) by minimizing the following weighted least squares function:
| (2.12) |
where hβ is a bandwidth.
Let Ã(s) be the minimizer of (2.12). Then, we have
where . We have
| (2.13) |
To select the bandwidth hβ, we pool the data from all n subjects and select a bandwidth hβ that minimizes the cross-validation score,
| (2.14) |
where β̃(s, hβ)(−i) is the local linear estimator of β(s) with the bandwidth hβ based on data excluding all the observations from the i-th subject.
Step (IV): We use the local linear regression method to smooth and then obtain an estimate of for each i and j. Since the local linear regression is a standard method (Fan and Gijbels, 1996; Wand and Jones, 1995), we omit the detailed steps for the approximation of uij,G(s). Furthermore, if there is an interest in recovering the subject-specific random effect bi(s), one could use the best linear unbiased predictors, which are commonly employed in linear mixed-effects models, to estimate bi(s) at each point s and then smooth over s.
Remark 1
To extend the estimation procedure to nonlinear mean functions μ(xij, β(s)), such as exponential functions or power functions, one needs to modify steps (I) and (III) by applying a Taylor expansion for μ(xij, β(sm)) at s,
where μ̇(xij, β(s)) = ∂μ(xij, β(s))/∂β(s) and μij(s) = (μ(xij, β(s)), μ̇(xij, β(s))β̇(s)h1). Then, one estimates A(s) by minimizing a nonlinear weighted least squares function:
In this general case, Â(s) does not have an explicit form, but it can be estimated by using optimization algorithms, such as the Gaussian Newton algorithm or Levenberg-Marquardt algorithm (Seber and Wild, 1989). Similar to Ln(A(s)), we can modify (2.12) in step (III).
2.3 Computational Complexity
The computational complexity of our estimation procedure is extremely important for high-dimensional neuroimaging data, which usually contain a large number of locations, especially when they correspond to the voxel locations of the image. For instance, M can have a magnitude of tens of thousands. For the linear mean function, the computational complexity of our estimation procedure in Section 2.2. is O(nh1T0M2 + nT0(R0M)2 + nT0hsM2). If we use leave-one-out cross-validation, then the computational effort increases by a factor of n.
We first discuss steps (I) and (III). In step (I), we need to calculate the local linear estimator of β(sm) at each grid point sm across 𝒮0 = {sm, m = 1, …, M}. The computational complexity of step (I) is almost the same as that in standard point-wise linear regression analysis. An alternative is to fit a linear mixed-effect model at each grid point sm using the maximum likelihood. However, this step is not necessary as it only applies to an initial estimate, which then is improved in step (III).
For step (III), we only need to calculate the weighted least squares estimators β̃(sm) in (2.13) across sm ∈ 𝒮0, which is computationally straightforward. The computational complexity is O(nT0h1M) for each sm, so overall it is O(nT0h1M2).
To improve computational efficiency, we standardize all covariates and then use a single tuning parameter h1 to smooth all the coefficient functions βj(s). Since this strategy works best for coefficient functions that exhibit similar degrees of smoothness, it may be necessary to use different tuning parameters for different coefficient functions (Fan and Zhang, 2008) when the coefficient functions have different level of smoothness.
Next, we discuss the computational complexity of step (II). First, estimating ûij(s) is computationally fast for all possible (i, j). Second, we do not need to calculate Σb(s, s′) and Σe,G(s, s′) for all possible (s, s′). As discussed in step (III) above, we only need the estimates of Σb(sm, sm) and Σe,G(sm, sm) for all sm ∈ 𝒮0. Therefore, in step (S2), we can focus on solving Σb(sm, sm) and Σe,G(sm, sm) with all (sm, sm′) in {(sm, sm′) ∈ 𝒮0 × 𝒮0 : |sm − sm′ | ≤ R0}, where R0 is a positive scalar. In this case, step (II) is computationally feasible even for large M when R0 is relatively small. The computational complexity is at most O(nT0(R0M)2) for (sm, sm′) ∈ 𝒮0 × 𝒮0.
A major computational hurdle is to calculate Σb(s, s′) and Σe,G(s, s′) for all possible (s, s′). If M is relatively large, it can be computationally challenging to estimate and across all possible . We take two different approaches. The first one is to estimate and for a small subset of S0 × S0. Specifically, we can bin the data to reduce the number of grid points substantially to a much smaller number M0 << M, and estimate Σb(s, s′) and Σe,G(s, s′) on those M0 points and interpolate the results elsewhere. The second approach is to apply the approaches proposed by Zipunnikov et al. (2014) and Xiao et al. (2016) to the estimation of Σb(s, s′) and Σe,G(s, s′). These methods include a fast implementation of the sandwich smoother for covariance smoothing and a two-step procedure where one first obtains the singular value decomposition of the data matrix and then smooths the eigenvectors.
Regarding the computational complexity of step (IV), we note that, similar to step (II), smoothing uij,P(s) for all possible (i, j) is computationally light. The overall computational complexity is approximately O(nT0hsM2), where hs is the bandwidth of the local linear method.
Remark 2
We discuss two possible extensions of (2.2). The first is to extend the estimation procedure from 𝒮 = [0, 1] to a D–dimensional compact subset of a Euclidean space. For this, we only need to modify steps (I) and (III) by changing β̇l(s) and sm − s into D × 1 vectors. The second extension is to assume that eij1,G(s) and eij2,G(s) for j1 ≠ j2 are dependent and have a separable covariance structure, cov(eij1,G(s), eij2,G(s)) = Σe,G(s, s′)ρ(tij1, tij2 ; θ), where ρ(tij1, tij2 ; θ) is usually a pre-specified correlation function of unknown parameter θ, such as the exponential correlation model with ρ(tij1, tij2 ; θ) = exp(−θ|tij1 − tij2|) (Diggle et al., 2002; Fitzmaurice et al., 2004). However, we found empirically that the use of the correlation function dramatically increases the computational complexity but does not lead to much efficiency gain for the estimation of β(·).
3. Theoretical Results
We systematically investigate the asymptotic properties of all estimators proposed in Section 2.2 and investigate several inference procedures based on the asymptotic properties. For any smooth function f(s), we use the notation ḟ(s) = df(s)/ds and f̈(s) = d2f(s)/ds2. We use uq = ∫ K(υ)υqdυ and υq = ∫ Kq(υ)dυ for q = 1 and 2, and ‖ · ‖2 for the Euclidean norm.
3.1 Assumptions
Throughout the paper, the following assumptions are used to facilitate the technical details. Some of the assumptions might be weakened but the current version simplifies the proof.
(A.1) The grid points in 𝒮0 = {sm, m = 1, …, M} are independently and identically distributed with a density function f(s), which has a continuous second-order derivative and bounded support 𝒮. Moreover, for some fl > 0 and fu < ∞, fl < f(s) < fu for all s ∈ 𝒮.
(A.1b) The grid points 𝒮0 = {sm, m = 1, …, M} are prefixed according to a design density function f(s) such that for m ≥ 1. Here f(s) has continuous second-order derivative and bounded support [0, 1], and fl < f(s) < fu for all s ∈ [0, 1], for some positive fl > 0 and fu < ∞.
(A.2) The covariate vectors xij = (xij1, …, xijpx)T and zij = zi(tij) = (zij1, …, zijpz)T, may or may not be time-dependent. Nevertheless, we use the notation xijl = xil(tij) for 1 ≤ l ≤ px, and zijl = zil(tij) for 1 ≤ l ≤ pz. We assume that supt∈𝒯 |xil(t)| and supt∈𝒯 |zil(t)| are almost surely bounded, where 𝒯 is a finite time domain.
(A.3) The kernel function K(t) is a symmetric density function with compact support [−1, 1], and is Lipschitz continuous.
(A.4) All components of β(s) have continuous second derivatives on 𝒮.
(A.5) With probability one, the sample paths of eij,G(·) and bi(·) are Lipschitz continuous.
(A.6) maxi Ti < T0, n, M → ∞, h → 0, Mh → ∞, nah → ∞ for some a > 0, where T0 is a fixed constant, and h could be h1, hβ, h2, and h3.
(A.7) E{sups∈[0,1] |eij,G(s)|2q} + E{sups∈S0 |eij,L(s)|2q} < ∞ for some q > 2.
(A.8) , for some q > 2.
(A.9) exists for any (s, s′).
(A.10) There is a positive fixed integer E < ∞ such that the eigenvalues of Σe,G satisfy , for some constant λ > 0, and analogously for the eigenvalues of Σb.
Remark 3
Our theoretical results hold for both random and fixed designs. Assumptions (A.1) is a standard condition on random design points s, while (A.1b) is for fixed designs. Assumption (A.2) is a condition on the boundedness of the covariate vectors. The bounded support restriction on K(·) in assumption (A.3) is not essential and can be removed if we put restrictions on the tail of K(·). Assumptions (A.4)–(A.5) are smoothness conditions on the coefficient functions, random functions and their covariances. The smoothness condition in assumption (A.5) can be relaxed with substantial additional efforts (Zhu et al., 2012). Assumption (A.6) is a weak condition on n, M and h, where h1 is the bandwidth used in Step (I) for the initial estimate of β. Assumptions (A.7) and (A.8) require uniform bounds on certain high-order moments of the random functions, which are standard assumptions in the literature (Zhu et al., 2012; Li and Hsing, 2010). Assumption (A.10) on simple multiplicity of the first E eigenvalues is only needed to investigate the asymptotic properties of the eigenfunctions. It is also a standard assumption in the literature.
3.2. Asymptotics of Estimation Procedure
We state the following theorems, for which detailed proofs can be found in the supplementary document. The first theorem tackles the theoretical properties of {β̃(s) : s ∈ 𝒮} obtained from step (III).
Theorem 1
Under (A.1) (or (A.1b)) and (A.2)–(A.9), we have the following results:
- The asymptotic bias and covariance of β̃(s) for s ∈ (0, 1) are
(3.1) If log M = o(Mhβ) and there exists γn → ∞ with and n−1/2γn log M = o(1) for some q > 2 that satisfies (A.7), then as n → ∞, converges weakly to a centered Gaussian process G(·) ~ 𝒢(0, R), where R(s, s′) = {Q*(s, s)}−1Q*(s, s′){Q*(s′, s′)}−1 with .
Theorem 1 (i) provides theoretical justification of steps (I)–(III) for the refined estimator β̃(s). It has several important implications. First, the estimator β̂(s) obtained in step I has asymptotic covariance
(details can be found in the proof of Theorem 1), which is larger than that of β̃(s). The improvement by the refined estimator β̃(s) is due to the incorporation of within-subject correlations among Ti longitudinal observations, and can lead to substantial efficiency gain in estimating {β(s) : s ∈ 𝒮}. Second, if we use the maximum likelihood (or the restricted maximum likelihood) estimators at each of the observed data at sm, the asymptotic covariance, given by , is larger than that of β̃(sm). The improvement achieved by β̃(sm) is due to incorporating the smoothness in the functional data. Therefore, one can construct more efficient estimators of β(s) by simultaneously accounting for the smoothness in functional data and the within subject covariance, since these functions are measured repeatedly and longitudinally. Moreover, the asymptotic bias of β̃(s) is of the order , which is similar to that of nonparametric regression for independent responses; whereas the asymptotic variance of β̃(s) is of the order n−1.
We note here that the efficiency gain discussed above is not in conict with the results in Lin and Carroll (2001), where they show that the most efficient estimator of the nonparametric function through kernel smoothing is achieved by ignoring the dependence structure among functional observations. In our setting, this means that kernel smoothing in the direction of s should be implemented as we did in Step (I) by ignoring the dependence structure among functional observations. However, in the FMEM setting of longitudinal functional data, it is possible to improve the β estimate as we did in Step (III) by incorporating the covariance structure Σyi,G(s, s). The analogy here is the standard linear mixed-effects model with just longitudinal data (i.e. no functional components), since FMEM is an extension of linear mixed-effects model. It is clear that in linear mixed-effects model one needs to do weighted least square to gain efficiency for the β estimator and this is what we did in Step (III) to refine the β estimator through a weighted least square estimator with weights from Σyi,G(s, s). We emphasize that we could implement Step (III) only after we have obtained a covariance estimate in Step (II), which relies on an initial unweighted least square estimator of β in Step (I). This explains why we need three steps to complete the estimation of β.
Theorem 1 (ii) establishes the weak convergence of the centered estimator β̃(s) − E(β̃(s)), which is essential to carry out the statistical inference for β(s) in Section 3.3 below. Let h = nα, M = nβ and γn = nγ. Anything that satisfies α < 0, α + β > 0 and will satisfy the assumptions, where q > 2 is a constant that satisfies the moment condition given in (A.7).
The second theorem provides the theoretical analysis of the estimators of Σe,G(s, s′) obtained from step (II). Similar results can be obtained for Σb,kk′(s, s′), 1 ≤ k, k′ ≤ pz and are provided in the online supplementary material.
Theorem 2
Under (A.1) (or (A.1b)) and (A.2)–(A.8), (A.10), if h1 = O((log n/n)1/4) and h3 = O(log n/n)1/4, then we have the following results:
sups,s′ |Σ̂e,G(s, s′) − Σe,G(s, s′)| = Op((log n/n)1/2);
For 1 ≤ l ≤ E, ;
For 1 ≤ l ≤ E, .
Theorem 2 characterizes the uniform convergence rates of Σ̂e,G(s, s′) and the associated eigenvalues and eigenfunctions. It can be regarded as an extension of Theorems 3.3–3.6 of Li and Hsing (2010), which established the strong uniform convergence rates of these estimates under a simpler model.
3.3. Asymptotics of Inference Procedure
In this subsection, we derive the asymptotic theory of a global test for testing linear hypotheses of β(·) and the theory for simultaneous confidence bands (SCB) for each component of β(·). These are key tools for statistical inference for the coefficient functions.
We first consider linear hypotheses for β(s),
| (3.2) |
where C is a q × px matrix with rank q, and β0(s) is a given q × 1 vector of functions. We define a global test statistic Sn as
| (3.3) |
where d(s) = Cβ̃(s) − bias(Cβ̃(s)) − β0(s). For simplicity and computational efficiency, we do not consider estimating the bias of Cβ̃(s), since it is negligible based on our simulation results reported below. It follows from Theorem 1 that under H0, we have
where ⇒ denotes weak convergence and GC(·) is a centered Gaussian process with covariance function {CQ*(s, s)CT}−1/2R(s, s′){CQ*(s′, s′)CT}−1/2. Thus, we can derive the asymptotic distribution of Sn under the null hypothesis and its asymptotic power under local alternative hypotheses.
Theorem 3
Under assumptions (A.1)–(A.9), if log M = o(Mhβ) and there exists γn → ∞ with and n−1/2γn log M = o(1) for some q > 2 that satisfies (A.7), we have the following results:
under the null hypothesis H0,
for a sequence of local alternatives H1n : Cβ(s)−β0(s) = n−τ/2d(s), where τ is any scalar in [0, 1), Sn,α is the upper 100α percentile of Sn under H0, and 0 < ∫𝒮 ‖d(s)‖2ds < ∞.
Theorem 3 can be regarded as a generalization of theorem 7 of Zhang and Chen (2007) and theorem 2 of Zhang (2011). The test statistic Sn has a weighted χ2-type asymptotic distribution under H0. Zhang and Chen (2007) (after theorem 7) provided a discussion of the estimation for the null distribution of Sn by χ2-approximation and bootstrapping, which also applies to the case we considered here. It is easy to see that part (ii) still holds when the critical value Sn,α is replaced by some estimated critical value.
Next, we construct simultaneous confidence bands for the coefficient functions, which can then be used for statistical inference for FMEM. For a given confidence level α, we construct a simultaneous confidence band for each βl(s), 1 ≤ l ≤ px, as
| (3.4) |
where and are the lower and upper limits of the SCB. Specifically, a 1 − α simultaneous confidence band for βl(s) is:
| (3.5) |
where Cl(α) is the critical value of sups∈𝒮 |G(s)| associated with β̂l(s) in Theorem 1.
To carry out the inference procedure developed above, we approximate both Cl(α) and Sn,α. Because the asymptotic distribution of Sn is quite complicated and it is difficult to directly approximate the percentiles of Sn under the null hypothesis, we use a wild bootstrap method to approximate the critical values of Sn. The wild bootstrap idea has been used by Zhu et al. (2012); details are presented in the Appendix. Let G(q)(·) be the bootstrapped samples for q = 1, ⋯, Q, where Q is the total number of wild bootstrap samples. The following theorem lays the ground for the wild bootstrap method to construct a simultaneous confidence band of β(s) and to approximate the null distribution of Sn.
Theorem 4
Under assumptions (A.1)–(A.9) and given the data, the bootstrapped process G(q)(s) converges in distribution to 𝒢(0, R), which is defined in part (ii) of Theorem 1, as n → ∞.
4. Simulation Studies
In this section, we present four sets of simulations to examine the finite-sample performance of the proposed estimation and inference procedures. In the first two simulations, we consider two competing methods, including wavelet-based functional mixed models (WFMM) (Morris and Carroll, 2006) and functional additive mixed models (FAMM) (Scheipl et al., 2015). All computations for these numerical examples were carried out using Windows 7, 3.60GHz quard-core Intel Core i7 CPU and 16GB DDR3 1066MHz memory. One can further reduce the computational time for FMEMs by using other computer languages, such as C++.
All simulated data sets were generated from the model:
| (4.1) |
where xij = (1, xij,1, xij,2)T, zij = (1, xij,2), , and eij,L(s) ~ N(0, Σe,L) for i = 1, …, n. Each subject was observed up to 3 times in this sample, among which 5%, 30% and 65% have only one, two and all three observations, respectively. We set sm = (m − 0.5)/M. The first covariate xij,1 was simulated from N(0, 1) and fixed across time for subject i and the second covariate xij,2 was assumed to vary with time, where the increments xij,2 − xi(j−1),2 were independently sampled from a uniform distribution on [0, 1]. Both covariates were standardized to have zero mean and unit variance. Moreover, we set for k = 1, 2, and Σe,L = 0.01. The functional coefficients and eigenfunctions were selected as
We fitted FMEM, WFMM, and FAMM to each simulated data set and calculated all the unknown quantities. The average computational times per simulated data set with n = 100 and M = 40 for FMEM, WFMM, and FAMM are, respectively, 19.6 seconds, 2.32 seconds, and 1.15 hours.
Simulation 1
The first simulation aims at evaluating the performance of the estimates for βj(·). We set n = 100 and M = 40 and 60 and then simulated 1,000 data sets from model (4.1) as described above. Table 1 summarizes the mean integrated absolute error (MIAE) and mean integrated squared error (MISE) of all estimated coefficient functions based on 1,000 simulations. The results in Table 1 indicate satisfactory performance of our estimators since all MIAE and MISE values are quite small. As expected, all the errors decrease as the number of grid points increases. Moreover, FMEM outperforms WFMM and FAMM in terms of both MIAE and MISE. However, this comparison may be unfair to WFMM, since it is designed for spiky data, not the intrinsically smooth functional data.
Table 1.
Simulation 1. MIAE×10−2 and MISE ×10−2 and their standard deviations ×10−2 are reported. MIAE denotes the mean integrated absolute error and MISE denotes the mean integrated square error. Standard deviations are in the parentheses. For each case, 100 simulated data sets were used.
| Method | MIAE×10−2 | MISE×10−2 | |||||
|---|---|---|---|---|---|---|---|
| M | β1(·) | β2(·) | β3(·) | β1(·) | β2(·) | β3(·) | |
| WFMM | 40 | 1.63 (0.73) | 1.67 (0.77) | 1.88 (0.78) | 0.04 (0.04) | 0.05 (0.04) | 0.06 (0.04) |
| 60 | 1.37 (0.61) | 1.39 (0.63) | 1.55 (0.64) | 0.03 (0.03) | 0.03 (0.03) | 0.04 (0.03) | |
|
| |||||||
| FAMM | 40 | 3.36 (2.11) | 2.84 (1.88) | 4.26 (3.27) | 0.23 (0.56) | 0.16 (0.35) | 0.38 (0.77) |
| 60 | 3.03 (1.93) | 2.51 (1.58) | 3.95 (3.29) | 0.18 (0.36) | 0.13 (0.21) | 0.34 (0.95) | |
|
| |||||||
| FMEM | 40 | 1.57 (0.72) | 1.44 (0.65) | 1.69 (0.70) | 0.04 (0.03) | 0.03 (0.03) | 0.05 (0.03) |
| 60 | 1.29 (0.60) | 1.23 (0.55) | 1.37 (0.53) | 0.03 (0.03) | 0.03 (0.01) | 0.03 (0.03) | |
Simulation 2
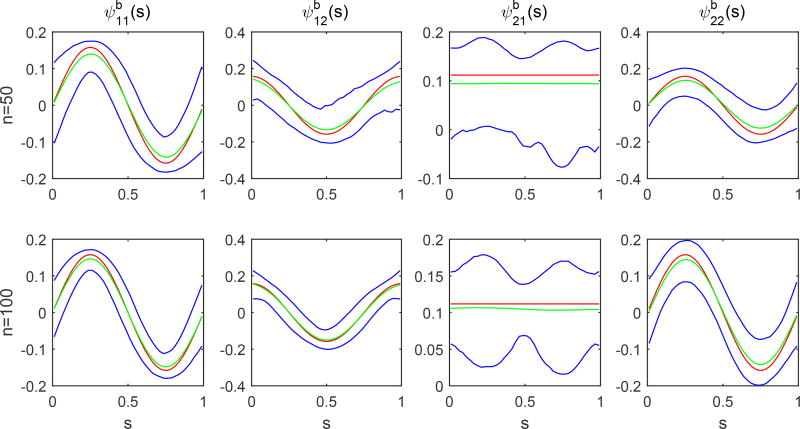
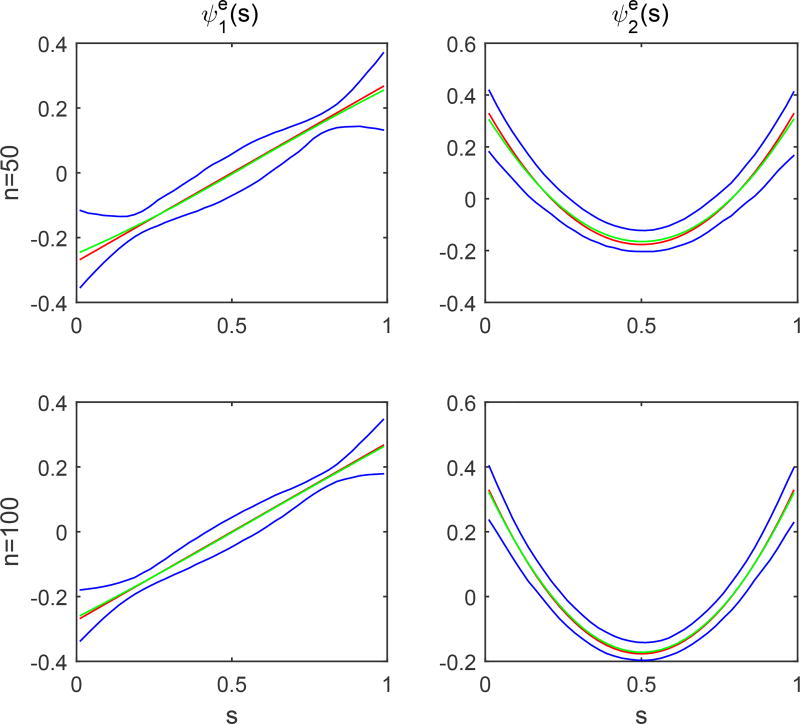
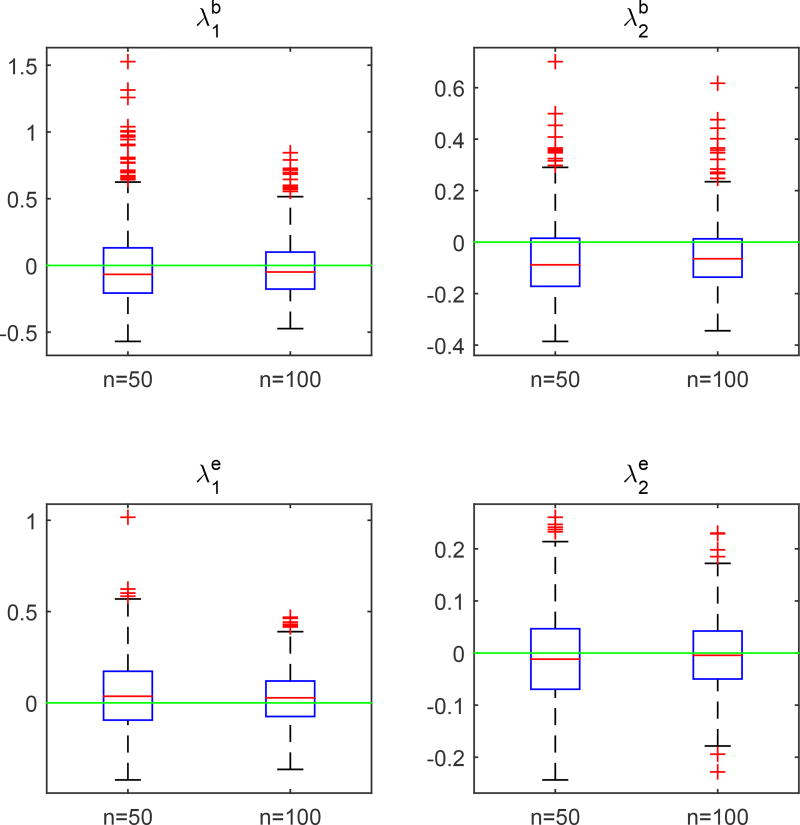
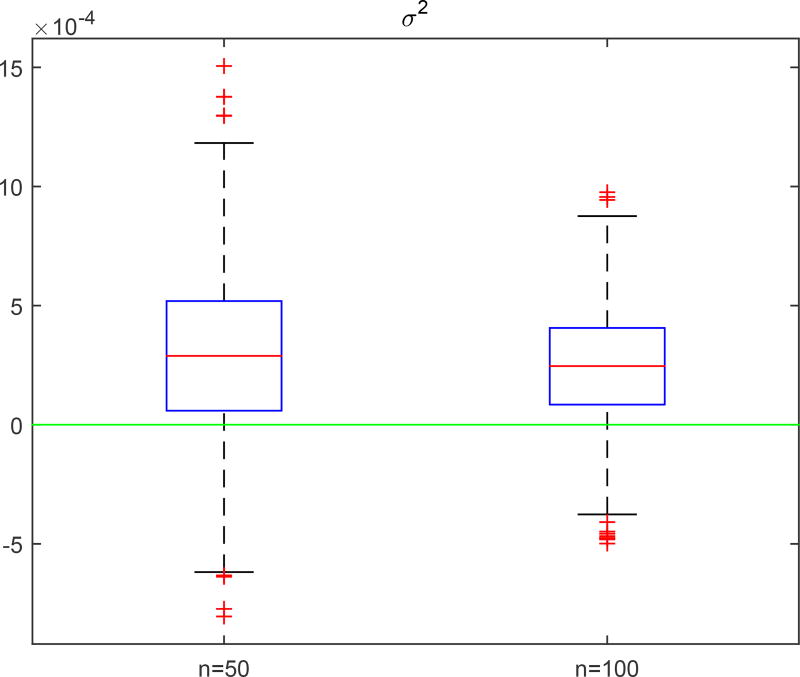
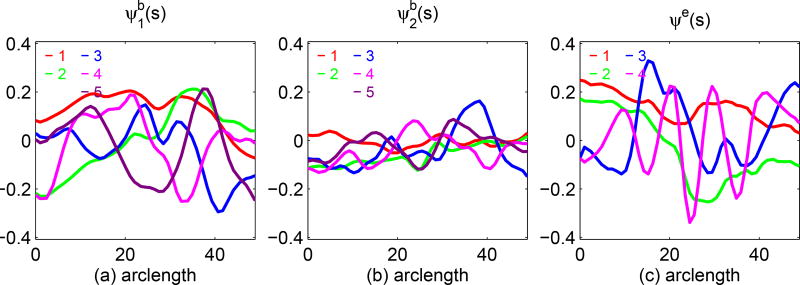
The second simulation is to evaluate the accuracy of the estimators of the eigenvalues and eigenfucntions of the covariance functions Σb(·, ·), Σe,G(·, ·) and Σe,L. We used the same parameter values as those in Simulation 1. We set c = 0.1 and n = 50 and 100, and generated 1,000 datasets for each combination. The accuracy of all kinds of estimators improves with the sample size. The estimated eigenfunctions were plotted in Figures 4.1 and 4.2, in which the mean and the pointwise 5th and 95th percentiles of the estimated functions were plotted along with the true eigenfunctions. Figures 4.3 and 4.4 show the boxplots for the estimates of the eigenvalues and σ2, which are quite close to their true values.
Figure 4.1.
Simulations 2: the estimates of the first two eigenfunctions for l, k = 1, 2 and their pointwise confidence intervals. The red solid, green dashed and blue solid, curves are, respectively, the true eigenfunctions, the pointwise means, and their pointwise 5th and 95th percentiles of estimated eigenfunctions based on 1,000 replications.
Figure 4.2.
Simulations 2: the estimates of the first two eigenfunctions and their pointwise confidence interval. The red solid, green dashed and blue solid, curves are, respectively, the true eigenfunctions, the pointwise means and their pointwise 5th and 95th percentiles of estimated eigenfunctions based on 1,000 replications.
Figure 4.3.
Simulation 2: boxplots of the differences between the estimated eigenvalues and , k = 1, 2 and their true values based on 1,000 replications.
Figure 4.4.
Simulation 2: boxplots of the differences between the estimated σ2 and its true values based on 1,000 replications.
Simulation 3
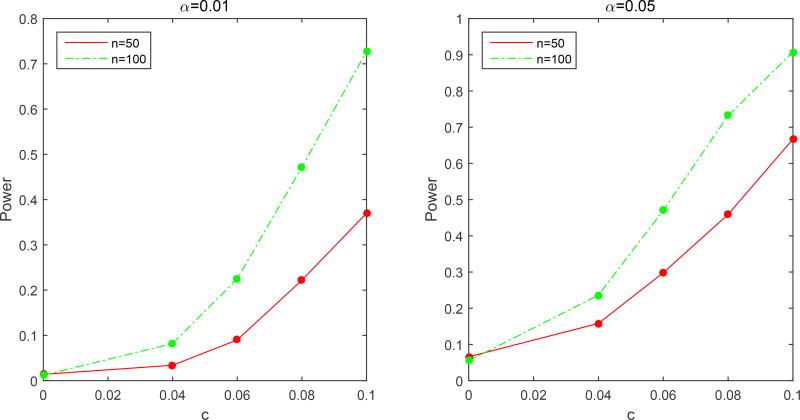
The third simulation is designed to evaluate the type I error rate and power of the global test statistics Sn. We are interested in testing H0 : β3(s) = 0 for all s, against H1 : β3(s) ≠ 0 for some s. All parameters in FMEM were specified as above except that β3(s) was set as 4cs(1 − s)−0.4c, where we first set c = 0 to assess the type I error rate of Sn and then c = 0.04, 0.06, 0.08, and 0.1 to examine the power of Sn at different effect sizes. Furthermore, we set n = 50 and 100 and used 1, 000 replications to estimate the rejection rate of Sn. The p-value of Sn was approximated by the wild bootstrap method with Q = 500 bootstrap samples.
Fig. 4.5 presents the rejection rates of Sn across all effect sizes at the two significance levels α = 0.05 and 0.01. Type I error rates are well maintained at the two significance levels for n = 100. Specifically, at α = 0.05 (or 0.01), the Type I error rates of Sn is 0.066 (or 0.014) for n = 50 and 0.055 (or 0.012) for n = 100, respectively. As expected, the statistical power for rejecting the null hypothesis increases with the sample size, the effect size c and the significance level.
Figure 4.5.
Simulation 3: Power curves as functions of c. Rejection rates of Sn using the wild bootstrap method are calculated at five different values of the effect size c (c = 0, 0.04, 0.06, 0.08 and 0.1) for two sample sizes (n = 50 and 100) at the 0.01 (a) and 0.05 (b) significance levels based on 1,000 replications.
Simulation 4
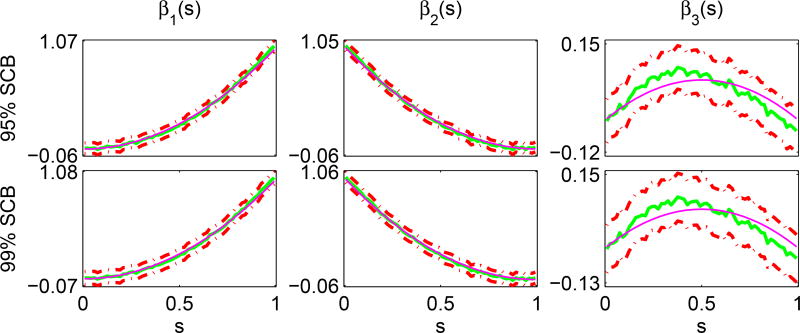
The fourth simulation aims at evaluating the coverage probability of the simultaneous confidence bands for βj(s). We use the same data generated from Simulation 1 above. Based on the 1,000 simulated data sets, we fitted FMEM, WFMM, and FAMM to each simulated data and then calculated SCB for each component in β(s). Table 2 presents the empirical coverage probabilities of all three methods for α = 0.01 and 0.05. The coverage probabilities improve with the number of grid points M. When M = 60, the coverage probabilities are quite close to the pre-specified confidence levels. Since FAMM only provides level (1 − α) confidence interval at each grid point, we use the Bonferroni method to approximate its simultaneous cover probabilities. Again, FMEM outperforms WFMM and FAMM in terms of the coverage probability. However, this comparison may be unfair to WFMM and FAMM, since they do not have any valid method to construct simultaneous confidence bands of βj(s) yet. Fig. 4.6 displays typical 95% and 99% simultaneous confidence bands for coefficient functions βl(s), l = 1, 2, 3 based on FMEM as M = 60.
Table 2.
Simulation 4: Coverage probabilities of estimated coefficient functions based on 1, 000 replications at simultaneous confidence levels 0.95 and 0.99. For each case, 1,000 simulated data sets were used.
| Method | 95% | 99% | |||||
|---|---|---|---|---|---|---|---|
| M | β1 | β2 | β3 | β1 | β2 | β3 | |
| WFMM | 40 | 0.787 | 0.807 | 0.710 | 0.913 | 0.900 | 0.872 |
| 60 | 0.784 | 0.767 | 0.719 | 0.897 | 0.895 | 0.875 | |
|
| |||||||
| FAMM | 40 | 0.991 | 1.000 | 0.993 | 0.996 | 1.000 | 0.996 |
| (Bonferroni) | 60 | 0.996 | 0.998 | 0.994 | 0.999 | 0.998 | 0.991 |
|
| |||||||
| FMEM | 40 | 0.945 | 0.948 | 0.924 | 0.989 | 0.992 | 0.992 |
| 60 | 0.933 | 0.920 | 0.938 | 0.984 | 0.985 | 0.987 | |
Figure 4.6.
Simulation 4: Typical 95% (the first row) and 99% (the second row) simultaneous confidence bands for functional coefficients . The magenta, green solid, and red dash-dotted curves are, respectively, the true curves, the estimated functional coefficients and their corresponding 95% and 99% confidence bands.
5. Data Analysis
The data set was taken from the national database for autism research (NDAR) (http: //http://ndar.nih.gov/), an NIH-funded research data repository that aims at accelerating progress in autism spectrum disorders (ASD) research through data sharing, data harmonization, and the reporting of research results. A total of 416 MRI scans are selected for 253 normal children (126 males and 127 females) following standard protocol. Table 3 contains demographic information and distribution of scan availability.
Table 3.
Autism spectrum disorder data analysis: demographic information for participants.
| Visit | Number of subjects | Age: mean(std) (years) | Age: range (years) |
|---|---|---|---|
| 1 | 58 | 10.53 (5.96) | [0, 18] |
| 2 | 148 | 12.25 (4.62) | [0, 21] |
| 3 | 160 | 12.29 (5.14) | [1, 22] |
| 4 | 19 | 1.84 (1.42) | [1, 6] |
| 5 | 7 | 1.57 (0.79) | [1, 3] |
| 6 | 10 | 2.70 (0.67) | [2, 4] |
| 7 | 6 | 3.17 (0.75) | [2, 4] |
| 8 | 5 | 3.40 (1.14) | [2, 5] |
| 9 | 3 | 3.67 (1.15) | [3, 5] |
|
| |||
| Gender | Male/Female | 126/127 | |
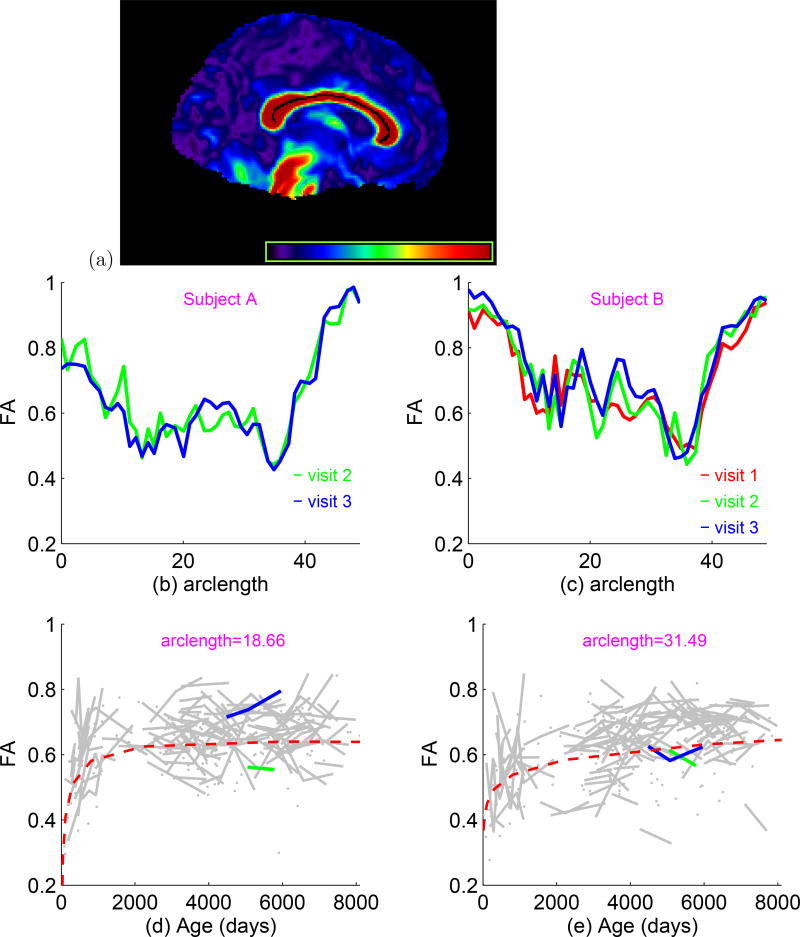
The diffusion tensor imaging (DTI) data were processed by two key steps including a weighted least squares estimation method (Basser et al., 1994) to construct the diffusion tensors and a pipeline for tract-based spatial statistics (TBSS) (Smith et al., 2006) to register DTIs from multiple subjects to create a mean image and a mean skeleton. Specifically, maps of fractional anisotropy (FA) were computed for all subjects from the DTI after Eddy current correction and automatic brain extraction using FMRIB software library. FA maps were then fed into the TBSS tool, which is also part of the FSL. In the TBSS analysis, the FA data for all subjects were aligned into a common space by a non-linear registration method and the mean FA images were created and thinned to obtain a mean FA skeleton, which represents the centers of all white matter tracts common to the group. Subsequently, each subject’s aligned FA data sets were projected onto this skeleton. While several DTI fiber tracts were tracked, we chose to focus in this paper on the corpus callosum (see Fig. 4.7 (a)) to illustrate the applicability of our method in assessing the effects of covariates of interest, such as patient age and gender. In this case, there are M = 45 grid points along each fiber tract. The FA values were extracted at each grid point across multiple times (1 to 9 times) along the selected fiber tracts for all 253 infants.
Figure 4.7.
Data analysis: (a) 3D visualization of the corpus callosum in the sagittal view, with the FA skeleton template overlaid on it. (b) and (c) FA’s along the corpus callosum obtained from 2 selected subjects A (b) and B (c) with 2 or 3 visits. Different visits for the same subjects are indicated by color. (d) and (e) FA values varying over age at selected locations: arclength=18.66 (d) and arclength=31.49 (e) along the corpus callosum for all 253 subjects, with green and blue lines corresponding to subjects A and B, respectively. Red dashed lines represent the fitted lines for the male group.
The goal of the data analysis is to delineate the development of skeleton diffusion properties across time. We fitted FMEM (2.1) and (2.2) with xi = (1, Gender, log(Age), {log(Age)}2)T and zi = (1, log(Age))T to the selected FA tracts obtained from all 253 subjects. The coefficient functions associated with log(Age) and {log(Age)}2 were included to detect age effect in FA changes. In addition, as shown in Fig. 4.7, there are random subject-to-subject variations in FA measures at each grid point along this tract as well as those in the age effect on FA measures. We included random intercept and age effects in the model in order to account for the inter-subject variations.
We applied FMEM, WFMM, and FAMM to this data set and estimated all unknown quantities but will only discuss the results based on FMEM below. The results for WFMM and FAMM are provided in the supplementary document. The computational times for FMEM, WFMM, and FAMM are, respectively, 55.8 seconds, 7.9 seconds, and 6.078 hours.
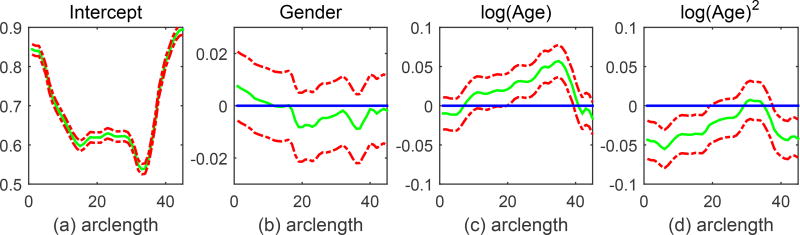
For FMEM, the estimated functional coefficients of β(s) and their 95% simultaneous confidence bands were constructed along with the global test statistic Sn to test for the significance of gender and age effects on FA values. The p-value of Sn was approximated using the resampling method with Q = 1, 000 replications. Figure 4.8 presents the estimated coefficient functions corresponding to intercept, gender, log(Age), and {log(Age)}2 along with their 95% simultaneous confidence bands. The intercept function describes the overall trend of FA along the corpus callosum. In general, the central regions of the corpus callosum show smaller FA values, whereas the peripheral regions show larger FA values. In Figure 4.8, the simultaneous confidence band contains the horizontal line crossing (0, 0) for the gender effect, whereas the horizontal line is out of the 95% simultaneous confidence band for the age effect, indicating a significant age effect. This agrees with our analysis results based on Sn for the gender and age effects. We obtained the p values of 0.215 and < 0.0001 for the gender and age effects, respectively, indicating significant age but no gender effect.
Figure 4.8.
95% simultaneous confidence bands for coefficient functions. The solid curves are the estimated coefficient functions and the dashed curves are the 95% simultaneous confidence bands. The thin horizontal line is the line crossing the origin (0, 0).
Table 4 displays the estimated eigenvalues and the percentage of total variability explained by different components in FMEM. It shows that 31.41% of the variability is explained by the first principal component for b and 18.22% by the first principal component for eG. Overall, the first 8 principal components for b explain 62.47% of the total variability, whereas the first 8 principal components for eG explain 32.18% of the total variability. This indicates that the random effects b capture most of the variation in the data. Within b, 53.57% and 8.90% of the total variation are explained by the random functional intercept and the subject-specific random slope, respectively. The within-curve measurement error explains only 5.35% of the total variation. Figure 4.9 shows the first five and four eigenfunctions for b and eG, respectively.
Table 4.
Autism spectrum disorder data analysis: Estimated eigenvalues and the percentage of the total variability explained by different components in the functional mixed effects model.
| k | (×10−2) | (%) | (%) | (×10−2) | (%) | σ2(%) |
|---|---|---|---|---|---|---|
| 1 | 7.96 | 31.41 | 0.71 | 4.51 | 18.22 | 5.35 |
| 2 | 3.08 | 9.34 | 3.08 | 1.31 | 5.28 | |
| 3 | 1.44 | 3.52 | 2.28 | 0.56 | 2.26 | |
| 4 | 1.15 | 3.53 | 1.09 | 0.43 | 1.72 | |
| 5 | 0.74 | 2.54 | 0.43 | 0.36 | 1.45 | |
| 6 | 0.59 | 1.45 | 0.93 | 0.34 | 1.38 | |
| 7 | 0.32 | 1.06 | 0.23 | 0.25 | 1.03 | |
| 8 | 0.22 | 0.74 | 0.15 | 0.21 | 0.85 | |
|
| ||||||
| 53.57 | 8.90 | 32.18 | 5.35 | |||
Figure 4.9.
(a) (b) The first five estimated eigenfunctions for the random intercept and slope processes. and correspond to the random functional intercept and random functional slope, respectively. (c) The first four estimated eigenfunctions for the visit specific deviation process.
Supplementary Material
Acknowledgments
The research of Dr. Zhu was supported by NSF grants SES-1357666 and DMS-1407655, NIH grant MH086633, a grant from the Cancer Prevention Research Institute of Texas, and the endowed Bao-Shan Jing Professorship in Diagnostic Imaging. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We would like to thank Drs. Morris and Herrick for helping with WFMM.
Appendix
Wild Bootstrap Method for Critical Values of Sn
We have shown that the asymptotic distribution of Sn is very complicated hence it is difficult to directly approximate the percentiles of Sn under the null hypothesis. Instead, we propose using a wild bootstrap method to obtain critical values of Sn. The wild bootstrap consists of the following three steps.
Step 1. Fit (2.1) and (2.2) under the null hypothesis H0, which yields β̂*(sm), and for all i, j and m = 1, …, M.
- Step 2. Generate a random sample and τij(sm)(q) from a N(0, 1) generator for all i, j and m = 1, …, M and then construct
Then, based on ŷij(sm)(q), we recalculate β̂(s)(q), and d(s)(q) = Cβ̂(s)(q) − β0(s). Subsequently, we compute Step 3. Repeat Step 2 Q times to obtain and then calculate . If p is smaller than a pre-specified significance level α, say 0.05, then one rejects the null hypothesis H0.
Wild Bootstrap Methods for Simultaneous Confidence Bands of β(·)
Although there are several methods of determining Cl(α) including random field theory (Worsley et al., 2004), we develop an efficient resampling method to approximate Cl(α) as follows (Kosorok, 2003).
We calculate for all i, j, and m.
- For q = 1, …, Q, we independently simulate from N(0, 1) and calculate a stochastic process G(s)(q) given by
We calculate sups∈[0,1] |elG(s)(q)| for all q, where el is a px × 1 vector with the l-th element 1 and 0 otherwise, and use their 1 − α empirical percentile to estimate Cl(α).
Footnotes
Supplementary materials available in the attached file include the proofs of Lemmas 1–13, Theorems 1–3, and Corollary 1.
Bibliography
- Basser PJ, Mattiello J, LeBihan D. Estimation of the effective self- diffusion tensor from the NMR spin echo. Journal of Magnetic Resonance Ser. B. 1994;103:247–254. doi: 10.1006/jmrb.1994.1037. [DOI] [PubMed] [Google Scholar]
- Cao G, Yang L, Todem D. Simultaneous inference for the mean function based on dense functional data. Journal of Nonparametric Statistics. 2012;24:359–377. doi: 10.1080/10485252.2011.638071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederbaum J, Pouplier M, Hoole P, Greven S. Functional linear mixed models for irregularly or sparsely sampled data. Statistical Modelling. 2016;16:67–88. [Google Scholar]
- Chen K, Müller H-G. Modeling repeated functional observations. Journal of the American Statistical Association. 2012;107:1599–1609. [Google Scholar]
- Di C, Crainiceanu CM, Caffo BS, Punjabi NM. Multilevel functional principal component analysis. Annals of Applied Statistics. 2009;3:458–488. doi: 10.1214/08-AOAS206SUPP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di C, Crainiceanu CM, Jank W. Multilevel sparse functional principal component analysis. Stat. 2014;3:126–143. doi: 10.1002/sta4.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle P, Heagerty P, Liang KY, Zeger S. Analysis of Longitudinal Data. 2. New York: Oxford University Press; 2002. [Google Scholar]
- Evans AC, Group BDC. The NIH MRI Study of Normal Brain Development. NeuroImage. 2006;30:184–202. doi: 10.1016/j.neuroimage.2005.09.068. [DOI] [PubMed] [Google Scholar]
- Fan J, Gijbels I. Local Polynomial Modelling and Its Applications. London: Chapman and Hall; 1996. [Google Scholar]
- Fan J, Zhang W. Statistical methods with varying coefficient models. Statistics and its Interface. 2008;1:179–195. doi: 10.4310/sii.2008.v1.n1.a15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzmaurice GM, Laird NM, Ware JH. Applied Longitudinal Analysis. New York: Wiley; 2004. [Google Scholar]
- Greven S, Crainiceanu S, Caffo BS, Reich D. Longitudinal functional principal component analysis. Electron. J. Statist. 2010;4:1022–1054. doi: 10.1214/10-EJS575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W. Functional mixed effects models. Biometrics. 2002;58:121–128. doi: 10.1111/j.0006-341x.2002.00121.x. [DOI] [PubMed] [Google Scholar]
- Horvath L, Kokoszka P. Inference for Functional Data with Applications. New York, N. Y: Springer; 2012. [Google Scholar]
- Kosorok MR. Bootstraps of sums of independent but not identically distributed stochastic processes. J. Multivariate Anal. 2003;84:299–318. [Google Scholar]
- Li Y, Hsing T. Uniform convergence rates for nonparametric regression and principal component analysis in functional/longitudinal data. The Annals of Statistics. 2010;38:3321–3351. [Google Scholar]
- Lin X, Carroll RJ. Semiparametric regression for clustered data using generalized estimating equations. Journal of the American Statistical Association. 2001;96:1045–1056. [Google Scholar]
- Meyer MJ, Coull BA, Versace F, Cinciripini P, Morris JS. Bayesian function-on-function regression for multilevel functional data. Biometrics. 2015;71:563–574. doi: 10.1111/biom.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JS, Carroll RJ. Wavelet-based functional mixed models. J. R. Stat. Soc. Ser. B Stat. Methodol. 2006;68:179–199. doi: 10.1111/j.1467-9868.2006.00539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L. Ways toward an early diagnosis in Alzheimer's disease: The Alzheimer's Disease Neuroimaging Initiative (ADNI) Alzheimer's & Dementia. 2005;1:55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay JO, Silverman BW. Functional Data Analysis. 2 Springer-Verlag; New York: 2005. [Google Scholar]
- Reiss PT, Huang L, Chen H, Colcombe S. Varying-smoother models for functional responses. arXiv preprint arXiv:1412.0778 2014 [Google Scholar]
- Scheipl F, Staicu A, Greven S. Additive mixed models for correlated functional data. Journal of Computational and Graphic Statistics. 2015;24:477–501. doi: 10.1080/10618600.2014.901914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seber GAF, Wild CJ. Nonlinear Regression. New York, N.Y: John Wiley & Sons; 1989. [Google Scholar]
- Shi JQ, Choi T. Gaussian Process Regression Analysis for Functional Data. Chapman & Hall/CRC; 2011. [Google Scholar]
- Smith SM, Jenkinson M, Johansen-Berg H, Rueckert D, Nichols TE, Mackay CE, Watkins KE, Ciccarelli O, Cader M, Matthews P, Behrens TE. Tractbased spatial statistics: voxelwise analysis of multi-subject diffusion data. NeuroImage. 2006;31:1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Staicu AM, Lahiri S, Carroll RJ. Significance tests for functional data with complex dependence structure. Journal of Statistical Planning and Inference. 2015;156:1–13. doi: 10.1016/j.jspi.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wand MP, Jones MC. Kernel Smoothing. London: Chapman and Hall; 1995. [Google Scholar]
- Worsley KJ, Taylor JE, Tomaiuolo F, Lerch J. Unified univariate and multivariate random field theory. NeuroImage. 2004;23:189–195. doi: 10.1016/j.neuroimage.2004.07.026. [DOI] [PubMed] [Google Scholar]
- Wu H, Zhang J. Local polynomial mixed-effects models for longitudinal data. Journal of the American Statistical Association. 2002;97:883–889. [Google Scholar]
- Wu H, Zhang J. Nonparametric Regression Methods for Longitudinal Data Analysis. Hoboken, New Jersey: John Wiley & Sons, Inc; 2006. [Google Scholar]
- Xiao L, Zipunnikov V, Ruppert D, Crainiceanu C. Fast covariance estimation for high-dimensional functional data. Stat. Computing. 2016;26:409–421. doi: 10.1007/s11222-014-9485-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao F, Müller H-G, Wang J-L. Functional data analysis for sparse longitudinal data. J. Amer. Statist. Assoc. 2005;100:577–590. [Google Scholar]
- Yuan Y, Gilmore JH, Geng X, Styner M, Chen K, Wang JL, Zhu H. FMEM: Functional mixed effects modeling for the analysis of longitudinal white matter tract data. NeuroImage. 2014;84:753–764. doi: 10.1016/j.neuroimage.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Statistical inferences for linear models with functional responses. Statistica Sinica. 2011;21:1431–1451. [Google Scholar]
- Zhang J, Chen J. Statistical inference for functional data. The Annals of Statistics. 2007;35:1052–1079. [Google Scholar]
- Zhou L, Huang JZ, Martinez JG, Maity A, Baladandayuthapani V, Carroll RJ. Reduced rank mixed effects models for spatially correlated hierarchical functional data. Journal of American Statistical Association. 2010;105:390–400. doi: 10.1198/jasa.2010.tm08737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Brown P, Morris J. Robust, adaptive functional regression in functional mixed model framework. Journal of the American Statistical Asssociation. 2011;106:1167–1179. doi: 10.1198/jasa.2011.tm10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu HT, Li R, Kong L. Multivariate varying coefficient model for functional responses. Annals of Statistics. 2012;40:2634–2666. doi: 10.1214/12-AOS1045SUPP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipunnikov V, Greven S, Shou H, Caffo B, Reich DS, Crainiceanu C. Longitudinal high-dimensional principal components analysis with application to diffusion tensor imaging of multiple sclerosis. Annals of Applied Statistics. 2014;8:2175–2202. doi: 10.1214/14-aoas748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.