Abstract
This is a review describing advances in CRISPR/Cas-mediated therapies for neuromuscular disorders (NMDs). We explore both CRISPR-mediated editing and dead Cas approaches as potential therapeutic strategies for multiple NMDs. Last, therapeutic considerations, including delivery and off-target effects, are also discussed.
Keywords: CRISPR/Cas, gene editing, neuromuscular disorders, muscular dystrophy, gene therapy
Introduction
Techniques to accomplish gene editing have been around for more than 20 years and include meganucleases, zinc finger nucleases, and transcription activator-like effector nucleases (TALENs); however, the newest approach using the clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein (Cas) system has revolutionized the field due to its ease of use and increased efficiency with wide-ranging potential applications (10). The CRISPR/Cas system was originally discovered in bacteria and was revealed to be a form of adaptive immunity. There are two main groups of CRISPR: class 1 (including types I, III, IV) utilizes multiple Cas enzymes, whereas class 2 (types II, V, VI) requires a single Cas protein (67). These Cas enzymes are often endonucleases that use RNA-guided targeting to cut nucleic acids and degrade foreign invading DNA (11, 20). For example, type II CRISPR systems utilize a CRISPR (cr) RNA complexed with a trans-activation CRISPR (tracr) RNA (referred to as a guide RNA), which targets the Cas9 protein to cut specific sites in DNA (34, 42).
The CRISPR/Cas system was recently adapted to enable gene editing in mammalian cells. In this application, a 20-bp sequence complementary to the target site in the genomic DNA [flanked by a protospacer adjacent motif (PAM)] is included in the guide RNA (gRNA), which targets a Cas protein to that site to create a double-stranded break (DSB) in the DNA (27, 56, 86). The most commonly used Cas enzymes for gene editing in mammalian cells include the type II Streptococcus pyogenes (Sp) Cas9 (27, 56, 86), the slightly smaller Staphylococcus aureus (Sa) Cas9 (109), or type V Cas12a/Cpf1 (62, 140). The PAM varies for each Cas enzyme: an NGG for SpCas9 (92), an NNGRRT for SaCas9 (109), and a TTTV for Cas12a (140). Cas9 creates a blunt-cut 3 bp upstream of the PAM (41), but Cas12a generates a staggered cut with a five nucleotide overhang 18–23 bp from the PAM (140). Cas9 contains two nuclease domains that cut each strand of DNA, or it can be converted into a nickase by mutation of one catalytic domain or into dead (d)Cas9 through mutation of both domains (56, 108).
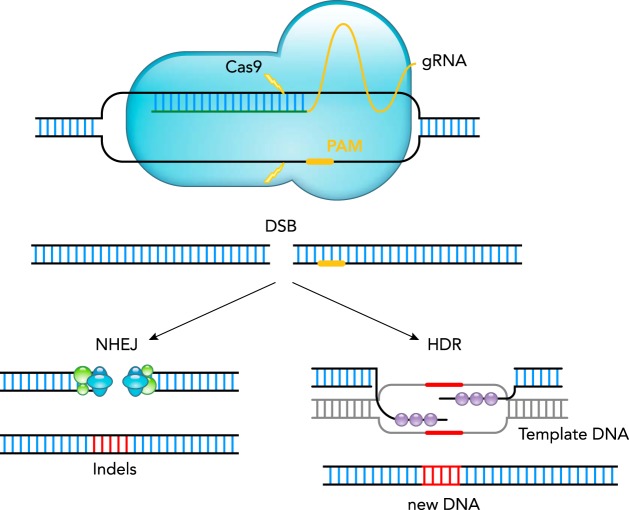
The DSB that occurs after Cas cutting can be repaired through two main mechanisms: either the cell’s endogenous repair machinery using non-homologous end joining (NHEJ), which rejoins the DSB and can result in small insertions or deletions (indels), or incorporation of new DNA through homology-directed repair (HDR) with the addition of an exogenous template (FIGURE 1). HDR will only occur during late S/G2 phase of the cell cycle (57).
FIGURE 1.
The CRISPR/Cas system for gene editing
The CRISPR/Cas9 gene editing system consists of two components: the Cas9 endonuclease (shown in blue) and a guide RNA (gRNA), which is composed of a target spacer region for DNA binding (shown in green) and a scaffold region for binding to Cas9 (in yellow). The gRNA spacer sequence is designed to be homologous to a target site in the genome, which must have a 3′ protospacer adjacent motif (PAM) sequence. gRNA targeting results in Cas9 creating a double-stranded break (DSB) in the DNA 3 bp upstream of the PAM. The DSB can be repaired either through the cells endogenous repair, non-homologous end joining (NHEJ), which can sometimes result in small insertions or deletions (indels), or through homology-directed repair (HDR) to incorporate new DNA by addition of template DNA containing homology arms.
Therapeutically, CRISPR can be used in vitro or in vivo to remove, edit, or add to a cell’s genome, or to activate or inactivate genes as a potential treatment for countless diseases (FIGURE 2). Strategies that rely on NHEJ can be applied through a single gRNA cut, leading to indels that cause a reading frame shift, disrupting or restoring gene expression, or through indels that remove a region of DNA such as a splice site (58, 86, 115). Using a pair of gRNAs, the intervening region of DNA can be deleted through NHEJ, and even seamless rejoining can occur (27, 86).
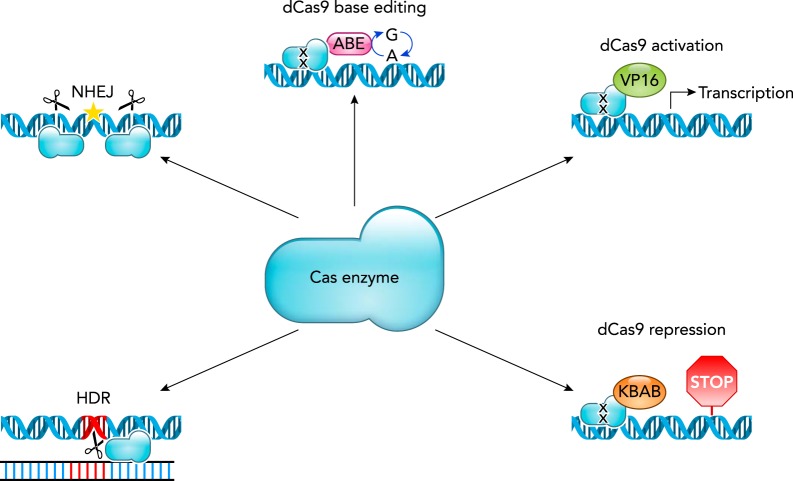
FIGURE 2.
Therapeutic strategies that exploit the CRISPR/Cas system
The CRISPR system has many applications. Some examples where it can be used to edit DNA, through NHEJ or HDR, or as an inactive targeting agent for base editing by fusion with an adenine base editor (ABE), with VP16 for gene activation, or with KRAB for gene repression are depicted. Inactivated Cas9 is represented by the black X on the protein.
CRISPR-mediated HDR can be utilized to precisely repair mutations or insert new DNA but requires an exogenous template containing homology arms around the cut site (27, 86). However non-cycling cells, such as post-mitotic myofibers or quiescent muscle stem cells, have very low levels of HDR (57). Alternatively, recent work has demonstrated methods to achieve targeted integration without the necessity for HDR. For example, microhomology-mediated end joining (e.g., CRIS-PITCh) is active in G1/early S phases (94), homology-independent targeted integration (HITI) can occur in post-mitotic cells (121), and other integration methods utilize the NHEJ pathway (8, 29, 63, 70, 77, 87, 113, 114).
CRISPR can also be adapted for non-DNA editing applications using a catalytically inactivated “dead” Cas protein. Here, dCas9 is used for specific targeting, such as to a promoter, and can be fused to an additional protein to induce activation or inactivation of genes, epigenome editing, imaging, base editing, or RNA targeting/degradation (2).
For therapeutic application to humans, CRISPR could be applied directly to the target tissue in vivo or to patient cells ex vivo through cell therapy. CRISPR/Cas editing in blood cells ex vivo was the first clinical application of CRISPR in humans. Clinical trials were initiated in 2016 in China (31) and for β-thalassemia in Germany (100). U.S. scientists plan to do in vivo editing in Leber’s congenital amaurosis type 10 in the near future (117). Thus CRISPR/Cas is transforming the field of precision medicine and gene therapy for human disease. In this review, we will discuss some applications of the CRISPR system for neuromuscular disorders (NMDs). Depending on the cause of disease, type of protein, and cycling state of the target cell, different CRISPR strategies may be more appropriate for certain diseases. Below, we have grouped by strategy different potential CRISPR-based approaches for various NMDs (Table 1).
Table 1.
Summary of CRISPR/Cas9 approaches for NMDs
| Approach | NMD | Model | References |
|---|---|---|---|
| NHEJ-mediated single exon deletion | DMD | Cells, mice | 37, 50, 66, 80, 84, 85, 97, 98, 101, 122 |
| NHEJ-mediated multi-exon deletion | DMD | Cells, mice | 15, 16, 36, 55, 69, 84, 85, 101, 110, 135, 137, 138 |
| NHEJ-mediated single cut | DMD | Cells, mice, dog | 5, 6, 66, 76, 81, 85, 90, 93, 141 |
| NHEJ-mediated duplication deletion | DMD | Cells | 71, 133 |
| NHEJ-mediated repeat region deletion | DM1 | Cells, mice | 3, 32, 105, 131 |
| NHEJ-mediated intronic deletion | MDC1A | Mice | 60 |
| UCMD | Cells | 17 | |
| HDR-mediated point mutation repair | DMD | Cells, mice | 15, 74, 82, 141, 142 |
| LGMD2B, 2D | Cells | 126 | |
| HDR-mediated exon replacement | DMD | Cells | 76 |
| HDR-mediated polyA insertion | DM1 | Cells | 131 |
| Base editing | DMD | Cells, mice | 112, 139 |
| dCas9 activation | DMD | Cells, mice | 78, 102, 133 |
| MDC1A | Mice | 59 | |
| dCas9 inactivation | FSHD | Cells | 52 |
| dCas9 interference | DM1 | Cells, mice | 13, 104 |
Non-Homologous End Joining
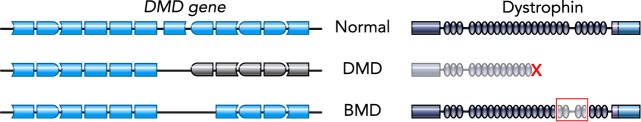
CRISPR-mediated NHEJ has potential uses for many NMDs. One of the most extensively studied applications is for Duchenne muscular dystrophy (DMD), which is often caused by out-of-frame mutations in the DMD gene encoding dystrophin. Through human and mouse studies, it has been demonstrated that not all of the large middle-rod domain of dystrophin is required for its function (35, 38). In fact, a milder, allelic disease, Becker muscular dystrophy (BMD), is typically caused by in-frame mutations in DMD resulting in production of an internally deleted but still at least somewhat functional dystrophin protein (FIGURE 3). Some Becker patients are asymptomatic until late in life, suggesting certain mutations lead to very functional proteins (88, 95, 123). Therefore, it is hypothesized that CRISPR can be applied to convert a DMD into a BMD mutation by deletion or permanent skipping of one or more exons in the DMD gene. Proof-of-principle that CRISPR gene editing by NHEJ can restore the reading frame for numerous DMD mutations has been validated in human-induced pluripotent stem cells (hiPSCs), DMD muscle cells, and dystrophic animal models. Additionally, NHEJ can be used to remove duplications, repeat regions, or intronic areas as therapeutic strategies for multiple NMDs.
FIGURE 3.
Schematics of normal, DMD, and BMD genes and protein
Representations of parts of the DMD gene, with blue boxes denoting exons (left) and corresponding dystrophin protein schematics (right). DMD mutations, often deletions of exons, disrupt the reading frame, preventing expression of dystrophin protein. BMD mutations are often in-frame, allowing for expression of an internally deleted but at least somewhat functional dystrophin.
NHEJ-Mediated Single Exonic Deletions or Single Cut Splice Site Disruptions
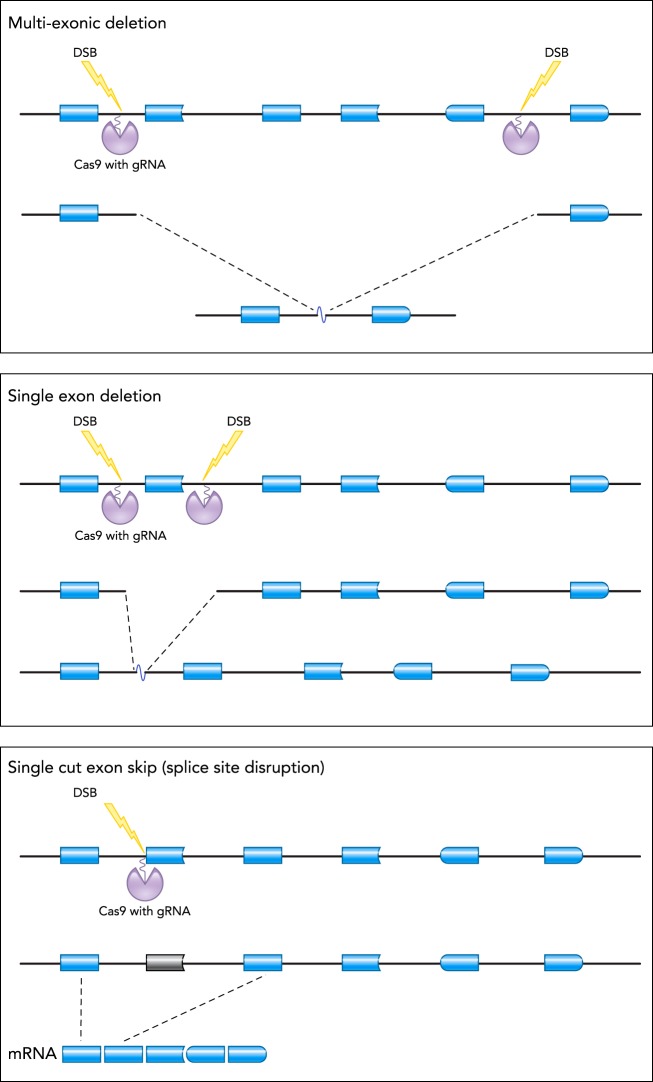
DMD reframing by NHEJ can be accomplished by a variety of strategies, including the use of two guides to achieve single- or multi-exon deletions or the use of a single guide to induce frameshifts or permanent exon skipping (FIGURE 4). Single-exon deletion or permanent skipping has been demonstrated for the murine Dmd exons 23 (50, 66, 80, 93, 97, 98, 122, 141), 45 (90), 51 (6), canine exon 51 (5), and human DMD exons 43 (90), 45 (76, 90), 51 (37, 81, 85, 101, 141), and 53 (84, 85). In vitro proof-of-principle has been demonstrated in human muscle cells (37, 84, 85, 101) and hiPSCs (76, 81, 90, 141). In vivo single-exon deletion or skipping by NHEJ has been accomplished in multiple dystrophic mouse models of Duchenne, including mdx mice with a stop codon in Dmd exon 23 (50, 66, 80, 97, 98, 122, 141), as well as in novel dystrophic mice with deletions of Dmd exons 44 (90) or 50 (6). One study tested single exon skipping by NHEJ in a dystrophic dog model (5). This study provided proof-of-principle that dystrophin can be successfully reframed in a large animal model.
FIGURE 4.
NHEJ-based CRISPR approaches for reframing DMD
Different CRISPR/Cas9-mediated strategies that could use NHEJ to reframe the DMD gene are depicted. Top: a multi-exon deletion. Middle: a single exon deletion. Bottom: a single cut permanent exon skipping approach. A region of the DMD gene is shown by blue boxes (representing exons); Cas9 and its associated gRNA are shown in purple; and the corresponding double-stranded break (DSB) is represented by the yellow lightning bolt.
Multiple Exonic Deletions and Rejoining by NHEJ
CRISPR/Cas9 can also accomplish large deletions encompassing multiple exons by using a pair of gRNAs and NHEJ. In DMD, removal of multiple exons provides the opportunity to create designer dystrophin species with high functionality and to encompass more patient mutations. It is important to keep in mind that more highly functional dystrophin species are known to often retain the proper repeat phasing in the rod domain of the protein. Researchers were able to achieve very large multi-exonic DMD deletions in vitro using human DMD myoblasts (36, 55, 84, 85, 101) and hiPSCs (69, 137). The feasibility of multi-exon Dmd deletion in vivo has been successfully demonstrated in the mdx mouse model of Duchenne by deletion of exons 21–23 (110, 135) and exons 52–53 in the mdx4CV mouse (15, 16). Two humanized DMD mouse models with out-of-frame deletions of exons 45 (138) or 52 (127) have been created to provide more relevant models for testing CRISPR strategies on the human DMD gene in vivo. These mice were used to demonstrate that functional human dystrophin proteins could be generated through multi-exon deletion and NHEJ of 450 or 708 kb (36, 138). Thus it is possible to accomplish NHEJ between two very distant parts of the large DMD gene to accomplish reframing to generate a functional protein that can improve the dystrophic phenotype.
Long-Term NHEJ Studies In Vivo
Two different studies used the mdx mouse model of DMD to assess the long-term consequence of restoring dystrophin through CRISPR/Cas9 gene editing using NHEJ. Mice were studied after 12 (98) or 18 mo (50) post a single adeno-associated virus (AAV)-CRISPR/Cas9 injection. Although both of these studies demonstrated loss of viral genomes and reduced editing in skeletal muscle over 12–18 mo, they also revealed that the heart was better able to retain edited nuclei. It is possible that this differential loss of viral genomes between heart and skeletal muscle relates to the extensive turnover that occurs in dystrophic skeletal (but not cardiac) muscle as it undergoes degeneration/regeneration cycles since co-delivery of micro-dystrophins prevents loss of edited skeletal myofibers (Bengtsson NE, Chamberlain JS, personal communication). Interestingly, both of these studies discovered a disproportionate loss of the AAV-gRNA vector compared with the AAV-Cas9 vector. It is difficult to establish the cause of the loss of AAV-gRNA vector; however, Hakim et al. demonstrated that increasing the ratio of AAV-gRNA to AAV-Cas9 led to an improvement in overall efficacy (50). Similar observations were made in a murine model lacking exon 44 and in the mdx4cv model, whereby it was demonstrated that increasing the AAV-gRNA-to-AAV-Cas9 ratio improved the level of dystrophin protein produced (14, 90).
NHEJ in Dystrophic Dogs
Only one study has tested the feasibility of CRISPR-mediated NHEJ in a large animal model. Investigators administered a CRISPR platform designed to induce permanent exon 51 skipping or reframing through disruption of an exon splicing enhancer to young dystrophic dogs (1 mo old) carrying an exon 50 deletion. Systemic administration of low-dose AAV showed very little dystrophin, whereas the single dog given the high dose (2 × 1014 vg/kg total virus) showed high dystrophin restoration in some muscles, with low dystrophin levels in other muscles (5). This study provided proof-of-concept that CRISPR gene editing could be accomplished to restore dystrophin protein production in a large animal model.
Removal of Duplications by NHEJ
Genetic duplications causing DMD (~10% of patients) can also be removed through NHEJ. Using a single gRNA targeted to a duplicated region, so that it binds twice, led to production of full-length dystrophin in patient cells containing an exon 18–30 duplication (133). A single guide or dual guides flanking a duplication were also designed to be able to remove an exon 2 duplication mutation, restoring dystrophin in vitro (71).
NHEJ-Mediated Deletion of Expanded Repeats
Myotonic dystrophy type 1 (DM1) is caused by a toxic CTG trinucleotide repeat expansion in the 3′ UTR of the dystrophia myotonica protein kinase gene. When the expanded repeats are expressed in RNA, the expansion sequesters RNA binding proteins, such as the muscle blind family of splicing factors, in ribonuclear foci, which prevents the correct splicing of many genes (72). One strategy for applying gene editing to these types of repeat expansion diseases is to remove the repeat region in the genomic DNA through NHEJ. Multiple groups have demonstrated effective deletion using gRNAs that flank the repeat region in DM1 patient cells (3, 32, 105, 131). However, this deletion was not always precise, since different types of events occurred, including partial deletions, inversions, or a single cut inducing loss of the entire repeat. Thus targeting repeat expansions may not lead to an exact or expected result.
NHEJ to Restore Correct Splicing
Congenital muscular dystrophy 1A (MDC1A) is due to a variety of mutations in the LAMA2 gene (encoding for the laminin α2 protein), where a large percentage (~40%) are caused by splice-site mutations (44). An NHEJ approach was utilized for a splice donor site mutation causing MDC1A. In this example, a DNA deletion spanning from the mutation into the intronic region in the dy2J/dy2J mouse model allowed for generation of a new splice donor site that could restore normal splicing (60).
A recently discovered, yet fairly common, mutation in COL6A1 that is associated with Ullrich congenital muscular dystrophy (UCMD) results in inclusion of a pseudoexon from an intron. It was demonstrated that a pair of gRNAs could cut and utilize NHEJ to delete both the mutation and the pseudoexon in the intronic region, and restore normal expression of collagen VI (17). Thus CRISPR-based NHEJ can be applied to introns to restore correct splicing for multiple diseases.
NHEJ for Other NMDs
Not all NMDs will be amenable to NHEJ-mediated gene editing. Based on successful exon skipping studies in limb girdle muscular dystrophies (LGMD) 2B and C (due to mutations in dysferlin or gamma-sarcoglycan respectively) (1, 12, 73, 134), it has been suggested that CRISPR could be used to generate functional proteins by NHEJ. Other NMDs caused by aberrant expression of toxic proteins, such as facioscapulohumeral muscular dystrophy (FSHD), could also in theory be targeted by NHEJ. On the other hand, many of the autosomal recessive LGMDs arise from mutations in enzymes, such as LGMD2A (CAPN3) or LGMD2H (TRIM32), and such disorders would not benefit from NHEJ-based approaches. Thus the potential to use gene editing for each NMD will need to be thoroughly explored on a case-by-case basis.
Homology-Directed Repair
HDR can be utilized for point mutation correction or insertion of new DNA. This strategy is especially relevant for diseases that have common point mutations or are enzymes, which cannot function with deletions. Additionally, incorporation of termination signals through HDR could prevent expression of toxic regions of DNA. However, challenges for applying HDR in post-mitotic tissues such as muscle and neurons still remain, since it is largely inefficient in non-diving cells.
HDR after CRISPR/Cas editing has been utilized for point-mutation correction of the mdx mouse, which contains a nonsense mutation in exon 23 of the Dmd gene. The first demonstration of HDR in mdx mice was through germline editing in zygotes with Cas9 (82) and later with Cas12a/Cpf1 (141), which showed proof-of-principle CRISPR-mediated restoration of dystrophin, but this avenue is currently not feasible for human patients. HDR was also utilized to correct the point mutation in mdx satellite cells ex vivo. Although the efficiency was low, they were able to demonstrate restored dystrophin after engraftment in vivo (142). Furthermore, HDR to knockin a deleted exon was used in DMD hiPSCs to restore full-length dystrophin (76). Finally, two groups observed low levels of HDR correction in vivo after either AAV-CRISPR delivery to mdx4Cv mice (15) or intramuscular injection of CRISPR-gold nanoparticles in mdx mice (74).
HDR was also utilized for specific point mutation correction of LGMD2B and 2D, caused by mutations in the dysferlin and alpha-sarcoglycan genes, respectively. CRISPR-mediated correction by HDR in patient hiPSCs was shown to rescue protein expression. One of these mutations is the most prevalent LGMD2D mutation found in ~32% of patients (126).
An effective HDR-based strategy to inhibit expression of the expanded repeats in DM1 cells was developed. Here, a polyadenylation (polyA) signal template is integrated before the repeat expansion. This termination signal prevented the repeat from being expressed in the RNA, thus inhibiting its toxic effects (131).
Dead CRISPR
As a potential therapeutic strategy for some neuromuscular disorders, a dead CRISPR system fused to another protein has been used for base editing or to activate compensatory proteins or inactivate toxic ones. Since dead CRISPR does not cause DSBs, it may be safer, although it would require continuous expression for gene activation or inactivation.
Base Editing
Base editing using a Cas9 nickase or dead Cas9 fused with a deaminase protein allows for C/G-to-T/A or A/T-to-G/C conversions in DNA (65). This approach could be applied to permanently fix certain point mutations or modulate splicing without creating a DSB or utilizing a homology template. Base editing for DMD was applied to a mouse model containing a nonsense mutation, which was converted to glutamine by an A-to-G base edit in vivo (112). This approach would only be effective for very specific mutations. An alternative approach used base editing to convert a splice site G to an A to induce permanent exon skipping and restore dystrophin in DMD hiPSCs (139). However, large amounts of off-target changes have recently been reported using base editors (47, 144).
Activation of Genes
For DMD, a transcriptional transactivator, dCas9-VP160, targeted to the utrophin promoter was used as a strategy to upregulate utrophin in patient cells (133). Utrophin has significant homology to dystrophin and can potentially be used to functionally replace it (48). Another system using short dead gRNAs with two MS2 RNA aptamer domains that recruit and bind the MS2:P65:HSF1 (MPH) transcriptional activation complex was used in the mdx mouse model of DMD to upregulate klotho to increase regenerative potential (132), utrophin to compensate for dystrophin, and/or follistatin to increase muscle mass (75) (78). Additionally, dCas9-VP160 was used in rag/mdx mice to upregulate the laminin subunit alpha 1 chain (Lama1) to increase laminin-111 to stabilize the muscle fiber membrane (102). For MDC1A, a non-mutation specific approach with SadCas9-2xVP64 was used to upregulate Lama1, which can compensate to rescue the dy2J/dy2J phenotype in vivo (59).
Inactivation of Genes
Inactivation strategies using dCas9 targeted to DNA or RNA have been applied to downregulate toxic repeat RNAs as a therapy for repeat expansion diseases, including DM1, DM2, and C9orf72 amyotrophic lateral sclerosis (ALS). dCas9 targeted to the DNA repeat could inhibit transcription (104), and dCas9 alone or fused to a PIN RNA endonuclease targeted to the repeat region of RNA could reduce toxic RNA levels (13).
FSHD is caused by contractions of the D4Z4 repeat and chromatin relaxation, leading to aberrant expression of DUX4 and its downstream targets, including genes involved in germline development, stem cells, and suppression of the innate immune response (33). Since chromatin dysregulation is correlated with disease, an inactivation strategy using dCas9-KRAB was developed to target the DUX4 promoter/exon 1 and was shown to cause chromatin repression and reduce DUX4 levels in patient cells (52).
Therapeutic Considerations: Translation of CRISPR Therapies to Patients With NMDs
Delivery of CRISPR
Although the promise of therapeutic gene editing is high for monogenic diseases, there are still many obstacles that need to be addressed before CRISPR therapies can be translated to NMD patients. Most importantly, it will be necessary to identify an appropriate vehicle that allows delivery of these platforms to all skeletal muscles and, for some NMDs, to cardiac muscle. Although delivery could be achieved by either viral or non-viral means, most in vivo studies have relied on viral delivery strategies. AAV is the most promising vector to translate CRISPR to patients; however, AAV presents serious concerns that could impact the safety and efficacy, which need to be addressed before AAV can be used clinically for CRISPR delivery. Concerns about the use of AAV-CRISPR relates to both the vector and the bacterially derived Cas9 cargo, especially regarding the predicted immune response, which could impact safety (25, 91, 98, 130). These potential issues include 1) the inability to re-dose AAV without additional procedures, which will reduce the efficacy of the CRISPR therapy (83); 2) the presence of pre-existing immunity to AAV, ranging in seroprevalence from 30 to 70% of people (19, 83) and to Cas9 (23, 119, 128); 3) sustained expression of Cas9 from the AAV episome or potential AAV integration, which could lead to long-term autoimmune consequences and increased off-target activity potential; and 4) functional demonstration of AAV-mediated targeting of quiescent muscle stem cells in vivo is still unclear (7, 46, 122), which means that corrected nuclei may become diluted over time. These issues could potentially be overcome with strategies to degrade Cas9, a new capsid generation that evades the immune system, new AAV serotypes that can target quiescent muscle stem cells, and other interventions that interfere with the ability of the immune system to reject AAV-infected cells (9, 26, 28, 89, 91). Considering that some strategies will likely necessitate continuous expression of dCas9, investigators will need to carefully assess the toxicities that arise from long-term expression of dCas9 in vivo.
A major challenge with AAV is determining the optimal dose, which involves counterbalancing the desire for high transduction with the problem of the immune response. Because AAV treatment is currently limited to a single dose, it is imperative to achieve the best transduction possible, especially if the patient is a child who will grow and dilute the amount of vector in their tissues. With regard to CRISPR, the issue of dose is compounded because of AAV’s small carrying capacity (~4.7 kb), which is not able to contain both SpCas9 plus multiple gRNAs in the same vector, thus requiring two different viral vectors to carry one platform. Using two vectors for one platform may effectively halve the effective dose. SaCas9 is an alternative, smaller nuclease that can be packaged along with a gRNA into a single AAV and has been successfully applied in some models of NMDs (15, 36, 59, 60, 97, 98, 104, 110, 122, 131). However, a higher percentage of the population is estimated to have pre-existing immunity to SaCas9 compared with SpCas9 (23, 119). Furthermore, loss of gRNA viral genomes has been shown to occur with time in murine models, and it was demonstrated that increasing the ratio of gRNA AAV to Cas9 AAV greatly increased the efficacy of the therapy (14, 50, 90). The necessity of increasing the gRNA vector may also adversely impact the dose of vector that is needed to be administered.
Engineered nanoparticles are one non-viral alternative that could facilitate delivery of CRISPR/Cas in vivo as DNA, RNA, or ribonucleoprotein (RNP) complexes. Nano-CRISPR studies for other diseases have targeted tissues such as the liver, lung, or tumors in vivo (22). However, delivery to muscle has its own challenges since it comprises such a large percentage of body mass, is post-mitotic, and has an extensive network of connective tissue that is a barrier to delivery. In the context of muscular dystrophy, where excessive connective tissue is increased as disease progresses, the challenges are further heightened. Only one in vivo study has attempted to use nanoparticles to deliver CRISPR locally in a NMD to correct the mdx mutation by HDR, and here they achieved an editing efficiency of 0.8% without cardiotoxin (74). This low efficiency is not surprising, since HDR requires cell cycling, and skeletal muscle is post-mitotic (57). It is likely that any HDR that was achieved arose from targeting activated myoblasts. Thus there is still much work to be done to create systemically compatible nanoparticles that will be efficient enough to be used as a therapeutic.
A second non-viral alternative would be a case where hiPSCs, muscle progenitors, or muscle stem cells are subjected to gene editing ex vivo, and then the cells are engrafted to muscles as a cell therapy. This approach could provide a corrected muscle stem cell with lifelong regenerative potential. Direct intramuscular injection of CRISPR-corrected muscle progenitor cells has been demonstrated in the mdx mouse (51, 101, 137, 142), but challenges still remain, including efficient systemic delivery of human skeletal muscle progenitors, loss of muscle stem cell potential when expanded ex vivo, and the inability to generate muscle stem cells from hiPSCs with long-term repopulation potential.
Level of Efficacy Needed
Another important consideration for CRISPR in NMDs is understanding the timing and efficiency necessary to achieve therapeutic levels of the restored protein. In the case of dystrophinopathies, where mutations lead to impaired membrane integrity, it will be necessary to restore the mutant protein to the threshold level needed to protect each individual muscle fiber. Although the overall level of dystrophin necessary for improved function has been approximated in DMD as between 3 and 40% (43, 45, 99, 106, 107, 116, 129), the level needed on a per fiber basis is still unclear. Tremblay and colleagues have estimated that the amount of dystrophin produced from a single nucleus is sufficient to cover ~549 µm along the length of the muscle fiber membrane (55). According to their estimates, dystrophin could be restored along the length of most muscle cells if ~4% of total myonuclei are edited. This estimation is only an approximation and will be influenced by many factors, including DMD mRNA and protein stability. Additionally, if muscle stem cells are also targeted (46), the impact of the therapy could be greatly amplified.
Restoring the reading frame by NHEJ is a strategy that will work for only a subset of neuromuscular disorders, and the functionality of each internally deleted protein will need to be assessed in the disease context. This point is especially important for DMD, where NHEJ will create a variety of internally deleted dystrophin species that could vary in their functionality and stability. In vivo studies are essential to establish the relative stability and functionality of each dystrophin species that is created by gene editing. It will also be important to conduct long-term in vivo assessments to determine the rate at which edited nuclei are lost over time and to gain insight into why gRNA AAVs are more unstable than AAVs carrying Cas9.
Many of the papers discussed here assessed functional outcomes after in vivo delivery. Skeletal muscle force and/or force drop after eccentric contraction were improved across multiple DMD studies, even with restoration of as little as 2% dystrophin (15, 50, 66, 90, 97, 122). Other studies showed improvement in grip strength and hanging time (6, 74, 78, 80, 82, 141). Heart function was assessed and demonstrated improvement in some DMD models (50, 110). Last, for other diseases, open-field and tetanic force were improved in a mouse model of MDC1A (59, 60), and less myotonia was observed in a model of DM1 (104) after CRISPR-based therapies were given. These studies are promising; however, the long-term benefit of gene editing on functional outcomes will still need to be fully explored.
Potential for Off-Target Activity
Although Watson-Crick base pairing makes the CRISPR/Cas system highly specific, it has potential for off-target activity since it has been to shown to be able to tolerate some mismatches within the gRNA target sequence (39). Most studies discussed here have assessed off-target activity at the predicted sites for each gRNA using T7 or Surveyor nuclease digestion, or deep sequencing of the target sites. A few studies applied additional unbiased assessments; for example, Li et al. used whole exome sequencing on corrected hiPSC clones (76) and Koo et al. performed Digenome-seq (66). Nelson et al. assessed genome-wide AAV integrations with Nextera sequencing, which was more sensitive than deep sequencing and may be a new approach for unbiased off-target assessment (98). For the dead Cas9 studies, assessment of off-target gene expression was performed using qRT-PCR or RNA sequencing (13, 52, 59, 102, 104). Across all these studies, only a few reported higher than background levels of editing but were still low percentages (36, 82, 97, 98, 101, 133). Unwanted on-target changes are also a possibility, which is discussed below. Additionally, after gene editing, there is potential for a p53 DNA damage response, which could lead to cell-cycle arrest or toxicity (49, 54); however, this is transient and may not be as big of a problem for post-mitotic muscle cells.
It is important to note that T7 and Surveyor assays are not very sensitive and cannot be used to demonstrate <1% or 3% editing, respectively (143). Additionally, it is preferred to use an unbiased method to capture all potential sites. These strategies include whole genome/exome sequencing in vitro [e.g., Digenome-seq (61)] or in vivo, such as genome-wide unbiased identification of DSBs enabled by sequencing (GUIDE-seq) (125), high-throughput genome-wide translocation sequencing (HTGTS) (53), circularization for in vitro reporting of cleavage effects by sequencing (CIRCLE-seq) (124), verification of in vivo off-targets (VIVO) (4), in situ breaks labeling enrichment and sequencing (BLESS) (30), or SITE-seq (21). Importantly, unbiased off-target assessment should be performed in the background genome of interest (e.g., human) and is critical to consider and test before clinical translation. However, there has been progress on ways to reduce CRISPR-mediated off targets, including the use of truncated gRNAs (40) and engineered Cas9s with higher specificity (24, 64, 120, 136).
Targeting Considerations for CRISPR Editing
With CRISPR/Cas9-mediated editing, there is also potential for unwanted on-target DNA changes, including inversions, translocations, or small or large indels. In the DMD gene, Ousterout et al. observed a low number of translocations in vitro (101), and Nelson et al. observed deletions, inversions, indels, and AAV integration in vivo (98). An unrelated paper also demonstrated large deletions or rearrangements at the on-target site in vitro (68). Since additional mutations can occur at the cut site, it is important to consider where gRNAs are targeted within the DNA.
CRISPR/Cas-mediated editing for NMDs can be achieved through various approaches where gRNAs can be targeted to introns, exons, intron/exon boundaries, or untranslated regions. Intronic targeting, such as to create exon deletions or remove psuedoexons, may be advantageous since unwanted small indels that arise at the rejoining site would not likely have a large effect, although unmapped regulatory elements could be affected. Targeting exons to precisely fix mutations would allow for full-length proteins to be expressed, but this approach requires HDR and complete precision. HDR is also currently less efficient than NHEJ, although strategies to improve efficiency or to use NHEJ-based integration pathways are ongoing (79, 96). Additionally, exons can be targeted to generate deletions and create hybrid exons; for example, a CinDel approach created hybrid exons in DMD that were designed to maintain the proper phasing of dystrophin (36, 55). However, for this approach, ideally seamless rejoining (or at least an indel of a multiple of three) should be achieved to maintain the reading frame. Although it may be simpler to use a single gRNA targeted to a splice site or enhancer to induce a deletion in the mRNA, this strategy relies on creation of indels, and not every DNA change will be the same, potentially leading to unwanted effects. Furthermore, this approach will require significant personalization, since each exon will need to be targeted individually. Targeting other noncoding regions, such as the 3′ UTR to insert a polyA or deleting repeats in the case of DM1, may also allow for on-target mistakes to be tolerated. Strategies using a single gRNA to remove duplications could be targeted to introns or exons, and thus may or may not require seamless rejoining, depending on where the gRNA binds. Importantly, there has been progress on ways to make CRISPR safer, for example, through the use of truncated gRNAs (40), engineered Cas9s with less off-target activity (24, 64, 120, 136), anti-CRISPR proteins to interfere with Cas9 activity (18, 118), or a self-inactivating strategy by targeting a gRNA against the Cas9 sequence (89, 103, 111).
Conclusions
In conclusion, CRISPR/Cas has wide-ranging uses for many NMDs and is showing much promise in pre-clinical studies. Efficacy has been demonstrated in models of DMD, congenital muscular dystrophy, LGMD, DM1, and FSHD. The field is ripe for further exploration; however, overcoming potential delivery, immune response, and off-target issues will need to be addressed before clinical translation can effectively occur.
Acknowledgments
The authors thank N.E. Bengtsson and J.S. Chamberlain for manuscript suggestions.
Funding was provided by a Ruth L. Kirschstein National Research Service Award T32AR065972 “Muscle Cell Biology, Pathophysiology, and Therapeutics” from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (to C.S.Y.).
C.S.Y., A.D.P., and M.J.S. are co-founders of MyoGene Bio.
C.S.Y. and M.J.S. conceived and designed research; C.S.Y. prepared figures; C.S.Y. and M.J.S. drafted manuscript; C.S.Y., A.D.P., and M.J.S. edited and revised manuscript; C.S.Y., A.D.P., and M.J.S. approved final version of manuscript.
References
- 1.Aartsma-Rus A, Singh KHK, Fokkema IFAC, Ginjaar IB, van Ommen G-J, den Dunnen JT, van der Maarel SM. Therapeutic exon skipping for dysferlinopathies? Eur J Hum Genet 18: 889–894, 2010. doi: 10.1038/ejhg.2010.4 . A corrigendum for this article is available at http://dx.doi.org/10.1038/ejhg.2010.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun 9: 1911, 2018. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Agtmaal EL, André LM, Willemse M, Cumming SA, van Kessel IDG, van den Broek WJAA, Gourdon G, Furling D, Mouly V, Monckton DG, Wansink DG, Wieringa B. CRISPR/Cas9-induced (CTG⋅CAG)n repeat instability in the myotonic dystrophy type 1 locus: implications for therapeutic genome editing. Mol Ther 25: 24–43, 2017. doi: 10.1016/j.ymthe.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akcakaya P, Bobbin ML, Guo JA, Malagon-Lopez J, Clement K, Garcia SP, Fellows MD, Porritt MJ, Firth MA, Carreras A, Baccega T, Seeliger F, Bjursell M, Tsai SQ, Nguyen NT, Nitsch R, Mayr LM, Pinello L, Bohlooly-Y M, Aryee MJ, Maresca M, Joung JK. In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature 561: 416–419, 2018. doi: 10.1038/s41586-018-0500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amoasii L, Hildyard JCW, Li H, Sanchez-Ortiz E, Mireault A, Caballero D, Harron R, Stathopoulou T-R, Massey C, Shelton JM, Bassel-Duby R, Piercy RJ, Olson EN. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science 362: 86–91, 2018. doi: 10.1126/science.aau1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amoasii L, Long C, Li H, Mireault AA, Shelton JM, Sanchez-Ortiz E, McAnally JR, Bhattacharyya S, Schmidt F, Grimm D, Hauschka SD, Bassel-Duby R, Olson EN. Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Sci Transl Med 9: eaan8081, 2017. doi: 10.1126/scitranslmed.aan8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnett AL, Konieczny P, Ramos JN, Hall J, Odom G, Yablonka-Reuveni Z, Chamberlain JR, Chamberlain JS. Adeno-associated viral (AAV) vectors do not efficiently target muscle satellite cells. Mol Ther Methods Clin Dev 1: 14038, 2014. doi: 10.1038/mtm.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Auer TO, Duroure K, De Cian A, Concordet J-P, Del Bene F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res 24: 142–153, 2014. doi: 10.1101/gr.161638.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnes C, Scheideler O, Schaffer D. Engineering the AAV capsid to evade immune responses. Curr Opin Biotechnol 60: 99–103, 2019. doi: 10.1016/j.copbio.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nat Biotechnol 34: 933–941, 2016. doi: 10.1038/nbt.3659. [DOI] [PubMed] [Google Scholar]
- 11.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712, 2007. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 12.Barthélémy F, Blouin C, Wein N, Mouly V, Courrier S, Dionnet E, Kergourlay V, Mathieu Y, Garcia L, Butler-Browne G, Lamaze C, Lévy N, Krahn M, Bartoli M. Exon 32 skipping of dysferlin rescues membrane repair in patients’ cells. J Neuromuscul Dis 2: 281–290, 2015. doi: 10.3233/JND-150109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Batra R, Nelles DA, Pirie E, Blue SM, Marina RJ, Wang H, Chaim IA, Thomas JD, Zhang N, Nguyen V, Aigner S, Markmiller S, Xia G, Corbett KD, Swanson MS, Yeo GW. Elimination of toxic microsatellite repeat expansion RNA by RNA-targeting Cas9. Cell 170: 899–912.e10, 2017. doi: 10.1016/j.cell.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bengtsson NE, Hall JK, Chamberlain JS. Systemic gene editing for muscular dystrophy using AAV-CRISPR/Cas9. In: ASGCT Annual Meeting Abstracts (Online). Milwaukee, WI: American Society of Gene & Cell Therapy, 2017. p. 16 https://kundoc.com/pdf-asgct-annual-meeting-abstracts-.html. [Google Scholar]
- 15.Bengtsson NE, Hall JK, Odom GL, Phelps MP, Andrus CR, Hawkins RD, Hauschka SD, Chamberlain JR, Chamberlain JS. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat Commun 8: 14454, 2017. doi: 10.1038/ncomms14454 . A corrigendum for this article is available at http://dx.doi.org/10.1038/ncomms16007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bengtsson NE, Seto JT, Hall JK, Chamberlain JS, Odom GL. Progress and prospects of gene therapy clinical trials for the muscular dystrophies. Hum Mol Genet 25, R1: R9–R17, 2016. doi: 10.1093/hmg/ddv420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bolduc V, Foley AR, Solomon-Degefa H, Sarathy A, Donkervoort S, Hu Y, Chen GS, Sizov K, Nalls M, Zhou H, Aguti S, Cummings BB, Lek M, Tukiainen T, Marshall JL, Regev O, Marek-Yagel D, Sarkozy A, Butterfield RJ, Jou C, Jimenez-Mallebrera C, Li Y, Gartioux C, Mamchaoui K, Allamand V, Gualandi F, Ferlini A, Hanssen E, Wilton SD, Lamandé SR, MacArthur DG, Wagener R, Muntoni F, Bönnemann CG; COL6A1 Intron 11 Study Group . A recurrent COL6A1 pseudoexon insertion causes muscular dystrophy and is effectively targeted by splice-correction therapies. JCI Insight 4: e124403, 2019. doi: 10.1172/jci.insight.124403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bondy-Denomy J, Garcia B, Strum S, Du M, Rollins MF, Hidalgo-Reyes Y, Wiedenheft B, Maxwell KL, Davidson AR. Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature 526: 136–139, 2015. doi: 10.1038/nature15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boutin S, Monteilhet V, Veron P, Leborgne C, Benveniste O, Montus MF, Masurier C. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum Gene Ther 21: 704–712, 2010. doi: 10.1089/hum.2009.182. [DOI] [PubMed] [Google Scholar]
- 20.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321: 960–964, 2008. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cameron P, Fuller CK, Donohoue PD, Jones BN, Thompson MS, Carter MM, Gradia S, Vidal B, Garner E, Slorach EM, Lau E, Banh LM, Lied AM, Edwards LS, Settle AH, Capurso D, Llaca V, Deschamps S, Cigan M, Young JK, May AP. Mapping the genomic landscape of CRISPR-Cas9 cleavage. Nat Methods 14: 600–606, 2017. doi: 10.1038/nmeth.4284. [DOI] [PubMed] [Google Scholar]
- 22.Carboni V, Maaliki C, Alyami M, Alsaiari S, Khashab N. Synthetic vehicles for encapsulation and delivery of CRISPR/Cas9 gene editing machinery. Adv Ther 2: 1800085, 2019. doi: 10.1002/adtp.201800085. [DOI] [Google Scholar]
- 23.Charlesworth CT, Deshpande PS, Dever DP, Dejene B, Gomez-Ospina N, Mantri S, Pavel-Dinu M, Camarena J, Weinberg KI, Porteus MH. Identification of pre-existing adaptive immunity to Cas9 proteins in humans. Nat Med 25: 249–254, 2019. doi: 10.1038/s41591-018-0326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, Sternberg SH, Joung JK, Yildiz A, Doudna JA. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550: 407–410, 2017. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chew WL, Tabebordbar M, Cheng JKW, Mali P, Wu EY, Ng AHM, Zhu K, Wagers AJ, Church GM. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat Methods 13: 868–874, 2016. doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chicoine LG, Montgomery CL, Bremer WG, Shontz KM, Griffin DA, Heller KN, Lewis S, Malik V, Grose WE, Shilling CJ, Campbell KJ, Preston TJ, Coley BD, Martin PT, Walker CM, Clark KR, Sahenk Z, Mendell JR, Rodino-Klapac LR. Plasmapheresis eliminates the negative impact of AAV antibodies on microdystrophin gene expression following vascular delivery. Mol Ther 22: 338–347, 2014. doi: 10.1038/mt.2013.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823, 2013. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corti M, Cleaver B, Clément N, Conlon TJ, Faris KJ, Wang G, Benson J, Tarantal AF, Fuller D, Herzog RW, Byrne BJ. Evaluation of readministration of a recombinant adeno-associated virus vector expressing acid alpha-glucosidase in pompe disease: preclinical to clinical planning. Hum Gene Ther Clin Dev 26: 185–193, 2015. doi: 10.1089/humc.2015.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cristea S, Freyvert Y, Santiago Y, Holmes MC, Urnov FD, Gregory PD, Cost GJ. In vivo cleavage of transgene donors promotes nuclease-mediated targeted integration. Biotechnol Bioeng 110: 871–880, 2013. doi: 10.1002/bit.24733. [DOI] [PubMed] [Google Scholar]
- 30.Crosetto N, Mitra A, Silva MJ, Bienko M, Dojer N, Wang Q, Karaca E, Chiarle R, Skrzypczak M, Ginalski K, Pasero P, Rowicka M, Dikic I. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat Methods 10: 361–365, 2013. doi: 10.1038/nmeth.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cyranoski D. CRISPR gene-editing tested in a person for the first time Nature 539: 479, 2016. doi: 10.1038/nature.2016.20988. [DOI] [PubMed] [Google Scholar]
- 32.Dastidar S, Ardui S, Singh K, Majumdar D, Nair N, Fu Y, Reyon D, Samara E, Gerli MFM, Klein AF, De Schrijver W, Tipanee J, Seneca S, Tulalamba W, Wang H, Chai YC, In’t Veld P, Furling D, Tedesco FS, Vermeesch JR, Joung JK, Chuah MK, VandenDriessche T. Efficient CRISPR/Cas9-mediated editing of trinucleotide repeat expansion in myotonic dystrophy patient-derived iPS and myogenic cells. Nucleic Acids Res 46: 8275–8298, 2018. doi: 10.1093/nar/gky548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daxinger L, Tapscott SJ, van der Maarel SM. Genetic and epigenetic contributors to FSHD. Curr Opin Genet Dev 33: 56–61, 2015. doi: 10.1016/j.gde.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471: 602–607, 2011. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duan D. Systemic AAV micro-dystrophin gene therapy for Duchenne muscular dystrophy. Mol Ther 26: 2337–2356, 2018. doi: 10.1016/j.ymthe.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duchêne BL, Cherif K, Iyombe-Engembe JP, Guyon A, Rousseau J, Ouellet DL, Barbeau X, Lague P, Tremblay JP. CRISPR-induced deletion with SaCas9 restores dystrophin expression in dystrophic models in vitro and in vivo. Mol Ther 26: 2604–2616, 2018. doi: 10.1016/j.ymthe.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ehrke-Schulz E, Schiwon M, Leitner T, Dávid S, Bergmann T, Liu J, Ehrhardt A. CRISPR/Cas9 delivery with one single adenoviral vector devoid of all viral genes. Sci Rep 7: 17113, 2017. doi: 10.1038/s41598-017-17180-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.England SB, Nicholson LV, Johnson MA, Forrest SM, Love DR, Zubrzycka-Gaarn EE, Bulman DE, Harris JB, Davies KE. Very mild muscular dystrophy associated with the deletion of 46% of dystrophin. Nature 343: 180–182, 1990. doi: 10.1038/343180a0. [DOI] [PubMed] [Google Scholar]
- 39.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31: 822–826, 2013. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32: 279–284, 2014. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garneau JE, Dupuis MÈ, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468: 67–71, 2010. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 42.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA 109: E2579–E2586, 2012. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gentil C, Le Guiner C, Falcone S, Hogrel J-Y, Peccate C, Lorain S, Benkhelifa-Ziyyat S, Guigand L, Montus M, Servais L, Voit T, Piétri-Rouxel F. Dystrophin threshold level necessary for normalization of neuronal nitric oxide synthase, inducible nitric oxide synthase, and ryanodine receptor-calcium release channel type 1 nitrosylation in golden retriever muscular dystrophy dystrophinopathy. Hum Gene Ther 27: 712–726, 2016. doi: 10.1089/hum.2016.041. [DOI] [PubMed] [Google Scholar]
- 44.Geranmayeh F, Clement E, Feng LH, Sewry C, Pagan J, Mein R, Abbs S, Brueton L, Childs A-M, Jungbluth H, De Goede CG, Lynch B, Lin J-P, Chow G, Sousa C, O’Mahony O, Majumdar A, Straub V, Bushby K, Muntoni F. Genotype-phenotype correlation in a large population of muscular dystrophy patients with LAMA2 mutations. Neuromuscul Disord 20: 241–250, 2010. doi: 10.1016/j.nmd.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Godfrey C, Muses S, McClorey G, Wells KE, Coursindel T, Terry RL, Betts C, Hammond S, O’Donovan L, Hildyard J, El Andaloussi S, Gait MJ, Wood MJ, Wells DJ. How much dystrophin is enough: the physiological consequences of different levels of dystrophin in the mdx mouse. Hum Mol Genet 24: 4225–4237, 2015. doi: 10.1093/hmg/ddv155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goldstein JM, Tabebordbar M, Zhu K, Wang LD, Messemer KA, Peacker B, Ashrafi Kakhki S, Gonzalez-Celeiro M, Shwartz Y, Cheng JKW, Xiao R, Barungi T, Albright C, Hsu YC, Vandenberghe LH, Wagers AJ. In situ modification of tissue stem and progenitor cell genomes. Cell Reports 27: 1254–1264.e7, 2019. doi: 10.1016/j.celrep.2019.03.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grünewald J, Zhou R, Garcia SP, Iyer S, Lareau CA, Aryee MJ, Joung JK. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 569: 433–437, 2019. doi: 10.1038/s41586-019-1161-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guiraud S, Roblin D, Kay DE. The potential of utrophin modulators for the treatment of Duchenne muscular dystrophy. Expert Opin Orphan Drugs 6: 179–192, 2018. doi: 10.1080/21678707.2018.1438261. [DOI] [Google Scholar]
- 49.Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med 24: 927–930, 2018. doi: 10.1038/s41591-018-0049-z. [DOI] [PubMed] [Google Scholar]
- 50.Hakim CH, Wasala NB, Nelson CE, Wasala LP, Yue Y, Louderman JA, Lessa TB, Dai A, Zhang K, Jenkins GJ, Nance ME, Pan X, Kodippili K, Yang NN, Chen SJ, Gersbach CA, Duan D. AAV CRISPR editing rescues cardiac and muscle function for 18 months in dystrophic mice. JCI Insight 3: e124297, 2018. doi: 10.1172/jci.insight.124297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hicks MR, Hiserodt J, Paras K, Fujiwara W, Eskin A, Jan M, Xi H, Young CS, Evseenko D, Nelson SF, Spencer MJ, Handel BV, Pyle AD. ERBB3 and NGFR mark a distinct skeletal muscle progenitor cell in human development and hPSCs. Nat Cell Biol 20: 46–57, 2018. doi: 10.1038/s41556-017-0010-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Himeda CL, Jones TI, Jones PL. CRISPR/dCas9-mediated transcriptional inhibition ameliorates the epigenetic dysregulation at D4Z4 and represses DUX4-fl in FSH muscular dystrophy. Mol Ther 24: 527–535, 2016. doi: 10.1038/mt.2015.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu J, Meyers RM, Dong J, Panchakshari RA, Alt FW, Frock RL. Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing. Nat Protoc 11: 853–871, 2016. doi: 10.1038/nprot.2016.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ihry RJ, Worringer KA, Salick MR, Frias E, Ho D, Theriault K, Kommineni S, Chen J, Sondey M, Ye C, Randhawa R, Kulkarni T, Yang Z, McAllister G, Russ C, Reece-Hoyes J, Forrester W, Hoffman GR, Dolmetsch R, Kaykas A. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med 24: 939–946, 2018. doi: 10.1038/s41591-018-0050-6. [DOI] [PubMed] [Google Scholar]
- 55.Iyombe-Engembe JP, Ouellet DL, Barbeau X, Rousseau J, Chapdelaine P, Lagüe P, Tremblay JP. Efficient restoration of the dystrophin gene reading frame and protein structure in DMD myoblasts using the CinDel method. Mol Ther Nucleic Acids 5: e283, 2016. doi: 10.1038/mtna.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821, 2012. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kakarougkas A, Jeggo PA. DNA DSB repair pathway choice: an orchestrated handover mechanism. Br J Radiol 87: 20130685, 2014. doi: 10.1259/bjr.20130685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kapahnke M, Banning A, Tikkanen R. Random splicing of several exons caused by a single base change in the target exon of CRISPR/Cas9 mediated gene knockout. Cells 5: 45, 2016. doi: 10.3390/cells5040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kemaladewi D, Bassi PS, Lindsay K, Erwood S, Hyatt E, Place KM, Marks R, Gawlik KI, Durbeej M, Ivakine EA, Cohn RD. A mutation-independent approach via transcriptional upregulation of a disease modifier gene rescues muscular dystrophy in vivo. bioRxiv. doi: 10.1101/286500. [DOI] [PubMed] [Google Scholar]
- 60.Kemaladewi DU, Maino E, Hyatt E, Hou H, Ding M, Place KM, Zhu X, Bassi P, Baghestani Z, Deshwar AG, Merico D, Xiong HY, Frey BJ, Wilson MD, Ivakine EA, Cohn RD. Correction of a splicing defect in a mouse model of congenital muscular dystrophy type 1A using a homology-directed-repair-independent mechanism. Nat Med 23: 984–989, 2017. doi: 10.1038/nm.4367. [DOI] [PubMed] [Google Scholar]
- 61.Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, Hwang J, Kim J-I, Kim J-S. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods 12: 237–243, 2015. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 62.Kim D, Kim J, Hur JK, Been KW, Yoon SH, Kim JS. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol 34: 863–868, 2016. doi: 10.1038/nbt.3609 . An erratum for this article is available at http://dx.doi.org/10.1038/nbt0816-888a. [DOI] [PubMed] [Google Scholar]
- 63.Kimura Y, Hisano Y, Kawahara A, Higashijima S. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci Rep 4: 6545, 2014. doi: 10.1038/srep06545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529: 490–495, 2016. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533: 420–424, 2016. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koo T, Lu-Nguyen NB, Malerba A, Kim E, Kim D, Cappellari O, Cho HY, Dickson G, Popplewell L, Kim J-S. Functional rescue of dystrophin deficiency in mice caused by frameshift mutations using campylobacter jejuni Cas9. Mol Ther 26: 1529–1538, 2018. doi: 10.1016/j.ymthe.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol 37: 67–78, 2017. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol 36: 765–771, 2018. doi: 10.1038/nbt0918-899c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kyrychenko V, Kyrychenko S, Tiburcy M, Shelton JM, Long C, Schneider JW, Zimmermann W-H, Bassel-Duby R, Olson EN. Functional correction of dystrophin actin binding domain mutations by genome editing. JCI Insight 2: e95918, 2017. doi: 10.1172/jci.insight.95918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lackner DH, Carré A, Guzzardo PM, Banning C, Mangena R, Henley T, Oberndorfer S, Gapp BV, Nijman SMB, Brummelkamp TR, Bürckstümmer T. A generic strategy for CRISPR-Cas9-mediated gene tagging. Nat Commun 6: 10237, 2015. doi: 10.1038/ncomms10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lattanzi A, Duguez S, Moiani A, Izmiryan A, Barbon E, Martin S, Mamchaoui K, Mouly V, Bernardi F, Mavilio F, Bovolenta M. Correction of the exon 2 duplication in DMD myoblasts by a single CRISPR/Cas9 system. Mol Ther Nucleic Acids 7: 11–19, 2017. doi: 10.1016/j.omtn.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee JE, Cooper TA. Pathogenic mechanisms of myotonic dystrophy. Biochem Soc Trans 37: 1281–1286, 2009. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee JJA, Maruyama R, Duddy W, Sakurai H, Yokota T. Identification of novel antisense-mediated exon skipping targets in DYSF for therapeutic treatment of dysferlinopathy. Mol Ther Nucleic Acids 13: 596–604, 2018. doi: 10.1016/j.omtn.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee K, Conboy M, Park HM, Jiang F, Kim HJ, Dewitt MA, Mackley VA, Chang K, Rao A, Skinner C, Shobha T, Mehdipour M, Liu H, Huang WC, Lan F, Bray NL, Li S, Corn JE, Kataoka K, Doudna JA, Conboy I, Murthy N. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat Biomed Eng 1: 889–901, 2017. doi: 10.1038/s41551-017-0137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee S-J, McPherron AC. Regulation of myostatin activity and muscle growth. Proc Natl Acad Sci USA 98: 9306–9311, 2001. doi: 10.1073/pnas.151270098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li HL, Fujimoto N, Sasakawa N, Shirai S, Ohkame T, Sakuma T, Tanaka M, Amano N, Watanabe A, Sakurai H, Yamamoto T, Yamanaka S, Hotta A. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports 4: 143–154, 2015. doi: 10.1016/j.stemcr.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li J, Meng X, Zong Y, Chen K, Zhang H, Liu J, Li J, Gao C. Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9. Nat Plants 2: 16139, 2016. doi: 10.1038/nplants.2016.139. [DOI] [PubMed] [Google Scholar]
- 78.Liao HK, Hatanaka F, Araoka T, Reddy P, Wu MZ, Sui Y, Yamauchi T, Sakurai M, O’Keefe DD, Núñez-Delicado E, Guillen P, Campistol JM, Wu CJ, Lu LF, Esteban CR, Izpisua Belmonte JC. In vivo target gene activation via CRISPR/Cas9-mediated trans-epigenetic modulation. Cell 171: 1495–1507.e15, 2017. doi: 10.1016/j.cell.2017.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu M, Rehman S, Tang X, Gu K, Fan Q, Chen D, Ma W. Methodologies for improving HDR efficiency. Front Genet 9: 691, 2019. doi: 10.3389/fgene.2018.00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351: 400–403, 2016. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Long C, Li H, Tiburcy M, Rodriguez-Caycedo C, Kyrychenko V, Zhou H, Zhang Y, Min Y-L, Shelton JM, Mammen PPA, Liaw NY, Zimmermann W-H, Bassel-Duby R, Schneider JW, Olson EN. Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Sci Adv 4: eaap9004, 2018. doi: 10.1126/sciadv.aap9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science 345: 1184–1188, 2014. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Louis Jeune V, Joergensen JA, Hajjar RJ, Weber T. Pre-existing anti-adeno-associated virus antibodies as a challenge in AAV gene therapy. Hum Gene Ther Methods 24: 59–67, 2013. doi: 10.1089/hgtb.2012.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maggio I, Liu J, Janssen JM, Chen X, Gonçalves MAFV. Adenoviral vectors encoding CRISPR/Cas9 multiplexes rescue dystrophin synthesis in unselected populations of DMD muscle cells. Sci Rep 6: 37051, 2016. doi: 10.1038/srep37051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maggio I, Stefanucci L, Janssen JM, Liu J, Chen X, Mouly V, Gonçalves MAFV. Selection-free gene repair after adenoviral vector transduction of designer nucleases: rescue of dystrophin synthesis in DMD muscle cell populations. Nucleic Acids Res 44: 1449–1470, 2016. doi: 10.1093/nar/gkv1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science 339: 823–826, 2013. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maresca M, Lin VG, Guo N, Yang Y. Obligate ligation-gated recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res 23: 539–546, 2013. doi: 10.1101/gr.145441.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Melis MA, Cau M, Muntoni F, Mateddu A, Galanello R, Boccone L, Deidda F, Loi D, Cao A. Elevation of serum creatine kinase as the only manifestation of an intragenic deletion of the dystrophin gene in three unrelated families. Eur J Paediatr Neurol 2: 255–261, 1998. doi: 10.1016/S1090-3798(98)80039-1. [DOI] [PubMed] [Google Scholar]
- 89.Merienne N, Vachey G, de Longprez L, Meunier C, Zimmer V, Perriard G, Canales M, Mathias A, Herrgott L, Beltraminelli T, Maulet A, Dequesne T, Pythoud C, Rey M, Pellerin L, Brouillet E, Perrier AL, du Pasquier R, Déglon N. The self-inactivating KamiCas9 system for the editing of CNS disease genes. Cell Reports 20: 2980–2991, 2017. doi: 10.1016/j.celrep.2017.08.075. [DOI] [PubMed] [Google Scholar]
- 90.Min Y, Li H, Rodriguez-Caycedo C, Mireault AA, Huang J, Shelton JM, Mcanally JR, Amoasii L, Mammen PPA, Bassel-Duby R, Olson EN. CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci Adv 5: eaav4324, 2019. doi: 10.1126/sciadv.aav4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mingozzi F, High KA. Overcoming the host immune response to adeno-associated virus gene delivery vectors: the race between clearance, tolerance, neutralization, and escape. Annu Rev Virol 4: 511–534, 2017. doi: 10.1146/annurev-virology-101416-041936. [DOI] [PubMed] [Google Scholar]
- 92.Mojica FJM, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155: 733–740, 2009. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 93.Mou H, Smith JL, Peng L, Yin H, Moore J, Zhang XO, Song CQ, Sheel A, Wu Q, Ozata DM, Li Y, Anderson DG, Emerson CP, Sontheimer EJ, Moore MJ, Weng Z, Xue W. CRISPR/Cas9-mediated genome editing induces exon skipping by alternative splicing or exon deletion. Genome Biol 18: 108, 2017. doi: 10.1186/s13059-017-1237-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nakade S, Tsubota T, Sakane Y, Kume S, Sakamoto N, Obara M, Daimon T, Sezutsu H, Yamamoto T, Sakuma T, Suzuki KT. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat Commun 5: 5560, 2014. doi: 10.1038/ncomms6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nakamura A, Yoshida K, Fukushima K, Ueda H, Urasawa N, Koyama J, Yazaki Y, Yazaki M, Sakai T, Haruta S, Takeda S, Ikeda S. Follow-up of three patients with a large in-frame deletion of exons 45-55 in the Duchenne muscular dystrophy (DMD) gene. J Clin Neurosci 15: 757–763, 2008. doi: 10.1016/j.jocn.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 96.Nami F, Basiri M, Satarian L, Curtiss C, Baharvand H, Verfaillie C. Strategies for in vivo genome editing in nondividing cells. Trends Biotechnol 36: 770–786, 2018. doi: 10.1016/j.tibtech.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 97.Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan X, Ran FA, Yan WX, Asokan A, Zhang F, Duan D, Gersbach CA. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351: 403–407, 2016. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nelson CE, Wu Y, Gemberling MP, Oliver ML, Waller MA, Bohning JD, Robinson-Hamm JN, Bulaklak K, Castellanos Rivera RM, Collier JH, Asokan A, Gersbach CA. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat Med 25: 427–432, 2019. doi: 10.1038/s41591-019-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Neri M, Torelli S, Brown S, Ugo I, Sabatelli P, Merlini L, Spitali P, Rimessi P, Gualandi F, Sewry C, Ferlini A, Muntoni F. Dystrophin levels as low as 30% are sufficient to avoid muscular dystrophy in the human. Neuromuscul Disord 17: 913–918, 2007. doi: 10.1016/j.nmd.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 100.Offord C. US Companies launch CRISPR clinical trial [Online]. The Scientist: 2018. https://www.the-scientist.com/news-opinion/us-companies-launch-crispr-clinical-trial-64746.
- 101.Ousterout DG, Kabadi AM, Thakore PI, Majoros WH, Reddy TE, Gersbach CA. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun 6: 6244, 2015. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Perrin A, Rousseau J, Tremblay JP. Increased expression of laminin subunit alpha 1 chain by dCas9-VP160. Mol Ther Nucleic Acids 6: 68–79, 2017. doi: 10.1016/j.omtn.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Petris G, Casini A, Montagna C, Lorenzin F, Prandi D, Romanel A, Zasso J, Conti L, Demichelis F, Cereseto A. Hit and go CAS9 delivered through a lentiviral based self-limiting circuit. Nat Commun 8: 15334, 2017. doi: 10.1038/ncomms15334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pinto BS, Saxena T, Oliveira R, Méndez-Gómez HR, Cleary JD, Denes LT, McConnell O, Arboleda J, Xia G, Swanson MS, Wang ET. Impeding transcription of expanded microsatellite repeats by deactivated Cas9. Mol Cell 68: 479–490.e5, 2017. doi: 10.1016/j.molcel.2017.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Provenzano C, Cappella M, Valaperta R, Cardani R, Meola G, Martelli F, Cardinali B, Falcone G. CRISPR/Cas9-mediated deletion of CTG expansions recovers normal phenotype in myogenic cells derived from myotonic dystrophy 1 patients. Mol Ther Nucleic Acids 9: 337–348, 2017. doi: 10.1016/j.omtn.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.van Putten M, Hulsker M, Nadarajah VD, van Heiningen SH, van Huizen E, van Iterson M, Admiraal P, Messemaker T, den Dunnen JT, ’t Hoen PAC, Aartsma-Rus A. The effects of low levels of dystrophin on mouse muscle function and pathology. PLoS One 7: e31937, 2012. doi: 10.1371/journal.pone.0031937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.van Putten M, Hulsker M, Young C, Nadarajah VD, Heemskerk H, van der Weerd L, ’t Hoen PA, van Ommen GJ, Aartsma-Rus AM. Low dystrophin levels increase survival and improve muscle pathology and function in dystrophin/utrophin double-knockout mice. FASEB J 27: 2484–2495, 2013. doi: 10.1096/fj.12-224170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183, 2013. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520: 186–191, 2015. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.El Refaey M, Xu L, Gao Y, Canan BD, Adesanya TMA, Warner SC, Akagi K, Symer DE, Mohler PJ, Ma J, Janssen PML, Han R. In vivo genome editing restores dystrophin expression and cardiac function in dystrophic mice. Circ Res 121: 923–929, 2017. doi: 10.1161/CIRCRESAHA.117.310996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ruan GX, Barry E, Yu D, Lukason M, Cheng SH, Scaria A. CRISPR/Cas9-mediated genome editing as a therapeutic approach for leber congenital amaurosis 10. Mol Ther 25: 331–341, 2017. doi: 10.1016/j.ymthe.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ryu SM, Koo T, Kim K, Lim K, Baek G, Kim ST, Kim HS, Kim DE, Lee H, Chung E, Kim JS. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat Biotechnol 36: 536–539, 2018. doi: 10.1038/nbt.4148. [DOI] [PubMed] [Google Scholar]
- 113.Sawatsubashi S, Joko Y, Fukumoto S, Matsumoto T, Sugano SS. Development of versatile non-homologous end joining-based knock-in module for genome editing. Sci Rep 8: 593, 2018. doi: 10.1038/s41598-017-18911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schmid-Burgk JL, Höning K, Ebert TS, Hornung V. CRISPaint allows modular base-specific gene tagging using a ligase-4-dependent mechanism. Nat Commun 7: 12338, 2016. doi: 10.1038/ncomms12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343: 84–87, 2014. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sharp PS, Bye-a-Jee H, Wells DJ. Physiological characterization of muscle strength with variable levels of dystrophin restoration in mdx mice following local antisense therapy. Mol Ther 19: 165–171, 2011. doi: 10.1038/mt.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sheridan C. Go-ahead for first in-body CRISPR medicine testing [Online]. Nature Biotechnology: 2018. https://www.nature.com/articles/d41587-018-00003-2.
- 118.Shin J, Jiang F, Liu JJ, Bray NL, Rauch BJ, Baik SH, Nogales E, Bondy-Denomy J, Corn JE, Doudna JA. Disabling Cas9 by an anti-CRISPR DNA mimic. Sci Adv 3: e1701620, 2017. doi: 10.1126/sciadv.1701620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Simhadri VL, McGill J, McMahon S, Wang J, Jiang H, Sauna ZE. Prevalence of pre-existing antibodies to CRISPR-associated nuclease Cas9 in the USA population. Mol Ther Methods Clin Dev 10: 105–112, 2018. doi: 10.1016/j.omtm.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science 351: 84–88, 2016. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, Hatanaka F, Yamamoto M, Araoka T, Li Z, Kurita M, Hishida T, Li M, Aizawa E, Guo S, Chen S, Goebl A, Soligalla RD, Qu J, Jiang T, Fu X, Jafari M, Esteban CR, Berggren WT, Lajara J, Nuñez-Delicado E, Guillen P, Campistol JM, Matsuzaki F, Liu G-H, Magistretti P, Zhang K, Callaway EM, Zhang K, Belmonte JCI. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540: 144–149, 2016. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tabebordbar M, Zhu K, Cheng JKW, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, Cong L, Zhang F, Vandenberghe LH, Church GM, Wagers AJ. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 351: 407–411, 2016. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Taglia A, Petillo R, D’Ambrosio P, Picillo E, Torella A, Orsini C, Ergoli M, Scutifero M, Passamano L, Palladino A, Nigro G, Politano L. Clinical features of patients with dystrophinopathy sharing the 45-55 exon deletion of DMD gene. Acta Myol 34: 9–13, 2015. [PMC free article] [PubMed] [Google Scholar]
- 124.Tsai SQ, Nguyen NT, Malagon-Lopez J, Topkar VV, Aryee MJ, Joung JK. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat Methods 14: 607–614, 2017. doi: 10.1038/nmeth.4278 . A corrigendum for this article is available at http://dx.doi.org/10.1038/nmeth0518-394c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, Aryee MJ, Joung JK. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 33: 187–197, 2015. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Turan S, Farruggio AP, Srifa W, Day JW, Calos MP. Precise correction of disease mutations in induced pluripotent stem cells derived from patients with limb girdle muscular dystrophy. Mol Ther 24: 685–696, 2016. doi: 10.1038/mt.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Veltrop M, van Vliet L, Hulsker M, Claassens J, Brouwers C, Breukel C, van der Kaa J, Linssen MM, den Dunnen JT, Verbeek S, Aartsma-Rus A, van Putten M. A dystrophic Duchenne mouse model for testing human antisense oligonucleotides. PLoS One 13: e0193289, 2018. doi: 10.1371/journal.pone.0193289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wagner DL, Amini L, Wendering DJ, Burkhardt LM, Akyüz L, Reinke P, Volk HD, Schmueck-Henneresse M. High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nat Med 25: 242–248, 2019. doi: 10.1038/s41591-018-0204-6. [DOI] [PubMed] [Google Scholar]
- 129.Waldrop MA, Gumienny F, El Husayni S, Frank DE, Weiss RB, Flanigan KM. Low-level dystrophin expression attenuating the dystrophinopathy phenotype. Neuromuscul Disord 28: 116–121, 2018. doi: 10.1016/j.nmd.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang D, Mou H, Li S, Li Y, Hough S, Tran K, Li J, Yin H, Anderson DG, Sontheimer EJ, Weng Z, Gao G, Xue W. Adenovirus-mediated somatic genome editing of Pten by CRISPR/Cas9 in mouse liver in spite of Cas9-specific immune responses. Hum Gene Ther 26: 432–442, 2015. doi: 10.1089/hum.2015.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang Y, Hao L, Wang H, Santostefano K, Thapa A, Cleary J, Li H, Guo X, Terada N, Ashizawa T, Xia G. Therapeutic genome editing for myotonic dystrophy type 1 using CRISPR/Cas9. Mol Ther 26: 2617–2630, 2018. doi: 10.1016/j.ymthe.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wehling-Henricks M, Welc SS, Samengo G, Rinaldi C, Lindsey C, Wang Y, Lee J, Kuro-O M, Tidball JG. Macrophages escape Klotho gene silencing in the mdx mouse model of Duchenne muscular dystrophy and promote muscle growth and increase satellite cell numbers through a Klotho-mediated pathway. Hum Mol Genet 27: 14–29, 2018. doi: 10.1093/hmg/ddx380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wojtal D, Kemaladewi DU, Malam Z, Abdullah S, Wong TWY, Hyatt E, Baghestani Z, Pereira S, Stavropoulos J, Mouly V, Mamchaoui K, Muntoni F, Voit T, Gonorazky HD, Dowling JJ, Wilson MD, Mendoza-Londono R, Ivakine EA, Cohn RD. Spell checking nature: versatility of CRISPR/Cas9 for developing treatments for inherited disorders. Am J Hum Genet 98: 90–101, 2016. doi: 10.1016/j.ajhg.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wyatt EJ, Demonbreun AR, Kim EY, Puckelwartz MJ, Vo AH, Dellefave-Castillo LM, Gao QQ, Vainzof M, Pavanello RCM, Zatz M, McNally EM. Efficient exon skipping of SGCG mutations mediated by phosphorodiamidate morpholino oligomers. JCI Insight 3: e99357, 2018. doi: 10.1172/jci.insight.99357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xu L, Park KH, Zhao L, Xu J, El Refaey M, Gao Y, Zhu H, Ma J, Han R. CRISPR-mediated genome editing restores dystrophin expression and function in mdx mice. Mol Ther 24: 564–569, 2016. doi: 10.1038/mt.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yin H, Song C-Q, Suresh S, Kwan S-Y, Wu Q, Walsh S, Ding J, Bogorad RL, Zhu LJ, Wolfe SA, Koteliansky V, Xue W, Langer R, Anderson DG. Partial DNA-guided Cas9 enables genome editing with reduced off-target activity. Nat Chem Biol 14: 311–316, 2018. doi: 10.1038/nchembio.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Young CS, Hicks MR, Ermolova NV, Nakano H, Jan M, Younesi S, Karumbayaram S, Kumagai-Cresse C, Wang D, Zack JA, Kohn DB, Nakano A, Nelson SF, Miceli MC, Spencer MJ, Pyle AD. A single CRISPR-Cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hiPSC-derived muscle cells. Cell Stem Cell 18: 533–540, 2016. doi: 10.1016/j.stem.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Young CS, Mokhonova E, Quinonez M, Pyle AD, Spencer MJ. Creation of a novel humanized dystrophic mouse model of duchenne muscular dystrophy and application of a CRISPR/Cas9 gene editing therapy. J Neuromuscul Dis 4: 139–145, 2017. doi: 10.3233/JND-170218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yuan J, Ma Y, Huang T, Chen Y, Peng Y, Li B, Li J, Zhang Y, Song B, Sun X, Ding Q, Song Y, Chang X. Genetic modulation of RNA splicing with a CRISPR-guided cytidine deaminase. Mol Cell 72: 380–394.e7, 2018. doi: 10.1016/j.molcel.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 140.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163: 759–771, 2015. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang Y, Long C, Li H, McAnally JR, Baskin KK, Shelton JM, Bassel-Duby R, Olson EN. CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice. Sci Adv 3: e1602814, 2017. doi: 10.1126/sciadv.1602814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhu P, Wu F, Mosenson J, Zhang H, He TC, Wu WS. CRISPR/Cas9-mediated genome editing corrects dystrophin mutation in skeletal muscle stem cells in a mouse model of muscle dystrophy. Mol Ther Nucleic Acids 7: 31–41, 2017. doi: 10.1016/j.omtn.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zischewski J, Fischer R, Bortesi L. Detection of on-target and off-target mutations generated by CRISPR/Cas9 and other sequence-specific nucleases. Biotechnol Adv 35: 95–104, 2017. doi: 10.1016/j.biotechadv.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 144.Zuo E, Sun Y, Wei W, Yuan T, Ying W, Sun H, Yuan L, Steinmetz LM, Li Y, Yang H. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 364: 289–292, 2019. doi: 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]