Abstract
The ubiquitin-proteasome offers novel targets for potential therapies with their specific activities and tissue localization. Recently, the expansion of our understanding of how ubiquitin ligases (E3s) specifically regulate transcription has demonstrated their roles in skeletal muscle, complementing their roles in protein quality control and protein degradation. This review focuses on skeletal muscle E3s that regulate transcription factors critical to myogenesis and the maintenance of skeletal muscle wasting diseases.
Keywords: ubiquitin ligase, transcription factors, skeletal muscle, differentiation, atrophy
Introduction
Skeletal muscle homeostasis is not only essential for day-to-day activities but also the healthy functioning of the body. Alterations in skeletal muscle strength and function are concomitant with several other disease pathophysiologies, including diabetes, COPD, Duchenne muscular dystrophy, and motor neuron diseases (54). The reduction in muscle mass, recognized commonly as muscle wasting or atrophy, results in loss of muscle function and is associated with increased morbidity and mortality (9). It occurs due to various physiological and pathological consequences. One of the most prevalent physiological causes of atrophy is aging and is termed sarcopenia (60), which involves several molecular, functional, and histological changes in the muscle leading to frailty and functional decline in older adults. The pathological causes of atrophy include cancer cachexia, burns, chronic heart failure, chronic kidney disease, AIDS, mechanical ventilation, chronic obstructive pulmonary disorder (COPD), sepsis, immune disorders, dystrophies, etc., which all contribute to muscle atrophy in unique ways (12).
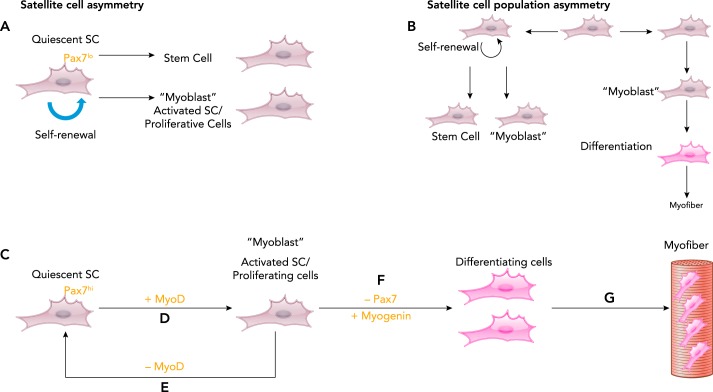
The regenerative capabilities of skeletal muscle in case of injury or damage is remarkable and is enabled by the presence of tissue-specific adult stem cells, termed satellite cells, which are located between the basal lamina and the sarcolemma of the muscle fibers (FIGURE 1) (27). Inherently quiescent, satellite cells get activated by a stimulus in response to injury, stretching, exercise, and denervation, among other pathological states such as myositis, muscular dystrophy, genetic mutations, and cancer cachexia, for example (10, 31, 43, 59). Quiescent satellite cells have a number of fates, including division to renew themselves and asymmetric division to both renew themselves and produce cells capable of differentiating (FIGURE 1A). Within the broader population of satellite cells, SCs can both divide asymmetrically and exist in asymmetric populations, some of which only differentiate into myocytes and do not self-renew (FIGURE 1B). In this review, we focus on these quiescent SC cells, which are Pax7 expressing (FIGURE 1, C–G). Quiescent SCs express the transcription factor paired box 7 (PAX7) (FIGURE 1C), whereas activated SCs coexpress PAX7 and muscle regulatory transcription factors MyoD and Myf5, and drive the proliferation of these cells (termed myoblasts) (FIGURE 1D) (68). On the other hand, the activated SCs can also go back to quiescence state by losing MyoD to maintain the progenitor pool (FIGURE 1E). The myoblasts, after a few rounds of proliferation, start to differentiate by losing PAX7 and expressing muscle regulatory transcription factor myogenin (FIGURE 1F). These mature myoblasts fuse to form the differentiated regenerated myofibers (FIGURE 1G) (60). These myofibers also express transcription factors like p53 and MyoD, whose specific function is unknown and may be related to the maintenance of neuromuscular junction support (35, 67).
FIGURE 1.
Stem cell renewal and the molecular regulation of muscle cell differentiation (myogenesis)
Skeletal muscle satellite (stem) cells can divide asymmetrically into differentiated cells and renew (A) with different stem cell populations asymmetrically dividing, with some satellite cells dividing asymmetrically, whereas others just differentiative (B). In generally, satellite cell levels of PAX7 are low in the asymmetrically dividing cells (A) and higher in the cells that differentiate (C). With myogenic stimuli (e.g., contractile activity, direct/indirect myocyte damage), PAX7-expressing quiescent cells (C) have increased expression of muscle regulatory gene MYOD (D), thus activating the cells and leading to proliferation (D). At this stage, the cells can return to the quiescent state by losing MYOD (E) or can differentiate (F) by losing PAX7, which increases expression of myogenin. G: the terminally differentiated cells fuse to form myofibers.
Muscle atrophy is the balance between protein synthesis and protein degradation, both of which regulate protein turnover (9). Autophagy-lysosome and ubiquitin-proteasome systems are the two major cell proteolytic systems that regulate the enhanced protein degradation in myocyte atrophy. Although the ubiquitin proteasome system (UPS) is primarily attributed to the protein degradation of sarcomere proteins in atrophy, its role in regulating transcription factors governing transcription is beginning to be appreciated in muscle physiology and pathophysiology. How the ubiquitin proteasome system regulates transcription is well established in embryonic stem cells (14), the tumor microenvironment (16), and the heart (13, 62). The ways in which the UPS posttranslationally modifies and regulates transcription by directing transcription factors by ubiquitination is emerging through multiple mechanisms. They include the degradation of transcription factors by enhancing poly-ubiquitination, leading to proteasome-dependent degradation. Other mechanisms include ubiquitin playing a role in directing the localization of transcription factors (in the case of mono-ubiquitination). Some of them regulate the transcription-factor part of the signaling pathways involved in the maintenance and regeneration of skeletal muscle, including those involved in skeletal muscle atrophy. This review focuses on the role of the skeletal muscle ubiquitin ligases, the enzymes giving the UPS specificity, in regulating transcription in skeletal muscle so that their broadening diverse roles are better appreciated and provide context to thinking about therapeutically targeting them in the future.
Ubiquitin Ligases and Their Degradation Mechanisms
The posttranslational modification of proteins via ubiquitination is mediated by three classes of proteins in a multistep process. Conjugation of proteins to ubiquitin (Ub) begins with an ATP-dependent step wherein there is a thioester linkage between the last residue of Ub, i.e., Gly 76, and Cys residue of the Ubiquitin-activating enzyme, E1. The activated Ub is then transferred to the Cys residue of the ubiquitin-conjugating enzyme (E2). The final step is the concurrent interaction of the Ub-loaded E2 and ubiquitin protein ligases (E3) resulting in the formation of Ub chains on the substrate. When E3s polyubiquitinate their substrate with Lys48-linked ubiquitin, the 26S proteasome binds the ubiquitin chains and degrades the substrate. Thus E3s are critical parts of this process, since they not only are responsible for substrate specificity but also regulate the efficiency of the process. Ubiquitination of proteins not only targets substrates for degradation but also controls their activity and localization, depending on the type of Ub chains added. On the other hand, there are also deubiquitinating enzymes (DUBs), which catalyze the removal of ubiquitin, cellular localization (e.g., mono-ubiquitin), or stability (e.g., non-lysine48 linked poly-ubiquitin-mediated modifications), and also balance the posttranslational modification of substrates, thus altering their susceptibility to proteasome-dependent degradation (3).
In the pathogenesis of skeletal muscle atrophy, satellite cells play an integral role in recovery and regeneration of muscle cells. One example highlighting the importance of the UPS system and the ubiquitin-mediated regulation of transcriptions factors has been described in the regulation of satellite cells in skeletal muscle atrophy. The RPT3 (regulatory protein of the 26S proteasome) protein is a putative ATPase integral to the 26S proteasome (18). Recent studies have identified that depleting RPT3 induced skeletal muscle atrophy and the loss of satellite cells in resting skeletal muscle (37). Mice with muscle-specific RPT3 depletion spontaneously develop skeletal muscle atrophy starting from 3 wk of age and predominant atrophy at 4 wk of age, evidenced by a decrease in myofiber cross-sectional area in gastrocnemius and soleus muscle (37). The depletion of RPT3 in mice led to decreased proteasome activity, specifically chymotrypsin-like and trypsin-like proteasome activity in the fast-twitch-dominant tibialis anterior muscle at 2 wk of age (37). Satellite cell-specific Rpt3 conditional knockout (Rpt3-scKO) mice were generated to deplete Rpt3 specifically in the satellite-cell population (36). These mice did not have a significant phenotype, as seen by no change in muscle weight or cross-sectional area in the tibialis anterior muscle upon induced genetic inactivation of Rpt3 in these mice (36). Nevertheless, these mice had impaired regeneration due to a defective proliferative stage and apoptosis in primary myoblasts isolated from these mice (36). This satellite cell-specific proliferation defect could be rectified on P53 depletion in primary myoblasts isolated from these mice (36).
The underlying mechanism of RPT3’s regulation of skeletal muscle atrophy pathogenesis was identified to be its regulation of the transcription p53, whereby RPT3-mediated p53 degradation through its ATPase activity is critical to proteasome activity (36). Depletion of RPT3 in satellite cells led to enhanced p53 expression (36), which then mediated defects in the satellite cell proliferative state and led to apoptosis (36) instead of the needed regeneration to counteract the atrophy (36). This example illustrates the critical role of the 26S proteasome’s degradatory functions and highlights the importance of upstream regulators of ubiquitination, specifically the ubiquitin ligases that give the process specificity. Ubiquitylation and degradation of many transcriptional activators at promoter sites (53) is one of the regulators of transcription among other regulators like activators, repressive complexes, and histones. This review expands this basic appreciation of the importance of the proteasome-mediated degradation of transcription factors (p53) in satellite cells in skeletal muscle atrophy and investigates how ubiquitin ligases specific for transcription factors regulate the body’s resistance to skeletal muscle atrophy.
TATA-Box Binding Protein and TATA-Box Binding Protein-Associated Factors
The TATA-box binding protein (TBP) and TATA-box binding protein-associated factors (TAF) form the RNA polymerase II preinitiation complex (PIC) that recognizes the transcription factor IID (TFIID) binding to the core promoter of the gene being transcribed. The transcriptional activity of MYOD in activated SCs in terms of inducing myogenin is essential for terminal differentiation. This mechanism involved functional interaction with the TFIID complex. First, the binding of MYOD to its recognition site is stabilized by TFIID complex/TBP. Eventually, MYOD mediates and stabilizes the association of TFIIB to the PIC (32). Thus TBP levels are vital in regulating the transcriptional activity of MYOD.
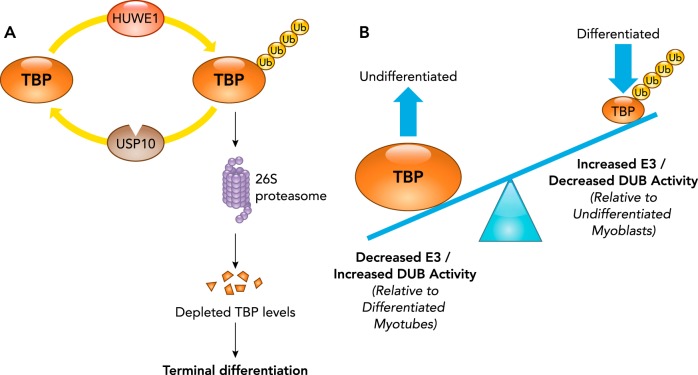
The TATA-box binding protein (TBP) protein is regulated posttranslationally by ubiquitination in recent studies. The ubiquitin ligase HUWE1 and deubiquitinase (DUB) USP10 coordinate to regulate TBP levels (FIGURE 2A) (41). Both HUWE1 (HECT, UBA, and WWE domain containing 1), a HECT ubiquitin ligase, and the USP10 DUB are found in proliferating undifferentiated myoblasts (41). HUWE1 was found to polyubiquitinate TBP and to drive its proteasome-dependent degradation (FIGURE 2A). In undifferentiated myoblasts, the USP10, a ubiquitin-specific protease, prevents proteasomal degradation of TBP by counteracting HUWE1 ubiquitination of TBP, thus maintaining stable TBP levels (FIGURE 2A) (41). The net result of this ubiquitin ligase/DUB pairing is the regulation of TBP levels during differentiation (FIGURE 2B). During differentiation, HUWE1 expression increases while USP10 expression decreases, favoring the polyubiquitination and subsequent degradation of TBP. The resulting myocyte depletion of TBP during differentiation by posttranslational degradation via the proteasome is an essential mechanism for differentiation (FIGURE 2B). In contrast, USP10 depletion was not sufficient to mediate differentiation (41). The importance of reduced TBP levels in differentiation has been reported to be related to TBP’s regulation of MyoD (45). By regulating the TBP levels through posttranslational ubiquitination and degradation, the ubiquitin ligase HUWE1 and deubiquitinase USP10 play a cohesive role in maintaining TBP levels during myogenesis (41). The HUWE ubiquitin ligase also has been reported to directly regulate MYOD protein level via mono-ubiquitination and proteasome-dependent degradation (50), which may parallel its indirection regulation of MYOD described above (41). Since the role of TBP in muscle regeneration has been established in previous studies, in-depth understanding of how TBP levels are modulated would be important to have a clear understanding of various pathways in muscle regeneration.
FIGURE 2.
Regulation of TATA-box binding protein levels by the ubiquitin proteasome system
A: the HUWE1 ubiquitin ligase polyubiquitinates TATA-box binding protein (TBP) 4 and targets it for recognition by the 26S proteasome, which degrades the protein. The deubiquitinase (DUB) ubiquitin proteasome (USP) 10 removes TBP polyubiquitin chains to counteract this targeted degradation. B: in proliferating myoblasts, HUWE1 ubiquitin ligase activity (polyubiquitination) is counteracted by USP10 DUB removing TBP polyubiquitin chains, resulting in the stabilization of TBP levels. C2C12 myoblasts induced to undergo differentiation have reduced TBP levels, resulting from an increase in HUWE1 levels and a decrease in USP10, resulting in the enhanced ubiquitin-mediated proteasome degradation of TBP.
Neural Precursor Cell-Expressed Developmentally Downregulated Gene 4
NEDD4 (neural precursor cell-expressed developmentally downregulated gene 4) has a role in cellular processes involving turnover of membrane-bound channels and receptors associated with protein trafficking, endocytosis, virus budding, and transcription. NEDD4, along with its family of proteins, has a phospholipid-binding C2 domain, two to four WW domains that recognize substrates, and a catalytic domain homologous to the E6-AP COOH-terminal (HECT) domain (69). NEDD4 associates with cellular membranes via its C2 lipid-binding domain, allowing for the interaction between the WW domains of NEDD4 and PPXY motifs of its membrane-associated protein substrates (38). WWP2 was shown to interact with and monoubiquitinate the membrane-anchored fragment of NOTCH3, leading to its degradation, possibly via the endosomal/lysosomal pathway. As a result, NOTCH signaling activity was reduced (73). NOTCH is a transmembrane receptor protein that is sequentially cleaved upon ligand binding, releasing its “activated” intracellular domain, which translocates from the cytosol to the nucleus to influence transcription.
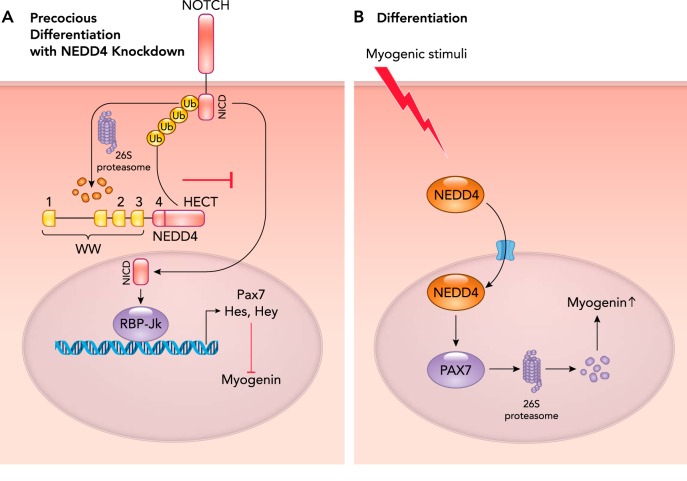
The ubiquitin ligase NEDD4 has recently been implicated in mediating denervation-induced atrophy, with NEDD4 depletion leading to muscle mass retention (FIGURE 3A) (49). Activation of NOTCH by its ligands leads to the release and translocation of its intracellular domain (NICD) to the nucleus, where it interacts with RBP-Jκ (recombination signal binding protein for immunoglobulin kappa J region), a DNA binding protein that activates transcription of Hes and Hey, which inhibit MyoD gene transcription (FIGURE 3A) (61). The NICD protein directly regulates PAX7 expression through RBP-Jκ to mediate self-renewal rather than differentiation. NEDD4 regulates the expression of the transcription factor PAX7, which in turn is responsible for myogenin induction (FIGURE 3A) (52, 68). Evidence for the regulation of Notch by NEDD4 comes from studies of unloading and denervation of the rat hindlimb, resulting in a significant increase in NEDD4 expression in the soleus, plantaris, and gastrocnemius in rats (38). Simultaneous with this increased NEDD4 expression, NOTCH1 expression decreased, suggesting that NOTCH1 is a NEDD4 substrate that is degraded by the UPS (38). To demonstrate this relationship, the expression of a dominant negative NEDD4 in soleus muscles was tested in vivo and found to completely reverse the unloading-induced decrease in NOTCH1 expression (38). When NEDD4 was conditionally increased in C2C12 cells, an increase in NOTCH1 ubiquitination was observed, indicating that NOTCH1 was a NEDD4 substrate in skeletal muscle atrophy (38).
FIGURE 3.
NEDD4-mediated transcriptional regulation in myogenesis
A: during the proliferation phase of myogenesis, NEDD4 regulates myogenesis by its effects on NOTCH signaling. NEDD4 polyubiquitinates NCID (NOTCH), preventing its nuclear translocation and activation of the RBP-Jκ (recombination signal binding protein for immunoglobulin kappa J region) transcription factor. By inhibiting RBP-Jκ, NEDD4 prevents the transcription of proteins critical to myocyte differentiation. Recent studies have demonstrated that increased NEDD4 expression leads to precocious myocyte differentiation by inhibiting Hes and Hey, allowing enhanced myogenin expression/activity. B: myogenic stimuli enhance NEDD4 translocation to the nucleus, where it ubiquitinates PAX7, targeting it for 26S-dependent proteasome degradation. Since PAX7 inhibits myogenin (A), increased myogenin and myocyte differentiation results from enhanced nuclear NEDD4 activity.
The mechanism by which PAX7 through NOTCH1 regulates skeletal muscle atrophy may be related to its regulation of muscle-derived stem cells (48). For example, NOTCH-activating factors from osteosarcoma cells repress myogenesis, thereby inducing atrophy (48). Thus NEDD4 mediated reduction in NOTCH signaling, leading to the precocious differentiation and depletion of SC reservoir; thus aggravating atrophy may be one mechanism to consider. PAX7 regulates skeletal muscle atrophy by regulating the regenerating mechanisms, as can be seen by cancer cachexia caused due to impaired regulation of PAX7 levels (63).
Subsequent studies reported that NEDD4 modulates PAX7 during early muscle differentiation by depleting PAX7 levels by subjecting it to ubiquitination; conversely, NEDD4 loss results in the accumulation of PAX7 (FIGURE 3B) (15). Transient nuclear accumulation of NEDD4 induced decrease in PAX7 and precocious muscle differentiation, demonstrating that NEDD4 functions as a novel PAX7 regulator temporally and spatially controlled to modulate PAX7 protein levels and satellite cell fate (15). PAX7 levels were found to be regulated by the proteasome system in cells initiating differentiation but not cells in the proliferating stage. Blocking the proteasome was found to increase the levels of PAX7 in differentiating myoblasts, consistent with NEDD4-mediated degradation of PAX7 (15).
Unexpectedly, the COOH-terminus domain of PAX7 in differentiating skeletal muscle cells was found to be mono-ubiquitinated by NEDD4 (15), which is not generally a signal for the proteasome-mediated degradation observed in these studies, since poly-ubiquitin chains are classically required to be recognized by the proteasome ubiquitin receptor subunits (20, 33). Interestingly, mono-ubiquitination is not an uncommon signal for degradation and has been demonstrated in other Pax family members, including PAX3 (11). Preventing the proteasome-dependent degradation of PAX3 in satellite cells blocks myogenic progression due to the impaired control of PAX levels (40). For myogenic progression to happen after satellite cell activation, PAX3 protein is depleted, whereas there is no change in PAX7 protein levels. The mechanism by which PAX3 is depleted during satellite cell activation has been reported to be through “mono-ubiquitination,” which facilitates subsequent poly-ubiquitination and degradation by the 26S proteasome (11). Although PAX7 protein levels do not change during this early stage of myogenesis, PAX7 degradation is a critical component in later stages of the myogenic process (i.e., differentiation) (26).
In addition to the proteasome-dependent mechanism, caspase-3-mediated cleavage also contributes to reduced PAX7 levels (29). Counteracting the proteasome-dependent PAX7 degradation is the casein kinase 2 (Ck-2) phosphorylation of PAX7, which prevents degradation. In addition to this, it was also found that Pax7 phosphorylation directed by CK-2 reduces caspase-mediated cleavage of PAX7 (21). When casein kinase 2 (Ck-2) phosphorylates PAX7 at S201, PAX7 degradation by the proteasome system and caspases is inhibited, thus stabilizing PAX7 levels (29), which is prominently seen in proliferating cells. The PAX7 protein is also regulated by the addition of small ubiquitin-like modifiers (SUMO). SUMOylation is a post-translational modification process paralleling ubiquitination involving the conjugation of SUMO instead of ubiquitin. SUMOlyation of PAX7 is also essential in myogenic differentiation as it mediates transactivation of PAX7 regulated genes before differentiation, as shown in C2C12 cells (44). Having a comprehensive understanding of various regulators of PAX7, including its modulation by the UPS system, should help in developing a holistic understanding of the regulators of PAX7.
Thyroid Hormone Receptor Interactor 1
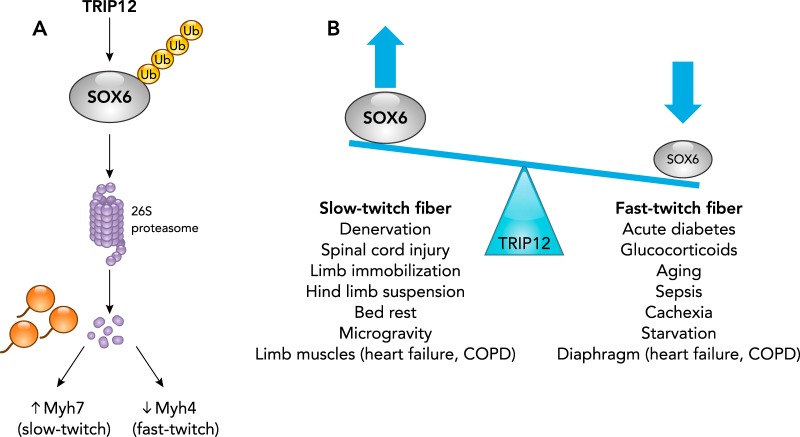
TRIP12 (thyroid hormone receptor interactor 1), a HECT domain encoding ubiquitin ligase, regulates the fiber-type switching in adult skeletal muscle myocytes (4). Adult skeletal muscles are composed of short-twitch fibers and fast-twitch fibers, based on various external and internal cues, and the composition varies in healthy and diseased states (55). At a molecular level, slow-twitch fibers express myosin heavy chain (MHC) isoform MHC-Iβ, Myh7, and use oxidative metabolism, whereas the fast-twitch fibers include those expressing MHC-IIA, MHC-IIX/D, and MHC-IIB, Myh4, which use glycolytic or mixed metabolism (51). The contraction speed of fast-twitch fibers is higher than that of slow-twitch fibers, and they also differ in their sensitivity to calcium and fatigue resistance, among other characteristics (55). Muscle atrophy caused by distinct pathophysiological conditions has a distinct effect on the composition of either slow- or fast-twitch fibers, based on the signaling mechanism by which the atrophic condition is induced. Muscle wasting induced by sepsis, cancer cachexia, AIDS, aging, glucocorticoid administration, starvation, acute diabetes in the diaphragm, heart failure, and COPD is characterized by the presence of fast-twitch fibers and fast-to-slow fiber-type shift (17). On the other hand, muscle wasting caused by hand denervation, spinal cord injury, limb immobilization, hindlimb suspension in a shortened position, spinal cord injury, bed rest, and microgravity, and in limb muscles during heart failure and COPD are characterized by a shift from slow- to fast-fiber types. The TRIP12 ubiquitin ligase regulates fiber-type switching by regulating the expression of the SOX6 transcription factor (FIGURE 4), which is critical in enhancing the transcription of fast-fiber genes such as Myh5 and suppressing the transcription of slow-fiber genes such as Myh7 and myogenin.
FIGURE 4.
The TRIP12 ubiquitin ligase regulates muscle fiber-type switching through its regulation of the SOX6 transcription factor
A: TRIP12 polyubiquitinates SOX6, which enhances SOX6 degradation via the 26S proteasome. By blocking SOX6 activity, enhanced expression of Myh7 (enhancing the slow-twitch muscle phenotype) and concurrent decrease in Myh5 expression (decreasing the fast-twitch muscle phenotype) results. In contrast, deletion of TRIP12 (leading to enhanced SOX6 expression) enhances a fast-twitch muscle phenotype. B: muscle fiber-type switches are typical in skeletal muscle atrophy. The significance of TRIP12’s regulation of SOX6 stems from its potential role in skeletal muscle atrophies characterized by slow-twitch fibers (left) or fast-twitch fibers (right).
The TRIP12 HECT domain is the SOX6 substrate recognition module and catalytic domain, which mediates SOX6 polyubiquitination, resulting in SOX6 targeting to the 26S proteasome for degradation (FIGURE 4A) (4). In C2C12 myotubes, TRIP12 or proteasome inhibition was found to increase SOX6 protein levels (4). Control of SOX6 by TRIP12 ubiquitination resulted in significant changes in the resulting myocyte phenotype. TRIP12 knockdown in C2C12 myotubes resulted in SOX6 upregulation, a decrease in slow fiber-specific Myh7 and myogenin expression, and an upregulation of the fast-fiber Myh4 (4). Together, these studies demonstrate that the TRIP12 ubiquitin ligase regulates the SOX6 protein level, which itself controls the fiber-type gene expression and phenotype critical to the alterations seen in different types of skeletal muscle atrophy (FIGURE 4B). In this way, Trip-12-mediated fiber-type switching could potentially alter the workload capacity and function in skeletal muscle atrophy, although this hypothesis has not directly been tested to date (4). The SOX6 protein is also regulated by the addition of small ubiquitin-like modifiers (SUMO). Recent studies have reported that SOX6 is modified in vivo by SUMO on two distinct lysine residues in a process dependent on the E2-conjugating UBC9 enzyme, involving the addition of the SUMO2 isoform, resulting in the enhancement of SOX6 activity (23). The significance of SOX6 SUMOlyation in skeletal muscle has not been tested directly to date but may offer complementary regulatory mechanisms to TRIP12 since both SUMO and Ubiquitin bind lysine residues and have been reported to compete in other biological systems.
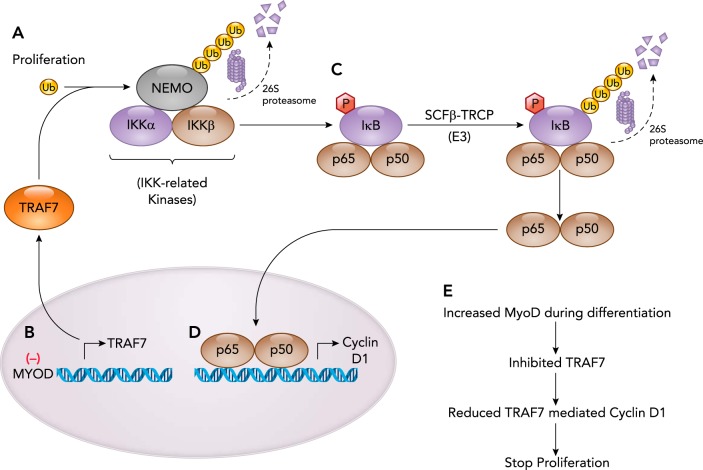
TNF Receptor-Associated Factor 7
Tumor necrosis factor (TNF)-associated factor (TRAF) proteins are cytosolic protein functioning as signal transduction pathways for receptors. TRAF7 is a member of the TRAFs. TRAF7 activates several signaling pathways and mediates cellular processes like proliferation, differentiation, and apoptosis. TRAF7 is unique among TRAFs as the only member to physically associate with IκB kinase/NF-κB essential modulator (NEMO) and the p65 (RelA/p65) member of the NF-κB transcription factor family (FIGURE 5) (72). TRAF7 is composed of an NH2-terminal RING domain with an adjacent zinc-domain and seven WD40 repeats at the carboxy terminal (71). Studies of TRAF7 in cancer cells identified that the TRAF7 ubiquitin ligase poly-ubiquitinates NEMO and p65 with lysine 29-linked chains, which promote their degradation (FIGURE 5A) (72). In addition to inhibiting NF-κB signaling, TRAF7 appears to promote AP-1 transcriptional activity (72) and the C/EBP-homologous protein (CHOP) transcriptional activity (64). Although considerable information about TRAF7’s regulation of NF-κB in cancer cells is known, its role in myocyte differentiation suggests that TRAF7’s role may go beyond NF-κB signaling.
FIGURE 5.
Proliferating myocytes exit the cell cycle during differentiation that is dependent on the TRAF7 ubiquitin ligase involved in downregulating Cyclin D1 expression/ activity
A: TRAF7 ubiquitin ligase polyubiquitinates NEMO and targets it for 26S degradation, when the NEMO component of the inhibitor of kB kinase (IKK) complex is degraded. B: in the studies described in the body of the text, the MYOD transcription factor depletes TRAF7 transcription and protein expression, leading to inactivation of the NF-κB activity and reduced cyclin D1, leading to exit from cell cycle and thereby driving the cells from proliferation to differentiation stage. C: the IKK complex is released from NEMO and can then phosphorylate the IκB protein (inhibitory kappa B protein) complexed with the p65 and p50 NF-κB subunits. This results in degradation of the IκB protein, allowing the p65/p50 NF-κB transcription factor to translocate to the nucleus (D) and regulate gene transcription, including Cyclin D1 critical to cell cycle and proliferation of cells. E: summary of TRAF7 activity in myocyte differentiation.
TRAF7 regulates myoblast differentiation through its activity as a ubiquitin ligase. The RING domain of TRAF7 promotes Lys-29-linked mono-ubiquitination of NEMO (IKKγ) (FIGURE 5C) (58). This leads to activation of the NF-κB signaling, which transcriptionally regulates cyclin D1 and mediates myoblast proliferation (FIGURE 5D) (30). MYOD1 is one of at least four transcription factors that regulate myocyte differentiation, in addition to MYF5, myogenin, and MRF4 (56). Chromatin IP (ChIP) studies in C2C12 myoblasts identified that MYOD directly regulates the expression of ubiquitin ligase, including Traf7 (58). When MYOD is depleted, a reduction of Traf7 gene expression results, implicating MYOD regulation of the NF-κB pathway via Traf7. In contrast, when TRAF7 is reduced in C2C12 myoblasts, significantly reduced NF-κB transcriptional activity is observed in luciferase reporter assays, consistent with TRAF7’s support of NF-κB activity by regulating the p65 subunit (58). Depletion of TRAF7 reduces p65 nuclear translocation, thereby diminishing NF-κB-mediated cyclin D1 transcription, leading to premature differentiation (FIGURE 5E) (58). However, the NF-κB-dependent cyclin D1 activation was reduced in the later stages of myogenesis, wherein the cells moved from a proliferating state to a differentiating state (47).
Up-Frameshift Suppressor 1 Homolog
Up-frameshift suppressor 1 homolog (UPF1) is an RNA helicase initially found to be a key component in the nonsense-mediated RNA decay (NMD) pathway and later reported to have a RING-like domain of UPF1, which has ubiquitin ligase activity (36). Interestingly, the UPF1 ubiquitin ligase activity was found to regulate myogenesis by regulating the MYOD transcription factor (22). UPF1 regulates MYOD levels by degrading mRNA encoding MYOD and also directly by ubiquitinating the MYOD protein and targeting it for proteasome-dependent degradation (22). This was confirmed by studies wherein depleting RING domain of MYOD did not reduce MYOD protein (22). Blocking UPF enhanced myogenesis in myoblasts due to sustained MYOD levels (22). UPF1 thus was found to play a critical role in myogenesis process by regulating MYOD’s targeted proteasome-mediated degradation, a key transcription factor in myogenesis.
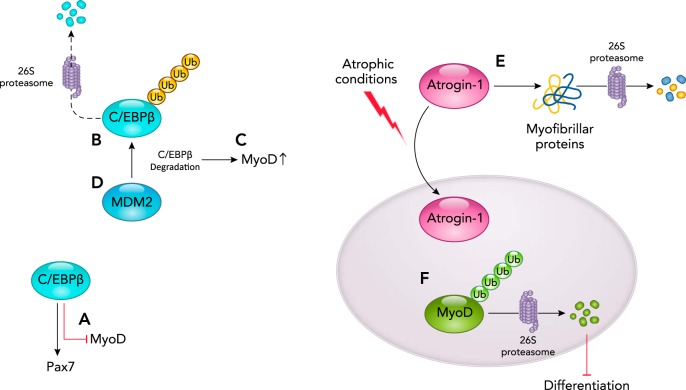
Mouse Double Minute 2 Homolog
Mouse double minute 2 homolog (MDM2) is a RING finger family ubiquitin ligase that regulates the differentiation of myoblasts via its posttranslational regulation of p53 via ubiquitination (26). The role of p53 in myocyte differentiation is well established in previous studies. Using an unbiased approach, studies of satellite cell-derived myoblast transcriptional regulation found that p53 is a key regulator of myoblast quiescence (24). Specifically, when cultured myoblasts are activated and proliferate, an upregulation of p53 is seen, and cells fail to differentiate (24). The sustained p53 increases in myoblast subpopulations of activated and proliferating cells not only hinders differentiation of these cells after their exit from cell cycle, it also leads to their quiescence (24). P53 binds to myogenin and represses its transcription, thus preventing the differentiation of myoblasts under genotoxic stress (65). It was also found that p53 expression is increased during atrophic conditions and plays an important role during atrophic conditions (25).
Satellite cells express CCAAT/enhancer-binding protein β (C/EBPβ), which maintains them in their quiescent state by stimulating PAX7 expression and by triggering decreased MYOD protein expression (FIGURE 6A). Recent studies have identified that increased MDM2 expression accompanies the loss of C/EBPβ proteins, which led to the hypothesis that MDM2 interacts with and ubiquitinates C/EBPβ (FIGURE 6B), leading to its degradation in a proteasome-dependent manner (FIGURE 6C) (26). Consistent with this mechanism of action, knockdown of MDM2 expression in myoblasts resulted in increased C/EBPβ and the blockade of myogenesis; similarly, knockdown of MDM2 in primary myoblasts resulted in the inability of muscle regeneration when grafted into cardiotoxin-injured muscle (26). Since the differentiation defect seen with increased MDM2 could be rescued by inhibiting the proteasome or by increasing C/EBPβ, a mechanism by which MDM2 regulates myogenesis by degrading C/EBPβ has been proposed (FIGURE 6D) (26). Studies demonstrating the interaction between MDM2 and C/EBPβ by co-IP and ubiquitination assays illustrate this mechanism as a pivotal regulator of myogenic differentiation and regeneration in vivo (26).
FIGURE 6.
The MDM2 ubiquitin ligase activity poly-ubiquitinates C/EBPβ, targeting it for proteasome-dependent degradation
A: in the undifferentiated state, C/EBPβ supports the expression of Pax7 and inhibits MYOD expression. B: the MDM2 ubiquitin ligase can poly-ubiquitinate C/EBPβ, targeting it for degradation by the 26S proteasome resulting in enhanced MYOD transcription secondary to the loss of C/EBPβ activity (C) and myogenic induction (differentiation) (D). E: during atrophic conditions, atrogin-1 expressed is increased, which in turn degrades the myofibrillar protein. F: in addition, atrogin-1 also translocates to the nucleus, wherein it ubiquitinates and degrades MYOD, thus blocking differentiation, as described in more detail in the text.
Atrogin-1/Muscle Atrophy F-Box (MAFBX)
During myocyte differentiation, the atrogin-1 ubiquitin ligase regulates differentiation by targeting MYOD1 for ubiquitination and 26S proteasome-dependent degradation, in addition to regulation of other transcription factors critical for muscle differentiation (39). Atrogin-1 is a muscle-specific ubiquitin ligase implicated in skeletal muscle atrophy (8). Atrogin-1 has been upregulated in skeletal muscle atrophy induced by glucocorticoids, denervation, spinal cord transection, hind limb suspension, immobilization, renal failure, diabetes, long-term mechanical ventilation, Cachexia, HIV, COPD, lipopolysaccharide (LPS)-induced changes in muscle catabolism, aging, alcohol, spinal muscle atrophy, heart failure, space flights, thermal injury, cytokine-mediated muscle loss, smoking, myositis, acute lung injury, statins, arthritis, hypoxia, knee arthroplasty, and pulmonary arterial hypertension (7). Previous studies have reported that increased atrogin-1 in atrophy contributes to the degradation of the myofibrillar proteins in the cytoplasm (FIGURE 6E) (28). In myotubes undergoing starvation-mediated atrophy, reduction in MyoD was observed on activation of atrogin-1 and its translocation to the nucleus (FIGURE 6F). Conversely, shRNA knockdown of atrogin-1 blocked MYOD proteolysis associated with muscle atrophy (39). Additionally, mutant MYOD (K113R), which lacked the atrogin-1 ubiquitination site, prevented atrophy of primary mouse myotubes and skeletal muscle fibers in vivo, illustrating that atrogin-1 plays a critical role in atrophy as a regulator of MYOD via poly-ubiquitin-mediated degradation during atrophic conditions.
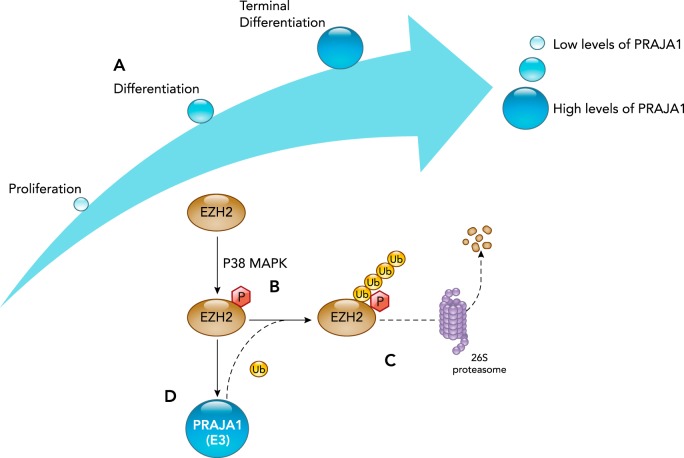
PRAJA1
The PRAJA1 ubiquitin ligase promotes skeletal muscle myogenesis through its regulation of the EZH2 transcription factor on P38α activation (FIGURE 7) (19). In myogenesis, PRAJA1 is first identified in the cytoplasm of MYOD-positive cells 3 days after cardiotoxin-induced muscle damage (19). The expression of PRAJA1 itself is increased during muscle cell differentiation (FIGURE 7A) (19). At later stages of the regeneration process, PRAJA1 re-localizes to the nucleus of the MYOD-positive cells, and the number of PJA1-positive cells begins to decline (19). The cytoplasm-to-nuclear changes suggest the PRAJA1 may have different targets in proliferating myoblasts when they are induced to differentiate (19). Recent bioinformatics analysis predicted that PRAJA1 would interact with enhancer of zeste homolog 2 (EZH2), which has been independently shown to be true experimentally (19). In skeletal muscle progenitor cells, EZH2 maintains muscle gene chromatin in a repressive conformation with the catalytic subunit of polycomb repressive complex 2 (PRC2) (19). EZH2’s gene repression plays a vital role in the satellite cell retention and proliferative phase of myogenesis, whereas in differentiating myoblasts the EZH2 levels are depleted (63). In C2C12 cells, depleting PRAJA1 blocks terminal differentiation, thus indicating the vital role of PRAJA1 in myogenesis (19).
FIGURE 7.
PRAJA1’s ubiquitin ligase activity regulates differentiation by targeting phosphorylated EZH2 for degradation by the 26S proteasome
A: PRAJA1 ubiquitin ligase expression is low during satellite cell proliferation and increases during differentiation. B: during differentiation, the P38 MAPK phosphorylates the EZH2 transcription factor, which is then recognized by the PRAJA1 ubiquitin ligase. C: PRAJA1 then poly-ubiquitinates EZH2, targeting it for degradation by the 26S proteasome. D: since EZH2 drives the transcription and expression of Praja1, PRAJA1-mediated degradation of EZH2 effectively forms a negative feedback, which decreases EZH2 expression.
A recent publication reported that the P38α kinase promotes the EZH2 by phosphorylating it at the threonine 372 position (FIGURE 7B) (19). In addition, both biochemical and genetic studies provided evidence that MYOD-induced PRAJA1 expression regulates EZH2 levels when P38α is activated (19). EZH2-associated proteins were then found to be targets of the PJA1-induced ubiquitination in muscle cells (19), consistent with previous studies using a cell-free system (70). The resulting EZH2 poly-ubiquitinated protein is then degraded in a proteasome-dependent manner (FIGURE 7C) (19). When EZH2 degradation is blocked in proliferating myocytes by decreasing PRAJA1 levels, EZH2 stays localized in the cytoplasm and has lower activity toward un-phosphorylated EZH2 (19). Together, these studies demonstrate that p38α signaling is necessary to induce the PJA1/EZH2 interaction, whereby EZH2 is ubiquitinated and degraded during satellite cell differentiation.
Summary
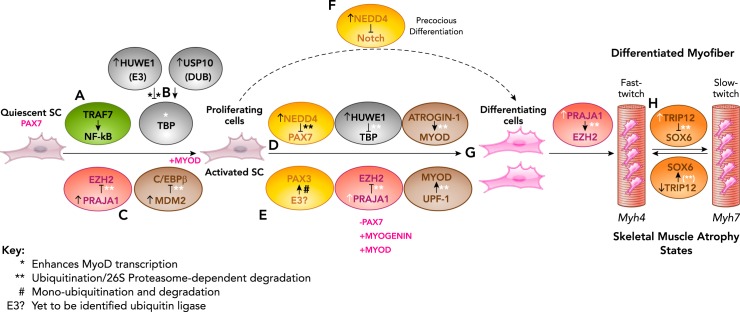
Posttranslational regulation is emerging as a critical regulator of skeletal muscle repair and regeneration in physiological and pathophysiological states. In this review, we discuss the role of both ubiquitin ligases (HUWE1, NEDD4, TRIP12, TRAF7, UPF1, MDM2, ATROGIN-1, PRAJA1) and deubiquitinase (USP10) in regulating multiple transcription factors known to control myogenesis, including the regulation of muscle fiber-type switching in differentiated myocytes in skeletal muscle atrophy (Table 1). TRAF7, HUWE1, PRAJA1, and MDM2 regulate the transition of quiescent satellite cells to activated, proliferating satellite cells by regulating the NF-κB, TBP, EZH2, and C/EBPβ transcription factors, respectively, via ubiquitination (FIGURE 8, A–C). Interestingly, the HUWE and PRAJA1 ubiquitin ligases further regulate SC differentiation in concert with NEDD4 and a yet-to-be-identified E3, which regulates the PAX7 and PAX3 transcription factors (FIGURE 8, D AND E). In addition, NEDD4 temporally and spatially mediates different stages of myogenesis by independently inhibiting NOTCH signaling (FIGURE 8F). Both atrogin-1, UPF-1, and HUWE1 ubiquitin ligases direct the posttranslational degradation of MYOD and are altered accordingly to allow MYOD expression during differentiation (FIGURE 8G). The TRIP12 ubiquitin ligase has an interesting role in serving as the focal point in slow- to fast-twitch fiber transition and vice-versa in different kinds of atrophies (FIGURE 8H). Together, these findings illustrate an array of examples by which ubiquitin ligases regulate transcription factors critical to myogenesis and likely are only a small sample of all the ubiquitin ligases involved.
Table 1.
Summary of ubiquitin ligases regulating transcription in skeletal muscle
| Ubiquitin Ligase | Substrate | Known E-Substrate Interaction | System E3/Substrate Tested | Role in Skeletal Muscle Atrophy | Reference(s) |
|---|---|---|---|---|---|
| HUWE1 (E3), USP10 (DUB) | TATA-binding protein (TBP) | HUWE1 targets TBP for K48-linkedin ubiquitination and proteasome-mediated degradation | C2C12 myoblasts and myotubes C2C12 myoblasts | Reduction in TBP levels, which regulate MyoD during terminal differentiation. | 41, 45 |
|
USP10 is a de-ubiquitinase (DUB) that removes poly-ubiquitin from TBP to counteract HUWE1 ubiquitination |
Increased HUWE1 (or decreased DUB) decrease TBP protein levels and block differentiation and myogenic progression. |
||||
|
Increase in HUWE1 and decrease in USP10 leads to depletion of TBP in differentiating cells. | |||||
| NEDD4 (E3) | PAX7, NOTCH | NEDD4 and PAX7 interact during early muscle differentiation. | Adult primary myoblasts and isolated myofibers. | Promotes differentiation. | 15, 38 |
|
PAX7 ubiquitinated and degraded. |
C2C12 cells. |
Promotes differentiation. |
|||
|
Intracellular domain of activated NOTCH translocates to the nucleus, wherein it upregulates PAX7. NEDD4 blocks translocation of NCID to the nucleus. |
Primary myoblasts from hindlimb skeletal muscles. |
Downregulated in denervation and unloading induced atrophy. |
|||
| TRIP12 (E3) | SOX6 | TRIP12 interacts with and polyubiquitinates SOX6 for proteasome-dependent degradation. | C2C12 myotubes. | Focal-point in mediating the transition between fast-slow fiber and slow-fast fiber. | 4 |
| TRAF7 (E3) | NEMO | TRAF7 binds IκB kinase/NF-κB essential modulator (NEMO) and the p65 NF-κB transcription factor to promote Lys-29 polyubiquitination of NEMO targeting lysosomal degradation of both proteins (72). | C2C12 myoblasts and myotubes. | Essential in myogenesis during the proliferation phase. | 58 |
|
Prevents precocious differentiation. | |||||
| UPF1 (E3) | MYOD | UPF1 poly-ubiquitinates MYOD and targets for proteasome-dependent degradation. | 54-1 and MB135 myoblasts. | Inhibits myogenesis. | 22 |
| MDM2 (E3) | C/EBPβ | MDM2 interacts with, ubiquitinates, and targets C/EBPβ for 26S proteasome-dependent degradation and can alter C/EBPβ-mediated satellite cells in the quiescent state. | C2C12 cells. | MDM2 degrades p53, thus enabling differentiation after exit from cell cycle. | 24, 26 |
|
Myoblasts. |
MDM2 knockdown in primary myoblasts results in the inability of muscle regeneration when grafted into cardiotoxin-injured muscle. |
||||
| ATROGIN-1 (E3) | MYOD | Atrogin-1 translocates to the nucleus during atrophic conditions and depletes MYOD levels. | C2C12 myotubes and in cellulo C2C12 myoblasts. | Mediates atrophic conditions by degrading MYOD proteins in a proteasome-dependent manner. | 39 |
| PRAJA1 (E3) | EZH2 | PRAJA1 poly-ubiquitinates EZH2, targeting it for proteasome-dependent degradation, resulting in a shift from proliferation to differentiation. | C2C12 cells. | Mediates terminal differentiation. | 19 |
HUWE1, HECT, UBA, and WWE domain containing 1, E3 ubiquitin protein ligase; MDM2, mouse double minute 2 homolog; NEDD4, neural precursor cell-expressed developmentally downregulated gene 4; TRIP12, thyroid hormone receptor interactor 12; TRAF7, TNF receptor-associated factor 7; UPF1, up-frameshift suppressor 1 homolog; USP10, ubiquitin specific peptidase 10.
FIGURE 8.
Summary of the role of ubiquitin ligase regulation of transcription factors during myocyte differentiation (myogenesis)
A: TRAF7’s ubiquitin ligase activity activates NF-κB signaling via NEMO ubiquitination to support the SC proliferation. B: the TBP transcription factor is regulated by the HUWE1 ubiquitination ligase and the USP1 deubiquitinase to maintain stable levels of TBP during proliferation stage. However, in the later stages, there is an increase in Huwe1 and a decrease in USP1 levels, thereby leading to reduced TBP levels, which is essential for differentiation. C: the PRAJA1 and MDM2 ubiquitin ligases are increased during proliferation, and poly-ubiquitinate and degrade the EZH2 and C/EBPβ, respectively, effectively releasing the inhibition on proliferation. PRAJA1 levels are tightly regulated throughout the myogenic lineage, which in a positive feedback loop mechanism degrades phosphorylated EZH2, a vital gene regulator in myogenesis. D: the NEDD4 ubiquitin ligase inhibits PAX7 protein levels during myogenesis while a yet to be identified ubiquitin ligase (E3?) inhibits the PAX3 transcription factor (E). NEDD4 leads to PAX7 depletion by ubiquitination and 26S proteasome degradation. F: NEDD4 also inhibits Notch signaling to accelerate differentiation in an unrelated mechanism. G: the ubiquitin ligases atrogin-1 and UPF-1 independently degrade MYOD, thereby modulating MYOD levels essential for differentiation. H: the TRIP12 ubiquitin ligase regulates SOX6 protein levels to control the transition of fast-twitch fibers to slow-twitch fibers. *Enhances MyoD transcription. **Ubiquitination/26S proteasome-dependent degradation. #Mono-ubiquitination and degradation. E3? , yet to be identified ubiquitin ligase.
Impaired myogenesis is seen in several diseases like cachexia, muscle dystrophy, and neuromuscular junction degeneration (1, 34, 42). Myogenesis capabilities are impaired on aging, and there have been several therapeutic options evolved to rejuvenate satellite cell function and activity to improve muscle regeneration. Apart from therapeutic strategies for dealing with aging, simple changes in lifestyle like regular exercise and low-calorie diet have been shown to have profound impact in improving the satellite cell number and activity (5). Similarly, atrophy has also been shown to be reversed by regular exercise and rehabilitation. Myogenesis is also blocked in rhabdomyosarcoma (RMS), a pediatric soft tissue sarcoma, and it involves modulation of several transcription factors like Pax3, EZH2, Notch, NF-κB, etc., which were discussed in this review (66). Differentiation therapy is one of the most possible therapeutic options being considered for RMS, in addition to targeting MYOD and MYF5 (57). There are several examples in the literature indicating how therapeutic targets are feasible at various stages of the myogenic lineage. The atrophy condition indicated by the extent of muscle mass present is an amalgamation of protein degradation and myogenesis. Several E3s, including atrogin-1, NEDD4, TRAF6, TRIM32, TRIM72, and USP10, either directly or indirectly impact the transcription factors, which are key to myogenic progression, in addition to regulating other signaling mechanisms that mediate myogenesis (6). Targeting the proteasome system has been a prominent therapeutic strategy for cancer cachexia (2), and further understanding of the intricate mechanisms regulated by the proteasome in mediating atrophy is critical. For example, MDM2, which degrades C/EBPβ as discussed in this paper, could be another possible therapeutic target in cancer cachexia, since it was shown earlier that C/EBPβ inhibits myogenesis (46). The possibility of specific E3 being assessed as the therapeutic target for the muscle-wasting diseases varies, since each of the diseases encompassing atrophy is regulated by distinct signaling mechanisms(s).
Acknowledgments
This work was supported by National Heart, Lung, and Blood Institute Grant R01 HL-104129 (to M.W.).
No conflicts of interest, financial or otherwise, are declared by the author(s).
V.S. and M.S.W. analyzed data; V.S. and M.S.W. interpreted results of experiments; V.S. and M.S.W. prepared figures; V.S. and M.S.W. drafted manuscript; V.S. and M.S.W. edited and revised manuscript; V.S. and M.S.W. approved final version of manuscript.
References
- 1.Abujarour R, Bennett M, Valamehr B, Lee TT, Robinson M, Robbins D, Le T, Lai K, Flynn P. Myogenic differentiation of muscular dystrophy-specific induced pluripotent stem cells for use in drug discovery. Stem Cells Transl Med 3: 149–160, 2014. doi: 10.5966/sctm.2013-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acharyya S, Guttridge DC. Cancer cachexia signaling pathways continue to emerge yet much still points to the proteasome. Clin Cancer Res 13: 1356–1361, 2007. doi: 10.1158/1078-0432.CCR-06-2307. [DOI] [PubMed] [Google Scholar]
- 3.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta 1695: 189–207, 2004. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 4.An CI, Ganio E, Hagiwara N. Trip12, a HECT domain E3 ubiquitin ligase, targets Sox6 for proteasomal degradation and affects fiber type-specific gene expression in muscle cells. Skelet Muscle 3: 11, 2013. doi: 10.1186/2044-5040-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bengal E, Perdiguero E, Serrano AL, Muñoz-Cánoves P. Rejuvenating stem cells to restore muscle regeneration in aging. F1000 Res 6: 76, 2017. doi: 10.12688/f1000research.9846.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bilodeau PA, Coyne ES, Wing SS. The ubiquitin proteasome system in atrophying skeletal muscle: roles and regulation. Am J Physiol Cell Physiol 311: C392–C403, 2016. doi: 10.1152/ajpcell.00125.2016. [DOI] [PubMed] [Google Scholar]
- 7.Bodine SC, Baehr LM. Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am J Physiol Endocrinol Metab 307: E469–E484, 2014. doi: 10.1152/ajpendo.00204.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294: 1704–1708, 2001. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 9.Bonaldo P, Sandri M. Cellular and molecular mechanisms of muscle atrophy. Dis Model Mech 6: 25–39, 2013. doi: 10.1242/dmm.010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bossola M, Marzetti E, Rosa F, Pacelli F. Skeletal muscle regeneration in cancer cachexia. Clin Exp Pharmacol Physiol 43: 522–527, 2016. doi: 10.1111/1440-1681.12559. [DOI] [PubMed] [Google Scholar]
- 11.Boutet SC, Disatnik MH, Chan LS, Iori K, Rando TA. Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell 130: 349–362, 2007. doi: 10.1016/j.cell.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 12.Brooks NE, Myburgh KH. Skeletal muscle wasting with disuse atrophy is multi-dimensional: the response and interaction of myonuclei, satellite cells and signaling pathways. Front Physiol 5: 99, 2014. doi: 10.3389/fphys.2014.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown DI, Parry TL, Willis MS. Ubiquitin ligases and posttranslational regulation of energy in the heart: the hand that feeds. Compr Physiol 7: 841–862, 2017. doi: 10.1002/cphy.c160024. [DOI] [PubMed] [Google Scholar]
- 14.Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, Moran-Crusio K, Farnsworth CL, Koller AA, Dasgupta R, Silva JC, Stadtfeld M, Hochedlinger K, Chen EI, Aifantis I. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell 11: 783–798, 2012. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bustos F, de la Vega E, Cabezas F, Thompson J, Cornelison DD, Olwin BB, Yates JR III, Olguín HC. NEDD4 regulates PAX7 levels promoting activation of the differentiation program in skeletal muscle precursors. Stem Cells 33: 3138–3151, 2015. doi: 10.1002/stem.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang SC, Ding JL. Ubiquitination and SUMOylation in the chronic inflammatory tumor microenvironment. Biochim Biophys Acta Rev Cancer 1870: 165–175, 2018. doi: 10.1016/j.bbcan.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Ciciliot S, Rossi AC, Dyar KA, Blaauw B, Schiaffino S. Muscle type and fiber type specificity in muscle wasting. Int J Biochem Cell Biol 45: 2191–2199, 2013. doi: 10.1016/j.biocel.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 18.Collins GA, Goldberg AL. The logic of the 26S proteasome. Cell 169: 792–806, 2017. doi: 10.1016/j.cell.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Consalvi S, Brancaccio A, Dall’Agnese A, Puri PL, Palacios D. Praja1 E3 ubiquitin ligase promotes skeletal myogenesis through degradation of EZH2 upon p38α activation. Nat Commun 8: 13956, 2017. doi: 10.1038/ncomms13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem 269: 7059–7061, 1994. [PubMed] [Google Scholar]
- 21.Dick SA, Chang NC, Dumont NA, Bell RA, Putinski C, Kawabe Y, Litchfield DW, Rudnicki MA, Megeney LA. Caspase 3 cleavage of Pax7 inhibits self-renewal of satellite cells. Proc Natl Acad Sci USA 112: E5246–E5252, 2015. doi: 10.1073/pnas.1512869112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng Q, Jagannathan S, Bradley RK. The RNA surveillance factor UPF1 represses myogenesis via its E3 ubiquitin ligase activity. Mol Cell 67: 239–251.e236, 2017. doi: 10.1016/j.molcel.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernández-Lloris R, Osses N, Jaffray E, Shen LN, Vaughan OA, Girwood D, Bartrons R, Rosa JL, Hay RT, Ventura F. Repression of SOX6 transcriptional activity by SUMO modification. FEBS Lett 580: 1215–1221, 2006. doi: 10.1016/j.febslet.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 24.Flamini V, Ghadiali RS, Antczak P, Rothwell A, Turnbull JE, Pisconti A. The satellite cell niche regulates the balance between myoblast differentiation and self-renewal via p53. Stem Cell Reports 10: 970–983, 2018. doi: 10.1016/j.stemcr.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fox DK, Ebert SM, Bongers KS, Dyle MC, Bullard SA, Dierdorff JM, Kunkel SD, Adams CM. p53 and ATF4 mediate distinct and additive pathways to skeletal muscle atrophy during limb immobilization. Am J Physiol Endocrinol Metab 307: E245–E261, 2014. doi: 10.1152/ajpendo.00010.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu D, Lala-Tabbert N, Lee H, Wiper-Bergeron N. Mdm2 promotes myogenesis through the ubiquitination and degradation of CCAAT/enhancer-binding protein β. J Biol Chem 290: 10200–10207, 2015. doi: 10.1074/jbc.M115.638577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fukada SI. The roles of muscle stem cells in muscle injury, atrophy and hypertrophy. J Biochem 163: 353–358, 2018. doi: 10.1093/jb/mvy019. [DOI] [PubMed] [Google Scholar]
- 28.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci USA 98: 14440–14445, 2001. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.González N, Moresco JJ, Cabezas F, de la Vega E, Bustos F, Yates JR III, Olguín HC. Ck2-dependent phosphorylation is required to maintain Pax7 protein levels in proliferating muscle progenitors. PLoS One 11: e0154919, 2016. doi: 10.1371/journal.pone.0154919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guttridge DC, Albanese C, Reuther JY, Pestell RG, Baldwin AS Jr. NF-kappaB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol Cell Biol 19: 5785–5799, 1999. doi: 10.1128/MCB.19.8.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hauerslev S, Vissing J, Krag TO. Muscle atrophy reversed by growth factor activation of satellite cells in a mouse muscle atrophy model. PLoS One 9: e100594, 2014. doi: 10.1371/journal.pone.0100594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heller H, Bengal E. TFIID (TBP) stabilizes the binding of MyoD to its DNA site at the promoter and MyoD facilitates the association of TFIIB with the preinitiation complex. Nucleic Acids Res 26: 2112–2119, 1998. doi: 10.1093/nar/26.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453: 481–488, 2008. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inaba S, Hinohara A, Tachibana M, Tsujikawa K, Fukada SI. Muscle regeneration is disrupted by cancer cachexia without loss of muscle stem cell potential. PLoS One 13: e0205467, 2018. doi: 10.1371/journal.pone.0205467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JA, Shon YH, Lim JO, Yoo JJ, Shin HI, Park EK. MYOD mediates skeletal myogenic differentiation of human amniotic fluid stem cells and regeneration of muscle injury. Stem Cell Res Ther 4: 147, 2013. doi: 10.1186/scrt358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitajima Y, Suzuki N, Nunomiya A, Osana S, Yoshioka K, Tashiro Y, Takahashi R, Ono Y, Aoki M, Nagatomi R. The ubiquitin-proteasome system is indispensable for the maintenance of muscle stem cells. Stem Cell Reports 11: 1523–1538, 2018. doi: 10.1016/j.stemcr.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kitajima Y, Tashiro Y, Suzuki N, Warita H, Kato M, Tateyama M, Ando R, Izumi R, Yamazaki M, Abe M, Sakimura K, Ito H, Urushitani M, Nagatomi R, Takahashi R, Aoki M. Proteasome dysfunction induces muscle growth defects and protein aggregation. J Cell Sci 127: 5204–5217, 2014. doi: 10.1242/jcs.150961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koncarevic A, Jackman RW, Kandarian SC. The ubiquitin-protein ligase Nedd4 targets Notch1 in skeletal muscle and distinguishes the subset of atrophies caused by reduced muscle tension. FASEB J 21: 427–437, 2007. doi: 10.1096/fj.06-6665com. [DOI] [PubMed] [Google Scholar]
- 39.Lagirand-Cantaloube J, Cornille K, Csibi A, Batonnet-Pichon S, Leibovitch MP, Leibovitch SA. Inhibition of atrogin-1/MAFbx mediated MyoD proteolysis prevents skeletal muscle atrophy in vivo. PLoS One 4: e4973, 2009. doi: 10.1371/journal.pone.0004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Grand F, Rudnicki MA. Skeletal muscle satellite cells and adult myogenesis. Curr Opin Cell Biol 19: 628–633, 2007. doi: 10.1016/j.ceb.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li L, Martinez SS, Hu W, Liu Z, Tjian R. A specific E3 ligase/deubiquitinase pair modulates TBP protein levels during muscle differentiation. eLife 4: e08536, 2015. doi: 10.7554/eLife.08536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu W, Klose A, Forman S, Paris ND, Wei-LaPierre L, Cortés-Lopéz M, Tan A, Flaherty M, Miura P, Dirksen RT, Chakkalakal JV. Loss of adult skeletal muscle stem cells drives age-related neuromuscular junction degeneration. eLife 6: e26464, 2017. doi: 10.7554/eLife.26464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loell I, Lundberg IE. Can muscle regeneration fail in chronic inflammation: a weakness in inflammatory myopathies? J Intern Med 269: 243–257, 2011. doi: 10.1111/j.1365-2796.2010.02334.x. [DOI] [PubMed] [Google Scholar]
- 44.Luan Z, Liu Y, Stuhlmiller TJ, Marquez J, García-Castro MI. SUMOylation of Pax7 is essential for neural crest and muscle development. Cell Mol Life Sci 70: 1793–1806, 2013. doi: 10.1007/s00018-012-1220-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malecova B, Dall’Agnese A, Madaro L, Gatto S, Coutinho Toto P, Albini S, Ryan T, Tora L, Puri PL. TBP/TFIID-dependent activation of MyoD target genes in skeletal muscle cells. eLife 5: e12534, 2016. doi: 10.7554/eLife.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marchildon F, Lamarche É, Lala-Tabbert N, St-Louis C, Wiper-Bergeron N. Expression of CCAAT/enhancer binding protein beta in muscle satellite cells inhibits myogenesis in cancer cachexia. PLoS One 10: e0145583, 2015. doi: 10.1371/journal.pone.0145583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mourkioti F, Rosenthal N. NF-kappaB signaling in skeletal muscle: prospects for intervention in muscle diseases. J Mol Med (Berl) 86: 747–759, 2008. doi: 10.1007/s00109-008-0308-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mu X, Agarwal R, March D, Rothenberg A, Voigt C, Tebbets J, Huard J, Weiss K. Notch signaling mediates skeletal muscle atrophy in cancer cachexia caused by osteosarcoma. Sarcoma 2016: 3758162, 2016. doi: 10.1155/2016/3758162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagpal P, Plant PJ, Correa J, Bain A, Takeda M, Kawabe H, Rotin D, Bain JR, Batt JA. The ubiquitin ligase Nedd4-1 participates in denervation-induced skeletal muscle atrophy in mice. PLoS One 7: e46427, 2012. doi: 10.1371/journal.pone.0046427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noy T, Suad O, Taglicht D, Ciechanover A. HUWE1 ubiquitinates MyoD and targets it for proteasomal degradation. Biochem Biophys Res Commun 418: 408–413, 2012. doi: 10.1016/j.bbrc.2012.01.045. [DOI] [PubMed] [Google Scholar]
- 51.Olguin HC, Olwin BB. Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: a potential mechanism for self-renewal. Dev Biol 275: 375–388, 2004. doi: 10.1016/j.ydbio.2004.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olguin HC, Yang Z, Tapscott SJ, Olwin BB. Reciprocal inhibition between Pax7 and muscle regulatory factors modulates myogenic cell fate determination. J Cell Biol 177: 769–779, 2007. doi: 10.1083/jcb.200608122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ouni I, Flick K, Kaiser P. Ubiquitin and transcription: the SCF/Met4 pathway, a (protein-) complex issue. Transcription 2: 135–139, 2011. doi: 10.4161/trns.2.3.15903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pinheiro CH, Guimarães-Ferreira L. Editorial: Frontiers in skeletal muscle wasting, regeneration and stem cells. Front Physiol 6: 141, 2015. doi: 10.3389/fphys.2015.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Talbot J, Maves L. Skeletal muscle fiber type: using insights from muscle developmental biology to dissect targets for susceptibility and resistance to muscle disease. Wiley Interdiscip Rev Dev Biol 5: 518–534, 2016. doi: 10.1002/wdev.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science 242: 405–411, 1988. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- 57.Tenente IM, Hayes MN, Ignatius MS, McCarthy K, Yohe M, Sindiri S, Gryder B, Oliveira ML, Ramakrishnan A, Tang Q, Chen EY, Petur Nielsen G, Khan J, Langenau DM. Myogenic regulatory transcription factors regulate growth in rhabdomyosarcoma. eLife 6: e19214, 2017. doi: 10.7554/eLife.19214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsikitis M, Acosta-Alvear D, Blais A, Campos EI, Lane WS, Sánchez I, Dynlacht BD. Traf7, a MyoD1 transcriptional target, regulates nuclear factor-κB activity during myogenesis. EMBO Rep 11: 969–976, 2010. doi: 10.1038/embor.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wallace GQ, McNally EM. Mechanisms of muscle degeneration, regeneration, and repair in the muscular dystrophies. Annu Rev Physiol 71: 37–57, 2009. doi: 10.1146/annurev.physiol.010908.163216. [DOI] [PubMed] [Google Scholar]
- 60.Walsh ME, Bhattacharya A, Sataranatarajan K, Qaisar R, Sloane L, Rahman MM, Kinter M, Van Remmen H. The histone deacetylase inhibitor butyrate improves metabolism and reduces muscle atrophy during aging. Aging Cell 14: 957–970, 2015. doi: 10.1111/acel.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wen Y, Bi P, Liu W, Asakura A, Keller C, Kuang S. Constitutive Notch activation upregulates Pax7 and promotes the self-renewal of skeletal muscle satellite cells. Mol Cell Biol 32: 2300–2311, 2012. doi: 10.1128/MCB.06753-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Willis MS, Patterson C. Into the heart: the emerging role of the ubiquitin-proteasome system. J Mol Cell Cardiol 41: 567–579, 2006. doi: 10.1016/j.yjmcc.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 63.Woodhouse S, Pugazhendhi D, Brien P, Pell JM. Ezh2 maintains a key phase of muscle satellite cell expansion but does not regulate terminal differentiation. J Cell Sci 126: 565–579, 2013. doi: 10.1242/jcs.114843. [DOI] [PubMed] [Google Scholar]
- 64.Xu LG, Li LY, Shu HB. TRAF7 potentiates MEKK3-induced AP1 and CHOP activation and induces apoptosis. J Biol Chem 279: 17278–17282, 2004. doi: 10.1074/jbc.C400063200. [DOI] [PubMed] [Google Scholar]
- 65.Yang ZJ, Broz DK, Noderer WL, Ferreira JP, Overton KW, Spencer SL, Meyer T, Tapscott SJ, Attardi LD, Wang CL. p53 suppresses muscle differentiation at the myogenin step in response to genotoxic stress. Cell Death Differ 22: 560–573, 2015. doi: 10.1038/cdd.2014.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu PY, Guttridge DC. Dysregulated myogenesis in rhabdomyosarcoma. Curr Top Dev Biol 126: 285–297, 2018. doi: 10.1016/bs.ctdb.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 67.Yun MH, Gates PB, Brockes JP. Regulation of p53 is critical for vertebrate limb regeneration. Proc Natl Acad Sci USA 110: 17392–17397, 2013. doi: 10.1073/pnas.1310519110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zammit PS, Relaix F, Nagata Y, Ruiz AP, Collins CA, Partridge TA, Beauchamp JR. Pax7 and myogenic progression in skeletal muscle satellite cells. J Cell Sci 119: 1824–1832, 2006. doi: 10.1242/jcs.02908. [DOI] [PubMed] [Google Scholar]
- 69.Zhu JY, Heidersbach A, Kathiriya IS, Garay BI, Ivey KN, Srivastava D, Han Z, King IN. The E3 ubiquitin ligase Nedd4/Nedd4L is directly regulated by microRNA 1. Development 144: 866–875, 2017. doi: 10.1242/dev.140368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zoabi M, Sadeh R, de Bie P, Marquez VE, Ciechanover A. PRAJA1 is a ubiquitin ligase for the polycomb repressive complex 2 proteins. Biochem Biophys Res Commun 408: 393–398, 2011. doi: 10.1016/j.bbrc.2011.04.025. [DOI] [PubMed] [Google Scholar]
- 71.Zotti T, Scudiero I, Vito P, Stilo R. The emerging role of TRAF7 in tumor development. J Cell Physiol 232: 1233–1238, 2017. doi: 10.1002/jcp.25676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zotti T, Uva A, Ferravante A, Vessichelli M, Scudiero I, Ceccarelli M, Vito P, Stilo R. TRAF7 protein promotes Lys-29-linked polyubiquitination of IkappaB kinase (IKKgamma)/NF-kappaB essential modulator (NEMO) and p65/RelA protein and represses NF-kappaB activation. J Biol Chem 286: 22924–22933, 2011. doi: 10.1074/jbc.M110.215426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zou X, Levy-Cohen G, Blank M. Molecular functions of NEDD4 E3 ubiquitin ligases in cancer. Biochim Biophys Acta 1856: 91–106, 2015. doi: 10.1016/j.bbcan.2015.06.005. [DOI] [PubMed] [Google Scholar]